Abstract
During neural development, complex organisms rely on progressive and regressive events whereby axons, synapses, and neurons are overproduced followed by selective elimination of a portion of these components. Tumor necrosis factor α (TNFα) together with its cognate receptor (Tumor necrosis factor receptor 1; TNFR1) have been shown to play both regressive (i.e. forward signaling from the receptor) and progressive (i.e. reverse signaling from the ligand) roles in sympathetic neuron development. In contrast, a paralog of TNFR1, p75 neurotrophic factor receptor (p75NTR) promotes mainly regressive developmental events in sympathetic neurons. Here we examine the interplay between these paralogous receptors in the regulation of axon branch elimination and arborization. We confirm previous reports that these TNFR1 family members are individually capable of promoting ligand-dependent suppression of axon growth and branching. Remarkably, p75NTR and TNFR1 physically interact and p75NTR requires TNFR1 for ligand-dependent axon suppression of axon branching but not vice versa. We also find that p75NTR forward signaling and TNFα reverse signaling are functionally antagonistic. Finally, we find that TNFα reverse signaling is necessary for nerve growth factor (NGF) dependent axon growth. Taken together these findings demonstrate several levels of synergistic and antagonistic interactions using very few signaling pathways and that the balance of these synergizing and opposing signals act to ensure proper axon growth and patterning.
Keywords: Sympathetic neuron, p75NTR, TNFR1, TNF, Reverse signaling, Axon arbor
1. Introduction
Assembly of a functional nervous system in complex organisms employs a strategy of trial and error, whereby dynamic and tightly controlled progressive and regressive developmental events pattern nascent circuits. For example, in the vertebrate nervous system, circuit components such as axons, synapses, and neurons are produced in excess and selectively eliminated to achieve appropriate connectivity and function (Hamburger and Levi-Montalcini, 1949; reviewed in Chao, 2003; Fitzsimonds and Poo, 1998). An emergent principle in developmental neuroscience is that pro-growth and pro-refinement signaling pathways compete with one another to promote or inhibit connectivity in order to define the architecture of the nervous system (Deppmann et al., 2008; Singh et al., 2008; Sharma et al., 2010). How cross-talk between progressive and regressive cues occurs to mediate nervous system wiring remains an open question.
During the development of the sympathetic nervous system, refinement of the excess neuronal population is mediated by competition between TrkA mediated pro-growth and p75-neurotrophin receptor (p75NTR) mediated regressive signaling (reviewed in Majdan and Miller, 1999; Deckwerth and Johnson, 1993; Majdan et al., 2001). Limiting amounts of target organ-derived nerve growth factor (NGF) bind axonal TrkA thereby transducing signaling cascades which promote target innervation, neuronal survival, synapse initiation, and a variety of other important developmental processes (McMahon et al., 1994; Glebova and Ginty, 2004; Sharma et al., 2010). On the other hand, p75NTR signaling represents a counterbalance to these TrkA-mediated pro-growth signals as it mediates neuronal death, axon degeneration, and synapse restriction (Bamji et al., 1998; Singh et al., 2008; Sharma et al., 2010). Neurons, axons, and synapses experiencing high levels of trophic signaling are refractory to p75NTR-mediated death signaling. However, neurons exposed to low levels of NGF-dependent survival signaling (termed suboptimal by Miller and colleagues, Majdan et al., 2001) are susceptible to ligand-induced regressive signaling by brain-derived neurotrophic factor (BDNF), neurotrophin 4/5 (NT4/5), or pro-neurotrophins through p75NTR (Deppmann et al., 2008; Deinhardt et al., 2011). During target innervation, neurons must traverse varying levels of NGF pro-growth signaling and suppress prodestructive cues to survive and stabilize components like axons and synapses (Deppmann et al., 2008; Deinhardt et al., 2011). Despite a reasonably good understanding of the signaling pathways mediating progressive and regressive signaling, we still do not understand how these antagonistic pathways interact with one another.
p75NTR is a member of the tumor necrosis factor receptor (TNFR) family and signaling related to several of these receptors has been reported to build or refine the sympathetic nervous system (O’Keeffe et al., 2008; Kisiswa et al., 2013; Barker et al., 2001; Nikolaev et al., 2009). Beyond p75NTR, other TNFR family members including TNFR1 and death receptor 6 (DR6) are expressed in the developing peripheral nervous system and play roles in regressive signaling (Wheeler et al., 2014; Barker et al., 2001, Gamage et al., 2017; Kisiswa et al., 2013; Kisiswa et al., 2017). TNFR1 and p75NTR in particular share several properties. For example, both receptors are capable of triggering common signaling pathways including c-Jun N-terminal kinase (JNK), NFkB (reviewed in Hempstead, 2002), and RhoA (Yamashita et al., 1999; Neumann et al., 2002). There are also several key differences between these family members, including variation in the number of ligands that each cognate receptor can bind to: TNFR1 is more limited in the ligands it prefers (e.g., TNFα and LTα), whereas p75NTR promiscuously binds to all immature and mature neurotrophins (NGF, BDNF, NT4/5, NT3) as well as non-neurotrophic factors secreted by glial cells such as Nogo, myelin-associated glycoprotein (MAG), and oligodendrocyte myelin glycoprotein (OMgp) (Bibel and Barde, 2000; Jansen et al., 2007; Wang et al., 2002). Additionally, while both TNFR1 and p75NTR promote apoptotic signaling, they do so via different mechanisms. TNFR1 ligand binding activates the extrinsic apoptotic pathway by causing the binding of adapter protein TNFR1-associated death domain protein (TRADD), promoting association and activation of procaspase-8 (Elmore, 2007). p75NTR, however, is capable of activating the caspase-9-dependent intrinsic cell pathway through interactions with a handful of its binding partners (Underwood and Coulson, 2008). There is also evidence that p75NTR can act as a dependence receptor, signaling without ligand binding (Rabizadeh et al., 1993; Bredesen et al., 1998). Whether TNFR family members like p75NTR and TNFR1 cooperate during refinement signaling to affect an individual neuron’s morphology is unknown.
However, TNFR family member-mediated signaling is not invariably regressive. For instance, TNFα, a major ligand for TNFR1, belongs to the TNF superfamily. Most TNF family members are type II transmembrane proteins and, in addition to being cleaved from the membrane and acting as ligands, may remain membrane-bound and function as receptors when interacting with their binding partners in a process called reverse signaling. Several examples of reverse signaling within the TNF superfamily exist in the context of immune system development and function (Sun and Fink, 2007). For example, TNFα reverse signaling on macrophages and monocytes reduces the production of pro-apoptotic cytokines upon stimulation with lipopolysaccharide (LPS) (Eissner et al., 2000). Within the nervous system, TNFR1, which induces pro-refinement signaling when acting as a membrane-bound receptor, can also be cleaved from the membrane and act as a ligand for membrane-bound TNFα (mTNFα), providing a potent pro-growth signal (Kisiswa et al., 2013). Intracellular signaling initiated from what the field views as classic receptors is referred to as forward signaling and signaling emanating from a membrane-bound ligand within the cell bearing that ligand is referred to as reverse signaling. While it remains unclear how TNFR1-TNFα forward and reverse signaling work together to govern nervous system patterning, this additional layer of control would likely provide a mechanism for juxtacrine regulation of progressive and regressive signaling.
In this study, we seek to uncover the interactions between each component of the bidirectional and antagonistic TNFR1-TNFα signaling axis and pro-refinement p75NTR. We report a unidirectional dependence between both ligand-dependent regressive signaling pathways: BDNF-p75NTR-mediated repression of axon growth requires the presence of TNFR1, while similar inhibition of axon growth through TNF-TNFR1 forward signaling does not require p75NTR. Further, we demonstrate a physical association between TNFR1 and p75NTR through co-immunoprecipitation and bimolecular fluorescence complementation (BiFC). However, we find that p75NTR forward and TNFR1-TNFα reverse signaling are functionally antagonistic, indicating that the primary role of each pathway is regressive and progressive, respectively. Moreover, we demonstrate that mTNFα reverse signaling is required for NGF-TrkA dependent axon growth. Taken together, these findings demonstrate that TNFR family signaling exhibit complex regulatory processes including co-dependence, compensation, and antagonism, all of which are required for proper elaboration of sympathetic axons.
2. Results
Soluble BDNF and TNFα reduced axon complexity in cultured sympathetic neurons.
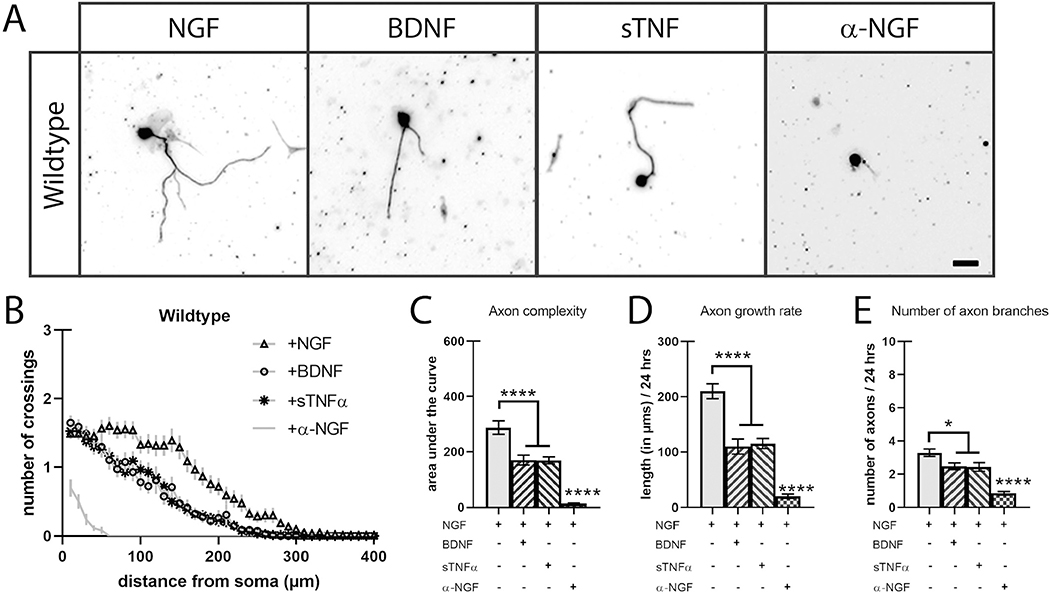
To compare the role of TNFR1, TNFα, and p75NTR activation on axon growth and branching, sympathetic neurons from the superior cervical ganglion (SCG) of wild-type (WT) P0 mice were cultured in the presence of 2 ng/ml of NGF and with 2 ng/ml of NGF with sTNFα (2 ng/ml), BDNF (250 ng/ml), or a function-blocking α-NGF antibody (1 μg/ml). After 24 h, the neurons were fixed and the complete axon arbor was visualized using immunocytochemistry for Class III β-tubulin antibody, TuJ1 (Fig. 1A). We then traced isolated neurons and analyzed axon complexity using Sholl analysis (Fig. 1B–C, STable 1). As expected, the presence of NGF promoted sympathetic axon complexity while blocking TrkA activation with α-NGF function neutralizing antibody reduced axon arborization drastically (Fig. 1A–C). Consistent with work from Davies and colleagues as well as Miller and colleagues, treatment with soluble BDNF or TNFα antagonizes NGF-TrkA signaling resulting in reduced axon complexity (Fig. 1A–C, STable 1) (Kisiswa et al., 2013; Singh et al., 2008). Axon complexity is the composite of multiple characteristics including axon growth rate, homeostasis between axon sprouting and axon pruning, hierarchical branching, and more. With this in mind, we assessed two of these characteristics: axon growth rate and the number of axon branches. We analyzed axon growth rate by measuring the length of the longest axon for each neuron providing a distance in microns over the 24 h growth period (Fig. 1D). The number of axon branches were quantified by counting axon endpoints (Fig. 1E). The reduction in axon complexity of both TNFR1 and p75NTR activation was due to a slowed axon growth rate (~50%) and a reduction (~20%) in branch number (Fig. 1D, E).
Fig. 1.
Soluble BDNF and TNFα reduces axon complexity in cultured sympathetic neurons.
A) SCG neurons were cultured in the presence of NGF (2 ng/mL) or a combination of NGF with p75NTR ligand BDNF (250 ng/mL), TNFR1 ligand sTNFα (2 ng/mL), or NGF-neutralizing antibody α-NGF (2 ng/mL) and stained with TUJ1 24 h after plating. Scale bar = 30 μm. B) Sholl analysis of P0 SCG neuron axon complexity was performed by counting trace intersections in 10 μm intervals. C) Quantification of the area under the curve from B). D) Axon growth rate was measured as the length of the longest axon segment per neuron after 24 h from B). E) Axon branching was measured as the number of axon branch endpoints per neuron after 24 h from B). Full statistical comparisons found in STable 1. Error bars represent s.e.m. n = 62 to 112 neurons per condition from 3 cultures of SCGs pooled from whole litters. *p < 0.05, ****p < 0.0001 using 2-way ANOVA with Tukey’s multiple comparisons test.
2.1. Loss of p75NTR increased axon number independently of TNFR1
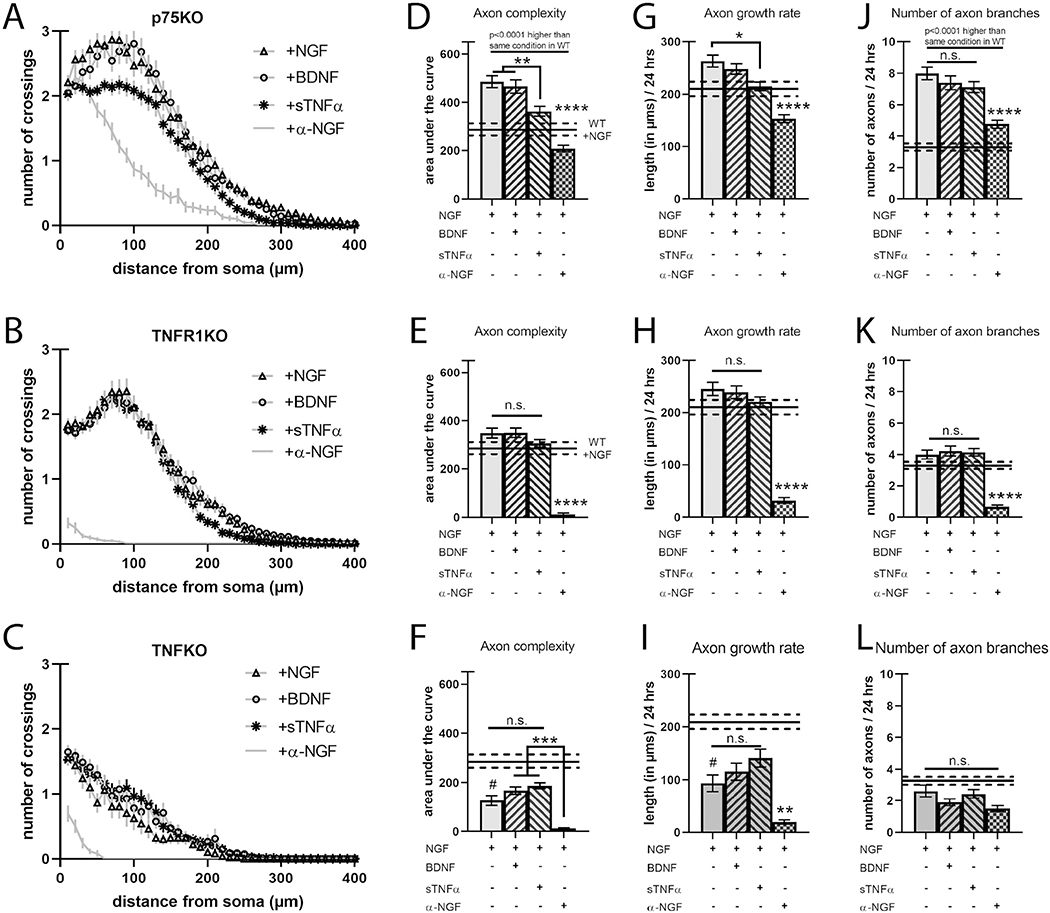
To further understand the role of TNFR1, TNFα, and p75NTR in axon complexity, we cultured sympathetic neurons from mutant mice deficient in TNFR1 (TNFR1KO), TNFα (TNFKO), or p75NTR (p75KO) and treated these cultures as in Fig. 1. p75NTR has been shown to promote axon pruning in sympathetic axons (Singh et al., 2008) and in its absence or when it is suppressed, nascent axon branches stabilize, resulting in elevated axon complexity. As expected, p75KO neurons exhibited exuberant axon complexity in all conditions compared to wild-type neurons (Fig. 2A, D, G, J, STable 1). Similarly, we find that these p75KO axon arbors had double the number of branches compared to wild-type axons exposed to the same ligands, including when exposed to α-NGF (Fig. 2J). Intriguingly, activation of p75NTR with BDNF caused a ~50% decrease in axon growth rate in wild-type neurons, however, loss of p75NTR only modestly potentiated axon growth rate (compare Figs. 1D and 2G). BDNF also stunted branch number by ~20% in wild-type neurons while loss of p75NTR more than doubled the number of axon branches (compare Figs. 1E and 2J). This suggests that p75NTR may provide regressive signals in the absence of BDNF stimulation. Finally, p75KO neurons still displayed dampened axon complexity due to reduced axon growth in response to sTNFα, implying that TNFR1 forward signaling does not depend on p75NTR (Fig. 2D, G).
Fig. 2.
Loss of TNFR1 does not alter axon complexity but is required for BDNF-dependent decrease in axon length.
A–C) Sholl analysis of SCG neurons cultured as in Fig. 1 but from p75KO, TNFR1KO, and TNFKO, respectively. D–F) Quantification of the area under the curve from A–C, respectively. G–I) Axon growth rate was measured as the length of the longest axon segment per neuron after 24 h from A–C, respectively. J–L) Axon branching was measured as the number of axon branch endpoints per neuron after 24 h from A–C, respectively. Solid black bars represent the mean axon characteristics displayed by neurons from wild-type animals treated with NGF with the dotted lines representing the s.e.m. shown in Fig. 1 and here for comparison. Full statistical comparisons found in STable 1. Error bars represent s.e.m. n = 50 to 120 neurons per condition from 3 cultures of SCGs pooled from whole litters for each genotype. n.s. = not significant, #p < 0.005 (between wild-type +NGF and TNFKO +NGF), *p < 0.05, ****p < 0.0001 using 2-way ANOVA with Tukey’s multiple comparisons test.
2.2. Loss of TNFR1 does not alter axon growth but is required for BDNF-p75NTR-dependent axon regression
Given that TNFR1 regulates axon growth by both acting as a ligand for pro-growth mTNFα and as a receptor for regressive sTNFα, one would expect its loss to affect axon complexity. Interestingly, TNFR1KO neurons grown in 2 ng/ml NGF or α-NGF have axon arbors indistinguishable from the corresponding wild-type axons in complexity, growth, and number (Fig. 2B, E, H, K, STable 1). As expected, these neurons do not respond to sTNFα as they lack the cognate receptor for this ligand (Fig. 2B, E, H, K). Surprisingly, TNFR1KO neurons also did not respond to application of BDNF, implying that BDNF-p75NTR regressive signaling requires TNFR1 (Fig. 2E, H, 2nd column).
2.3. Loss of TNFα reduces axon complexity by slowing axon growth
We next sought to uncover the contribution of mTNFα as a receptor. Consistent with its role as a pro-growth receptor in developing sympathetic neurons and its ability to act as a regressive ligand when solubilized (Kisiswa et al., 2013), loss of TNFα results in reduced axon development due to slower outgrowth (Fig. 2C, F, I, L, STable 1). In order to examine the role of mTNFα alone, we added back sTNFα to compensate for its loss in TNFKO neurons. However, these neurons do not exhibit an additive loss of axon complexity when exposed to either sTNFα or BDNF despite retaining receptors for both of these ligands, indicating that mTNFα is critical for the proper growth of axons (Fig. 2C, F, I, L). It may be that mTNFα signaling is a foundational pro-growth pathway and other pathways are recruited at higher concentrations of NGF (i.e. higher levels of TrkA activity).
2.4. TNFR1 and p75NTR physically interact
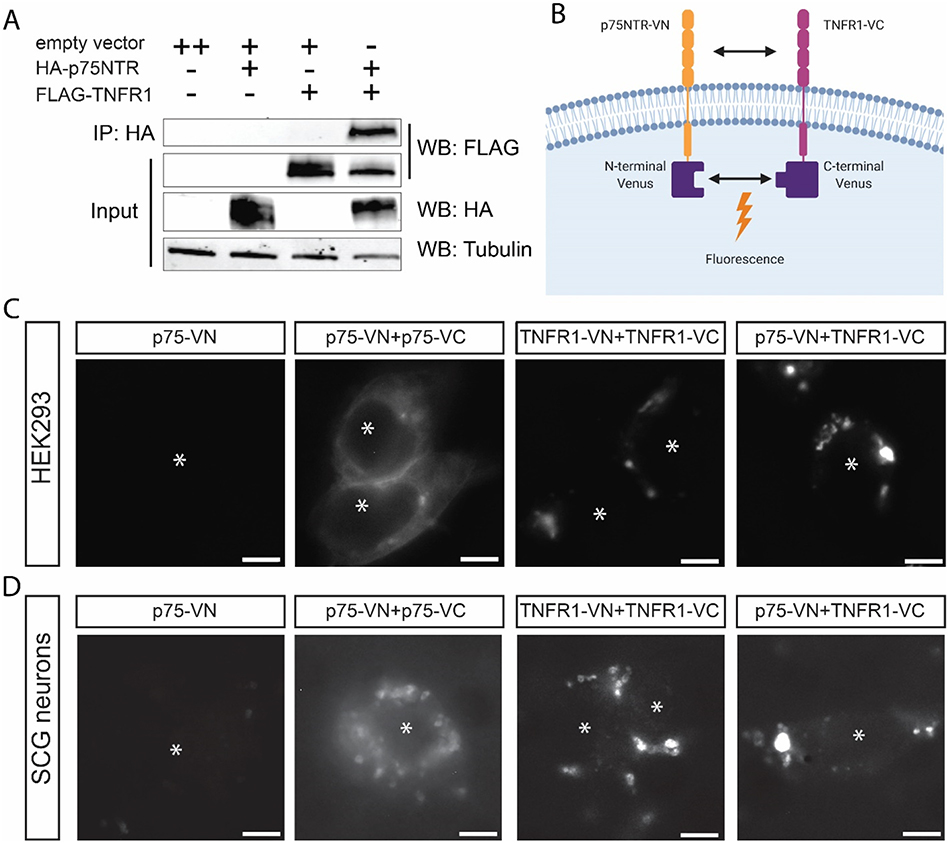
To better understand the observed unidirectional dependence of p75NTR on TNFR1, we asked whether the receptors are physically associated. We generated HA-tagged p75NTR and FLAG-tagged TNFR1 expression constructs (SFig. 1A). These constructs were expressed in HEK293 cells followed by analysis of interaction by co-immunoprecipitation and immunoblot analysis. In the immunoprecipitated lanes, we detected FLAG-TNFR1 only when HA-p75NTR and TNFR1 were expressed, suggesting association of p75NTR and TNFR1 (Fig. 3A).
Fig. 3.
TNFR1 interacts with p75NTR and changes its subcellular localization.
A) Co-immunoprecipitation (co-IP) of p75NTR and TNFR1 constructs using HEK293 cells. The experiment was performed 4 times (n = 4). B) Schematic of bimolecular fluorescence complementation (BiFC). This figure was created with Biorender.com and exported under a paid subscription. C, D) Bimolecular fluorescence complementation (BiFC) of p75-NTR and TNFR1 constructs using HEK293 cells (C) or sympathetic neurons (D). Transfected cells were imaged using fluorescence excitation at 514 nm. Asterisks indicate the cells in each image. Images are representative of experiments performed at least 3 times (n = 3). Scale bar = 5 μm.
We next sought to determine the subcellular localization of p75NTR/TNFR1 heteromeric complexes compared to their respective homomeric interactions. To visualize the association of p75NTR and TNFR1, we used bimolecular fluorescence complementation (BiFC), where p75NTR and TNFR1 are cloned in frame with split-portions of the Venus fluorescent protein (N-terminus or C-terminus) (Hu and Kerppola, 2003) (Fig. 3B). To avoid overexpression, which could lead to inappropriate interaction, these fusion constructs were expressed in the pTRE2hyg vector backbone, a tetracycline-regulated expression system (SFig. 1B). We expressed the constructs in HEK293 cells cultured in tetracycline-free media in order to maintain minimal “leaky” heterologous expression (SFig. 1C). Cells expressing the homomeric pair of p75NTR exhibited uniform fluorescence in the cytoplasm and plasma membrane as well as a few intracellular puncta; whereas cells expressing homomeric TNFR1 displayed fluorescent patches suggesting that these receptors have distinct subcellular distributions in the cell (Fig. 3C). Each homomeric pair - p75NTR or TNFR1-exhibited fluorescence in the absence of its cognate ligand, which is consistent with previous reports (Vilar et al., 2009; Chan et al., 2000) (Fig. 3C, SFig. 1C). Interestingly, in cells expressing the heteromeric pair of p75NTR and TNFR1, punctate fluorescence was similar to homomeric TNFR1 (Fig. 3C, SFig. 1C). To ensure that these results were not particular to HEK293 cells, we next expressed the same constructs in primary mouse sympathetic cultures. In all pairings of BiFC constructs, fluorescence expression was similar to expression in the HEK293 cells (Fig. 3D, SFig. 1C). Homomeric expression of p75NTR still exhibited uniform expression although more puncta were evident (Fig. 3D, SFig. 1C). Homomeric TNFR1 BiFC pairs also displayed punctate membrane localization (Fig. 3D, SFig. 1C). Finally, the p75NTR and TNFR1 heteromeric BiFC pair displayed a similar distribution in homomeric TNFR1 (Fig. 3D, SFig. 1C). This indicates that the subcellular localization of the p75NTR:TNFR1 complex is principally influenced by TNFR1 across cell types.
2.5. Loss of TNFR1 or TNFα in a p75NTR-deficient background rescues exuberant axon arborization
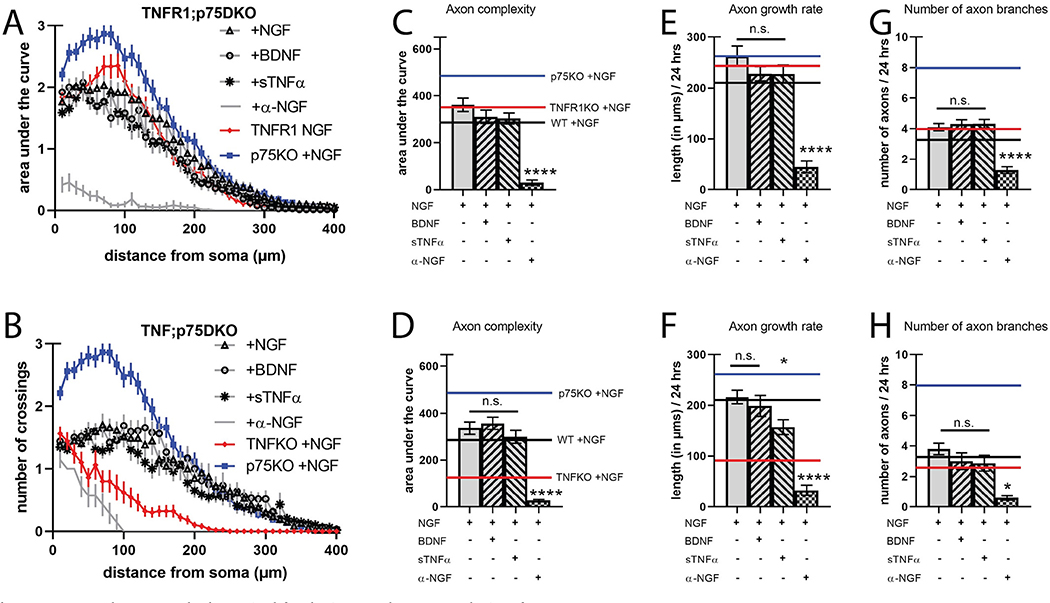
The physical and functional interactions of TNFR1 and p75NTR led us to investigate axon branching in TNFR1−/−;p75NTR−/− (TNFR1;p75DKO) neurons. Based on observations in single null mutants, we predicted that TNFR1;p75DKO neurons would exhibit axon complexity at least as exuberant as p75KO. Surprisingly, TNFR1;p75DKO axon arbors were identical to wild-type, even though activation of either of these receptors represses axon growth and branching (Fig. 4A, C, E, G, STable 1). Neither BDNF or sTNFα elicited an effect as both cognate receptors were absent (Fig. 4A, C, E, G). To explain the lack of exuberant branching observed in the TNFR1;p75DKO compared to p75KO (compare Figs. 2D and 4C), we speculated that loss of TNFR1 ablates a critical pro-growth signal provided by mTNFα activation that we failed to uncover in the presence of p75NTR (Fig. 2C, F, I, L). Indeed, work from our lab and others has shown that TNFR1-TNFα reverse signaling is a potent inducer of axon growth and branching in sympathetic (Kisiswa et al., 2013) and sensory (Wheeler et al., 2014) neurons. This led us to hypothesize that TNFR1 and mTNFα pro-growth reverse signaling and p75NTR pro-refinement signaling are coupled to ensure proper axon development.
Fig. 4.
TNFR1 and TNFα are both required for the increased axon complexity of p75KO neurons.
A, B) Sholl analysis of SCG neurons cultured as in Fig. 1 but from TNFR1;p75DKO, and TNF;p75DKO, respectively. C, D) Quantification of the area under the curve from A, B, respectively. E, F) Axon growth rate was measured as the length of the longest axon segment per neuron after 24 h from A–B, respectively. G, H) Axon branching was measured as the number of axon branch endpoints per neuron after 24 h from A–B, respectively. Solid black bars represent axon characteristics displayed by neurons from wild-type animals treated with NGF shown in Fig. 1 for comparison. The red line indicates TNFR1KO (in A, C, E, G), and TNFKO (in B, D, F, H), both +NGF, and the blue line indicates p75KO + NGF for comparison. For clarity, the s.e.m. was not represented. Full statistical comparisons found in STable 1. Error bars represent s.e.m. n = 27 to 82 neurons per condition from 3 cultures of SCGs pooled from whole litters for each genotype. n.s. = not significant, *p < 0.05, ****p < 0.0001 using 2-way ANOVA with Tukey’s multiple comparisons test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To test whether p75NTR counterbalances TNFR1-TNFα pro-growth signaling we assessed axon growth in TNFα−/−;p75NTR−/− (TNF;p75DKO) neurons. If the primary output of TNFR1-TNFα signaling is pro-growth reverse signaling with TNFR1 as ligand and mTNFα as receptor, then loss of either protein should have a negative effect on axon complexity. In this case, we would expect loss of TNFα in a p75NTR-deficient background to result in wild-type levels of axon complexity, similarly to loss of TNFR1. Indeed, TNF;p75DKO neurons show identical axon complexity as TNFR1;p75DKO and wild-type in all conditions, suggesting that deletion of either member of the reverse signaling pathway has a similar effect on axon dynamics (Fig. 4B, D, F, H, STable 1). Interestingly, while application of sTNFα did not reduce overall axon complexity in TNF;p75DKO neurons as it previously had in neurons containing TNFR1, it did slow axon growth rate compared to both TNF;p75DKO with NGF and TNFR1;p75DKO with sTNFα (comparison not shown), similar to the effect of sTNFα in wild-type neurons (Fig. 4D, F). This suggests that exogenous and endogenous sTNFα combines to diminish both axon growth rate and complexity, and that exogenous sTNFα alone is insufficient to produce this effect. These results indicate that by losing either TNFα or TNFR1 in addition to p75NTR, neurons have lost both a regressive and progressive signal, returning axon complexity to wild-type levels. Thus, p75NTR-dependent axon refinement functionally antagonizes TNFR1-TNFα reverse signaling to maintain an appropriate range of axon complexity. Activation of either pathway pushes axon arbors towards upper and lower boundaries of complexity by specifically titering growth or branch stability.
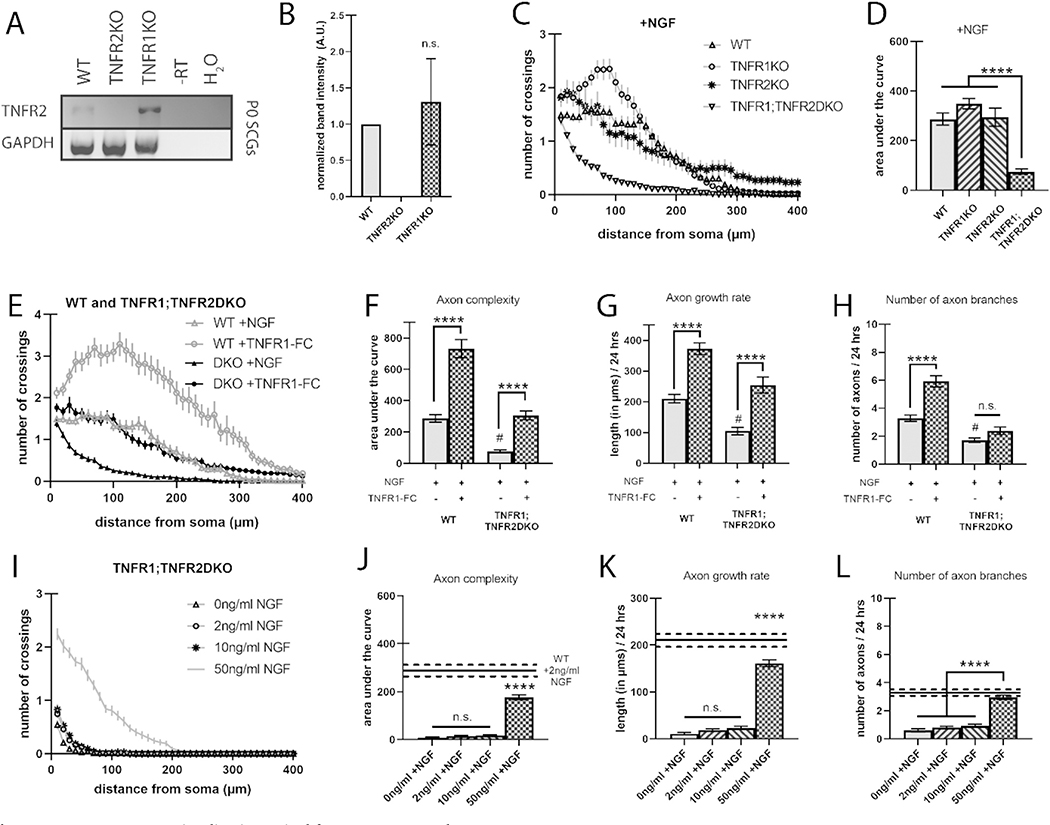
2.6. TNFR2 compensates for TNFR1 and loss of both TNFR1 and TNFR2 results in catastrophic loss of axon complexity
If TNFR1 and TNFα are essential for a pro-growth signal during axon development, then loss of TNFR1 should result in a dramatic loss of axon arbors. However, we found that TNFR1KO neurons were identical to wild-type conditions (Fig. 2B, E). This could be explained by another ligand for mTNFα compensating when TNFR1 is lost. TNFR2, a TNFR superfamily member closely related to TNFR1 with extensive sequence and functional similarity, is considered the canonical ligand for mTNFα in the mediation of proliferation, differentiation, and survival of diverse tissues (Grell et al., 1995; Qu et al., 2017; Juhász et al., 2014). Davies and colleagues report weak immunohistochemical staining of TNFR2 in cultured mouse sympathetic neurons (Kisiswa et al., 2013). We hypothesized that in the absence of TNFR1, TNFR2 may be expressed to compensate. We therefore pooled SCGs from litters of P0 wild-type, TNFR1KO, and TNFR2−/− (TNFR2KO) mice, and performed RT-PCR using 2 primer sets against the coding region of TNFR2 (Fig. 5A, SFig. 2A, B). We found that TNFR2 transcript is expressed in both wild-type and TNFR1KO, but not in TNFR2KO SCGs (Fig. 5A, B). Furthermore, transcript levels do not seem to increase in the absence of TNFR1, indicating that if compensation occurs, it may be at the protein and not expression level (Fig. 5B). To determine whether TNFR2 functionally compensates for loss of TNFR1, we assessed axon complexity in wild-type, TNFR1KO, TNFR2KO, and TNFR1−/−;TNFR2−/− (TNFR1;TNFR2DKO) neurons. Similar to TNFR1KO neurons, axon complexity of TNFR2KO was identical to wild-type controls, however, TNFR1;TNFR2DKO neurons exhibited a striking loss of axon complexity, to the levels of wild-type neurons treated with α-NGF (Fig. 5C, D, STable 2). Thus, the TNFR1;TNFR2DKO suggests that TNFR1 and TNFR2 work in parallel to activate progressive signaling through mTNFα. To ensure that the loss of axon branching in TNFR1;TNFR2DKO neurons was due to deficient reverse signaling, we attempted to rescue this phenotype using a soluble extracellular peptide of TNFR1, TNFR1-Fc (10 ng/ml). We confirmed that addition of TNFR1-Fc to wild-type cultures elicited a robust increase in axon complexity and found that TNFR1-Fc rescued the loss of axon complexity observed in the TNFR1;TNFR2DKO neurons (Fig. 5E–H, STable 3).
Fig. 5.
TNFR-TNFα reverse signaling is required for NGF axon growth.
A) Representative RT-PCR showing expression of TNFR2 mRNA in SCG neurons derived from pooled P0 wild-type, TNFR1KO, and TNFR2KO litters. B) Relative TNFR2 mRNA expression as measured by RT-PCR from 3 separate pooled litters per genotype; SFig. 2 shows the RT-PCR from all 3 blots. C) Sholl analysis of TNFR2KO SCG neurons grown with 2 ng/ml NGF and compared to wild-type and TNFR1KO neurons from Figs. 1 and 2, respectively. D) Quantification of the area under the curve from C. E) Sholl analysis of wild-type or TNFR1;TNFR2DKO SCG neurons grown with 2 ng/ml NGF and in the presence or absence of 10 ng/ml TNFR1-Fc, soluble extracellular TNFR1 peptide. F) Quantification of the area under the curve from E. G) Axon growth rate was measured as the length of the longest axon segment per neuron after 24 h from E. H) Axon branching was measured as the number of axon branch endpoints per neuron after 24 h from E. I) Sholl analysis of TNFR1;TNFR2DKO SCG neurons grown in 0, 2, 10, and 50 ng/ml NGF. J) Quantification of the area under the curve from I. K) Axon growth rate was measured as the length of the longest axon segment per neuron after 24 h from I. L) Axon branching was measured as the number of axon branch endpoints per neuron after 24 h from I. Full statistical comparisons found in STables 2, 3, and 4. Error bars represent s.e.m. n = 52 to 107 neurons per condition from 3 cultures of SCGs pooled from whole litters for each genotype. n.s. = not significant, #p < 0.0001 (between wild-type +NGF and TNFR1;TNFR2DKO + NGF), ****p < 0.0001 using 1-way ANOVA for B-D and I-L and 2-way ANOVA for E–H all with Tukey’s multiple comparisons test.
Further, the TNFR1;TNFR2DKO reveals that NGF pro-growth signaling requires TNFR-TNFα reverse signaling since loss of both TNFR ligands for mTNFα abrogates axon growth even in the presence of NGF (Fig. 5D). To determine if this requirement only exists in suboptimal levels of NGF or if it is a general requirement, we assessed axon complexity in TNFR1;TNFR2DKO neurons grown in 0, 2, 10, and 50 ng/ml NGF. We found that neurons in no (0 ng/ml NGF) or suboptimal NGF (2 ng/ml and 10 ng/ml) displayed similar loss of axon complexity but optimal NGF (50 ng/ml) exhibited a partial rescue compared to wild-type neurons grown in suboptimal NGF (Fig. 5I–L, STable 4). Although TNFR1;TNFR2DKO neurons treated with 50 ng/ml NGF had the same number of axon branches as wild-type neurons in suboptimal NGF, the DKO axons grew significantly slower (Fig. 5K, L). This suggests that optimal NGF compensates for the loss of TNFR1 and TNFR2 for axon branching but not for axon growth. Together, these data suggest that TNFR1 and TNFR2 work in concert as ligands for mTNFα reverse pro-growth signaling.
3. Discussion
In this study, we have examined how p75NTR, TNFR1 and TNFα signaling interact to control axon growth (Fig. 6). First, we show that ligand-dependent p75NTR activation requires TNFR1 to slow axon growth and that regressive TNFR1 forward signaling occurs independently of p75NTR. Second, we demonstrate that TNFR1 and p75NTR physically interact and colocalize. Third, we show that TNFR1 fails to compensate for loss of p75NTR, resulting in exuberant axon branching, while loss of TNFR1 does not affect axon complexity due to compensation by TNFR2. Fourth, concurrent loss of TNFR1 or TNFα with p75NTR rescues axon arborization to wild-type levels compared to the overgrowth observed in the absence of p75NTR. Finally, we demonstrate that TNFR-TNFα reverse signaling is required for suboptimal NGF to promote axon growth. Thus, our work indicates that p75NTR suppression of axon branching is a critical balance to the pro-growth TNFR1-TNFα reverse signaling.
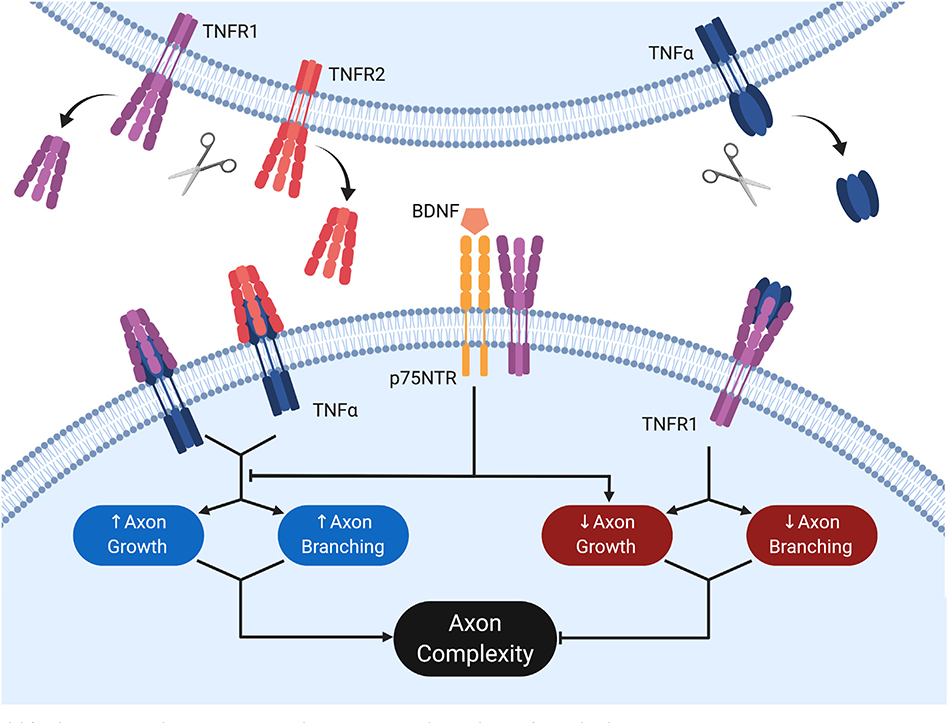
Fig. 6.
Model for the interactions between p75NTR and TNFR1-TNFα in the regulation of axon development.
Activation of membrane-bound TNFα by either sTNFR1 or sTNFR2 promotes both axon growth rate and branching, leading to an increase in overall axon complexity. Conversely, membrane-bound TNFR1 may be activated by soluble TNFα, suppressing axon growth and branching, thereby reducing axon complexity. Pro-growth signaling through the former pathway is dominant, such that loss of either TNFα or both TNFR1 and TNFR2 results in catastrophic loss of axon complexity. p75NTR mediates a reduction in axon complexity and functionally antagonizes the pro-growth TNFR1-TNFα signaling axis. As such, loss of p75NTR results in exuberant axon branching, which is abrogated upon subsequent loss of either TNFR1 or TNFα. While p75NTR requires the presence of TNFR1 to suppress axon growth and branching in response to BDNF, TNFR1 displays no such dependence. This figure was created with Biorender.com and exported under a paid subscription.
We observe that p75NTR appears to be regulating axon complexity through two distinct mechanisms: 1) BDNF-dependent reduction of axon complexity through inhibition of axon growth rate, and 2) suppression of progressive mTNFα reverse signaling. The mechanism by which p75NTR suppressed pro-growth mTNFα reverse signaling requires further investigation. While we had thought it likely that TNFR1 and p75NTR had either a compensatory or a co-dependent relationship, we were surprised to discover that p75NTR was overall antagonistic to TNFR1-TNFα reverse signaling while requiring TNFR1 forward signaling for ligand-dependent regressive events.
How is it that p75NTR might be dependent on TNFR1 forward signaling to respond to BDNF? In general, death receptors are thought to initiate similar pathways such as JNK, NFkB, Rho, and Caspase 8/10 cascades (Locksley et al., 2001; Hempstead, 2002). It is possible that individual TNFR family members induce subtle variations in distinct pathways, the combinations of which may be sufficient to create diversified outcomes. Alternatively, interactions between family members may be permissive for particular signaling functionality through a shift in subcellular locale. A portion of p75NTR-dependent signaling is thought to require hierarchical cleavage by BACE followed by gamma secretase in order to liberate the intracellular domain (Kenchappa et al., 2010; Kanning et al., 2003). Importantly, BACE and gamma secretase activity are thought to be most active in endosomes and lipid rafts, respectively (Osenkowski et al., 2008; Das et al., 2013). Further, a recent study demonstrated that p75NTR, upon ligand binding, preferentially associates with lipid raft microdomains, facilitating pro-apoptotic signaling (Marchetti et al., 2019). It is known that TNFR1 is enriched in lipid rafts whereas p75NTR is detected in both raft and non-raft compartments (Cottin et al., 2002; Zhang et al., 2013). Our finding that homomeric TNFR1 as well as p75NTR:TNFR1 complexes exist in punctate microdomains suggests that, upon ligand binding, TNFR1 may deliver p75NTR to microenvironment(s) necessary for p75NTR cleavage and signaling, providing an explanation for the observed reliance of p75NTR on TNFR1 for its function.
A similar explanation may be provided for how p75NTR regulates the relative strengths of TNFR1-TNFα forward and reverse signaling. Perhaps this TNFR1-p75NTR complex sequesters TNFR1 from TNFα. This could either be through occlusion of TNFR1 cleavage by ADAM17 (Giai et al., 2013) or the binding site between TNFR1 and mTNFα. Another possibility is that p75NTR and mTNFα reverse signaling are independent pathways, both pruning and sprouting axons simultaneously and resulting in a function sum balance when unperturbed.
Regardless of the mechanism of p75NTR regulation of TNFα signaling, it is intimately linked to axon growth in suboptimal NGF environments. Especially intriguing is how mTNFα reverse signaling is necessary for low levels of NGF to promote axon growth (Fig. 5E). These results differ from Kisiswa and colleagues, where neurons were treated with slightly higher levels of NGF and show no such requirement. This in turn indicates that the 2 ng/ml NGF used in our experiments is capable of supporting axon growth, but sensitizes neurons to the effect of regressive signaling pathways. TNFα reverse signaling, however, is only necessary for NGF-promoted growth when p75NTR is present, as loss of p75NTR rescues the dramatic loss of axon arbors in the TNFα-deficient background. Thus, a single trophic signal, here NGF, is able to influence the refinement activity not just of p75NTR, but also that of related TNFR family members. Given that these proteins have distinct spatial distributions and functional roles, this would allow the cell to regulate progressive and regressive signals not just at the cell body in ways particular to p75NTR, but to diversify this response across the cell and in varying contexts. Further investigation is required to determine precisely how NGF influences pathways that control axon regression, especially the extent that NGF concentration or spatial distribution plays a role.
Several other examples of signaling systems, where either bidirectional or non-canonical reverse signaling in developmental patterning, have been well described. Eph-ephrin signaling is one such example and, like TNFR1-TNFα, is able to modulate different cellular events depending on the context in which the two proteins are expressed, including cell migration, diverse morphogenetic processes, and axon guidance. Indeed, a number of receptor/ligand pairs from the TNFR/TNF superfamily have recently been shown to participate in bidirectional signaling in the context of neural development. CD40-induced reverse signaling through CD40L is important for dendritic elaboration for striatal neurons (Carriba and Davies, 2017) and promotes innervation density by sympathetic neurons in an NGF-dependent manner (McWilliams et al., 2015). Activation of TWE-PRIL receptor, a fusion protein between two TNF superfamily members TWEAK and APRIL, opposes NGF-induced axon growth whereas soluble APRIL promotes axon growth (Howard et al., 2018). Recent work indicates that the critical period for TNFR/TNF superfamily signaling in the peripheral nervous system is narrow and defined by the expression of the signaling components (Kisiswa et al., 2013; Nolan et al., 2014). For example, sTNFR1 application induces reverse signaling at P0 and P5 in cultured rat neurons, but has no effect on axon growth at E18 or P10 (Nolan et al., 2014). Thus, TNFR-TNFα reverse signaling activity coincides with the developmental window for sympathetic innervation of target organs (Davies, 2009).
What is the purpose of juxtaposing two opposing and mutually activating signaling pathways present in the same cell? One would imagine that either the two would counterbalance one another leading to no net effect, or that one would become dominant over the other resulting in branch growth or elimination. In either case, the need for both members to be bi-directional signaling complex would seem to be obviated. However, the localization of TNFR superfamily bidirectional partners may give us further clues about the purpose of this arrangement. Davies and colleagues showed that while TNFR1 was restricted to the somas of sympathetic neurons, TNFα was broadly expressed across axons as well. Since TNFα, but not TNFR1, is localized to these growing axons (Kisiswa et al., 2013; Erice et al., 2019), it is likely that TNFα is a major contributor to pro-growth regulation of axon patterning. Thus, TNFα is positioned to receive and relay progressive signals from target tissue promoting axon elaboration whereas TNFR1 sharpens the response to competition as a pro-destruction cue at the cell body similar to p75NTR (Deppmann et al., 2008), In fact, due to where the ligand is derived in each signaling direction, we might consider TNFR1-TNFα bidirectional signaling predominantly as a divide between autocrine and paracrine signaling; TNFR1 activation occurs from autocrine sTNFα when NGF availability is low and paracrine mTNFα activation occurs distally from tissue rich with TNFR1 or TNFR2. Thus, the utility in having both systems may be derived from spatially separating the two. In culture, this spatial separation is perturbed, disrupting proper signaling by providing ligands globally. Thus the roles of forward and reverse TNFR-TNFα signaling may be obscured by the loss of each pathway’s spatial restriction.
Our work, in sympathetic neurons cultured in suboptimal NGF, shows that p75NTR and mTNFα signaling are the dominant regressive and progressive pathways, respectively. To appreciate the dynamic interaction of regressive and progressive pathways, full characterization of both temporal and spatial expression across the sympathetic chain and target tissues will be necessary. Given the size of the TNFR family, as well as the number of co-receptors reported to play a role in their function, we suggest that interactions between p75NTR and TNFR1 merely scratch the surface of how many heteromeric receptor combinations are possible. For example, p75-NTR has been reported to interact with Sortilin which mediates pro-neurotrophin signaling (Nykjaer et al., 2004), Nogo Receptor (NgR) which mediates Nogo, MAG, and OMGp signaling (Wang et al., 2002), and DR6 which mediates beta amyloid signaling (Hu et al., 2013). Given the wide range of cellular processes these proteins mediate and the plethora of ligands they respond to, a wide range of developmental responses may be derived from expressing them in different combinations. In this way, a complex combinatorial code of TNFR family interactions may act as a Rosetta stone to interpret patterns of relatively simple yet highly mobile developmental cues.
4. Materials and methods
4.1. Animals
All animals were maintained in a c57bl6 background. Transgenic lines were purchased from The Jackson Laboratory: p75NTR−/− (Lee et al., 1992; JAX stock #002213), TNFR1−/− (Peschon et al., 1998; JAX stock #003243), TNFR2−/− (Erickson et al., 1994; JAX #002620), and TNFα−/− (Pasparakis et al., 1996; JAX #005540). The following double knockouts were bred by crossing the previously mentioned lines: TNFR1−/−;p75NTR−/−, TNFα−/−;p75NTR−/−, and TNFR1−/−;TNFR2−/−. Wild-type controls were c57bl6. Genotyping was done on DNA extracted from tail as per REDExtract-N-Amp Tissue PCR Kit (Sigma, E7526, T3073, N3910, and R4775) using the following primer sets recommended by Jackson Laboratory for each line.
p75NTR−/−
Common: GCT CAG GAC TCG TGT TCT CC
Mutant: TGG ATG TGG AAT GTG TGC GAG
Wild-type: CCA AAG AAG GAA TTG GTG GA
TNFR1−/−
Common: AGA GCT CCA GGC ACA AGG GC
Mutant: CCG GTG GAT GTG GAA TGT GTG
Wild-type: AAC GGG CCA GAC CTC GGG T
TNFR2−/−
Common: TAG AGC TCC AGG CAC AAG G
Mutant: AGA CTG CCT TGG GAA AAG CG
Wild-type: AGC CAC TGG GTA TTT CTG GA
TNFα−/−
Common: TAG CCA GGA GGG AGA ACA GA
Mutant: CGT TGG CTA CCC GTG ATA TT
Wild-type: AGT GCC TCT TCT GCC AGT TC
All experiments were carried out in compliance with the Association for Assessment of Laboratory Animal Care policies and approved by the University of Virginia Animal Care and Use Committee.
4.2. Primary neuronal culture
Superior cervical ganglia (SCG) dissected from postnatal day 0 (P0) mice were serially enzymatically dissociated, first with 4 mg/ml Collagenase type II (Worthington, LS004176), 0.2 mg/ml Deoxyribonuclease I from bovine pancreas (Sigma, DN25), and 0.2 mg/ml Hyaluronidase from bovine testes (Sigma, H3884) in BSA (Sigma, A9647) for 20 min, then with 3 mg/ml Trypsin (Sigma, T4799) for 15 min (Keeler et al., 2017). Following enzymes, the SCGs were triturated gently with a fire-polished glass pipette. After the ganglia were completely dissociated, they were grown in 4.5 g/L D-Glucose/L-Glutamine DMEM (Gibco; 11965–092) supplemented with 10% fetal bovine serum (FBS; Gibco; 16000–044), 1 U/ml penicillin-streptomycin (Gibco; 15140–122), and either 2 ng/ml NGF (purified in-house from mouse salivary glands at an amount that is sufficient to maintain survival but unable to block death receptor signaling (Deppmann et al., 2008, Kisiswa et al., 2013)), or a combination of 2 ng/ml NGF and one of the following: 2 ng/ml TNFα (Peprotech, 315–01A), 250 ng/ml BDNF (Novus, NBP199674), 10 ng/ml TNFR1-Fc (R&D Systems, 430-RI-050), or 1 μg/ml α-NGF (Sigma, AB1528SP) neutralizing antibody (Kisiswa et al., 2013; Suo et al., 2015). Neurons were plated on cover glass coated with poly-D-lysine (PDK;Sigma, P7886) and Laminin (Fisher, 23017015) and hereafter incubated at 37 °C in 10% CO2. After 24 h, the neuronal cultures were fixed with 4% PFA (diluted from 16%, Fisher, PI 28908) for 10 min.
4.3. In vitro axon complexity measurement
Fixed neurons were blocked in 0.1% TritonX-100 (Sigma, X100) and 3% Normal Donkey Serum (Sigma, S30) stained overnight with a Class III β-tubulin antibody, TuJ1 (generous gift from A. Spano, 1:1000 for immunohistochemistry). Cells were washed and stained for 1 h at room temperature with goat anti-mouse Alexa 488 antibody (Life Technologies, A-11001, 1:500 for immunofluorescence), mounted with DAPI Fluoromount G (Fisher, OB010020) then imaged. Only isolated neurons where TuJ1 staining did not overlap with other neurons and where DAPI indicated that a cell body contained only a single nuclei were imaged. Axon complexity data was generated by Sholl analysis (Sholl, 1953) in increments of 10 μm with both Neuron Studio (Wearne et al., 2005) and FIJI’s Simple Neurite Tracer plugin (Longair et al., 2011). Briefly, Sholl analysis measures the number of intersections between neuritic processes and superimposed circles of increasing radius centered on the soma. Scorers were blinded to the conditions they were imaging for. Axon growth rate data was calculated by measurement of the longest axon length and number of branches were the number of complete segments traced, both using FIJI’s Simple Neurite Tracer plugin.
4.4. Construction of plasmids
HA-p75NTR and FLAG-TNFR1: The full-length coding sequence of wild-type rat p75NTR (Gift from J. Tuttle) and TNFR1 (Obtained from the mammalian gene collection) were expressed from the pcDNA 3.1 backbone and pExpress1 backbone, respectively. N-terminal hemagglutinin (HA) epitope and triple repeats of FLAG epitope was inserted between signal sequence and mature peptide sequence of p75NTR and TNFR1, respectively. p75-VN, p75-VC, TNFR1-VN, TNFR-VC: The full length coding sequence of wild-type rat p75NTR and TNFR1 were expressed from the pTREhyg backbone. Triple repeats of the FLAG epitope were inserted between the signal sequence and mature peptide sequence of p75-NGFR and TNFR1. In the C-terminus of p75NTR and TNFR1, Venus N- or C-terminal fragments were inserted.
4.5. HEK293 cell culture
Cells were kept at 37 °C in 5% CO2. HEK293FT cells were cultured in DMEM containing 20% FBS and 1 U/ml penicillin-streptomycin and split at 1:20 to 1:10 dilution every 72–96 h following a PBS wash and 0.05% trypsin-EDTA treatment (Gibco; 25200–056).
4.6. Immunoblot analysis
HEK293 cells were transfected with the constructs of HA-p75-NGFR and/or FLAG-TNFR1 using Lipofectamine 2000 overnight and then lysed in ice-cold lysis buffer containing 1% Triton X-100, 60 mM Octyl β-D-glucopyranoside (Sigma, O8001) and cOmplete, mini, EDTA-free protease inhibitor cocktail (Sigma, 11836153001). For immunoprecipitation, lysed sample was incubated with agarose-conjugated chicken anti-HA (Aves Lab, ET-HA100, 1:500 immunoprecipitation) overnight at 4 °C. After electrophoresis, gels were blotted to PVDF membranes. The blots were immunostained with anti-FLAG (Sigma, F1804), anti-HA, and anti-Tubulin (Cell Signaling Technologies, 86298) overnight at 4 °C, and then imaged using Odyssey Infrared system (LI-COR).
4.7. BiFC visualization
BiFC was performed as previously described (Hu and Kerppola, 2003). HEK293 cells were transfected with the BiFC constructs using Lipofectamine 2000 (Fisher, 11668019) according to manufacturer’s instructions for 8–10 h and then imaged using fluorescence microscopy (514 nm excitation, 8 s exposure). SCG cultures were grown for 24 h, then transfected with the BiFC constructs using Lipofectamine LTX with Plus Reagent (Fisher, A12621) according to manufacturer’s instructions. SCG cells were then imaged after 24 and 48 h using fluorescence microscopy (514 nm excitation, 8 s exposure).
4.8. Reverse transcription PCR of TNFR2
SCGs were pooled from whole litters of postnatal day 0 mice. RNA purification was performed using an RNeasy Mini Kit (Qiagen, 74104) according to manufacturer’s guidelines. The Superscript III First-Strand Kit (Fisher, 18080051) was used for cDNA synthesis and RT-PCR according to manufacturer’s guidelines. The same primers were used to synthesize TNFR2 cDNA as used for RT-PCR. See SFig. 2 for RT-PCR primers.
Supplementary Material
Acknowledgments
The authors are grateful to Barry Condron, Ali Güler, Nikki Watson, Sushanth Kumar, Vitaly Zimyanin, and Amrita Pathak for helpful discussions. The authors thank Martin Hetzer. The authors also thank Pam Neff for technical assistance. Rat p75NTR coding sequence was a generous gift from Jeremy Tuttle. This work was supported by NIH_NINDS grants F32NS103770 and R01NS091617 award to A.B.K. and C.D.D, respectively, and the NSF-Career Award 1453242, the Sloan Foundation, and UVa Fund for excellence in science and technology awarded to C.D.D.
Abbreviations:
- BDNF
brain-derived neurotrophic factor
- BiFC
bimolecular fluorescence complementation
- DR6
death receptor 6
- JNK
c-Jun N-terminal kinase
- MAG
myelin-associated glycoprotein
- mTNFα
membrane-bounded tumor necrosis factor α
- NGF
nerve growth factor
- p75NTR
p75 neurotrophic factor receptor
- NgR
Nogo receptor
- OMgp
oligodendrocyte myelin glycoprotein
- sTNFα
soluble tumor necrosis factor α
- TNFα
tumor necrosis factor α
- TNFR1
tumor necrosis factor receptor 1
- TNFR2
tumor necrosis factor receptor 2
- WT
wild-type
Footnotes
Declaration of competing interest
The authors declare no competing interests.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mcn.2020.103467.
References
- Bamji SX, Majdan M, Pozniak CD, Belliveau DJ, Aloyz R, Kohn J, Causing CG, Miller FD, 1998. The p75 neurotrophin receptor mediates neuronal apoptosis and is essential for naturally occurring sympathetic neuron death. J. Cell Biol 140, 911–923. 10.1083/jcb.140.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker V, Middleton G, Davey F, Davies AM, 2001. TNFalpha contributes to the death of NGF-dependent neurons during development. Nat. Neurosci 4, 1194–1198. 10.1038/nn755. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA, 2000. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 14, 2919–2937. 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Bredesen DE, Ye X, Tasinato A, Sperandio S, Wang JJ, Assa-Munt N, Rabizadeh S, 1998. p75NTR and the concept of cellular dependence: seeing how the other half die. Cell Death Differ. 5, 365–371. 10.1038/sj.cdd.4400378. [DOI] [PubMed] [Google Scholar]
- Carriba P, Davies AM, 2017. CD40 is a major regulator of dendrite growth from developing excitatory and inhibitory neurons. Elife 6 10.7554/eLife.30442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ, 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288, 2351–2354. 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- Chao MV, 2003. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat. Rev. Neurosci 4, 299–309. 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Cottin V, Doan JES, Riches DWH, 2002. Restricted localization of the TNF receptor CD120a to lipid rafts: a novel role for the death domain. J. Immunol 168, 4095–4102. 10.4049/jimmunol.168.8.4095. [DOI] [PubMed] [Google Scholar]
- Das U, Scott DA, Ganguly A, Koo EH, Tang Y, Roy S, 2013. Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron 79, 447–460. 10.1016/j.neuron.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AM, 2009. Extracellular signals regulating sympathetic neuron survival and target innervation during development. Auton. Neurosci. 151, 39–45. 10.1016/j.autneu.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Deckwerth TL, Johnson EM, 1993. Temporal analysis of events associated with programmed cell death (apoptosis) of sympathetic neurons deprived of nerve growth factor. J. Cell Biol 123, 1207–1222. 10.1083/jcb.123.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinhardt K, Kim T, Spellman DS, Mains RE, Eipper BA, Neubert TA, Chao MV, Hempstead BL, 2011. Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Sci. Signal. 4, ra82 10.1126/scisignal.2002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppmann CD, Mihalas S, Sharma N, Lonze BE, Niebur E, Ginty DD, 2008. A model for neuronal competition during development. Science 320, 369–373. https://oi.org/10.1126/science.1152677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissner G, Kirchner S, Lindner H, Kolch W, Janosch P, Grell M, Scheurich P, Andreesen R, Holler E, 2000. Reverse signaling through transmembrane TNF confers resistance to lipopolysaccharide in human monocytes and macrophages. J. Immunol. 164, 6193–6198. 10.4049/jimmunol.164.12.6193. [DOI] [PubMed] [Google Scholar]
- Elmore S, 2007. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol. 35, 495–516. 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erice C, Calhan OY, Kisiswa L, Wyatt S, Davies AM, 2019. Regional differences in the contributions of TNF reverse and forward signaling to the establishment of sympathetic innervation. Dev. Neurobiol. 79, 317–334. 10.1002/dneu.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson SL, de Sauvage FJ, Kikly K, Carver-Moore K, Pitts-Meek S, Gillett N, Sheehan KC, Schreiber RD, Goeddel DV, Moore MW, 1994. Decreased sensitivity to tumour-necrosis factor but normal T-cell development in TNF receptor-2-deficient mice. Nature 372, 560–563. 10.1038/372560a0. [DOI] [PubMed] [Google Scholar]
- Fitzsimonds RM, Poo MM, 1998. Retrograde signaling in the development and modification of synapses. Physiol. Rev. 78, 143–170. 10.1152/physrev.1998.78.1.143. [DOI] [PubMed] [Google Scholar]
- Gamage KK, Cheng I, Park RE, Karim MS, Edamura K, Hughes C, Spano AJ, Erisir A, Deppmann CD, 2017. Death receptor 6 promotes wallerian degeneration in peripheral axons. Curr. Biol. 27, 890–896. 10.1016/j.cub.2017.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giai C, Gonzalez C, Ledo C, Garofalo A, Di Genaro MS, Sordelli DO, Gomez MI, 2013. Shedding of tumor necrosis factor receptor 1 induced by protein A decreases tumor necrosis factor alpha availability and inflammation during systemic Staphylococcus aureus infection. Infect. Immun 81, 4200–4207. 10.1128/IAI.00593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebova NO, Ginty DD, 2004. Heterogeneous requirement of NGF for sympathetic target innervation in vivo. J. Neurosci 24, 743–751. 10.1523/JNEUROSCI.4523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell M, Douni E, Wajant H, Löhden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, Scheurich P, 1995. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 83, 793–802. 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Levi-Montalcini R, 1949. Proliferation, differentiation and degeneration in the spinal ganglia of the chick embryo under normal and experimental conditions. J. Exp. Zool 111, 457–501. 10.1002/jez.1401110308. [DOI] [PubMed] [Google Scholar]
- Hempstead BL, 2002. The many faces of p75NTR. Curr. Opin. Neurobiol 12, 260–267. 10.1016/S0959-4388(02)00321-5. [DOI] [PubMed] [Google Scholar]
- Howard L, Wosnitzka E, Okakpu D, White MA, Wyatt S, Davies AM, 2018. TWE-PRIL reverse signalling suppresses sympathetic axon growth and tissue innervation. Development 145 10.1242/dev.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C-D, Kerppola TK, 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539–545. 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Lee X, Shao Z, Apicco D, Huang G, Gong BJ, Pepinsky RB, Mi S, 2013. A DR6/p75(NTR) complex is responsible for β-amyloid-induced cortical neuron death. Cell Death Dis. 4, e579 10.1038/cddis.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen P, Giehl K, Nyengaard JR, Teng K, Lioubinski O, Sjoegaard SS, Breiderhoff T, Gotthardt M, Lin F, Eilers A, Petersen CM, Lewin GR, Hempstead BL, Willnow TE, Nykjaer A, 2007. Roles for the pro-neurotrophin receptor sortilin in neuronal development, aging and brain injury. Nat. Neurosci 10, 1449–1457. 10.1038/nn2000. [DOI] [PubMed] [Google Scholar]
- Juhász P, Mester A, Biró A-J, Héjj G, Poór G, 2014. Clinical and radiological dissociation of anti-TNF plus methotrexate treatment in early rheumatoid arthritis in routine care: results from the ABRAB study. BMC Musculoskelet. Disord. 15, 251 10.1186/1471-2474-15-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanning KC, Hudson M, Amieux PS, Wiley JC, Bothwell M, Schecterson LC, 2003. Proteolytic processing of the p75 neurotrophin receptor and two homologs generates C-terminal fragments with signaling capability. J. Neurosci 23, 5425–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeler AB, Suo D, Park J, Deppmann CD, 2017. Delineating neurotrophin-3 dependent signaling pathways underlying sympathetic axon growth along intermediate targets. Mol. Cell. Neurosci 82, 66–75. 10.1016/j.mcn.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchappa RS, Tep C, Korade Z, Urra S, Bronfman FC, Yoon SO, Carter BD, 2010. p75 neurotrophin receptor-mediated apoptosis in sympathetic neurons involves a biphasic activation of JNK and up-regulation of tumor necrosis factor-alpha-converting enzyme/ADAM17. J. Biol. Chem 285, 20358–20368. 10.1074/jbc.M109.082834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiswa L, Osório C, Erice C, Vizard T, Wyatt S, Davies AM, 2013. TNFα reverse signaling promotes sympathetic axon growth and target innervation. Nat. Neurosci 16, 865–873. 10.1038/nn.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiswa L, Erice C, Ferron L, Wyatt S, Osório C, Dolphin AC, Davies AM, 2017. T-type Ca2+ channels are required for enhanced sympathetic axon growth by TNFα reverse signalling. Open Biol 7 10.1098/rsob.160288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Li E, Huber LJ, Landis SC, Sharpe AH, Chao MV, Jaenisch R, 1992. Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell 69, 737–749. 10.1016/0092-8674(92)90286-l. [DOI] [PubMed] [Google Scholar]
- Locksley RM, Killeen N, Lenardo MJ, 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104, 487–501. 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD, 2011. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454. 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Majdan M, Miller FD, 1999. Neuronal life and death decisions functional antagonism between the Trk and p75 neurotrophin receptors. Int. J. Dev. Neurosci 17, 153–161. 10.1016/S0736-5748(99)00016-7. [DOI] [PubMed] [Google Scholar]
- Majdan M, Walsh GS, Aloyz R, Miller FD, 2001. TrkA mediates developmental sympathetic neuron survival in vivo by silencing an ongoing p75NTR-mediated death signal. J. Cell Biol 155, 1275–1285. 10.1083/jcb.200110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti L, Bonsignore F, Gobbo F, Amodeo R, Calvello M, Jacob A, Signore G, Schirripa Spagnolo C, Porciani D, Mainardi M, Beltram F, Luin S, Cattaneo A, 2019. Fast-diffusing p75NTR monomers support apoptosis and growth cone collapse by neurotrophin ligands. Proc. Natl. Acad. Sci. U. S. A 10.1073/pnas.1902790116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Armanini MP, Ling LH, Phillips HS, 1994. Expression and coexpression of Trk receptors in subpopulations of adult primary sensory neurons projecting to identified peripheral targets. Neuron 12, 1161–1171. 10.1016/0896-6273(94)90323-9. [DOI] [PubMed] [Google Scholar]
- McWilliams TG, Howard L, Wyatt S, Davies AM, 2015. Regulation of autocrine signaling in subsets of sympathetic neurons has regional effects on tissue innervation. Cell Rep. 10, 1443–1449. 10.1016/j.celrep.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Schweigreiter R, Yamashita T, Rosenkranz K, Wekerle H, Barde Y-A, 2002. Tumor necrosis factor inhibits neurite outgrowth and branching of hippocampal neurons by a rho-dependent mechanism. J. Neurosci 22, 854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DDM, Tessier-Lavigne M, 2009. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457, 981–989. 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nolan AM, Collins LM, Wyatt SL, Gutierrez H, O’Keeffe GW, 2014. The neurite growth inhibitory effects of soluble TNFα on developing sympathetic neurons are dependent on developmental age. Differentiation 88, 124–130. 10.1016/j.diff.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Lee R, Teng KK, Jansen P, Madsen P, Nielsen MS, Jacobsen C, Kliemannel M, Schwarz E, Willnow TE, Hempstead BL, Petersen CM, 2004. Sortilin is essential for proNGF-induced neuronal cell death. Nature 427, 843–848. 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- O’Keeffe GW, Gutierrez H, Pandolfi PP, Riccardi C, Davies AM, 2008. NGF-promoted axon growth and target innervation requires GITRL-GITR signaling. Nat. Neurosci 11, 135–142. 10.1038/nn2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenkowski P, Ye W, Wang R, Wolfe MS, Selkoe DJ, 2008. Direct and potent regulation of gamma-secretase by its lipid microenvironment. J. Biol. Chem 283, 22529–22540. 10.1074/jbc.M801925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Alexopoulou L, Episkopou V, Kollias G, 1996. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J. Exp. Med 184, 1397–1411. 10.1084/jem.184.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon JJ, Torrance DS, Stocking KL, Glaccum MB, Otten C, Willis CR, Charrier K, Morrissey PJ, Ware CB, Mohler KM, 1998. TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J. Immunol. 160, 943–952. [PubMed] [Google Scholar]
- Qu Y, Zhao G, Li H, 2017. Forward and reverse signaling mediated by transmembrane tumor necrosis factor-alpha and TNF receptor 2: potential roles in an immunosuppressive tumor microenvironment. Front. Immunol 8, 1675 10.3389/fimmu.2017.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabizadeh S, Oh J, Zhong LT, Yang J, Bitler CM, Butcher LL, Bredesen DE, 1993. Induction of apoptosis by the low-affinity NGF receptor. Science 261, 345–348. 10.1126/science.8332899. [DOI] [PubMed] [Google Scholar]
- Sharma N, Deppmann CD, Harrington AW, St Hillaire C, Chen Z-Y, Lee FS, Ginty DD, 2010. Long-distance control of synapse assembly by target-derived NGF. Neuron 67, 422–434. 10.1016/j.neuron.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA, 1953. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat 87, 387–406. [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Park KJ, Hong EJ, Kramer BM, Greenberg ME, Kaplan DR, Miller FD, 2008. Developmental axon pruning mediated by BDNF-p75NTR-dependent axon degeneration. Nat. Neurosci 11, 649–658. 10.1038/nn.2114. [DOI] [PubMed] [Google Scholar]
- Sun M, Fink PJ, 2007. A new class of reverse signaling costimulators belongs to the TNF family. J. Immunol 179, 4307–4312. 10.4049/jimmunol.179.7.4307. [DOI] [PubMed] [Google Scholar]
- Suo D, Park J, Young S, Makita T, Deppmann CD, 2015. Coronin-1 and calcium signaling governs sympathetic final target innervation. J. Neurosci 35, 3893–3902. 10.1523/JNEUROSCI.4402-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood CK, Coulson EJ, 2008. The p75 neurotrophin receptor. Int J Biochem Cell Biol 40, 1664–1668. 10.1016/j.biocel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Vilar M, Charalampopoulos I, Kenchappa RS, Simi A, Karaca E, Reversi A, Choi S, Bothwell M, Mingarro I, Friedman WJ, Schiavo G, Bastiaens PIH, Verveer PJ, Carter BD, Ibáñez CF, 2009. Activation of the p75 neurotrophin receptor through conformational rearrangement of disulphide-linked receptor dimers. Neuron 62, 72–83. 10.1016/j.neuron.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chun S-J, Treloar H, Vartanian T, Greer CA, Strittmatter SM, 2002. Localization of Nogo-A and Nogo-66 receptor proteins at sites of axon-myelin and synaptic contact. J. Neurosci. 22, 5505–5515 (doi:20026582). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR, 2005. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience 136, 661–680. 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- Wheeler MA, Heffner DL, Kim S, Espy SM, Spano AJ, Cleland CL, Deppmann CD, 2014. TNF-α/TNFR1 signaling is required for the development and function of primary nociceptors. Neuron 82, 587–602. 10.1016/j.neuron.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Tucker KL, Barde YA, 1999. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron 24, 585–593. 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Zhang YH, Khanna R, Nicol GD, 2013. Nerve growth factor/p75 neurotrophin receptor-mediated sensitization of rat sensory neurons depends on membrane cholesterol. Neuroscience 248, 562–570. 10.1016/j.neuroscience.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.