Abstract
Objective
To explore the causal relationships between sleep, major depressive disorder (MDD), and Alzheimer disease (AD).
Methods
We conducted bidirectional 2-sample Mendelian randomization analyses. Genetic associations were obtained from the largest genome-wide association studies currently available in UK Biobank (n = 446,118), Psychiatric Genomics Consortium (n = 18,759), and International Genomics of Alzheimer's Project (n = 63,926). We used the inverse variance–weighted Mendelian randomization method to estimate causal effects and weighted median and Mendelian randomization–Egger for sensitivity analyses to test for pleiotropic effects.
Results
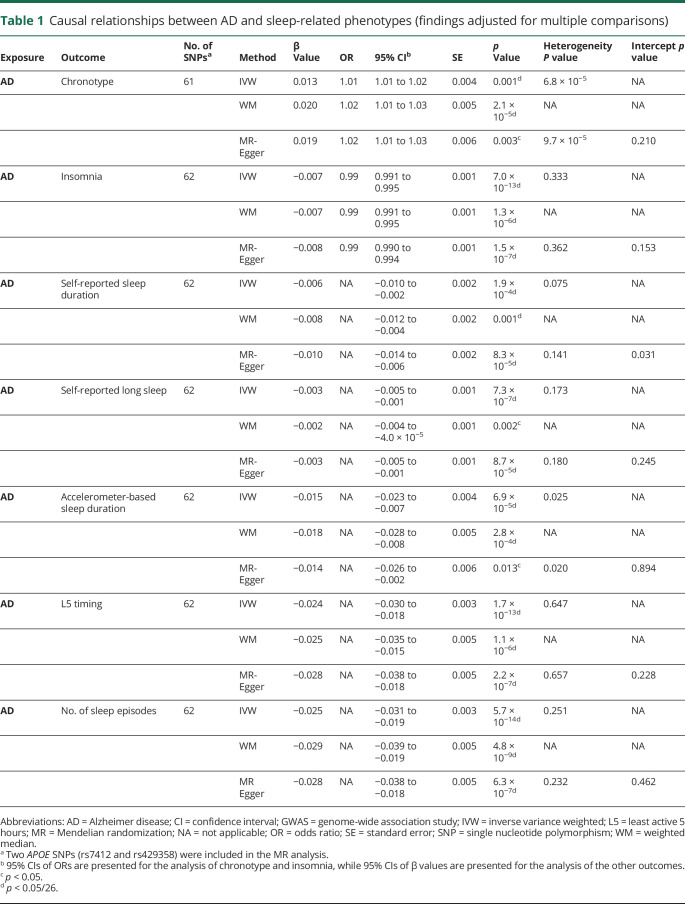
We found that higher risk of AD was significantly associated with being a “morning person” (odds ratio [OR] 1.01, p = 0.001), shorter sleep duration (self-reported: β = −0.006, p = 1.9 × 10−4; accelerometer based: β = −0.015, p = 6.9 × 10−5), less likely to report long sleep (β = −0.003, p = 7.3 × 10−7), earlier timing of the least active 5 hours (β = −0.024, p = 1.7 × 10−13), and a smaller number of sleep episodes (β = −0.025, p = 5.7 × 10−14) after adjustment for multiple comparisons. We also found that higher risk of AD was associated with lower risk of insomnia (OR 0.99, p = 7 × 10−13). However, we did not find evidence that these abnormal sleep patterns were causally related to AD or for a significant causal relationship between MDD and risk of AD.
Conclusion
We found that AD may causally influence sleep patterns. However, we did not find evidence supporting a causal role of disturbed sleep patterns for AD or evidence for a causal relationship between MDD and AD.
Aging populations across the world have led to an increasing prevalence of both Alzheimer disease (AD) and depression.1–3 The comorbidity of depressive disorders and late-life neurodegenerative diseases, including AD, has been widely reported.4–6 However, it is not known if any causal relationship exists between them or, alternatively, whether their co-occurrence is due to confounding or common risk factors such as aging. A genome-wide association study (GWAS) did not find evidence for a shared genetic architecture between depression and AD,7 supporting roles for common nongenetic risk factors. Sleep habits are important aspects of lifestyle, and abnormal sleep patterns are among the clinical signs and symptoms of both depression and AD. Sleep deprivation affects brain function and is associated with cognitive decline, anxiety, and depression.8 Clinical associations have been reported between major depressive disorder (MDD) and AD and are supported indirectly by their common effects on hippocampal atrophy and the involvement of molecular pathways related to oxidative stress in the progression of both diseases.9 However, potential causal relationships between sleep habits and MDD or AD have not been explored directly at a population level to the best of our knowledge.
We hypothesized that sleep causally affects MDD and AD but that there is no causal relationship between MDD and AD. To test these hypotheses, we investigated causal relationships in the triad of sleep, MDD, and AD using Mendelian randomization (MR) analyses. The MR approach overcomes unmeasured confounding and reverse causation in observational studies.10 Understanding these causal relationships will elucidate the mechanisms responsible for development of these diseases, better describe potential interactions between depression and AD, and further test the hypothesis that interventions to improve sleep patterns could modify disease risks.
Methods
Standard protocol approvals, registrations, and patient consents
This study used summary data published by multiple GWAS; patient consents were obtained by corresponding studies. This study is reported following the Strengthening the Reporting of Observational Studies in Epidemiology reporting guideline.
Study design
We conducted a bidirectional 2-sample MR study to investigate causal relationships between 6 sleep-related phenotypes, MDD, and AD.
Data sources
Sleep-related phenotypes
GWAS of sleep-related phenotypes were based on data from the UK Biobank, a population-based prospective study.11 We investigated sleep using a number of sleep-related phenotypes. The phenotypes either were self-reported (chronotype, insomnia, sleep duration) or were estimated with the accelerometer (the number of sleep episodes, sleep duration, least active 5 hours [L5] timing, and sleep efficiency). We chose instrumental variables and extracted summary statistics for the association of genetic variants with sleep-related phenotypes using a number of recent GWAS as listed in supplementary table e-1, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt.
Chronotype
Chronotype is considered the proclivity for earlier or later timing of sleep.12 An individual who prefers going to bed and waking earlier is considered a “morning person,” while a person who prefer going to bed and waking later is considered an “evening person.”12 We obtained the genetic associations of self-reported chronotype from a GWAS among 403,195 individuals of European ancestry from the UK Biobank.12 Chronotype was defined by 1 question (“Do you consider yourself to be?”). Participants who considered themselves “definitely a ‘morning’ person” or “more a ‘morning’ than ‘evening’ person” were included as morning persons (nmorning = 252,287); participants who considered themselves “definitely an ‘evening’ person” or “more an ‘evening’ than a ‘morning’ person” were included as evening persons as the reference (nevening = 150,908). Participants who responded with “do not know” or “prefer not to answer” were excluded.
Insomnia
Insomnia refers to the difficulty in falling asleep or maintaining sleep.13 We obtained the genetic associations of insomnia from the GWAS among 237,627 individuals of European ancestry from the UK Biobank.13 Insomnia was assessed by self-reports with the question, “do you have trouble falling asleep at night, or do you wake up in the middle of the night?” Individuals who reported “usually” were considered to have frequent insomnia (ncase = 129,270), and those who reported “never/rarely” were considered controls (ncontrol = 237,627).
Self-reported sleep duration
Self-reported sleep duration refers to the number of hours of sleep an individual get in every 24 hours (including naps).14 Genetic associations of self-reported sleep duration were obtained from a GWAS among 446,118 individuals of UK Biobank.14 Participants responded to the question “about how many hours sleep do you get in every 24 hours? (please include nap)” with hour increments. Extreme responses of <3 hours or >18 hours were not included in the GWAS analysis. Self-reported sleep duration was used as a continuous variable. Two binary variables were derived from self-reported sleep duration, i.e., short sleep and long sleep. Short sleep was defined as a sleep duration <7 hours, while long sleep was defined as a sleep duration ≥9 hours. Similar cutoffs were used in previous prospective studies on the effect of sleep duration.15 The GWAS on short sleep compared the short sleep group (nShortSleep = 106,192) to the reference group (≥7 and <9 hours of sleep, nreference = 305,742). Accordingly, the GWAS on long sleep compared the long sleep group (nLongSleep = 34,184) to the same reference group as in the GWAS on short sleep.
Accelerometer-based sleep-related phenotypes
A triaxial accelerometer device (Axivity AX3, Axivity, Newcastle Upon Tyne, UK) was used continuously for up to 7 days by 103,711 UK Biobank participants.16 After the removal of individuals with low measurement quality or poor wear time (<72 hours), data were available for ≈85,000 UK Biobank participants. Genetic associations of accelerometer-based sleep-related phenotypes were obtained from a GWAS among 85,670 individuals of the UK Biobank.16 We focused on 4 accelerometer-based sleep-related phenotypes in this study: number of sleep episodes, sleep duration, L5 timing, and sleep efficiency. Individual activity levels based on wrist-worn accelerometer were used to distinguish movement from nonmovement and to estimate the sleep period time (SPT) window. During the SPT window, a period of at least 5 minutes with a change <5° on the z-axis (the dorsal-ventral direction in the anatomic position) was considered 1 sleep episode. A number of sleep episodes <5 or >30 was excluded. Duration of each sleep episode was added up to determine the total sleep duration. Sleep durations <3 hours or >12 hours were excluded. The L5 timing was defined as the 5-hour period with the minimum average acceleration starting from the previous midnight, which indicates the timing when the individuals were asleep. The midpoint of the L5 was used in the GWAS analysis. Sleep efficiency was calculated as sleep duration divided by the duration of the SPT window.
Major depressive disorder
Genetic associations with MDD were obtained from the publicly available GWAS among individuals of European ancestry (ncase = 9,240 and ncontrol = 9,519) contributed from the Psychiatric Genomics Consortium database (supplementary table e-2, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt).17 Cases of MDD were defined as clinically diagnosed MDD according to the DSM-IV.
Alzheimer disease
Genetic associations with AD were obtained from the meta-analysis of GWAS on individuals of European ancestry (ncase = 21,982 and ncontrol = 41,944) contributed by the International Genomics of Alzheimer's Project (IGAP) (supplementary table e-3, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt).18 Details of the IGAP were published elsewhere.18 In brief, the meta-analysis comprises 4 GWAS, namely the Alzheimer's Disease Genetics Consortium, the Cohort for Heart and Ageing Research in Genomic Epidemiology consortium, the European Alzheimer's Disease Initiative consortium, and the Genetic and Environmental Risk in Alzheimer's Disease consortium. Either postmortem autopsy or clinical examination was used to evaluate the participants for AD in the individual studies.18 Across all cohorts included in the meta-analysis, the mean age at onset of AD in cases ranged from 71.1 to 82.6 years, and the mean age at examination of controls ranged from 51.0 to 78.9 years.18
We performed an additional analyses using a larger meta-analysis of AD/AD-by-proxy in individuals of European ancestry (ncase = 71,880 and ncontrol = 383,378) (supplementary table e-3, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt).19 The meta-analysis comprises GWAS on cases with AD and GWAS on AD-by-proxy cases. GWAS on cases with AD were contributed by the IGAP, Alzheimer work group initiative of the Psychiatric Genomic Consortium, and Alzheimer's Disease Sequencing Project (ncase = 24,087 and ncontrol = 55,058). Cases of AD were diagnosed by physician examination or autopsy confirmation. GWAS of AD-by-proxy cases was conducted with UK Biobank data. AD-by-proxy status was based on self-reported parents' diagnoses of AD. A total of 47,793 participants had 1 or both parents affected. AD-by-proxy cases with 2 affected parents were up-weighted. Unaffected parents were weighted by the age or age at death to account for late-onset AD. The author reported a genetic correlation of 0.81 between AD status and AD-by-proxy status.19 We used the publicly available summary statistics for the meta-analysis of GWAS on AD and AD-by-proxy cases.
Statistical analysis
To assess the causal relationship between sleep-related phenotypes, MDD, and AD, we performed bidirectional MR analysis for each pair of traits in this triad. For sleep-related phenotypes and AD, we chose the genetic instruments using genome-wide significant threshold (p < 5 × 10−8). For MDD, we used an arbitrary threshold of 1 × 10−4. We removed correlated single nucleotide polymorphisms (SNPs) (r2 > 0.1) by keeping the SNP with the smallest p value for the association with the exposure. For all exposures, we filtered the instruments for F statistics >10 to mitigate potential effects of weak instrument bias using the genotype data.17 SNPs were aligned on the basis of their presumed effect allele.
We estimated SNP-specific associations using the Wald ratio (the ratio of the genetic association with the outcome to the genetic association with the exposure).20 We combined the SNP-specific associations for MR using inverse variance weighting (IVW).21 We also performed sensitivity analyses using weighted median and MR-Egger regression methods to assess the validity of instruments and to investigate influences of potential pleiotropic effects.21–23 The MR–pleiotropy residual sum and outlier method was used to identify potential outlier SNPs, which were excluded in the sensitivity analysis.24 We accounted for multiple comparisons of 26 groups of associations using a Bonferroni correction (p value threshold of 0.05/26 = 0.002).
All statistical analyses were performed with the TwoSampleMR package25 in R 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria).
Data availability
All data used for the analyses are publicly available. Supplementary data are available from Dryad (doi.org/10.5061/dryad.ffbg79cqt).
Results
In this study, we investigated the causal relationships between sleep-related phenotypes, MDD, and AD by conducting bidirectional MR analysis between each pair of traits. The numbers of instrumental variables (i.e., SNPs associated with exposure) varied for sleep-related phenotypes, MDD, and for AD (supplementary table e-4, doi.org/10.5061/dryad.ffbg79cqt). The full results are available in supplementary table e-5 and supplementary figures e-1 through e-38.
Sleep and MDD
We did not find causal relationships between sleep-related phenotypes and MDD in either direction (supplementary figures e-1–e-18, doi.org/10.5061/dryad.ffbg79cqt).
MDD and AD
The MR estimates of the causal relationships between MDD and AD were positive in both directions using MR with IVW, but neither was statistically significant (supplementary figures e-19 and e-20, doi.org/10.5061/dryad.ffbg79cqt).
Sleep and AD
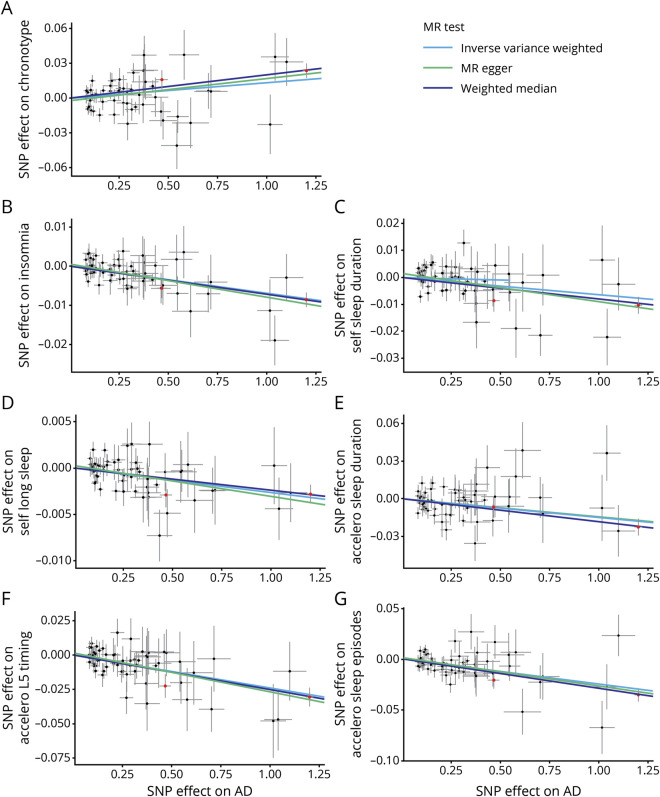
Genetically higher risks of AD were associated with being a morning person, a lower risk of insomnia, a shorter self-reported and accelerometer-based sleep duration, being less likely to report long sleep, an earlier L5 timing, and a smaller number of sleep episodes after adjustment for multiple comparisons (table 1 and figure 1 and supplementary figures e-21–e-29, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt). MR findings using IVW, weighted median, and MR-Egger were consistent. Results based on the GWAS of AD/AD-by-proxy cases also were similar except that accelerometer-based sleep duration was the only significant sleep phenotype that appeared to be potentially caused by AD when IVW was used after adjustment for multiple comparisons. None of the MR analyses supported a causal effect of sleep-related phenotypes on AD risk (supplementary figures e-30–e-38, available from Dryad).
Table 1.
Causal relationships between AD and sleep-related phenotypes (findings adjusted for multiple comparisons)
Figure 1. Causal relationships between AD and sleep-related phenotypes in scatterplots (findings adjusted for multiple comparisons).
(A) Alzheimer disease (AD) → chronotype; (B) AD → insomnia; (C) AD → self-reported sleep duration; (D) AD → self-reported long sleep; (E) AD → accelerometer-based sleep duration; (F) AD → least active 5 hours (L5) timing; (G) AD → number of sleep episodes (APOE single nucleotide polymorphisms [SNPs] [rs7412 and rs429358] are labeled in red). MR = Mendelian randomization.
Because of the larger independent contribution of APOE ε4, a gene with pleiotropic effects influencing multiple disease processes that might contribute to AD risk,26 we performed a sensitivity analysis excluding APOE SNPs (rs7412 and rs429358). Without APOE ε4, the causal associations of AD with self-reported and accelerometer-based sleep duration were not significant (supplementary figures e-21–e-29, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt). However, all other causal associations of AD on sleep phenotypes explored here were supported.
Discussion
A systematic review (supplementary table e-6, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt) found an association between self-reported low sleep quality and depression or depressive symptoms among college students.27 Among older adults, the prevalence of coexisting sleep disturbances (defined as insomnia, poor sleep quality, and complaints of insomnia) and depressive symptoms was 10.6%.28 In prospective studies among older adults, a bidirectional relationship between sleep disturbances and depression also was reported (supplementary table e-6, available from Dryad).28 However, in our study, we did not find evidence for a causal relationship between sleep-related phenotypes and MDD. Apparent inconsistences between our finding and previous observations may be due in part to the different definitions of sleep-related phenotypes. Previous studies considered multiple sleep-related phenotypes together as a global concept of sleep disturbances, while we investigated a number of the commonly characterized sleep-related phenotypes separately. Our concern was that, given the complexity of sleep, the different sleep-related phenotypes provide measures of different elements of sleep behavior; none of the individual phenotypes alone nor their aggregation into a global measure has been validated as a surrogate for the full clinical concept of sleep. The different sleep-related phenotypes also may have individually distinct impacts on human health. Moreover, considered as individual measures, they have a precision that may increase analysis sensitivity.
The comorbidity of depression and AD has been reported widely.4,5 Previous systematic reviews (supplementary table e-7, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt) consistently suggested simple associations of depression with both all-cause dementia and AD.29,30 In addition, only a trajectory of increasing depressive symptoms was associated with higher risk of dementia.31 These findings suggest that depressive symptoms may be a prodrome of AD reflecting preclinical disease progression.32 Evidence for associations of MDD and AD at the clinical and molecular levels also has been reported.9 Reports have argued that oxidative stress leading to neuronal dysfunction could contribute to the development of both MDD and AD.9 The upregulation of brain nuclear factor-kβ and inflammatory cytokine production found in both MDD and AD potentially decreases neuronal proliferation in both diseases.9 However, we did not find epidemiologic evidence supporting a causal relationship between MDD and AD. As appropriate genetic instruments become available for late-onset depression, future work should revisit its causal role in AD33 because late-onset depression is related more proximately in time to clinical symptoms of AD.
Our study found that a genetically higher risk of AD is associated with several sleep-related phenotypes. Specifically, we found that those who are genetically at higher risk of AD are likely to have lower risk of insomnia and earlier L5 timing. Both the lower risk of insomnia and earlier L5 timing suggest that the individuals were less active in the first half of the night. These results from the study of people at high genetic risk of AD are not consistent with the observational findings of sleep disturbances in people who express AD clinically; e.g., previous studies (supplementary table e-8, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt) describe sleep disturbances as typical symptoms of AD and sleep disordered breathing, excessive daytime sleepiness, and insomnia as frequent sleep disorders in patients with dementia or mild cognitive impairment.34,35 Thus, disruption of the sleep-wake cycle—rather than insomnia—is prevalent in patients with AD.36 Sleep disturbances also can precede clinical expression of AD.34 Both short sleep duration and long sleep duration are associated with a higher risk of a broad range of cognitive disorders, including mild cognitive impairment, dementia, AD, and cognitive decline, suggesting a nonlinear relationship.37–39 In our study, we found causal associations of genetically higher risk of AD with shorter self-reported sleep duration, as did the earlier observational studies. While the evidence is not strong, on the basis of these data and our observations, we hypothesize that preclinical stages of AD and more severe, clinically expressed AD may have different effects on sleep phenotypes. These relationships highlight the potential importance of sleep management to improve the quality of life in patients with all stages of AD.
APOE is a particularly important genetic contributor to AD risk, alone accounting for ≈13% of the phenotypic variance of AD.40 While this may appear to make APOE SNPs strong instruments for assessing risk of AD with MR,10 because APOE is associated with multiple disorders besides AD,26 the use of APOE SNPs as instruments in an MR analysis for AD violates exclusion restriction assumptions for MR (that genetic instruments affect only the outcome of interest through the risk factor).10 While the pleiotropy test from MR-Egger regression suggested little pleiotropy (table 1), we performed an additional sensitivity analysis excluding the APOE SNPs. All causal associations of higher risk of AD with sleep patterns were sustained except for the observation of an association with shorter sleep duration. A possible interpretation of these results is that the MR estimate with inclusion of APOE may be influenced both by its impact on the genetic risk of AD and through its associations with other disorders.41
In this study, we explored the bidirectional relationship in the triad of sleep-related phenotypes, MDD, and AD. The application of MR circumvents the conundrum of unmeasured confounding and reverse causation in observational studies and thus can provide some insights into causal relationships. We have examined the assumptions of MR carefully and selected appropriate data sources for each association under investigation. For example, all SNPs used as instruments have an F statistic >10, indicating that weak instrument bias is unlikely. We also performed sensitivity analyses to explore the potential impact of pleiotropic effects of the SNPs used as instruments.
Our study has notable strengths. Specifically, we used data from the largest GWAS available for each trait or disorder under investigation, and we explored a wide range of sleep-related phenotypes, including those objectively estimated with an accelerometer. The objective estimates of sleep may be more precise and are less prone to bias and misclassification than self-reports.42 Nevertheless, there are several potential limitations of our study. (1) Overall, we investigated different exposure traits and used different sets of predictive SNPs, but the power of our analyses with the different instruments varies. (2) MR analysis assumes a lifetime exposure to the risk factor and is likely to overestimate the effect of clinical intervention on the outcome,43,44 so it cannot be assumed to suggest that an intervention to modify an associated factor will bring clinical benefits (although characterizing any causal relationships can direct hypothesis generation for discovery of biological mechanisms). (3) The most relevant critical exposure period cannot be distinguished with MR because the data available to us for the GWAS for sleep and incidence of AD were obtained only from middle-aged or older individuals (precluding tests of effects of exposure to abnormal sleep patterns in childhood or adolescence). (4) Biological mechanisms underlying different forms of dementia resembling AD may differ,45 and only ≈19% of the AD cases used for the GWAS used were autopsy confirmed (moreover, AD subtypes were not taken into account).18 (5) APOE SNPs may have both direct and indirect (e.g., through their influence on risks of other diseases) effects on risk of AD. (6) Sleep-related phenotypes were either self-reported or estimated with accelerometers rather than measured directly with polysomnography. (7) We used data from individuals of mainly European ancestry, although sleep habits may vary between cultural and ethnic groups. (8) Finally, null findings in our study could reflect lack of power; however, the 2-sample MR design increases statistical power by using multiple data sources,10 and our estimates of statistical power (assuming the observed effect and confidence interval in this study represent the true effect sizes)46 (supplementary table e-5, available from Dryad, doi.org/10.5061/dryad.ffbg79cqt) suggested that the lack of an effect of long sleep on AD is unlikely to be due to lack of power.
In this study, we found evidence supporting a potential causal influence of AD on sleep disturbances. However, we did not find evidence supporting a causal role of disturbed sleep patterns on AD, suggesting that observed associations between sleep disorders and AD may be due to reverse causation. We did not find evidence for significant causal relationships between MDD and AD. As a first step toward better understanding the basis for observed associations between depression and AD, future work could explore the genetic heterogeneity of depression syndromes to test for causal relationships between potentially etiologically distinct subtypes of depression (e.g., late-onset depression33) and AD.
Glossary
- AD
Alzheimer disease
- DSM-IV
Diagnostic and Statistical Manual of Mental Disorders, 4th edition
- GWAS
genome-wide association study
- IGAP
International Genomics of Alzheimer's Project
- IVW
inverse variance weighting
- L5
least active 5 hours
- MDD
major depressive disorder
- MR
Mendelian randomization
- SNP
single nucleotide polymorphism
- SPT
sleep period time
Appendix. Authors
Study funding
Study funded by the UK Dementia Research Institute.
Disclosure
J. Huang, and V. Zuber report no disclosures relevant to the manuscript. P.M. Matthews acknowledges consultancy fees from Adelphi Communications, Biogen, Celgene, and Roche. He has received honoraria or speakers' honoraria from Biogen, Novartis, and Roche, and has received research or educational funds from Biogen, GlaxoSmithKline, Nodthera, and Novartis. He is a paid member of the Scientific Advisory Board for Ipsen Pharmaceuticals. P. Elliott, J. Tzoulaki, and A. Dehghan report no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Global Burden of Disease Study Collaborators. Global, regional, and national burden of Alzheimer's disease and other dementias, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 2019;18:88–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whiteford HA, Degenhardt L, Rehm J, et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet 2013;382:1575–1586. [DOI] [PubMed] [Google Scholar]
- 3.Thielke SM, Diehr P, Unutzer J. Prevalence, incidence, and persistence of major depressive symptoms in the Cardiovascular Health Study. Aging Ment Health 2010;14:168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anisman H, Merali Z, Hayley S. Neurotransmitter, peptide and cytokine processes in relation to depressive disorder: comorbidity between depression and neurodegenerative disorders. Prog Neurobiol 2008;85:1–74. [DOI] [PubMed] [Google Scholar]
- 5.Hurley LL, Tizabi Y. Neuroinflammation, neurodegeneration, and depression. Neurotox Res 2013;23:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haaksma ML, Vilela LR, Marengoni A, et al. Comorbidity and progression of late onset Alzheimer's disease: a systematic review. PLoS One 2017;12:e0177044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson J, Russ TC, Adams MJ, et al. Assessing the presence of shared genetic architecture between Alzheimer's disease and major depressive disorder using genome-wide association data. Transl Psychiatry 2017;7:e1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhai L, Zhang H, Zhang D. Sleep duration and depression among adults: a meta-analysis of prospective studies. Depress Anxiety 2015;32:664–670. [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues R, Petersen RB, Perry G. Parallels between major depressive disorder and Alzheimer's disease: role of oxidative stress and genetic vulnerability. Cell Mol Neurobiol 2014;34:925–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies NM, Holmes MV, Davey Smith G. Reading mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones SE, Lane JM, Wood AR, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun 2019;10:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane JM, Jones SE, Dashti HS, et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet 2019;51:387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dashti HS, Jones SE, Wood AR, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun 2019;10:1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cappuccio FP, D'Elia L, Strazzullo P, Miller MA. Sleep duration and all-cause mortality: a systematic review and meta-analysis of prospective studies. Sleep 2010;33:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones SE, van Hees VT, Mazzotti DR, et al. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat Commun 2019;10:1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013;18:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkle BW, Grenier-Boley B, Sims R, et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 2019;51:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen IE, Savage JE, Watanabe K, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet 2019;51:404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for mendelian randomization. Stat Methods Med Res 2017;26:2333–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burgess S, Thompson SG. Interpreting findings from mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers AJ, Nemeroff CB. APOE: a risk factor for multiple disorders. Am J Geriatr Psychiatry 2012;20:545–548. [DOI] [PubMed] [Google Scholar]
- 27.Dinis J, Braganca M. Quality of sleep and depression in college students: a systematic review. Sleep Sci 2018;11:290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao YP, Han Y, Ma J, et al. Cooccurrence and bidirectional prediction of sleep disturbances and depression in older adults: meta-analysis and systematic review. Neurosci Biobehav Rev 2017;75:257–273. [DOI] [PubMed] [Google Scholar]
- 29.Cherbuin N, Kim S, Anstey KJ. Dementia risk estimates associated with measures of depression: a systematic review and meta-analysis. BMJ Open 2015;5:e008853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diniz BS, Butters MA, Albert SM, Dew MA, Reynolds CF III. Late-life depression and risk of vascular dementia and Alzheimer's disease: systematic review and meta-analysis of community-based cohort studies. Br J Psychiatry 2013;202:329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirza SS, Wolters FJ, Swanson SA, et al. 10-year trajectories of depressive symptoms and risk of dementia: a population-based study. Lancet Psychiatry 2016;3:628–635. [DOI] [PubMed] [Google Scholar]
- 32.Elsworthy RJ, Aldred S. Depression in Alzheimer's disease: an alternative role for selective serotonin reuptake inhibitors? J Alzheimers Dis 2019;69:651–661. [DOI] [PubMed] [Google Scholar]
- 33.Grayson L, Thomas A. A systematic review comparing clinical features in early age at onset and late age at onset late-life depression. J Affect Disord 2013;150:161–170. [DOI] [PubMed] [Google Scholar]
- 34.Brzecka A, Leszek J, Ashraf GM, et al. Sleep disorders associated with Alzheimer's disease: a perspective. Front Neurosci 2018;12:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guarnieri B, Adorni F, Musicco M, et al. Prevalence of sleep disturbances in mild cognitive impairment and dementing disorders: a multicenter Italian clinical cross-sectional study on 431 patients. Dement Geriatr Cogn Disord 2012;33:50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim MM, Gerstner JR, Holtzman DM. The sleep-wake cycle and Alzheimer's disease: what do we know? Neurodegener Dis Manag 2014;4:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu L, Sun D, Tan Y. A systematic review and dose-response meta-analysis of sleep duration and the occurrence of cognitive disorders. Sleep Breath 2018;22:805–814. [DOI] [PubMed] [Google Scholar]
- 38.Bokenberger K, Strom P, Dahl Aslan AK, et al. Association between sleep characteristics and incident dementia accounting for baseline cognitive status: a prospective population-based study. J Gerontol A Biol Sci Med Sci 2017;72:134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohara T, Honda T, Hata J, et al. Association between daily sleep duration and risk of dementia and mortality in a Japanese community. J Am Geriatr Soc 2018;66:1911–1918. [DOI] [PubMed] [Google Scholar]
- 40.Ridge PG, Hoyt KB, Boehme K, et al. Assessment of the genetic variance of late-onset Alzheimer's disease. Neurobiol Aging 2016;41:200.e213–200.e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satizabal CL, Samieri C, Davis-Plourde KL, et al. APOE and the association of fatty acids with the risk of stroke, coronary heart disease, and mortality. Stroke 2018;49:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doherty A, Jackson D, Hammerla N, et al. Large scale population assessment of physical activity using wrist worn accelerometers: the UK Biobank Study. PLoS One 2017;12:e0169649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol 2017;14:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess S, Butterworth AS, Thompson JR. Beyond Mendelian randomization: how to interpret evidence of shared genetic predictors. J Clin Epidemiol 2016;69:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira D, Pereira JB, Volpe G, Westman E. Subtypes of Alzheimer's disease display distinct network abnormalities extending beyond their pattern of brain atrophy. Front Neurol 2019;10:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess S. Sample size and power calculations in mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 2014;43:922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used for the analyses are publicly available. Supplementary data are available from Dryad (doi.org/10.5061/dryad.ffbg79cqt).