Abstract
Objective
To determine whether naturally occurring autoantibodies against the prion protein are present in individuals with genetic prion disease mutations and controls, and if so, whether they are protective against prion disease.
Methods
In this case–control study, we collected 124 blood samples from individuals with a variety of pathogenic PRNP mutations and 78 control individuals with a positive family history of genetic prion disease but lacking disease-associated PRNP mutations. Antibody reactivity was measured using an indirect ELISA for the detection of human immunoglobulin G1–4 antibodies against wild-type human prion protein. Multivariate linear regression models were constructed to analyze differences in autoantibody reactivity between (1) PRNP mutation carriers vs controls and (2) asymptomatic vs symptomatic PRNP mutation carriers. Robustness of results was examined in matched cohorts.
Results
We found that antibody reactivity was present in a subset of both PRNP mutation carriers and controls. Autoantibody levels were not influenced by PRNP mutation status or clinical manifestation of prion disease. Post hoc analyses showed anti-PrPC autoantibody titers to be independent of personal history of autoimmune disease and other immunologic disorders, as well as PRNP codon 129 polymorphism.
Conclusions
Pathogenic PRNP variants do not notably stimulate antibody-mediated anti-PrPC immunity. Anti-PrPC immunoglobulin G autoantibodies are not associated with the onset of prion disease. The presence of anti-PrPC autoantibodies in the general population without any disease-specific association suggests that relatively high titers of naturally occurring antibodies are well-tolerated.
Clinicaltrials.gov identifier
Prion diseases are diseases of the CNS that not only occur as sporadic and transmissible forms, but can also be transmitted through the germ line as autosomal dominant traits.1 Genetic prion diseases (gPrDs) account for ∼10%–15% of all prion diseases and are characterized by pathogenic, nonsynonymous mutations of the human prion protein gene PRNP.2 The most prevalent human prion disease, sporadic Creutzfeldt-Jakob disease (sCJD), is characterized by a rapidly progressive dementia and a short survival time (usually <1 year) from clinical onset.3 In contrast, PRNP mutation carriers often present with atypical phenotypes; for example, long survival rates can be observed in Gerstmann-Sträussler-Scheinker (GSS) disease.4
The cellular prion protein PrPC consists of an unstructured flexible tail (FT) on its N-terminal end and a C-terminal globular domain (GD).5 We showed in 2001 that humoral immunity against PrPC can protect against prion neuroinvasion.6 Antibodies against the FT of PrPC, or removal of amino acid residues from the FT, abrogate the neurotoxic effects of anti-PrPC-GD antibodies and reduce the toxicity of bona fide prions.7,8 Naturally occurring PrP antibodies may exist in the general population: for instance, reactivity against a 21-residue PrP peptide was observed in commercial pooled immunoglobulin,9 and a unique blood group has been observed in individuals homozygous for the E219K polymorphism.10
Clinical trials have yet to deliver an effective antiprion agent.11–14 An ongoing clinical study involves the administration of PRN100, a humanized antibody against PrPC-GD, to individuals with Creutzfeldt-Jakob disease (CJD).15 There is much hope that this trial will be successful, but the murine counterpart of PRN100, ICSM18, exhibits an on-target, dose-dependent toxicity, and whether a therapeutic window exists has not yet been established.16–18
The frequency of PRNP missense variants exceeds the reported gPrD prevalence, suggesting a spectrum of disease penetrance in gPrDs rather than complete penetrance of nonsynonymous PRNP mutations.19 The mechanisms by which these mutations induce disease are largely unclear. The majority of structural studies on human PrPC variants linked to gPrD failed to identify consistent effects on global protein stability.20 Age at onset in gPrD is highly variable, and typically middle age or older, which might suggest that a protective mechanism guards some individuals against the prion protein–induced toxicity.2 We hypothesized that subtle conformational alterations of pathogenic PrPC variants could stochastically generate immunogenic neoepitopes, which in turn might elicit a protective humoral anti-PrPC immune response. We therefore conducted an extensive search for such autoantibodies in individuals carrying pathogenic PRNP mutations, and in unaffected relatives as controls.
Methods
Standard protocol approvals, registrations, and patient consents
The Cantonal Ethics Committee of the Canton of Zurich approved this study (permit no. KEK-ZH Nr.2015-0514). This trial was registered at clinicaltrials.gov (no. NCT02837705). The protocol for this study was approved by the institutional review board at each participating institution with the University of Zurich being the lead regulatory site. Written informed patient consent was received by all individuals participating in this study.
Human participants and study design
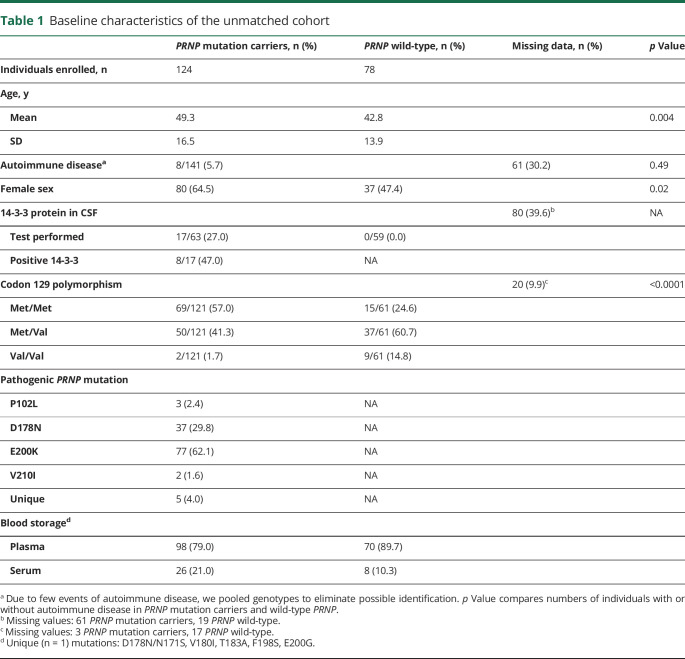
We defined PRNP mutation carriers as individuals with a nonsynonymous mutation in the open reading frame of the PRNP gene that was previously reported to be pathogenic.2 Between September 2015 and October 2018, we contacted both international patient organizations as well as national prion disease reference centers for further reuse of existing blood samples. Individuals at any age with a confirmed PRNP mutation were considered eligible for this study. Individuals with a confirmed PRNP mutation in a blood relative who did not undergo PRNP sequencing prior to enrollment in this study were also considered eligible if they gave consent for PRNP sequencing. Blood samples without information on age or sex were excluded from further analysis. PRNP wild-type individuals with neurologic or psychiatric symptoms indicative of gPrD were excluded from the study.21 Clinical manifestation of gPrD was defined as presence of both a pathogenic PRNP mutation and PrD-typical symptoms.21 The latter were assessed by clinical examination and neuropsychological assessment, in some cases complemented by ancillary tests such as presence of 14-3-3 proteins in CSF, real-time quaking-induced conversion assays, EEG, and MRI.22 Personal history of autoimmune disease and other immunologic disorders could be obtained in 141 participants. A detailed description of the patient cohort is given in table 1. For sensitivity analysis, cases and controls were matched on age (±5 years), sex, and blood sample type (i.e., serum or plasma).
Table 1.
Baseline characteristics of the unmatched cohort
PRNP genotyping
PRNP genotyping was performed using a modified version of the DNeasy Blood & Tissue Kit (Qiagen, Venlo, the Netherlands). Twenty microliters of proteinase K (600 mAU/mL) and 200 μL of 5 M guanidine hydrochloride with 1% Triton-X100 at pH = 5.0 were added to 200 μL of anticoagulated blood, vortexed thoroughly, and incubated for 24 hours at room temperature. Two hundred microliters EtOH (96%–100%) was added to the reaction and the rest of the DNA purification was performed according to the manufacturer's guidelines. The primer pair PRNP_up and PRNP_low (table e-1, doi.org/10.5061/dryad.08kprr4xk) was used in combination with Q5 high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA) to amplify the open reading frame from exon 2 of PRNP. Sanger sequencing was performed at the Department of Molecular Pathology (Institute of Surgical Pathology, University Hospital Zurich) using 4 different sequencing primers (PRNP_up, PRNP_up2, PRNP_low, PRNP_low2; table e-1, doi.org/10.5061/dryad.08kprr4xk). Sequencing traces were aligned to reference DNA from the Reference Sequence Database (RefSeq at National Center of Biotechnology Information, Bethesda, MD) using CLC Main Workbench (Qiagen) and packages sangerseqr23 and DECIPHER24 for Bioconductor25 in R.
Statistical analyses
We performed a priori testing of anti-PrPC autoantibody reactivity for the following hypotheses: (1) differences in anti-PrPC autoantibody reactivity between PRNP mutation carriers and PRNP wild-type individuals and (2) differences in anti-PrPC autoantibody reactivity between PRNP mutation carriers showing clinical signs of prion diseases and those without. All other analyses were performed post hoc. We used already established predictors of autoimmune disease such as age26 and sex27 as well as storage conditions known to affect antibody responses such as presence of coagulation factors28 as covariates in our multivariate regression model. Using the purposeful selection of covariates method as described previously,29 effects of covariates on autoantibody titers were tested by bivariate linear regression analyses using the Wald test and included for multivariate testing at a p value cutoff point of 0.25. In the multivariate model, covariates were removed if they were nonsignificant at the 0.1 α level or not a confounder, as determined by a change in the remaining parameter estimate greater than 20% as compared to the full model. PRNP mutation status, clinical signs of prion disease, and PRNP codon 129 polymorphism were added after establishment of significant confounders. In matched cohorts, multivariate models were adjusted for matching factors.
All values are given as average ± SD unless mentioned otherwise. For analysis, autoantibody titers were log10-transformed, and reported β coefficients and confidence intervals represent back-transformed values. Normality was tested using the D'Agostino-Pearson normality test. For values following a Gaussian distribution, differences between 2 groups were compared using 2-tailed Student t test. For not normally distributed values, Mann-Whitney U test was used for comparison of 2 groups. For comparison of categorical variables, Fisher exact test and χ2 test were used for comparison of 2 and more than 2 groups, respectively. Pearson correlation coefficient was computed for data sampled from Gaussian distributions and Spearman ρ for those sampled from non-Gaussian distributions. Matching of cases and controls was done using the find.matches function from the Hmisc package in R. We used lm for R for linear regression analysis. Python and R were used for statistical analysis and data visualization was performed using Prism 7 (GraphPad).
Data availability
The study participants, if they have not undergone predictive testing themselves, participated under the condition of not knowing their PRNP genotype. Due to the relatively small sample size and risk of de-identification, all raw study data involving human participants were made available to the editors and reviewers but will not be made available publicly. Supplementary data, as well as DNA sequences of gene blocks used for construction of humanized antibodies and human PrPC-AviTag, are available at Dryad (doi.org/10.5061/dryad.08kprr4xk).
Results
Description of the cohort
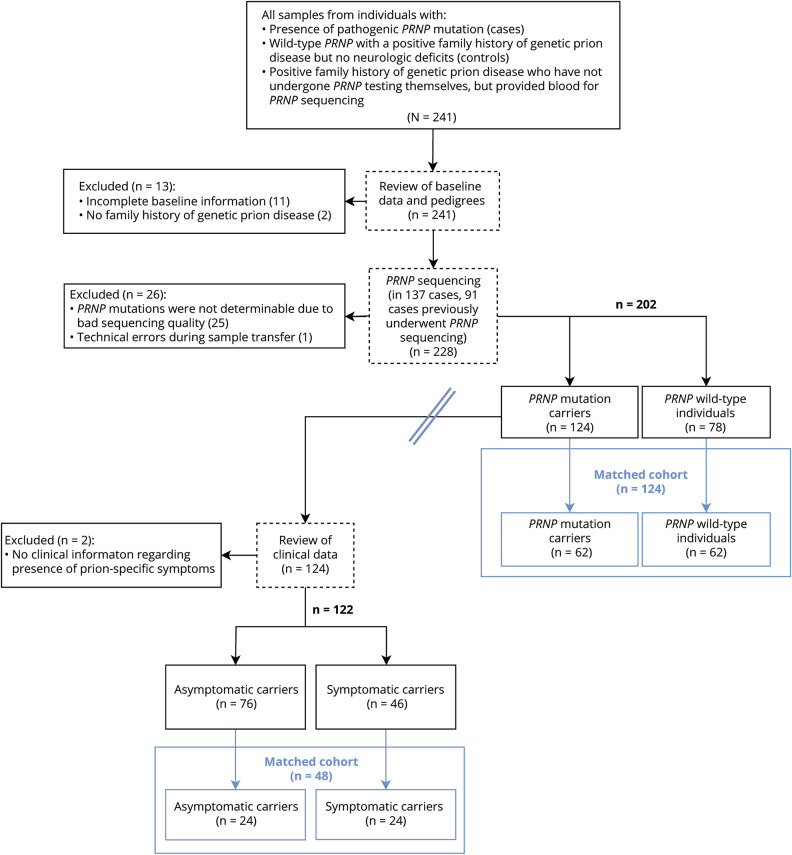
We received blood samples and clinical information from a total of 241 individuals and selected 202 unmatched cases and controls for this analysis (figure 1). To test the robustness of our results, we matched 64 cases on 64 controls based on age (±5 years), sex, and blood storage conditions (i.e., serum/plasma; table e-2, doi.org/10.5061/dryad.08kprr4xk). Anti-PrPC autoantibody reactivity was measured by a sandwich ELISA; a description of the assay is provided in extended text and figures e-1 and e-2 (doi.org/10.5061/dryad.08kprr4xk). Briefly, blood samples were diluted over a range of >2 logs and bound autoantibodies were detected with anti-human IgG antibodies. Antibody titers are expressed as negative common logarithm of the half-maximal effective concentration (figure e-1E). Anti-PrPC antibody reactivity was independent of serum IgG levels (Spearman ρ = 0.07, p = 0.69; figure 2A). The age of probands did not influence the IgG levels (Pearson r = 0.33, p = 0.16). To confirm our ability to detect human antibodies against specific targets, we tested a subset of individuals for the presence of IgG against EBNA. A total of 4/5 PRNPWT and 16/16 PRNPMut individuals tested positive (corresponding to 95% positive individuals), in line with anti-EBNA IgG seroprevalence in the general population (figure 2B).30
Figure 1. Flowchart of patient selection.
Double line indicates cohorts selected for comparison of anti-PrPC autoantibody titers from individuals carrying wild-type or mutated PRNP alleles (right of double line) and cohort selected for comparing anti-PrPC autoantibody titers of symptomatic vs asymptomatic mutation carriers (left of double line). Blue boxes indicate matched cohorts.
Figure 2. Correlation of anti-PrPC autoantibody reactivity with total immunoglobulin G (IgG) levels, IgG anti–Epstein-Barr virus (EBV) autoantibodies, and change of autoantibody titers over time.
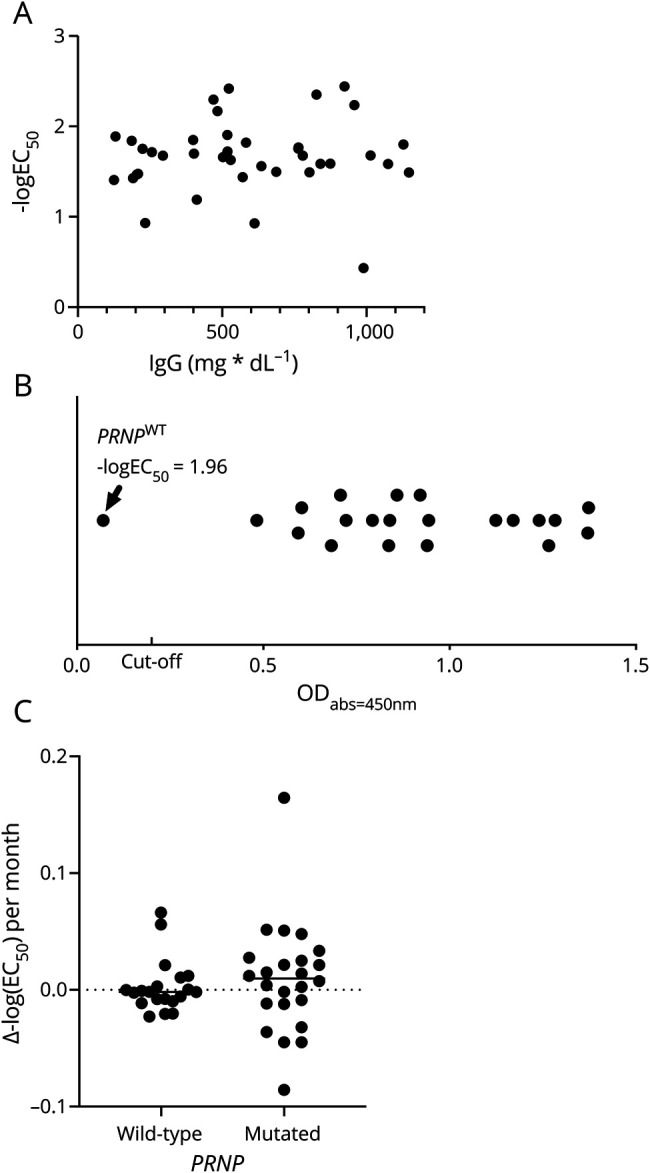
(A) Correlation of total IgG with anti-PrPC autoantibody titers. (B) Qualitative assessment of anti–Epstein-Barr nuclear antigen (EBNA) IgG antibodies in blood shows 1 PRNPWT individual without detectable anti-EBNA IgG antibodies. Cutoff: ODabs = 450 nm (optical density at absorbance λ = 450 nm) = 0.2 according to the manufacturer's guidelines. (C) In 2 subsequent blood drawings, mean change in antibody titers per year is stable and similar between PRNP mutation and wild-type carriers, but variance is larger in PRNP mutation carriers.
Prevalence of anti-PrPC autoantibodies in PRNP mutation carriers
The presence of coagulation factors (e.g., plasma instead of serum), and possibly age, but not female sex were associated with anti-PrPC autoantibody reactivity in bivariate and multivariate analyses (table 2).29 We henceforth adjusted all analyses for age and presence of coagulation factors. Presence or absence of a pathogenic PRNP mutation was not associated with significant changes in anti-PrPC autoantibody reactivity (table 3). In addition, we matched 62 cases and controls on age (±5 years), sex, and blood sample type26–28 (table e-2, doi.org/10.5061/dryad.08kprr4xk). As with the unmatched cohort, PRNP mutation did not significantly influence anti-PrPC autoantibody titers in multivariate linear regression adjusted for matching factors (table e-2).
Table 2.
Age and lack of coagulation factors in blood (e.g., serum probes), but not sex, are significantly associated with anti-PrPC autoantibody reactivity
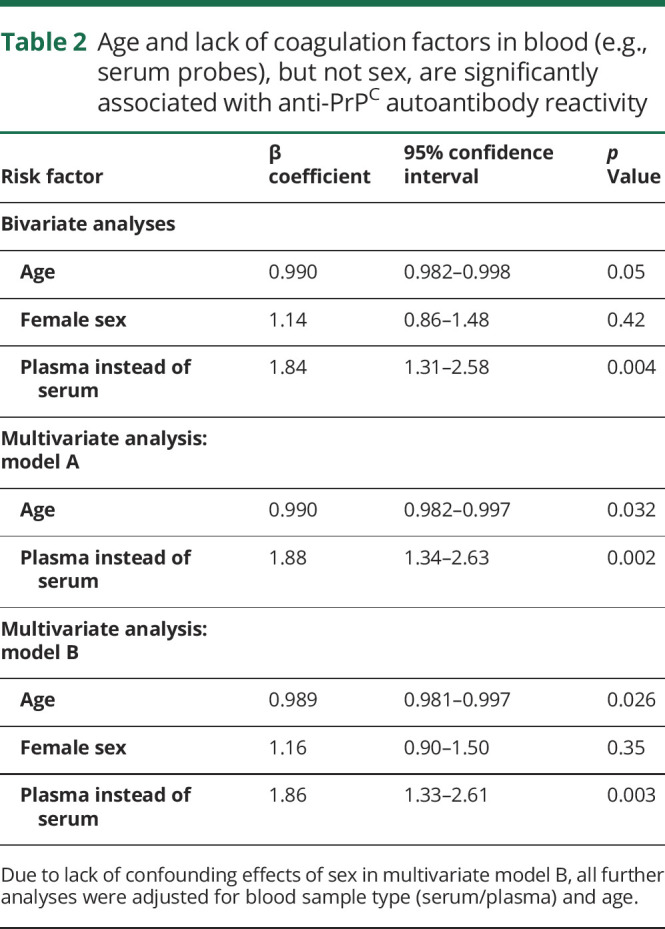
Table 3.
Effect of PRNP mutation status on anti-PrPC autoantibody reactivity
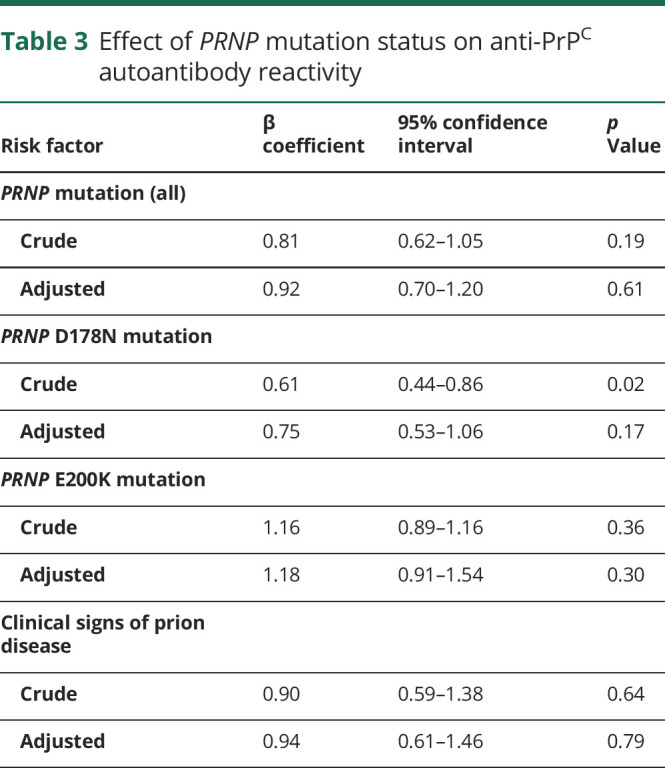
We then tested whether anti-PrPC autoantibody response was associated with symptoms of prion disease. Presence or absence of clinical signs was reported by 122 PRNP mutation carriers (out of a total of 124 enrolled): 76 (62.3%) were asymptomatic carriers whereas 46 (37.7%) presented with clinically apparent disease. Detailed clinical data were available in 14 cases. The most common clinical presentations entailed cerebellar signs (n = 12 [85.7%]) and dementia (n = 11 [78.6%]). Status of 14-3-3 protein in CSF, albeit a poor predictor of gPrD,31 was provided by 121 study participants. Seventeen individuals (all PRNP mutation carriers with clinically apparent disease) were tested, with 8 (47.1%) testing positive, in line with previous findings.31 Presence of prion-specific symptoms was not associated with alterations in anti-PrPC autoantibodies in an unmatched cohort (table 3). This was confirmed in an analysis of a cohort consisting of 24 symptomatic PRNP mutation carriers and 24 asymptomatic PRNP mutation carriers matched on PRNP mutation, age, and sample type (table e-2, doi.org/10.5061/dryad.08kprr4xk).
Post hoc subgroup analyses on the association of anti-PrPC autoantibodies with specific PrPC mutations, PrPC p.129 polymorphism, and autoimmune disease and other immunologic disorders
We analyzed the effects of PRNP mutations that were present at least 5 times in the study population, namely D178N and E200K, on anti-PrPC autoantibody titers: individuals with D178N mutations showed a significant trend towards lower autoantibody titers in bivariate analysis (table 3). This finding, however, was not significant after adjusting for age and sample type (table 3). E200K mutation carriers did not show significant changes in autoantibody reactivity (table 3). The methionine/valine polymorphism at codon 129 of the human PRNP gene was reported to affect the susceptibility to prion diseases.32 Information on p.129 polymorphism was available in 182 study participants: 84 (46.2%) were homozygous for methionine (p.129MM), 87 (47.8%) p.129MV, and 11 (6.0%) p.129VV. None of the polymorphisms significantly altered autoantibody response to PrPC in a post hoc analysis (table 4). In D178N carriers, the clinical phenotype was suggested to be dependent on the PRNP cis c.129 polymorphism: methionine was associated with fatal familial insomnia and valine with familial CJD,2 although this association may not be universal.33,34 In our cohort, 28 patients could be unambiguously identified as D178N_cis129M and 5 patients as D178N_cis129V. No differences in mean antibody reactivity were seen between those 2 groups (table 4).
Table 4.
Effect of PRNP codon 129 polymorphism and history of autoimmune disease on anti-PrPC autoantibody reactivity
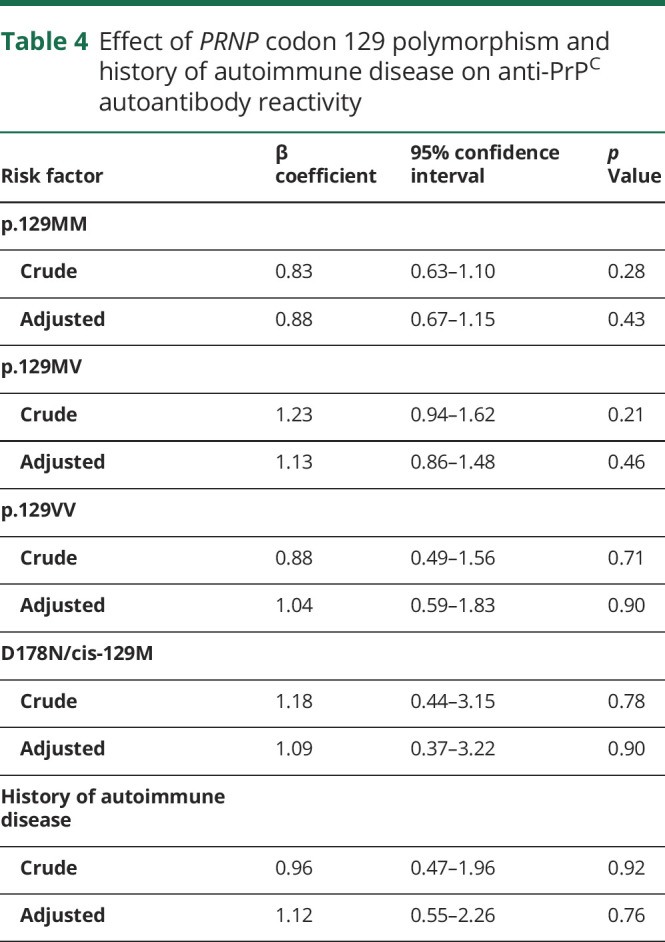
Co-occurrence of multiple autoimmune diseases is a commonly observed phenomenon.35 In order to test the influence of preexisting autoimmune diseases on anti-PrPC autoantibody titers, we searched clinical reports of study participants for presence of autoimmune disease and other immunologic disorders. We were able to retrieve this information in 141 (69.8%) cases: 8 individuals were diagnosed with autoimmune disease, namely Hashimoto thyroiditis (n = 3), Graves disease (n = 1), monoclonal gammopathy of unknown significance (n = 1), multiple sclerosis (n = 1), psoriasis (n = 1), and rheumatoid arthritis (n = 1). Multivariate linear regression analysis adjusted for age, sex, and type of blood sample did not show a significant association of autoimmune disease with anti-PrPC autoantibody titers (table 4).
Temporal evolution of anti-PrPC autoantibodies
Forty-four individuals (21.8%) donated blood multiple times, several months apart, on which we performed a post hoc time course analysis. PRNP wild-type individuals were observed over a longer time period compared to PRNP mutation carriers (17 ± 1.78 months vs 10 ± 6.21 months, p = 1.42 × 10−5). PRNP mutation carriers showed larger variability in autoantibody titers; mean proportional change per year, however, was similar across groups (p = 0.23) and was overall negligible between 2 blood drawings (113.2 ± 61.44% per year in PRNP mutation carriers vs 99.95 ± 17.22% per year in PRNP wild-type individuals, figure 2C). None of the PRNP mutation carriers tested in this time course analysis exhibited clinical signs of prion disease.
Discussion
The diagnosis of a disease-associated PRNP mutation is a fateful and often devastating event for individuals carrying such mutations. The clinical penetrance of PRNP mutations can be very high, and no disease-modifying therapy is available.2 Clinical signs of familial prion disease typically erupt in late adulthood, although carriers arguably produce the mutated protein from the first day of their life.5 There are at least 2 scenarios that may account for this phenomenon: (1) the pathogenic mutations may slightly destabilize PrPC, thereby infinitesimally increasing the probability of pathologic aggregation; or (2) the pathogenic conformation of PrPC is attained early on, but the body's defenses stave off its consequences for decades.
In the case of scenario 1, extensive structural studies on pathogenic PrPC variants failed to reveal major structural alterations.20 We hypothesized that under scenario 2, the stochastic generation of PrPSc in mutation carriers might engender neoantigens, which in turn might lead to protective humoral responses. Remarkably, however, we found no evidence of induction of humoral antibody-mediated immunity against PrPC by pathogenic PRNP variants. Instead, our study suggests the prevalence of naturally occurring anti-PrPC antibodies in the general population independent of clinical signs of prion disease, PRNP variant, or PRNP p.129 polymorphism. Although reactivity to wild-type PrP has been reported in the serum of E219K homozygotes,10 and reactivity to a non-naturally occurring PrP peptide was reported in commercial IgG,9 the present report is to our knowledge the first observation of the PRNP genotype-independent presence of autoantibodies to full-length, wild-type PrP in humans. Without disease-specific antibodies, one might speculate that PRNP mutations accumulate subclinical levels of prions to a point when clinical symptoms become evident.
In a subset of individuals, anti-PrPC autoantibody reactivity was tested in multiple blood drawings up to 1.5 years apart: the mean change of autoantibody titers was similar across PRNP genotypes, in line with previous reports that showed stable autoantibody levels at least over several years.36,37
Matching in case–control studies is a controversial topic.38 In our study, initial analyses were performed on unmatched cohorts adjusted for known confounders of blood autoantibody levels; this approach was described to increase statistical power.39 To strengthen our arguments, we compared anti-PrPC autoantibody levels in cases and matched controls. These results are in line with findings from the unmatched cohorts.
An increasing number of autoantibodies against neurodegenerative targets are being explored as biomarkers and as potential therapeutics. Naturally occurring autoantibodies against hyperphosphorylated tau protein have been isolated from several asymptomatic blood donors.40 Researchers from Neurimmune (Schlieren, Switzerland) recently reported the development of a fully human antibody against amyotrophic lateral sclerosis targeting pathologically misfolded SOD1, α-miSOD1, from a memory B-cell library from healthy elderly individuals.41 Phase III trials involving aducanumab, a bona fide human antibody with potent β-amyloid clearing capabilities, were stopped prematurely.42
In previous works, we found that anti-PrPC antibodies can efficaciously counteract prions,6 a finding later confirmed by several other researchers.43 We speculated that anti-PrPC autoantibodies from the general population could represent a reservoir of potential therapeutic agents against prion diseases. We find, however, that the distribution of titers appears similar between mutation carriers and controls, and between symptomatic and presymptomatic mutation carriers, arguing against the possibility that these autoantibodies are broadly beneficial. This is at variance with a previous preclinical report claiming neuroprotective effects for naturally occurring antibodies to a PrP peptide.9 Similarly, naturally occurring anti-β-amyloid autoantibodies with neuroprotective effects were reported in mice, but did not meet primary cognitive endpoints when tested in a phase III clinical trial.44
Nonetheless, our work does not rule out the possibility of protective anti-PrP autoantibodies in the general population or in PRNP mutation carriers specifically. Our study was restricted to the assessment of autoantibody levels against full-length, wild-type, recombinant human PrPC. We did not evaluate the presence of antibodies specific to pathogenic PRNP mutations or to neoepitopes created by those mutations. Moreover, it is possible that humans develop antibodies specific to PrPSc, the aggregated form of the prion protein. In our experience, such anti-PrPSc antibodies tend to cross-react, at least to some level, with PrPC.45 Another difficulty is that PrPSc structure is very heterogenous in gPrDs: while brains from patients with genetic CJD and sCJD show similar patterns of PrPSc, PrPSc is fragmented and of low molecular weight in brains from patients with GSS and can show marked variation in individuals with the D178N mutation.2,46 Future studies will focus on the detection of rare, low-titer anti-PrPSc antibodies, which may possess unique prion-clearing properties.
Acknowledgment
The authors thank the individuals who participated in this study, the participating patients, the patients' families, the CJD Foundation, referring clinicians, all the members of the National Prion Disease Pathology Surveillance Center for technical help, and Anne Kerschenmeyer, Tina Kottarathil, and Rita Moos at the University Hospital of Zurich for technical assistance.
Glossary
- CJD
Creutzfeldt-Jakob disease
- EBNA
Epstein-Barr nuclear antigen
- FT
flexible tail
- GD
globular domain
- gPrD
genetic prion diseases
- GSS
Gerstmann-Sträussler-Scheinker
- sCJD
sporadic Creutzfeldt-Jakob disease
Appendix 1. Authors
Appendix 2. Coinvestigators
Study funding
The authors received financial support from the EPSRC, BBSRC, ERC, and the Frances and Augustus Newman Foundation. This work was supported by the programs “Investissements d'avenir” ANR-10-IAIHU-06, “Santé Publique France,” and grants from NIH (R01NS103848) and CDC (UR8/CCU515004). Collection of samples at Massachusetts General Hospital was funded by Prion Alliance. The funders played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
K. Frontzek received an unrestricted grant by Ono Pharmaceuticals and was funded by the Theodor Ida Herzog-Egli Stiftung. M. Carta, M. Losa, and M. Epskamp report no disclosures. G. Meisl is funded by a Ramon Jenkins Research Fellowship at Sidney Sussex College. A. Anane, J.-P. Brandel, U. Camenisch, J. Castillas, and S. Haïk report no disclosures. T. Knowles received financial support by the EPSRC, BBSRC, ERC, and the Frances and Augustus Newman Foundation. E. Lindner was funded by the National Organization for Rare Diseases. A. Lutterotti reports no disclosures. E.V. Minikel has received research support in the form of charitable contributions from Charles River Laboratories and Ionis Pharmaceuticals and has consulted for Deerfield Management. I. Roiter, J.G. Safar, R. Sanchez-Valle, and D. Žáková report no disclosures. S. Hornemann is the recipient of grants from SystemsX.ch (SynucleiX) and the innovations commission of the University Hospital of Zurich. A. Aguzzi is the recipient of an Advanced Grant of the European Research Council (ERC 250356) and is supported by grants from the Swiss National Foundation (SNF, including a Sinergia grant), the Swiss Initiative in Systems Biology, SystemsX.ch (PrionX, SynucleiX), the Klinische Forschungsschwerpunkte (KFSPs) “small RNAs” and “Human Hemato-Lymphatic Diseases,” and a Distinguished Investigator Award of the Nomis Foundation. Go to Neurology.org/N for full disclosures.
References
- 1.Aguzzi A, Lakkaraju AKK, Frontzek K. Toward therapy of human prion diseases. Annu Rev Pharmacol Toxicol 2018;58:331–351. [DOI] [PubMed] [Google Scholar]
- 2.Kim MO, Takada LT, Wong K, Forner SA, Geschwind MD. Genetic PrP prion diseases. Cold Spring Harbor Perspect Biol 2018;10:pii: a033134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Will RG, Ironside JW. Sporadic and infectious human prion diseases. Cold Spring Harb Perspect Med 2017;7:pii: a024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minikel EV, Vallabh SM, Orseth MC, et al. Age at onset in genetic prion disease and the design of preventive clinical trials. Neurology 2019;93:e125–e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheckel C, Aguzzi A. Prions, prionoids and protein misfolding disorders. Nat Rev Genet 2018;19:405–418. [DOI] [PubMed] [Google Scholar]
- 6.Heppner FL, Musahl C, Arrighi I, et al. Prevention of scrapie pathogenesis by transgenic expression of anti-prion protein antibodies. Science 2001;294:178–182. [DOI] [PubMed] [Google Scholar]
- 7.Sonati T, Reimann RR, Falsig J, et al. The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 2013;501:102–106. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann US, Sonati T, Falsig J, et al. Prion infections and anti-PrP antibodies trigger converging neurotoxic pathways. PLoS Pathog 2015;11:e1004662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei X, Roettger Y, Tan B, et al. Human anti-prion antibodies block prion peptide fibril formation and neurotoxicity. J Biol Chem 2012;287:12858–12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omae Y, Ito S, Takeuchi M, et al. Integrative genome analysis identified the KANNO blood group antigen as prion protein. Transfusion 2019;59:2429–2435. [DOI] [PubMed] [Google Scholar]
- 11.Haik S, Marcon G, Mallet A, et al. Doxycycline in Creutzfeldt-Jakob disease: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol 2014;13:150–158. [DOI] [PubMed] [Google Scholar]
- 12.Haïk S, Brandel JP, Salomon D, et al. Compassionate use of quinacrine in Creutzfeldt–Jakob disease fails to show significant effects. Neurology 2004;63:2413–2415. [DOI] [PubMed] [Google Scholar]
- 13.Collinge J, Gorham M, Hudson F, et al. Safety and efficacy of quinacrine in human prion disease (PRION-1 study): a patient-preference trial. Lancet Neurol 2009;8:334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto M, Cepek L, Ratzka P, et al. Efficacy of flupirtine on cognitive function in patients with CJD: a double-blind study. Neurology 2004;62:714–718. [DOI] [PubMed] [Google Scholar]
- 15.Killworth H. Fourth patient to be given innovative treatment for CJD [online]. Available at: ucl.ac.uk/news/2019/jan/fourth-patient-be-given-innovative-treatment-cjd. Accessed September 13, 2019.
- 16.Reimann RR, Aguzzi A. Intrinsic toxicity of antibodies to the globular domain of the prion protein. Biol Psychiatry 2018;84:e51–e52. [DOI] [PubMed] [Google Scholar]
- 17.Reimann RR, Sonati T, Hornemann S, et al. Differential toxicity of antibodies to the prion protein. PLoS Pathog 2016;12:e1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu B, McDonald AJ, Markham K, et al. The N-terminus of the prion protein is a toxic effector regulated by the C-terminus. Elife 2017;6:pii: e23473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minikel EV, Vallabh SM, Lek M, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med 2016;8:322ra329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biljan I, Ilc G, Plavec J. Understanding the effect of disease-related mutations on human prion protein structure: insights from NMR spectroscopy. Prog Mol Biol Transl Sci 2017;150:83–103. [DOI] [PubMed] [Google Scholar]
- 21.WHO Manual for Surveillance of Human Transmissible Spongiform Encephalopathies, Including Variant Creutzfeldt-Jakob Disease. Geneva: World Health Organization; 2003. [Google Scholar]
- 22.Ironside JW, Ritchie DL, Head MW. Prion diseases. Handb Clin Neurol 2017;145:393–403. [DOI] [PubMed] [Google Scholar]
- 23.Hill JT, Demarest BL, Bisgrove BW, Su YC, Smith M, Yost HJ. Poly peak parser: method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev Dyn 2014;243:1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright ES. Using DECIPHER v2.0 to analyze big biological sequence data in R. R J 2016;8:352–359. [Google Scholar]
- 25.Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015;12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watad A, Bragazzi NL, Adawi M, et al. Autoimmunity in the elderly: insights from basic science and clinics: a mini-review. Gerontology 2017;63:515–523. [DOI] [PubMed] [Google Scholar]
- 27.Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Front Neuroendocrinol 2014;35:347–369. [DOI] [PubMed] [Google Scholar]
- 28.Kifude CM, Rajasekariah HG, Sullivan DJ Jr, et al. Enzyme-linked immunosorbent assay for detection of Plasmodium falciparum histidine-rich protein 2 in blood, plasma, and serum. Clin Vaccin Immunol 2008;15:1012–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bursac Z, Gauss CH, Williams DK, Hosmer DW. Purposeful selection of variables in logistic regression. Source Code Biol Med 2008;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess RD. Routine Epstein-Barr virus diagnostics from the laboratory perspective: still challenging after 35 years. J Clin Microbiol 2004;42:3381–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zerr I, Bodemer M, Gefeller O, et al. Detection of 14-3-3 protein in the cerebrospinal fluid supports the diagnosis of Creutzfeldt-Jakob disease. Ann Neurol 1998;43:32–40. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi A, Teruya K, Matsuura Y, et al. The influence of PRNP polymorphisms on human prion disease susceptibility: an update. Acta Neuropathol 2015;130:159–170. [DOI] [PubMed] [Google Scholar]
- 33.Sun L, Li X, Lin X, Yan F, Chen K, Xiao S. Familial fatal insomnia with atypical clinical features in a patient with D178N mutation and homozygosity for Met at codon 129 of the prion protein gene. Prion 2015;9:228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, He S, Shi XH, et al. The clinical features in Chinese patients with PRNP D178N mutation. Acta Neurol Scand 2018;138:151–155. [DOI] [PubMed] [Google Scholar]
- 35.Matusiewicz A, Strozynska-Byrska J, Olesinska M. Polyautoimmunity in rheumatological conditions. Int J Rheum Dis 2019;22:386–391. [DOI] [PubMed] [Google Scholar]
- 36.Nagele EP, Han M, Acharya NK, DeMarshall C, Kosciuk MC, Nagele RG. Natural IgG autoantibodies are abundant and ubiquitous in human sera, and their number is influenced by age, gender, and disease. PLoS One 2013;8:e60726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lacroix-Desmazes S, Mouthon L, Kaveri SV, Kazatchkine MD, Weksler ME. Stability of natural self-reactive antibody repertoires during aging. J Clin Immunol 1999;19:26–34. [DOI] [PubMed] [Google Scholar]
- 38.Mansournia MA, Jewell NP, Greenland S. Case-control matching: effects, misconceptions, and recommendations. Eur J Epidemiol 2018;33:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faresjo T, Faresjo A. To match or not to match in epidemiological studies: same outcome but less power. Int J Environ Res Public Health 2010;7:325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pascual G, Wadia JS, Zhu X, et al. Immunological memory to hyperphosphorylated tau in asymptomatic individuals. Acta Neuropathol 2017;133:767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier M, Welt T, Wirth F, et al. A human-derived antibody targets misfolded SOD1 and ameliorates motor symptoms in mouse models of amyotrophic lateral sclerosis. Sci Transl Med 2018;10. [DOI] [PubMed] [Google Scholar]
- 42.Selkoe DJ. Alzheimer disease and aducanumab: adjusting our approach. Nat Rev Neurol 2019;15:365–366. [DOI] [PubMed] [Google Scholar]
- 43.White AR, Enever P, Tayebi M, et al. Monoclonal antibodies inhibit prion replication and delay the development of prion disease. Nature 2003;422:80–83. [DOI] [PubMed] [Google Scholar]
- 44.Relkin NR, Thomas RG, Rissman RA, et al. A phase 3 trial of IV immunoglobulin for Alzheimer disease. Neurology 2017;88:1768–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol 2005;4:805–814. [DOI] [PubMed] [Google Scholar]
- 46.Haik S, Peoc'h K, Brandel JP, et al. Striking PrPsc heterogeneity in inherited prion diseases with the D178N mutation. Ann Neurol 2004;56:909–910; author reply 910–911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The study participants, if they have not undergone predictive testing themselves, participated under the condition of not knowing their PRNP genotype. Due to the relatively small sample size and risk of de-identification, all raw study data involving human participants were made available to the editors and reviewers but will not be made available publicly. Supplementary data, as well as DNA sequences of gene blocks used for construction of humanized antibodies and human PrPC-AviTag, are available at Dryad (doi.org/10.5061/dryad.08kprr4xk).