Abstract
Aim:
The present study aimed to identify human protein–host protein interactions of SARS-CoV-2 infection in the small intestine to discern the potential mechanisms and gain insights into the associated biomarkers and treatment strategies.
Background:
Deciphering the tissue and organ interactions of the SARS-CoV-2 infection can be important to discern the potential underlying mechanisms. In the present study, we investigated the human protein–host protein interactions in the small intestine.
Methods:
Public databases and published works were used to collect data related to small intestine tissue and SARS-CoV-2 infection. We constructed a human protein-protein interaction (PPI) network and showed interactions of host proteins in the small intestine. Associated modules, biological processes, functional pathways, regulatory transcription factors, disease ontology categories, and possible drug candidates for therapeutic targets were identified.
Results:
Thirteen primary protein neighbors were found for the SARS-CoV-2 receptor ACE2. ACE2 and its four partners were observed in a highly clustered module; moreover, 8 host proteins belonged to this module. The protein digestion and absorption as a significant pathway was highlighted with enriched genes of ACE2, MEP1A, MEP1B, DPP4, and XPNPEP2. The HNF4A, HNF1A, and HNF1B transcription factors were found to be regulating the expression of ACE2. A significant association with 12 diseases was deciphered and 116 drug-target interactions were identified.
Conclusion:
The protein-host protein interactome revealed the important elements and interactions for SARS-CoV-2 infection in the small intestine, which can be useful in clarifying the mechanisms of gastrointestinal symptoms and inflammation. The results suggest that antiviral targeting of these interactions may improve the condition of COVID-19 patients.
Key Words: SARS-CoV-2, Small intestine, Interactome, Protein interaction network, Regulatory network, Drug targets
Introduction
COVID-19 disease as a new type of beta-coronavirus disease first emerged in Wuhan, China during December 2019, and is currently spreading rapidly throughout the world. The 2019-nCOV has caused cases of COVID-19 disease throughout the world today (1). Until August 24, 2020, a total of 23.4 million COVID-19 cases have been diagnosed, with 809,000 deaths worldwide. The World Health Organization (WHO) has identified this disease as a global health threat and has supported every research in finding a vaccine (2). The major way of cause infection is human-to-human transmission (3), and despite the rapid spread of this virus, the detailed molecular mechanism and pathogenesis of it remain unclear. The main clinical symptoms of COVID-19 cases are fever, cough, and sore throat (4), however, gastrointestinal symptoms appear in some patients (2%-10%) especially in younger COVID-19 patients, including diarrhea, vomiting, and abdominal pain, proposing that not only does COVID-19 involve lung tissue but that the digestive system can also be affected by SARS-CoV-2 (5, 6). The almost 86.9% similarity of the nucleotide sequence of SARS-COV2 to the SARS-COV and also the pairwise protein sequence analysis results suggest that COVID-19 uses a similar mechanism to enter the host cell (7, 8). In addition, previous studies suggested that angiotensin-converting enzyme 2 (ACE2) is the major receptor of SARS-COV; playing an important role in the entry of the SARS-CoV-2 virus into the host cell (9). So, it can be concluded that ACE2 can be a potential vaccine target for COVID-19 disease therapy. ACE2 greatly express in human organs including lung epithelial cells, kidney, heart, blood, and the small intestine (10). The widespread expression of ACE2 in different tissues might indicate the multi-organ dysfunction in COVID-19 patients (11). Interestingly, the expression of ACE2 protein is downregulated after host infection with SARS-COV (12). Decreased ACE2 expression levels can increase susceptibility to intestinal inflammation due to damage to epithelial cells and its expression on surface cells of the small intestine might participate in the virus invasion, amplification, and also activation of gastrointestinal inflammation (12). Due to the genomic similarity of SARS-CoV-2 to SARS-CoV, SARS-CoV-2 may also impair ACE2 expression, and this reduction or inhibition of ACE2 expression can lead to dysfunction in the gastrointestinal tract, resulting in digestive symptoms in COVID-19 patients, and can also be a possible molecular pathway for adverse digestive symptoms emerging in patients with COVID-19. On the other hand, ACE2 also plays a key role in the intestine including maintaining amino acid homeostasis (13). Constant expression of ACE2 on the small intestine may relate to gastrointestinal symptoms, impact the infection route, and virus pathogenicity (11). So far, there are no effective antiviral drugs or vaccines against SARS-CoV-2, which is due to the limited understanding of the molecular pathways of coronavirus pathogenesis (14). Understanding COVID-19 disease pathogenesis and also how the COVID-19 causes gastrointestinal symptoms in our body are crucial points to be considered (15), and the detection of host protein interactions with ACE2 in the small intestine could help to discover coronavirus pathology pathways and present potential drug and vaccine targets. The viral entry can affect the interaction of the ACE2 with host cellular proteins. Accordingly, protein interaction analyses may provide further insight into understanding the molecular basis of COVID-19 pathogenesis. Besides, identification and knowledge of the distribution of ACE2 receptor in various human tissues are of particular importance in discovering treatment strategies for COVID-19 infection. Small intestine enterocytes show a high expression of ACE2 mRNA and protein (16, 17). In this study, to provide a view into the biological processes and pathways in the host cell by COVID-19 infection, and gastrointestinal inflammation, we analyzed the human protein-host protein interactome of the SARS-CoV-2 infection in the small intestine using protein interaction and a transcription factor regulatory network. Furthermore, we extracted possible drug-target interactions.
Methods
This study aimed to identify possible human protein–host protein interactions of SARS-CoV-2 infection in the small intestine and attempted to discern the potential mechanisms of gastrointestinal symptoms in the COVID-19 disease.
Data related to small intestine tissue and SARS-CoV-2 infection
The small intestine-specific genes list was retrieved from the Tissue-specific Gene Expression and Regulation (TiGER) database (18). Various recent studies have presented interaction networks of ACE2 (as a major host receptor for SARS-CoV-2) and some reports have exhibited specific expression of it in small intestinal enterocytes after virus infection (17, 19-21). Hence, in this study, the first interacting protein neighbors of ACE2 were gathered from the UniProt (22) and GeneCards databases (23) based on the interaction records in the STRING database (24). The serine protease TMPRSS2 as a host factor that is required for SARS-CoV-2 entry (25) was also added to the list. The human host proteins of SARS-CoV-2 were compiled from two different studies by Gordon et al. and Kumar et al (26, 27). After obtaining these lists of proteins, the tissue specific- expression analysis of the protein list was performed by the Database for Annotation, Visualization and Integrated Discovery (DAVID) webserver using the “UNIGENE_EST_QUARTILE” categories (28). The significant tissue-specific genes for the small intestine were selected with an FDR corrected p-value < 0.05. The resultant gene list was used for further analysis.
Network generation, modules detection, and functional analyses
Small intestine tissue-specific genes were queried into the STRING database to mine the related protein-protein interactions (PPI) (24). STRING is a web resource for exploiting the known and predicted PPI information. The combined score > 0.4 was considered as a criterion for the selection of protein interactions. These connections were then imported into the Cytoscape software to build a PPI network (29). The host proteins were determined using a different color in the resulting network. ACE2 interactome network, in the resultant network of small intestine tissue, was presented using ACE2, the first interacting protein neighbors of ACE2, and second neighbor host proteins. A module is a cluster of highly interacting proteins. The molecular complex detection (MCODE) algorithm was employed to screen the high modularity clusters of the PPI network in the Cytoscape software (30). Moreover, cross-talk and interactions between the detected modules and ACE2 were extracted. Functional roles of the high score clusters were evaluated through gene ontology biological processes enrichment, and KEGG (Kyoto Encyclopedia of Genes and Genomes) (31) pathway analysis, with the aid of the DAVID database (28).
Regulatory network construction
Possible upstream regulating transcription factors of the 176 small intestine-specific proteins were enriched using the GeneTrail2 web service (32). GeneTrail2 as a web-interface provides access to different tools for biological analyses and extraction of molecular signatures. The validated transcription factor-target gene interactions with q-value < 0.05 were retrieved from the TRANSFAC database through the GeneTrail2 web service (33). The transcription factor-target gene interactions were imported into Cytoscape to generate the regulatory network.
Disease Ontology Annotation and disease-gene network construction
Overlapping genes across the 176 genes of small intestine tissue and different diseases were mined using the GAD (Genetic Association Database)_DISEASE category with the aid of the DAVID database (28). P-value < 0.05 was considered a specifying parameter for the identification of GAD_DISEASE ontology terms (34). Disease-gene associations were imported into Cytoscape to generate a disease-gene network.
Prediction of drug-target interactions and generation of drug-protein interaction network
Drugs for the enriched KEGG pathways were obtained from the KEGG database (31). Moreover, overlapping diseases with the 176 genes list were queried in repoDB drug repositioning database (35). The resultant drugs list from the KEGG and repoDB databases with the 176 genes list were merged and compiled into one list and mapped into the STITCH database to identify possible drug-target interactions (36). The drug-target interactions were imported into Cytoscape to construct a drug-protein interaction network.
Results
Data preparation for analysis
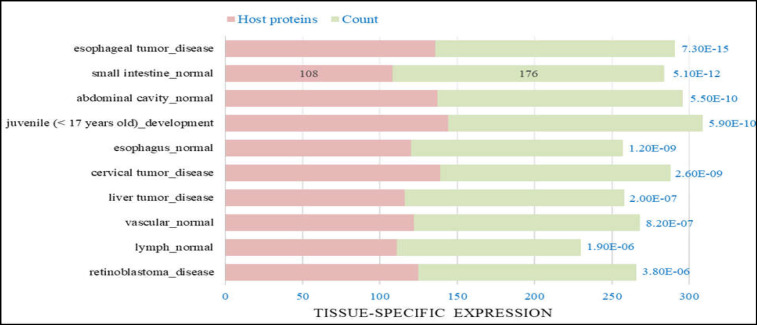
A list of 101 small intestine-specific genes was retrieved from TiGER. ACE2, 17 proteins as the first interacting protein neighbors of ACE2, and TMPRSS2 were added to the gene list. Lists of 508 host proteins were compiled from studies of Gordon et al. (332 host proteins) and Kumar et al. (390 host proteins) (26, 27). Finally, a comprehensive list of 625 genes was compiled. Subsequently, the gene list was queried to the DAVID database to identify tissue-specific expression categories, which showed 176 significant small intestine-specific genes (108 host proteins) with FDR-corrected p-value = 5.10E-12 as shown in Figure 1. The 176 genes list was used to construct a PPI network.
Figure 1.
Top 10 tissue-specific expression categories with FDR-corrected p-value <. 05. 176 genes for small intestine tissue were enriched with FDR-corrected p-value = 5.10E-12, of which 108 genes were common with the host protein list
Network generation, modules detection, and functional analyses
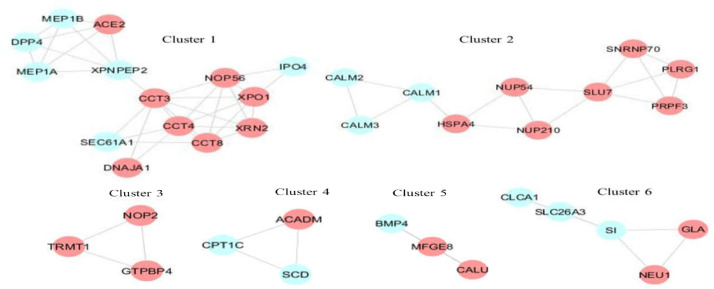
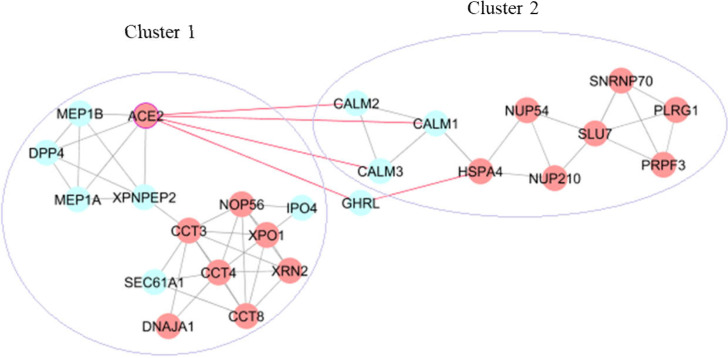
Human protein interactions with a medium confidence score for the 176 small intestine-specific genes were mined from the STRING database. These associations were imported into Cytoscape and a scale-free PPI network with an r-squared value of 0.764 was constructed that showed 146 nodes (91 host proteins) and 366 edges (Figure 2. (A)). Thirteen interactions were found for ACE2 with small intestine-specific proteins. Figure 2 (B) presents possible ACE2 interactome in the small intestine including ACE2, the first interacting protein neighbors of ACE2 (DPP4, MEP1A, MEP1B, AGT, NTS, TFRC, GHRL, NPEPPS, TMPRSS2, CALM1, CALM2, CALM3, and XPNPEP2), and the second interacting host protein neighbors (PLAT, ACTR3, HSPA4, OS9, CCT3, FAR2, CALU, SLC9A3R1, LOX, FKBP10, FAF2, ATP6AP1, HMOX1, DOX41, TPM3, and GOLGA2). Six modules were detected with MCODE scores > 3. Figure 3 demonstrates the skeletal structure of the 6 clusters. The first cluster with the highest MCODE score of 5.077 showed 14 nodes and 33 edges. ACE2 and 4 partners of ACE2 (DPP4, MEP1A, MEP1B, and XPNPEP2) were seen in Cluster1. Moreover, 8 host proteins (ACE2, XPO1, DNAJA1, XRN2, CCT3, CCT8, CCT4, and NOP56) belonged to Cluster1. The results revealed Cluster2 with an MCODE score of 3.333, 10 nodes (7 host proteins), and 15 edges. Cross-talk among Cluster1 and other clusters through ACE2 was investigated that showed a direct interaction of ACE2 with CALM1, CALM2, and CALM3 in Cluster2. An indirect interaction was also seen through GHRL that connects ACE2 in Cluster1 to the HSPA4 host protein in Cluster2 (Figure 4). Functional analysis of Cluster1 enriched the protein digestion and absorption KEGG pathway with FDR corrected p-value < 0.05.
Figure 2.
(A). PPI Network of the 176 small intestine-specific proteins (146 nodes and 366 edges). Red nodes: host proteins (91 proteins). Blue edges: ACE2-first neighbor interactions. Red edges: ACE2-second neighbor interactions. (B). ACE2 interaction network in the small intestine including ACE2, the first interacting protein neighbors of ACE2 (13 proteins), and second neighbor host proteins (16 proteins)
Figure 3.
Skeletal structure of 6 clusters. Red nodes: host proteins
Figure 4.
Cross-talk among the clusters through ACE2 in the network. Red nodes: host proteins
ACE2 and MEP1B, MEP1A, DPP4, XPNPEP2 proteins were significantly enriched in this KEGG pathway (Table 1). Additionally, gene ontology (GO) biological processes terms were detected, for example, toxin transport, regulation of protein localization to Cajal body, and protein localization to nuclear body.
Table 1.
Gene ontology and KEGG pathway analysis of Cluster1
| Category | Term | FDR | Genes |
|---|---|---|---|
| KEGG_PATHWAY | protein digestion and absorption | 0.0018 | MEP1A, ACE2, MEP1B, DPP4, XPNPEP2 |
| GOTERM_BP_ALL | toxin transport | 2.36E-05 | CCT4, CCT8, DNAJA1, MEP1B, CCT3 |
| GOTERM_BP_ALL | regulation of protein localization to Cajal body | 0.0232 | CCT4, CCT8, CCT3 |
| GOTERM_BP_ALL | positive regulation of protein localization to Cajal body | 0.0232 | CCT4, CCT8, CCT3 |
| GOTERM_BP_ALL | protein localization to Cajal body | 0.0298 | CCT4, CCT8, CCT3 |
| GOTERM_BP_ALL | protein localization to nuclear body | 0.0298 | CCT4, CCT8, CCT3 |
Regulatory network construction
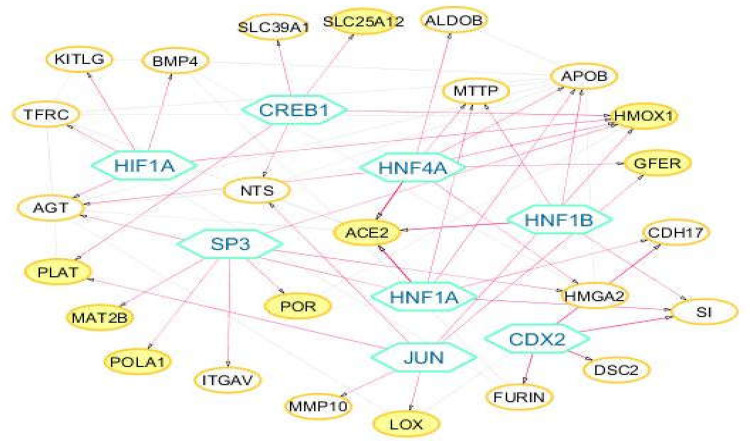
Eight transcription factors (TFs) (HNF4A, HNF1A, HNF1B, CDX2, JUN, HIF1A, CREB1, and SP3) with q-value < 0.05 were enriched for 25 target genes from the 176 genes list. The regulatory network showed 45 validated transcription factor-target gene interactions between 8 TFs and 25 target genes (Figure 5). HNF factors including HNF4A, HNF1A, and HNF1B were found to be regulating the expression of the ACE2 target gene (Table 2).
Figure 5.
Transcription factor-target gene regulatory network. Hexagon nodes represent the 8 enriched transcription factors. Yellow nodes: host proteins
Table 2.
Enriched transcription factors for the 176 genes list
| N | Transcription factor | q-value | Target genes (of the 176 small intestine-specific proteins) |
|---|---|---|---|
| 1 | HNF4A | 9.66E-07 | ACE2, AGT, ALDOB, APOB, GFER, HMGA2, HMOX1, MTTP |
| 2 | HNF1A | 3.14E-05 | ACE2, AGT, APOB, CDH17, MTTP, SI |
| 3 | HNF1B | 1.13E-04 | ACE2, APOB, MTTP, SI |
| 4 | CDX2 | 0.0018 | CDH17, DSC2, FURIN, SI |
| 5 | JUN | 0.0027 | GFER, HMOX1, LOX, MMP10, NTS, PLAT |
| 6 | HIF1A | 0.0092 | AGT, BMP4, HMOX1, KITLG, TFRC |
| 7 | CREB1 | 0.0208 | HMOX1, NTS, PLAT, SLC25A12, SLC39A1 |
| 8 | SP3 | 0.0365 | HMGA2, HMOX1, ITGAV, MAT2B, POLA1, POR |
Disease-gene network construction
GAD disease category associations were scanned to further classify the 176 genes and annotating their function. Twelve GAD_DISEASE categories were found with a p-value < 0.05 (Table 3). A disease-gene network was constructed using 7 GAD_DISEASE categories that have highlighted their importance in the COVID-19 response (diabetes, immunodeficiency, cardiovascular diseases, hypertension, blood pressure, colorectal cancer, and nephropathy (Figure 6). Seven GAD_DISEASE categories, 64 genes, and 91 associated-gene interactions are presented in the disease-gene network. Subsequently, these overlapping diseases were queried in repoDB to identify drug candidates.
Table 3.
Disease ontology of 176 genes
| Category | Term | Count | P-Value |
|---|---|---|---|
| GAD_DISEASE | Type 2 Diabetes| edema | rosiglitazone | 45 | 7.60E-06 |
| GAD_DISEASE | blood pressure, arterial | 5 | 2.80E-03 |
| GAD_DISEASE | Acquired Immunodeficiency Syndrome|Disease Progression | 20 | 3.00E-03 |
| GAD_DISEASE | Cardiovascular Diseases| | 5 | 5.30E-03 |
| GAD_DISEASE | Cleft Lip|Cleft Palate | 11 | 8.70E-03 |
| GAD_DISEASE | Hyperlipidemias|Hypertension | 3 | 1.00E-02 |
| GAD_DISEASE | Spinal Dysraphism | 4 | 2.50E-02 |
| GAD_DISEASE | protein C protein S | 2 | 3.10E-02 |
| GAD_DISEASE | restenosis | 4 | 3.20E-02 |
| GAD_DISEASE | longevity | 10 | 3.50E-02 |
| GAD_DISEASE | Colorectal Cancer | 10 | 3.50E-02 |
| GAD_DISEASE | nephropathy, diabetic | 3 | 4.70E-02 |
Figure 6.
Disease-gene network. Yellow nodes represent the 7 overlapping diseases with 176 genes. Red nodes: host proteins
Drug-protein interaction network
A list was prepared of 24 drugs that are in the KEGG database for protein digestion and absorption KEGG pathway (the most important pathway was found in our analysis approach).
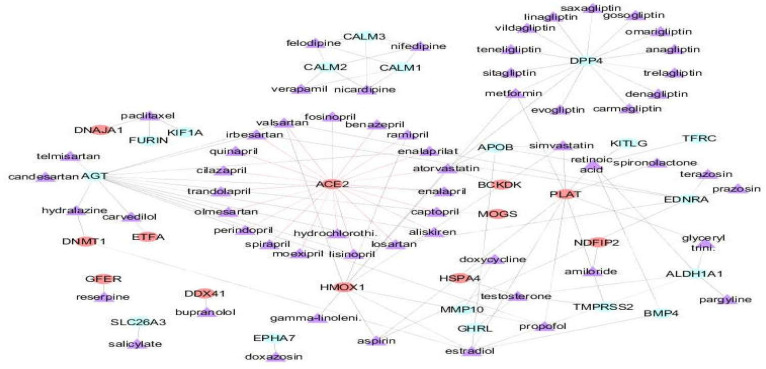
Additionally, 110 drugs were mined from repoDB for overlapping diseases with the 176 genes. Subsequently, the resultant drugs along with the 176 genes list were mapped into the STITCH database. Three new drugs were also revealed in the data list that was predicted by STITCH, including gamma-linolenic acid, retinoic acid, and glyceryl trinitrate (nitroglycerin). A Drug-protein interaction network was constructed using the STITCH outcomes in Cytoscape that revealed 116 drug-protein interactions, including 30 proteins (12 host proteins) and 60 drugs. The drug candidates mostly belong to the drug classes of ACE inhibitors, Angiotensin receptor blockers (ARBs), calcium channel blockers, gliptins, and statins (Figure 7, Table 4). ACE2 showed 20 drug interactions.
Figure 7.
Drug-protein interaction network. Violet nodes represent the 60 drugs. Red nodes: host proteins
Table 4.
This table presents the drugs list and associated drug classes
| Drug classes | Drug names | Drug targets |
|---|---|---|
| ACE inhibitors | benazepril, spirapril, trandolapril, cilazapril, fosinopril, moexipril, quinapril | ACE2 |
| captopril | ACE2, AGT, MOGS | |
| enalapril, enalaprilat, perindopril, lisinopril | ACE2, AGT | |
| ramipril | ACE2, HMOX1 | |
| Gliptins | anagliptin, carmegliptin, denagliptin, evogliptin, gosogliptin, linagliptin, omarigliptin, saxagliptin, sitagliptin, teneligliptin, trelagliptin, vildagliptin | DPP4 |
| Statins | atorvastatin | ACE2, APOB, BCKDK, DPP4, HMOX1 |
| simvastatin | APOB, HMOX1, PLAT | |
| Calcium channel blockers | carvedilol | AGT, ETFA |
| felodipine, nicardipine, nifedipine | CALM1, CALM2, CALM3 | |
| verapamil | CALM1, CALM2 | |
| Angiotensin receptor blockers (ARBs) | valsartan | ACE2, AGT, EDNRA, HMOX1 |
| irbesartan, losartan | ACE2, AGT, EDNRA | |
| telmisartan, candesartan, olmesartan | AGT | |
| olmesartan, hydrochlorothiazide | ACE2 | |
| Alpha-blockers | doxazosin | EPHA7 |
| prazosin, terazosin | EDNRA | |
| bupranolol | DDX41 | |
| Aldosterone receptor antagonists | spironolactone | PLAT |
| Intravenous anesthetic | propofol | AGT, PLAT, HMOX1 |
| Vasodilators | nitroglycerin | ALDH1A1, PLAT |
| hydralazine | AGT, DNMT1 | |
| Adrenergic blocking agent | reserpine | GFER |
| Antibiotic | doxycycline | MMP10, HSPA4 |
| Estrogens | estradiol | ALDH1A1, APOB, BMP4, DNMT1, HMOX1, PLAT |
| Fatty acid, Omega-6 fatty acid | gamma-linolenic | HMOX1 |
| Antihyperglycemic agent | metformin | DPP4, HMOX1, PLAT |
| non-steroidal anti-inflammatory drug | aspirin (acetylsalicylic acid) | EDNRA, HMOX1, PLAT |
| salicylate | SLC26A3 | |
| Monoamine Oxidase Inhibitors | pargyline | ALDH1A1 |
| potassium-sparing diuretic family | amiloride | TMPRSS2, NDFIP2, PLAT |
| renin inhibitor | aliskiren | ACE2, AGT |
| Chemotherapy | paclitaxel | FURIN, DNAJA1, KIF1A |
| Thiazide diuretics | hydrochlorothiazide | ACE2 |
| Retinoids | retinoic acid | ALDH1A1, APOB, BMP4, KITLG, PLAT, TFRC |
| Androgens | testosterone | ALDH1A1, GHRL, HSPA4, TMPRSS2 |
Discussion
As recent studies have suggested, SARS-CoV-2 may infect the host cells by the virus binding to the ACE2 receptor. It could be assumed, then, that ACE2 may be a potential therapeutic target against COVID-19 (37). Previous studies have reported the expression pattern of the ACE2 gene as a host cell receptor of SARS-CoV-2 in the small intestine (17). They have demonstrated ACE2 expression on the surface enterocytes that potently facilitates the entrance of SARS-CoV-2 and results in host infection and gastrointestinal symptoms (17). In this study, we aimed to identify the SARS-CoV-2–host protein interactions in the small intestine and understand the possible mechanisms of gastrointestinal symptoms in the COVID-19 disease to gain insights about the associated biomarkers. PPI network of the small intestine-specific proteins revealed the 13 ACE2 interacting partners (DPP4, MEP1A, MEP1B, AGT, NTS, TFRC, GHRL, NPEPPS, TMPRSS2, CALM1, CALM2, CALM3, and XPNPEP2) and their associated functional pathways in the small intestine. ACE2 and four of its primary protein neighbors (DPP4, MEP1A, MEP1B, and XPNPEP2) were seen in the highly clustered module enriched in the protein digestion and absorption KEGG pathway. ACE2 is abundantly presented on the luminal surface of intestinal epithelial cells and functions as a co-receptor for amino acid and nutrient uptake from food (38). The deficiency of ACE2 in the intestine, due to SARS-CoV-2 infection, may further impair the nutrition of the host and have abilities to mount a balanced immune response (39). Moreover, several studies have demonstrated that ACE2 also functions as the chaperone for the membrane trafficking of the amino acid transporter B0AT1 (40). Therefore, when SARS-CoV-2 blocks ACE2 it also blocks B0AT1, thus shutting down intestinal amino acid transport (40). It was shown that mutations in B0AT1 led to Hartnup disease with defective adsorption of amino acids through the kidneys and small intestine. Patients with Hartnup disease develop pellagra-like symptoms under stress conditions, such as rash, cerebellar ataxia, and diarrhea (39). In ACE2 deficient mice, B0AT1 is completely absent from the small intestine and, therefore, displays a dramatically decreased level of tryptophan which also causes colitis (13, 40). Since tryptophan-rich peptides are a subset of antimicrobial peptides, the interaction of SARS-CoV-2 with the enterocytic ACE2/B0AT1 receptors can cause an abnormal composition of the gut microbiome followed by a massive inflammatory response and cytokine storm in the small bowel (39). Meprins (MEP1A, MEP1B), as shown in the results, are the first interacting protein neighbors of ACE2. They are extracellular proteases involved in connective tissue homeostasis, intestinal barrier function, and immunological processes (41). MEP1A has been identified as a genetic susceptibility factor for inflammatory bowel disease. It encodes meprin α, an astacin-like metalloprotease, which is secreted into the intestinal lumen or it is preserved at the brush border membrane in relation to transmembrane meprin β (42). Therefore, any reduction in meprin α or β expression can cause similar defects in the host. Various substrates including extracellular matrix proteins, growth factors, and cytokines are cleaved by meprins. Meprin β on the apical side of intestinal epithelial cells detaches the constitutive mucus due to the prevention of bacterial overgrowth (41). Meprin α deficient mice were more susceptible to dextran sulfate sodium-induced experimental colitis and underwent larger colon disorders and inflammation than wild-type mice (42). Meprins may function as a mucosal defense mechanism that maintains the intestinal epithelium against potential toxic peptides and also against enteric commensal and pathogenic bacteria by regulating the interaction between microbes and the host mucosa (42). Another primary protein neighbor of ACE2; namely Dipeptidyl peptidase 4 (DPP4) (also known as cluster of differentiation 26 (CD26)), is a transmembrane glycoprotein presented ubiquitously in several tissues such as lung, kidney, liver, gut, and immune cells (43). DPP4 as a serine exo-peptidase is involved in various physiological processes and can cleave a wide range of substrates including growth factors, chemokines, neuropeptides, and vasoactive peptides. DPP4 modulates immune responses by functioning as a costimulatory molecule on T-cells and regulates glucose homeostasis by degrading incretin hormones (43). The DPP-4 activity, as one of the peptidases at the surface of enterocytes, participates in the last step of protein digestion (44). According to recent studies, it was suggested that SARS–CoV-2, like MERS–Co-V, may use the DPP4/CD26 receptor as a co-receptor when virus enters the cells via ACE2. The co-expression of ACE2 and DPP4/CD26, as receptors of S glycoproteins, could hypothesize that verity human CoVs target similar cell types across various human tissues and explain the emergence of similar clinical characteristics in patients infected with verity CoVs (45). In our results, ghrelin (GHRL) as a primary protein neighbor of ACE2 links together two of the detected highly clustered modules. Ghrelin is a peptide hormone mostly secreted by X/A-like gastric cells, functions through the growth hormone secretagogue receptor (GHSR), and exhibits a modulatory role in the immune system (46). Ghrelin also induces gastric motility and emptying as well as motility in the small and large intestine. Ghrelin has been shown to be affected in multiple gastrointestinal diseases and disorders such as inflammatory bowel disease, coeliac disease, infectious diseases, functional disorders and diabetes gastroenteropathy. This demonstrates that ghrelin is implicated in the pathophysiology of gastrointestinal diseases and disorders (47).
In this study, we also enriched HNF proteins (HNF4A, HNF1A, and HNF1B) as transcription factors which regulate expression of the ACE2 target gene. HNF4 factors were identified in one recent study as key transcriptional regulators of SARS-CoV-2 entry genes in the intestine (48). Chen et al., using epigenomic approaches and mouse genetic models, reported that HNF4 factors bind to the loci of the ACE2 gene, alter chromatin looping, shape epigenetic modifications, and ultimately show a dramatic impact on its expression upon transcription factor knockout (48). Our predictions, for HNF1A and HNF1B factors, have been formerly reported experimentally to drive ACE2 expression in pancreatic islet cells and insulinoma cells, respectively (49). Recently, Barker et al., using bioinformatics tools, also predicted that the HNF1A transcription factor in colon tissue has high positive correlation with the expression of ACE2 (50). CDX2 was found to be another important transcription factor that was enriched in this study. It was shown that this factor regulates furin expression during intestinal epithelial cell differentiation (51). The small intestine is rich in furin, a serine protease, that can cleave the S protein of the coronavirus into two “pinchers” (S1 and 2). The cleaving of the S protein into S1 and S2 is important for the binding of the virion to both the ACE2 receptor and the cell membrane (39). Indeed, furin is a broadly distributed enzyme in the small intestine and is the main enzyme in the process of activation of other enteric toxins (39).
Moreover, in this study, we extracted a list of 60 drugs that interact with the 176 gene list. Some of these reported drugs can be beneficial in use against COVID-19 infection.
One preliminary clinical study reported that treatment with renin inhibitor aliskiren, as an antihypertensive agent, is effective and safe for severe COVID-19 patients complicated with hypertension (52). In another study, it was reported that aliskiren, by molecular docking, showed higher energies of binding than that of the co-crystallized ligand N3 with COVID-19 main protease Mpro (53). They suggested that aliskiren can be a repurposing drug for the treatment of COVID-19. Gliptins are antidiabetic drugs controlling glucose homeostasis by the prevention of DPP4 enzymatic activity. Gliptins can protect endothelial function by their reported anti-inflammatory, anti-oxidant, and potentially protective effects on the vascular system, which are beneficial aspects in the fight against COVID-19 (54). Gliptins might be also used to restrain SARS-CoV-2 binding to host cells. DPP4 inhibitor ability to inhibit coronavirus entry into host cells has also been investigated (54). Another noteworthy point is that a recent study reported that anagliptin might exert a cholesterol-lowering action through DPP4-dependent and glucagon-like peptide 1-independent suppression of intestinal cholesterol transport (55).
Candesartan, as an angiotensin receptor blocker (ARB), reduces inflammation and protects lung and brain functions, and it has recently been reported that candesartan can be therapeutically effective in COVID-19 patients and ameliorate the cytokine storm (56). One recent first-time study demonstrated that candesartan treatment alleviates hypertension-associated pathophysiological alterations in the gut, enhances microbial production of short-chain fatty acids, and protects gut Lactobacillus under hypertensive conditions. This information sheds novel light on the pharmacological implications of candesartan (57). Other ARBs such as losartan and telmisartan were enriched in our study as therapeutics for SARS-CoV-2 virus infections. These drugs are widely used in the clinic since the 1990s for control of hypertension and kidney disorders and are known as safe drugs that are rarely implicated in adverse drug events (58). Telmisartan, which is well absorbed after oral administration, has the longest plasma half-life (24 hr), and achieves the highest tissue concentrations because of its high lipid solubility and high levels of dissemination (500 L). This drug separates more slowly after attachment to the Ang receptor I, leading to an apparently irreversible block (59). Recently, a randomized open-label controlled trial began enrollment of patients in Hospital de Clínicas José de San Martín (School of Medicine, University of Buenos Aires, Argentina). Clinical studies to investigate the safety of Telmisartan in healthy cases, or in hypertensive patients receiving daily doses of up to 160 mg, indicated no distinction among those treated with Telmisartan and the placebo group in frequency and severity of detrimental effects (59). Another recent study reported that Telmisartan can attenuate colon inflammation, oxidative perturbations, and apoptosis in a rat model with experimental inflammatory bowel disease (60). Valsartan is another ARB that was enriched in this study. The PRAETORIAN-COVID trial has been begun as a project to provide the much-expected evidence regarding the controversy on the acts of Valsartan in patients with SARS-CoV-2 infectious disease (61).
Carvedilol is a drug with vasodilating properties initially designed for managing hypertension and coronary artery disease. Unlike ACE inhibitors that enhance the expression of ACE2, carvedilol reduces its expression, therefore, this drug can be useful for all COVID-19 patients (62). Carvedilol can fight COVID-19 through another mechanism because it has interleukin 6 (IL-6) suppressing properties and which plays a major role in the inflammatory cascade of COVID-19 (62).
Gamma-linolenic acid, as a bioactive lipid, inhibits the production of pro-inflammatory IL-6 and TNF-α and could be employed to treat cytokine storm that is seen in COVID-19 patients. Reports show that gamma-linolenic acid inactivates enveloped viruses including COVID-19, thus, infusions of appropriate amounts of this acid are of significant therapeutic benefit in treating COVID-19 (63). According to these data, a prospective double-blinded controlled 14-day trial on 30 SARS-CoV-2 positive cases has been conducted (Anti-inflammatory/Antioxidant Oral Nutrition Supplementation in COVID-19 [ONSCOVID19]; NCT04323228). The participants were randomly assigned to two groups (n = 15/each); intervention (IG) and placebo (PG). The IG group received an anti-inflammatory and antioxidant oral nutrition supplement (ONS) enriched in eicosapentaenoic acid, gamma-linolenic acid, and antioxidants daily, while the PG group received an isocaloric placebo. It can be concluded that with more maintenance of the nutritional status of infected patients the anti-inflammatory-antioxidant ONS might contribute to the decrease of COVID-19 severity.
Studies have shown that Verapamil can interfere with coronavirus entry and amplification by blocking ion channels. It was proven that Verapamil is effective against RNA viruses in vitro and inhibited filovirus infection in cell cultures and mouse models (64). Interestingly, a randomized trial has been commenced comparing Verapamil and amiodarone with usual care in hospitalized patients with confirmed COVID-19 (Amiodarone or Verapamil in COVID-19 Hospitalized Patients with Symptoms [ReCOVery-SIRIO]; NCT04351763).
The human protein-host protein interactome of SARS-CoV-2 infection in the small intestine was investigated. The outcomes indicate that ACE2 and its interacting proteins in the small intestine may explain gastrointestinal symptoms and intestinal inflammation. The results suggest that antiviral targeting these interactions may improve the condition of COVID-19 patients; however, more research is needed to find possible mechanisms of the gastrointestinal symptoms and confirm associated biomarkers..
Acknowledgment
The authors gratefully acknowledge the support of the Shahid Beheshti University of Medical Sciences (code No. 24895).
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Cao W. Clinical features and laboratory inspection of novel coronavirus pneumonia (COVID-19) in Xiangyang, Hubei. medRxiv. 2020 [Google Scholar]
- 2.Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199–207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Z, Yi F, Wu K, Lai K, Sun X, Zhong N, et al. Clinical characteristics of 2019 coronavirus pneumonia (COVID-19): an updated systematic review. MedRxiv. 2020 [Google Scholar]
- 6.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395:507–13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Yu C, Qu J, Zhang L, Jiang S, Huang D, et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47:1275–80. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The Lancet. 2020;395:514–23. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamming I, Timens W, Bulthuis M, Lely A, Navis Gv, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–7. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikhra V. The Agent and Host Factors in Covid-19: Exploring Pathogenesis and Therapeutic Implications. Biomed J Sci Techn Res. 2020;27:20669–20681. [Google Scholar]
- 12.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nature Med. 2005;11:875–9. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–81. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Bai T, Yang L, Hou X. Discovery of potential drugs for COVID-19 based on the connectivity map. 2020 doi: 10.3389/fgene.2020.558557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Z, Wang Y, Qi W. The small intestine, an underestimated site of SARS-CoV-2 infection: from red queen effect to probiotics. Preprints. 2020:2020030161. [Google Scholar]
- 16.Liang W, Feng Z, Rao S, Xiao C, Xue X, Lin Z, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–3. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Li HB, Lyu JR, Lei XM, Li W, Wu G, et al. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int J Infect Dis. 2020;96:19–24. doi: 10.1016/j.ijid.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinf. 2008;9:1–7. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circul Res. 2020;126:1456–74. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wicik Z, Eyileten C, Jakubik D, Pavao R, Siller-Matula JM, Postula M. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors. BioRxiv. 2020 doi: 10.3390/jcm9113743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karunakaran KB, Balakrishnan N, Ganapathiraju MK. Interactome of SARS-CoV-2/nCoV19 modulated host proteins with computationally predicted PPIs. 2020 [Google Scholar]
- 22.Consortium U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–15. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinf. 2016;54:1–30. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon D, Jang G, Bouhaddou M, Xu J, Obernier K, O’Meara M, et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug Repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar R, Verma H, Singhvi N, Sood U, Gupta V, Singh M, et al. Comparative Genomic Analysis of Rapidly Evolving SARS-CoV-2 Reveals Mosaic Pattern of Phylogeographical Distribution. Msystems. 2020:5. doi: 10.1128/mSystems.00505-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:1–11. [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stöckel D, Kehl T, Trampert P, Schneider L, Backes C, Ludwig N, et al. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics. 2016;32:1502–8. doi: 10.1093/bioinformatics/btv770. [DOI] [PubMed] [Google Scholar]
- 33.Matys V, Fricke E, Geffers R, Gößling E, Haubrock M, Hehl R, et al. TRANSFAC®: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–8. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–2. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 35.Brown AS, Patel CJ. A standard database for drug repositioning. Sci Data. 2017;4:1–7. doi: 10.1038/sdata.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–4. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Deng W, Li S, Yang X. Advances in research on ACE2 as a receptor for 2019-nCoV. Cell Mol Life Sci. 2020:1–14. doi: 10.1007/s00018-020-03611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–90. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mönkemüller K, Fry L, Rickes S. Covid-19, Coronavirus, SARS-CoV-2 and the small bowel. Rev Esp Enferm Dig. 2020;112:383–8. doi: 10.17235/reed.2020.7137/2020. [DOI] [PubMed] [Google Scholar]
- 40.Camargo SM, Singer D, Makrides V, Huggel K, Pos KM, Wagner CA, et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009;136:872–82. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold P, Otte A, Becker-Pauly C. Meprin metalloproteases: molecular regulation and function in inflammation and fibrosis. Biochim Biophys Acta Mol Cell Res. 2017;1864:2096–104. doi: 10.1016/j.bbamcr.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Vazeille E, Bringer MA, Gardarin A, Chambon C, Becker-Pauly C, Pender SL, et al. Role of meprins to protect ileal mucosa of Crohn's disease patients from colonization by adherent-invasive E. coli. PLoS One. 2011;6:e21199. doi: 10.1371/journal.pone.0021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strollo R, Pozzilli P. DPP4 inhibition: preventing SARS‐CoV‐2 infection and/or progression of COVID‐19? Diabetes Metab Res Rev. 2020 doi: 10.1002/dmrr.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivares M, Schüppel V, Hassan AM, Beaumont M, Neyrinck AM, Bindels LB, et al. The potential role of the dipeptidyl peptidase-4-like activity from the gut microbiota on the host health. Front Microbiol. 2018;9:1900. doi: 10.3389/fmicb.2018.01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solerte SB, Di Sabatino A, Galli M, Fiorina P. Dipeptidyl peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetol. 2020:1. doi: 10.1007/s00592-020-01539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira JAdS, Silva FCd, de Moraes-Vieira PMM. The impact of ghrelin in metabolic diseases: an immune perspective. J Diabetes Res. 2017 doi: 10.1155/2017/4527980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Salhy M. Ghrelin in gastrointestinal diseases and disorders: a possible role in the pathophysiology and clinical implications. Int J Mol Med. 2009;24:727–32. doi: 10.3892/ijmm_00000285. [DOI] [PubMed] [Google Scholar]
- 48.Chen L, Marishta A, Ellison CE, Verzi MP. Identification of transcription factors regulating SARS-CoV-2 entry genes in the intestine. Cell Mol Gastroenterol Hepatol. 2020;6:S2352–30129. doi: 10.1016/j.jcmgh.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen KB, Chhabra KH, Nguyen VK, Xia H, Lazartigues E. The transcription factor HNF1α induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim Biophys Acta Mol Cell Res. 2013;1829:1225–35. doi: 10.1016/j.bbagrm.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barker H, Parkkila S. Bioinformatic characterization of angiotensin-converting enzyme 2, the entry receptor for SARS-CoV-2. BioRxiv. 2020 doi: 10.1371/journal.pone.0240647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gendron F-P, Mongrain S, Laprise P, McMahon S, Dubois CM, Blais M, et al. The CDX2 transcription factor regulates furin expression during intestinal epithelial cell differentiation. Am J Physiol Gastrointest Liver Physiol. 2006;290:G310–8. doi: 10.1152/ajpgi.00217.2005. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y, Zeng J, Li Q, Li P, Luo F, Zhang W, et al. Preliminary clinical study of direct renin inhibitor aliskiren in the treatment of severe COVID-19 patients with hypertension. Zhonghua Nei Ke Za Zhi. 2020;59:E011. doi: 10.3760/cma.j.cn112138-20200328-00310. [DOI] [PubMed] [Google Scholar]
- 53.Aly OM. Molecular docking reveals the potential of aliskiren, dipyridamole, mopidamol, rosuvastatin, rolitetracycline and metamizole to inhibit COVID-19 virus main protease. chemrxiv. 2020 doi: 10.1039/d0ra03582c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valencia I, Peiró C, Lorenzo Ó, Sánchez-Ferrer CF, Eckel J, Romacho T. DPP4 and ACE2 in diabetes and COVID-19: therapeutic targets for cardiovascular complications? Front Pharmacol. 2020;11:1161. doi: 10.3389/fphar.2020.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goto M, Furuta S, Yamashita S, Hashimoto H, Yano W, Inoue N, et al. Dipeptidyl peptidase 4 inhibitor anagliptin ameliorates hypercholesterolemia in hypercholesterolemic mice through inhibition of intestinal cholesterol transport. J Diabetes Invest. 2018;9:1261–9. doi: 10.1111/jdi.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elkahloun AG, Saavedra JM. Candesartan could ameliorate the COVID-19 cytokine storm. Biomed Pharmacother. 2020:110653. doi: 10.1016/j.biopha.2020.110653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu D, Tang X, Ding L, Cui J, Wang P, Du X, et al. Candesartan attenuates hypertension-associated pathophysiological alterations in the gut. Biomed Pharmacother. 2019;116:109040. doi: 10.1016/j.biopha.2019.109040. [DOI] [PubMed] [Google Scholar]
- 58.Gurwitz D. Angiotensin receptor blockers as tentative SARS‐CoV‐2 therapeutics. Drug Dev Res. 2020;81:537–40. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothlin RP, Vetulli HM, Duarte M, Pelorosso FG. Telmisartan as tentative angiotensin receptor blocker therapeutic for COVID‐19. Drug Dev Res. 2020:10. doi: 10.1002/ddr.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arab HH, Al-Shorbagy MY, Abdallah DM, Nassar NN. Telmisartan attenuates colon inflammation, oxidative perturbations and apoptosis in a rat model of experimental inflammatory bowel disease. PLoS One. 2014;9:e97193. doi: 10.1371/journal.pone.0097193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gommans DF, Nas J, Pinto-Sietsma SJ, Koop Y, Konst RE, Mensink F, et al. Rationale and design of the PRAETORIAN-COVID trial: A double-blind, placebo-controlled randomized clinical trial with valsartan for PRevention of Acute rEspiraTORy dIstress syndrome in hospitAlized patieNts with SARS-COV-2 Infection Disease. Am Heart J. 2020;226:60–68. doi: 10.1016/j.ahj.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skayem C, Ayoub N. Carvedilol and COVID-19: A Potential Role in Reducing Infectivity and Infection Severity. Am J Med Sci. 2020;360:300. doi: 10.1016/j.amjms.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Das UN. Bioactive Lipids as Mediators of the Beneficial Action (s) of Mesenchymal Stem Cells in COVID-19. Aging Dis. 2020;11:746–55. doi: 10.14336/AD.2020.0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castaldo N, Aimo A, Castiglione V, Padalino C, Emdin M, Tascini C. Safety and efficacy of amiodarone in a patient with COVID-19. JACC Case Rep. 2020;2:1307–10. doi: 10.1016/j.jaccas.2020.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]