Adult tissue-derived organoids allow for the expansion and maintenance of primary epithelial cells in a near-native state. These 3D and self-organizing organotypic cultures derived from adult tissues have been increasingly used in fundamental and translational research. A key feature of this organoid system is that it recapitulates the stem cell lineage and thus, the differentiated cell-type heterogeneity of the in vivo tissue of origin. Importantly, we and others have shown that organoids can be manipulated to expand different cell lineages, allowing for the study of rare cell types that would otherwise be very difficult to analyze. Here, focusing specifically on organoids of the small intestine, we discuss recent advances and future directions of this new avenue of organoid research. We highlight methods used to enrich specific cell types including stem cells, enterocytes, Paneth cells, goblet cells, micro-fold (M)-cells, tuft cells, and enteroendocrine cells (EECs) in intestinal organoids and focus on what each of these methods has taught us about the differentiation of adult intestinal stem cells (ISCs) to specific cell fates. Furthermore, we highlight how these new cell type-enriched intestinal organoids can be used to answer a diversity of questions relevant to human biology and disease.
The intestinal cellular landscape
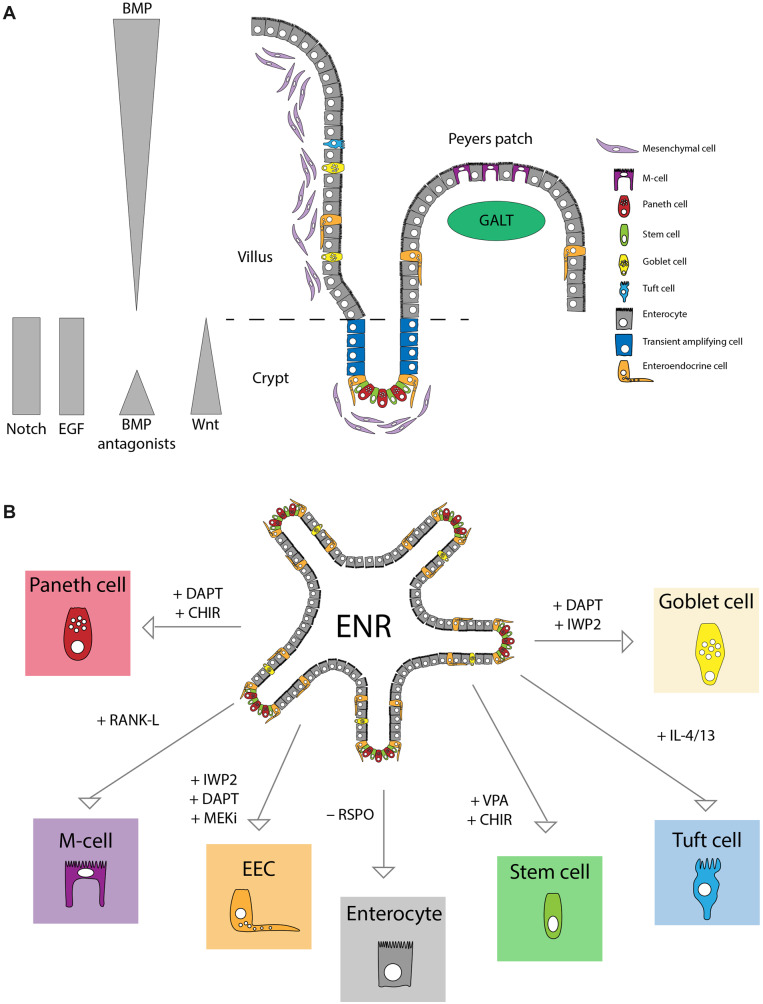
The small intestine is part of the gastrointestinal tract and functions mainly in the digestion of food and absorption of nutrients. The inner lining of the intestine is made up of finger-like protrusions called villi, which maximize the absorptive surface. At the base of the villi are invaginations called crypts, which are home to the intestinal crypt columnar base cells, the ISCs that contribute to self-renewal of the small intestinal epithelium and give rise to all the differentiated epithelial cell types in this tissue. There are six main cell types in the absorptive epithelium of the small intestine, which differ in their relative abundance and their location along the crypt‒villus axis (Figure 1A). The cells with the highest abundance are the absorptive enterocytes, which make up ∼80% of all epithelial cells. Paneth cells, which are secretory cells in the intestinal crypts, and mucus-producing goblet cells, which are found in both the crypt and villus account for ∼3%‒8% and 4%‒12% of all intestinal epithelial cells, respectively. The cells with the lowest abundance include environment sensing M-cells (<1%), chemosensory tuft cells (0.4%‒2%), and hormone-producing EECs (<1%) (Sternini et al., 2008; van der Flier and Clevers, 2009; Gerbe et al., 2012; Haber et al., 2017).
Figure 1.
Mouse intestinal cell types and enrichment in organoids. (A) Schematic overview of the small intestinal tissue. Indicated are the different cell types and niche signals. (B) Indicated are modulations of mouse intestinal organoid medium that result in enrichment for specific cell types.
The small intestinal epithelium is the fastest self-renewing tissue in mammals and it has served as the principle model for the study of adult stem cell biology (van der Flier and Clevers, 2009). Strict regulation of ISC maintenance and differentiation toward specific lineages is the result of different signaling microenvironments along the crypt‒villus axis. While the signals that promote self-renewal and the stem cell state are restricted to the crypt, signals that drive differentiation can be found in different gradients along the villus axis. The primary signaling pathways involved in these processes include Wnt, Notch, bone morphogenic protein (BMP), and epidermal growth factor (EGF) signaling (Figure 1A). These signals are tightly regulated and changes in the abundance or source of one or more often directly result in a disturbed balance in cell type abundance and/or intestinal homeostasis. Our knowledge of these signaling pathways and their role in ISC self-renewal have contributed to the development of adult tissue-derived intestinal organoids (discussed in more detail later). In turn, recent advances in our understanding of the essential minimal signals required for ISC fate specification have been made possible by the organoid system.
Wnt
Wnt signaling is essential for homeostatic ISC renewal in the intestine, and activating mutations of Wnt pathway components are often found in colorectal cancer (CRC; The Cancer Genome Atlas, 2012; Zhan et al., 2017). Canonical Wnt signaling results in the transcription of β-catenin/TCF/LEF target genes. In the absence of Wnt ligands, β-catenin is targeted for degradation by a destruction complex. Binding of Wnt ligands to Frizzled receptors and LRP5/6 at the plasma membrane inactivates the destruction complex, leading to accumulation of β-catenin and its translocation to the nucleus, where it binds to TCF/LEF transcription factors and activates target gene expression.
The importance of Wnt target gene expression to ISCs was first demonstrated by the finding that knockout of Tcf4 in mice resulted in loss of nearly all proliferative crypts (Korinek et al., 1998). Since then, it has been shown that Wnt signaling regulates the expression of a number of genes involved in stem cell maintenance and proliferation. Some of these Wnt target genes include genes that act as positive and negative feedback regulators of the pathway (Zhan et al., 2017). The Wnt target genes, Rnf43 and Znrf3, for example, encode for ubiquitin ligases that downregulate Wnt pathway activity by enabling downregulation of the Frizzled receptors at the plasma membrane (Koo et al., 2012). Lgr5, a cell surface receptor involved in positive regulation of Wnt signaling, is also a Wnt target gene. Secreted R-spondin proteins bind to Lgr5 and together they sequester Rnf43/Znrf3, thereby potentiating Wnt signaling by removing inhibitory signals (de Lau et al., 2011).
Wnt3 ligands secreted by Paneth cells and Wnt2B and R-spondin1 ligands secreted by the subepithelial mesenchyme surrounding the crypt restrict Wnt pathway activation to the crypts (Sato et al., 2011b; Farin et al., 2012; Moor et al., 2016). Importantly, these ligands act in a non-redundant, cooperative manner to regulate ISC maintenance and self-renewal. Wnt ligands are required to prime ISCs to express Lgr5 and thus are responsive to potentiation of the pathway by R-spondin (Yan et al., 2017). The cooperative action of these two ligands further ensures tight regulation of Wnt pathway activation in the intestine.
BMP
BMP signals have also been shown to be important for the control of intestinal tissue homeostasis, and mutations in BMP pathway components are highly associated with CRC (The Cancer Genome Atlas, 2012). Upon binding of BMP ligands to BMP receptors (BMPRs) at the cell surface, an intracellular signaling cascade is enabled resulting in SMAD phosphorylation and transcription of BMP target genes. In the intestine, the main BMP ligands, BMP2 and BMP4, are expressed in the mesenchyme and epithelium of the villus. Likewise, BMPRs are also expressed in both the epithelium and mesenchyme of the small intestine, suggesting the importance of this pathway to both of these cellular compartments. The main BMPR in the intestinal epithelium is BMPR1A. BMP inhibitors Noggin, Gremlin 1/2, and Chordin-like 1 are produced in the mesenchyme underlying the crypt (Wang and Chen, 2018). As a result, BMP pathway activation is highest at the villus tip and decreases along the villus axis toward the crypt. The first indication that BMP signaling inhibits the ISC identity was the finding that transgenic mice expressing Xenopus Noggin formed ectopic crypts in the villi (Haramis et al., 2004). Similarly, loss of BMPR1A in the intestinal epithelium resulted in expansion of the ISC compartment in mice (Qi et al., 2017).
Notch
Notch signaling has been shown to both promote ISC self-renewal and homeostasis and regulate cell fate decisions. Furthermore, activated Notch signaling has been observed in CRC (Noah and Shroyer, 2013). Notch signaling occurs via cell‒cell contact. Binding of any of the five canonical Notch ligands to any of the four Notch receptors results in γ-secretase-mediated proteolytic cleavage of the receptor and release of the Notch intracellular domain (NICD). The NICD then translocates to the nucleus where it binds to the DNA-binding protein, RBPjκ, and activates transcription of Notch target genes. Direct Notch target genes include the hairy/enhancer of split (HES) class of transcriptional repressors, which, upon activation, act as Notch signaling effectors.
While Notch ligands DLL1 and DLL4 are presented by Paneth cells and secretory progenitor cells, Notch receptors such as NOTCH1 are expressed by ISCs, and their interactions in the crypt contribute to stem cell maintenance. Notch signaling is also involved in cell-type specification in the intestine. Whereas active Notch signaling in cells undergoing specification promotes the absorptive cell fate, lack of Notch signals leads to induction of a secretory cell fate via expression of Math1, the main transcription factor involved in determining secretory lineages (Noah and Shroyer, 2013). Once specified, secretory progenitors upregulate the expression of Notch ligands and thus promote the absorptive cell fate in surrounding cells. Inactivation of the Notch effectors, Hes1, Hes3, and Hes5, in mice resulted in decreased proliferation and increased differentiation toward the secretory lineage (Ueo et al., 2012).
EGF
EGF signaling is one of the major contributors to ISC proliferation, and mutations in EGF pathway components, including EGF receptor (EGFR) and the downstream effector, KRAS, are frequent in CRC (The Cancer Genome Atlas, 2012). Binding of EGF to its receptors EGFR/ErbB1 and other ErbB family members induces tyrosine kinase activity and activates downstream pro-proliferative and pro-survival signaling cascades, such as the MAPK and PI3K pathways. In the intestine, EGF ligands are produced by Paneth cells and the mesenchyme underlying the crypt. The ISCs, in turn, co-express EGFRs (ErbB family) and their negative regulator, leucine-rich repeats and immunoglobulin-like domains protein 1 (LRIG1). The importance of this co-expression to maintaining ISC homeostasis is highlighted by the fact that Lrig1 knockout mice show both increased EGFR activation and crypt hyperplasia (Wong et al., 2012).
In vitro modeling of the intestine in organoid cultures
In 2009, Sato and colleagues applied what was known about the ISC niche to grow small intestinal epithelium from isolated ISCs. The resulting culture system, which was based on growing isolated ISCs in extracellular matrix and a specific growth factor cocktail, was referred to as intestinal adult stem cell organoid culture technology (Sato et al., 2009). The specific growth factor cocktail consisted of EGF, the BMP antagonist Noggin, and R-spondin, and was thus termed ENR. Modification of this medium by providing additional components allowed for the expansion of human small intestinal and colon organoids (Sato et al., 2011a). Under basal growth conditions, mouse and human intestinal organoids show cell-type heterogeneity similar to that of the tissue of origin. Over the years, through further modification of the system, a diverse array of adult tissue-derived cultures from other epithelial tissues and from many different kinds of tumors have been successfully established (Kretzschmar and Clevers, 2016). It is important to note that similar organoid structures can also be derived from induced pluripotent stem cells (iPSCs), and these culture systems have contributed significantly to our understanding of stem cell biology and disease (Kretzschmar and Clevers, 2016). For the purposes of this perspective article, we will focus on adult tissue-derived intestinal organoids.
Mouse and human small intestinal organoids have been used in a wide variety of applications including disease modeling, regenerative medicine, drug screening, and proof of concept correction of genetic defects in vitro. An exciting new application for adult tissue-derived organoids is the study of rare cell types and how these contribute to human biology and disease.
In vitro cell-type enrichment in intestinal organoids
Under basic conditions, murine intestinal organoids contain ISCs, Paneth cells, goblet cells, EECs, and enterocytes with the abundance similar to that in the in vivo tissue. To allow for research into specific (and rare) cell types, intestinal organoid protocols have been adapted to push cellular homeostasis within the organoid toward specific cell fates. Stem cell-enriched organoid cultures are a prime example of this (Figure 1B).
Primary epithelial cell types of the intestine: stem cells, enterocytes, Paneth cells, and goblet cells
Addition of Wnt3A-conditioned medium to the standard ENR medium (WENR medium) results in stem cell-enriched organoids that homogenously proliferate (Sato et al., 2011a). Intestinal organoid cultures can be even further enriched for stem cells by simultaneous stimulation of Wnt signaling and activation of the Notch pathway through addition of the GSK3β-inhibitor CHIR990221 (CHIR) and the HDAC inhibitor valproic acid (ENR-CV medium) (Yin et al., 2014). Similarly, organoid cellular homeostasis can be pushed in the opposite direction, by simply removing the Wnt potentiator, R-spondin, from the standard ENR medium (EN medium), thereby recapitulating the absence of Wnt signaling in the villi in vivo (Yin et al., 2014). These stem cell-enriched and stem cell-depleted intestinal organoid cultures have been exploited to identify defining features of stem cells vs. differentiated cells and to further understand the molecular mechanisms that drive differentiation. For example, through multi-omic analysis of standard ENR, stem cell-enriched CV-ENR, and enterocyte-enriched EN organoids, researchers identified the transcription factor hepatocyte nuclear factor 4 gamma (Hnf4g) as a driver of differentiation toward the enterocyte lineage (Lindeboom et al., 2018).
Paneth and goblet cells are two related cell types of the secretory lineage with different functions. In addition to acting as anti-microbial cells that produce lysozyme and various defensins, Paneth cells function as niche cells that surround ISCs in the crypts and provide growth signaling molecules that are required for ISC proliferation and maintenance (Sato et al., 2011b). Goblet cells produce mucus, which lubricates the intestinal lining and serves as a protective layer against mechanical and biological damage and pathogen entry. As mentioned above, absence of Notch signaling during cell-type specification leads to differentiation toward a secretory cell fate (Noah and Shroyer, 2013). The Wnt signaling context under which a cell experiences absence of Notch signaling determines which of these specific secretory cell types, Paneth cell or goblet cell, is generated. Consistent with their location along the crypt‒villus axis in vivo, Paneth cells, found in the crypt, are dependent on high Wnt signaling, while goblet cells, found mostly, though not exclusively, along the villus are not (Farin et al., 2012; Yin et al., 2014).
Addition of the NOTCH inhibitor DAPT to stem cell-enriched murine intestinal organoids promotes secretory cell differentiation but does not appear to favor one secretory lineage vs. the other beyond what would be expected given the natural abundance of each lineage in vivo. However, simultaneous addition of DAPT and CHIR (ENR+DC), to stimulate the Wnt pathway in these cultures, results in a clear preference for Paneth cell differentiation (Figure 1B; Yin et al., 2014). Simultaneous addition of DAPT and the Wnt pathway inhibitor IWP2 (ENR+DI), on the other hand, results in intestinal organoid cultures in which goblet cell differentiation is favored (Figure 1B; Yin et al., 2014). This system has been used to identify cell type-specific regulators of Paneth and goblet cells through transcriptomic analysis and comparison of ENR organoids to Paneth and goblet cell-enriched organoids (Treveil et al., 2019). In a separate study, using single-cell RNA-sequencing (scRNA-seq) of ENR organoids, Paneth cell-enriched organoids, and freshly isolated primary mouse intestine tissue, researchers identified Nupr1 as a potentially important transcription factor for the survival and development of Paneth cells (Mead et al., 2018).
Rare intestinal epithelial cell types: M-cells, tuft cells, and EECs
M-cells are specialized epithelial cells located in the follicle-associated epithelium (FAE) overlaying gut-associated lymphoid tissue (GALT), including Peyer’s patches and isolated lymphoid follicles. In the FAE, M-cells function as gatekeeper cells, sampling antigens from the intestinal lumen and transferring them to lymphocytes in the GALT where immune surveillance takes place (Randall et al., 2008). The direct lineage relationship between Lgr5+ ISCs and M-cells was first established in 2012 through genetic lineage tracing of Lgr5+ cells in mice (de Lau et al., 2012). In the same study, researchers showed that treatment of intestinal organoids with RANKL, a cytokine that is expressed by the reticular cells below the FAE, resulted in increased M-cell differentiation in these cultures. This was the first example of an intestinal organoid culture system enriched for a very rare cell type.
The authors of this study further showed that RANKL-induced M-cell differentiation of ISCs in organoids was dependent on expression of the Ets transcription factor SpiB—a finding consistent with their observation in vivo that SpiB−/− mice lack M-cells (de Lau et al., 2012). This differentiation protocol was later applied in combination with scRNA-seq to identify an M-cell-specific gene expression signature, which was previously not possible due to scarcity of these cells in vitro (Haber et al., 2017).
Chemosensory tuft cells are rare cells in the small intestinal epithelium with an abundance of <2%. In mice, these cells have been shown to orchestrate type-2 immune responses to a variety of stimuli, including infection with the parasite Helminth (Gerbe et al., 2016; Howitt et al., 2016). The tuft cell-mediated type-2 immune response to Helminth infection was accompanied by an expansion of tuft cell number, implicating a positive feedback loop in which tuft cell activation resulted in further differentiation of stem cells toward the tuft cell fate. Interleukin-4 receptor α (IL-4Rα) signaling is important for type-2 immune responses to Helminth infection, and treatment of mice with recombinant IL-4 and IL-13, two cytokines that signal through IL-4Rα, was sufficient to induce expansion of both goblet and tuft cells in vivo (Gerbe et al., 2016). Stimulation of intestinal organoid cultures with recombinant IL-4 and/or IL-13 led to an increase of goblet and tuft cells in vitro, suggesting that tuft cell responses to immune signaling can be recapitulated in organoids. These experiments in organoids confirmed the data from in vivo experiments in mice and concurrently provided a new ISC differentiation protocol that can be used as a tool to enrich and study tuft cells in vitro. Through scRNA-seq profiling of mouse small intestinal epithelium, Haber et al. (2017) identified two subtypes of tuft cells, tuft-1 and tuft-2, which were characterized by a neuronal-like signature and an immune-like signature, respectively. Enrichment for tuft cells in human small intestinal organoids by stimulation with IL-4 and IL-13 might allow researchers to not only identify similar heterogeneity in human tuft cells but also perform functional analyses of these cells in a defined in vitro system.
Among the rare cell types of the intestinal epithelium, EECs show the highest degree of complexity. Based on their hormone expression patterns, EECs can be divided into seven major lineages, K-cells, L-cells, delta-cells, X-cells, I-cells S-cells, and N-cells. Classification based on combinatorial expression patterns of different EEC-produced hormones, however, can result in up to 20 subtypes (Habib et al., 2012; Haber et al., 2017). These numbers support the need for a method to enrich the full diversity of EECs in vitro. Studies in intestinal organoids have provided fundamental insights into the cellular signals and transcriptional programs that drive EEC differentiation.
In particular, shRNA-mediated knockdown of the transcription factor Neurogenin 3 (NEUROG3) in human iPSC-derived intestinal organoids confirmed previous studies in mice showing that Neurog3 is required for EEC development (Spence et al., 2011). Furthermore, overexpression of NEUROG3 in the same system showed that it is also sufficient to induce EEC differentiation in human cells.
EEC differentiation in murine adult tissue-derived organoids has been achieved by applying an EEC differentiation cocktail for combined inhibition of Notch, Wnt, and MAPK signaling (Figure 1B; Basak et al., 2014). Importantly, this system recapitulates the EEC subtype distribution observed in vivo across the length of the small intestine, i.e. ileum-derived organoids favor ileal EEC subtypes and duodenum-derived organoids favor duodenal EEC subtypes. Likewise, a follow-up study showed that while the original EEC differentiation cocktail gave rise to EEC subtypes found in the crypt region including GLP-1-producing L-cells, stimulation of organoids with the EEC differentiation cocktail and BMP4 resulted in EECs consistent with those found in the villus, such as Secretin (Sct)-producing S-cells (Beumer et al., 2018). Treatment of mice with an inhibitor of the BMP receptor, BMPR1Α, resulted in the loss of S-cells concomitant with an increase in L-cells specifically in the villus.
This EEC differentiation protocol for intestinal organoids has also proven useful for assessing gene function in the context of EEC differentiation. Using scRNA-seq of sorted EECs at different stages of their differentiation trajectory from early NEUROG3-expressing cells to mature EECs, researchers characterized spatiotemporal gene expression in the EEC lineage and identified potential regulators of the process. CRISPR-based knockout of these candidate regulators of EEC differentiation in mouse intestinal organoids followed by induction of EEC differentiation in knockout organoids allowed researchers to assess their function. Knockout of Rfx6 and Tox3 in mouse intestinal organoids, for example, resulted in skewed EEC subtype specification upon EEC differentiation (Gehart et al., 2019).
Future perspectives
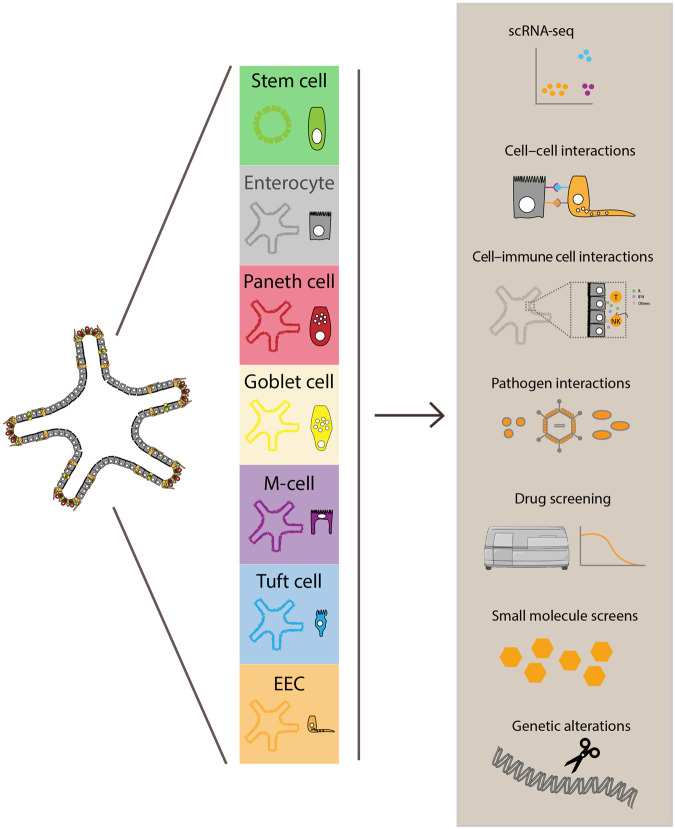
The ability to easily modify in vitro organoid cultures by modulating the media composition and thereby influencing their differentiation state has made it possible to perform experiments on cells that are too sparse in vivo to study (Figure 2). Enrichment for specific cell types in organoids has increased the feasibility of performing large-scale single-cell transcriptomic analysis of specific cell types along their differentiation trajectory from stem cell to mature differentiated cell. Furthermore, organoid differentiation protocols provide a platform for the study of cell type-specific responses to external soluble stimuli such as small molecules and drugs. As certain pathogens are known to infect only specific cell types that harbor expression of certain membrane proteins, it is easy to imagine how organoids enriched for specific cell types might serve as a platform to study mechanisms of infection as well as cell type-specific responses to infection in vitro. Some of the examples we have provided here highlight how organoids enriched for specific cell types can be used for faster assessment of gene function and phenotypes in rare cell types than is possible in vivo.
Figure 2.
Overview of the generation and use of cell type-enriched intestinal organoids. Stimulation of organoids with pathogens, specific niche factors, certain drugs, or nutrients results in cell-type enrichment in organoids. These cell type-enriched organoids can be used for a wide variety of different kinds of studies including single-cell scRNA-seq analysis, cell‒cell or cell‒pathogen interaction studies, screens with drugs or small molecules, and studies involving analysis of specific genetic alterations. T, T-cells; NK, natural killer cells; IL, interleukins; IFN, interferons.
Small intestinal organoids enriched for EECs could, for example, be challenged with panels of small molecules to identify those that stimulate the production and secretion of specific hormones. In vitro insights into the molecular mechanisms that drive hormone production and secretion could potentially be translated to the clinic for patients in need of more hormone production. Small molecules that enrich and activate L-cells, which produce GLP-1, a hormone that induces the release of insulin, could be used to treat diabetic patients. Similar scenarios can be envisioned for tuft cell-enriched organoids, which could be used in combination with type-2 immune response stimuli to elucidate tuft cell-involved intrinsic mechanisms of type-2 immune responses or to model how these responses might be overactivated in immune-mediated diseases.
Altogether, the studies emphasized here exemplify how in vivo systems can be used to inform the development of in vitro organoid models of specific differentiation states to support the study of rare cell types. The ability to drive ISC differentiation toward specific rare cell-type lineages by modulating the medium components of organoid cultures has made the study of these specific cell types more accessible. Whereas previously, the primary means of studying rare cell types was the use of model organisms or fresh human tissue in combination with surface marker or genetic lineage label-based isolation, rare cell type-enriched organoids have made it possible to study these cell types sometimes even at the level of bulk organoid cultures. Furthermore, while still more labor intensive than 2D cell lines, organoids are more easily manipulated than an entire organism, and the system opens the door to studying rare cell types in the context of a more simplified cellular milieu allowing researchers to distill specific cell‒cell interactions and dynamics. In the future, the complexity of this cellular milieu could be increased in a stepwise manner. Cell type-enriched organoids could be co-cultured with specific cellular components of the non-epithelial microenvironment, such as fibroblasts or specific immune cell types, to study the crosstalk of specific epithelial cell types with their microenvironment. Moreover, many of the differentiation protocols highlighted here have not yet been exploited to their full potential. In the next few years, we envisage rapidly growing implementation of these systems and others in both basic and clinical research.
[This work was supported by the European Research Council under ERC Advanced Grant Agreement no. 67013 (H.C. and K.B.) and the Neuroendocrine Tumor Research Foundation under a Petersen Accelerator Award (H.C. and T.D.). T.D. was supported by an EMBO postdoctoral fellowship and a Marie Curie Individual Fellowship. H.C. is an inventor of several patents related to organoid technology. His full disclosure is given at https://www.uu.nl/staff/JCClevers/Additional functions.]
References
- Basak O., van den Born M., Korving J., et al. (2014). Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 33, 2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer J., Artegiani B., Post Y., et al. (2018). Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 20, 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., et al. (2011). Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–298. [DOI] [PubMed] [Google Scholar]
- de Lau W., Kujala P., Schneeberger K., et al. (2012). Peyer’s patch M cells derived from Lgr5+ stem cells require SpiB and are induced by RankL in cultured 'miniguts'. Mol. Cell. Biol. 32, 3639–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin H.F., van Es J.H., Clevers H. (. 2012). Redundant sources of Wnt regulate intestinal stem cells and promote formation of paneth cells. Gastroenterology 143, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Gehart H., van Es J.H., Hamer K., et al. (2019). Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell 176, 1158–1173. [DOI] [PubMed] [Google Scholar]
- Gerbe F., Legraverend C., Jay P. (. 2012). The intestinal epithelium tuft cells: specification and function. Cell. Mol. Life Sci. 69, 2907–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F., Sidot E., Smyth D.J., et al. (2016). Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber A.L., Biton M., Rogel N., et al. (2017). A single-cell survey of the small intestinal epithelium. Nat. Publ. Gr. 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib A.M., Richards P., Cairns L.S., et al. (2012). Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology 153, 3054–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramis A.-P.G., Begthel H., van den Born M., et al. (2004). De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686. [DOI] [PubMed] [Google Scholar]
- Howitt M.R., Lavoie S., Michaud M., et al. (2016). Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.-K., Spit M., Jordens I., et al. (2012). Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Moerer P., et al. (1998). Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19, 379–383. [DOI] [PubMed] [Google Scholar]
- Kretzschmar K., Clevers H. (. 2016). Review organoids: modeling development and the stem cell niche in a dish. Dev. Cell 38, 590–600. [DOI] [PubMed] [Google Scholar]
- Lindeboom R.G.H., van Voorthuijsen L., Oost K.C., et al. (2018). Integrative multi-omics analysis of intestinal organoid differentiation. Mol. Syst. Biol. 14, e8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B.E., Ordovas-Montanes J., Braun A.P., et al. (2018). Harnessing single-cell genomics to improve the physiological fidelity of organoid-derived cell types. BMC Biol. 16, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor A.E., Cantu C., Aguet M., et al. (2016). Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 15, 911–918. [DOI] [PubMed] [Google Scholar]
- Noah T.K., Shroyer N.F. (2013). Notch in the intestine: regulation of homeostasis and pathogenesis. Annu. Rev. Physiol. 75, 263–288. [DOI] [PubMed] [Google Scholar]
- Qi Z., Li Y., Zhao B., et al. (2017). BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall T.D., Carragher D.M., Rangel-Moreno J. (2008). Development of secondary lymphoid organs. Annu. Rev. Immunol. 26, 627–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., et al. (2011. a). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772. [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., et al. (2011. b). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., et al. (2009). Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–266. [DOI] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternini C., Anselmi L., Rozengurt E. (2008). Enteroendocrine cells: a site of 'taste' in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 15, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treveil A., Sudhakar P., Matthews Z.J., et al. (2019). Regulatory network analysis of Paneth cell and goblet cell enriched gut organoids using transcriptomics approaches. Mol. Omics 16, 39–58. [DOI] [PubMed] [Google Scholar]
- Ueo T., Imayoshi I., Kobayashi T., et al. (2012). The role of Hes genes in intestinal development, homeostasis and tumor formation. Development 139, 1071–1082. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., Clevers H. (2009). Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260. [DOI] [PubMed] [Google Scholar]
- Wang S., Chen Y.-G. (2018). BMP signaling in homeostasis, transformation and inflammatory response of intestinal epithelium. Sci. China Life Sci. 61, 800–807. [DOI] [PubMed] [Google Scholar]
- Wong V.W.Y., Stange D.E., Page M.E., et al. (2012). Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 14, 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K.S., Janda C.Y., Chang J., et al. (2017). Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature 545, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Farin H.F., van Es J.H., et al. (2014). Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T., Rindtorff N., Boutros M. (2017). Wnt signaling in cancer. Oncogene 36, 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]