Abstract
In 2020, as part of a diagnosis attempt at IHU Méditerranée Infection in Marseille (France), a blood specimen was obtained from a 59-year-old man with chronic obstructive pulmonary disease symptoms, from which we isolated the new bacterial Corynebacterium haemomassiliense strain Marseille-Q3615T. Matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) failed to identify this isolate. Analysis of the 16S ribosomal RNA gene and genome-to-Genome comparison suggested that this taxon belongs to a novel bacterial species within the family Corynebacteriaceae in the phylum Actinobacteria. We describe the main phenotypic characteristics, genome sequence and annotation of Corynebacterium haemomassiliense strain Marseille-Q3615T, a new member of the Corynebacterium genus, which we propose as the type strain.
Keywords: Bacteria, corynebacterium, genome, human, sp. nov., species, taxonogenomics
Introduction
The genus Corynebacterium comprises 173 species [1], some of which are of medical, veterinary or biotechnological interest [2]. Corynebacterium haemomassiliense strain Marseille-Q3615T was isolated as part of a diagnosis attempt at IHU Méditerranée Infection in Marseille (France). A taxonogenomics approach—including matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS), phylogenetic analysis, main phenotypic description and genome sequencing—was used to describe this species [3,4]. The genome of Corynebacterium haemomassiliense strain Marseille-Q3615T is 2.578.128 bp long with 65.28% G+C content. This new bacterium is most closely related to Corynebacterium pilbarense strain DSM 45350, with a 16S ribosomal RNA (rRNA) sequence identity value of 99.12%. Furthermore, genomic comparison using the OrthoANI parameter provided a value of 73.65% with Corynebacterium striatum strain KC-NA01 (NZ_CP014634.1) and a digital DNA-DNA hybridization (dDDH) value of 43.3% with Corynebacterium afermentans strain DSM 44280, the closest species with standing in nomenclature.
Material and method
Strain isolation and phenotypic tests
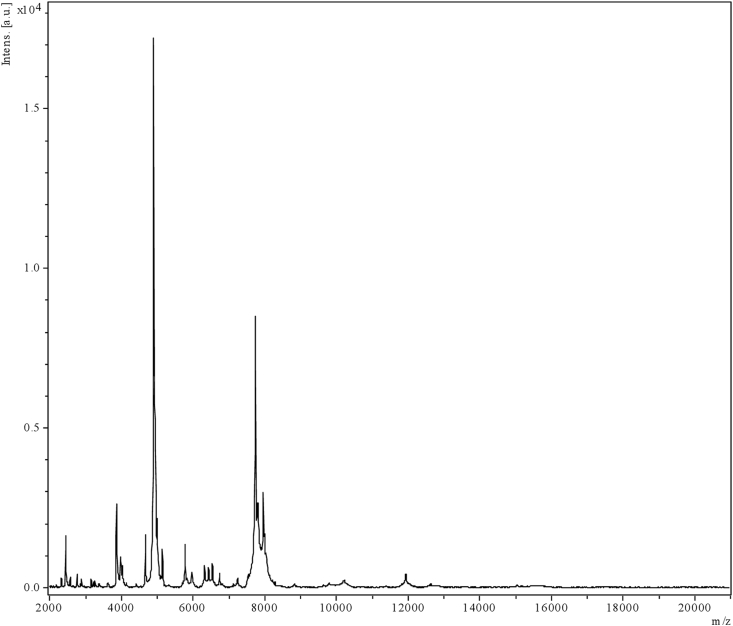
Corynebacterium haemomassiliense strain Marseille-Q3615T was initially isolated from a liquid aerobic haemoculture bottle (BACT/ALERT; bioMérieux, Marcy l’Etoile, France) incubated for 24 hours at 37°C and is routinely cultivated on Columbia agar with 5% sheep's blood media (bioMérieux) incubated in in the presence of oxygen at 37°C. MALDI-TOF MS protein analysis was carried out with a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [5]. Spectra from strain Marseille-Q3615T (Fig. 1) were imported into Biotyper 3 software (Bruker) and analysed by standard pattern matching using the default parameter settings. Different growth temperatures (20, 31.5, 37, 45, 56°C), atmosphere conditions, anaerobic, aerobic and microaerophilic (CampyGEN; Oxoid, Basingstoke, UK) and pH (5.5, 6.5, 7.5, 8.5) were tested. API ZYM, API Coryne and API 50 CH strips (bioMérieux) were used to evaluate the biochemical properties of the strain according to the manufacturer's instructions. For scanning electronic microscopy, a colony was collected from agar and immersed into a 2.5% glutaraldehyde fixative solution. The slide was gently washed in water, air dried and a sample approximately 60 cm in height and 33 cm in width examined to evaluate bacterial structure on a TM4000Plus microscope (Hitachi High-Tech, Tokyo, Japan). A motility test was performed using the semisolid 2,3,5‐triphenyltetrazolium chloride (TTC) media as described by Tittsler and Sandholzer [6].
Fig. 1.
MALDI-TOF MS reference mass spectrum. Spectra from 12 individual colonies of Corynebacterium haemomassiliense strain Marseille-Q3615T were compared and reference spectrum generated.
The study was validated by the ethics committee of the Institut Fédératif de Recherche IFR48.
Genome sequencing
Genomic DNA of Corynebacterium haemomassiliense strain Marseille-Q3615T was extracted using the EZ1 biorobot (Qiagen, Germantown, MD, USA) with the EZ1 DNA tissue kit. After mechanical and enzymatic pretreatment (respectively by glass bead and acid washing; G4649-500g; Sigma-Aldrich, St Louis, MO, USA) using a FastPrep-24 5G grinder (mBio, Santa Ana, CA, USA) and lysozyme incubation at 37°C. Genomic DNA was next sequenced on the MiSeq Technology device (Illumina, San Diego, CA, USA) with the paired end strategy using the Nextera XT DNA sample prep kit (Illumina). The purification step was performed using AMPure XP beads (Beckman Coulter, Brea, CA, USA), and libraries were normalized according the Nextera XT protocol (Illumina). They were pooled into a single library for sequencing via MiSeq Automated cluster generation. Paired end sequencing with dual index reads was performed in a single 39-hour run at a 2 × 250 bp read length. Total information (4.8 Gb) was obtained from a 511/mm2 cluster density with a cluster passing quality-control filters of 90.7%. Within this run, the index representation for Corynebacterium haemomassiliense Marseille-Q3615T was determined to index 4.8%. The 9 843 335 paired end reads were filtered according to the read qualities.
Phylogeny, genome annotation and genome comparison
Assembly was performed by SPAdes software v3.10 using the default parameters [7]. Genome annotation was obtain through the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline [8]. By extracting the sequence, a 16S rRNA–based phylogenetic tree was obtained using the Maximum Likelihood method parameter within MEGA 7 software [9]. The Genome-to Genome Distance Calculator (GGDC) web server (http://ggdc.dsmz.de) was used to estimate the overall identity among compared genomes and to replace wet-lab DNA-DNA hybridization (DDH) with dDDH. The degree of genomic identity of Corynebacterium haemomassiliense strain Marseille-Q3615T with closely related species was estimated by OrthoANI software [10]. Antibiotic resistance genes and presence of pathogenesis-related proteins was investigated using the ABRicate tool and CARD, Resfinder, Virulence Factor Database (VFDB) and PlasmidFinder databases of the Online Galaxy platform [11].
Results
Strain identification and classification
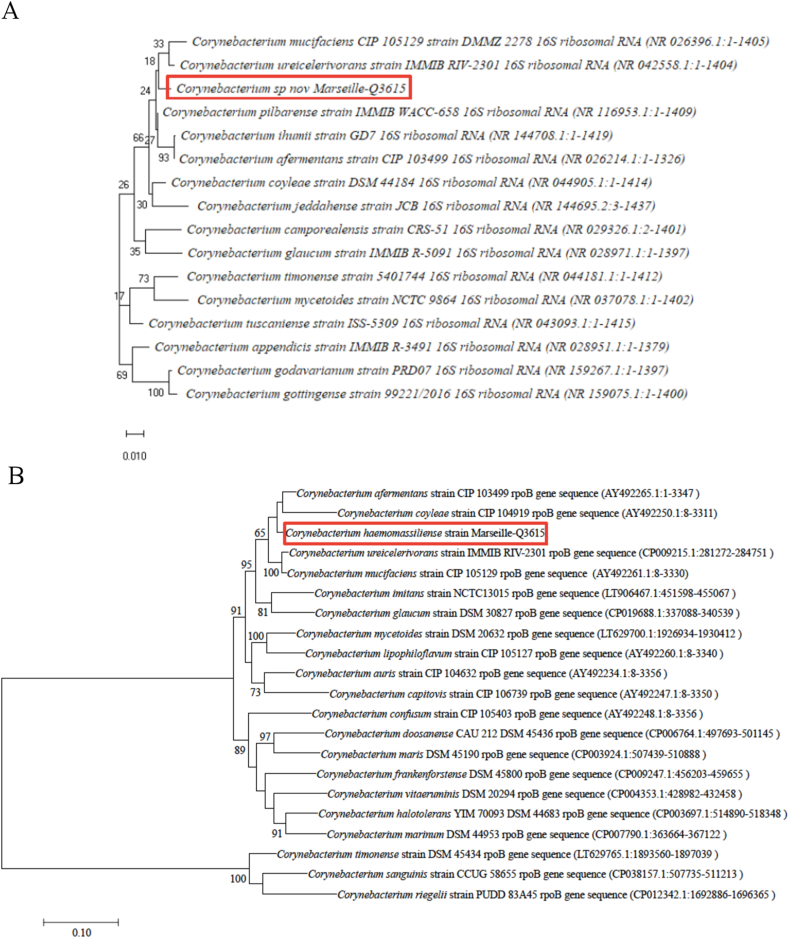
Corynebacterium haemomassiliense strain Marseille-Q3615T was isolated from a blood specimen of a 59-year-old man with chronic obstructive pulmonary disease symptoms. This strain failed to be identified by our systematic MALDI-TOF MS screening, suggesting that the corresponding species was not in the database (https://www.mediterranee-infection.com/acces-ressources/base-de-donnees/urms-data-base/). By analysing its conserved sequences, Corynebacterium haemomassiliense strain Marseille-Q3615T exhibited a 99.12% 16S gene sequence identity with Corynebacterium pilbarense strain DSM 45350 (NR_116953.1), the phylogenetically closest bacterium with standing in nomenclature, and a 95.60% rpoB gene identity, which was shown to be more discriminant for Corynebacterium species, thereby validating the <96.6% identity cutoff described by Khamis et al. [12], with sequence identity with Corynebacterium ureicelerivorans strain DSM 45051 (CP_009215.1) (Fig. 2). The dDDH analysis between the novel organism with the Corynebacterium afermentans strain DSM 44280 type strain revealed an identity of only 43.3%, and the OrthoANI parameter provided a value of 73.65% with Corynebacterium aurismucosum strain ATCC 700975. These both values are below the species delineation cutoff [13].
Fig. 2.
16S rRNA gene (A) and rpoB gene (B) based phylogenetic trees highlighting position of Corynebacterium haemomassiliense sp. nov. strain Marseille-Q3615 (red) relative to other closely related bacterial taxa. Sequences were aligned by Muscle v3.8.31 with default parameters; phylogenetic relationship was inferred by maximum likelihood method, with 1000 bootstrap replicates, within MEGA 7 software.
Phenotypic characteristics
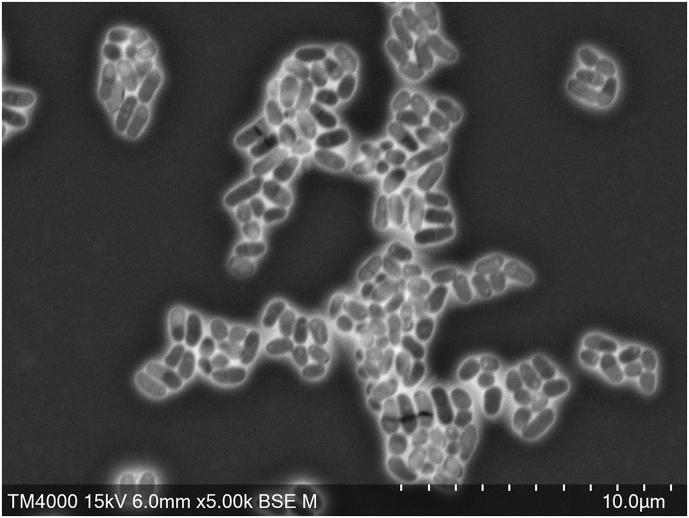
Colonies from strain Marseille-Q3615T showed white pigmentation and no haemolysis. Bacterial cells were Gram-positive, nonmotile, rod-shaped bacilli with a size of 1.8 × 0.2 μm determined by scanning electron microscopy (Fig. 3). Strain Marseille-Q3615T is a facultative aerobe. Optimal growth medium pH and NaCl concentration are 5.5–8.5 and 10–15 g/L respectively. The sporulation test (20 minutes at 80°C) was negative. Using API (analytical profile index) strips (bioMérieux), positive reactions were obtained for pyrazinecarboxamide, 2-naphthyl-phosphate, d-glucose, d-ribose, d-saccharose (sucrose), alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase and N-acetyl-β-glucosaminidase. All other reactions tested were negative. In addition, this bacterium was catalase positive and oxidase negative. Table 1 provides the main characteristic of the strain compared with relative species with standing in nomenclature.
Fig. 3.
Scanning electron micrograph of Corynebacterium haemomassiliense sp. nov. strain Marseille-Q3615T using TM4000Plus tabletop microscope (Hitachi High-Tech, Tokyo, Japan). Scale bar represents 5 μm.
Table 1.
Characteristics permitting discrimination of Corynebacterium haemomassiliense strain Marseille-Q3615T from closest species with standing in nomenclature
| Characteristic | C. haemomassiliense | C. pilbarense | C. ureicelerivorans | C. mucifaciens | C. coyleae | C. ihumii |
|---|---|---|---|---|---|---|
| Strain | Marseille-Q3615 | IMMIB WACC 658 | IMMIB RIV-2301 | CCUG 36878 | DSM 44184 | GD7 |
| Cell diameter | 1.8 × 0.2 μm | 0.5–2.0 μm | 1 mm | 0.7 μm | ||
| Oxygen requirement | Facultative | Facultative | Facultative | Facultative | Facultative | Facultative |
| Gram strain | − | + | + | + | + | + |
| Motility | − | − | − | − | − | − |
| Endospore formation | NA | − | − | − | − | − |
| Optimum temperature for growth | 31.5–56°C | NA | NA | NA | NA | 37°C |
| Production of: | ||||||
| Alkaline phosphatase | + | + | + | + | + | + |
| Catalase | + | + | + | + | + | + |
| Oxidase | − | − | − | − | NA | − |
| α-Glucosidase | − | − | − | − | − | − |
| β-Galactosidase | − | − | − | − | − | − |
| Acid from: | ||||||
| N-Acetylglucosamine | + | − | − | − | − | + |
| l-Arabinose | − | − | + | − | − | + |
| d-Ribose | + | + | + | v | + | + |
| d-Mannose | − | NA | NA | + | + | + |
| d-Mannitol | − | − | − | − | − | + |
| d-Glucose | + | + | + | + | + | + |
| d-Fructose | − | NA | NA | + | + | + |
| d-Maltose | − | − | − | − | − | + |
| d-Lactose | − | − | NA | − | − | + |
| G+C content | 65.28% | NA | NA | 64 mol% | 62 mol% | 65.1 mol% |
| Habitat | Human healthy skin | Ankle aspirate from male patient | Blood culture | Human clinical material | Human clinical specimens | Faecal flora from 62-year-old man |
+, positive result; −, negative result; v, variable result; NA, data not available.
Genome properties
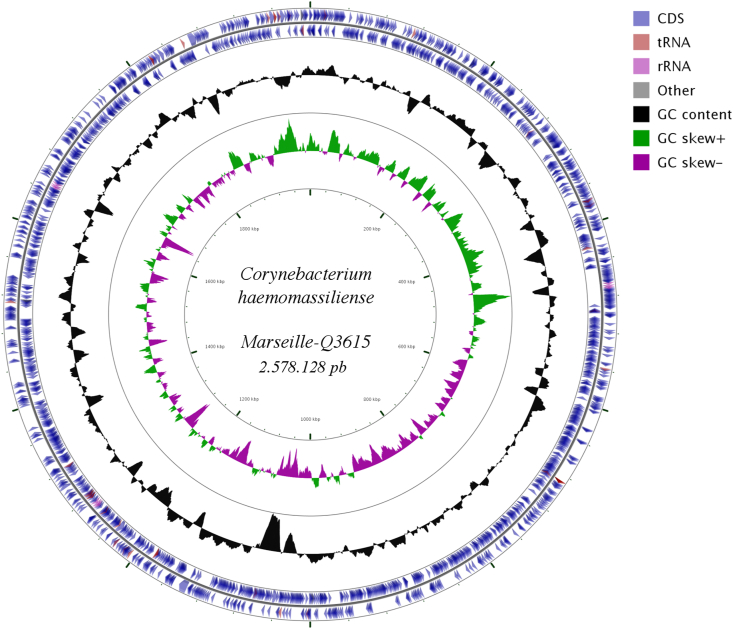
The genome size of strain Marseille-Q3615T is 2 578 128 bp long with a 65.28% G+C content. The genome de novo assembly of this strain was achieved on four contigs (Fig. 4). Of the 2431 predicted genes, 2365 were protein-coding genes and 66 were RNAs (four 16S rRNA, four additional 5S rRNAs, four additional 23S rRNAs, three noncoding RNAs, 51 transfer RNAs). The genome properties and distribution of genes into Clusters of Orthologous Groups (COGs) functional categories are detailed in Table 2. The in silico resistome of the strain Marseille-Q3615T shows three genes—erm(X)_4, tet(W)_4 and cmx_1—with high percentage identity (94.85%, 99.01% and 99.83% respectively) that could be involved in tetracycline and chloramphenicol resistance. Neither associated plasmid nor virulence factor was found. The OrthoANI parameter provided a value of 73.65% with Corynebacterium striatum strain KC-NA01 (Fig. 5) and a dDDH value of 43.3% with Corynebacterium afermentans strain DSM 44280.
Fig. 4.
Graphical circular map of genome from strain Marseille-Q3615T obtained by CGView Server tool [14].
Table 2.
Corynebacterium haemomassiliense strain Marseille-Q3615T genes associated with 25 general COGs functional categories
| Code | Description | N |
|---|---|---|
| Information storage and processing | ||
| J | Translation, ribosomal structure and biogenesis | 175 |
| A | RNA processing and modification | 1 |
| K | Transcription | 135 |
| L | Replication, recombination and repair | 113 |
| B | Chromatin structure and dynamics | 0 |
| Cellular processes and signaling | ||
| D | Cell-cycle control, cell division, chromosome partitioning | 31 |
| Y | Nuclear structure | 0 |
| V | Defense mechanisms | 61 |
| T | Signal transduction mechanisms | 73 |
| M | Cell wall/membrane/envelope biogenesis | 99 |
| N | Cell motility | 8 |
| Z | Cytoskeleton | 0 |
| W | Extracellular structures | 1 |
| U | Intracellular trafficking, secretion, and vesicular transport | 15 |
| O | Posttranslational modification, protein turnover, chaperones | 82 |
| X | Mobilome: prophages, transposons | 35 |
| Metabolism | ||
| C | Energy production and conversion | 95 |
| G | Carbohydrate transport and metabolism | 117 |
| E | Amino acid transport and metabolism | 169 |
| F | Nucleotide transport and metabolism | 72 |
| H | Coenzyme transport and metabolism | 107 |
| I | Lipid transport and metabolism | 68 |
| P | Inorganic ion transport and metabolism | 131 |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 34 |
| Poorly characterized | ||
| R | General function prediction only | 143 |
| S | Function unknown | 98 |
COGs, Clusters of Orthologous Groups database.
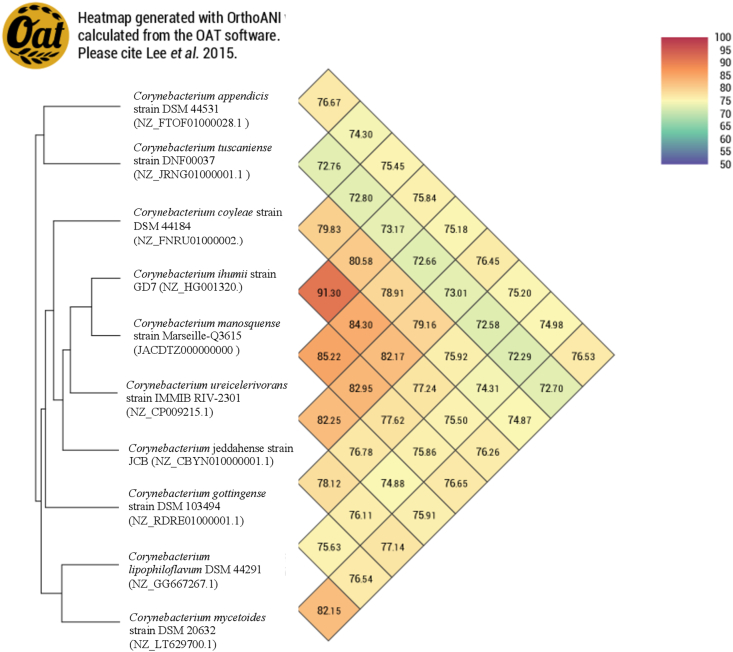
Fig. 5.
Heat map generated with OrthoANI values calculated by OAT software between Corynebacterium haemomassiliense sp. nov. strain Marseille-Q3615T and other closely related species with standing in nomenclature.
Discussion and conclusion
Using the taxonogenomics concept (i.e. the combination of genomic and phenotypic properties of a putative new taxon), we have characterized a new bacterial species representing a new species within the family Corynebacteriaceae found in humans (Table 3). It was named Corynebacterium haemomassiliense strain Marseille-Q3615T, from Gr. fem. n. korynê, ‘club’; L. neut. n. bacterium, ‘rod’, and in biology a bacterium (so called because the first ones observed were rod shaped); N.L. neut. n. Corynebacterium, ‘club bacterium’; Haemomassiliense, ‘blood’ (L. transliteration haema), referring to the nature of the specimen; and massiliense, ‘to Massilia’, the antic name of Marseille, France, where the strain was isolated.
Table 3.
Description of Corynebacterium haemomassiliense sp. nov. strain Marseille-Q3615T
| Type of description | New description |
|---|---|
| Species name | haemomassiliense |
| Genus name | Corynebacterium |
| Specific epithet | Corynebacterium |
| Species status | sp. nov. |
| Species etymology | Corynebacterium haemomassiliense strain Marseille-Q3615T, from Gr. fem. n. korynê, ‘club’; L. neut. n. bacterium, ‘rod’, and in biology a bacterium (so called because the first ones observed were rod shaped); N.L. neut. n. Corynebacterium, ‘club bacterium’; Haemomassiliense, ‘blood’ (L. transliteration haema), referring to the nature of the specimen; and massiliense, ‘to Massilia’, the antic name of Marseille, France, where the strain was isolated |
| Authors | Manon Boxberger, Angéline Antezack, Sibylle Magnien, Nadim Cassir, Bernard La Scola |
| Designation of the type strain | Marseille-Q3615 |
| Strain collection number | CSUR |
| 16S rRNA gene accession number | MT772001 |
| Genome accession number | JACDTZ000000000 |
| Genome status | Whole genome |
| Genome size | 2 578 128 bp |
| GC% | 65.28% |
| Country of origin | France |
| Date of isolation | 2019 |
| Source of isolation | Human healthy skin |
| Conditions used for standard cultivation | Columbia agar with 5% sheep's blood (bioMérieux, Marcy l’Etoile, France) |
| Gram stain | + |
| Cell shape | Irregular rods |
| Cell size | 1.8 × 0.2 μm |
| Motility | − |
| Sporulation | − |
| Colony morphology | White, smooth |
| Temperature range | 20–56°C |
| Temperature optimum | 31.5–56°C |
| Relationship to O2 | Facultative |
| O2 for strain testing | + |
| Oxidase | − |
| Catalase | + |
Deposit in culture collections and sequence databases
Corynebacterium haemomassiliense strain Marseille-Q3615T has been deposited in the Collection de Souches de l’Unité des Rickettsies under accession number CSUR-Q3615. This Whole Genome Shotgun project has been deposited at GenBank under accession number JACDTZ000000000. The 16S gene sequence has been deposited under accession number MT772001.
Conflicts of interest
None declared.
Acknowledgements
MB PhD grant is supported by the collaboration between M&L Laboratories and Aix-Marseille Université (reference PVM 2018-200). This study was supported by the French state, managed by the National Research Agency under the ‘Investissements d'avenir’ programme (reference ANR-10-IAHU-03, Méditerranée Infection) and by the Région Provence-Alpes-Côte-d’Azur and European funding via FEDER PRIMI. The authors are indebted to L. Brechard, Institut Hospitalo-Universitaire Méditerranée Infection for sequencing the genome and help with the electron microscopy IHU-MI platform for the electron micrographs.
References
- 1.Parte A.C. LPSN—list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol. 2018;68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira A., Oliveira L.C., Aburjaile F., Benevides L., Tiwari S., Jamal S.B. Insight of genus Corynebacterium: ascertaining the role of pathogenic and nonpathogenic species. Front Microbiol. 2017;8:1937. doi: 10.3389/fmicb.2017.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J.C., Dubourg G., Million M., Cadoret F., Bilen M., Fenollar F. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 4.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 5.Carbonnelle E., Mesquita C., Bille E., Day N., Dauphin B., Beretti J.L. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin Biochem. 2011;44(1):104–109. doi: 10.1016/j.clinbiochem.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Tittsler R.P., Sandholzer L.A. The use of semisolid agar for the detection of bacterial motility. J Bacteriol. 1936;31:575–580. doi: 10.1128/jb.31.6.575-580.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee I., Ouk Kim Y., Park S.C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 11.Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Čech M. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khamis A., Raoult D., La Scola B. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol. 2004;42:3925–3931. doi: 10.1128/JCM.42.9.3925-3931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdallah R.A., Beye M., Diop A., Bakour S., Raoult D., Fournier P.E. The impact of culturomics on taxonomy in clinical microbiology. Antonie Van Leeuwenhoek. 2017;110:1327–1337. doi: 10.1007/s10482-017-0871-1. [DOI] [PubMed] [Google Scholar]
- 14.Grant J.R., Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]