Abstract
The COVID-19 caused by a novel coronavirus, named Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) has taken a great toll of life affecting lakhs of people around the globe. It was detected initially in Wuhan, China and has spread rapidly to more than 208 countries to date. A range of molecular and immunoassay-based techniques ranging from central laboratory testing to point-of-care tests is urgently needed for the diagnosis and management of COVID-19 patients. Intensive research is going on for the rapid and highly sensitive detection of COVID 19 using varied approach. Hence, this review will focus on the structure of SARS-CoV-2 and recent progress of different detection tool for the detection of COVID-19. This review will also stimulate academics and researcher to update their current technology. Additionally, we also state about the future revolving around the detection of the novel coronavirus. Lately, the way ahead for better management are also put forward.
Keywords: SARS-CoV-2, Point of care, RT-PCR, CRISPR, Isothermal amplification, Biosensors
Introduction
The year 2019 ended with a fatal outbreak of a novel coronavirus, Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2), preliminarily identified as a causative agent of a series of unusual pneumonia cases in Wuhan City, Hubei Province, China. The International Committee on Taxonomy of Viruses (ICTV) permanently named the pathogen SARS-CoV-2 and the disease it caused coronavirus disease 2019 (COVID-19) and declared the crisis a pandemic (Enserink 2020). As per the World Health Organisation (WHO), latest situation report (Sep 14, 2020) at the time of this writing, more than 28.9 million confirmed cases with more than millions of deaths have been reported in 208 countries (Organisation 2020). The first reported case of COVID-19 was in Wuhan, Hubei province with symptoms such as fever, cold, shortness in breath very similar to the common cold in few cases and within a week had a reported cluster cases of acute pneumonia (Nishiura et al. 2020). The Computerized Tomography (CT) scan of the patients revealed slight opacity and difference from the healthy scan of the lungs (Ai et al. 2020). It was concluded as a possible case of pneumonia but the speculation was rejected as soon as PCR results revealed it belongs to coronavirus family using the bronchoalveolar lavage samples of the patients (Ai et al. 2020). It was speculated that the viral disease caused had a zoonotic source. Genome sequencing of SARS-CoV-2 has revealed that it is 96.2% identical to the bat coronavirus and 79.6% identical to SARS-CoV, while 50% similarity to MERS-CoV (Zhou et al. 2020; Ai et al. 2020). The USA can be regarded as the new epicenter of the diseases more than 6.7 million reported cases with 198,520 death followed by India and Brazil having more than 450,000 cases as of Sep 14, 2020.The infection spread is increasing at an alarming rate all around the globe as no great breakthrough has been made in vaccine development to treat the virus. Although, structural analysis and molecular characterization has illuminated a lot of advancement to fight the COVID-19 outbreak even though there is a lot to be uncovered for better disease detection and prevention.
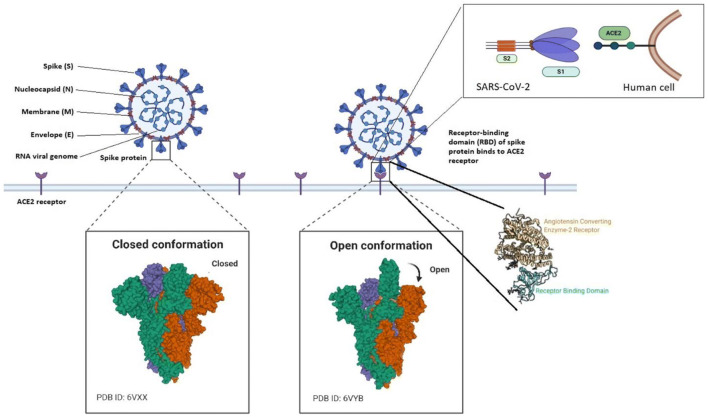
SARS-CoV-2 belongs to beta lineage of subgenus Sarbecovirus, genus Betacoronavirus, the Coronaviridae family, and the Orthocoronavirinae subfamily (Huids et al. 2020; Weiss and Leibowitz 2011). It is an enveloped single-stranded RNA virus (+ve ssRNA) with a genome size of 29.9 kb that spreads widely among humans and other mammals, causing a wide range of symptoms from those of the common cold to fatal diseases such as SARS (Zhu et al. 2020; Zhou et al. 2020). Based on their antigenic properties, they were classified into three main groups (Schoeman and Fielding 2019): i) alpha-CoVs, responsible for gastrointestinal disorders; ii) beta-CoVs, which includes: (a) Bat coronavirus (BCoV), and mammals (b) the human Severe Acute Respiratory Syndrome (SARS) virus, (c) Middle Eastern Respiratory Syndrome (MERS) virus; and iii) gamma-CoVs, that mainly infect avian species (iv) delta-CoVs mostly in pigs. Severe disease has been caused previously to humans by corona family of viruses like MERS-CoV, SARS-CoV and very recent SARS-CoV-2 being the leading cause of death around the globe falls in beta type of coronavirus (Schoeman and Fielding 2019). The four major structural proteins of the Coronaviridae family consists of the spike surface glycoprotein (S), a small envelope protein (E), matrix protein (M), and nucleocapsid protein (N) as shown in Fig. 1.
Fig. 1.
Structure of COVID-19
The spike protein has great importance for entry into the host and can greatly act as a major point to focus for therapeutics and vaccine development as it has better binding as compared to SARS-CoV (Wrapp et al. 2020; Yuan et al. 2017).The M protein has two conformations elongated and compact hence aids in viral assembly (Neuman et al. 2011). The E protein act as an ion-channeling viroporin and interacts with host cell protein (Schoeman and Fielding 2019). The transmission from animal to human lead to the occurrence of the diseases while the spread of the disease is attributed mostly to human transmission making it impossible to constrain the spread of the diseases without distancing (Li et al. 2020a; Chan et al. 2020b).The symptoms for SARS-CoV-2 vary from patient to patient. The symptoms are highly influenced by factors such as sex, age and the attributed diseases underlying a person like hypertension, cardiovascular diseases or even diabetes. The symptoms can range from mild fever, cough, fatigue in certain cases symptoms like diarrhea, hemoptysis and even headache are also reported. In certain cases, complications like acute respiratory distress syndrome, anemia, acute cardiac injury and other secondary infection are also reported (Huang et al. 2020). The case-fatality rate of COVID-19 ranges from 0.6% to 7.2% by region and seem to be substantially higher than the 0.1% mortality rate of seasonal influenza (Onder et al. 2020; Dong et al. 2020; Centers For Disease Control and Prevention 2020a). Few medications are highlighted to treat the COVID-19 which includes of Lopinavir /Ritonavir (Yao et al. 2020; Lu 2020). Chloroquine has also been described to be effective in the treatment of the novel virus (Gao et al. 2020). Vaccine development for treatment is under extensive research and is under rigorous screening before that it can be made available for human use. Currently, the vaccine is in the developmental stages but research is progressing rapidly to find a cure. To constrain the further spread until a vaccine is available for treatment constant efforts also needs to be made in the direction of accurate and point of care detection technique. Point of care testing is a bed side testing which helps to take quick medical decisions by the individuals based on the proximate result. In recent scenario, point of care testing can act as an armor by mass screening the population as it provides result in minutes. The virus can be the next great pandemic in the history of diseases caused by viruses if not managed appropriately. The death toll is increasing day by day with no potential vaccine in hand. Although the field of disease detection has revolutionized in a short time like never before.
The prospect related to COVID-19 detection is reforming at great pace. In regard to the amount of people screened as of Sep 14,2020 for detection with highest testing been done by China having a count of one hundred sixty million followed by USA and India having ninety-two million and fifty seven million testing done respectively (Elflein 2020).Pertaining to the number of test performed per 1000 people is highest in countries like Australia, Canada, USA,Russia, Saudi Arabia and Spain. Whereas relatively fewer tests are performed per 1000 people in countries like Brazil, Peru, Colombia and South Africa to name a few (Hannah Ritchie et al. 2020). Case Fatality Rate (CFR) is high in countries like Mexico, Italy, UK and Yemen (Hannah Ritchie et al. 2020).This review is a holistic resource of all the information currently necessary to be considered for COVID-19 diagnosis. The review will act as an aid in field of COVID-19 detection and diagnosis. In a systemic way it also put forward various efforts across the globe taken to tackle the deadly virus. Moreover, it provides information in what way nanotechnology interventions could possibly be linked in a better and more accurate detection apart from the available technology. Initially it sheds lights on the current scenario and rate at which the virus has affected the globe followed by the structure of the virus and later the detection of the virus. Finally, it also states various pioneering commercial names at present in the field of detection. The goal of this review is to present an overall idea pertaining to SARS-CoV-2 detection.
Current diagnostic tests for COVID-19
COVID-19 has spread massively all over the world since its breakthrough making the field of disease detection and testing to revolutionize (Udugama et al. 2020). Enormous biological examination is done attributed to the structure of virus this is help in development of both the vaccine and the diagnostics measures further to constrain the spread of virus. Here in we elaborate specifically on Nucleic acid and protein based diagnostic techniques developed over time for detection of COVID-19.
Nucleic acid based detection
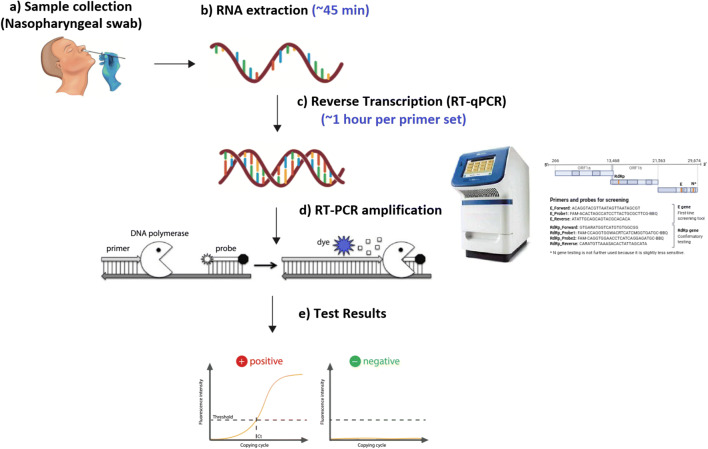
With increasing in several cases each day there is a constant need for the diagnosis of the diseases. Currently, the diagnosis of the COVID-19 is done using nucleic acid-based testing method. In nucleic acid-based testing usually, RT-PCR is used for detection (Corman et al. 2020). A nasopharyngeal samples taken from the respiratory tract to assess the presence of one or several Nucleic Acid (NA) targets specific to SARS–CoV-2 is preferred choice at the moment, but samples from oropharyngeal, mid-turbinate, or anteriornares are also acceptable (Centers for Disease Control and Prevention 2020b; Zou et al. 2020). For better sensitivity than upper respiratory tract samples, sputum, endotracheal aspirates, and bronchoalveolar lavage are also preferred after microbiological processing (Zhu et al. 2020). Inadequate or improper sample collection may result in a false-negative test. For setting up the reaction initially a reverse transcription of SARS-CoV-2 virus RNA is done which converts the RNA into complementary DNA (cDNA) strand which is then followed by amplification of specific regions of the cDNA (Fig. 2). Three regions that are usually targeted to carry out the detections using RT-PCR are (a) the RDRP gene (RNA-dependent RNA polymerase gene) in the open reading frame ORF1ab region, (b) the E gene (envelope protein gene), and (c) the N gene (nucleocapsid protein gene) (Corman et al. 2012a, b). Two types of RT-PCR formats are currently available in the market one known as the singleplex format and other the multiplexed format. In both the format one primer and probe set is used to detect human RNase P (RP) to ensure the RNA extraction was successful. Singleplex format uses three primers whereas the multiplexed make use of only two primers (Authorization 2020). Basic steps for setting up the RT-PCR for detection of COVID-19 are simple although needs trained personnel to carry it out. The major drawback in using lone RT-PCR test for detection is the occurrence of false-negative detection of the specimens making it difficult to take the necessary steps for taking the right measures for the treatment of the patient. In the case of asymptomatic patients if becomes difficult to identify such individuals on the bases of RT-PCR results and take the necessary actions to stop the further spread of the disease (Lee et al. 2020). Hence it was reported that it was beneficial to make use of CT scans to complement the RT-PCR results for enhanced and better diagnosis of the diseases (Ai et al. 2020; Feng et al. 2020; Fang et al. 2020). Leo L M Poon et al. reported making of RT-PCR based test for detection of COVID-19 which specifically used two different regions (ORF1b and N) of the virus (CHU et al. 2020). Another group reported the development of RT-PCR based testing targeting RNA-dependent RNA polymerase (RdRp)/helicase (Hel), spike (S), and nucleocapsid (N) genes of SARS-CoV-2 and compared it with RdRp-P2 assay, and concluded that COVID-19-RdRp/Hel assay was reported as the most effective test for detection of the COVID-19 as it did not cross-react with other coronavirus species (Chan et al. 2020a). The major downfall in diagnosis using the traditional RT-PCR in the testing of COVID-19 includes of unavailability of enough test reagent for testing, requirement of trained personnel for carrying out the testing, availability of developed and well-equipped lab for the entire process to occur in a critical manner. Lastly, the time required for completion of the test is too large to produce results. The ability of RT-PCR assays to rule out COVID-19 on the basis of upper respiratory tract samples obtained at a single time point remains unclear. To overcome some of the problems encountered efforts are made around the globe to transverse this problem at the earliest.
Fig. 2.
Traditional RT-PCR based nucleic acid testing
Recently the development of straightforward detection technique which made use of mini PCR and well plate reader was reported which can be possibly used in a remote area as well provide early results after the detection. Samples were combined with a DNA intercalating reagent (Evagreen Dye) which was then detected after amplification using the fluorescence indicator in a 96 well plate format (Gonzalez-Gonzalez 2020). Another progress in the field is the development of direct rapid extraction-free PCR (DIRECT-PCR). The single-tube homogeneous reaction was developed which included all Viral lysis, reverse transcription, amplification and detection within 36 min in just a tube for regions where laboratory competency is a major constrain. It would be helpful for early screening of mass population (Wee et al. 2020). To overcome the problem faced by using nucleic acid-based detection using RT-PCR technique which included of laborious system, a large amount of time required to process the sample, necessity of developed facilities to carry out the detection, trained personnel to carry out the test and occurrence of false-positive results (Gallagher 2020). Lastly, the test is not point of care hence there is a need to find some alternatives that are easy to use and provides accurate results instantaneously.
Aptamer based testing
Aptamer are single stranded nucleic acid molecule which can be readily used for detection of specific disease in an accurate manner as they have property to be modified at random position and could act as a great advantage to tackle the COVID-19 crisis at hand. Exploiting aptamers as they are small oligonucleotide which can be used for detection of COVID-19 as they are stable and easy to modify (Chen and Yang 2015). Again, synthesis of aptamers is easy in large quantity and at a lower price when compared to RT-PCR based nucleic acid testing. As many strategies are already put forward in line for commending the use of aptamers one such strategy was used was put forward by Ahn et al. in 2009 for detection of SARS-CoV by finding application in the preparation of nanoaptamer chip in detection by making use of the nucleocapsid protein which acted as a selective marker for detection a chemiluminescence based immunosorbent assay was developed and a detection limit of 2 pg/ml was achieved (Ahn et al. 2009). Another possibility can be making use of aptamers to develop anti-corona virus agent (Jang et al. 2008). Group of scientist from Aptamer Group Ltd. are working extensively to develop aptamers based point of care test for COVID-19 using spike protein of the SARS-CoV2 (Group 2020). Selex (systemic evolution of ligands through exponential enrichment) process is another potential way of creating aptamers and finding its application in virus detection in conjugation with the microfluidics system (Liebich 2020; Lai et al. 2012). COVID-19 being an RNA virus recently reported MANGO a fluorogenic dye tactic in the detection of the virus as the dye has background-free technology for live-cell RNA imaging upon ligand binding (Cawte et al. 2020). The group of scientists from Simon Fraser University (SFU) proposed making use of MANGO system which consists of an RNA Mango aptamer bounded by fluorescent dye and glows brightly when bounded by the target. The team has proposed developing Mango NABSA (nucleic acid sequence-based amplification) kit for detection of the COVID-19 (News 2020).
CRISPR-Cas based testing
Another aspect to be explored for detecting COVID-19 is making use of CRISPR-Cas gene editing tool which can create wonders in the field of detection (Chekani-Azar et al. 2020). Making use of CRISPR-Cas has been reported for use in medicine and diagnostics over the years. Success in making use of the CRISPR-Cas has enabled to ascertain new systems for targeting and manipulating nucleic acids including those from Cas9, Cas12, Cascade and Cas13 orthologues (Pickar-Oliver and Gersbach 2019; Li et al. 2019). Possibilities of developing a diagnostic tool for detection of TB is already known using CRISP-Cas technology which lay the foundation for making its use in COVID-19 detection as well (Ai et al. 2019). Very recently, developed SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) for detection of COVID-19 using CRISPR-Cas12. This developed lateral flow strip could decrease the detection time required by the ongoing RT-PCR testing and aid in better disease management in this crucial time (Broughton et al. 2020). Exploiting the potential of CRISPR-Cas technology fluorescence and colorimetric based detection of COVID-19 was made possible. The technology is named as specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) by making use of recombinase-mediated polymerase pre-amplification of DNA or RNA for detection. The reported technology takes less than an hour to provide results making it one of the potent applicants in the field of diagnosis of COVID-19 (Kellner et al. 2019). An improved version of SHERLOCK was put forward which overcome major challenges of SHERLOCK and is named as STOP (SHERLOCK testing in one pot). The mentioned assay is capable of solely performing extraction, amplification and CRISPR-mediated detection of the viral RNA in a go on a single temperature. Two version are presented as STOP covid version 1 and 2 respectively. STOP covid version 1 is compatible with lateral-flow and fluorescence readouts whereas STOP covid version 2 makes use of magnetic beads concentrated SARS-CoV-2 RNA for detection (Joung et al. 2020). CREST is another pioneering discovery in the field of diagnosis for detection of COVID-19. The CREST stands for (Cas13-based, Rugged, Equitable, Scalable Testing) and has overcome major drawbacks associated with the current diagnostic system of COVID-19. The Cas13 can be used in point of care setting and it would be low cost when compared to traditional lateral flow assay as it doesn’t require antibody conjugates for the detection (Rauch et al. 2020). Another effective use of CRISPR Cas 12 was seen by integrating into a lateral flow system named as iSCAN having potential to be used as effective management system for point of care detection(Ali et al. 2020). AIOD CRISPR assay, made use of a pair of crRNAs to initiate the detection using CRISPR-Cas12a for improved sensitivity. The assay have potential in detecting nucleic acids (DNA and RNA) of SARS-CoV-2 and HIV (Ding and Yin 2020). Integration of lateral flow system along with nucleic acid as a substrate for quick and point of care results can prove to be fruitful amid this pandemic as shown in Fig. 3a. Another perspective to be explored is pairing CRISPR-Cas with electrochemical based sensing as it can help in achieving even lower limit of detection. Can Dincer et.al, reported (CRISPR)/Cas13a-powered microfluidic based electrochemical sensor for detection of cancer reaching a limit of 10 pM on site (Bruch et al. 2019). Such a system should be made available at this crucial time in detection of COVID-19. Development of CRISPR-Cas based on-chip electrical detection of nucleic acid could be the future of detection in COVID-19 screening as it can detect even in femto Molar levels (Hajian et al. 2019).
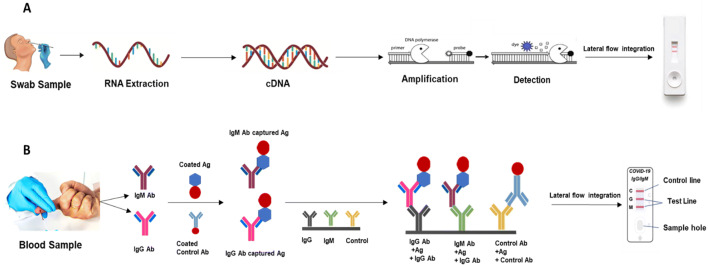
Fig. 3.
Lateral flow integration for a Nucleic acid-based testing for COVID 19 detection, b Protein based testing for COVID 19 detection
Protein based testing
Along with the on-going nucleic acid-based testing for detection of COVID-19 protein-based detection can also act as an excellent supplement alongside for better mass screening for early measures in a point of care setting. Although nucleic acid, CRISPR-Cas and aptamer-based testing are currently been employed for detection protein-based avenues needs to be explored. Antigen and antibody are the among the major protein-based entities which are already used for detection of several diseases over a long time. Protein-based detection system mostly includes ELISA, chromogenic based or fluorescence-based detection using dye, lateral flow-based detection. Not much was developed until recently since the viral load has reported fluctuating with weeks after detection providing results which are not possibly reproducible when test was made using saliva sample from infected patients. Viral load was highest at the start and subsequently declined at the end (To et al. 2020). Although it can be speculated that antibody-based test can provide more time since antibody can be detected even after a long time in the patient.
Antigen based testing
An antigen test has ability to reveals if a person is presently infected with a pathogen such as the SARS-CoV-2 virus. Antigen tests mostly detect proteins or glycans, such as the spike proteins found on the surface of the SARS-CoV-2 virus. Sona nanotech is one of the pioneers in the filed leading the way of COVID-19 testing by developing antigen-based test kit for detection. The gold nanorod particles used are CTAB (cetyltrimethylammonium) free hence limiting the toxic risks associated with the use of other gold nanorod technologies in medical applications. It was reported recently that the test attained a positive response to a recombinant whole spike protein control reagent specific to SARS-CoV2 and neither had false positives (Nanotech 2020). Another milestone is reached by E25Bio’s rapid diagnostic which provide results in 15 min by detecting the presence of the virus in the patient sample (Misra 2020). Iceni Diagnostics run by Prof Rob Field, has developed a diagnostic technique that make use of an artificial glycan receptor to detect the novel virus (Iceni Diagnostics Ltd 2020). OraSure envisions is another established name being working on developing antigen based testing kit (OraSure Technologies 2020). Roche has reported of developing test having sensitivity of 96.52% and a specificity of 99.68%, based on 426 samples from two independent study centers the test helps in identifying both symptomatic and asymptomatic people with in 15 min (Roche Ltd 2020a). Lately special consideration is being given to point of care antigen testing in order to detect infected person at the earliest and decreasing the chances of mass transfer of the virus (Dinnes et al. 2020). However, it was reported that antigen based detection alone cannot be effective in detection of SARS-CoV-2 (Mak et al. 2020).
Antibody based detection
Major class of immunoglobins includes of IgG, IgM, IgA, IgE and IgD which can contribute in detection of diseases. IgG been the first line of defense initiated as part of the immune response when infected by a pathogen and creates immunological memory to further fight the infection is mostly targeted in detection of COVID-19. Also reported is that IgM appears in the blood of the patient when infected by SARS after 3 to 6 days and IgG can be detected 8 days after the infection occurred (Racine and Winslow 2009; Wan et al. 2003).
IgG and IgA ELISAs in conjunction with the EURO Lab workstation (Euroimmun, Lübeck, Germany). EUROIMMUN enzyme-linked immunosorbent assay (ELISA) for semi-quantitative detection of IgA and IgG antibodies in serum and plasma samples using recombinant S1 domain of the SARS-CoV-2 spike protein as antigen is reported demonstrates excellent sensitivity for detection of IgA and IgG antibodies from samples collected ≥3 days and ≥ 4 days, respectively, after COVID-19 diagnosis by PCR (Beavis et al. 2020). Other ELISA based IgG and IgA showed specificity of 91.9% and 73.0% 12 and 11 days after symptom onset (Jääskeläinen et al. 2020). Many industries as well have developed ELISA based detection kit for COVID-19 like Epitope diagnostic, Inc. and Eagle bioscience although tests are yet to be approved by FDA for use. Eagle bioscience test kits are now available for use in the US for detection (Epitope Diagnostics 2020; Bioscience 2020). Making use of protein for detection is known and adopted for development over decades but recently making use of neutralizing antibody had been adopted to fight against the novel coronavirus. The antibody targets the spike protein of the virus which is made up of two subunits the S1 and S2. The S1 subunit is necessary for entry into the host cell while S2 subunit helps in fusion of the cellular and viral membrane. SARS-CoV-2 binds the receptor human angiotensin-converting enzyme 2 (ACE2) (Wang et al. 2020a).Considering the fact that neutralizing antibodies have great potential to fight the diseases making its use in disease diagnosis could be the near future. On evaluation of IgM/IgG based detection kit the highest specificity was reported for the Wantai SARS-CoV-2 Total Antibody ELISA followed by 93% for the Euroimmun IgA ELISA and 96% for the Euroimmun IgG ELISA with sensitivities of 90%, 90%, and 65%, respectively (Lassaunière et al. 2020).
Development of lateral flow strip for detection makes use of antigen-antibody reaction. Here the reaction occurs on the nitrocellulose membrane. Once the sample is added into the strip the antigen interacts with the already loaded capture antibody for detection through capillary action. The gold nanoparticle-conjugated antibody is present near the sample port to interact with the sample for the formation of the visible color change after formation of the immunocomplex. Making use of lateral flow assay for diseases detection which makes use of antigen-antibody for detection can be a better alternative for COVID-19 diseases management. Lateral flow assay(LFA) is well established in terms of diseases detection and been used for more than a decade as shown in Fig. 3b (Bahadir and Sezgintürk 2016).
Building on this concept various lateral flow strip has been developed lately (Li et al. 2020b).Development of a rapid and simple point-of-care lateral flow immunoassay that can detect immunoglobulin M (IgM) and IgG antibodies simultaneously against SARS-CoV-2 virus in human blood within 15 min having an overall testing sensitivity of 88.66% and specificity of 90.63%. This IgM-IgG combined assay would result in better utility and sensitivity on contrary to single IgM or IgG test. (Li et al. 2020b). Lateral flow immunoassay strip with various optimizing parameters such as concentration of coating antigen, BSA blocking concentration and pH value for conjugation was also developed (Wen et al. 2020). IgG based NG-Test® based lateral flow immunoassay strip showed sensitivity (around 80%) for CLIA, ELISA and LFIA (Nicol et al. 2020). IgG and IgA ELISAs in conjunction with the EURO Lab workstation (Euroimmun, Lübeck, Germany).
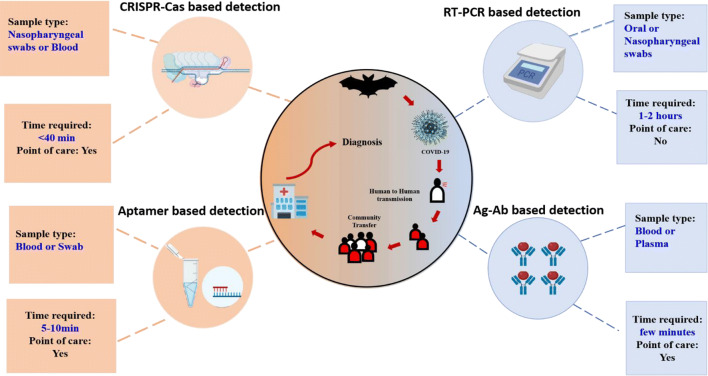
Many companies have developed LFA which is ready to use and provide results within minutes. Roche diagnostic reported making a serological test for COVID-19 named as Elecsys which make use of cobas-e analyser for analysis of the results and provides results in 18 min with detection at a rate of 300 test/h can by targeting the nucleocapsid (N) antigen for the determination of antibodies against SARS-CoV-2 (Roche Ltd 2020b). Abbott’s, on the other hand, developed another platform knowns as ARCHITECT i1000SR and i2000SR laboratory-based instruments which run 100–200 tests an hour for effective detection using antibodies (Abbott 2020). Although IgG and IgM based test strip are claimed to provide results which are not accurate and reliable hence now are having limited use in the field of diagnosis of COVID-19 due to the drawbacks (Burog et al. 2020). Milenia Biotec a Germany based company has already intervened in the development of dipstick based COVID-19 detection system named as HybriDetect which uses gene amplification system or protein for detection of the novel virus (Biotec 2020). Although a lot has developed within a short time in testing related to COVID-19 multiple detection of close related biomarker or multiple analyte could be the next step in early and timely detection of the virus (Dincer et al. 2017). Concurrently, it was reported recently that alone RT-PCR had few limitation which could be overcome by making simultaneous use of antibody based tests resulting in an overall sensitivity and specificity of 95.7% and 98.7% respectively (Wang 2020). Concluding, we represent the current tool of diagnosis for COVID- 19 Diagnosis as shown in Fig. 4.
Fig. 4.
Current diagnostics tool of COVID-19 diagnosis
Chemiluminescence based COVID-19 detection
Chemiluminescent based immunoassay is one the effective modification to overcome traditional immunoassay drawbacks which can be used in order to detect COVID-19 effectively. Peptide-based luminescent immunoassay to detect IgG and IgM is been proposed for COVID-19. Cut-off value of this assay was determined by the detection of 200 healthy sera and 167 sera from patients infected with other pathogens than SARS-CoV-2. To evaluate the performance of this assay, detection of IgG and IgM in the 276 sera from confirmed patients were done. The positive rate of IgG and IgM were 71.4% (197/276) and 57.2% (158/276) respectively. By combining with real time RT-PCR detection, this assay might help to enhance the accuracy of diagnosis of SARS-CoV-2 infection(Cai et al. 2020). Highly purified receptor-binding domain (RBD) of the SARS-CoV-2 spike protein was expressed in human 293F cells and used to make a set of chemical luminescence kits for detecting the presence of RBD-specific IgA, IgM, and IgG, respectively The RBD-specific IgA, IgM, and IgG kits showed diagnostic sensitivities of 98.6%, 96.8%, and 96.8%, and specificities of 98.1%, 92.3%, and 99.8%, respectively (Ma et al. 2020).
Nanotechnology interventions
Nanotechnology intervention is disease detection, vaccine development and therapeutics is known for a long time. Advanced research has been done for hand in hand application of nanotechnology along with disease management. The recent pandemic can put forward more ways to tackle the pandemic by making use of nanotechnology. Few advancements are already done is a field of COVID-19 diagnosis using nanotechnology (Weiss et al. 2020; Mujawar et al. 2020). In recent time, the field-effect transistor (FET)-based biosensing device for detecting SARS-CoV-2 was developed. The developed biosensing platform made use of graphene-coated FET against specific spike protein against COVID-19. One major advantage of the biosensing platform is that does not require any pre-treatment of the sample for detection (Seo et al. 2020). Another use of nanotechnology-based COVID-19 detection is in making use of innovative nanoparticles to simplify the existing available technologies for efficient results. One such developed nanoparticle is of poly (amino ester) with carboxyl groups (PC)-coated magnetic nanoparticles (pcMNPs) which aided in viral RNA extraction. pcMNPs-RNA complex can directly integrate into the RT-PCR reactions and help in detection. The proposed pcMNPs can be a great help in this decisive time to reduce the turnaround time which is obligatory to take early measure as fast as possible (Zhao et al. 2020). Recent improvement and novel strategy were integrated in making LFA for detection benefiting the advantage of glyconanoparticles. It was accomplished by exploiting the sialic acids affinity with SARS-CoV-2 spike protein by using glyconanoparticle platform rather than the traditional IgG and IgM based testing. It novel strategy helped in having a detection limit of 5 μg·mL−1 (Baker et al. 2020).
Another report is in development of naked eye detection test for SARS-CoV-2 by making use of AuNPs capped with thiol-modified antisense oligonucleotides (ASOs) specific for N-gene (nucleocapsid phosphoprotein). Results appears in 10 min from the isolated RNA samples showing agglomeration in the presence of its target RNA sequence of SARS-CoV-2, moreover RNaseH helps in cleaving the RNA strand from the RNA–DNA hybrid aiding in visually detectable precipitate. The limit of detection is 0.18 ng/μL of RNA having SARS-CoV-2 viral load (Moitra et al. 2020). Development of lanthanide-doped polysterene nanoparticles (LNPs) used as a fluorescent reporter to detect anti-SARV-CoV-2 IgG in human serum using Lateral flow immunoassay. The developed assay can be of great benefit in positive identification in doubtful cases and evaluating patients response to treatment (Chen et al. 2020). Wuhan Dgensee Clinical Laboratory Co., Ltd. reportedly developed nanopore target sequencing (NTS) to detect SARS-CoV-2 and other respiratory viruses simultaneously within 6–10 h. The NTS is not constrained to detect only COVID-19 but can be used in the application of diseases detection of other pathogenic viruses as well for improved results (Wang et al. 2020b).
Lately, smart electronics has emerged as a potential field in the development and detection of pathogenic infections. Diseases like cancer, H1N1, HIV and many more have been detected by making use of biosensors. Point-of-care based biosensors are reported to have high selectivity and sensitivity and works on bio affinity receptors (Morales-Narváez and Dincer 2020). All kinds of biomolecules like aptamers, antibody nucleic acid and lipids are reported to be detected by making use of biosensors (Tu et al. 2019). TriSilix is a recent advancement in the field of detection which makes use of silicon and have three modes of working on a single chip starting from electrical heater followed by temperature sensor and lastly electrochemical sensor when tested using cDNA from SARS-Cov-2 the system could detected up to 1 pg (Nunez-Bajo et al. 2020). Further, development of exhaled breathe analyzer based on nanomaterial hybrid array was developed and showed an accuracy of 90% and 95% in differentiating COVID-19 patients or other related lung infection if any, such an analyzer will help in better screening of patient in point of care setting (Shan et al. 2020). Integration of biosensors into a mobile technology can be the future of disease management and detection which can also be adapted for detection of COVID-19 (Zhang and Liu 2016). Multiplexing using smart and flexible electronics can also be adopted at this crucial time for detection of the novel virus as it has been reported for accurate and timely detection of many diseases (Konstantinov et al. 2009). Microfluidics can be used in detection as they have the capability of been point of care also been small in size it becomes user friendly in diseases management (Myers and Lee 2008; (Chin et al. 2011; Martinez et al. 2008). Table 1 represents the available list of commercial SARS-CoV-2 in-vitro diagnostic assays.
Table 1.
List of commercial SARS-CoV-2 in vitro diagnostic assays given an EUA from the FDA as of 9 May 2020
| No. | Manufacturer | Diagnostic test name | Technology/Platform | Description |
|---|---|---|---|---|
| Molecular | ||||
| 1 | Rutgers Clinical Genomics Laboratory at RUCDR Infinite Biologics - Rutgers University | Rutgers Clinical Genomics Laboratory TaqPath SARS-CoV-2-Assay | rRT-PCR | Real-time Reverse Transcription Polymerase Chain Reaction (rRT-PCR) test intended for the qualitative detection of nucleic acid from SARS-CoV-2 in oropharyngeal (throat) swab, nasopharyngeal swab, anterior nasal swab, mid-turbinate nasal swab, and bronchoalveolar lavage (BAL) fluid from individuals suspected of COVID-19. |
| 2 | Zymo Research Corporation | Quick SARS-CoV-2rRT-PCR Kit | rRT-PCR | Real-time RT-PCR test intended for the qualitative detection of nucleic acids from SARS-CoV-2 in upper respiratory specimens (such as nasal, nasopharyngeal, mid-turbinate or oropharyngeal swabs), and lower respiratory specimens (such as sputum, tracheal aspirates, and bronchoalveolar lavage) from patients suspected of COVID-19. |
| 3 | OPTI Medical Systems, Inc. | OPTI SARS-CoV-2 RT PCR Test | rRT-PCR | The OPTI SARS-CoV-2 RT-PCR test kit is based on real-time reverse transcription polymerase chain reaction (RT-PCR), which provides detection of the viral RNA in the sample. It is designed for the detection of SARS-CoV-2 RNA extracted from nasopharyngeal swabs, oropharyngeal swabs, bronchoalveolar lavage and sputum samples. |
| 4 | Sherlock BioSciences, Inc. | Sherlock CRISPR SARS-CoV-2 Kit | CRISPR/RT-LAMP | The kit works by programming a CRISPR molecule to detect the presence of a specific genetic signature – in this case, the genetic signature for SARS-CoV-2 – in a nasal swab, nasopharyngeal swab, oropharyngeal swab or bronchoalveolar lavage (BAL) specimen. When the signature is found, the CRISPR enzyme is activated and releases a detectable signal. |
| 5 | BioMérieux SA | SARS-COV-2 R-GENE | rRT-PCR | The SARS-COV-2 R-GENE® kit is a molecular detection kit, using real-time PCR after viral nucleic acids extraction. Assay includes all necessary reagents optimized to detect SARS-COV-2 for in vitro diagnostic use |
| 6 | Fast Track Diagnostics Luxembourg S.á.r.l. (a Siemens Healthineers Company) | FTD SARS-CoV-2 | rRT-PCR | The FTD SARS-CoV-2 Assay is a real-time PCR test used to detect the new coronavirus SARS-CoV-2 causing COVID-19. The FTD SARS-CoV-2 Assay uses the same protocol, including PCR cycling profile, as other FTD respiratory assays. |
| 7 | Sansure BioTech Inc. | Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR-Fluorescence Probing) | rRT-PCR | The kit is a real-time RT-PCR test intended for the qualitative detection of RNA from SARSCoV-2 in nasopharyngeal swabs, oropharyngeal (throat) swabs, anterior nasal swabs, midturbinate swabs, nasal washes and nasal aspirates from individuals who are suspected of COVID19. |
| 8 | Bio-Rad Laboratories, Inc | Bio-Rad SARS-CoV-2 ddPCR Test | digital droplet rRT-PCR | The test is a partition-based endpoint RT-PCR test intended for the qualitative detection of nucleic acids from SARS-CoV-2 in nasopharyngeal anterior nasal and mid-turbinate nasal swab specimens, as well as nasopharyngeal wash/aspirate and nasal aspirate specimens from patients suspected of having COVID-19 |
| 9 | BioFire Diagnostics, LLC | BioFire Respiratory Panel 2.1 (RP2.1) | rRT-PCR | The BIOFIRE® RP2.1 panel takes approximately 45 min and tests nasopharyngeal swab samples in transport media which includes 22 pathogens that cause respiratory infections, including SARS-CoV-2 |
| 10 | LabGenomics Co., Ltd. | LabGun COVID-19 RT-PCR Kit | rRT-PCR | Qualitative detection of nucleic acid from SARS-CoV-2 in nasopharyngeal, or oropharyngeal, anterior nasal and mid-turbinate nasal swabs, as well as nasopharyngeal wash/aspirate or nasal aspirate specimens and sputum, from individuals who are suspected of COVID-19 |
| 11 | Rheonix, Inc. | Rheonix COVID-19 MDx Assay | rRT-PCR | The Rheonix COVID-19™ MDx Assay is an endpoint RT-PCR assay intended for the qualitative detection of nucleic acid from SARS-CoV-2 in respiratory samples from individuals who are suspected of COVID-19 |
| 12 | SEASUN BIOMATERIALS | U-TOP COVID-19 Detection Kit | rRT-PCR | The kit is an in-vitro diagnostic product intended for Real-time PCR based detection of new Coronavirus (COVID-19) infection via simultaneous amplification of ORF1ab and N-gene in patients with suspect COVID-19 infection. |
| 13 | SD Biosensor, Inc. | STANDARD M nCoV Real-Time Detection Kit | rRT-PCR | Real-Time Detection kit is used for rapid identification and detection of novel coronavirus (2019-nCoV) nucleic acids in human nasopharyngeal swabs and throat swab samples targeting ORF1ab (RdRp) gene and E gene. |
| 14 | altona Diagnostics GmbH | RealStar SARS-CoV02 RT-PCR Kits U.S. | rRT-PCR | The kit is a reagent system, based on real time PCR technology, for the qualitative detection and differentiation of lineage B-betacoronavirus (B-βCoV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) specific RNA (targets E and S gene). |
| 15 | Covance/Seegene, Inc. | Allplex 2019-nCoV Assay | rRT-PCR | Assay is multiplex Real-time PCR assay for simultaneous detection of 3 target genes of SARS-CoV-2 in a single tube. The assay is designed to detect RdRP and N genes specific for SARS-CoV-2, and E gene for all of Sarbecovirus including SARS-CoV-2. |
| 16 | Trax Management Services Inc. | PhoenixDx 2019-CoV | rRT-PCR | The test should be run on the BIO-RAD CFX96-IVD platform, with RNA extracted using a separate kit. Sensitivity was determined using 30 positive samples, and specificity from 10 negative samples. |
| 17 | OSANG Healthcare | GeneFinder COVID-19 Plus RealAmp Kit | rRT-PCR | This is a one-step RT-PCR test, with each kit having reagents for 100 tests. |
| 18 | Fosun Pharma USA Inc. | Fosun COVID-19 RT-PCR Detection Kit | rRT-PCR | This test uses TaqMan chemistry. Sensitivity was determined from 50 positive samples, and specificity from 100 negative samples. Their results were compared to the CDC RT-PCR protocol. |
| 19 | KorvaLabs Inc. | Curative-Korva SARS-Cov-2 Assay | rRT-PCR | The test should be run on the BioRad CFX 96 Touch, BioRad CFX Connect Real-Time PCR systems and Roche LightCycler 480 II Real-Time PCR systems. Sensitivity was determined from 34 positive samples, and specificity from 18 samples. |
| 20 | GenoSensor, LLC | GS™ COVID-19 RT-PCR KIT | rRT-PCR | The test should be run on the Applied Biosystems 7500 Fast Dx Real-Time PCR system, and can be run in a 96-well or 384-well format. The test takes about 1.5 h. Sensitivity and specificity were determined using 32 positive and negative samples. |
| 21 | Maccura Biotechnology (USA) LLC | SARS-CoV-2 Fluorescent PCR Kit | rRT-PCR | The kit includes reagents for RNA extraction and for PCR. The test takes about 2 h, and requires the Applied Biosystems 7500 Real-Time PCR Systems with v2.3 software. Sensitivity and specificity were determined from 30 positive and negative samples. There was a false positive rate of 3.3% in the N gene. |
| 22 | Atila BioSystems, Inc. | iAMP COVID-19 Detection Kit | Isothermal amplification (OMEGA), patented | There is no extraction process necessary for this kit, and the amplification is isothermal. The test takes about 1 h, and 94 samples can be processed in a single run. |
| 23 | DiaCarta, Inc | QuantiVirus SARS-CoV-2 Test kit | rRT-PCR | This is a multiplex rRT-PCR diagnostic, called the QuantiVirus SARS-CoV-2 Multiplex Test. The test can be run in under 2 h. The test should be run on the Thermo Fisher (ABI) QuantStudio 5, Thermo Fisher (ABI) 7500 Fast Dx, and Bio-Rad CFX 384 platforms. Sensitivity was determined from 41 positive clinical samples, and specificity from 52 negative clinical samples. |
| 24 | Becton, Dickinson & Company | BD SARS-CoV-2Reagents for BD MAX System | Antigen (chromatographic digital immunoassay) | The nasal swab specimen is directly placed in the extraction reagent tube, during which time virus particles present in the specimen are disrupted, exposing internal viral nucleoproteins. After processing the swab in the extraction reagent, the specimen is added to the BD Veritor System test device. SARS-CoV-2 antigens present in the specimen then bind to antibodies conjugated to detector particles in the test strip. The antigen-conjugate complexes migrate across the test strip to the reaction area and are captured by a line of antibodies bound on the membrane. Test results of the BD Veritor System test device are read using the BD Veritor Plus Analyzer Instrument, or other authorized instrument, when the 15-min assay development time is complete. |
| 25 | InBios International, Inc | Smart Detect SARS-CoV-2 rRT-PCR Kit | rRT-PCR | RNA extraction should be performed before using this kit. The test should be run with the 7500 Fast Dx Real-Time PCR Instrument (Applied Biosystems) with Sequence Detection System (SDS) Software, version 1.4 (Applied Biosystems) or the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with CFX Maestro Software (Bio-Rad). |
| 26 | Gnomegen LLC | Gnomegen COVID-19 RT-Digital PCR Detection Kit | rRT-PCR | The oligonucleotide primers and probes for detection of SARS-CoV-2 were selected CV03030005 R01 Gnomegen 4 from regions of the virus nucleocapsid (N) gene. The panel is designed for specific detection of the 2019-nCoV (two primer/probe sets). An additional primer/probe set to detect the human RNase P gene (RP) in control samples and clinical specimens is also included in the panel. RNA isolated and purified from upper and lower respiratory specimens is reverse transcribed to cDNA and subsequently amplified in the Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument with SDS version 1.4 software. |
| 27 | Co-Diagnostics, Inc | Logix Smart Coronavirus Disease 2019 (COVID-19) Kit | rRT-PCR | The test is a one-step, RT-PCR with CoPrimer technology. The test takes 1–1.5 h, for use with CoDx Box, MIC qPCR Cycler, and Eco48 cycler. |
| 28 | ScienCell Research Laboratories | ScienCell SARS-CoV-2 Coronavirus Real-time RT-PCR (RT-qPCR) Detection Kit | rRT-PCR | The test should be performed on the AB 7500 Fast DX system, and extraction of RNA is required before use of this kit. |
| 29 | Luminex Corporation | ARIES SARS-CoV-2 Assay | rRT-PCR | The test must be performed on an ARIES unit with an ARIES cassette, which can hold up to six samples. Sample lysis, RNA extraction, and RT-PCR are all completed in the ARIES unit. |
| 30 | Becton, Dickinson & Company (BD) | BioGX SARS-CoV-2 Reagents for BD MAX System | RT-PCR | A combination of lytic and extraction reagents is used to perform cell lysis and DNA/RNA extraction. Nucleic acids released from the target organisms are captured on magnetic affinity beads. The beads, together with the bound nucleic acids, are washed and the nucleic acids are eluted by a combination of heat and pH variation. The final eluate is used to rehydrate BioGX SARS-CoV-2 Reagents, which contains all reagents necessary for RT-PCR including primers and probes. After reconstitution, the BD MAX System dispenses a fixed volume of RT-PCR-ready solution containing extracted nucleic acids into the PCR Cartridge. Microvalves on the cartridge are sealed by the system prior to initiating PCR in order to contain the amplification mixture and thus prevent evaporation and contamination. |
| 31 | Ipsum Diagnostics, LLC | COV-19 IDx assay | rRT-PCR | The test should be used with Applied Biosystems QuantStudio12 Flex (QS12) instrument with software version 1.2.2. Clinical sensitivity and specificity were determined using 36 and 30 samples, respectively. |
| 32 | QIAGEN GmbH | QIAstat-Dx Respiratory SARS-CoV-2 Panel | rRT-PCR | The Bio-Speedy Direct RT-qPCR SARS-CoV-2 is a one-step molecular test. The test should be run on the Roche LightCycler 96, Bio-Rad CFX96 Touch, or Qiagen Rotor-Gene 5 Plex platforms. They did a very extensive study of the limit of detection using various sample types and collection methods. Sensitivity was determined from 347 positive clinical samples, and negativity from 94 negative samples. |
| 33 | NeuMoDx Molecular, Inc. | NeuMoDx SARS-CoV-2 Assay | rRT-PCR | The test relies on a one-step RT-qPCR with TaqMan chemistry. The kit should be used with the NeuMoDx Molecular System, and swabs should be collected with the Copan UTM-RT® System or BD UVT System. For sensitivity and specificity, they used 87 and 82 samples, respectively. |
| 34 | Luminex Molecular Diagnostics, Inc. | NxTAG CoV Extended Panel Assay | rRT-PCR | The test incorporates multiplex Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) with the Luminex® proprietary universal tag sorting system on the Luminex platform to easily detect SARS-CoV-2 s. Extracted total nucleic acid is added to pre-plated, Lyophilized Bead Reagents (LBRs) and mixed to resuspend the reaction reagents. The reaction is amplified via RT-PCR and the reaction product undergoes near simultaneous bead hybridization within the sealed reaction well. The hybridized, tagged beads are then sorted and read on the MAGPIX® instrument. The generated signals are analyzed using the NxTAG CoV Extended Panel Assay File for SYNCT™ |
| 35 | Abbott Diagnostics Scarborough, Inc. | ID NOW COVID-19 | rRT-PCR | Assay utilizes isothermal nucleic acid amplification technology for the qualitative detection of SARS-CoV-2 viral nucleic acids. It is comprised of a Sample Receiver, containing elution/lysis buffer, a Test Base, comprising two sealed reaction tubes, each containing a lyophilized pellet, a Transfer Cartridge for transfer of the eluted sample to the Test Base, and the ID NOW Instrument. The reaction tubes in the Test Base contain the reagents required for amplification of SARS-CoV-2, as well as an internal control. The templates (similar to primers) designed to target SARS-CoV-2 RNA amplify a unique region of the RdRp segment. Fluorescently-labeled molecular beacons are used to specifically identify each of the amplified RNA targets |
| 36 | BGI Genomics Co. Ltd | Real-Time Fluorescent RT-PCR Kit for Detecting SARS-CoV-2 | rRT-PCR | The limit of detection (for 95% sensitivity) was 100–150 virions/mL. They also tested cross reactivity with other 54 other pathogens, such as influenza A and SARS-CoV, and found no cross-reactivity. The sensitivity in 384 clinical samples was 88.1%, and specificity was 99.6%. |
| 37 | Avellino Lab USA, Inc. | AvellinoCoV2 test | rRT-PCR | The test should be run on Applied Biosystems 7500 Fast Real-Time PCR System with software version 2.3. |
| 38 | PerkinElmer, Inc. | PerkinElmer New Coronavirus Nucleic Acid Detection Kit | rRT-PCR | The test should be run on PerkinElmer’s chemagic 360 instrument, with each kit having reagents for 48 tests. |
| 39 | Mesa Biotech Inc. | Accula SARS-Cov-2 Test | PCR and lateral flow | The amplification of the viral RNA is based on PCR, the detection is based on visualization of the nucleic acid on a lateral flow strip. The readout is qualitative. The test must be run on the Accula Dock or the Silaris Dock, and takes about 30 min. The system is an “all in one,” where sample lysis, RNA extraction, and RT-PCR are performed within the dock. |
| 40 | BioFire Defense, LLC | BioFire COVID-19 Test | rRT-PCR | The test takes about 45 min, and runs on the FilmArray 2.0 and FilmArrayTorch systems. |
| 41 | Cepheid | Xpert Xpress SARS-CoV-2 test | rRT-PCR | The test should be run on the GeneXpert Dx and GeneXpert Infinity systems. This is an “all in one” system which includes sample preparation, nucleic acid extraction and amplification, and detection of the target sequences. |
| 42 | Primerdesign Ltd. | Primerdesign Ltd. COVID-19 genesig Real-Time PCR assay | rRT-PCR | The kit uses TaqMan technology with fluorescent probes for detection of viral RNA, and it must be coupled with an approved extraction kit. The test can be used with the following platforms: Applied Biosystem® 7500 Real-Time PCR System, Bio-Rad CFX96, Roche® LightCycler 480 II. |
| 43 | GenMark Diagnostics, Inc. | ePlex SARS-CoV-2 Test | rRT-PCR | The test must be run on the ePlex system, and provides qualtiative diagnosis of SARS-CoV-2 viral RNA in the sample. Rather than using fluorescent probes, the test uses the eSensor technology to detect amplification based on electrochemical detection. 96 tests can be run in an 8-h shift, with each run taking 2 h. Clinical sensitivity was 94.4%, and there was cross-reactivity with SARS CoV-1 virus. |
| 44 | DiaSorin Molecular LLC | Simplexa COVID-19 Direct assay | rRT-PCR | The test is an “all in one” system, with extraction, reverse transcription, and PCR all performed in the Liaison MDX system. In 52 positive and 56 negative samples, sensitivity and specificity were 100%. There are 24 reactions/kit. |
| 45 | Abbott Molecular | Abbott RealTime SARS-CoV-2 assay | rRT-PCR | The target sequences for the Abbott RealTime SARS-CoV-2 assay are in the SARS-CoV-2 RdRp and N genes of the SARS-CoV-2 genome. The selected target sequences are highly conserved and also specific to this strain of coronavirus. The IC target sequence is derived from the hydroxypyruvate reductase gene from the pumpkin plant, Cucurbita pepo, and is delivered in an Armored RNA® particle that has been diluted in negative human plasma. |
| 46 | Quest Diagnostics Infectious Disease, Inc. | Quest SARS-CoV-2 rRT-PCR | rRT-PCR | The test is a real-time RT-PCR test intended for the qualitative detection of nucleic acid from the SARS-CoV-2 in upper respiratory specimens (for example, nasopharyngeal swabs, oropharyngeal swabs, sputum, BAL, and tracheal aspirates). The assay is composed of two principal steps: (1) extraction of RNA from patient specimens, (2) one-step reverse transcription and PCR amplification with SARS-CoV-2 specific primers and real-time detection with 2019-nCoV specific probes. The assay targets regions of the virus nucleocapsid gene (N1 & N3) and is designed for the detection of SARSCoV-2. Amplification and detection are accomplished using TaqMan chemistry on the ABI 7500. |
| 47 | Quidel Corporation | Lyra SARS-CoV-2 Assay | rRT-PCR | This test procedure received EUA status on 3/17/2020, but this revised EUA replaces the RNA extraction step with a single-step heat extraction in order to decrease reagent bottlenecks. The kit now contains a “process buffer” and there are some differences in volumes of positive and negative controls. The rest of the procedure is approximately the same as the entry on 3/17/2020, with changes made to incorporate the new RNA extraction procedure. |
| 48 | Laboratory Corporation of America (LabCorp) | COVID-19 RT-PCR Test | rRT-PCR | The test is an rRT-qPCR test, and therefore detects viral RNA. LabCorp does not collect specimens, only running the samples. Samples may be collected by the healthcare provider, or at home using the Pixel by LabCorp COVID-19 test home collection kit. |
| 49 | Hologic, Inc. | Panther Fusion SARS-CoV-2 Assay | rRT-PCR | The Assay amplifies and detects two conserved regions of the ORF1ab gene in the same fluorescence channel. The two regions are not differentiated and amplification of either or both regions leads to a fluorescence signal. The Panther Fusion system compares the fluorescence signal to a predetermined cut-off to produce a qualitative result for the presence or absence of the analyte. |
| 50 | Thermo Fisher Scientific, Inc. | TaqPath COVID-19 Combo Kit | rRT-PCR | Performed using TaqPath COVID-19 Combo Kit on the Applied Biosystems 7500 Fast Dx Real-Time PCR instrument, or other authorized instruments. |
| 51 | Roche Molecular Systems, Inc. (RMS) | cobas SARS-CoV-2 | rRT-PCR | The cobas SARS-CoV-2 test is based on fully automated sample preparation (nucleic acid extraction and purification) followed by reverse transcription, PCR amplification and detection on the cobas 6800/8800 system, or other authorized instruments. Designed to be a single-well, dual target qualitative assay |
| 52 | Wadsworth Center, New York State Department of Public Health’s (CDC) | New York SARS-CoV-2 Real-time Reverse Transcriptase (RT)-PCR Diagnostic Panel | RT-PCR | Test and analysis performed on Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument with SDS 1.4 software. |
| 53 | Centers for Disease Control and Prevention’s (CDC) | CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel (CDC) | RT-PCR | It is a real-time RT-PCR test intended for the qualitative detection of nucleic acid from the 2019-nCoV in upper and lower respiratory specimens (such as nasopharyngeal or oropharyngeal swabs, sputum, lower respiratory tract aspirates, bronchoalveolar lavage, and nasopharyngeal wash/aspirate or nasal aspirate) collected from individuals who meet 2019-nCoV clinical and/or epidemiological criteria |
| Serological | ||||
| 1 | EUROIMMUN US Inc. | Anti-SARS-CoV-2 ELISA (IgG) | Serology IgG | The test kit contains microplate strips each with 8 break-off reagent wells coated with recombinant structural protein of SARS-CoV-2. In the first reaction step, diluted patient samples are incubated in the wells. In the case of positive samples, specific IgG (also IgA and IgM) antibodies will bind to the antigens. To detect the bound antibodies, a second incubation is carried out using an enzyme-labelled anti-human IgG (enzyme conjugate) catalyzing a color reaction. |
| 2 | Roche Diagnostics | Elecsys Anti-SARS-CoV-2 | Serology Antibody | It is an immunoassay based test for the in vitro qualitative detection of antibodies (including IgG) to SARS-CoV-2 in human serum and plasma. The test is intended as an aid in the determination of the immune reaction to SARS-CoV-2. |
| 3 | Wadsworth Center, New York State Department of Health | New York SARS-CoV Microsphere Immunoassay for Antibody Detection | Serology Total Antibody | It is to be used as an aid in identifying individuals who may have high levels of SARS-CoV-2-reactive antibodies in their blood that reflect an adaptive immune response to SARS-CoV-2 indicating recent or prior infection. |
| 4 | Bio-Rad Laboratories, Inc | Platelia SARS-CoV-2 Total Ab assay | Serology Total Antibody | The Platelia SARS-CoV-2 Total Ab is a one-step antigen capture format, enzyme-linked immunosorbent assay (ELISA), intended for the qualitative detection of total antibodies (IgM/IgG/IgA) to SARS-CoV-2 in human serum and plasma (dipotassium EDTA, tripotassium EDTA, lithium heparin, ACD, or sodium citrate). |
| 5 | Abbott Laboratories Inc. | SARS-CoV-2 IgG assay | Serology IgG only | The SARS-CoV-2 IgG assay is a chemiluminescent microparticle immunoassay (CMIA) used for the qualitative detection of IgG antibodies to SARS-CoV-2 in human serum and plasma on the Alinity i and ARCHITECT i Systems. |
| 6 | DiaSorin Inc. | LIAISON SARS-CoV-2 S1/S2 IgG | Serology IgG only | The tests make use of magnetic beads coated with S1 & S2 Antigens |
| 7 | Ortho-Clinical Diagnostics, Inc. | VITROS Immunodiagnostic Products Anti-SARS-CoV-2 IgG Reagent Pack | Serology IgG only | Qualitative detection of IgG antibodies to SARS-CoV-2 in human serum. Intended for use as an aid in identifying individuals with an adaptive immune response to SARS-CoV-2, indicating recent or prior infection |
| 8 | Autobio Diagnostics Co. Ltd. | Anti-SARS-CoV-2 Rapid Test | Serology IgM and IgG | Qualitative detection and differentiation of IgM and IgG antibodies to SARS-CoV-2 in human plasma from anticoagulated blood (Heparin/ EDTA/ sodium citrate) or serum. Intended for use as an aid in identifying individuals with an adaptive immune response to SARS-CoV-2, indicating recent or prior infection |
| 9 | Cellex Inc. | qSARS-CoV-2 IgG/IgM Rapid Test | Serology IgM and IgG | A lateral flow immunoassay intended for the qualitative detection and differentiation of IgM and IgG antibodies to SARS-CoV-2 in serum, plasma (EDTA, citrate) or venipuncture whole blood specimens from patients suspected of COVID-19 infection by a healthcare provider |
| 10 | Mount Sinai Laboratory | COVID-19 ELISA IgG Antibody Test | Serology IgG | ELISA IgG Antibody Test consists of two serial direct Enzyme-Linked Immunosorbent Assays (ELISA) for the qualitative detection of human IgG antibodies in serum and plasma specimens collected from individuals suspected of prior infection with the virus that causes COVID-19 by their healthcare provider. An initial ELISA is performed against recombinant Receptor Binding Domain of SARS-CoV-2 in serum and plasma, followed for positive specimen by a confirmatory ELISA against full length SARS-CoV-2 Spike protein in serum and plasma. |
| 11 | Chembio Diagnostic System, Inc | DPP COVID-19 IgM/IgG System | Serology IgM and IgG | A single-use rapid immunochromatographic test for the qualitative detection and differentiation of Immunoglobulin M (IgM) and Immunoglobulin G (IgG) antibodies to SARS-CoV-2 in fingerstick whole blood, venous whole blood, serum, or plasma (lithium heparin or EDTA) samples from patients suspected of COVID-19 infection by a healthcare provider |
EUA Emergency Use Authorization, FDA U.S. Food and Drug Administration)
Conclusion and future prospect
The development and research have ramped up at a greater speed than ever in the field of diseases diagnosis due to COVID-19 outbreak leading to an increase in the mortality rate each day in an unpredictable manner. Still countries are struggling to speed up testing due to the laborious and expensive RT-PCR lab technique, which often require centralized services. Few RT-PCR based innovative diagnostic methods are in the developmental stage which detect viral materials in different ways such as gene editing tool CRISPR. But, need of the hour in this pandemic is the development and establishing innovative diagnostic tool which would be easy to use, accurate, sensitive and point of care. Although point of care serological tests (lateral flow immunoassay based) is readily available in the market for COVID-19 detection but still the quality of commercial kits needs to be re-evaluated due to occurrence of false positive results. The lateral flow-based immunoassay has advantages over RT-PCR in developing nations were the facilities for RT-PCR set up is scare. However, the lateral flow immunoassay does not give an explicit picture of the severity of the disease but can help as an aid the detection alongside with RT-PCR technique.
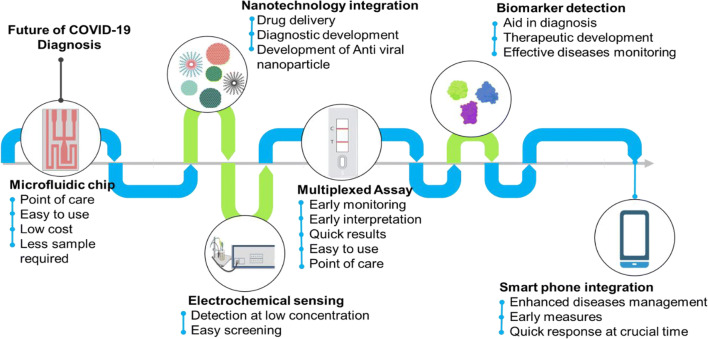
With the available technology, the diagnosing of the disease could be a menace shortly. Although the process of technology development is hurtling up day by day integrating them for a better output is essential. In our view, following developments can make an advanced COVID -19 detection tools (Fig. 5): 1) Making isothermal amplification-based LFA could be one of the alternatives in hand to overcome the problem with current lateral flow assay of giving false negative results. 2) Paper-based device with real-rime simultaneous detection of multiple DNA targets can be developed for highly sensitive and specific detection, 3) Nanotechnology intervention in the pipeline of disease detection can be potentially fruitful, 4) Development needs to be made in making use of microfluidic devices for development of new prototype for early point of care detection of the novel virus. Even screening of close biomarkers for development of accurate and precise diagnostics kit is necessary, 5) Efforts need to be made for technology forward approach by integrating it to smartphone mobile devices which are omnipresent in nature, 6) Making CRISPR-Chip based electrical detection can help us to detect the analyte even at lower concentration, 7) Development of novel face mask which in turn is integrated with an sensor can help in detecting even lower limit of virus 8) Making use of electrochemical based device as it can detect even narrow antigenic load. There is also possibility to explore the development of point-of-care tests with multiplex assays and breathe analyzer. With integration with technology it will be easy not only to identify the people suffering and also take measure as early as possible for better diseases management.
Fig. 5.
Future prospects of COVID-19 diagnosis
Research in the field of COVID-19 is evolving quickly and new data are generated each day. Some referenced manuscripts are preprints and have not been peer-reviewed.
Acknowledgements
We gratefully acknowledge Gujarat State Biotechnology Mission (GSBTM Project ID: AJJV48) and SHODH: ScHeme of Developing High Quality Research Scholarship (Ref No: 201901440009) for financial support.
Authors’ contributions
All the authors have contributed equally.
Data availability
Not applicable.
Compliance with ethical standards
Conflicts of interest/competing interests
Authors declare no conflict of interest.
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbott: Abbott launches Covid-19 antibody test (2020). https://www.abbott.com/corpnewsroom/product-andinnovation/abbott-launches-covid-19-antibody-test.html. Accessed 15 May 2020.
- Ahn D-G, Jeon I-J, Kim JD, Song M-S, Han S-R, Lee S-W, Jung H, Oh J-W. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst. 2009;134:1896–1901. doi: 10.1039/b906788d. [DOI] [PubMed] [Google Scholar]
- Ai J-W, Zhou X, Xu T, Yang M, Chen Y, He G-Q, Pan N, Cai Y, Li Y, Wang X. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 2019;8:1361–1369. doi: 10.1080/22221751.2019.1664939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;200:642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;198:129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Authorization, E. U.: LabCorp COVID-19 RT-PCR test EUA Summary (2020). https://www.labcorp.com/coronavirusdisease-covid-19. Accessed 15 May 2020
- Bahadir EB, Sezgintürk MK. Lateral flow assays: Principles, designs and labels. TrAC Trends in Analytical Chemistry. 2016;82:286–306. [Google Scholar]
- Baker AN, Richards S-J, Guy CS, Congdon TR, Hasan M, Zwetsloot AJ, Gallo A, Lewandowski JZR, Stansfeld PJ, Straube A. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis KG, Matushek SM, Abeleda APF, Bethel C, Hunt C, Gillen S, Moran A, Tesic V. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020;104:468. doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus COVID-19 IgM ELISA Assay Kit (2020)
- Milenia Biotec, COVID-19 test development with lateral flow & CRISPR “Hybridetect” (2020). https://www.milenia-biotec.com/en/covid-19-rapid-test-development. Accessed 15 May 2020.
- J.P. Broughton, X. Deng, G. Yu, C.L. Fasching, V. Servellita, J. Singh, X. Miao, J.A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 1–5 (2020) [DOI] [PMC free article] [PubMed]
- Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W, Dincer C, Urban GA. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 2019;31:1905311. doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- A.I.L.D. Burog, C.P.R.C. Yacapin, R.R.O. Maglente, A.A. Macalalad-Josue, E.J.B. Uy, Should IgM/IgG rapid test kit be used in the diagnosis of COVID-19? Pacific Center for Evidence Based Healthcare 4, 1–12 (2020)
- Cai X, Chen J, Hu J, Long Q, Deng H, Fan K, Liao P, Liu B, Wu G, Chen Y. A Peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J. Infect. Dis. 2020;222(2):189–193. doi: 10.1093/infdis/jiaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawte AD, Unrau PJ, Rueda DS. Live cell imaging of single RNA molecules with fluorogenic Mango II arrays. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-14932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention: Burden Of Influenza (2020a). https://www.cdc.gov/flu/about/burden/index.html. Accessed 15 May 2020
- Centers for Disease Control and Prevention: Coronavirus Disease 2019 COVID-19 (2020b). https://www.cdc.gov/coronavirus/2019-ncov/index.html. Accessed 15 May 2020
- J.F.-W. Chan, C.C.-Y. Yip, K.K.-W. To, T.H.-C. Tang, S.C.-Y. Wong, K.-H. Leung, A.Y.-F. Fung, A.C.-K. Ng, Z. Zou, H.-W. Tsoi, Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 58 (2020a) [DOI] [PMC free article] [PubMed]
- Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, Poon RW-S. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekani-Azar S, Gharib Mombeni E, Birhan M, Yousefi M. CRISPR/Cas9 gene editing technology and its application to the coronavirus disease (COVID-19), a review. J. Life Sci. Biomed. 2020;10:01–09. [Google Scholar]
- Chen A, Yang S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015;71:230–242. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang Z, Zhai X, Li Y, Lin L, Zhao H, Bian L, Li P, Yu L, Wu Y. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, Using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 2011;17:1015. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, Liu Y, NG DY, Wan CK, Yang P, Wang Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V. Corman, I. Eckerle, T. Bleicker, A. Zaki, O. Landt, M. Eschbach-Bludau, S. van Boheemen, R. Gopal, M. Ballhause, T. Bestebroer, Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance 17 (2012a) [DOI] [PubMed]
- V. Corman, M. Müller, U. Costabel, J. Timm, T. Binger, B. Meyer, P. Kreher, E. Lattwein, M. Eschbach-Bludau, A. Nitsche, Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance 17 (2012b) [DOI] [PubMed]
- V. Corman, T. Bleicker, S. Brünink, C. Drosten, M. Zambon, Diagnostic detection of 2019-nCoV by real-time RT-PCR (World Health Organization, 2020)
- Dincer C, Bruch R, Kling A, Dittrich PS, Urban GA. Multiplexed point-of-care testing–xPOCT. Trends Biotechnol. 2017;35:728–742. doi: 10.1016/j.tibtech.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- X. Ding, K. Yin, Z. Li, C. Liu, Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 11(1) (2020) [DOI] [PMC free article] [PubMed]
- J. Dinnes, J.J. Deeks, A. Adriano, S. Berhane, C. Davenport, S. Dittrich, D. Emperador, Y. Takwoingi, J. Cunningham, S. Beese, Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. (2020) [DOI] [PMC free article] [PubMed]
- Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z, Tong S. Epidemiological characteristics of 2143 pediatric patients with 2019 coronavirus disease in China. Pediatrics. 2020;145(6):e20200702. [Google Scholar]
- J. Elflein, (2020). Available: https://www.statista.com/statistics/1028731/covid19-tests-select-countries-worldwide/
- M. Enserink, Update: ‘A bit chaotic.’ Christening of new coronavirus and its disease name create confusion [Online] (2020). Available: https://www.sciencemag.org/news/2020/02/bit-chaotic-christening-new-coronavirus-and-its-disease-name-create-confusion.
- Epitope Diagnostics, Inc: ELISA For Novel Coronavirus (2019-Ncov, SARS-Cov-2) Causing an outbreak of pneumonia (COVID-19) — Epitope Diagnostics, Inc. http://www.epitopediagnostics.com/covid-19-elisa (2020). Accessed 15 May 2020
- Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;200:432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H. Feng, Y. Liu, M. Lv, J. Zhong, A case report of COVID-19 with false negative RT-PCR test: Necessity of chest CT. Jpn. J. Radiol. 1–2 (2020) [DOI] [PMC free article] [PubMed]
- J. Gallagher, Are coronavirus tests flawed. BBC News, https://www.bbccom/news/health-51491763, 13 (2020)
- J. Gao, Z. Tian, X. Yang, Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends (2020) [DOI] [PubMed]
- E. Gonzalez-Gonzalez, G. Trujillo-De Santiago, I.M. Lara-Mayorga, S.O. Martinez-Chapa, M.M. Alvarez, Portable and accurate diagnostics for COVID-19: Combined use of the miniPCR thermocycler and a well-plate reader for SARS-CoV-2 virus detection. PLOS One 15(8), e0237418 (2020) [DOI] [PMC free article] [PubMed]
- Group, A.:Aptamer Group. 2020. Applications of aptamers in detection and therapeutics of SARS-Cov - Aptamer Group (2020). https://www.aptamergroup.co.uk/applications-of-aptamers-in-detection-and-therapeutics-of-sars-cov. Accessed 15 May 2020
- Hajian R, Balderston S, Tran T, Deboer T, Etienne J, Sandhu M, Wauford NA, Chung J-Y, Nokes J, Athaiya M. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E. Ortiz-Ospina, H. Ritchie, D. Beltekian, E. Mathieu, J. Hasell, B. Macdonald, C. Giattino, M. Roser, (2020). Available: https://ourworldindata.org/coronavirus-testing#the-scale-of-testing-compared-to-the-scale-of-the-outbreak [DOI] [PMC free article] [PubMed]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huids IAE, Madani T, Ntoumi F, Koch R, Dar O. The continuing 2019-nCoV epidemicthreatof novel coronaviruses to global health: the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A.J. Jääskeläinen, E. Kekäläinen, H. Kallio-Kokko, L. Mannonen, E. Kortela, O. Vapalahti, S. Kurkela, M. Lappalainen, Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Eurosurveillance. 25, 2000603 (2020) [DOI] [PMC free article] [PubMed]
- Jang KJ, Lee N-R, Yeo W-S, Jeong Y-J, Kim D-E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J. Joung, A. Ladha, M. Saito, N.-G. Kim, A.E. Woolley, M. Segel, R.P. Barretto, A. Ranu, R.K. Macrae, G. Faure, Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. (2020) [DOI] [PMC free article] [PubMed]
- Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov KN, Sitdikov RA, Lopez GP, Atanassov P, Rubin RL. Rapid detection of anti-chromatin autoantibodies in human serum using a portable electrochemical biosensor. Biosens. Bioelectron. 2009;24:1949–1954. doi: 10.1016/j.bios.2008.09.032. [DOI] [PubMed] [Google Scholar]
- H. Lai, C. Wang, C. Weng, T. Liou, G. Lee, An integrated SELEX microfluidic system for rapid screening of influenza virus specific aptamers. Proceedings of the 16th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Okinawa, Japan, 14021404 (2012)
- R. Lassaunière, A. Frische, Z.B. Harboe, A.C. Nielsen, A. Fomsgaard, K.A. Krogfelt, C.S. Jørgensen, Evaluation of nine commercial SARS-CoV-2 immunoassays (Medrxiv, 2020)
- Lee EY, Ng M-Y, Khong P-L. COVID-19 pneumonia: what has CT taught us? Lancet Infect. Dis. 2020;20:384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li S, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Li X, Zai J, Wang X, Li Y. Potential of large “first generation” human-to-human transmission of 2019-nCoV. J. Med. Virol. 2020;92:448–454. doi: 10.1002/jmv.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S. Liebich, Potent SELEX aptamer-based therapeutic method for novel SARS-CoV2 virus disease (COVID-19) (2020)
- F. Hoffmann-La Roche Ltd, Roche to launch SARS-CoV-2 Rapid Antigen Test in countries accepting CE mark, allowing fast triage decisions at point of care [Online] ( 2020a). Available: https://www.roche.com/media/releases/med-cor-2020-09-01b.htm
- F. Hoffmann-La Roche Ltd, Elecsys Anti-SARS-CoV-2 (2020b)
- Iceni Diagnostics Ltd, Rapid COVID-19 detection – Iceni Diagnostics offers new approach [Online] (n.d.). Available: https://www.icenidiagnostics.com/news/rapid-covid-19-detection-iceni-diagnostics-offers-new-approach/
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- H. Ma, W. Zeng, H. He, D. Zhao, D. Jiang, P. Zhou, L. Cheng, Y. Li, X. Ma, T. Jin, Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 1–3 (2020) [DOI] [PMC free article] [PubMed]
- Mak GC, Cheng PK, Lau SS, Wong KK, Lau C, Lam ET, Chan RC, Tsang DN. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;104:500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AW, Phillips ST, Carrilho E, Thomas SW, III, Sindi H, Whitesides GM. Simple telemedicine for developing regions: camera phones and paper-based microfluidic devices for real-time, off-site diagnosis. Anal. Chem. 2008;80:3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S. Misra, Biotech startup with MIT Tata Center roots produces 15-min tests for Covid-19 [Online] (2020). Available: http://energy.mit.edu/news/mit-tata-center-startup-e25bio-produces-15-minute-tests-for-covid-19/
- Moitra P, Alafeef M, Dighe K, Frieman M, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E, Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;112:274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujawar MA, Gohel H, Bhardwaj SK, Srinivasan S, Hickman N, Kaushik A. Aspects of nano-enabling biosensing systems for intelligent healthcare; towards COVID-19 management. Mater. Today Chem. 2020;100:306. doi: 10.1016/j.mtchem.2020.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers FB, Lee LP. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab Chip. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- S. Nanotech, Sona Nanotech provides Covid-19 antigen test progress update [Online] (2020). Available: https://sonanano.com/sona-nanotech-provides-covid-19-antigen-test-progress-update/
- Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Engineering and Biotechnology News: COVID-19 test kits being developed using RNA imaging (2020). https://www.genengnews.com/news/covid-19-test-kits-being-developed-using-rna-imaging. Accessed 15 May 2020.
- Nicol T, Lefeuvre C, Serri O, Pivert A, Joubaud F, Dubée V, Kouatchet A, Ducancelle A, Lunel-Fabiani F, Le Guillou-Guillemette H. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech) J. Clin. Virol. 2020;129:104511. doi: 10.1016/j.jcv.2020.104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H. Nishiura, H. Oshitani, T. Kobayashi, T. Saito, T. Sunagawa, T. Matsui, T. Wakita, M. Covid, M. Suzuki, Closed environments facilitate secondary transmission of coronavirus disease 2019 (COVID-19) (MedRxiv, 2020)
- E. Nunez-Bajo, M. Kasimatis, Y. Cotur, T. Asfour, A. Collins, U. Tanriverdi, M. Grell, M. Kaisti, G. Senesi, K. Stevenson, Ultra-low-cost integrated silicon-based transducer for on-site, genetic detection of pathogens (bioRxiv, 2020)
- G. Onder, G. Rezza, S. Brusaferro, Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA (2020) [DOI] [PubMed]
- World Health Organization (WHO), Home (2020). https://www.who.int. Accessed 15 May 2020.
- OraSure Technologies, I.: Orasure Technologies, Inc. Announces 2020 Second quarter financial results and update On COVID-19 testing programs. GlobeNewswire News Room (2020). https://www.globenewswire.com/news-release/2020/08/05/2073582/0/en/OraSure-Technologies-Inc-Announces-2020-Second-Quarter-Financial-Resultsand-Update-on-COVID-19-Testing-Programs.html. Accessed 15 May 2020.
- Pickar-Oliver A, Gersbach CA. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine R, Winslow GM. IgM in microbial infections: taken for granted? Immunol. Lett. 2009;125:79–85. doi: 10.1016/j.imlet.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J.N. Rauch, E. Valois, S.C. Solley, F. Braig, R.S. Lach, N.J. Baxter, K.S. Kosik, C. Arias, D. Acosta-Alvear, M.Z. Wilson, A scalable, easy-to-deploy, protocol for Cas13-based detection of SARS-CoV-2 genetic material (bioRxiv, 2020) [DOI] [PMC free article] [PubMed]
- Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG. Rapid detection of Covid-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shan B, Broza YY, Li W, Wang Y, Wu S, Liu Z, Wang J, Gui S, Wang L, Zhang Z. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020;14:12125–12,132. doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- To KK-W, Tsang OT-Y, Leung W-S, Tam AR, Wu T-C, Lung DC, Yip CC-Y, Cai J-P, Chan JM-C, Chik TS-H. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Torrente-Rodríguez RM, Wang M, Gao W. The era of digital health: A review of portable and wearable affinity biosensors. Adv. Funct. Mater. 2019;1:906713. [Google Scholar]
- Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VY, Chen H, Mubareka S, Gubbay JB, Chan WC. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Wan Z, Zhang X, Yan X. IFA in testing specific antibody of SARS coronavirus. South China J Prev. Med. 2003;29:36–37. [Google Scholar]
- Wang P. Combination of serological total antibody and RT-PCR test for detection of SARS-COV-2 infections. J. Virol. Methods. 2020;283:113919. doi: 10.1016/j.jviromet.2020.113919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. Wang, W. Li, D. Drabek, N.M. Okba, R. Van Haperen, A.D. Osterhaus, F.J. Van Kuppeveld, B.L. Haagmans, F. Grosveld, B.-J. Bosch, A human monoclonal 1 antibody blocking SARS-CoV-2 infection (Biorxiv, 2020a) [DOI] [PMC free article] [PubMed]
- M. Wang, A. Fu, B. Hu, Y. Tong, R. Liu, J. Gu, J. Liu, W. Jiang, G. Shen, W. Zhao, Nanopore target sequencing for accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses (medRxiv, 2020b) [DOI] [PMC free article] [PubMed]
- S.K. Wee, S.P. Sivalingam, E.P.H. Yap, Rapid Direct Nucleic Acid Amplification Test without RNA Extraction for SARS-CoV-2 using a Portable PCR Thermocycler (bioRxiv, 2020) [DOI] [PMC free article] [PubMed]
- S.R. Weiss, J.L. Leibowitz, Coronavirus pathogenesis. Adv. Virus Res. (Elsevier, 2011) [DOI] [PMC free article] [PubMed]
- Weiss C, Carriere M, Fusco L, Capua I, Regla-Nava JA, Pasquali M, Scott JA, Vitale F, Unal MA, Mattevi C. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano. 2020;14(6):6383–6406. doi: 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
- Wen T, Huang C, Shi F-J, Zeng X-Y, Lu T, Ding S-N, Jiao Y-J. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst. 2020;145:5345–5352. doi: 10.1039/d0an00629g. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, Mclellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus—A possible reference for coronavirus disease-19 treatment option. J. Med. Virol. 2020;92(6):556–563. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu Q. Biosensors and bioelectronics on smartphone for portable biochemical detection. Biosens. Bioelectron. 2016;75:273–284. doi: 10.1016/j.bios.2015.08.037. [DOI] [PubMed] [Google Scholar]
- Z. Zhao, H. Cui, W. Song, X. Ru, W. Zhou, X. Yu, A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2 (bioRxiv, 2020) [DOI] [PMC free article] [PubMed]
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382(8), 727-733 (2020) [DOI] [PMC free article] [PubMed]
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.