Abstract
Introduction
The therapy of patients with advanced phase gastric cancer is still a huge threat, with extremely imperfect therapies authorized. Even if the amassed indications have validated the significance of lncRNA in gastric cancer, few understandings are stated concerning nuclear paraspeckle assembly transcript 1 (NEAT1) practical functions and molecular mechanisms.
Methods
In this research, the expression of NEAT1 and miR-1224-5p in gastric cancer tissues was measured by qRT-PCR analysis, and the expression of remodeling and spacing factor 1 (RSF1) was measured by IHC assay. Then, the bioinformatics prediction software ENCORI was applied to envisage the assumed binding sites. The monitoring roles of NEAT1 or miR-1224-5p on the cell proliferation and migration capacity were verified by CCK-8, wound healing and transwell assay, correspondingly. The interactions among NEAT1, miR-1224-5p and RSF1 were investigated via luciferase analysis.
Results
Our findings revealed high expression levels of NEAT1, RSF1 and a decreased expression level of miR-1224-5p in gastric cancer. Upregulation of NEAT1 or knockdown of miR-1224-5p elevated gastric cancer cell proliferation, and migration. Bioinformatics and luciferase analyses simplified that NEAT1 directly cooperated with miR-1224-5p to weaken miR-1224-5p binding to the RSF1 3ʹ-UTR region. Likewise, the mechanical inquiries ratified that initiation of the miR-1224-5p/RSF1 regulatory loop by miR-1224-5p knockdown or overexpressed RSF1 validated the functions of NEAT1 in endorsing gastric cancer cell malignancy.
Discussion
Our research initially validated that NEAT1 may regulate the expression of RSF1 competitive sponge to miR-1224-5p, contributed to the supervision of gastric cancer evolution, which exposed new brightness for diagnosis and therapy of gastric cancer.
Keywords: competes endogenous RNAs, remodeling and spacing factor 1, long non-coding RNAs, microRNAs, gastric cancer, nuclear paraspeckle assembly transcript 1
Introduction
Gastric cancer is the fifth most common global cause of cancer-related death, and is characterized with delayed diagnosis and high metastatic probability.1,2 Most patients with early gastric cancer have no overt clinical symptoms,3 although some people may have nausea and vomiting or upper gastrointestinal symptoms similar to ulcer disease, all of which lack specificity for the diagnosis of gastric cancer.4 Most gastric cancer patients are at an advanced stage when diagnosis is confirmed.5 At present, there is a lack of effective diagnostic indicators, and hence it is vital to look for effective diagnostic or therapeutic targets for early detection and treatment of gastric cancer.6 There are a large number of RNA molecules in human life, most of which are non-protein-coding RNAs.7 Non-coding RNAs (ncRNAs) refer to RNAs that can be transcribed from the genome but with no protein-coding capability, so they can function at their respective RNA levels.8 Most ncRNAs are functional, including antisense RNAs (asRNAs), microRNAs (miRNAs), small interfering RNAs (siRNAs), and long non-coding RNAs (lncRNAs).9 LncRNA is one of a type of endogenous RNA molecules greater than 200 nucleotides in length.10 LncRNA has a wide range of functions and can participate in many vital physiological procedures including transcriptional regulation and chromatin modification.11 More notably, lncRNA has a vital impact in abnormal cellular regulation and tumorigenesis induction, and some studies have found that lncRNA is closely related to tumors, and its abnormal expression and regulation are found in liver, breast, colon and stomach cancers.12 Nuclear paraspeckle assembly transcript 1 (NEAT1) is a novel lncRNA with potential carcinogenic activity screened by lncRNA microarray, which has the function of promoting tumor metastasis and has a vital impact on the progression of prostate, breast and laryngeal squamous cell cancers.13–15 However, the detailed molecular role of the impact of NEAT1 in gastric cancer needs to be further studied.15
Remodeling and remodeling factor 1 (RSF1), a member of ATP-dependent chromatin remodeling factor, was also identified as hepatitis B x-antigen associated protein (HBXAP).16 RSF1 binds with sucrose non-ferment protein 2 homolog (hSNF2H) to produce the RSF1/hSNF2H composite, which sequentially plays a role in various biological processes by regulating nucleosome remodeling, including transcription, DNA replication, and the cell cycle.17 Recently, RSF1 has been found to have abnormal expression and dysfunction in solid tumors.18 Studies have shown that RSF1 is evaluated and is distinctly associated with TNM stage and poor prognosis in renal and prostate cancers.19 However, the character and potential mechanism of RSF1 in gastric cancer remain uncertain. Therefore, it is of vital clinical meaning to investigate the expression of RSF1 in gastric cancer, its upstream genetic factor, and potential supervisory methods. Previously, to uncover the potential target genes mediated by RSF1 in gastric cancer cell, we identified RSF1 as one of the target gene of NEAT1/miR-1224-5p loop through the bioinformatics ENCORI software. The data revealed that the NEAT1 can regulate the expression level of RSF1 via a competing endogenous RNA (ceRNA) method. In this study, we aimed to explore the biological character of RSF1 on gastric cancer progression and its potential regulatory mechanisms. We indicated that RSF1 was evaluated in gastric cancer and the NEAT1/miR-1224-5p/RSF1 regulatory loop axis seems to be a possible therapeutic object in gastric cancer therapy.
Materials and Methods
Human Specimen
Seventy-nine cases of gastric cancer together with 78 cases of non-neoplastic gastric mucosal specimens were obtained from patients having received resection surgeries at the First Affiliated Hospital of Jilin University from May 2010 to March 2014. No patients had received chemotherapeutic or radiotherapeutic treatment before surgeries. Each patient had signed an informed consent in the written form. Our research was reviewed and approved by the Research Ethics Committee of Jilin University (approval number: JLU-2013157) and was conducted strictly in compliance with the ethics of the Declaration of Helsinki.
qRT-PCR
Isolation of total RNA was conducted using RNeasy Mini Kit (Qiagen). Reverse transcription was conducted via PrimeScriptTM RT reagent Kit (Takara, Shiga, Japan) as per the protocol specified by the manufacturer. Quantification of relative gene expression was conducted through qRT-PCR with SYBR Premix ExTaq kit (Takara, Shiga, Japan). One microgram of RNA was reverse-transcribed using an M-MLV Kit (Takara, Shiga, Japan) and random 9-mer primers (Takara, Shiga, Japan). Semiquantitative PCR was conducted using 50 ng of reverse-transcribed cDNA and 0.4 μM of each primer in a final reaction volume of 20 µL containing 1× PCR master mix (Takara, Shiga, Japan). GAPDH was used for expression normalization via the 2−ΔΔCt method. The used primer sequences are itemized in Table 1.
Table 1.
Primer Sequences for Quantitative Real-Time PCR
| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| NEAT1 | 5ʹ-CAGACTAGATACAAGCGAGAAG-3’ | 5ʹ-GTTTCAACAGATTGGCCAAAGA-3’ |
| hsa-miR-1224-5p | 5ʹ- GATGTAAGATCCGCCGTATATAC −3’ | 5ʹ- TGCAGTGGTGGGCAGGAGT −3’ |
| RSF1 | 5ʹ- CTAGCGACGAGATACCAGGG-3’ | 5ʹ- CATCTGAGTGCTCCAACACC-3’ |
| U6 | 5ʹ-CTTCAGCCGGCACAGCT-3’ | 5ʹ-CGCTAATTTGCGTTCAAACG-3’ |
| GAPDH | 5ʹ-CAGGCCTGTACGTTCGCT-3’ | 5ʹ-TAGGTCTGCTAAGTACTGC-3’ |
Western Blot Assay
Cells lysis was carried out in RIPA buffer (Beyotime, China) with inhibitor cocktail (Roche, USA). Twenty micrograms of total proteins were subjected to 10% SDS–PAGE and were later electro transferred onto PVDF membranes (Millipore, USA). After being blocked by skim milk, membranes were then subjected to incubation overnight by primary antibodies. The following antibodies were used: anti-RSF1 (1:10,000, ab109002, Abcam) at 4°C overnight. After washing 3 times with TBST including 0.1% Tween 20, the membrane was incubated with secondary antibodies and examined with ECL Western Blotting Substrate (Millipore, USA) and analyzed with Image Lab 6.0.1 Software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Immunohistochemistry (IHC) Analysis
Routine hematoxylin–eosin staining and antigen retrieval prior to immunohistochemistry were performed as formerly described.20 An immunohistochemical analysis was performed according to the manufacturer’s protocol of UltraSensitiveTM SP (Mouse/Rabbit) IHC Kit (Maixin Biological Technology Development Company, Fujian, China). Following proteolytic digestion and BCA blocking, the slices were incubated with polyclonal RSF1 antibody (Cat. 109002, Abcam, USA) at 4°C overnight, and the HRP/Fab polymer conjugated second antibody (Cat. PV-6000-D, Zhongshan Goldenbridge Biotechnology Co. Ltd., China) was served as secondary antibody. The ultimate immunoreactivity scores (IRS) were calculated consistent with the scores of the ratio of positive dyed cells as formerly described.21 For negative controls, the tissue sections were incubated with isotype antibodies (diluted at same concentration with primary antibodies) the at 4°C overnight.
Cell Culture
The gastric cancer cell lines (SNU-1, SNU-5, SNU-16 and AGS) and human gastric epithelium cell line GES-1 were acquired from Shanghai Institute of Biological Science. All cells were regularly cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS, Gibco) in an incubator at constant 37°C in a 5% CO2 atmosphere.
Cells Transfection
The plasmids of mimics NC, inhibitor NC, miR-1224-5p mimics and miR-1224-5p inhibitor, NEAT1 expression plasmids pcDNA-NEAT1, NEAT1 knockdown plasmids siRNA (si-NEAT1), and the negative control siRNA (si-NC) were synthesized by GenePharma (Shanghai, China). The Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was chosen to perform the cell transfection following the protocol.
Cell Growth Assay
The CCK-8 assay was utilized to evaluate the cell growth together with colony formation assays. For detecting cell growth, cell seeding was conducted in 96-well culture plates (1000 cells for each well), and was added with 10 μL CCK-8 reagent (Dojindo, Japan). The plates were then incubated for one extra hour at 37°C. The OD value at 450 nm wavelength was detected. For colony formation assay, 1000 cells were plated in six-well plates. After 15 days, cells were fixed with 10% paraformaldehyde and stained with 0.2% crystal violet.
Wound Healing Assay
Transfected cells were plated and then cultured up to the time of confluency. The wound was formed via scraping the cell coating using a 10-μL pipette tip. The streaked cells were further cultured for an additional 36 hours. Cell migration was recorded at the 0, 12 and 24th hours with an inverted optical microscope (Olympus, Japan).
Transwell Assay
In brief, 1 × 104 cells were plated into the superior layer of transwell membrane (Corning, USA) layered with Matrigel Matrix in 100 μL serum-free culture media. The lower layer was added with complete medium with 100 mL/L FBS, followed by 36-hour incubation. Cells drifted to the inferior chamber were coloured with 0.5% crystal violet and quantified via an inverted light microscope.
Dual-Luciferase Reporter Assays
Wild and mutant reporter plasmids of NEAT1 (NEAT1-WT-luc, NEAT1-MUT-luc) containing a wild or mutant miR-1224-5p or mimics NC binding sites were produced by GenePharma (Shanghai, China). These reporter plasmids were co-currently transfected into 293t cells via Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. The activities of the luciferase were evaluated via the dual-luciferase reporter assay system (Promega, USA) after 48 h.
Statistical Analysis
GraphPad Prism 6 was adopted for analyzing data. Chi-square test, ANOVA test, two-tailed Student’s t test and Kaplan–Meier analysis were used as appropriate. The data were represented as the mean ± standard error mean deviations and a P value < 0.05 was deemed to have statistical significance.
Results
NEAT1 and RSF1 Was Evaluated While miR-1224-5p Was Reduced in Gastric Cancer Tissues and Cell Lines
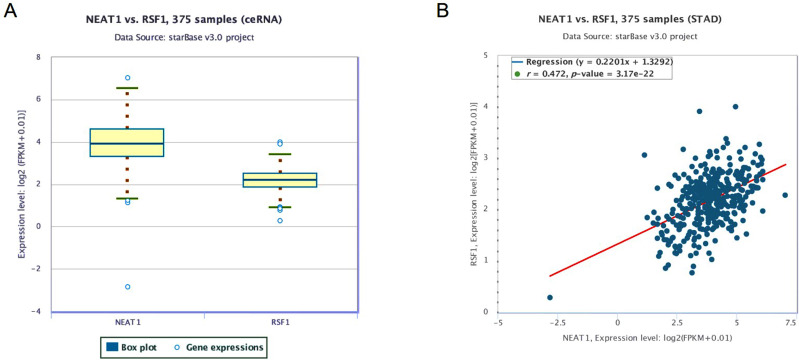
To explore the regulatory impact of NEAT1 in gastric cancer, we first speculated whether NEAT1 and miR-1224-5p were dysregulated in gastric cancer. To uncover the potential target genes mediated by miR-1224-5p in gastric cancer cell, we identified RSF1 as one of the target gene of miR-1224-5p through the bioinformatics ENCORI software. The Pan-Cancer analysis of miRNA-targets and RBP-RNAs in 375 cases of gastric cancer and the expression data of cancers were downloaded from TCGA project via Genomic Data Commons Data Portal. The data revealed that the NEAT1 can regulate the expression level of RSF1 via a ceRNA method, and the expression values of NEAT1 and RSF1 from RNA-seq data revealed that the expression level of NEAT1 is positively related with RSF1 in 375 cases of gastric cancer (Figure 1A and B).
Figure 1.
The association between the expression values of NEAT1 and RSF1. (A) The expression values of NEAT1 and RSF1 from RNA-seq data. (B) The association between the expression values of NEAT1 and RSF1. The expression values of genes from RNA-seq data were scaled with log2 (FPKM + 0.01).
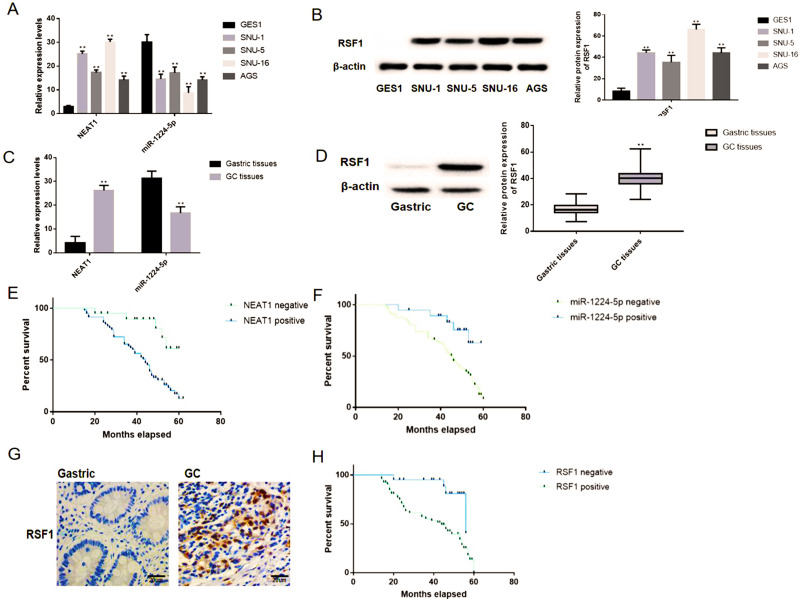
The qRT-PCR and western blotting observations showed the substantial enhancements of NEAT1 and RSF1 expression in four gastric cancer cell lines (SNU-1, SNU-5, SNU-16 and AGS) than a human gastric epithelium cell line (GES-1), while the miR-1224-5p was expressed in an inverse form (Figure 2A and B). To further evaluate the prognostic role of the NEAT1, miR-1224-5p and RSF1 in gastric cancer tissues, we similarly accessed the NEAT1, miR-1224-5p and RSF1 expression levels in gastric cancer tissues and non-neoplastic gastric tissues. As expected, NEAT1 and RSF1 had the elevated expression while miR-1224-5p had a reduced expression in gastric cancer tissues compared to gastric tissues (Figure 2C and D). Furthermore, the associations between the NEAT1, RSF1 and miR-1224-5p expression and prognosis of gastric cancer patients were explored via Kaplan–Meier survival curves with the Log rank test. As it shown in Figure 2E, F and H, the gastric cancer patient with positive miR-1224-5p expression had a longer overall survival time than a patient with negative miR-1224-5p expression. Meanwhile, the gastric cancer patient with positive NEAT1 or RSF1 expression had a shorter overall survival time than a patient with negative NEAT1 or RSF1 expression. Furthermore, the RSF1 expression in gastric cancer tissues was explored via IHC analysis, and it is shown in Figure 2G and Table 2 that the RSF1 has an elevated expression in gastric cancer tissues (56/79) versus gastric tissues (14/78), and the RSF1 was disclosed to be associated with the TNM stage (AJCC) (P < 0.01) and distant metastasis (P < 0.01). These evidences supplied that NEAT1, miR-1224-5p and RSF1 may have vital impacts in the advancement of gastric cancer.
Figure 2.
NEAT1 and RSF1 were upregulated while miR-1224-5p was downregulated in gastric cancer tissues and cell lines. (A) The expression levels of NEAT1 and miR-1224-5p were tested in gastric cancer cell lines (SNU-1, SNU-5, SNU-16 and AGS) and gastric epithelium cell line (GES-1) by qRT-PCR. (B) The western blotting observations showed that RSF1 was expressed at a higher level in four gastric cancer cell lines (SNU-1, SNU-5, SNU-16 and AGS) than a normal human gastric epithelium cell line (GES-1). (C) The expression patterns of NEAT1 and miR-1224-5p were measured in gastric cancer samples and normal tissue via qRT-PCR. (D) The expression patterns of RSF1 were measured in gastric cancer samples and normal tissue via western blotting. (E) The associations between the NEAT1 expression and prognosis of gastric cancer patients were explored. (F) The associations between the miR-1224-5p expression and prognosis of gastric cancer patients. (G) The expression patterns of RSF1 were measured in gastric cancer samples and normal tissue via IHC (magnification ×400). (H) The associations between the RSF1 expression and prognosis of gastric cancer patients. Data are shown as mean ± SD. **P < 0.01.
Table 2.
The Expression Levels of RSF1 and Summary of the Clinicopathological Characteristics in Gastric Cancer Patients
| Variables | Patients, N | RSF1 (+) |
RSF1(–) | P-value |
|---|---|---|---|---|
| Gastric cancer tissues | 79 | 56 | 23 | <0.01 |
| Gastric mucosal tissues | 78 | 14 | 64 | |
| Age (years) | ||||
| ≤60 | 44 | 31 | 13 | 0.718* |
| >60 | 35 | 25 | 10 | |
| Sex | ||||
| Male | 45 | 32 | 13 | 0.672* |
| Female | 34 | 24 | 10 | |
| Distant metastasis | ||||
| + | 47 | 38 | 9 | <0.01 |
| − | 32 | 18 | 14 | |
| TNM stage (AJCC) | ||||
| I–II | 42 | 25 | 17 | <0.01 |
| III–IV | 37 | 31 | 6 |
Note: *No statistical significance was found with the χ2 test/χ2 Goodness-of-Fit Test.
Abbreviation: TNM, tumor-node-metastasis.
NEAT1 Reduction Inhibited the Malignant Behaviors of Gastric Cancer Cells In Vitro
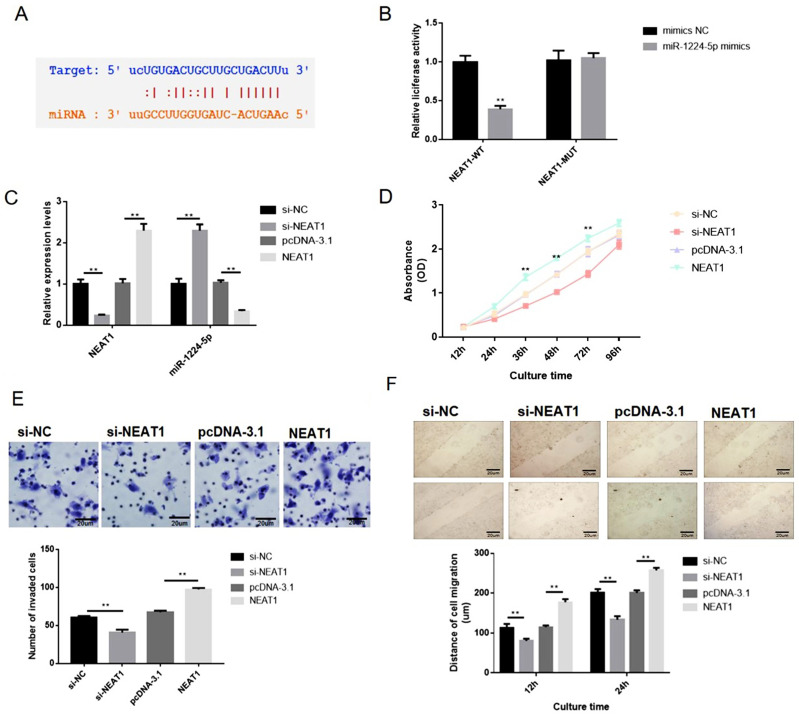
As aforementioned, the NEAT1 and miR-1224-5p were negatively interrelated in gastric cancer cells and specimens (Figure 2), implying the inhibitory impact of NEAT1 on miR-1224-5p. Bioinformatics elucidated that NEAT1 can directly cooperated with miR-1224-5p (Figure 3A). Dual-luciferase reporter assays observed that miR-1224-5p mimics transfection distinctly declining the luciferase signal of reporters comprising NEAT1-WT, but no impact was detected on the luciferase signal of reporters comprising NEAT1-MUT (Figure 3B). Next, we transfected SNU-16 cells with NEAT1 siRNA (si-NEAT1) or pcDNA3.1-NEAT1 overexpression vector (NEAT1). The siRNA (si-NC) or pcDNA-3.1 plasmid were taken as knockdown or overexpression control. Moreover, the expression of NEAT1 and miR-1224-5p were explored via RT-qPCR (Figure 3C). The qRT-PCR observations indicated that the expression level of NEAT1 in NEAT1 overexpression group is 2.2-fold changes higher that vector group and the expression level of NEAT1 in NEAT1 siRNA group is 0.21-fold changes lower that vector group. The expression level of miR-1224-5p in NEAT1 overexpression group is 0.27-fold changes lower that vector group and the expression level of miR-1224-5p in NEAT1 siRNA group is 2.4-fold changes higher that vector group. These observations revealed a negatively regulative association between NEAT1 and miR-1224-5p (Figure 3C). In summary, these observations confirmed that NEAT1 served as a ceRNA for miR-1224-5p to constrain miR-1224-5p expression. Moreover, the knockdown of NEAT1 radically reduce the proliferation, invasion, and migration of SNU-16 cells, which was nonetheless increased by NEAT1 upregulation, as demonstrated by CCK-8 assay, transwell assay and wound-healing assay, respectively (Figure 3D–F). In contrast, overexpression of NEAT1 speeded the proliferation, invasion, and migration of SNU-16 cells (Figure 3D–F).
Figure 3.
NEAT1 overexpression enhanced the proliferation, migration and invasion of gastric cancer SNU-16 cells in vitro. SNU-16 cells were transfected with NEAT1 targeted siRNA (si-NEAT1), negative control siRNA (si-NC), pcDNA3.1-NEAT1 overexpression vector (pcDNA-NEAT1) and blank vector plasmid (pcDNA-3.1), respectively. (A) Bioinformatics clarified that NEAT1 can directly interacted with miR-1224-5p. (B) Dual-luciferase reporter assays were performed to verify the impact of miR-1224-5p on the luciferase signal of reporters containing NEAT1. (C) The expression level of NEAT1 and miR-1224-5p was explored via RT-qPCR. (D) Cell proliferation was tested by CCK-8 assay. (E) Cell invasion was detected by transwell assay. (F) Cell migration was detected by wound healing assay. Data are shown as mean ± SD. **P < 0.01.
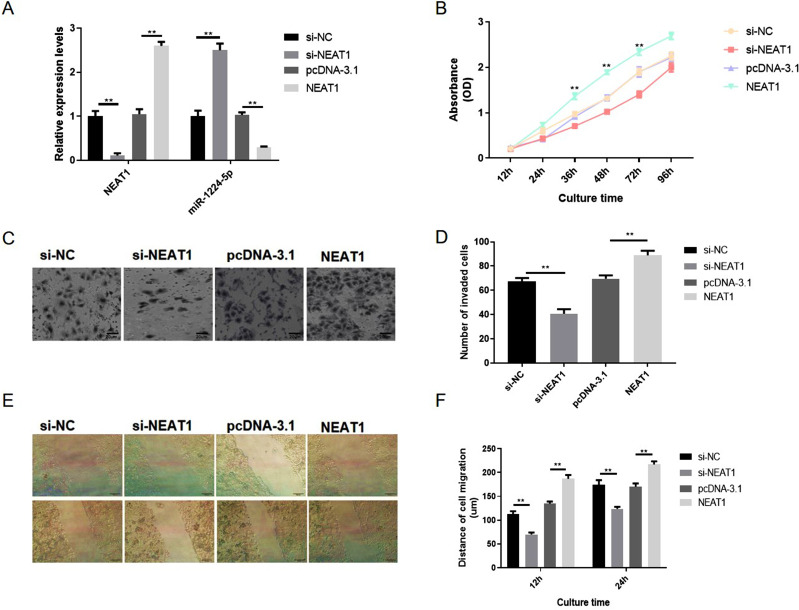
Moreover, the influence of NEAT1 reduction on the malignant behaviors of gastric cancer cells in vitro was also explored in another gastric cell line AGS. The qRT-PCR observations indicated that the expression level of NEAT1 in NEAT1 overexpression group is 2.3-fold changes higher than the vector group and the expression level of NEAT1 in NEAT1 siRNA group is 0.14-fold changes lower than the vector group. The expression level of miR-1224-5p in NEAT1 overexpression group is 0.34-fold changes lower than the vector group and the expression level of miR-1224-5p in NEAT1 siRNA group is 2.5-fold changes higher than the vector group. These observations also revealed a negatively regulative association between NEAT1 and miR-1224-5p (Figure 4A). Similarly, our observations revealed that the NEAT1 reduction inhibited the malignant behaviors of gastric cancer cells and the NEAT1 overexpression enhanced the malignant behaviors of gastric cancer cells in vitro (Figure 4B–F). Hence, these observations implied NEAT1 has an oncogenic potential to stimulate gastric cancer cell malignancy.
Figure 4.
NEAT1 knockdown inhibited the proliferation, migration and invasion of gastric cancer AGS cells in vitro. AGS cells were also transfected with NEAT1 targeted siRNA (si-NEAT1), negative control siRNA (si-NC), pcDNA3.1-NEAT1 overexpression vector (pcDNA-NEAT1) and blank vector plasmid (pcDNA-3.1), respectively. (A) The expression levels of NEAT1 and miR-1224-5p were explored via RT-qPCR. (B) Cell proliferation was tested by CCK-8 assay. (C and D) Cell invasion was detected by transwell assay. (D–F) Cell migration were detected by wound healing assay. Data are shown as mean ± SD. **P < 0.01.
miR-1224-5p Constrained the Malignancy of Gastric Cancer Cells via Down-Regulating RSF1 In Vitro
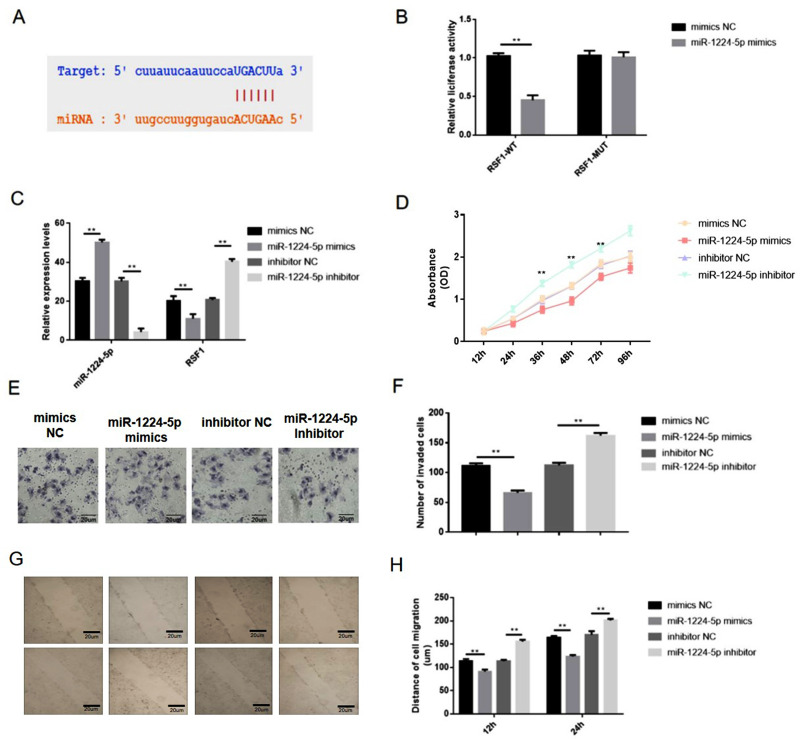
However, the impacts of RSF1 on gastric cancer and its interaction with miR-1224-5p have not yet been analyzed. Bioinformatics and luciferase assay elucidated that miR-1224-5p can directly interact with the RSF1 3ʹ-UTR region (Figure 5A). Next, a dual-luciferase reporter gene assay implied that miR-1224-5p probably interacts with the 3ʹ-UTR of RSF1 (Figure 5B). Additional qRT-PCR authorized that upregulation of miR-1224-5p (1.6-fold changes) distinctly abridged the expression level of RSF1 (0.6-fold changes), whereas reduction of miR-1224-5p (0.13-fold changes) distinctly evaluated the mRNA levels expression of RSF1 (1.7-fold changes) in SNU-16 cells (Figure 5C). Hence, these observations implied that miR-1224-5p functioned as the upstream regulator to change RSF1 expression. CCK‐8 assays displayed an apparently reduced cell viability in miR-1224-5p mimics group, while a substantially augmented cell viability in miR-1224-5p inhibitor group than control group (Figure 5D). Moreover, the migration or invasion ability of gastric cancer cells were restrained when carrying miR-1224-5p mimics transfection, whereas strengthened when carrying inhibitor transfection (Figure 5E–H).
Figure 5.
miR-1224-5p inhibited the proliferation, migration and invasion of gastric cancer cells via downregulating RSF1. (A) Bioinformatics and luciferase assay clarified that miR-1224-5p can directly binding to the RSF1 3ʹ-UTR region. (B) The miR-1224-5p mimics and luciferase reporter plasmids with wild-type or mutant RSF1 3ʹ-UTR were co-transfected into SNU-16 cells. Dual luciferase reporter gene assay was performed to verify the direct binding relationship between miR-1224-5p and RSF1. (C) SNU-16 was transfected with mimics NC, miR-1224-5p mimics, inhibitor NC, miR-1224-5p inhibitor, and then qRT-PCR was taken to evaluate relative expression levels of miR-1224-5p and RSF1. (D) Cell proliferation was tested by CCK-8 assay. (E) Cell invasion was detected by transwell assay. (F) The number of invaded cells. (G) Cell migration was detected by wound healing assay. (H) The distance of the cell migration detected by wound healing assay. Data are shown as mean ± SD. **P < 0.01.
NEAT1 Controlled Gastric Cancer Cells via miR-1224-5p/RSF1 Pathway
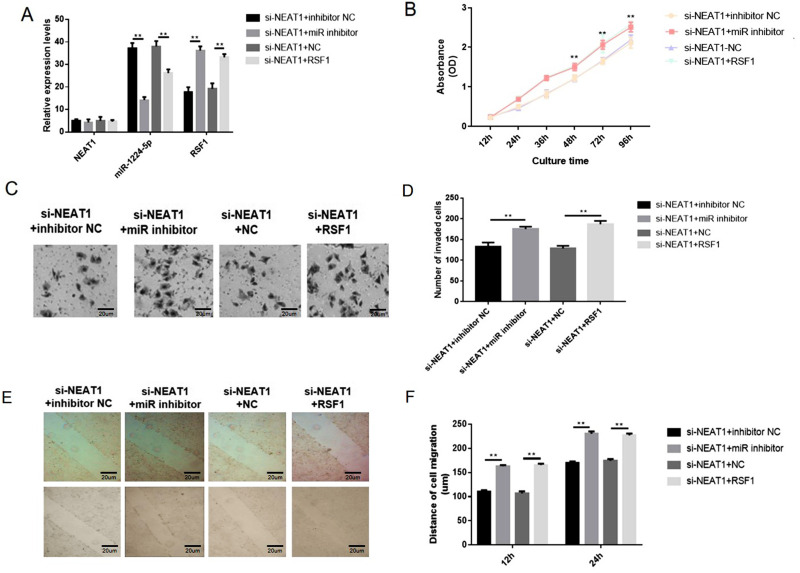
The rescue assays were performed to evaluate the impacts of NEAT1-miR-1224-5p-RSF1 pathway on SNU-16 cell behaviors. Overexpression of RSF1 was realized via transfecting the cells with pcDNA3.1‐RSF1 or miR-1224-5p inhibitor (Figure 6A). The observations exposed that loss of NEAT1 distinctly decreased cell proliferation, migration, and invasion. The co-transfection with si-NEAT1 and miR-1224-5p inhibitor or RSF1 distinctly enhanced cell malignancy in contrast cells transfected with si-NEAT1 alone (Figure 6B–F). Correspondingly, the data indicated the regulatory function of NEAT1 was achieved through miR-1224-5p/RSF1 axis to stimulate the advancement of gastric cancer cells.
Figure 6.
NEAT1 regulated gastric cancer cells via miR-1224-5p/RSF1 pathway. Overexpression of RSF1 was achieved via transfecting the cells with pcDNA3.1‐RSF1 or miR-1224-5p inhibitor (Figure 6A). The knockdown of NEAT1 distinctly reduced cell proliferation, migration and invasion. However, co-transfection with si-NEAT1 and miR-1224-5p inhibitor or RSF1 distinctly increased cell proliferation, migration and invasion in contrast cells transfected with si-NEAT1 alone (Figure 6B–F). Data are shown as mean ± SD. **P < 0.01.
Discussion
In recent years, more and more research has been devoted to elucidate the genetic and epigenetic mechanisms involved in the progression of gastric cancer, but the pathogenesis of gastric cancer is still not fully understood.22 Sequence studies confirm that abnormal expression of lncRNA, an essential part of the human transcriptome, in different cancers is apparent after modification, transcription and its regulation and regulating tumor-related gene expression of the tumor cell malignancy and radiation sensitivity have a vital impact in the process of development.23,24 It has been reported that lncRNA can be used as an anti-tumor target for the therapy of cancer and has certain clinical application value.25 Meanwhile, lncRNA can become a potential prognostic marker of tumors by promoting tumor recurrence and metastasis. NEAT1 is a newly discovered lncRNA, and the mechanism of its action in tumor is occasionally studied.26,27 Studies have found that NEAT1 overexpression predicted prostate cancer progression, indicating that lncRNA-NEAT1 may be a potential oncogene.28 Therefore, we speculate that NEAT1 may also impact the development of gastric cancer. In this study, the expression of NEAT1 in gastric cancer tissues and cells were explored via real-time fluorescent qPCR, and it was found that NEAT1 showed abnormally high expression in gastric cancer.
The miRNAs monitor gene translation via binding with the 3′-UTR of the target gene, which trigger translational repression or mRNA cleavage. Investigations have demonstrated that they could competitively sponge miRNAs by binding to mRNA response components in miRNAs, and ultimately adjust the surface of the basis, thus playing a ceRNA role.29 Bioinformatic analysis suggested that NEAT1 may be associated with the regulation of gastric cancer cell viability and apoptosis as a ceRNA with miR-1224-5p/RSF1. Thus, we assumed that NEAT1 could influence miR-1224-5p expression via ceRNA mechanism. As an inspiring observation, we authorized that miR-1224-5p did have a reciprocal inhibitive impact on NEAT1 expression and knockdown of miR-1224-5p decreased the cell malignancy of gastric cancer cells. Prominently, the dual-luciferase assay additionally presented NEAT1 can directly binds with miR-1224-5p to diminish its expression, revealing NEAT1 role as a miRNA sponge that interact and regulate the expression of miR-1224-5p.
RSF1 was revealed to participate in the regulation of chromatin remodeling and exemplifies a potential therapeutic target in various human cancer.30 The evaluated expression level of RSF1 has been related to adverse tumor characteristics in diverse tumor types,31 but its role in gastric cancer is uncertain. Here we revealed that RSF1 was regulated via the EBLN3P/miR-144-3p axis through bioinformatics analyses. Further rescue assays indicated that EBLN3P enhanced cell malignancy in gastric cancer cells via regulating the miR-144-3p/RSF1 signaling axis. Although we primarily disclosed an original downstream regulated mechanism of the gastric cancer cells caused by NEAT1 in vitro, to profoundly comprehend this complicated mechanism, there is still a great deal to be executed. As it is more complex and has more ambiguity in the animal body than cell models, future mice model research is necessary to further validate the innovative molecular mechanism and demonstrate its possibility of therapeutic objects.
In conclusion, the current study initially validated that NEAT1 acts as an innovative oncogene in gastric cancer. Furthermore, NEAT1 performs as a ceRNA to sponge miR-1224-5p, thereby regulating RSF1 expression and modifying the evolution of gastric cancer. Consequently, our discoveries gave practical evidence to detect new-found target for premature diagnosis and therapeutic treatment in gastric cancer evolution.
Abbreviations
ceRNA, competinh endogenous RNAs; lncRNA, long non-coding RNAs; RSF1, remodeling and spacing factor 1.
Disclosure
The authors declare that there is no conflict of interest.
References
- 1.Tan P, Yeoh KG. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149(5):1153–1162.e1153. doi: 10.1053/j.gastro.2015.05.059 [DOI] [PubMed] [Google Scholar]
- 2.Aichler M, Luber B, Lordick F, Walch A. Proteomic and metabolic prediction of response to therapy in gastric cancer. World J Gastroenterol. 2014;20(38):13648–13657. doi: 10.3748/wjg.v20.i38.13648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barreto SG, Windsor JA. Redefining early gastric cancer. Surg Endosc. 2016;30(1):24–37. doi: 10.1007/s00464-015-4184-z [DOI] [PubMed] [Google Scholar]
- 4.Choi JH, Kim ES, Lee YJ, et al. Comparison of quality of life and worry of cancer recurrence between endoscopic and surgical treatment for early gastric cancer. Gastrointest Endosc. 2015;82(2):299–307. doi: 10.1016/j.gie.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 5.Antonio P, Natale F. Screening for gastric cancer. Am J Gastroenterol. 2019. [Google Scholar]
- 6.Chen P, Lin Y, Zheng K, et al. Risk factors of gastric cancer in high-risk region of China: a population-based case-control study. Asian Pac J Cancer Prev. 2019;20(3):775–781. doi: 10.31557/APJCP.2019.20.3.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun X, Jing H, Wenkang Y. Systematic Identification of Non-coding RNAs. Adv Exp Med Biol. 2018. [DOI] [PubMed] [Google Scholar]
- 8.Deng G, Sui G. Noncoding RNA in oncogenesis: a new era of identifying key players. Int J Mol Sci. 2013;14(9):18319–18349. doi: 10.3390/ijms140918319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W-T, Han C, Sun Y-M, Chen T-Q, Chen Y-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J Hematol Oncol. 2019;12(1):55. doi: 10.1186/s13045-019-0748-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long B, Zhan H, Du H, Wang Z, Jiao Z. The association between long non-coding RNA overexpression and poor prognostic of gastric cancer: a meta-analysis. Chin J Evid-Based Med. 2018;18(3):286–293. [Google Scholar]
- 11.Liu H, Wu Y. Long non-coding RNA gastric carcinoma highly expressed transcript 1 promotes cell proliferation and invasion in human head and neck cancer. Oncol Lett. 2018;15(5):6941–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amodio N, Raimondi L, Juli G, et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou K, Zhang C, Yao H, et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17(1):105. doi: 10.1186/s12943-018-0849-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin MA, Chakravarty D, Sboner A, Nair SS, Elemento O The Estrogen receptor alpha regulated NEAT1 long non-coding RNA promotes prostate cancer progression. Paper presented at: Epigenetics; December 3–6, 2013, Shoal Bay, NSW, Australia; 2013. [Google Scholar]
- 15.Ma Y, Liu L, Yan F, Wei W, Deng J, Sun J. Enhanced expression of long non-coding RNA NEAT1 is associated with the progression of gastric adenocarcinomas. World J Surg Oncol. 2016;14(1). doi: 10.1186/s12957-016-0799-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang TL, Park JT. RSF1 (remodeling and spacing factor 1). Atlas Genet Cytogenet Oncol Haematol. 2011;6. doi: 10.4267/2042/44492. [DOI] [Google Scholar]
- 17.Helfricht A, Wiegant W, Thijssen P, Vertegaal A, Luijsterburg M, Van Attikum H. Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell Cycle. 2013;12(18):3070–3082. doi: 10.4161/cc.26033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Xue D, Hao F, et al. Remodeling and spacing factor 1 overexpression is associated with poor prognosis in renal cell carcinoma. Oncol Lett. 2018;15(3):3852–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hflmayer D, Hamuda M, Schroeder C, Hube-Magg C, Büscheck F. High RSF1 protein expression is an independent prognostic feature in prostate cancer. Acta Oncol (Madr). 2019;59(2):1–6. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Wang H, Li Q, et al. CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int. 2018;18(1):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Wang X, Wang A, Li Q, Zhou M, Li T. CLDN10 promotes a malignant phenotype of osteosarcoma cells via JAK1/Stat1 signaling. J Cell Commun Signal. 2019;13(3):395–405. doi: 10.1007/s12079-019-00509-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alessandra R, Federico N, Giorgia P, Di BM, De BF, Filippo P. Genomic markers of resistance to targeted treatments in gastric cancer: potential new treatment strategies. Pharmacogenomics. 2018;2018–2077. [DOI] [PubMed] [Google Scholar]
- 23.Qin QH, Yin ZQ, Li Y, Wang BG, Zhang MF. Long intergenic noncoding RNA 01296 aggravates gastric cancer cells progress through miR-122/MMP-9. Retour Au Numéro. 2018;97:450–457. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205–217. doi: 10.3727/096504016X14549667334007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niknafs YS, Han S, Ma T, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7:12791. doi: 10.1038/ncomms12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;20(16):3373–3377. [PubMed] [Google Scholar]
- 27.Zhang M, Wu WB, Wang ZW, et al. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur Rev Med Pharmacol Sci. 2017;21(5):1020–1026. [PubMed] [Google Scholar]
- 28.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Rubin MA. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z, Zhou Z, Guo H, et al. Long noncoding RNA gastric cancer-related lncRNA1 mediates gastric malignancy through miRNA-885-3p and cyclin-dependent kinase 4. Cell Death Dis. 2018;9(6):607. doi: 10.1038/s41419-018-0643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, Fu L, Li Q. Rsf-1 regulates malignant melanoma cell viability and chemoresistance via N-κB/Bcl-2 signaling. Mol Med Rep. 2019;20(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang YI, Ahn JH, Lee KT, et al. RSF1 is a positive regulator of NF-κB-induced gene expression required for ovarian cancer chemoresistance. Cancer Res. 2014;74(8):2258–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]