All multicellular organisms are associated with a diverse and specific community of microorganisms; consequently, the microbiome is of fundamental importance for health and fitness of the multicellular host. However, studies on microbiome contribution to host fitness are in their infancy, in particular, for less well-established hosts such as the moon jellyfish Aurelia aurita. Here, we studied the impact of the native microbiome on the asexual reproduction and on further fitness traits (health, growth, and feeding) of the basal metazoan due to induced changes in its microbiome. We observed significant impact on all fitness traits analyzed, in particular, in the absence of the protective microbial shield and when challenged with marine potentially pathogenic bacterial isolates. Notable is the identified crucial importance of the native microbiome for the generation of offspring, consequently affecting life cycle decisions. Thus, we conclude that the microbiome is essential for the maintenance of a healthy metaorganism.
KEYWORDS: Aurelia aurita, host, host fitness, microbiome, microbiota, reproduction
ABSTRACT
All multicellular organisms are associated with microbial communities, ultimately forming a metaorganism. Several studies conducted on well-established model organisms point to immunological, metabolic, and behavioral benefits of the associated microbiota for the host. Consequently, a microbiome can influence the physiology of a host; moreover, microbial community shifts can affect host health and fitness. The present study aimed to evaluate the significance and functional role of the native microbiota for life cycle transitions and fitness of the cnidarian moon jellyfish Aurelia aurita. A comprehensive host fitness experiment was conducted studying the polyp life stage and integrating 12 combinations of treatments with microbiota modification (sterile conditions, foreign food bacteria, and potential pathogens). Asexual reproduction, e.g., generation of daughter polyps, and the formation and release of ephyrae were highly affected in the absence of the native microbiota, ultimately resulting in a halt of strobilation and ephyra release. Assessment of further fitness traits showed that health, growth, and feeding rate were decreased in the absence and upon community changes of the native microbiota, e.g., when challenged with selected bacteria. Moreover, changes in microbial community patterns were detected by 16S rRNA amplicon sequencing during the course of the experiment. This demonstrated that six operational taxonomic units (OTUs) significantly correlated and explained up to 97% of fitness data variability, strongly supporting the association of impaired fitness with the absence/presence of specific bacteria. Conclusively, our study provides new insights into the importance and function of the microbiome for asexual reproduction, health, and fitness of the basal metazoan A. aurita.
INTRODUCTION
In recent years, a new conceptual framework—the metaorganism concept—was established, which is considering a metaorganism as the collective interactions among a multicellular host and its associated microbial species (1). The metaorganism concept focuses on the function and contribution (beneficial or detrimental) of the host-associated microbiota in a given environment, which depend on the identity, abundance, and activity of the microbes (2–4). The function of a specific microbe or microbial consortia is dynamic and depends on the developmental stage of the host, its age, reproductive state, or physiological condition (4–6). Several studies have recently found specific host-associated microbiota to contribute to host metabolism (7, 8), development (9), organ morphogenesis (10), pathogen protection and immunity (11, 12), behavior (13), environmental sensing and adaptation (5, 14–19), developmental transitions (20–23), and reproduction (24, 25). Conversely, in the absence of a proper microbial community, the functioning of the metaorganism may be compromised, which can result in various diseases; in humans, for instance, anxiety, depression, diabetes, cancer, obesity, and chronic inflammations are linked to microbiome imbalances (6, 26, 27).
Complex relationships and dependencies within a metaorganism are best investigated in controlled experiments that manipulate host microbiota composition (4), thereby enabling insights into the functional contributions of microbes to metaorganism function. The simplest and most drastic experimental setup is to compare hosts that are largely or completely lacking microbes (gnotobiotic or germfree, respectively) with those harboring their native microbiota (11, 28–31). Such studies on the well-documented Euprymna scolopes-Vibrio symbiosis demonstrated that only in the presence of their native microbiota (comprising the symbiont Vibrio fischeri), juvenile squid underwent a tissue remodeling of the external epithelial cell layer of the light organ (32, 33). In contrast, in squid raised in the absence of V. fischeri, tissue regression did not occur (34). Moreover, recolonization experiments using sterile animals in combination with available microbial isolates further enable assigning a certain function to specific microbes (11, 30, 35–37), determining colonization dynamics of microbes (30, 38, 39), and considering bacteria-bacteria interactions (11, 31, 40, 41). Until now, most studies on host-microbiota interactions were performed with well-established model organisms, such as the model plant Arabidopsis thaliana, the fruit fly Drosophila melanogaster, the nematode Caenorhabditis elegans, or mice (42, 43). However, their complexity and/or long generation times often limit studies on host-microbiota interactions (44). Novel model systems might lack some of the features making model organisms easy to investigate, such as low-cost and easy husbandry, short life cycles, high fertility, and genetic manipulability; however, they might enable to gain new insights into host-microbe interactions, as demonstrated by recent studies comparing the microbiota of a wide range of different metaorganisms (45, 46).
Here, we develop a member of the phylum Cnidaria as a new model system for metaorganism research among the basal metazoans. Our chosen model is the moon jellyfish Aurelia aurita, representing one of the most widely distributed Scyphozoa (47, 48). This species plays an influential role in the marine ecosystem, since they affect the structure of the planktonic food web and cause jellyfish blooms around the world due to their enormous stress tolerance, regeneration potential, and high reproduction output (49–53). Furthermore, A. aurita has a simple body plan with only two tissue layers, where host-microbe interactions can take place—the ectoderm and endoderm, separated by a jelly-like layer called the mesoglea. A. aurita possesses a complex life cycle. After sexual reproduction, A. aurita releases planula larvae, which settle on a suitable substratum and develop to the sessile, benthic polyp stage. A. aurita shows two forms of asexual reproduction—budding and strobilation. During budding, at one specific polyp site, a bud develops as an outgrowth due to repeated cell division to clonally form new individuals, which detach from the parent body when fully mature (54, 55). During strobilation, induced by environmental triggers, a transverse segmentation of the polyp body takes place. This mode of reproduction produces many offspring at a high rate that are key to understanding frequent jellyfish blooms (56). The process starts with preliminary morphological changes; in particular, the tentacles are reabsorbed. First transverse constrictions appear near the upper stalk; furthermore, the number of constriction sites increases and migrates down the body length, transforming the body into a sequence of segments. Those segments are released as ephyrae, the preform of juvenile medusa (52). The polyp foot remains adhered to the substrate and regenerates to a new polyp (52). In the present study, we were particularly interested in the role and impact of microbiota for regulating A. aurita complicated reproductive cycle involving very different life stages—mobile medusa, as a member of the marine plankton, and sessile polyp, as a member of the benthos (52, 57). Previously, we demonstrated specific bacterial community patterns for A. aurita life stages, which undergo significant restructuring during the polyp-jellyfish transition, strongly arguing for an important functional role of the associated microbiota, particularly in the context of the life cycle (29). A crucial microbial impact on animal development has already been demonstrated for the bacterial induction of settlement and metamorphosis of many marine invertebrate larvae (58), such as sponges (59), cnidarians (60), ascidians (61), and bryozoans (62). The settlement of planula larvae of the scyphozoans A. aurita (60) and Cassiopea andromeda (63) or the hydrozoan Hydractinia echinata (20) is prevented in the absence of microbes; in contrast, the presence of bacteria induces the settlement and development of each of those animals (64). A comprehensive host fitness experiment was designed and conducted with the benthic polyp life stage of A. aurita to evaluate the importance and function of the microbiome on fitness traits such as asexual reproduction and survival. The bacterial communities of polyps were manipulated in 12 different treatments that included polyps with native microbiota or sterile ones kept in artificial seawater (sterile or with native microbiota), feeding with Artemia salina (unmanipulated or sterile), and experimental infection with potentially pathogenic bacteria correlating with the concomitant changes in microbial community composition. Our overall aim was to assess the importance of the native microbiome for health and, particularly, for the generation of offspring, ultimately proposing that the microbiome is essential for the maintenance of a healthy metaorganism.
RESULTS
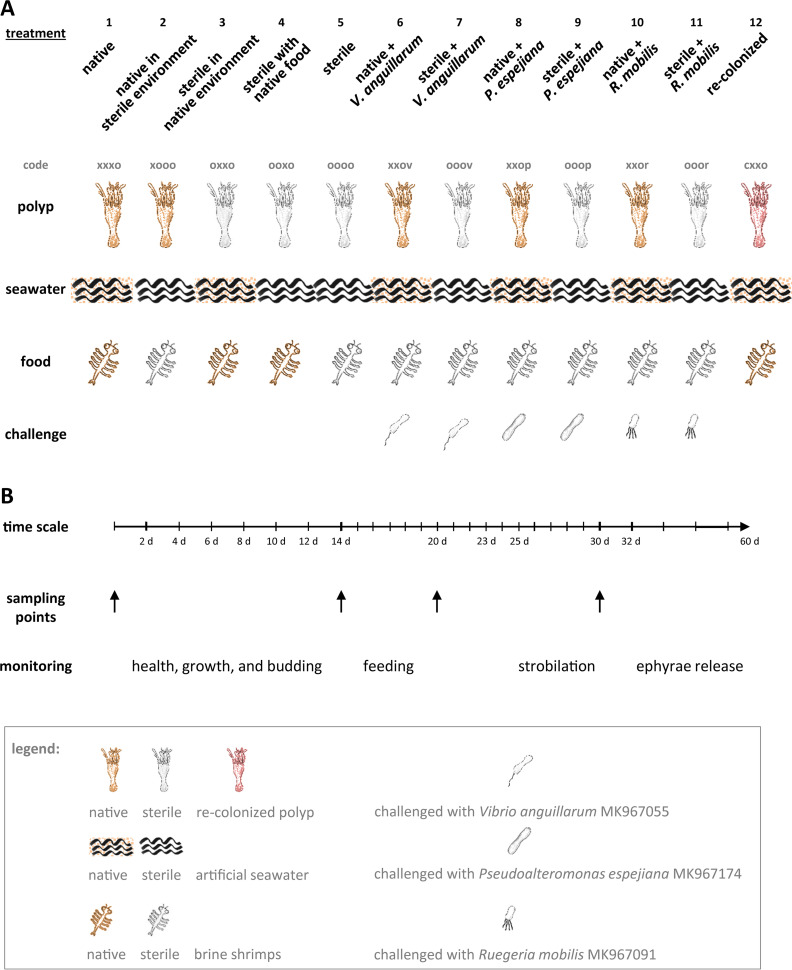
A comprehensive host fitness experiment with A. aurita polyps with high numbers of replicates (96) for each treatment was conducted in 48-well plates, where polyps were kept in 1 ml artificial seawater, to evaluate the importance and function of the microbiome on host fitness traits, i.e., asexual reproduction, survival, growth, and feeding (the experimental design and abbreviations are summarized in Fig. 1). To examine the general effects of microbiota, native polyps that harbor a native diverse microbial community (polyps were kept under artificial lab conditions) were compared with sterile polyps generated by using an antibiotic mixture and recolonized polyps. As major treatment factors, native as well as sterile polyps were kept in artificial seawater (sterile or with native microbiota), fed with Artemia salina (sterile or unmanipulated), and challenged with potentially pathogenic bacteria previously isolated from A. aurita polyps and ambient seawater (Vibrio anguillarum, Pseudoalteromonas espejiana, and Ruegeria mobilis) to evaluate the impact of microbiota on the resistance of the metaorganism against bacterial infections (Fig. 1).
FIG 1.
Study design for host fitness experiment. (A) Each treatment comprises a certain combination of polyp, seawater, and food regarding their microbial composition. Selected potential pathogenic bacteria were used for the microbial challenge. A code is introduced for each treatment describing the presence “x” or absence “o” of microbes in the order polyp, ambient water, and food; microbial challenge is labeled with “v” for Vibrio anguillarum, “p” for Pseudoalteromonas espejiana, and “r” for Ruegeria mobilis; recolonized polyps are labeled with” c.” (B) An experimental schedule comprising the time scale of the experiment with monitoring events and sampling points for subsequent 16S rRNA amplicon sequencing is illustrated.
Influence of the microbiota on asexual reproduction of A. aurita.
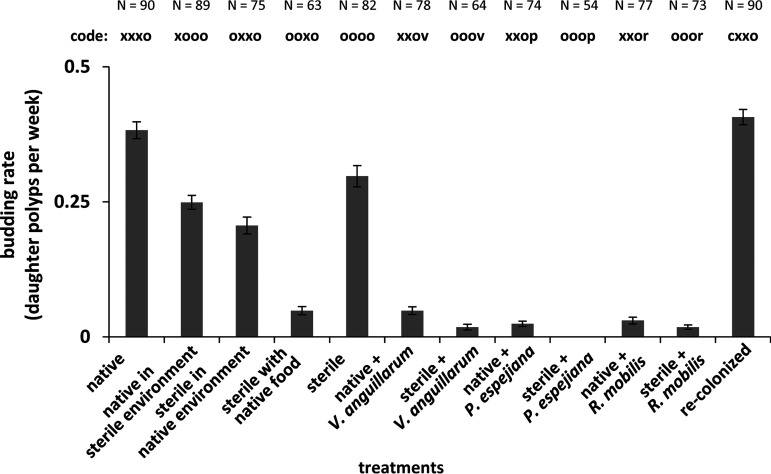
The generation of daughter polyps was monitored every 48 h over 14 days. Native as well as recolonized polyps showed a similar budding rate of 0.4 daughter polyps per week (0.38 ± 0.02 and 0.41 ± 0.01, respectively), whereas sterile polyps generated 0.3 ± 0.02 daughter polyps/week, representing a statistically significant decrease of 21% (P value of 0.0003) (Fig. 2; see also Fig. S1 and Table S1C in the supplemental material). Environmentally challenged polyps (treatments 3 and 4) had further decreases in budding rates, with decreases for sterile polyps in native environment by 45% and sterile polyps incubated with unmanipulated food by 87% (Fig. 2 and S1; Table S1D) (treatment 3: t = 4.0, P value < 0.0002; treatment 4: t = 6.0, P value < 0.0001). Budding rates also decreased remarkably when polyps were infected with potential pathogens, resulting in almost completely impaired generation of daughter polyps, in particular, when challenged with P. espejiana (Fig. 2 and S1; Table S1E) (F = 71.3, P value for permutational multivariate analysis of variance [PPERMANOVA] < 0.0001).
FIG 2.
Asexual reproduction of A. aurita polyps as the daughter polyp generation. Budding was followed every 48 h for 14 days by monitoring the generation of daughter polyps of healthy and harmed polyps. Budding rate was calculated as daughter polyp generation per week.
Daughter polyp generation of A. aurita polyps. Budding was followed every 48 h for 14 days by monitoring the generation of daughter polyps of healthy and harmed polyps. Numbers of generated daughter polyps are shown as percentages after 14 days. Download FIG S1, DOCX file, 0.1 MB (104.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PERMANOVA tests. (A) Overview of PERMANOVA study design. (B) All variables were tested under three test designs, A, B, and C. (C) PERMANOVA test A with full data set (4-factorial, partly nested). (D) PERMANOVA test B with polyp-seawater-food data set (3-factorial, partly nested). (E) PERMANOVA test C with bacterial challenge data set (2-factorial, fully crossed). ns, not significant. Download Table S1, DOCX file, 0.1 MB (56.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
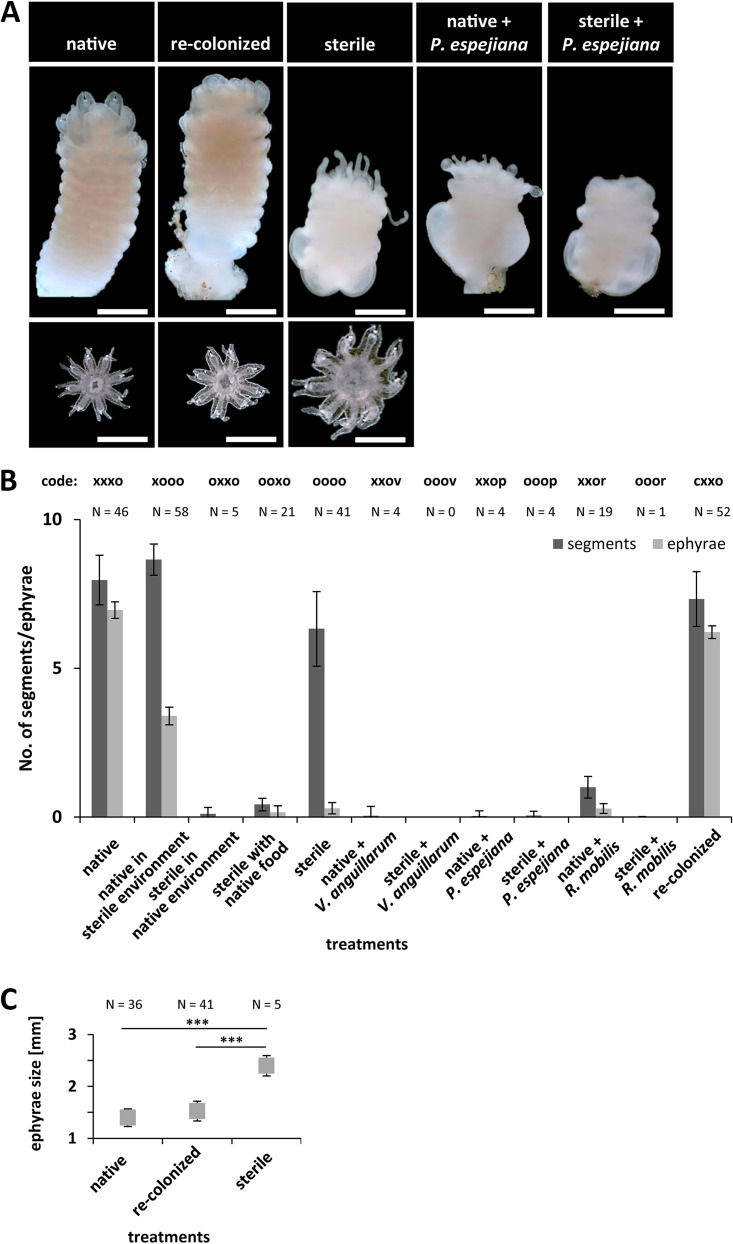
In a parallel experimental setup, strobilation of polyps was induced by adding the synthetic strobilation inducer 5 μM 5-methoxy-2-methyl indole (Fig. 1) (58). Strobilation induction was monitored each day, and strobila phenotypes as well as number of segments were monitored and detected beginning on day 5, when native polyps initiated segmentation (early strobila) (Fig. 3A; Table S2). Native polyps showed full segmentation and shaping as late strobila on day 9 (Fig. 3A; Table S2). Specifically, 51% of native and 58% of recolonized polyps showed initiation of segmentation on day 5 after induction (Fig. 3B; Table S2). Even 64% of native polyps in the sterile environment formed strobilae, which showed typical segmentation (Fig. 3B). However, the formation of strobilae was delayed, with early strobilae appearing after 8 days and late strobilae after 17 days. In contrast, 45% of sterile polyps showed segmentation (Fig. 3B; Table S1C) (t = 10.7, P value < 0.0001) (Table S2), but remarkably, phenotypically abnormal strobilae were formed (Fig. 4A). Phenotypical abnormalities were manifested by colorless, minimized, and thickened strobilae without absorbed tentacles (see examples in Fig. 4A). Polyps, which were environmentally challenged (treatments 3 and 4) showed massively reduced formation of strobilae, and the few ones observed were phenotypically abnormal (colorless, minimized, and thickened strobilae with inflated calyx) (Fig. 4). Likewise, infected polyps (treatments 6 to 11) showed almost no formation of strobilae (Fig. 3B; Table S1E) (F = 121.7, PPERMANOVA < 0.0001), resulting in almost completely halted segmentation (Fig. 4A). Furthermore, subsequent ephyra release was detected for all treatments, beginning on day 12 after induction of strobilation, when native strobilae began to release ephyrae (Fig. 3A and 4A; Table S2). Native and recolonized strobilae formed approximately eight segments and finally released seven ephyrae with a size of 1.5 ± 0.1 mm in diameter after 12 to 14 days after induction (Fig. 4B; Table S2). Native polyps in a sterile environment formed nine segments but deferred release of only three ephyrae (Fig. 4B; Table S2). Sterile polyps were only slightly affected in induced segmentation (45% strobilae with a mean of six segments); however, an abnormal strobila phenotype was continuously detected, resulting in massively impaired ephyra release (Fig. 4; Table S1C) (F = 171.9, PPERMANOVA < 0.0001). Moreover, only 15% of the formed sterile strobilae were able to release only a single ephyra of increased size (2.4 ± 0.2 mm, diameter increase of 70%) (Fig. 4C). Ultimately, infected polyps showed almost no segmentation; consequently, no ephyrae were released (Fig. 4; Table S1C) (F = 10.2, PPERMANOVA < 0.0001). These findings strongly argue that strobilation and consequently ephyra release are massively interrupted in the absence of the native polyp microbiota as well as when infected with potential pathogens, ultimately resulting in halted generation of offspring.
FIG 3.
Asexual reproduction of A. aurita polyps as the generation of strobilae. Strobilation of polyps was induced using 5 μM 5-methoxy-2-methyl indole. (A) Original photographs show progress of segmentation for polyps under native conditions. Scale bars correspond to 1 mm. (B) Segmentation of polyps was monitored each day, and the numbers of generated strobilae were determined.
FIG 4.
Segmentation of polyps and ephyra release. Strobilation of polyps was induced using 5 μM 5-methoxy-2-methyl indole. (A) Original photographs show phenotypical appearance and segmentation of strobilae under selected representative treatments. Photographs of released ephyrae are shown at the bottom at 15 days postinduction. Scale bars correspond to 1 mm. (B) Numbers of segments per strobila and numbers of released ephyrae were monitored each day for a maximum of 4 weeks. (C) Diameters of released ephyrae under native, sterile, and recolonized conditions were quantified. ***, P < 0.001.
Summary of mean fitness status of A. aurita polyps and strobilae for all treatments. Heat map summarizes the significant decreases measured within the treatment groups compared to that in the native treatment. Based on individual results after host fitness experiments, monitored fitness parameter survival, growth, budding, feeding, strobilation, and ephyra release are categorized as unaffected (orange), slightly affected (purple), and crucially affected (red) for each treatment. Additionally, timelines for generation of strobilae and ephyra release are depicted as days of first appearance after strobilation induction with the synthetic inducer. na, not applicable. Download Table S2, DOCX file, 0.1 MB (25.7KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial impact on the fitness traits survival, growth, and feeding.
In parallel with asexual reproduction (budding), further important fitness-correlated traits, in particular, survival and growth of A. aurita polyps, were monitored every 48 h over 14 days. Based on the overall phenotypical appearance of polyps and their tentacles, three health conditions were differentiated, namely, “healthy” (normal polyp phenotype, contracting tentacles), “harmed” (polyp phenotype impaired, tentacles degenerated), and “dead” (degenerated or no polyp) (see “Host fitness experiment” for detailed definition on phenotype characteristics), which are exemplified in Fig. 5A. Recolonized polyps showed similar survival rates as native polyps without an impaired phenotype (99%, corresponding to 89 ± 1 polyps, and 97%, corresponding to 87 ± 3 polyps, respectively), whereas 37% of sterile polyps were affected in health after 14 days (tend) (Fig. 5B; Table S1C) (t = 3.8, P value < 0.0001). Native polyps were able to maintain their associated native microbiota (see below and Fig. 5) when kept in a sterile environment at least over the short period during the experiment (code xooo, only 5% polyps were affected) (Fig. 5B). However, sterile polyps were affected in survival or at least showed malformation when kept under native environmental conditions (39% affected) (Table S1D) (t = 2.8, P value < 0.0008) and were affected even more when fed with A. salina harboring their native microbiota (78% affected) (Fig. 5B; Table S1D) (t = 5.6, P value < 0.0001). Environmentally challenged polyps (treatments 3 and 4) (Fig. 1) showed significantly increased malformation and mortality already at the onset of the experiment, which worsened over time. Remarkable health loss, with up to 76% impaired polyps, was also detected when challenging with potentially pathogenic bacteria which were previously isolated from A. aurita and ambient water (65) (Fig. 5B; Table S1C) (P value < 0.0001). The survival rate of polyps was significantly affected by each bacterial challenge (Table S1E) (F = 15.9, PPERMANOVA < 0.0001), with the exception of R. mobilis infection (Table S1E) (pairwise tests, P value was not significant). In more detail, we observed that unmanipulated native polyps were affected by infection with potentially pathogenic bacterial isolates during the course of the experiment, whereas sterile polyps were affected early on. Furthermore, R. mobilis and V. anguillarum (belonging to A. aurita native microbiota) affected the health/survival of native polyps only slightly (28%, of which 16% were dead, and 29%, of which 14% were dead, respectively) (Table S1E) (P value < 0.0001) compared to the effect of P. espejiana (56%, of which 19% were dead) (Table S1E) (P value < 0.0001) (Fig. 5B). A significant interaction of the type of ambient water (unmanipulated or sterile) as well as Artemia supplied as food (unmanipulated or sterile) was further identified (Table S1E) (F = 7.6, PPERMANOVA < 0.0005 and F = 34.5, PPERMANOVA < 0.0001, respectively), indicating that specific bacteria differed in their challenge depending on both water and food quality.
FIG 5.
Impact of microbes on health of A. aurita polyps. (A) Original photographs show examples of healthy, harmed, and dead polyps after 14 days for representative selected treatments. Scale bars correspond to 0.5 mm. (B) Percentages of healthy, harmed, and dead polyps monitored based on phenotypical appearance of polyps and presence of tentacles are shown for all treatments monitored after 48 h (t1) and 14 days (tend) comprising 90 biological replicates each.
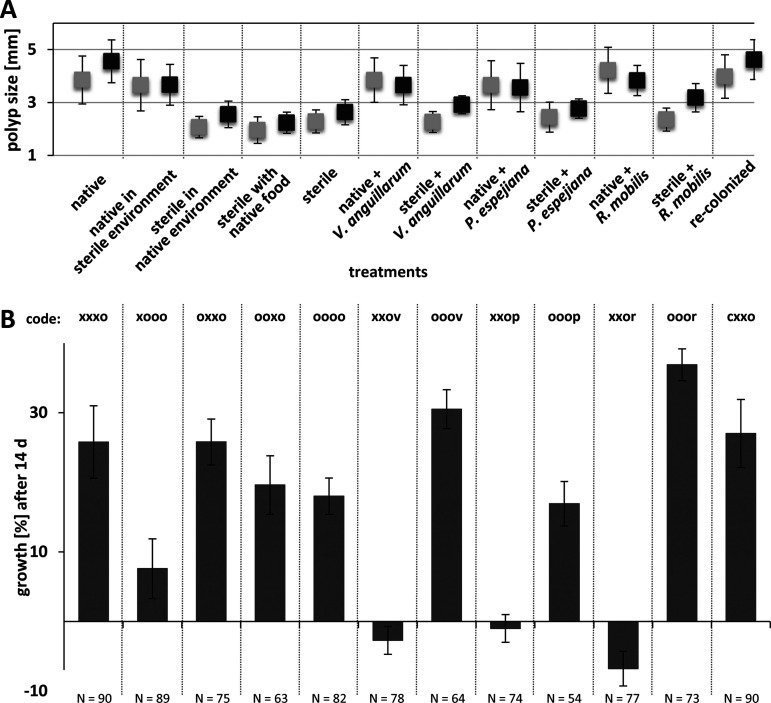
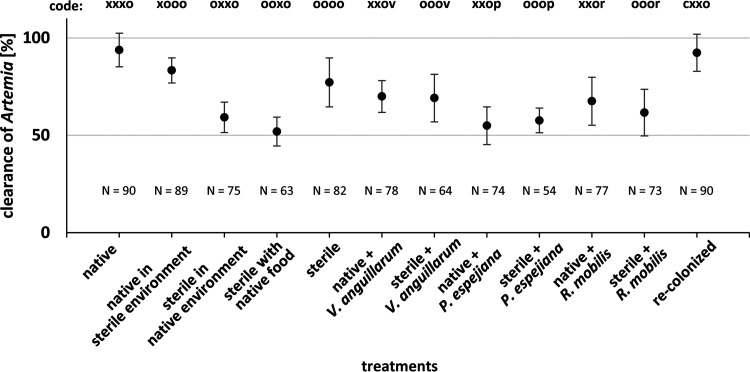
Healthy and harmed polyps were also analyzed with respect to growth and feeding rates. Native polyps had a mean polyp size (length plus width) of 3.85 ± 0.91 mm, whereas sterile polyps showed reduced start sizes of 2.3 ± 0.43 mm (Fig. 6). Native and recolonized polyps showed similar initial sizes and further similar growth rates (Fig. 6). Analyses indicated that growth was primarily affected in sterile artificial seawater and when infected with Vibrio anguillarum and Ruegeria mobilis (Fig. 6; Table S1). Feeding rates of healthy and harmed polyps were assessed in week three of the experiment on the five following days (Fig. 7 and S2). Native polyps had a mean clearance rate of 93.8% ± 8.6%, thus, similar to recolonized polyps (92.4% ± 9.5%), whereas sterile polyps showed a 77.2% ± 12.5% clearance rate (Fig. 7 and S2; Table S1C) (t = 12.5, P value <0.0001). Feeding rates were significantly reduced when treatments affected the overall health of polyps (Fig. S2). Analyses further disclosed a significant effect on the presence of the associated polyp microbiota as well as an effect of bacterial infection on feeding rates (Fig. 7 and S2). Native polyps were further affected in effective feeding when kept in sterile artificial seawater (Table S1) (P values < 0.0001).
FIG 6.
Growth rate of A. aurita polyps. Growth of polyps was followed every 48 h for 14 days by measuring the polyp size (length plus width) of healthy and harmed polyps. (A) Mean polyp start sizes (▪) and end sizes after 14 days (▪) per treatment. (B) Growth rates were calculated as percentages after 14 days.
FIG 7.
Clearance of Artemia salina by A. aurita polyps. Feeding rate as clearance of A. salina in percentages. Single polyps were incubated for 1 h with 20 A. salina. After incubation, remaining A. salina were counted to calculate the feeding rate of healthy and harmed polyps for all treatments. Feeding rates are displayed as means of all monitoring days (Σ5).
Clearance of Artemia salina by A. aurita polyps. Feeding rate as clearance of A. salina in percentages. Single polyps were incubated for 1 h with 20 A. salina. After incubation, remaining A. salina were counted to calculate the feeding rate of healthy as well as harmed polyps for all treatments (corresponding codes in parentheses). Compared are the results from day 1 (I1) and day 5 (tend). Download FIG S2, DOCX file, 0.2 MB (242.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial community shifts correlate with impaired fitness traits.
To correlate the impaired asexual reproduction, survival rates, growth, and feeding with changes in the microbial community of the native A. aurita microbiota, 16S V1-V2 amplicon sequencing was performed during the course of the experiment (see Table S3). Antibiotic-treated polyps and strobilae, developed from sterile polyps, were indeed shown to be sterile (Fig. 8 and S3, S4, and S5). In line with our previous report (29), the present analysis further showed restructuring of the microbial community during polyp-jellyfish transition (Fig. S3, S4, and S5). Native and recolonized polyps showed similar bacterial community patterns due to successful reconstitution (Fig. 8A and S4; Table S3). However, relative abundances of community members were different, resulting also in identification of specific indicator operational taxonomic units (iOTUs) explaining the microbial communities (Fig. S4; Table 1). Moreover, changes in the native microbial community composition induced by the experimental treatments were identified (Fig. 8 and S3). Notable are the higher abundances of unclassified (uncl.) Gammaproteobacteria, uncl. Cyanobacteria, and Melitea as well as the lower abundance of Arcobacter in the native microbiota of the animals (Fig. S3). Redundancy analysis revealed four distinct microbial community clusters resulting from the respective manipulation (native, environmental challenged, native plus challenged, and sterile plus challenged) for polyps as well as strobilae (Fig. 8). Indicator OTU analyses further revealed bacterial taxa, which significantly correlated either positively or negatively with fitness traits (Tables 1 and 2). For instance, the presence of OTU0142 (uncl. Prevotella) was linked to essentially impaired fitness traits, whereas OTU0015 (uncl. Neptuniibacter) and OTU0003 (uncl. Alteromonas) might be beneficial bacteria (Tables S4 and S5). Moreover, OTU0057, OTU0178, and OTU0336 (Olleya, Rothia, and Gemella, respectively) were exclusively beneficial for strobilation or, in contrast, the presence of OTU0110 and OTU0144 (Corynebacterium and Balneola, respectively) resulted in halted transverse constriction (Table S5). Finally, the multivariate ordination analysis visually summarizes the observed fitness effects (Fig. 9), which mirror the identified microbial community clusters from redundancy analysis (RDA) (Fig. 8), ultimately correlating the observed fitness effects with microbial community shifts.
FIG 8.
Microbial composition of A. aurita polyps and strobilae under selected microbial challenges. Composition of microbiota associated with A. aurita polyps after 14 days of microbial challenge (12 different treatments) during the host fitness experiment. Microbial communities were analyzed by sequencing the amplified V1-V2 region of 16S bacterial rRNA genes. Redundancy analysis plots of Hellinger-transformed OTU abundances. Each point represents the whole microbial community of the respective sample; replicates of one sample group are framed by polygons. Microbial community patterns of polyps (A) and strobilae (B) are shown.
TABLE 1.
Indicator operational taxonomic units
| Indicator OTUa | Taxonomic classificationb | P value |
|---|---|---|
| Polyps | ||
| Native | ||
| OTU0016 | Melitea salexigens | 0.001 |
| OTU0083 | Uncl. Glaciecola | 0.041 |
| OTU0120 | Melitea salexigens | 0.005 |
| OTU0220 | Uncl. Sinobacteraceae | 0.007 |
| Recolonized | ||
| OTU0015 | Neptuniibacter caesariensis | 0.003 |
| OTU0121 | Uncl. Oceanospirillaceae | 0.001 |
| OTU0130 | Uncl. Bacteriovorax | 0.001 |
| OTU0131 | Uncl. Flavobacteriales | 0.001 |
| OTU0134 | Uncl. Pseudoalteromonas | 0.001 |
| OTU0179 | Uncl. Ruegeria | 0.001 |
| Native microbiota | ||
| OTU0009 | Uncl. Cyanobacteria | 0.004 |
| OTU0073 | Uncl. Gammaproteobacteria | 0.001 |
| OTU0126 | Uncl. Gammaproteobacteria | 0.001 |
| Native in sterile environment | ||
| OTU0078 | Uncl. Vibrio | 0.009 |
| OTU0267 | Uncl. Gammaproteobacteria | 0.020 |
| Sterile in native environment | ||
| OTU0054 | Lewinella cohaerens | 0.006 |
| OTU0082 | Uncl. Flavobacteriaceae | 0.001 |
| OTU0128 | Uncl. Polaribacter | 0.003 |
| OTU0141 | Uncl. Balneola | 0.005 |
| OTU0218 | Uncl. Fluviicola | 0.001 |
| OTU0241 | Uncl. Polaribacter | 0.004 |
| Sterile with native food | ||
| OTU0048 | Uncl. Plesiocystis | 0.008 |
| OTU0065 | Uncl. Arcobacter | 0.002 |
| OTU0097 | Uncl. Saprospiraceae | 0.001 |
| OTU0099 | Uncl. Oleibacter | 0.005 |
| OTU0116 | Uncl. Oleibacter | 0.005 |
| OTU0138 | Uncl. Gammaproteobacteria | 0.007 |
| OTU0145 | Uncl. Lactococcus | 0.005 |
| OTU0163 | Microbacterium aurum | 0.011 |
| Sterile | ||
| Not detected | ||
| Native plus V. anguillarum | ||
| OTU0107 | Uncl. Chromobacterium | 0.001 |
| OTU0155 | Uncl. Flavobacteriaceae | 0.009 |
| OTU0258 | Uncl. Proteobacteria | 0.040 |
| OTU0355 | Uncl. Proteobacteria | 0.016 |
| OTU0203 | Uncl. Vibrio | 0.001 |
| Sterile plus V. anguillarum | ||
| OTU0203 | Uncl. Vibrio | 0.003 |
| Native plus P. espejiana | ||
| OTU0021 | Uncl. Bizionia | 0.002 |
| OTU0052 | Uncl. Proteobacteria | 0.001 |
| OTU0166 | Micrococcus luteus | 0.001 |
| OTU0175 | Rothia mucilaginosa | 0.003 |
| OTU0200 | Uncl. Bacteria | 0.001 |
| OTU0098 | Uncl. Pseudoalteromonas | 0.004 |
| Sterile plus P. espejiana | ||
| OTU0098 | Uncl. Pseudoalteromonas | 0.004 |
| Native plus R. mobilis | ||
| OTU0077 | Uncl. Nannocystis | 0.005 |
| OTU0093 | Uncl. Cytophagales | 0.003 |
| OTU0142 | Uncl. Prevotella | 0.001 |
| OTU0169 | Uncl. Mogibacteriaceae | 0.015 |
| OTU0179 | Uncl. Ruegeria | 0.002 |
| Sterile plus R. mobilis | ||
| OTU0179 | Uncl. Ruegeria | 0.002 |
| Strobilae | ||
| Native | ||
| OTU0029 | Uncl. Shewanella | 0.007 |
| OTU0142 | Uncl. Prevotella | 0.006 |
| OTU0169 | Uncl. Mogibacteriaceae | 0.002 |
| Recolonized | ||
| OTU0108 | Uncl. Gammaproteobacteria | 0.019 |
| OTU0167 | Uncl. Gammaproteobacteria | 0.002 |
| OTU0192 | Uncl. HTTC | 0.005 |
| Native in sterile environment | ||
| OTU0162 | Uncl. Staphylococcus | 0.034 |
| OTU0260 | Uncl. Neptuniibacter | 0.005 |
| OTU0268 | Uncl. Lewinella | 0.001 |
| Sterile in native environment | ||
| OTU0051 | Uncl. Proteobacteria | 0.033 |
| OTU0052 | Uncl. Proteobacteria | 0.032 |
| Sterile with native food | ||
| OTU0094 | Uncl. Alteromonas | 0.016 |
| OTU0099 | Uncl. Oleibacter | 0.026 |
| OTU0343 | Uncl. Oceanospirillaceae | 0.003 |
| Sterile | ||
| Not detected | ||
| Native plus V. anguillarum | ||
| OTU0138 | Uncl. Gammaproteobacteria | 0.043 |
| OTU0163 | Uncl. Microbacterium | 0.018 |
| OTU0203 | Uncl. Vibrio | 0.048 |
| OTU0252 | Uncl. Alteromonas | 0.005 |
| OTU0324 | Uncl. Arcobacter | 0.009 |
| Sterile plus V. anguillarum | ||
| OTU0203 | Uncl. Vibrio | 0.004 |
| Native plus P. espejiana | ||
| OTU0098 | Uncl. Pseudoalteromonas | 0.003 |
| OTU0173 | Uncl. Haliangiaceae (Kofleriaceae) | 0.017 |
| Sterile plus P. espejiana | ||
| OTU0098 | Uncl. Pseudoalteromonas | 0.004 |
| Native plus R. mobilis | ||
| OTU0077 | Uncl. Nannocystis | 0.013 |
| OTU0082 | Uncl. Flavobacteriaceae | 0.002 |
| OTU0103 | Uncl. Streptococcus | 0.005 |
| OTU0175 | Uncl. Rothia | 0.009 |
| OTU0179 | Uncl. Ruegeria | 0.006 |
| Sterile plus R. mobilis | ||
| OTU0179 | Uncl. Ruegeria | 0.022 |
Indicator OTUs identified as key operational taxonomic units for the respective treatments.
Uncl., unclassified.
TABLE 2.
Correlation of operational taxonomic units with fitness dataa
| OTU | Taxonomic classification | Adj. R2 | SS(trace) | Pseudo-F | P value | Prop | Cumul | Res df |
|---|---|---|---|---|---|---|---|---|
| OTU0142 | Uncl. Prevotella | 0.55 | 41.51 | 13.11 | 0.00 | 0.59 | 0.59 | 9 |
| OTU0167 | Uncl. Gammaproteobacteria | 0.65 | 8.83 | 3.59 | 0.01 | 0.13 | 0.72 | 8 |
| OTU0015 | Neptuniibacter caesariensis | 0.76 | 7.99 | 4.79 | 0.01 | 0.11 | 0.83 | 7 |
| OTU0106 | Uncl. Corynebacterium | 0.84 | 5.00 | 4.49 | 0.01 | 0.07 | 0.90 | 6 |
| OTU0065 | Uncl. Arcobacter | 0.90 | 3.10 | 4.34 | 0.01 | 0.04 | 0.95 | 5 |
| OTU0267 | Uncl. Gammaproteobacteria | 0.93 | 1.70 | 3.64 | 0.03 | 0.02 | 0.97 | 4 |
| OTU0003 | Uncl. Alteromonas | 0.95 | 0.91 | 2.87 | 0.08 | 0.01 | 0.99 | 3 |
| OTU0110 | Uncl. Gammaproteobacteria | 0.97 | 0.58 | 3.04 | 0.12 | 0.01 | 0.99 | 2 |
| OTU0085 | SC3-56 | 0.99 | 0.29 | 3.23 | 0.25 | 0.00 | 1.00 | 1 |
| OTU0355 | Uncl. Proteobacteria | 0.99 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 | 1 |
Distance based linear model (DistLM) analysis and sequential tests show predictors (operational taxonomic units [OTUs]) explaining the host fitness data structure in the best model (adjusted R2 = 0.99, R2 = 1.00, RSS [residual sums of squares] = 0.09, number of fitted variables = 10 OTUs) with a significance level of 0.05, which are marked in bold. Uncl., unclassified; Adj. R2, adjusted R2; SS(trace), sum of squares; Pseudo-F, multivariate analogue to Fisher’s F test; Prop, proportion of variance explained; Res df, residual degrees.
FIG 9.
Multivariate ordination results (nonmetric multidimensional scaling [NMDS] based on Bray-Curtis) of average fitness traits.
Microbial community composition of A. aurita in the experimental treatment groups. Microbial communities were analyzed by sequencing the V1-V2 region of 16S bacterial rRNA genes. OTU abundances were summarized at the genus level and normalized by the total number of reads per sample. Bar plots are grouped according to sample type, each group including 2 to 6 replicates. Microbial community patterns of polyps (A) and strobilae (B) are shown. Download FIG S3, DOCX file, 0.6 MB (581.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial composition of recolonized polyps. Compositions of microbiota associated with native and recolonized A. aurita polyps as well as of the generated native microbiota for recolonization of sterile polyps. Microbial communities were analyzed by sequencing the V1-V2 region of 16S bacterial rRNA genes. (A) OTU abundances were summarized at the genus level and normalized by the total number of reads per sample. Bar plots are grouped according to sample type, each group including 6 replicates. (B) Redundancy analysis plots of Hellinger-transformed OTU abundances. Each point represents the whole microbial community of the respective sample, replicates of one sample group are framed by polygons. (C) Results of pairwise tests in beta diversity analysis. Tests were conducted for specified comparisons. df, degree of freedom; v, variance. Download FIG S4, DOCX file, 0.1 MB (150KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of pairwise tests in beta diversity analysis. Permutation test for RDA under reduced model was conducted with permutations (number of permutations, 9,999) for all comparisons, but only specified comparisons are shown. Model: RDA (formula = OTUhel ∼ group, data = metaDataSub); df = 0.321, v = 6.663, P value = 0.00001. Download Table S3, DOCX file, 0.1 MB (26.5KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Correlation of operational taxonomic units (OTUs) with the fitness status of A. aurita polyps. OTUs listed with their taxonomic classification are correlated with the fitness status of unaffected/healthy (orange), slightly affected/marginally harmed (purple), and crucially affected/essentially harmed (red) of A. aurita polyps. At least 50% of the biological replicates analyzed were associated with the respective OTU. Download Table S4, DOCX file, 0.1 MB (34.9KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Correlation of operational taxonomic units (OTUs) with the segmentation of A. aurita polyps and ephyra release. Unaffected (orange), slightly affected (purple), and crucially affected (red) formation of strobilae and further ephyra release are correlated with respective listed OTUs. At least 50% of the biological replicates analyzed were associated with the respective OTU. Download Table S5, DOCX file, 0.1 MB (39.7KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
The findings of the present study clearly demonstrate that the native microbiota is particularly crucial for asexual reproduction of A. aurita as well as for survival, growth, and feeding (Fig. 1 and 7; see also Table S2 in the supplemental material). While the absence of the microbiota clearly impairs A. aurita fitness traits, they can be restored when sterile polyps are recolonized with the native microbiota, strongly arguing for a significant bacterial impact on host fitness. Although we cannot completely exclude off-target impacts of antibiotics on cell division or metabolism, recolonized polyps showed a normal phenotype despite antibiotic treatment; further indications are obtained from transcriptomic data (unpublished data). Moreover, the associated native microbiota of native polyps appears to be stably maintained even in a sterile environment. The decrease of growth rates in the absence of the microbiota and when additionally infected with potentially pathogenic bacteria confirms our assumption of the importance of a balanced microbiota (Fig. 6). However, the finding that growth rates did not decrease or even increase when bacteria were present in the environment (unmanipulated environment and infection with bacteria, treatments 3, 7, 9, and 11) compared to rates for polyps in a sterile environment (treatment 2) indicates that bacteria from the environment most likely act as an additional food source for the polyps (Fig. 6) (29, 66, 67). Sterile polyps in a completely sterile environment already showed reduced fitness traits, but infection with foreign (treatment 4) or potentially pathogenic bacteria (treatments 6 to 11) caused a strong inhibition of asexual reproduction as well as an increase in mortality/malformation (Fig. 9; Table S2). The results of the present study demonstrate that the native microbiota is crucially required for asexual reproduction of the moon jellyfish by budding. Budding rates were correlated with the observed health status of polyps and thus diminished in absence and after manipulation of the native microbiota (Fig. 2). Likewise, bacteria have been found to have profound effects on budding of the fresh water polyp Hydra viridissima (68). Sterile animals were also unable to produce buds and multiply asexually, which is the exclusive type of asexual reproduction for Hydra, because they lack the process of strobilation (69). Generation of daughter polyps was rescued by inoculation with native bacterial associates of Hydra (68). Furthermore, our study revealed that the native microbiota is fundamental for correct transverse constriction of A. aurita during the process of strobilation, which ultimately results in the release of ephyrae as offspring (Fig. 4). Generation and release of offspring are halted in the absence of the associated microbiota and when infected with potential pathogens. The requirement of an associated microbiota for reproduction was recently also demonstrated for the fruit fly Drosophila (70). It was shown that the fly gut stimulated bacterial diversity, which, in turn, enhanced development, aging, and reproduction of the host. At the current stage, we can only speculate on the bacterial contribution to A. aurita reproduction at the molecular level, but a bacterial metabolite or small molecule appears likely to be fundamental. As hypothesized, the native microbiota most likely acts as a protective shield and supports defense against facing potentially pathogenic bacteria, since polyp fitness was impaired in the absence of the microbiome and was even more crucial when additionally infected with potentially pathogenic bacteria. Multivariate ordination analysis (Fig. 9) of observed fitness effects perfectly matches with microbial community patterns identified in RDAs (Fig. 2). Indicator OTU analysis points to key bacteria, which are correlated with health or impaired fitness of A. aurita (Tables 1, S4, and S5).
The role of microbes in pathogen defense is exemplified for various invertebrates. In corals, commensal bacteria defend the attachment of a pathogen on the mucus and protect against the bleaching-causative Vibrio shiloi (71, 72). Moreover, the endosymbiont Spiroplasma protects Drosophila against nematodes by producing RIP toxins (73). The ability of associated microbes to interfere with pathogen colonization and growth is known as colonization resistance (74–76) and has also been documented across vertebrates and plants (77, 78). While the concept of colonization resistance is not new, its underlying molecular mechanisms, ranging from direct microbe-microbe competition (e.g., niche competition or direct antagonism) to indirect induction or priming of host metabolism or immunity, are less understood (79). Nevertheless, one prominent example from vertebrates clearly demonstrates the role of bacteria in combating infectious diseases. Nowadays, patients suffering from severe diarrhea caused by Clostridium difficile infection can be successfully treated using fecal transplants from healthy donors, which harbor beneficial microbes required for gut homeostasis (80).
In conclusion, our study contributes to the growing body of evidence that associated microbes are an essential part of the host phenotype, influencing fitness, and are thus ecologically important traits of their hosts (81–84). The functional contributions of native microbiomes to host fitness are manifold, comprising, for instance, pathogen protection, nutrient allocation, development, behavior, adaptation, detoxification, and mating selection (reviewed in reference 85). The observed decline in reproductive output and fitness of the jellyfish A. aurita upon disturbance of its natural microbiota in the present study clearly points to the importance of an organism-specific associated microbiota, ultimately facilitating a healthy metaorganism. Our results further emphasize the importance of using novel model organisms to study host-microbe interactions and highlight the importance of the microbiome for health and fitness of a host in general. We further assume that bacterial diversity and species abundances within the microbiome are balanced on the polyp, most likely due to interbacterial (11, 40, 41, 83) and host-microbe interactions (31, 86) as well as host genotype (87, 88). However, the molecular mechanism of the microbial contribution to A. aurita fitness, in particular, to asexual reproduction, has to be elucidated in the future. Impending identification of the underlying mechanism and involved molecules also has a potential for application. Concisely, asexual reproduction ensures enormous multiplication of polyps in an environment by budding or massive production of juvenile medusae via strobilation (56). This fast and frequent multiplication can lead to jellyfish blooms, which significantly impact ecological community composition and structure by altering carbon, nitrogen, and phosphorus cycling and reducing available prey for higher predators (89, 90). In the future, the knowledge of microbial molecules triggering massive offspring generation might lead to the development of compounds interfering with the asexual reproduction of A. aurita, which will ultimately be useful to control jellyfish blooms.
MATERIALS AND METHODS
Aurelia aurita polyp husbandry and generation of sterile polyps.
Husbandry and generation of sterile polyps are described in detail by Weiland-Bräuer et al. (29). Briefly, polyps of subpopulation North Atlantic (Roscoff, France) were kept more than 10 years in the lab in 2-liter plastic tanks in 3% artificial seawater (ASW) (tropical sea salts; Tropic Marin). Polyps were fed twice a week with freshly hatched Artemia salina (HOBBY, Grafschaft-Gelsdorf, Germany). Generation of sterile polyps was performed with antibiotic mixture added to sterile (filtered through 0.22-μm Durapore membrane filters; Merck Millipore, Darmstadt) artificial seawater (Provasoli’s antibiotic mixture with final concentrations of 360,000 U/liter penicillin G, 1.5 mg/liter chloramphenicol, 1.8 mg/liter neomycin, and 9,000 U/liter polymyxin B; all components from Carl Roth, Karlsruhe, Germany). Generation of sterile Artemia salina was similarly conducted. Eggs were hatched in sterile ASW with Provasoli’s antibiotic mixture (complemented with 3.5 mg/liter nystatin and 3.5 mg/liter amphotericin B). The absence of bacteria was confirmed by plating homogenized animals on marine bouillon agar plates (Carl Roth). After incubation at 19°C for 5 days, the CFU were determined. Absence of CFU indicated successful antibiotic treatment. Additionally, the full-length bacterial 16S rRNA gene was amplified using GoTaq polymerase (Promega, Madison, WI, USA) and primer set 27F (5′-AGAGTTTGATCCTGGCTCAG-3′)/1492R (5′-GGTTACCTTGTTACGACTT-3′) with genomic DNA isolated from putative sterile polyps and A. salina using a Wizard genomic DNA purification kit according to the manufacturer’s instructions (Promega). Samples without successful amplification were graded as sterile.
Bacterial growth conditions.
Bacteria (Vibrio anguillarum, Pseudoalteromonas espejiana, and Ruegeria mobilis; GenBank accession numbers MK967055, MK967174, and MK967091, respectively) for microbial challenge during host fitness experiments were isolated from A. aurita polyps of subpopulations from the Baltic Sea and North Atlantic as well as 3% ASW (30 practical salinity units [PSU]), respectively, as described in reference 65. Strains were grown in marine bouillon (MB; Carl Roth, Karlsruhe, Germany) at 30°C and 120 rpm to a turbidity optical density at 600 nm (OD600) of 0.8. Bacterial cell numbers were detected using a Neubauer count chamber (91), and 105 cells/ml were complemented with 5% dimethyl sulfoxide (DMSO) (1-ml aliquots) to store at −80°C before using 10 μl of 105 cells/ml in 1 ml ASW per well during host fitness experiments. Concisely, Vibrio anguillarum isolated from A. aurita Baltic Sea polyps is widely distributed in marine and estuarine environments around the world (92–94). It is a global causative agent of vibriosis in marine fish and shellfish species, because it can efficiently grow and proliferate under environmental stress (95). Pseudoalteromonas espejiana isolated from artificial seawater is a ubiquitous bacterium abundantly found in marine environments and often associated with marine multicellular organisms (96). P. espejiana is able to degrade polymers and induces metamorphosis of Hydractinia echinata hydroid larvae (20, 97). Ruegeria mobilis was exclusively isolated from the polyp life stage (65). R. mobilis has primarily been isolated from marine aquaculture, where the tropodithietic acid (TDA)-producing strains have probiotic potential due to the inhibition of fish pathogens (98). V. anguillarum and R. mobilis belong to A. aurita’s native microbiota but are known as a potential pathogen and beneficial bacterium, respectively. P. espejiana is a nonnative bacterium.
Recolonization of sterile polyps.
Sterile polyps were used for recolonization with generated native A. aurita polyp microbiota. For generation of native microbiota, native polyps were transferred from 2-liter husbandry tanks into 1.5 ml reaction tubes (polyps were not fed for at least 3 days before transposition) and washed three times with 1 ml sterile ASW. Polyps (10 per reaction tube) were homogenized with a motorized pestle (KONTES; DWK Life Sciences, Wertheim, Germany) and resuspended in 1 ml sterile ASW. The homogenate was subsequently filtered through 3.1-μm filters (Lab Logistic Group, Meckenheim, Germany) to remove eukaryotic cells. Prior to conducting the host fitness experiment, 10 μl of the filtrate (approximately 2.5 × 107 cells/ml) was added to a single polyp in 1 ml ASW (48-well plate) and incubated for 48 h at 19°C. Polyps were washed three times with 1 ml sterile ASW to remove noncolonized bacteria. Additionally, prepared native microbiota (filtrate) and recolonized polyps (after 48 h of incubation) were used for bacterial 16S rRNA amplicon sequencing.
Host fitness experiment.
Single native and sterile polyps of the subpopulation from the North Atlantic were transferred from 2-liter husbandry tanks to 48-well plates with 1 ml 3% ASW per well. Polyps settled to the bottom of the wells prior to the experiment start. For each treatment, 96 replicates were used. Twelve different treatments comprising combinations of polyp, food, and ambient seawater in terms of microbial composition (native, polyps maintained in ASW in the lab; sterile, antibiotic [AB] treated, challenged) were conducted to elucidate the impact of microbes on the fitness of the host (Fig. 1). Briefly, polyps either harbored their native microbiota (lab conditions) or were made sterile using an antibiotic mixture (see above). Moreover, one of the 12 treatments comprised sterile polyps, which were recolonized with generated native A. aurita polyp microbiota. Native or sterile artificial seawater (30 PSU, filtered through 0.22-μm filters) was used as ambient water, and Artemia salina with its native microbiota or sterile brine shrimps were applied as food. Native as well as sterile polyps were additionally infected with selected bacteria Vibrio anguillarum, Pseudoalteromonas espejiana, and Ruegeria mobilis. Six different fitness traits (survival, growth, feeding, budding, strobilation, and ephyra release) were analyzed using the stereomicroscope Novex Binokulares RZB-PL Zoom-Mikroskop 65.500 (Novex, Arnhem, Netherlands) with a high-definition multimedia interface (HDMI)/HD camera as follows. The survival/malformation of polyps was assessed every 48 h for 14 days based on overall phenotypical appearance of polyps and presence of tentacles and categorized as healthy, harmed, and dead (see Fig. 5). Healthy polyps showed typical morphology comprising an attached stalk and a stretched calyx with fully developed and extended tentacles. Polyps were rated as harmed when they lost at least two of those criteria, mostly showing a shrunken calyx and contracted or even absorbed tentacles. Dead polyps were of a shrunken roundish morphology or even dispersed. The health of polyps was compared between the first monitoring after 48 h (t1) and at the end of experiment after 14 days (tend). The growth of polyps was documented every 48 h for 14 days by measuring the length and width of the polyps, which were still alive (healthy and harmed). Mean start (t1) sizes (length plus width) and end sizes after 14 days (tend) of polyps were compared per treatment, and growth rates were calculated in percentages after 14 days for the remaining replicates. Moreover, budding as one form of asexual reproduction of A. aurita polyps, where daughter polyps are generated, was monitored for 14 days (in parallel to health and growth). Numbers of daughter polyps were counted for all alive (healthy and harmed) polyps, and the corresponding budding rate was calculated as daughter polyp generation per week. In addition, on days 15 to 19, feeding of alive polyps (healthy and harmed) was monitored each day. Therefore, freshly hatched Artemia salina were harvested from 500-ml brood containers in a harvesting container with an Artemia sieve (0.15-mm mesh). A. salina were washed three times with sterile ASW on the mesh and concentrated to 50 ml. A single polyp was fed with 20 A. salina (in 50 μl of sterile ASW) and incubated for 1 h before counting remaining A. salina. A clearance rate of A. salina was calculated per treatment in percentages for each day (day 1 [t1] up to day 5 [tend]). Finally, a new set of polyps (96 replicates per treatment) was prepared as mentioned above in 48-well plates, with a single polyp in 1 ml ASW corresponding to treatments shown in Fig. 1. Strobilation of polyps was induced by adding 5 μM 5-methoxy-2-methyl indole to the ambient water on the three following days (involving daily washing). Afterwards, polyps were washed on day 4 with the respective ambient water. Strobilation induction was monitored each day, and strobila phenotypes as well as number of segments were detected beginning on day 5, when native polyps began segmentation (early strobila). Native polyps showed full segmentation and shaping as late strobila on day 9. Recently, 5-methoxy-2-methyl indole was demonstrated to act as the temperature-dependent synthetic activator of strobilation in A. aurita (58). The selected procedure of chemical induction enables the reduction of the complexity of the strobilation process to a minimum, particularly, for investigating the impact of the microbiome and not the environmental triggers during reproduction. This procedure is more time efficient than the natural induction by lowering the temperature (approximately 1 month at 8°C). Ephyra release was monitored each day after first appearance, and numbers of released ephyrae were detected beginning on day 12, when native strobilae began to release ephyrae. Ephyra release was monitored for the next 4 weeks.
In addition to the monitoring of fitness traits, 16S rRNA amplicon sequencing was performed to analyze potential changes in the microbial community composition based on the treatments, ultimately correlating observed fitness traits with changes in microbial community patterns. Therefore, six polyps were removed from the 48-well plates before the experimental start (t0) for DNA isolation and subsequent 16S rRNA amplicon sequencing (90 replicates remaining for each treatment at t0). After 14 days (tend) of experiment, further six polyps were removed for 16S rRNA analysis. Furthermore, six polyps were also removed from the second set of polyps before induction of strobilation (t0). Likewise, six late strobila (removed on day 10 after strobilation induction) per treatment were removed for sequence analysis.
Data analysis of host fitness parameters.
Factor effects were assessed for each fitness variable (i.e., counts of harmed and dead polyps, growth rate in percent, clearance rate in percent, budding rate in percent, strobilae count, and ephyra production in percent) using univariate permutational analysis of variance (99). All variables were tested under three test designs as outlined in Table S1. PERMANOVAs were performed on Euclidian resemblance matrices calculated from log2 (x + 1)-transformed data (100) and were based on 9,999 permutations of residuals under a reduced model and type III partial sums of squares using Primer-E V6. (100). Within each significant factor, pairwise post hoc tests followed, providing insight into differences between the treatment groups (Table S1).
Nucleic acid isolation.
Eukaryotic as well as bacterial DNA from single A. aurita polyps was isolated using a Wizard genomic DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
Illumina sequencing.
Bacterial DNA was isolated as described above and used for the generation of PCR amplicon libraries using uniquely barcoded primers flanking the V1-V2 hypervariable regions (27F–338R; V1_A_Pyro_27F [5′-CGTATCGCCTCCCTCGCGCCATCAGTCAGAGTTTGATCCTGGCTCAG-3′] and V2_B_Pyro_27F [5′-CTATGCGCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3′]) with fused MiSeq adapters and heterogeneity spacers in a 20-μl PCR using Phusion high-fidelity DNA polymerase (New England Biolabs) (101). Reactions included the following components: 4 μl of HF buffer, 0.4 μl deoxynucleoside triphosphates (dNTPs; 200 μM each), 0.8 μl each of forward and reverse primers (2 μM), 0.2 μl Phusion Hot Start II high-fidelity DNA polymerase, and 2 μl DNA (20 ng). PCRs were conducted with the following cycling conditions (98°C, 30 s; 30 × [98°C, 9 s; 55°C, 60 s; 72°C, 90 s]; 72°C, 10 min; 10°C, infinity). Amplicons were size checked and purified using a MinElute gel extraction kit (Qiagen). Purified amplicons were quantified using a Quant-iT PicoGreen kit (Invitrogen), and equal amounts of the purified PCR products were pooled for subsequent Illumina sequencing as indicated by band intensity and measured with Quant-iT PicoGreen kit. Amplicon sequencing was performed on the Illumina MiSeq platform with v3 chemistry (2 × 300 cycle kit) according to the manufacturer’s instructions at Max-Planck Institute for Evolutionary Biology in cooperation with S. Künzel.
16S rRNA data processing and bioinformatics.
All steps of sequence processing were conducted with the program mothur v1.39.5 according to the MiSeq standard operating procedure (SOP). In brief, 16S rRNA amplicon reads were aligned against the bacterial SILVA database, which was previously trimmed to the 16S V1-V2 rRNA gene regions to improve accuracy of the alignment (align.seqs). Subsequently, a random subset of 1,000 sequences per sample was generated to eliminate bias due to unequal sampling effort (sub.sample). The aligned and subsampled sequences were then classified using the Greengenes database v13_8_99 (classify.seqs) and reads assigned to the lineages Archaea, Chloroplast, Mitochondria, and unknown, were removed. The remaining sequences were used to compute a distance matrix (dist.seqs) for binning sequences into operational taxonomic units (OTUs) by average neighbor clustering (cluster.split). Five hundred eleven OTUs at a 97% similarity threshold (roughly corresponding to species level) distinction were considered. All downstream computations were performed in R v2.15.1 (102) as described in reference 29. Indicator OTU analysis was also performed in R. Mothur-shared OTU tables were imported into R using the phyloseq package (103). Subsequently, the multipatt function of the indicspecies package (104) was used to identify OTUs that were significantly associated with the replicate groups. For this analysis, only the original replicate groups, as provided in the group file, were considered, and 500 permutations were conducted. Significantly associated OTUs were summarized for each group, and their taxonomic classification was assigned. Furthermore, relationships of bacterial OTU abundance with host fitness variables in the treatment groups were evaluated using distance based linear models (DistLM) as implemented in Primer-E V6. Prior to analysis, host fitness data were averaged across the treatment groups, normalized, and aligned with square root-transformed OTU abundance data (100). The DistLM routine was performed using Euclidian distances and implementing a forward selection procedure with 999 permutations and adjusted R2 criterion. Additionally, Spearman rank correlation coefficients were obtained for the predictors.
Data availability.
Sequence data were deposited under the NCBI BioProject PRJNA633008 comprising locus tag prefixes SAMN14930335 to SAMN14930451.
Microbial composition of strobilae. Compositions of microbiota associated with native A. aurita polyps before strobilation induction and native late strobilae 9 days after induction of strobilation with 5 μM 5-methoxy-2-methyl indole. Microbial communities were analyzed by sequencing the V1-V2 region of 16S bacterial rRNA genes. (A) OTU abundances were summarized at the genus level. (B) Redundancy analysis plots of Hellinger-transformed OTU abundances (df = 0.136, v = 9.652, P value = 0.005). Download FIG S5, DOCX file, 0.2 MB (160.4KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
This work was conducted with financial support of the excellence initiative “The Future Ocean,” Kiel Marine Science (KMS), and the DFG as part of the CRC1182 “Origin and function of metaorganisms.”
We thank Sven Künzel and colleagues from the Department for Evolutionary Genetics of the Max Planck Institute for Evolutionary Biology for next-generation deep sequencing.
N.W.-B. and R.A.S. designed the research; N.W.-B., D.L., and N.P. performed the research; S.G. and C.M.C. analyzed the sequence data; N.W.-B. and A.R. analyzed the fitness data; N.W.-B. and R.A.S. wrote the paper.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Citation Weiland-Bräuer N, Pinnow N, Langfeldt D, Roik A, Güllert S, Chibani CM, Reusch TBH, Schmitz RA. 2020. The native microbiome is crucial for offspring generation and fitness of Aurelia aurita. mBio 11:e02336-20. https://doi.org/10.1128/mBio.02336-20.
REFERENCES
- 1.Bosch TC, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology (Jena) 114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg E, Zilber-Rosenberg I. 2012. The hologenome concept, p 323–340, Beneficial microorganisms in multicellular life forms. Springer Science & Business Media, Berlin, Germany. [Google Scholar]
- 3.Rosenberg E, Zilber-Rosenberg I. 2011. Symbiosis and development: the hologenome concept. Birth Defects Res C Embryo Today 93:56–66. doi: 10.1002/bdrc.20196. [DOI] [PubMed] [Google Scholar]
- 4.Jaspers C, Fraune S, Arnold AE, Miller DJ, Bosch T, Voolstra CR. 2019. Resolving structure and function of metaorganisms through a holistic framework combining reductionist and integrative approaches. Zoology (Jena) 133:81–87. doi: 10.1016/j.zool.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Bang C, Dagan T, Deines P, Dubilier N, Duschl WJ, Fraune S, Hentschel U, Hirt H, Hülter N, Lachnit T, Picazo D, Pita L, Pogoreutz C, Rädecker N, Saad MM, Schmitz RA, Schulenburg H, Voolstra CR, Weiland-Bräuer N, Ziegler M, Bosch TCG. 2018. Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology (Jena) 127:1–19. doi: 10.1016/j.zool.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Esser D, Lange J, Marinos G, Sieber M, Best L, Prasse D, Bathia J, Ruhlemann MC, Boersch K, Jaspers C, Sommer F. 2019. Functions of the microbiota for the physiology of animal metaorganisms. J Innate Immun 11:393–312. doi: 10.1159/000495115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochsenkühn MA, Röthig T, D'Angelo C, Wiedenmann J, Voolstra CR. 2017. The role of floridoside in osmoadaptation of coral-associated algal endosymbionts to high-salinity conditions. Sci Adv 3:e1602047. doi: 10.1126/sciadv.1602047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaffer JP, U'Ren JM, Gallery RE, Baltrus DA, Arnold AE. 2017. An endohyphal bacterium (Chitinophaga, Bacteroidetes) alters carbon source use by Fusarium keratoplasticum (F. solani species complex, Nectriaceae). Front Microbiol 8:350. doi: 10.3389/fmicb.2017.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rook G, Bäckhed F, Levin BR, McFall-Ngai MJ, McLean AR. 2017. Evolution, human-microbe interactions, and life history plasticity. Lancet 390:521–530. doi: 10.1016/S0140-6736(17)30566-4. [DOI] [PubMed] [Google Scholar]
- 10.Sommer F, Bäckhed F. 2013. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 11.Fraune S, Anton-Erxleben F, Augustin R, Franzenburg S, Knop M, Schröder K, Willoweit-Ohl D, Bosch TC. 2015. Bacteria–bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J 9:1543–1556. doi: 10.1038/ismej.2014.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran NA, Yun Y. 2015. Experimental replacement of an obligate insect symbiont. Proc Natl Acad Sci U S A 112:2093–2096. doi: 10.1073/pnas.1420037112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. 2012. Animal behavior and the microbiome. Science 338:198–199. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- 14.Roder C, Bayer T, Aranda M, Kruse M, Voolstra CR. 2015. Microbiome structure of the fungid coral Ctenactis echinata aligns with environmental differences. Mol Ecol 24:3501–3511. doi: 10.1111/mec.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hume BC, Voolstra CR, Arif C, D’Angelo C, Burt JA, Eyal G, Loya Y, Wiedenmann J. 2016. Ancestral genetic diversity associated with the rapid spread of stress-tolerant coral symbionts in response to Holocene climate change. Proc Natl Acad Sci U S A 113:4416–4421. doi: 10.1073/pnas.1601910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortzfeld BM, Urbanski S, Reitzel AM, Künzel S, Technau U, Fraune S. 2016. Response of bacterial colonization in Nematostella vectensis to development, environment and biogeography. Environ Microbiol 18:1764–1781. vol p doi: 10.1111/1462-2920.12926. [DOI] [PubMed] [Google Scholar]
- 17.Röthig T, Costa RM, Simona F, Baumgarten S, Torres AF, Radhakrishnan A, Aranda M, Voolstra CR. 2016. Distinct bacterial communities associated with the coral model Aiptasia in aposymbiotic and symbiotic states with Symbiodinium. Front Mar Sci 3:234. doi: 10.3389/fmars.2016.00234. [DOI] [Google Scholar]
- 18.Araldi-Brondolo SJ, Spraker J, Shaffer JP, Woytenko EH, Baltrus DA, Gallery RE, Arnold AE. 2017. Bacterial endosymbionts: master modulators of fungal phenotypes. Microbiol Spectr 5:FUNK-00256-2016. doi: 10.1128/microbiolspec.FUNK-0056-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. 2017. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:14213–14218. doi: 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leitz T, Wagner T. 1993. The marine bacterium Alteromonas espejiana induces metamorphosis of the hydroid Hydractinia echinata. Mar Biol 115:173–178. doi: 10.1007/BF00346332. [DOI] [Google Scholar]
- 21.Unabia C, Hadfield M. 1999. Role of bacteria in larval settlement and metamorphosis of the polychaete Hydroides elegans. Mar Biol 133:55–64. doi: 10.1007/s002270050442. [DOI] [Google Scholar]
- 22.Woznica A, Gerdt JP, Hulett RE, Clardy J, King N. 2017. Mating in the closest living relatives of animals is induced by a bacterial chondroitinase. Cell 170:1175.e11–1183.e11. doi: 10.1016/j.cell.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster NS, Smith LD, Heyward AJ, Watts JEM, Webb RI, Blackall LL, Negri AP. 2004. Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl Environ Microbiol 70:1213–1221. doi: 10.1128/aem.70.2.1213-1221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chilton SN, Enos MK, Burton JP, Reid G. 2014. The effects of diet and the microbiome on reproduction and longevity: a comparative review across 5 continents. J Nutr Food Sci 5:3. doi: 10.4172/2155-9600.1000364. [DOI] [Google Scholar]
- 25.Jacob S, Parthuisot N, Vallat A, Ramon-Portugal F, Helfenstein F, Heeb P. 2015. Microbiome affects egg carotenoid investment, nestling development and adult oxidative costs of reproduction in Great tits. Funct Ecol 29:1048–1058. doi: 10.1111/1365-2435.12404. [DOI] [Google Scholar]
- 26.DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. 2016. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis 22:1137–1150. doi: 10.1097/MIB.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding R-x, Goh W-R, Wu R-n, Yue X-q, Luo X, Khine WWT, Wu J-r, Lee Y-K. 2019. Revisit gut microbiota and its impact on human health and disease. J Food Drug Anal 27:623–631. doi: 10.1016/j.jfda.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredricks DN. 2013. The human microbiota: how microbial communities affect health and disease. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 29.Weiland-Bräuer N, Neulinger SC, Pinnow N, Kunzel S, Baines JF, Schmitz RA. 2015. Composition of bacterial communities associated with Aurelia aurita changes with compartment, life stage, and population. Appl Environ Microbiol 81:6038–6052. doi: 10.1128/AEM.01601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domin H, Zurita-Gutiérrez YH, Scotti M, Buttlar J, Hentschel Humeida U, Fraune S. 2018. Predicted bacterial interactions affect in vivo microbial colonization dynamics in Nematostella. Front Microbiol 9:728. doi: 10.3389/fmicb.2018.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiland-Bräuer N, Fischer MA, Pinnow N, Schmitz RA. 2019. Potential role of host-derived quorum quenching in modulating bacterial colonization in the moon jellyfish Aurelia aurita. Sci Rep 9:34. doi: 10.1038/s41598-018-37321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFall-Ngai M. 2014. Divining the essence of symbiosis: insights from the squid-Vibrio model. PLoS Biol 12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyholm SV, McFall-Ngai M. 2004. The winnowing: establishing the squid–Vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 34.Kremer N, Philipp EER, Carpentier M-C, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EAC, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voolstra CR. 2013. A journey into the wild of the cnidarian model system Aiptasia and its symbionts. Mol Ecol 22:4366–4368. doi: 10.1111/mec.12464. [DOI] [PubMed] [Google Scholar]
- 36.Heintz-Buschart A, Wilmes P. 2018. Human gut microbiome: function matters. Trends Microbiol 26:563–574. doi: 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Barnes EM, Carter EL, Lewis J. 2020. Predicting microbiome function across space is confounded by strain-level differences and functional redundancy across taxa. Front Microbiol 11:101. doi: 10.3389/fmicb.2020.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wein T, Dagan T, Fraune S, Bosch TC, Reusch TB, Hülter NF. 2018. Carrying capacity and colonization dynamics of Curvibacter in the Hydra host habitat. Front Microbiol 9:443. doi: 10.3389/fmicb.2018.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X-Y, Pietschke C, Fraune S, Altrock PM, Bosch TC, Traulsen A. 2015. Which games are growing bacterial populations playing? J R Soc Interface 12:20150121. doi: 10.1098/rsif.2015.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longford SR, Campbell AH, Nielsen S, Case RJ, Kjelleberg S, Steinberg PD. 2019. Interactions within the microbiome alter microbial interactions with host chemical defences and affect disease in a marine holobiont. Sci Rep 9:1363. doi: 10.1038/s41598-018-37062-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller B, Grossniklaus U. 2010. Model organisms—a historical perspective. J Proteomics 73:2054–2063. doi: 10.1016/j.jprot.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Bosch TC, Guillemin K, McFall-Ngai M. 2019. Evolutionary “experiments” in symbiosis: the study of model animals provides insights into the mechanisms underlying the diversity of host–microbe interactions. BioEssays 41:1800256. doi: 10.1002/bies.201800256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kostic AD, Howitt MR, Garrett WS. 2013. Exploring host-microbiota interactions in animal models and humans. Genes Dev 27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell JJ, Theriot JA, Sood P, Marshall WF, Landweber LF, Fritz-Laylin L, Polka JK, Oliferenko S, Gerbich T, Gladfelter A, Umen J, Bezanilla M, Lancaster MA, He S, Gibson MC, Goldstein B, Tanaka EM, Hu C-K, Brunet A. 2017. Non-model model organisms. BMC Biol 15:55. doi: 10.1186/s12915-017-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taormina G, Ferrante F, Vieni S, Grassi N, Russo A, Mirisola MG. 2019. Longevity: lesson from model organisms. Genes 10:518. doi: 10.3390/genes10070518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mutlu E. 2001. Distribution and abundance of moon jellyfish (Aurelia aurita) and its zooplankton food in the Black Sea. Mar Biol 138:329–339. doi: 10.1007/s002270000459. [DOI] [Google Scholar]
- 48.Lucas CH, Graham WM, Widmer C. 2012. Jellyfish life histories: role of polyps in forming and maintaining scyphomedusa populations. Adv Mar Biol 63:133–196. doi: 10.1016/B978-0-12-394282-1.00003-X. [DOI] [PubMed] [Google Scholar]
- 49.Brodeur RD, Sugisaki H, Hunt GL, Jr.. 2002. Increases in jellyfish biomass in the Bering Sea: implications for the ecosystem. Mar Ecol Prog Ser 233:89–103. doi: 10.3354/meps233089. [DOI] [Google Scholar]
- 50.Purcell JE. 2005. Climate effects on formation of jellyfish and ctenophore blooms: a review. J Mar Biol Assoc U K 85:461–476. doi: 10.1017/S0025315405011409. [DOI] [Google Scholar]
- 51.Sommer U, Lengfellner K. 2008. Climate change and the timing, magnitude, and composition of the phytoplankton spring bloom. Glob Chang Biol 14:1199–1208. doi: 10.1111/j.1365-2486.2008.01571.x. [DOI] [Google Scholar]
- 52.Goldstein J, Steiner UK. 2020. Ecological drivers of jellyfish blooms–The complex life history of a ‘well-known’ medusa (Aurelia aurita). J Anim Ecol 89:910–920. doi: 10.1111/1365-2656.13147. [DOI] [PubMed] [Google Scholar]
- 53.Dong Z. 2019. Blooms of the moon jellyfish Aurelia: causes, consequences and controls, p 163–171. World seas: an environmental evaluation. Elsevier, Philadelphia, PA. [Google Scholar]
- 54.Ishii H, Watanabe T. 2003. Experimental study of growth and asexual reproduction in Aurelia aurita polyps. Sessile Org 20:69–73. doi: 10.4282/sosj.20.69. [DOI] [Google Scholar]
- 55.Han C-H, Uye S-I. 2010. Combined effects of food supply and temperature on asexual reproduction and somatic growth of polyps of the common jellyfish Aurelia aurita. Plankton Benthos Res 5:98–105. doi: 10.3800/pbr.5.98. [DOI] [Google Scholar]
- 56.Lucas CH. 2001. Reproduction and life history strategies of the common jellyfish, Aurelia aurita, in relation to its ambient environment. Hydrobiologia 451:229–246. doi: 10.1023/A:1011836326717. [DOI] [Google Scholar]
- 57.Brekhman V, Malik A, Haas B, Sher N, Lotan T. 2015. Transcriptome profiling of the dynamic life cycle of the scypohozoan jellyfish Aurelia aurita. BMC Genomics 16:74. doi: 10.1186/s12864-015-1320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst EM, Dirksen P, Bohm AM, Hemmrich G, Sommer F, Domazet-Loso T, Klostermeier UC, Anton-Erxleben F, Rosenstiel P, Bosch TC, Khalturin K. 2014. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr Biol 24:263–273. doi: 10.1016/j.cub.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Woollacott RM, Hadfield MG. 1996. Induction of metamorphosis in larvae of a sponge. Invertebr Biol 115:257–262. doi: 10.2307/3227015. [DOI] [Google Scholar]
- 60.Schmahl G. 1985. Induction of stolon settlement in the scyphopolyps of Aurelia aurita (Cnidaria, Scyphozoa, Semaeostomeae) by glycolipids of marine bacteria. Helgol Mar Res 39:117–127. doi: 10.1007/BF01997446. [DOI] [Google Scholar]
- 61.Schuett C, Doepke H, Groepler W, Wichels A. 2005. Diversity of intratunical bacteria in the tunic matrix of the colonial ascidian Diplosoma migrans. Helgol Mar Res 59:136–140. doi: 10.1007/s10152-004-0212-4. [DOI] [Google Scholar]
- 62.Kittelmann S, Harder T. 2005. Species-and site-specific bacterial communities associated with four encrusting bryozoans from the North Sea. Germany J Experimental Marine Biology and Ecology 327:201–209. doi: 10.1016/j.jembe.2005.06.020. [DOI] [Google Scholar]
- 63.Hofmann DK, Fitt WK, Fleck J. 1996. Checkpoints in the life-cycle of Cassiopea spp.: control of metagenesis and metamorphosis in a tropical jellyfish. Int J Dev Biol 40:331–338. [PubMed] [Google Scholar]
- 64.Müller WA, Leitz T. 2002. Metamorphosis in the Cnidaria. Can J Zool 80:1755–1771. doi: 10.1139/z02-130. [DOI] [Google Scholar]
- 65.Prasse D, Weiland-Bräuer N, Jaspers C, Reusch TBH, Schmitz RA. 9 April 2019. Evaluating the quorum quenching potential of bacteria associated to Aurelia aurita and Mnemiopsis leidyi. bioRxiv doi: 10.1101/602268. [DOI]
- 66.Tinta T, Kogovšek T, Klun K, Malej A, Herndl GJ, Turk V. 2019. Jellyfish-associated microbiome in the marine environment: exploring its biotechnological potential. Mar Drugs 17:94. doi: 10.3390/md17020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kramar MK, Tinta T, Lučić D, Malej A, Turk V. 2019. Bacteria associated with moon jellyfish during bloom and post-bloom periods in the Gulf of Trieste (northern Adriatic). PLoS One 14:e0198056. doi: 10.1371/journal.pone.0198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rahat M, Dimentman C. 1982. Cultivation of bacteria-free Hydra viridis: missing budding factor in nonsymbiotic hydra. Science 216:67–68. doi: 10.1126/science.7063873. [DOI] [PubMed] [Google Scholar]
- 69.Schaible R, Ringelhan F, Kramer BH, Scheuerlein A. 2017. Hydra, p 238–254. In Shefferson R, Jones O, Salguero-Gomez R. (eds), The evolution of senescence in the tree of life. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 70.Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Korasidis N, Gavryushkin A, Carlson JM, Beerenwinkel N, Ludington WB. 2018. Microbiome interactions shape host fitness. Proc Natl Acad Sci U S A 115:E11951–E11960. doi: 10.1073/pnas.1809349115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krediet CJ, Ritchie KB, Alagely A, Teplitski M. 2013. Members of native coral microbiota inhibit glycosidases and thwart colonization of coral mucus by an opportunistic pathogen. ISME J 7:980–990. doi: 10.1038/ismej.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mills E, Shechtman K, Loya Y, Rosenberg E. 2013. Bacteria appear to play important roles in both causing and preventing the bleaching of the coral Oculina patagonica. Mar Ecol Prog Ser 489:155–162. doi: 10.3354/meps10391. [DOI] [Google Scholar]
- 73.Hamilton PT, Peng F, Boulanger MJ, Perlman SJ. 2016. A ribosome-inactivating protein in a Drosophila defensive symbiont. Proc Natl Acad Sci U S A 113:350–355. doi: 10.1073/pnas.1518648113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pamer EG. 2016. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science 352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawley TD, Walker AW. 2013. Intestinal colonization resistance. Immunology 138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stecher B, Berry D, Loy A. 2013. Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol Rev 37:793–829. doi: 10.1111/1574-6976.12024. [DOI] [PubMed] [Google Scholar]
- 77.Masson F, Lemaitre B. 2017. Symbiosis: protection from within. Elife 6:e24111. doi: 10.7554/eLife.24111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oldroyd GE. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 79.Chiu L, Bazin T, Truchetet M-E, Schaeverbeke T, Delhaes L, Pradeu T. 2017. Protective microbiota: from localized to long-reaching co-immunity. Front Immunol 8:1678. doi: 10.3389/fimmu.2017.01678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Drekonja D, Reich J, Gezahegn S, Greer N, Shaukat A, MacDonald R, Rutks I, Wilt TJ. 2015. Fecal microbiota transplantation for Clostridium difficile infection: a systematic review. Ann Intern Med 162:630–638. doi: 10.7326/M14-2693. [DOI] [PubMed] [Google Scholar]
- 81.Fraune S, Bosch TC. 2010. Why bacteria matter in animal development and evolution. Bioessays 32:571–580. doi: 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- 82.McFall-Ngai M. 2007. Care for the community. Nature 445:153–153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 83.Nyholm SV, Graf J. 2012. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol 10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bosch TC. 2013. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol 67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 85.Rosenberg E, Zilber-Rosenberg I. 2014. The hologenome concept: human, animal and plant microbiota. Springer Science & Business Media, Berlin, Germany. [Google Scholar]
- 86.Hoye BJ, Fenton A. 2018. Animal host-microbe interactions. J Anim Ecol 87:315–319. doi: 10.1111/1365-2656.12788. [DOI] [PubMed] [Google Scholar]
- 87.Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 88.Deines P, Lachnit T, Bosch TC. 2017. Competing forces maintain the Hydra metaorganism. Immunol Rev 279:123–136. doi: 10.1111/imr.12564. [DOI] [PubMed] [Google Scholar]
- 89.Condon RH, Steinberg DK, Del Giorgio PA, Bouvier TC, Bronk DA, Graham WM, Ducklow HW. 2011. Jellyfish blooms result in a major microbial respiratory sink of carbon in marine systems. Proc Natl Acad Sci U S A 108:10225–10230. doi: 10.1073/pnas.1015782108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pitt KA, Welsh DT, Condon RH. 2009. Influence of jellyfish blooms on carbon, nitrogen and phosphorus cycling and plankton production. Hydrobiologia 616:133–149. doi: 10.1007/s10750-008-9584-9. [DOI] [Google Scholar]
- 91.Phelan MC, Lawler G. 1997. Cell counting. Curr Protoc Cytom 00:A.3A.1–A.3A.4. doi: 10.1002/0471142956.cya03as00. [DOI] [PubMed] [Google Scholar]
- 92.Toranzo AE, Magariños B, Romalde JL. 2005. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246:37–61. doi: 10.1016/j.aquaculture.2005.01.002. [DOI] [Google Scholar]
- 93.Kim E-Y, Kim Y-R, Kim D-G, Kong I-S. 2012. A susceptible protein by proteomic analysis from Vibrio anguillarum under various environmental conditions. Bioprocess Biosyst Eng 35:273–282. doi: 10.1007/s00449-011-0636-6. [DOI] [PubMed] [Google Scholar]
- 94.Ma Y, Wang L, Shao Z. 2006. Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ Microbiol 8:455–465. doi: 10.1111/j.1462-2920.2005.00911.x. [DOI] [PubMed] [Google Scholar]
- 95.Gao X, Pi D, Chen N, Li X, Liu X, Yang H, Wei W, Zhang X. 2018. Survival, virulent characteristics, and transcriptomic analyses of the pathogenic Vibrio anguillarum under starvation stress. Front Cell Infect Microbiol 8:389. doi: 10.3389/fcimb.2018.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holmstrom C, Kjelleberg S. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol 30:285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 97.Uchida M, Nakata K, Maeda M. 1997. Conversion of Ulva fronds to a hatchery diet for Artemia nauplii utilizing the degrading and attaching abilities of Pseudoalteromonas espejiana. J Appl Phycol 9:541–549. [Google Scholar]
- 98.Sonnenschein EC, Nielsen KF, D'Alvise P, Porsby CH, Melchiorsen J, Heilmann J, Kalatzis PG, López-Pérez M, Bunk B, Spröer C, Middelboe M, Gram L. 2017. Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis. ISME J 11:569–583. doi: 10.1038/ismej.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 100.Anderson M, Gorley R, Clarke K, Anderson M, Gorley R, Clarke K, Anderson M, Gorley R, Andersom M. 2008. PERMANOVA+ for PRIMER. Guide to software and statistical methods. PRIMER-E Ltd., Plymouth, United Kingdom. [Google Scholar]
- 101.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. 2014. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.R Core Team 2012. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 103.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.De Cáceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Daughter polyp generation of A. aurita polyps. Budding was followed every 48 h for 14 days by monitoring the generation of daughter polyps of healthy and harmed polyps. Numbers of generated daughter polyps are shown as percentages after 14 days. Download FIG S1, DOCX file, 0.1 MB (104.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PERMANOVA tests. (A) Overview of PERMANOVA study design. (B) All variables were tested under three test designs, A, B, and C. (C) PERMANOVA test A with full data set (4-factorial, partly nested). (D) PERMANOVA test B with polyp-seawater-food data set (3-factorial, partly nested). (E) PERMANOVA test C with bacterial challenge data set (2-factorial, fully crossed). ns, not significant. Download Table S1, DOCX file, 0.1 MB (56.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of mean fitness status of A. aurita polyps and strobilae for all treatments. Heat map summarizes the significant decreases measured within the treatment groups compared to that in the native treatment. Based on individual results after host fitness experiments, monitored fitness parameter survival, growth, budding, feeding, strobilation, and ephyra release are categorized as unaffected (orange), slightly affected (purple), and crucially affected (red) for each treatment. Additionally, timelines for generation of strobilae and ephyra release are depicted as days of first appearance after strobilation induction with the synthetic inducer. na, not applicable. Download Table S2, DOCX file, 0.1 MB (25.7KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clearance of Artemia salina by A. aurita polyps. Feeding rate as clearance of A. salina in percentages. Single polyps were incubated for 1 h with 20 A. salina. After incubation, remaining A. salina were counted to calculate the feeding rate of healthy as well as harmed polyps for all treatments (corresponding codes in parentheses). Compared are the results from day 1 (I1) and day 5 (tend). Download FIG S2, DOCX file, 0.2 MB (242.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial community composition of A. aurita in the experimental treatment groups. Microbial communities were analyzed by sequencing the V1-V2 region of 16S bacterial rRNA genes. OTU abundances were summarized at the genus level and normalized by the total number of reads per sample. Bar plots are grouped according to sample type, each group including 2 to 6 replicates. Microbial community patterns of polyps (A) and strobilae (B) are shown. Download FIG S3, DOCX file, 0.6 MB (581.6KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial composition of recolonized polyps. Compositions of microbiota associated with native and recolonized A. aurita polyps as well as of the generated native microbiota for recolonization of sterile polyps. Microbial communities were analyzed by sequencing the V1-V2 region of 16S bacterial rRNA genes. (A) OTU abundances were summarized at the genus level and normalized by the total number of reads per sample. Bar plots are grouped according to sample type, each group including 6 replicates. (B) Redundancy analysis plots of Hellinger-transformed OTU abundances. Each point represents the whole microbial community of the respective sample, replicates of one sample group are framed by polygons. (C) Results of pairwise tests in beta diversity analysis. Tests were conducted for specified comparisons. df, degree of freedom; v, variance. Download FIG S4, DOCX file, 0.1 MB (150KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of pairwise tests in beta diversity analysis. Permutation test for RDA under reduced model was conducted with permutations (number of permutations, 9,999) for all comparisons, but only specified comparisons are shown. Model: RDA (formula = OTUhel ∼ group, data = metaDataSub); df = 0.321, v = 6.663, P value = 0.00001. Download Table S3, DOCX file, 0.1 MB (26.5KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Correlation of operational taxonomic units (OTUs) with the fitness status of A. aurita polyps. OTUs listed with their taxonomic classification are correlated with the fitness status of unaffected/healthy (orange), slightly affected/marginally harmed (purple), and crucially affected/essentially harmed (red) of A. aurita polyps. At least 50% of the biological replicates analyzed were associated with the respective OTU. Download Table S4, DOCX file, 0.1 MB (34.9KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Correlation of operational taxonomic units (OTUs) with the segmentation of A. aurita polyps and ephyra release. Unaffected (orange), slightly affected (purple), and crucially affected (red) formation of strobilae and further ephyra release are correlated with respective listed OTUs. At least 50% of the biological replicates analyzed were associated with the respective OTU. Download Table S5, DOCX file, 0.1 MB (39.7KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial composition of strobilae. Compositions of microbiota associated with native A. aurita polyps before strobilation induction and native late strobilae 9 days after induction of strobilation with 5 μM 5-methoxy-2-methyl indole. Microbial communities were analyzed by sequencing the V1-V2 region of 16S bacterial rRNA genes. (A) OTU abundances were summarized at the genus level. (B) Redundancy analysis plots of Hellinger-transformed OTU abundances (df = 0.136, v = 9.652, P value = 0.005). Download FIG S5, DOCX file, 0.2 MB (160.4KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequence data were deposited under the NCBI BioProject PRJNA633008 comprising locus tag prefixes SAMN14930335 to SAMN14930451.
Microbial composition of strobilae. Compositions of microbiota associated with native A. aurita polyps before strobilation induction and native late strobilae 9 days after induction of strobilation with 5 μM 5-methoxy-2-methyl indole. Microbial communities were analyzed by sequencing the V1-V2 region of 16S bacterial rRNA genes. (A) OTU abundances were summarized at the genus level. (B) Redundancy analysis plots of Hellinger-transformed OTU abundances (df = 0.136, v = 9.652, P value = 0.005). Download FIG S5, DOCX file, 0.2 MB (160.4KB, docx) .
Copyright © 2020 Weiland-Bräuer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.