Abstract
Histone posttranslational modifications are key regulators of chromatin-associated processes including gene expression, DNA replication and DNA repair. Monoubiquitinated histone H2A, H2Aub (K118 in Drosophila or K119 in vertebrates) is catalyzed by the Polycomb group (PcG) repressive complex 1 (PRC1) and reversed by the PcG-repressive deubiquitinase (PR-DUB)/BAP1 complex. Here we critically assess the current knowledge regarding H2Aub deposition and removal, its crosstalk with PcG repressive complex 2 (PRC2)-mediated histone H3K27 methylation, and the recent attempts toward discovering its readers and solving its enigmatic functions. We also discuss mounting evidence of the involvement of H2A ubiquitination in human pathologies including cancer, while highlighting some knowledge gaps that remain to be addressed.
Subject terms: Biochemistry, Cancer, Molecular biology, Epigenetics
Histone H2A monoubiquitination on lysine 119 in vertebrate and lysine 118 in Drosophila (H2Aub) is an epigenomic mark usually associated with gene repression by Polycomb group factors. Here the authors review the current knowledge on the deposition and removal of H2Aub, its function in transcription and other DNA-associated processes as well as its relevance to human disease.
Introduction
Eukaryotic DNA is organized into a highly ordered structure called chromatin that can be broadly classified as either euchromatin (relaxed form) or heterochromatin (compacted form). The building blocks of this structure are nucleosomes, formed by the association between chromosomal DNA and histone octamers (dimers of H2A-H2B and H3-H4) to form arrays of nucleosomes connected by a shorter DNA linker with variable lengths. The crystal structure of the nucleosome revealed an organization of the 146 base pair DNA fragment around the histone octamer, with the histone N-terminal and C-terminal tails extending away from the nucleosome core1. Importantly, the histone tails are targeted by several posttranslational modifications (PTMs), which, in addition to giving an epigenetic dimension to chromatin, are involved in the regulation of virtually all DNA-dependent processes. These PTMs are catalyzed by a wide spectrum of chromatin modifying factors, including Polycomb group (PcG) and Trithorax group (TrxG) complexes, that act as molecular machines to orchestrate chromatin-associated processes2–6. Indeed, many PTMs are found on specific conserved residues of histone tails, including phosphorylation, methylation, acetylation, and ubiquitination where they regulate DNA replication, DNA damage/repair, and gene transcription2–6. Histone PTMs play important roles in a wide spectrum of biological processes including cell proliferation, cell fate determination, and differentiation as well as stress responses. Many studies have associated distinct profiles of histone modifications with human pathologies and it is increasingly recognized that alterations of chromatin function are causally linked to disease development and progression6–8.
Polycomb group (PcG)-mediated H2A ubiquitination occurs on K119 in vertebrates, K118 in Drosophila and K121 in Arabidopsis and will be hereafter referred to as H2Aub9,10. The function and genomic localization of H2Aub remained a mystery for a long time, but this situation has changed with the identification of its E3 ubiquitin ligase, the PRC1 complex, and its obligate components RING1A or RING1B. Indeed, inactivation of mammalian RING1A/RING1B or their Drosophila orthologue Sce completely abolishes H2A ubiquitination11–19. In this review, we will outline the functions of the complexes and factors that attach ubiquitin on H2A (writers), decode the ubiquitination signal (readers) or remove ubiquitin from this histone (erasers) (Fig. 1). We discuss our current understanding of the link between H2Aub and chromatin-associated processes and provide an update on the potential role of H2Aub deregulation in the development of human pathologies (additional definitions are listed in Box 1).
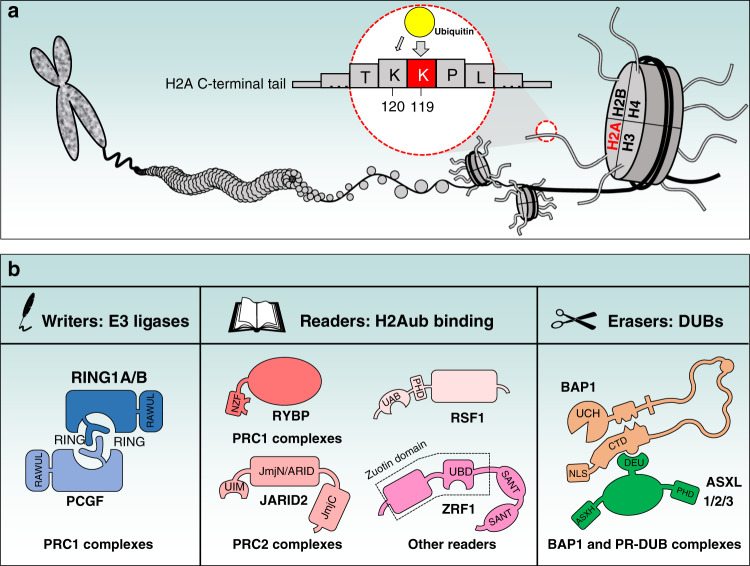
Fig. 1. Mammalian H2AK119ub (H2Aub) regulation by writers (ubiquitin E3 ligases), readers (ubiquitin-binding proteins) and erasers (deubiquitinases).
a Schematic representation of chromatin and the mammalian H2AK119 ubiquitination site by PRC1 E3 ligase. Note that the mammalian H2AK120 site is a minor site for PRC1-mediated ubiquitination. b Schematic representation of known modulators of H2Aub. Important functional domains are indicated. RING: Really Interesting New Gene, UBD: Ubiquitin-Binding Domain, UIM: Ubiquitin Interaction Motif, NZF: Nuclear protein localization 4 Zinc Finger, UAB: Ubiquitin-Associated Domain, RAWUL: Ring-finger and WD40 associated Ubiquitin-Like, PHD: Plant Homeodomain, UCH: Ubiquitin C-terminal Hydrolase, CTD: C-Terminal Domain, NLS: Nuclear Localization Signal, DEUBAD (DEU): DEUBiquitinase Activating Domain, ASXH: HARE-HTH of ASXLs.
Box 1 Definitions.
CpG: DNA regions rich in CG nucleotides found in the genome and are often protected from cytosine methylation.
CUBI: Composite Ubiquitin-Binding Interface is a protein interaction interface formed by the catalytic domain of BAP1, its C-terminal domain and the DEUBAD of ASXLs.
DEUBAD: DEUBiquitinase ADaptor is a conserved domain found in Asx/ASXLs, RPN13 and INO80G and is responsible for regulating DUB activity.
EZH: Enhancer of Zeste Homolog 2 is the methyltransferase enzyme of the PRC2 complex that catalyzes histone H3K27 methylation and corresponds to E(z) in Drosophila. Mammalians have two paralogues of EZH (EZH1 and EZH2).
H3K36me3: A histone PTM found on the gene body of actively transcribed genes.
H3K4me1: A histone PTM found on active enhancers.
H3K4me3: A histone PTM found at gene promoters and is a landmark of activate genes.
Homeobox (Hox): Originally identified in Drosophila and correspond to a group of genes that are responsible for defining the identity of body segments along the anterior/posterior axis.
PBAF: Polybromo-associated BAF complex is a SWI/SNF family chromatin-remodeling complex that regulates transcription in ATP-dependent manner.
PHD: Plant homeodomain (PHD) is a small zinc finger domain with similarities to RING fingers. PHD can bind histone PTMs such as methylated lysine. Some PHD domains can also bind an unmodified lysine.
PTEN: Phosphatase and Tensin Homolog deleted on Chromosome 10 (PTEN) is a dual phosphatase that uses protein and lipid as substrates.
RING: Really Interesting New Gene domain is a distinct type of zinc finger domain found in E3 ubiquitin ligases.
TrxG: Transcriptional activators that counteract PcG action. TrxG can be histone-modifying complexes or ATP-dependent chromatin-remodeling complexes.
UCH: Ubiquitin C-terminal Hydrolase is the catalytic domain found in the UCH family of deubiquitinases that include BAP1.
Mechanisms of H2Aub deposition
Genome-wide distribution of H2Aub on chromatin
H2Aub is a highly conserved histone modification found in eukaryotic organisms including animals and plants. Although it is still unclear when this modification was established during the course of evolution, H2Aub seems to be present in certain unicellular eukaryotes including diatoms and ciliates20,21. In mammalian cells, it is estimated that H2A is the most ubiquitinated protein, with up to 10% of total H2A harboring this PTM22. Early studies relied on visualizing the global levels of H2Aub in mammalian cells indicated that this modification is enriched in heterochromatic X and Y chromosomes (XY bodies) in spermatocytes and inactive X chromosome in female cells and was thus linked to gene silencing11,23–25. ChIP-seq studies in Drosophila cell lines revealed that H2Aub is broadly distributed on the genome, and is found in transcription start sites (TSS), gene body, and intergenic regions26. Moreover, H2Aub colocalizes with H3K27me3, a transcription repressive mark catalyzed by the PcG PRC2 complex (section “Positioning PRC1 complexes within the PcG landscape”)26. However, H2Aub can also exist in chromatin regions independently of H3K27 methylation26. In the plant Arabidopsis, H2Aub marks are also widespread with a significant colocalization with H3K27me3 on repressed genes, although about half H2Aub signals are not associated with detectable H3K27me327. ChIP-seq studies in mammalian cells also revealed that H2Aub is mostly associated with repressed genes14–16,28–32. For instance, in mouse embryonic stem cells (mESC), strong H2Aub signals are associated with promoters of repressed genes, notably PcG targets, which are often found within high-density CpG regions15,16,31–34. In addition, low levels of H2Aub are observed throughout the genome32. Importantly, a strong overlap between H2Aub and H3K27me3 is observed in stem cells and other cell types14–16,28,29,31,32. Of note, in several systems, low H2Aub signals could also be detected on active chromatin14,27,29,31. However, it is important to note that several factors can result in artificial H2Aub chromatin occupancy. These include the widespread nature of H2Aub, the chemical crosslinking conditions, and the stringency on ChIP-seq peak calling. Moreover, the presence of heterogeneous cell populations such as mixed differentiation states or asynchronous cell populations, can also complicate interpretation of H2Aub distribution. Thus, it is currently difficult to ascertain a definitive link between H2Aub and active chromatin.
In summary, the deposition of H2Aub is associated with repressed genes, notably PcG targets, and strongly overlaps with H3K27me3. H2Aub might also be associated with other chromatin contexts, possibly defining subcategories of H2Aub targets with specific histone PTMs and transcriptional states. Clearly, how the interplay between H2Aub and other histone PTMs orchestrates chromatin function is still not fully understood. Quantitative analysis of the genomic distribution of H2Aub in diverse cellular systems would shed light on how this PTM regulates chromatin-dependent processes in response to pathophysiological signaling.
Positioning PRC1 complexes within the PcG landscape
The roots of PcG genes date back to early studies characterizing genes that regulate development and anterior/posterior body segments identity using the Drosophila model2,5,35. It was found that defects in the regulation of Homeobox (Hox) genes induces distinct morphological phenotypes characterized by their ectopic expression in other body segments, a phenomenon known as homeotic transformation2,5,35. We know now that the expression of Hox genes is regulated by two groups of genes, the PcG and the Trithorax group (TrxG), with opposing roles as repressors and activators, respectively2,5,35. Moreover, it is well established that the PcG protein family are epigenetic regulators that play critical roles in body patterning during development and in maintaining cellular identity2,5,35.
In Drosophila, PcG factors are divided into five multi-protein repressor complexes including the canonical Polycomb repressive complex 1 (cPRC1), dRing-associated factors (dRAF) complex (vPRC1), Polycomb repressive complex 2 (PRC2), Polycomb repressive DUB complex (PR-DUB) and Pho-repressive complex (PhoRC)5,35–38 (Fig. 2a). PRC1 and PRC2 are the most studied complexes and both have a wide range of variants. The PRC1 complex components are Polycomb (Pc), Sex Comb Extra (Sce or dRing), Posterior Sex Combs (Psc), Polyhomeotic (Ph) and Sex Comb on Midleg (Scm), whereas PRC2 core components are Enhancer of Zeste (E(z)), Suppressor of Zeste 12 (Su(Z)12), Extra Sex Combs (Esc) and Nucleosome Remodelling Factor 55 (Nurf55). PRC1 and PRC2 complexes ensure ubiquitination of H2AK118 and methylation of H3K27 through the E3 ubiquitin ligase Sce and the methyltransferase E(z), respectively2,5,35. dRAF contains Sce, Psc and the lysine demethylase dKDM2 and is also associated with H2Aub deposition37. PhoRC is composed by Pleiohomeotic (Pho) and the Scm-related protein containing four MBT domains (Sfmbt) protein which confer sequence-specific DNA recognition and chromatin binding36,39 (section “Deposition of H2Aub and coordination with H3K27me3”). Finally, the PR-DUB complex which deubiquitinates H2Aub contains the DUB Calypso and Additional Sex Comb Asx38. Asx and it mammalian orthologue ASX-Like 1 (ASXL1) possess dual roles of enhancing PcG and Trithorax functions, and belongs to a group of factors termed Enhancers of Trithorax and Polycomb (ETP)40,41 (section “Deubiquitination of H2Aub by PR-DUB and BAP1”). Overall, the diversity of Drosophila PcG complexes suggests that multiple mechanisms of gene repression can take place to ensure proper gene silencing during development and homeostasis.
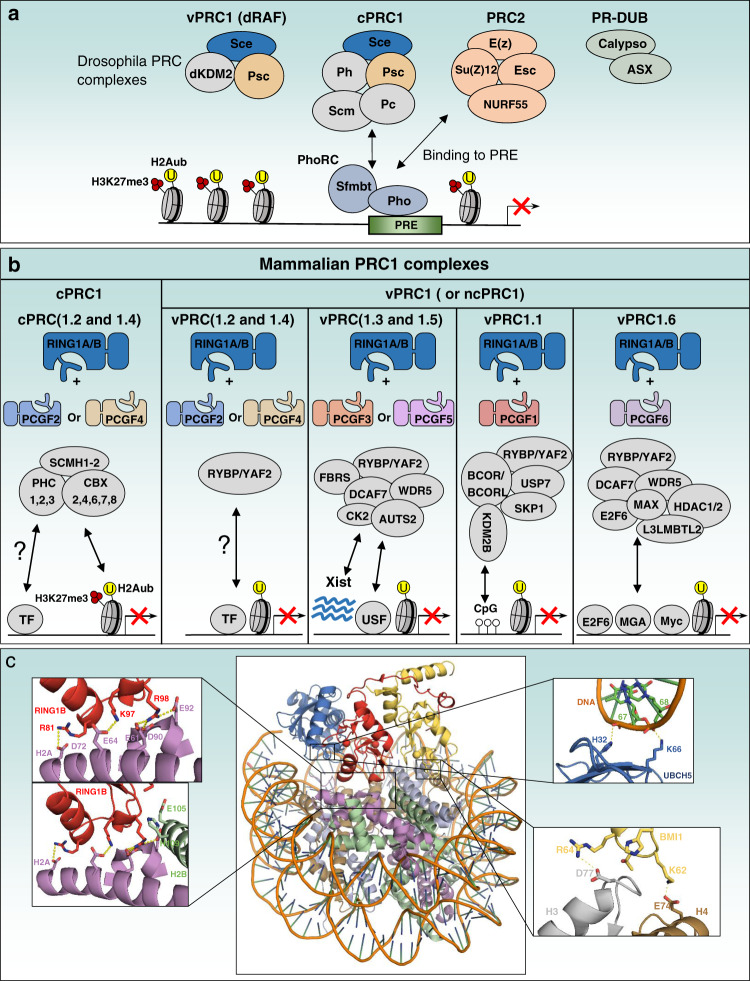
Fig. 2. PcG complexes in Drosophila and PRC1 complexes in mammals.
a Schematic representation of the five PcG complexes in Drosophila: canonical (cPRC1) and variant (vPRC1:dRAF) PRC1 complexes, PRC2 complex, PR-DUB complex and PhoRC complex. PhoRC binds to gene regulatory regions via the interaction of its subunit Pho with specific DNA sequences, PRE (PcG Responsive Elements). Pho can also directly bind to components of cPRC1 complexes to mediate their recruitment even in the absence of H3K27me3. The arrows indicate protein interactions responsible for the recruitment of PcG complexes to the PRE. b Schematic representation of the canonical (cPRC1) and variant (vPRC1) PRC1 complexes in mammalians. RING1A or RING1B are obligate components of the PRC1 complexes. Their association with either PCGF2/PCGF4 or other PCGFs (PCGF1, PCGF3, PCGF5, PCGF6) lead to the formation of cPRC1 or variants vPRC1 complexes, respectively. CBXs/HPHs and RYBP/YAF2 associate in mutually exclusive manner with RING1A/RING1B for the formation of cPRC1 and vPRC1 complexes, respectively. H3K27me3 and CpG islands recruit cPRC1 and vPRC1/PCGF1 complexes respectively. Transcription factors (TF), USF and other chromatin factors (E2F6, MGA, MYC) might also be involved in the recruitment of PRC1 to chromatin. The arrows indicate protein-protein or protein DNA/RNA interactions responsible for the recruitment of PRC1 complexes to chromatin. Ubiquitination is shown as the yellow circle and H3K27me3 is shown by the three red circles (a, b). c In the center, crystal structure of a minimal human PRC1 complex (RING1B /BMI1-UbcH5c) interacting with the nucleosome core particle (NCP) (PDB: 4R8P). The left panels show the contact between RING1B and H2A/H2B acidic patch. The top right panel shows the DNA-UBCH5 interactions. BMI1 also makes polar contacts with Histone H3 and H4 as shown in the bottom right panel. Interaction of RING1B with BMI1 on one hand and histone H2A on the other hand is essential for establishing H2Aub on the latter.
The PcG complexes are much more diverse in mammals. Focusing on PRC1, recent studies provided a classification of canonical (cPRC1) and variant PRC1 (vPRC1) complexes depending on their subunit composition13,14,32,42 (Fig. 2b). The two Sce orthologues RING1A (RNF1) or RING1B (RNF2) are obligate and mutually exclusive components of PRC1 complexes and their depletion results in a complete loss of H2Aub chromatin signals11–16. The mammalian orthologues of Pc, Psc, and Ph are respectively the H3K27me3-binding chromobox family (CBX2/4/6/7/8), the PcG RING fingers family (PCGF1/2/3/4/5/6) and the human Ph family (PHC1/2/3) proteins13,14,32,42. Two other factors, RING1-Yin Yang1 (YY1) Binding Protein (RYBP) and YY1-Associated Factor 2 (YAF2), are also associated with a subset of PRC1 complexes in a mutually exclusive manner with CBX and HPH factors13,14,32,42. Thus, cPRC1 complexes are described as the association of RING1A or RING1B, CBXs, HPHs and either PCGF4 (BMI1) or PCGF2 (MEL18) and vPRC1 variants are composed of RING1A or RING1B, RYBP or YAF2 and one of the six PCGF factors13,14,32,42–47 (Fig. 2b). The wide diversity of PRC1 complexes in higher eukaryotes could be explained by a functional specialization that has been imposed during evolution to fine-tune gene silencing. Combinatorial interactions within PRC1 complexes and between PRC1 complexes and other factors would tailor repression mechanisms to specific cell fate decisions. It will be interesting to determine the dynamics of PRC1 complexes with a focus on transient interactions that take place at gene regulatory regions, in response to signaling events and cell fate transitions, and determine their relevance to H2Aub deposition and function.
Deposition of H2Aub and coordination with H3K27me3
Role of DNA-binding proteins in H2Aub deposition: Pioneering studies in Drosophila indicated that PcG-mediated repression relies on specific DNA sequences called PRE (or PcG Responsive Elements)48,49. PRE-mediated gene silencing was later found to involve DNA-binding PcG proteins such as pleiohomeotic Pho and Pho-like, whose vertebrate orthologue is YY136,50–53. For instance, Pho can bind to components of the PRC1 complex and recruit its subunits directly to DNA, in the absence of nucleosomes53,54 (Fig. 2a). In addition, Sfmbt subunit of PhoRC can directly interact with PRC1 and promote its recruitment at PRE55. This might explain the presence of H2Aub in chromatin regions in the absence H3K27me326. Thus, PhoRC is possibly the first PcG to be recruited to DNA and to nucleate repression56. Nonetheless, further work is needed to define the exact contribution of PhoRC complexes in coordinating H2Aub genomic distribution during Drosophila development and how Pho and other transcription factors orchestrate the function of PcG complexes to ensure gene silencing. The dRAF complex contains dKDM2 which possesses a CXXC-type zinc finger capable of direct DNA binding57. dRAF was shown to promote both ubiquitination of histone H2AK118 and demethylation of histone H3K36me237. However, the exact role of DNA binding by dKDM2 in H2Aub deposition has not been established. In addition, dKDM2 mutant flies are viable58,59, arguing against a major role of this factor in H2Aub deposition and PcG repression.
Studies in high-order organisms failed to define true PREs, suggesting divergent modes of H2Aub deposition and regulation35. Recent biochemical and genome-wide studies indicated that PRC1 complex composition and DNA-binding factors are important in defining its recruitment and H2A ubiquitination13–16,32,42,45,60 (Fig. 2b). For instance, PCGF factors exert specific and overlapping roles in the establishment of H2Aub and H3K27me313,14,32. In particular, recruitment of vPRC1 complexes harboring KDM2B, which binds DNA on unmethylated CpGs, through its CXXC zinc finger domain, plays an important role in H2A ubiquitination13,33,61–63. Several transcription factors can also be involved in the recruitment of canonical or vPRC1 complexes to chromatin, including REST, USF1/2, MGA, and E2F614,64–69 (Fig. 2b). It is important to note that the exact contribution of many transcription factors to the deposition of H2Aub at specific target genes remains incompletely defined and further studies are needed to establish how transcription factors orchestrate H2Aub deposition depending on cell type and chromatin contexts.
Impact of H3K27me3 on H2Aub deposition: An initial model involved PRC2 recruitment at PcG target genes and recognition of H3K27me3 by Pc through its trimethyl-lysine binding domain known as chromodomain49,53,70–73. While this implied that PRC2 and PRC1 are sequentially recruited to the same regions to deposit H3K27me3 and subsequently H2Aub, respectively, many studies provided evidence for different mechanisms of H2Aub deposition53–55. First, as mentioned above, in Drosophila, PRC1 can be directly recruited to chromatin through PhoRC55. Second, mutation of Drosophila E(z), which strongly reduces H3K27me3, had only a minimal effect on the global levels of H2Aub26. Interestingly, while a significant reduction of H2Aub was observed around PREs following inactivation of PRC2, only a marginal decrease of global H2Aub levels was observed74. In mammals, several studies indicated that the bulk of genomic H2Aub does not depend on PRC2 and H3K27me345,75–78. For example, in mESC which lacks the PRC2 component EED, global H2Aub levels remain unchanged in the absence of H3K27me345,75. Moreover, it was found that deposition of H2Aub on inactivated X chromosome is independent of PRC2 and precedes H3K27me3 deposition79,80. However, other studies in mESC and somatic cells showed that inactivation of PRC2 can result in significantly decreased levels H2Aub both globally and on gene regulatory regions81,82. The reasons behind these discrepancies are unclear, but are likely to be associated with variations in cell systems and experimental conditions used. Thus, while a possible general model seems to point toward H2Aub being largely independent of H3K27me3, definitive answers on the role of H3K27 methylation in shaping H2A ubiquitination are still needed.
Role of H2Aub in H3K27 methylation: Inactivation of PRC1 in Drosophila, which results in the global depletion of H2Aub, also leads to partially decreased levels of H3K27me3 in mutant embryos19,26. Interestingly, H2Aub creates a docking site for PRC2 complex containing Jumonji And AT-Rich Interaction Domain Containing 2 (JARID2), and this leads to H3K27 trimethylation83 (section “Readers of H2Aub”). This provides a molecular link between H2Aub and H3K27me3 that might also explain the co-existence of both marks on PcG target genes.
In mammalian systems, notably mESC, inhibition of H2Aub deposition leads to a substantial reduction of PRC2 recruitment and concomitant deposition of H3K27me3 at PcG targets13–16,32,84. In addition, inhibition of PRC1 and subsequent H2Aub deposition prevents the recruitment of the PRC2 component EED (Embryonic Ectoderm Development) and subsequent H3K27 trimethylation during X chromosome inactivation85. This feedback loop between these two histone marks seems also to be mediated by JARID2 in mammals83,86. Consistent with this model, depletion of H2Aub results in a strong reduction in the chromatin occupancy of JARID215,16,87. However, inactivation of JARID2 in mESC results in a marginal reduction of the global levels of H3K27me3, although this can be explained by functional redundancy within PRC2 complexes87. Strikingly, other studies showed that ablation of PRC1 does not impact H3K27me330,87,88, which is difficult to reconcile with the findings mentioned above. While discrepancies can be explained by differences in experimental settings or tissue-specific regulation, additional studies are needed to further define how deposition of H2Aub impacts H3K27me3 distribution.
In summary, PRC1 recruitment to gene regulatory regions and subsequent H2Aub deposition is highly influenced by DNA-binding proteins, PRC1 subunit composition and the state of chromatin. While several scenarios of crosstalk between H2Aub and H3K27me3 can be envisioned, one emerging model involves the deposition of H2Aub by vPRC1 leading to the recruitment of PRC2 and subsequent deposition of H3K27me3. Methylation of H3K27, in turn, leads to the recruitment of cPRC1 through methyl binding by the CBX subunits. This model also implies different functions between vPRC1 and cPRC1 complexes (discussed later). These findings also raise some interesting questions of what determines the composition and stoichiometry of PcG complexes and how different complexes dynamically direct target gene expression outcomes. The use of quantitative approaches to analyze H2Aub, H3K27me3 and other histone PTMs throughout the genome in well-defined cell models and experimental settings would help unify conclusions.
Structural insights into the mechanism of H2A ubiquitination by PRC1
While the PRC1 complex was originally purified from Drosophila cells and shown to antagonize the chromatin-remodeling activity of a SWI/SNF factor89, it was later purified from human cells by virtue of its ability to ubiquitinate H2Aub9. Subsequently, a mammalian PRC1 complex was reconstituted in vitro, which revealed that BMI1 (PCGF4) strongly enhances the E3 ligase activity of RING1B and is essential for complex assembly17. In addition, early structural studies indicated that RING1B and BMI1 form a heterodimer via their RING domains while the E2 ubiquitin conjugating enzyme UBCH5 interacts directly with RING1B90,91. The crystal structure of PRC1 ubiquitination module (composed of the E2 UBCH5C and the minimal RING1B/BMI1 RING heterodimer) indicates that the N-terminal tail of RING1B is wrapped around BMI1 stabilizing the RING-RING interaction and simultaneously promoting E2 binding90,91. (Fig. 2c). On the other hand, the positioning of the RING1B/BMI1 heterodimer on the nucleosome core particle (NCP) involves an interface composed of basic residues that interact with the nucleosome acidic patch, an electronegative cleft formed by H2A/H2B histones dimer that constitutes a protein interaction interface92,93. Interestingly, all four histones contribute to the interaction with the RING1B/BMI1 complex. Of note, while the RING1B/BMI1-NCP interaction is independent of the DNA sequence, the proper orientation of RING1B/BMI1 heterodimer towards H2A involves an additional interaction of the E2 enzyme with nucleosomal DNA93. This multiple surface binding of the RING1B/BMI1/UBCH5C results in an enhanced catalytic activity of the complex while positioning the E2 enzyme very closely to H2A93. The fact that RING1B/BMI1-UBCH5C interacts with H2A-H2B, H3-H4, and DNA explains why PRC1 requires a fully assembled nucleosome for ubiquitination93. Overall, these studies provided important insights into the mechanism of H2A ubiquitination that involve the additive effects of multiple interactions that altogether increase the overall affinity and specificity of PRC1 towards nucleosomal H2Aub92,93.
Deubiquitination of H2Aub by PR-DUB and BAP1
The H2Aub DUB Calypso was originally identified as a gene whose mutation induces a PcG phenotype in Drosophila94. BAP1, the mammalian orthologue of Calypso, is found in the cytoplasm and nucleus95,96. Nuclear BAP1 associates with ASXLs and also catalyzes H2Aub deubiquitination38,97 (Fig. 3a). Of note, depletion of ASXL1 and ASXL2 strongly reduces BAP1 protein levels indicating the importance of these factors in regulating BAP1 function97. In addition, nuclear BAP1 assembles, with different stoichiometries, large complexes containing several transcriptional regulators including, the Host Cell Factor-1 (HCF-1), the O-Linked N-Acetylglucosamine Transferase (OGT), the Lysine demethylase KDM1B (LSD2/AOF1), the Histone acetyltransferase HAT1 as well as FOXK1/2, YY1 transcription factors and the E2/E3 hybrid enzyme UBE2O98–100 (Fig. 3a). While BAP1, HCF-1, OGT, ASXLs, and possibly FOXK1/2, appear to form the core complex95,97,100–102, the composition of BAP1 subcomplexes remains to be rigorously determined. In addition, how BAP1 complexes catalyze H2Aub deubiquitination in different physiological contexts and during development remains to be established.
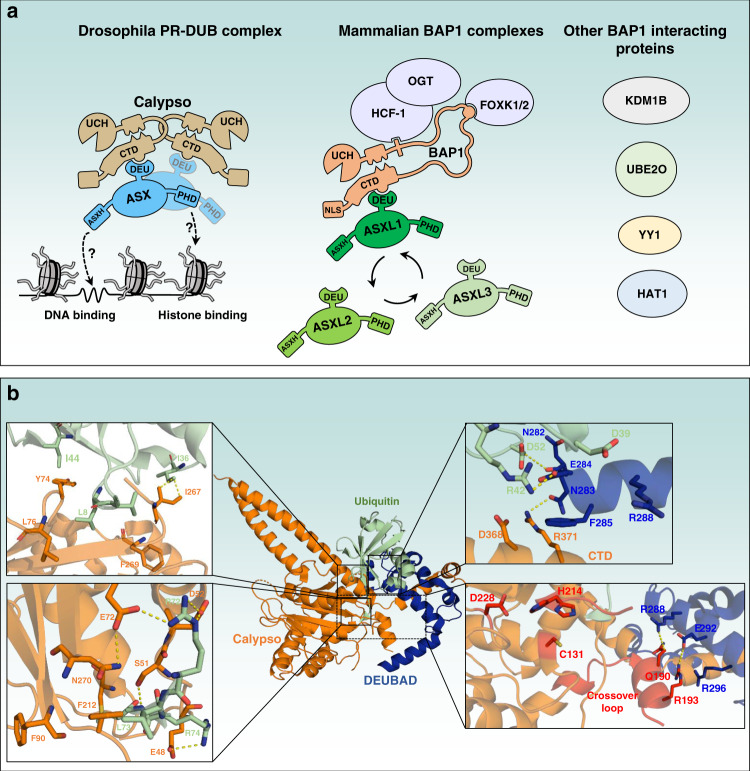
Fig. 3. PR-DUB and BAP1 as major deubiquitinase complexes for H2Aub.
a Schematic representation of the Drosophila PR-DUB (Calypso/ASX) and mammalian BAP1 complexes (BAP1/ASXLs). The dotted arrows indicate potential interactions of ASX with chromatin. Calypso and ASX form an oligomer of 2:2 molecules. This specific arrangement is important for the recruitment to chromatin and H2Aub deubiquitination. Note that the mammalian PR-DUB complex (BAP1 complexes) is composed of many additional subunits including HCF-1, OGT, and FOXK1/2. Other BAP1-interacting partners, KDM1B, UBE2O, YY1 and HAT1 are shown. The arrows in circle indicate the mutually exclusive binding of ASXLs to BAP1. b Structural model of the Drosophila core PR-DUB complex (Calypso/DEUBAD (DEUBAD of Asx)) bound to ubiquitin. The model was obtained by superimposing the crystal structure of Calypso/DEUBAD (PDB: 6HGC) with the UCH-L5-RPN13-Ub crystal structure (PDB: 4UEL). Hydrogen bound interactions between calypso catalytic domain and ubiquitin are shown (left panels). DEUBAD also makes contact with the CTD domain of Calypso and ubiquitin (right top panel). The DEUBAD interaction with Calypso stabilizes Calypso’s crossover loop (shown in red, right bottom panel) which promotes Calypso’s catalytic activity towards ubiquitin. The triad catalytic sites: C131, H214, D228 are shown in red (right bottom panel).
Mechanistically, Calypso/BAP1 DUB activity requires a stable interaction with a conserved domain, termed DEUBAD (DEUBiquitinase ADaptor) in Asx and ASXLs38,97,103,104. BAP1 adopts a conformational configuration through its association with the DEUBAD forming a Composite Ubiquitin-Binding Interface (CUBI). This interface is essential for BAP1 binding and deubiquitination of H2Aub97.
Structural studies of ubiquitin bound to a minimal PR-DUB complex indicated that BAP1/Calypso catalytic domain engages with ubiquitin through the hydrophobic patch and the C-terminal tail of ubiquitin103,105 (Fig. 3b). The overall model for PR-DUB-ubiquitin also revealed a third binding mode between BAP1/Calypso and ubiquitin, which involves the DEUBAD. The DEUBAD increases Calypso-ubiquitin binding by favouring a suitable arrangement between the C-terminal domain (CTD) and the UCH (Ubiquitin C-terminal Hydrolase) catalytic domain103,105. This includes a stabilization of the crossover loop (an amino acid extension that limits substrate access to the active site) and direct interactions between the DEUBAD and ubiquitin103,105 (Fig. 3b). Another layer of stabilization consists of the interaction between the DEUBAD and the crossover loop of BAP1/Calypso. Overall, the proposed model suggests that DEUBAD enhances BAP1/Calypso activation by increasing the latter’s affinity for ubiquitin, through conformational changes. Remarkably, Calypso-DEUBAD are arranged in a specific oligomerization state formed by 2:2 Calypso:DEUBAD molecules103,106. This oligomerization requires the coiled-coil domain of both Calypso molecules and involves the formation of an extended cleft separating the UCH domains. This arrangement facilitates the recruitment on nucleosomes, as dimerization defective mutants of Calypso exhibit reduced PR-DUB interaction with nucleosomes106. Importantly, oligomerization of Calypso is only required for its activity towards nucleosomal histone H2A, as the intrinsic DUB activity is not altered by mutation of the dimerization interface106.
Interestingly, the DEUBAD domain itself is predominately monoubiquitinated and this event is conserved from Drosophila to human104,107. DEUBAD monoubiquitination enhances H2Aub deubiquitination by BAP1/Calypso, providing another layer of regulation of H2Aub deubiquitination104,107. Conversely, it was recently reported that methylation of adenine N6 in DNA (N6-methyladenine) is recognized by the ASXH domain of ASXL1, triggering the degradation of BAP1/ASXL1 and preventing H2Aub deubiquitination and gene silencing108. Hence, it will be interesting to decipher how deubiquitination of H2Aub is orchestrated in response to stimuli and signaling pathways.
Readers of H2Aub
Several factors including RYBP, JARID2, the Zuotin-Related Factor (ZRF1) and the Remodeling and Spacing Factor 1 (RSF1) have been proposed as readers for H2Aub85,86,109–112 (Fig. 4). RYBP plays an important role in the deposition of H2Aub and genome-wide binding of RYBP is strongly reduced following depletion of RING1A/RING1B in mESCs16,42,45,112. In defining how RYBP interacts with chromatin, it was initially found that RYBP can bind ubiquitin and H2Aub through a NZF (Nuclear protein localization 4 Zinc Finger) ubiquitin-binding domain84,109 (Fig. 4a). On the other hand, RYBP was also shown to act as an activator of PRC142,84,109. However, in the latter studies, the impact of RYBP-H2Aub binding was not clearly separated from its E3 ligase stimulatory effect42,84. Interestingly, vPRC1 complexes containing PCGF3/5 promote Xist RNA-dependent X chromosome inactivation through RYBP-H2Aub binding85. Notably, following Xist induction in RYBP knockout mESCs, RYBP, but not its H2Aub-binding defective mutant (T31A/F32A), rescues PcG-mediated deposition of H2Aub and H3K27me3 on inactive X chromosome85. More recently, it was revealed that RYBP-mediated H2A ubiquitination is strongly increased on partially ubiquitinated versus unmodified nucleosome arrays112. This stimulatory effect is dependent on the ability of RYBP to bind H2Aub and is increased by nucleosome array compaction with histone H1. Moreover, using a gene reporter assay, it was observed that H2Aub is propagated in vivo through the ability of RYBP to bind H2Aub, thus providing an epigenetic mechanism for H2Aub maintenance during cell division112. In summary, RYBP appears to ensure a positive-feedback mechanism by promoting H2Aub propagation and spreading. Moreover, the role of this factor in allosteric regulation of PRC1 might also be orchestrated with its H2Aub reader function. Structural studies of PRC1-RYBP bound to nucleosomes should provide insights into H2Aub deposition and propagation by RYBP. On the other hand, taking into account that H2Aub is erased during mitosis and deposited in G1113, and that the half-life of H2Aub is relatively short114,115, the mechanism by which RYBP would contribute to the global maintenance of H2Aub is perhaps more complex than anticipated and further studies are required to better define the dynamics of H2Aub deposition and removal.
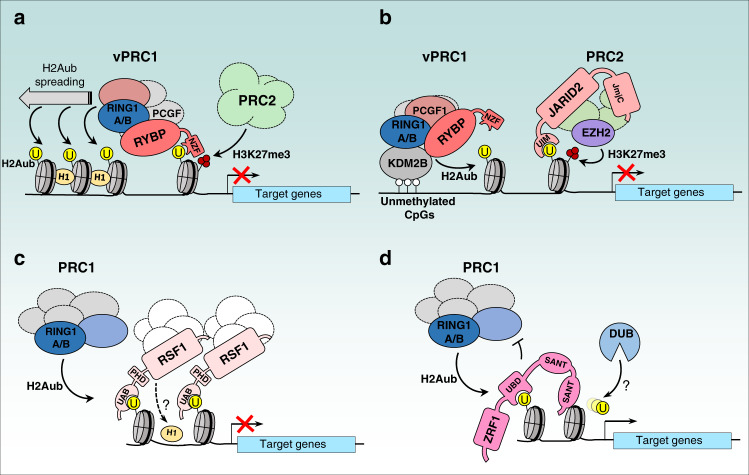
Fig. 4. H2Aub readers in transcription regulation.
a The vPRC1 component RYBP uses its NZF-UBD to bind H2Aub. This binding is increased with nucleosome compaction with histone H1 to promote H2Aub spreading and a repressive state of chromatin. b Positive-feedback loop between PRC1 and PRC2 complexes is ensured by the binding of vPRC1 component, KDM2B to unmethylated CpGs island promoting H2Aub deposition. This results in a specific binding of nucleosomal H2Aub by the PRC2 component JARID2 through its UIM domain. JARID2 binding promotes the recruitment of the PRC2 complex and eventually the establishment of H3K27me3 by EZH2 and maintenance of gene repression. c RSF1 binds H2Aub through its UAB domain and promotes the recruitment of the RSF1 chromatin-remodeling complex. This ensures proper nucleosome arrangement at target gene promoters with maintenance of histone H1 positioning, ensuring gene transcription repression in collaboration with PRC1 complexes. d ZRF1 uses its UBD Ubiquitin-Binding Domain to bind H2Aub, thus blocking the access of PRC1 to the nucleosomes, and likely promoting recruitment of DUBs, allowing ubiquitin removal and transcription activation. Ubiquitin is shown as a yellow circle and H3K27me3 is shown by the three red circles.
JARID2 promotes PRC2-EZH2 enzymatic activity through its N-terminal domain116. Insights into the mechanism of activation were provided by structural studies showing that JARID2 stabilizes the PRC2 complex and allosterically promote PRC2-mediated H3K27 methylation117–120. It was also found that JARID2 uses its N-terminal ubiquitin interaction motif (UIM) to specifically bind nucleosomal H2Aub and this results in increased H3K27 trimethylation, thus defining a positive-feedback loop between PRC1 and PRC2 (Fig. 4b)83,86. These findings are in agreement with several reports, demonstrating the dependency of PRC2 recruitment on H2A ubiquitination13,15,63, although further studies on the link between H2Aub binding by JARID2 and H3K27me3 deposition by PRC2 are needed. Notably genome-wide studies of both histone PTMs, following gene replacement in vivo, using discrete mutations of JARID2 in its UIM will help elucidating the link between H2Aub and H3K27me3. It will also be interesting to further dissect, at the structural level, how JARID2 binding to H2Aub is linked to its stimulatory effect of EZH2 methyltransferase activity.
The remodeling and spacing factor RSF1 was also recently proposed as a reader of H2Aub111. ChIP-seq analysis showed an overlap between RSF1 and H2Aub peaks and depletion of RING1B appears to reduce RSF1 chromatin recruitment. Knockdown of RSF1 induces target gene derepression without alteration of RING1B protein levels or H2A ubiquitination state111. A model was proposed that implicates the binding of RSF1 to H2Aub in order to promote nucleosomal stability and facilitate transcriptional silencing111 (Fig. 4c). However, it remains unclear how RSF1 stabilizes chromatin through H2Aub binding and how RSF1 interacts with PcG factors to ensure repression.
In contrast to the transcription silencing effects of the factors described above, ZRF1 was found as a potential reader of H2Aub with the opposite outcome110. ZRF1 binds directly to H2Aub, through its conserved zuotin domain, and appear to compete with PRC1 on many of its target genes110. Following retinoic acid induction, over half of ZRF1 genomic targets overlap with those of RING1B and H2Aub. The transcriptional activation of the overlapping targets was shown to be dependent on the presence of ZRF1, suggesting that the recruitment of this factor facilitates derepression of PRC1 targets (Fig. 4d)110. It will be interesting to further dissect the mechanism that orchestrates ZRF1 recruitment and its link with H2Aub deubiquitination and gene derepression. ZRF1 would likely rely on additional interacting factors to efficiently bind H2Aub and compete with PRC1. The potential identification of these factors as well as structural studies on ZRF1-chromatin interaction might provide insights into the mechanisms of gene activation by ZRF1.
In summary, several factors were proposed as readers of ubiquitinated H2A. However, a deeper understanding of their contributions in PcG silencing or gene activation will require further studies. In particular, additional questions relate to the chromatin contexts and the signaling events that might dictate which potential H2Aub reader needs to be recruited to coordinate gene expression.
Functions of H2Aub in chromatin-associated processes
Roles of H2Aub in transcriptional repression
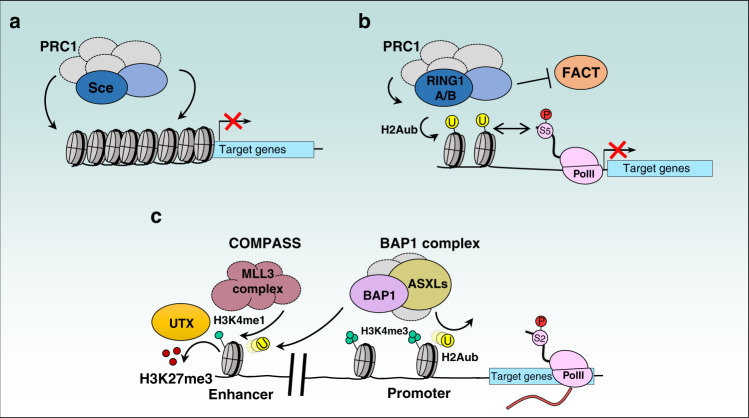
Inactivation of PRC1 E3 ligase activity in Drosophila, through the use of a Sce-E2 ubiquitin conjugating-interaction defective mutant (I48A), results in the depletion of H2Aub, but has no visible effects on the expression of the Hox genes Antennapedia (Antp), Ultrabithorax (Ubx), and Abdominal-B (Abd-B)19. However, the complete absence of Sce induces major embryotic homeotic transformations associated with deregulation of Hox gene expression19. In addition, replacing histone H2A gene cluster with an ubiquitination-defective mutant allele leads to similar conclusions that absence of H2Aub does not impact Hox gene expression19. These results are consistent with previous studies showing that PRC1 promotes chromatin compaction independently of its catalytic activity89,121. Interestingly, loss of PRC1 during Drosophila embryonic development results in higher order opening of chromatin at Hox loci122. Overall, while H2Aub is still required for viability of Drosophila embryos, the presence of Sce, but not its catalytic activity, is required to maintain Hox gene repression (Fig. 5a), indicating that PRC1 possess H2Aub-dependent and independent chromatin-associated activities.
Fig. 5. Regulation of gene transcription by H2Aub ubiquitination and deubiquitination.
a Recruitment of PRC1 complexes to compact chromatin regions in H2Aub-independent manner. These chromatin domains become inaccessible to the transcriptional machinery and are thus repressed. b H2A ubiquitination could contribute to gene repression by blocking the FACT histone chaperone. This would block nucleosome rearrangement resulting in maintenance of poised RNA PolII at promoters and inhibition of transcription elongation of specific target genes. Poised PolII is phosphorylated on serine 5. c BAP1 DUB complexes act at enhancers to recruit MLL3 and UTX to deposit H3K4me1 and demethylate H3K27me3, respectively. These events along with deubiquitination of H2Aub promote gene activation. Ubiquitination is shown in yellow circle, H3K27me3 is shown by the three red circles, H3K4me1/3 is shown by green circles and phosphorylation by bigger red circle.
In mammalian cells, initial studies using E2-interaction defective mutant of RING1B (e.g., I53A), provided initial evidence that enzymatic activity of RING1B is important for PcG gene silencing and maintenance of mESC identity28. However, RING1B I53A mutation does not completely abolish the E3 ligase activity15,16,123. Using RING1B mutants completely defective in E3 ligase activity (e.g., I53A/D56K), it was shown that PRC1 catalytic activity is required for neuronal gene silencing in neural stem or progenitor cells123. Using similar approaches in mESCs, it was later shown that PRC1-mediated ubiquitination is critical for repression of PcG target genes15,16. Thus, PRC1 catalytic activity plays an important role in transcriptional repression in higher organisms. Mechanistically, H2Aub may interfere with the function of RNA Polymerase II (RNA Pol II). In mESC, depletion of both RING1A/RING1B induces a loss of H2Aub and release of poised RNA Pol II, and this was associated with transcription elongation and increased gene expression124 (Fig. 5b). On the other hand, H2A ubiquitination appears to inhibit the recruitment of the histone chaperone FACT (Facilitates Chromatin Transcription), thus blocking nucleosomal rearrangement and subsequent early gene transcription elongation125 (Fig. 5b). However, it remains largely unclear how H2Aub inhibits FACT recruitment and whether this involves intermediate factors harboring ubiquitin-binding domains.
On the other hand, depletion of the PR-DUB complex in Drosophila results in the global increase of H2Aub levels and this was unexpectedly associated with transcriptional derepression of Ubx, in a DUB activity-dependent manner38. While the exact mechanism by which Calypso represses transcription remains elusive, these results support the notion that ubiquitination and deubiquitination of H2A might be dynamically involved in regulating Hox gene expression38. Moreover, this raises the question of whether mammalian BAP1 can also possess transcription repression properties. In investigating its potential functions in gene expression, BAP1 was initially described as a transcription activator100. BAP1 co-activator function correlated with increased H3K4me3 and concomitant depletion of H2Aub and H3K27me3 at target genes126–129. Consistent with this, BAP1 was also shown to promote the recruitment of KMT2C (MLL3) and KDM6A (UTX) to enhancers, thus ensuring H3K4me1 deposition and H3K27me3 removal, respectively130 (Fig. 5c). Of note, BAP1, but not Calypso, possesses a larger insertion in the middle of the protein that is responsible for interaction with HCF-1, OGT, and FOXKs38,97,131 (Fig. 3a). Notably HCF-1 is part of TrxG complexes2, and this might explain BAP1 transcription activation properties that would be coordinated with H2A deubiquitination. Moreover, FOXKs were shown to play an important role in BAP1-mediated H2Aub deubiquitination and transcriptional regulation129,131,132. Overall, while the general consensus is that BAP1-mediated H2A deubiquitination in mammalian systems promotes transcriptional activation127–129, there is also evidence for BAP1-mediated H2Aub deubiquitination promoting transcriptional repression126,132,133. Further studies are clearly needed to determine whether mammalian BAP1 has dual transcriptional regulatory functions and what are the exact cellular and chromatin contexts that involve one function versus another.
Does H2Aub play a role in transcriptional activation?
Intriguingly, gene repression was not the only outcome associated with the presence of H2Aub, as recent studies implicated PRC1 in gene activation134–136. For instance, during mESC differentiation, a transient swap of CBX7 with CBX8 subunits of PRC1 complex was shown to promote transcription activation of genes that appear to maintain a certain level of H2Aub135. However, mESCs undergoing differentiation are heterogeneous cell populations and RING1B or H2Aub ChIP signals detected at target genes might be contributed by non-differentiated cells. vPRC1 (PRC1.5) containing the casein kinase 2 (CK2), AUTS2 and PCGF5 subunits was also recently implicated in gene activation134. AUTS2 appears to block H2A ubiquitination by recruiting CK2 which was shown to phosphorylate and inactivate RING1B. In parallel, AUTS2 could also interact with and recruit the acetyltransferase p300 to promote gene expression134. However, these studies relied on transcription reporter systems and further validation is needed to evaluate the function of PRC1.5 in the context of natural recruitment to target genes134. In addition, it is unclear how the proposed function of PRC1.5 can be reconciled with the recently described PCGF5 function in H2Aub deposition and X chromosome inactivation32,85. Similarly, transcription activation in quiescent B and T lymphocytes also appears to be a manifestation of PRC1’s counterintuitive activity. Both the Aurora B kinase and the PRC1 complex appear to be recruited to actively transcribed genes136. In this case, Aurora B was shown to phosphorylate the E2 ubiquitin-conjugating enzyme UBCH5C to prevent RING1B-mediated H2A ubiquitination. However, the exact molecular mechanism of gene activation remains unclear136. In addition, the H2Aub-independent function of PRC1 was mostly associated with gene repression and chromatin compaction121,137,138. Thus, it remains to be determined, at the mechanistic level, how PRC1 complex composition can influence gene activation.
Clearly, additional work is needed to better define how H2Aub exactly regulates transcription and which chromatin contexts and factors act in concert or antagonism with this modification in the regulation of gene expression.
Role of H2Aub in DNA damage signaling and repair
The cellular response to genotoxic stress involves multiple signaling and repair factors, many of which are associated with chromatin function. Notably, the early responders, ATM (Ataxia Telangiectasia-Mutated) and ATR (ATM- and Rad3-related) kinases, phosphorylate several factors, including the histone variant H2AX, leading to the ordered assembly of multi-protein complexes that ensure DNA damage recognition and execution of the repair process139–142. It was initially found that UV-induced helix-distorting DNA lesions promote the local recruitment of RING1B and subsequent ubiquitination of H2A in ATR-dependent manner143. The initial incision at the DNA damage site by the nucleotide excision repair (NER) machinery appears to be required for H2Aub deposition at DNA damage sites, although the precise impact of H2Aub on the execution of the NER process remains unknown143.
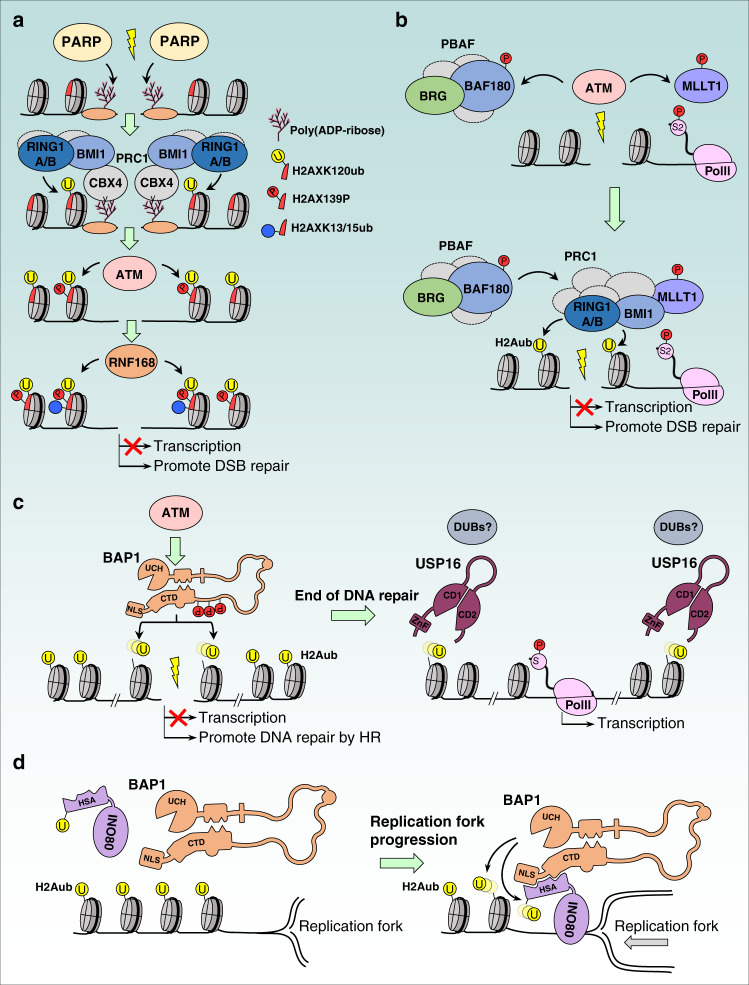
The role of H2Aub in DNA damage signaling has been mostly examined in the context of DNA double strand breaks (DSBs). Following induction of DSBs and phosphorylation of H2AX (γH2AX), an orchestrated ubiquitin-signaling cascade involving the ubiquitin ligases RNF8 and RNF168 leads to the monoubiquitination of H2AK13/K15 followed by the recruitment of 53BP1 and BRCA1 repair factors and DSB repair141,144. Using laser micro-irradiation-induced localized DNA damage, it was found that BMI1 and RING1B are rapidly and transiently recruited to DSB sites to promote the ubiquitination of γH2AXK120ub (γH2AXK120 is the corresponding ubiquitination site of H2AK119) and subsequent assembly of DNA repair factors145. These events are dependent on PARP-mediated poly(ADP-ribosyl)ation, which occurs rapidly following DNA break formation145–147. The initial recruitment of BMI1 and RING1B occurs through the binding of CBX4 to poly(ADP-ribose) prior to noticeable phosphorylation of H2AX and RNF8/RNF168 signaling145,147 (Fig. 6a). Thus, DNA damage induces a rapid recruitment of PcG factors and γH2AXK120 ubiquitination, which facilitates H2AK13/K15 ubiquitination and DSB signaling145,147. Of note, in many studies on DNA damage signaling, H2AK119ub (H2Aub) and H2AXK120ub (H2AXub) cannot be distinguished by immunostaining or ChIP, as both PTMs are recognized by antibodies against these PTMs148–151. Therefore, the specific contribution of each of these histone marks in DNA damage signaling and repair remains unclear.
Fig. 6. Involvement of H2Aub in DNA double strand break signaling and replication fork progression.
a Induction of DNA double strand breaks (DSBs) induces a rapid recruitment of PARP to DNA damage sites and subsequent protein poly(ADP-ribosyl)ation. Poly(ADP-ribose) serves as an anchor for the PRC1 complex, through its CBX4 subunit which leads to the recruitment of PRC1 to DNA damage site and ubiquitination of the histone variant H2AX. This ubiquitination event induces a more efficient recruitment of ATM and phosphorylation of H2AX (γH2AX), a hallmark of the DNA damage response. This in turn leads to the recruitment of RNF168 with catalyzes H2AXK13/K15 ubiquitination. Altogether, this cascade of posttranslational modifications leads to effective recruitment of the DNA repair machinery and inhibition of transcription. b Specific recruitment of the PRC1 complex to DNA DSB sites could be enhanced through interaction between BMI1 and the phosphorylated form of the transcription elongation factor MLLT1. Phosphorylation of PBAF also promotes the recruitment of the PRC1 complex to the site of DNA damage. c BAP1 is phosphorylated following DNA damage and is also recruited to DSB site to promote homologous recombination (HR). BAP1 appears to deubiquitinate H2Aub only near the DSB site, likely promoting DNA-end resection. Once the repair process is completed, the deubiquitinase USP16 or other DUBs could remove H2Aub mark more widely, allowing normal gene transcription. CD1 and CD2: the two parts of the catalytic domain. d During DNA synthesis, the chromatin-remodeling factor INO80 is protected from proteasomal degradation by BAP1. On the other hand, BAP1 deubiquitinates H2A and promotes coordinated DNA replication. HSA, Helicase-SANT associated domain. Ubiquitination is shown by the yellow circle, and phosphorylation by the red circle.
Mechanistically, it is still not well-defined how γH2AXub or H2Aub promotes DNA repair, but an attractive hypothesis that was originally proposed consists of the involvement of H2Aub/γH2AXub in transcription repression of regions neighboring DNA damage in order to facilitate the repair process145,146. Indeed, genomic regions around DSB sites become silenced in an ATM-dependent manner (Fig. 6a)148. When BMI1/RING1B is inhibited, ATM-dependent transcriptional silencing no longer occurs115. Interestingly, the chromatin-remodeling complex PBAF, which also acts downstream of ATM, appears to mediate transcriptional silencing in response to DSBs through PRC1/PRC2 recruitment149. The presence of the PBAF complex and ATM-mediated phosphorylation of its subunit BAF180 appears to be required for efficient deposition of H2Aub/γH2AXub and rapid DNA repair (Fig. 6b). On the other hand, phosphorylation of the transcriptional elongation factor MLLT1, component of the Super Elongation Complex (SEC), by ATM enhances its interaction with BMI1150. MLLT1 would normally promote transcription elongation. Instead, its phosphorylation promotes the subsequent recruitment of the PRC1 complex to DNA damage sites, H2Aub/γH2AXub deposition and transcriptional silencing (Fig. 6b)150. How PBAF and MLLT1 functions are coordinated at the DSB sites remains an interesting question to address.
The roles of H2Aub/γH2AXub in the DNA damage response were also revealed through investigating the contribution of DUBs in DSB repair. BAP1 promotes DSB repair, likely as a part of its tumor suppressor function96,151,152. BAP1 is phosphorylated by ATM, which promotes its recruitment to the DSB site. BAP1 might subsequently remove ubiquitin from H2Aub/γH2AXub, but only at the vicinity of the DSB site151. BAP1 might facilitate chromatin organization near DSB sites, and this might lead to the initiation of DNA-end resection and subsequent DSB repair by homologous recombination151,152 (Fig. 6c). USP16 would remove H2Aub/γH2AXub following the accomplishment of DNA repair promoting the recovery of gene transcription148 (Fig. 6c), although a direct role of USP16 in DSB signaling has been recently debated153. Despite these findings, the exact mechanism behind DUB recruitment and removal of ubiquitin from H2Aub/γH2AXub at DSB sites is not known. In addition, the temporal order of DUB recruitment at DSB sites and their specific roles in DNA damage signaling and repair require further studies.
In summary, while the importance of H2Aub/γH2AXub in DNA damage signaling and repair processes is becoming increasingly established, a mechanistic understanding is needed to explain how this histone PTM acts in concert with other histone modifications and chromatin-associated complexes to orchestrate transcription and DNA repair at DNA damage sites.
Roles of H2Aub in other DNA-associated processes
H2Aub was also implicated in other DNA-associated processes including DNA replication and chromosome segregation. Early observations revealed that H2Aub could form distinct foci which partially colocalized with Proliferating Cell Nuclear Antigen (PCNA)154. This colocalization was enhanced following inhibition of DNA replication, suggesting a potential regulatory mechanism involving H2Aub at replication forks154. It was also shown that the major satellite regions at pericentric heterochromatin—a condensed and silenced form of chromatin found near centromeres and is characterized by hypoacetylated histones and the presence of H3K9me3 ― are decorated with H2Aub12. It was proposed that H2Aub might be involved in the progression of the replication fork in these heterochromatic regions12. Recently, it was found that INO80 chromatin-remodeling factor associates with BAP1, which appears to promote INO80 recruitment to replication forks155. In addition, BAP1 also targets INO80 for deubiquitination thus promoting its stability at the replication fork (Fig. 6d)155,156. It was proposed that the presence of H2Aub provides an anchor-like function to recruit BAP1/INO80 to ensure proper replication fork progression and genomic stability155,156. However, how the dynamics of H2Aub deposition and removal impacts the progression of the replication fork remains largely unknown.
Changes in H2Aub levels were also associated with mitotic progression. It was previously observed that H2Aub levels are strongly reduced at the onset of M phase and re-established in early G1113,157. Depletion of USP16 was shown to prevent the mitotic decrease of H2Aub levels and inhibit cell cycle progression158. Moreover, H2Aub levels were reported to be inversely correlated with histone H3S10 phosphorylation by Aurora B kinase hinting to a potential crosstalk between these two histone modifications158. Mechanistically, USP16 appears to directly deubiquitinate H2Aub and this was shown to influence histone H3S10 phosphorylation (H3S10P), chromosome segregation and mitotic progression158. However, it was recently shown that H2Aub deubiquitination does not precede H3S10P deposition153. Thus, the link between phosphorylation of H3S10 and deubiquitination of H2Aub remains unclear.
Deregulation of H2Aub in human pathologies
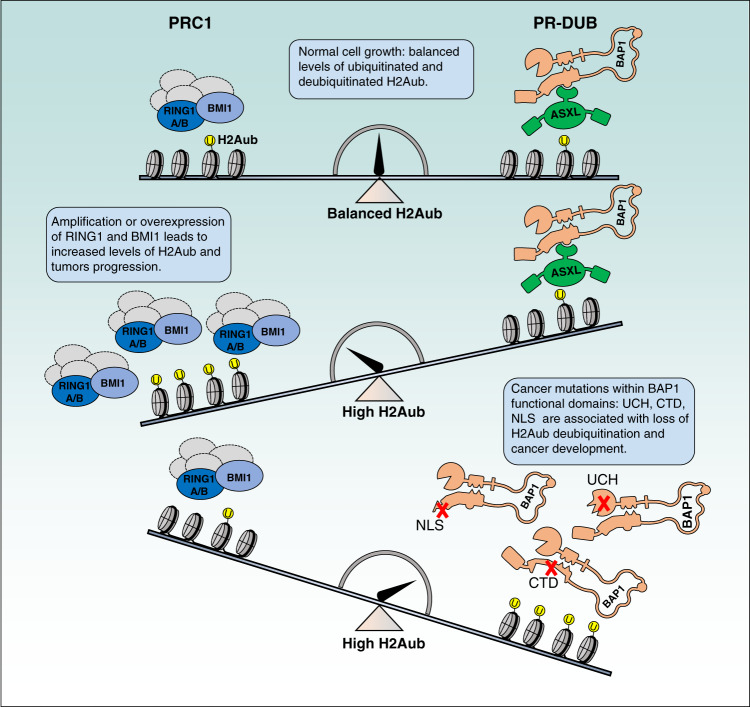
Several components of the PRC1 complexes, including RING1B and BMI1, are overexpressed in cancer159–163. Upregulation of BMI1 is associated with increased proliferation, reduced apoptosis and increased capacity of stem cell self-renewal163–166. The oncogenic properties of BMI1 appear to involve its transcriptional repressor function. BMI1 is a negative regulator of several target genes controlling cell cycle progression such as p16Ink4a, p19Arf, p21WAF1/CIP1, in addition to the tumor suppressor PTEN160,163,167–169. However, oncogenic functions of BMI1 that are independent of H2Aub have also been proposed162,170. On the other hand, there are also evidence for tumor suppressor activities for some PRC1 components including PHC3 and KDM2B171,172. Importantly, the relationship between the function of several PRC1 proteins and H2Aub in tumorigenesis remains unclear.
The potential role of H2Aub and cancer is also provided by the DUB BAP1 and its co-factors ASXLs. BAP1 is a tumor suppressor mutated in multiple cancers101,173–182. Cancer mutations of the catalytic domain compromise the DUB activity of BAP1181. Interestingly, a cancer mutation consisting of a small in-frame deletion in the CTD domain of BAP1 results in its selective dissociation from ASXLs and inability to deubiquitinate H2Aub and regulate cell proliferation97. In further linking BAP1-mediated H2A deubiquitination and tumor suppression, it was found that loss of BAP1 results in the repression or activation of genes that regulate cell death126,128. While the role of H2Aub in repression versus activation has not been directly tested in these studies, the data suggest that catalytic activity of BAP1 regulates cell death and tumor suppression126,128. BAP1 inactivation also compromises its ability to promote DNA repair and genomic integrity96,151,152,156, although it is currently unclear what is the contribution of BAP1 function in DNA repair for tumor suppression.
Overall, an emerging model of H2Aub-associated tumorigenesis involves the gain of function of PRC1 or the loss of function of PR-DUB with the resulting increase of H2Aub levels on chromatin. Uncontrolled H2Aub deposition on chromatin might be a key determinant in cancer development (Fig. 7). However, despite the number of studies indicating an important role of BAP1 in tumor suppression, recent studies also provided evidence for a potential gain of function of BAP1/ASXLs in myeloid malignancies29,107,183. Thus, further studies are needed to firmly determine how altered deposition of H2Aub exactly alters chromatin function and promotes oncogenic signaling.
Fig. 7. Model for the potential role of H2Aub in cancer development.
PRC1 mediates H2Aub deposition and represses target genes that are negative regulators of cell cycle progression and cell death. PRC1 also promotes stem cell self-renewal. PRC1 overexpression results in increased cell ability to overcome cell cycle control, reduced apoptosis as well as increased capacity of stem cell self-renewal. BAP1 regulates cell proliferation and promotes cell death. Inactivation of BAP1 results in increased H2Aub deposition at target genes and modulation of cell cycle and resistance to cell death. PRC1 and BAP1 also regulate DNA repair, defect of which can promote carcinogenesis. BAP1 cancer-associated mutations disrupting BAP1 functional domains are shown by the red cross. Ubiquitination is shown by the yellow circle.
Mutations in several components implicated in modulation of H2Aub levels have been associated with several syndromes and brain pathologies. For instance, mutations in AUTS2 are associated with autism and intellectual disability184. Similarly, defects in PHC1 are linked to brain development disorders such as autosomal recessive primary microcephaly. PHC1 mutations are associated with impaired H2A ubiquitination, cell cycle progression and DNA repair185. PR-DUB complex is also associated with rare developmental syndromes. For example, germline mutations of ASXL1 are found in patients with congenital disorders such as the Bohring-Opitz Syndrome, a very rare syndrome characterized by multiple congenital malformations and severe intellectual disabilities186. ASXL2 and ASXL3 mutations are also linked to developmental disorders187,188. Notably, Bainbridge-Ropers syndrome patients with mutated ASXL3 exhibit higher levels of H2Aub which correlated with differential gene expression outcomes189. Nonetheless, the exact mechanisms that mediate the effects of ASXLs mutations in the pathogenesis of these diseases are largely unknown. Overall, defects in either components that establish or erase H2Aub are observed in disease conditions, suggesting an important role of this PTM in normal physiology. The possibility to conduct genome-wide mapping for this modification, in conjunction with gene expression analysis and functional assays following gene targeting, would help to elucidate how deregulation of H2Aub underlies human disease.
Concluding remarks
Almost half a century of research on H2Aub revealed important aspects of its function. However, how H2Aub coordinates cellular processes and how its deregulation contributes to human disease remains incompletely defined. At the mechanistic level, how H2Aub deposition is translated into changes of chromatin function that can further impact transcription or DNA repair remains to be fully established. A major knowledge gap also remains regarding the regulation of H2Aub. In particular, PRC1 complexes are targeted by PTMs and it is unclear how these PTM translate signaling pathways into changes in H2Aub levels and chromatin function. On the other hand, recent studies provided possible mechanisms of H2Aub propagation. However, additional studies are needed to determine the mechanisms of H2Aub propagation in the context of natural recruitment and how its spreading is orchestrated by PRC1 and PR-DUB/BAP1. Clearly, future work is needed to better position H2Aub in the epigenomic landscape and determine how deregulation of this modification underlies human disease.
Acknowledgements
This work was supported by grants from the Natural Sciences and Engineering; Research Council of Canada (2015-2021) to E.B.A. E.B.A. is a senior scholar of the Fonds de la Recherche du Québec-Santé (FRQ-S). H.B. received a PhD scholarship from the Ministry of Higher Education and Scientific Research of Tunisia and the Cole Foundation. SD is a recipient of the Banting Postdoctoral Fellowship.
Author contributions
H.B. and E.B.A. proposed the topic for this review. H.B., S.D., M.H., and E.B.A. collected previous references and wrote the manuscript. H.B., S.D., and E.B.A. realized the illustrations.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 3.Poynter ST, Kadoch C. Polycomb and trithorax opposition in development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:659–688. doi: 10.1002/wdev.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Valencia AM, Kadoch C. Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. cell Biol. 2019;21:152–161. doi: 10.1038/s41556-018-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marazzi I, Greenbaum BD, Low DHP, Guccione E. Chromatin dependencies in cancer and inflammation. Nat. Rev. Mol. Cell Biol. 2018;19:245–261. doi: 10.1038/nrm.2017.113. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 10.Bratzel F, Lopez-Torrejon G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol.: CB. 2010;20:1853–1859. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 11.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Bravo M, et al. Polycomb RING1A- and RING1B-dependent histone H2A monoubiquitylation at pericentromeric regions promotes S-phase progression. J. Cell Sci. 2015;128:3660–3671. doi: 10.1242/jcs.173021. [DOI] [PubMed] [Google Scholar]
- 13.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scelfo A, et al. Functional landscape of PCGF proteins reveals both RING1A/B-dependent-and RING1A/B-independent-specific activities. Mol. Cell. 2019;74:1037–1052. doi: 10.1016/j.molcel.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackledge NP, et al. PRC1 catalytic activity is central to polycomb system function. Mol. Cell. 2020;77:857–874. doi: 10.1016/j.molcel.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamburri S, et al. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol. Cell. 2020;77:840–856. doi: 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez L, et al. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development. 2012;139:117–127. doi: 10.1242/dev.074450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pengelly AR, Kalb R, Finkl K, Muller J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015;29:1487–1492. doi: 10.1101/gad.265439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davie JR, Lin R, Allis CD. Timing of the appearance of ubiquitinated histones in developing new macronuclei of Tetrahymena thermophila. Biochem. Cell Biol. = Biochim. Biologie Cellulaire. 1991;69:66–71. doi: 10.1139/o91-009. [DOI] [PubMed] [Google Scholar]
- 21.Veluchamy A, et al. An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum. Genome Biol. 2015;16:102. doi: 10.1186/s13059-015-0671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldknopf IL, et al. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 23.Baarends WM, et al. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 1999;207:322–333. doi: 10.1006/dbio.1998.9155. [DOI] [PubMed] [Google Scholar]
- 24.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J. Biol. Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 25.Baarends WM, et al. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell. Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HG, Kahn TG, Simcox A, Schwartz YB, Pirrotta V. Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res. 2015;25:1170–1181. doi: 10.1101/gr.188920.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Romero-Campero FJ, Gomez-Zambrano A, Turck F, Calonje M. H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 2017;18:69. doi: 10.1186/s13059-017-1197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endoh M, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balasubramani A, et al. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat. Commun. 2015;6:7307. doi: 10.1038/ncomms8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen I, et al. PRC1 fine-tunes gene repression and activation to safeguard skin development and stem cell specification. Cell Stem Cell. 2018;22:726–739. doi: 10.1016/j.stem.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pivetti S, et al. Loss of PRC1 activity in different stem cell compartments activates a common transcriptional program with cell type-dependent outcomes. Sci. Adv. 2019;5:eaav1594. doi: 10.1126/sciadv.aav1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fursova NA, et al. Synergy between variant PRC1 complexes defines polycomb-mediated gene repression. Mol. Cell. 2019;74:1020–1036. doi: 10.1016/j.molcel.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z, et al. Kdm2b regulates somatic reprogramming through variant PRC1 complex-dependent function. Cell Rep. 2017;21:2160–2170. doi: 10.1016/j.celrep.2017.10.091. [DOI] [PubMed] [Google Scholar]
- 35.Kuroda MI, Kang H, De S, Kassis JA. Dynamic competition of polycomb and trithorax in transcriptional programming. Annu. Rev. Biochem. 2020;89:1–19. doi: 10.1146/annurev-biochem-120219-103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagarou A, et al. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheuermann JC, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfieri C, et al. Structural basis for targeting the chromatin repressor Sfmbt to Polycomb response elements. Genes Dev. 2013;27:2367–2379. doi: 10.1101/gad.226621.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milne TA, Sinclair DA, Brock HW. The Additional sex combs gene of Drosophila is required for activation and repression of homeotic loci, and interacts specifically with Polycomb and super sex combs. Mol. Gen. Genet. 1999;261:753–761. doi: 10.1007/s004380050018. [DOI] [PubMed] [Google Scholar]
- 41.Fisher CL, et al. Additional sex combs-like 1 belongs to the enhancer of trithorax and polycomb group and genetically interacts with Cbx2 in mice. Dev. Biol. 2010;337:9–15. doi: 10.1016/j.ydbio.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R, et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure. 2010;18:966–975. doi: 10.1016/j.str.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junco SE, et al. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure. 2013;21:665–671. doi: 10.1016/j.str.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, et al. A Non-canonical BCOR-PRC1.1 complex represses differentiation programs in human ESCs. Cell Stem Cell. 2018;22:235–251. doi: 10.1016/j.stem.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon J, Chiang A, Bender W, Shimell MJ, O’Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- 49.Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell. 1998;1:1057–1064. doi: 10.1016/S1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 51.Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- 52.Brown JL, Fritsch C, Mueller J, Kassis JA. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development. 2003;130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 53.Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frey F, et al. Molecular basis of PRC1 targeting to Polycomb response elements by PhoRC. Genes Dev. 2016;30:1116–1127. doi: 10.1101/gad.279141.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alhaj Abed J. et al. De novo recruitment of Polycomb-group proteins in Drosophila embryos. Development145, dev165027 (2018). [DOI] [PMC free article] [PubMed]
- 57.Holowatyj A, Yang ZQ, Pile LA. Histone lysine demethylases in Drosophila melanogaster. Fly (Austin) 2015;9:36–44. doi: 10.1080/19336934.2015.1074787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng Y, et al. The lysine demethylase dKDM2 is non-essential for viability, but regulates circadian rhythms in Drosophila. Front. Genet. 2018;9:354. doi: 10.3389/fgene.2018.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng Y, et al. A developmental genetic analysis of the lysine demethylase KDM2 mutations in Drosophila melanogaster. Mech. Dev. 2014;133:36–53. doi: 10.1016/j.mod.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernstein E, et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farcas AM, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He J, et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cooper S, et al. Targeting polycomb to pericentric heterochrom; atin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 65.Ren X, Kerppola TK. REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol. Cell. Biol. 2011;31:2100–2110. doi: 10.1128/MCB.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trojer P, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell. 2011;42:438–450. doi: 10.1016/j.molcel.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu M, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol. Cell. 2012;45:330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dietrich N, et al. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012;8:e1002494. doi: 10.1371/journal.pgen.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 2018;14:e1007193. doi: 10.1371/journal.pgen.1007193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poux S, Melfi R, Pirrotta V. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 2001;15:2509–2514. doi: 10.1101/gad.208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 72.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Kahn TG, et al. Interdependence of PRC1 and PRC2 for recruitment to Polycomb response elements. Nucleic Acids Res. 2016;44:10132–10149. doi: 10.1093/nar/gkw701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leeb M, et al. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang C, et al. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol.: CB. 2013;23:1324–1329. doi: 10.1016/j.cub.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 77.Aoyama K, et al. Ezh1 targets bivalent genes to maintain self-renewing stem cells in Ezh2-insufficient myelodysplastic syndrome. iScience. 2018;9:161–174. doi: 10.1016/j.isci.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat. Commun. 2019;10:1679. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zylicz JJ, et al. The implication of early chromatin changes in X chromosome inactivation. Cell. 2019;176:182–197. doi: 10.1016/j.cell.2018.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu X, et al. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 2008;36:3590–3599. doi: 10.1093/nar/gkn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cao Q, et al. The central role of EED in the orchestration of polycomb group complexes. Nat. Commun. 2014;5:3127. doi: 10.1038/ncomms4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 84.Rose N. R. et al. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. Elife5, e18591 (2016). [DOI] [PMC free article] [PubMed]
- 85.Almeida M, et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science. 2017;356:1081–1084. doi: 10.1126/science.aal2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cooper S, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016;7:13661. doi: 10.1038/ncomms13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Healy E, et al. PRC2.1 and PRC2.2 synergize to coordinate H3K27 trimethylation. Mol. Cell. 2019;76:437–452. doi: 10.1016/j.molcel.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 88.Chiacchiera F, et al. Polycomb complex PRC1 preserves intestinal stem cell identity by sustaining Wnt/beta-catenin transcriptional activity. Cell Stem Cell. 2016;18:91–103. doi: 10.1016/j.stem.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 89.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 90.Li Z, et al. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J. Biol. Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 91.Buchwald G, et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bentley ML, et al. Recognition of Marazzi et al. (2018) UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaytan de Ayala Alonso A, et al. A genetic screen identifies novel polycomb group genes in Drosophila. Genetics. 2007;176:2099–2108. doi: 10.1534/genetics.107.075739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mashtalir N, et al. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell. 2014;54:392–406. doi: 10.1016/j.molcel.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 96.Bononi A, et al. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature. 2017;546:549–553. doi: 10.1038/nature22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Daou S, et al. The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. J. Biol. Chem. 2015;290:28643–28663. doi: 10.1074/jbc.M115.661553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sowa ME, Bennett EJ, Gygi S;P, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu H, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dey A, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hauri S, et al. A high-density map for navigating the human Polycomb complexome. Cell Rep. 2016;17:583–595. doi: 10.1016/j.celrep.2016.08.096. [DOI] [PubMed] [Google Scholar]
- 103.Sahtoe DD, van Dijk WJ, Ekkebus R, Ovaa H, Sixma TK. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 2016;7:10292. doi: 10.1038/ncomms10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Daou S, et al. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nat. Commun. 2018;9:4385. doi: 10.1038/s41467-018-06854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De I, et al. Structural basis for the activation of the deubiquitinase Calypso by the Polycomb protein ASX. Structure. 2019;27:528–536. doi: 10.1016/j.str.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 106.Foglizzo M, et al. A bidentate Polycomb repressive-deubiquitinase complex is required for efficient activity on nucleosomes. Nat. Commun. 2018;9:3932. doi: 10.1038/s41467-018-06186-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Asada S, et al. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat. Commun. 2018;9:2733. doi: 10.1038/s41467-018-05085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kweon SM, et al. An adversarial DNA N(6)-methyladenine-sensor network preserves Polycomb silencing. Mol. Cell. 2019;74:1138–1147. doi: 10.1016/j.molcel.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arrigoni R, et al. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006;580:6233–6241. doi: 10.1016/j.febslet.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 110.Richly H, et al. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature. 2010;468:1124–1128. doi: 10.1038/nature09574. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Z, et al. Role of remodeling and spacing factor 1 in histone H2A ubiquitination-mediated gene silencing. Proc. Natl Acad. Sci. USA. 2017;114:7949–7958. doi: 10.1073/pnas.1711158114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao J, et al. RYBP/YAF2-PRC1 complexes and histone H1-dependent chromatin compaction mediate propagation of H2AK119ub1 during cell division. Nat. Cell Biol. 2020;22:439–452. doi: 10.1038/s41556-020-0484-1. [DOI] [PubMed] [Google Scholar]
- 113.Matsui SI, Seon BK, Sandberg AA. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc. Natl Acad. Sci. USA. 1979;76:6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seale RL. Rapid turnover of the histone-ubiquitin conjugate, protein A24. Nucleic Acids Res. 1981;9:3151–3158. doi: 10.1093/nar/9.13.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ismail IH, McDonald D, Strickfaden H, Xu Z, Hendzel MJ. A small molecule inhibitor of polycomb repressive complex 1 inhibits ubiquitin signaling at DNA double-strand breaks. J. Biol. Chem. 2013;288:26944–26954. doi: 10.1074/jbc.M113.461699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanulli S, et al. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell. 2015;57:769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Justin N, et al. Structural basis of oncogenicBratzel et al., (2010) histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kasinath V, et al. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018;359:940–944. doi: 10.1126/science.aar5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 122.Cheutin T, Cavalli G. Loss of PRC1 induces higher-order opening of Hox loci independently of transcription during Drosophila embryogenesis. Nat. Commun. 2018;9:3898. doi: 10.1038/s41467-018-05945-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsuboi M, et al. Ubiquitination-independent repression of PRC1 targets during neuronal fate restriction in the developing mouse neocortex. Dev. Cell. 2018;47:758–772. doi: 10.1016/j.devcel.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 124.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 125.Zhou W, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymede Napoles et al., (2004)rase II transcriptional elongation. Mol. Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 2018;20:1181–1192. doi: 10.1038/s41556-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Campagne A, et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat. Commun. 2019;10:348. doi: 10.1038/s41467-018-08255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He M, et al. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science. 2019;364:283–285. doi: 10.1126/science.364.6439.506. [DOI] [PubMed] [Google Scholar]
- 129.Kolovos, P. et al. PR-DUB maintains the expression of critical genes through FOXK1/2- and ASXL1/2/3-dependent recruitment to chromatin and H2AK119ub1 deubiquitination. Genome Res.30, 1119–1130 (2020). [DOI] [PMC free article] [PubMed]
- 130.Wang L, et al. Resetting the epigenetic balance of Pol; ycomb and COMPASS function at enhancers for cancer therapy. Nat. Med. 2018;24:758–769. doi: 10.1038/s41591-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ji Z, et al. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res. 2014;42:6232–6242. doi: 10.1093/nar/gku274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Okino Y, Machida Y, Frankland-Searby S, Machida YJ. BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J. Biol. Chem. 2015;290:1580–1591. doi: 10.1074/jbc.M114.609834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dai F, et al. BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc. Natl Acad. Sci. USA. 2017;114:3192–3197. doi: 10.1073/pnas.1619588114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao Z, et al. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–354. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M. A Cbx8-containing polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLoS Genet. 2014;10:e1004851. doi: 10.1371/journal.pgen.1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Frangini A, et al. The aurora B kinase and the polycomb protein ring1B combine to regulate active promoters in quiescent lymphocytes. Mol. Cell. 2013;51:647–661. doi: 10.1016/j.molcel.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 137.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol. Cell. 2001;8:545–556. doi: 10.1016/S1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 138.Eskeland R, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 140.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 142.Uckelmann M, Sixma TK. Histone ubiquitination in the DNA damage response. DNA repair. 2017;56:92–101. doi: 10.1016/j.dnarep.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 143.Bergink S, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 145.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chou DM, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl Acad. Sci. USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ismail IH, et al. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40:5497–5510. doi: 10.1093/nar/gks222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kakarougkas A, et al. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell. 2014;55:723–732. doi: 10.1016/j.molcel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ui A, Nagaura Y, Yasui A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol. Cell. 2015;58:468–482. doi: 10.1016/j.molcel.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 151.Yu H, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc. Natl Acad. Sci. USA. 2014;111:285–290. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ismail IH, et al. Germ-line mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 2014;74:4282–4294. doi: 10.1158/0008-5472.CAN-13-3109. [DOI] [PubMed] [Google Scholar]
- 153.Sen Nkwe N, et al. A potent nuclear export mechanism imposes USP16 cytoplasmic localization during interphase. J. Cell Sci. 2020;133:1–19. doi: 10.1242/jcs.239236. [DOI] [PubMed] [Google Scholar]
- 154.Vassilev AP, Rasmussen HH, Christensen EI, Nielsen S, Celis JE. The levels of ubiquitinated histone H2A are highly upregulated in transformed human cells: partial colocalization of uH2A clusters and PCNA/cyclin foci in a fraction of cells in S-phase. J. Cell Sci. 1995;108:1205–1215. doi: 10.1242/jcs.108.3.1205. [DOI] [PubMed] [Google Scholar]
- 155.Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat. Commun. 2014;5:5128. doi: 10.1038/ncomms6128. [DOI] [PubMed] [Google Scholar]
- 156.Lee HS, et al. BAP1 promotes stalled fork restart and cell survival via INO80 in response to replication stress. Biochem. J. 2019;476:3053–3066. doi: 10.1042/BCJ20190622. [DOI] [PubMed] [Google Scholar]
- 157.Mueller RD, Yasuda H, Hatch CL, Bonner WM, Bradbury EM. Identification of ubiquitinated histones 2A and 2B in Physarum polycephalum. Disappearance of these proteins at metaphase and reappearance at anaphase. J. Biol. Chem. 1985;260:5147–5153. [PubMed] [Google Scholar]
- 158.Joo HY, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 159.Bea S, et al. BMI-1 gene amplification and overexpression in hematological malignancies occur mainly in mantle cell lymphomas. Cancer Res. 2001;61:2409–2412. [PubMed] [Google Scholar]
- 160.Martinez-Romero C, et al. The epigenetic regulators Bmi1 and Ring1B are differentially regulated in pancreatitis and pancreatic ductal adenocarcinoma. J. Pathol. 2009;219:205–213. doi: 10.1002/path.2585. [DOI] [PubMed] [Google Scholar]
- 161.Bosch A, et al. The Polycomb group proteinTamburri (840-) RING1B is overexpressed in ductal breast carcinoma and is required to sustain FAK steady state levels in breast cancer epithelial cells. Oncotarget. 2014;5:2065–2076. doi: 10.18632/oncotarget.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chan HL, et al. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 2018;9:3377. doi: 10.1038/s41467-018-05728-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Su W, et al. The Polycomb Repressor Complex 1 drives double-negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell. 2019;36:139–155. doi: 10.1016/j.ccell.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 165.Abdouh M, et al. BMI1 sustains human glioblastoma multiforme stem cell renewal. J. Neurosci. 2009;29:8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 2010;30:10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 168.Song LB, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J. Clin. Investig. 2009;119:3626–3636. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fasano CA, et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 170.Zhu S, et al. BMI1 regulates androgen receptor in prostate cancer independently of the polycomb repressive complex 1. Nat. Commun. 2018;9:500. doi: 10.1038/s41467-018-02863-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Deshpande AM, et al. PHC3, a component of the hPRC-H complex, associates with E2F6 during G0 and is lost in osteosarcoma tumors. Oncogene. 2007;26:1714–1722. doi: 10.1038/sj.onc.1209988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Isshiki Y, et al. KDM2B in polycomb repressive complex 1.1 functions as a tumor suppressor in the initiation of T-cell leukemogenesis. Blood Adv. 2019;3:2537–2549. doi: 10.1182/bloodadvances.2018028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wiesner T, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Testa JR, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat. Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Goldstein AM. Germline BAP1 mutations and tumor susceptibility. Nat. Genet. 2011;43:925–926. doi: 10.1038/ng.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bott M, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pena-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jiao Y, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Popova T, et al. Ger; mline BAP1 mutations predispose to renal cell carcinomas. Am. J. Hum. Genet. 2013;92:974–980. doi: 10.1016/j.ajhg.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Carbone M, et al. BAP1 and cancer. Nat. Rev. Cancer. 2013;13:153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ohar JA, et al. Germline BAP1 mutational landscape of asbestos-exposed malignant mesothelioma patients with family history of cancer. Cancer Res. 2016;76:206–215. doi: 10.1158/0008-5472.CAN-15-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yang H, et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131:328–341. doi: 10.1182/blood-2017-06-789669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Oksenberg N, Ahituv N. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet.: TIG. 2013;29:600–608. doi: 10.1016/j.tig.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Awad S, et al. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum. Mol. Genet. 2013;22:2200–2213. doi: 10.1093/hmg/ddt072. [DOI] [PubMed] [Google Scholar]
- 186.Hoischen A, et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 2011;43:729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- 187.Bainbridge MN, et al. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5:11. doi: 10.1186/gm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shashi V, et al. De novo truncating variants in ASXL2 are associated with a unique and recognizable clinical phenotype. Am. J. Hum. Genet. 2016;99:991–999. doi: 10.1016/j.ajhg.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Srivastava A, et al. De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge-Ropers syndrome. Hum. Mol. Genet. 2016;25:597–608. doi: 10.1093/hmg/ddv499. [DOI] [PMC free article] [PubMed] [Google Scholar]