Abstract
Inhibition of neuropeptide Y1 receptor stimulates osteogenesis in vitro and in vivo. However, the underlying mechanisms involved in these effects remain poorly understood. Here we identify the effects of Y1 receptor deficiency on osteogenic differentiation in human bone marrow stromal cells (BMSCs) by using genetic and pharmacological regulation, and to explore the pathways mediating these effects. In BMSCs, inhibition of Y1 receptor stimulates osteogenesis and upregulates the expression levels of the master transcriptional factor RUNX2. Mechanistically, Y1 receptor deficiency increases the levels of intracellular cAMP, which via protein kinase A (PKA) mediated pathways results in activation of phospho-CREB (p-CREB). We find RUNX2 activation induced by Y1 receptor deficiency is reversed by H-89, a PKA inhibitor. These results indicate Y1 receptor deficiency activates PKA-mediated phosphorylation of CREB, leading to activation of RUNX2 and enhances osteogenic differentiation in BMSCs. In conclusion, these data indicate that Y1 receptor deficiency promotes osteogenic differentiation by RUNX2 stimulation through cAMP/PKA/CREB pathway.
Keywords: neuropeptide Y1 receptor, RUNX2, osteogenesis, bone marrow stromal cells (BMSCs), protein kinase A
Introduction
Neuropeptide Y receptors belong to the G protein-coupled receptor (GPCR) family and are widely expressed in the central nervous system and peripheral tissues (1, 2). Among the five NPY receptors (Y1, Y2, Y4, Y5, and Y6 receptors), Y1 is the first characterized receptor which plays important roles in cancer (1), energy homeostasis (3), and bone metabolism (4). In vivo, Y1 receptor ablation enhances osteoblast differentiation and bone formation, thus results in a high bone mass phenotype of mice (5). Pharmacological blockade of Y1 receptors by antagonists also improves bone mass in mice (6). In vitro, bone marrow-derived mesenchymal stem cells (BMSCs) isolated from Y1 receptor deletion mice show greater potentials to differentiated into mature osteoblast under the conditioned medium (7). Furthermore, We found that Y1 receptor knockdown rescued osteogenesis inhibition induced by glucocorticoid in preosteoblastic MC3T3-E1 cells (8). These findings together suggest Y1 receptor deficiency promotes osteogenesis. However, the underlying mechanisms involved in these effects remain elusive.
Human bone marrow stromal cells (hBMSCs) have the potential to differentiate into the osteogenic lineage given the appropriate conditions (9). This process is regulated by numerous osteogenesis factors including RUNX2, Osterix, Wnt-β-catenin, parathyroid hormone (PTH), and bone morphogenetic protein (BMP) (10). As the master transcriptional factor of osteogenesis, RUNX2 regulates osteogenic differentiation and is essential for normal skeletal formation as loss of RUNX2 in mice results in impaired postnatal skeletogenesis (11). RUNX2 regulates osteogenic differentiation by induction of specific osteoblast differentiation markers, namely, Osterix (12), ALP (alkaline phosphatase), OPN (osteopontin), BSP (bone sialoprotein), and OCN (osteocalcin) (13, 14).
Meanwhile, studies have showed that protein kinase A (PKA) signaling plays a critical role in osteogenic differentiation of BMSCs (15, 16). PKA is preferentially activated by increased level of cyclic adenosine monophosphate (cAMP), which then translocates into the nucleus and activates the cAMP response element-binding protein (CREB) at Ser-133. Activation of phospho-PKA (p-PKA) and phospho-CREB (p-CREB) contributes to osteogenic differentiation of BMSCs (15). The pro-osteogenic effects of cAMP/PKA/CREB signaling in BMSCs are associated with the induction of bone-related cytokines such as BMP2, TGF-β, IGF-1 (15), osteogenic markers including RUNX2 (17), and Osterix (18).
Interestingly, the association between NPY receptors and cAMP/PKA/CREB signaling has been widely reported. Activated by the ligands (neuropeptide Y, peptide YY, pancreatic polypeptide), Y receptors preferentially couple to Gi/o G-proteins, and thus inhibit adenylyl cyclase and lead to reduced cAMP level in islet, cancer, and fat tissues (1, 19, 20). Whereas Y1 receptor deficiency increases cAMP levels, which via PKA/CREB pathway results in improved outcome of islet transplantation (21). In osteoblastic cells, NPY is reported to reduce cAMP production and inhibit osteoblast differentiation (22, 23). Particularly, Y1 receptors are highly expressed in BMSCs. However, the interaction between cAMP/PKA/CREB signaling and Y1 receptor in the regulation of osteogenic differentiation in BMSCs remains unknown. Here we test whether the pro-osteogenic effects of Y1 receptor deficiency are associated with cAMP/PKA/CREB signaling.
In the present study, we hypothesize that inhibition of Y1 receptor signaling might increase RUNX2 levels and stimulate osteogenic differentiation due to the activation of cAMP/PKA pathway in human BMSCs.
Materials and Methods
Isolation and Culture of hBMSCs
Bone marrow aspirates (5 ml) were isolated from the posterior iliac crest from five healthy volunteers (3 males and 2 female; aged 20–40 years). The volunteers had similar lifestyles (low-moderate level of physical activity; no smoking habit), and all of them denied the history of long-term intake of calcium, vitamin D or other potentially nutrients influencing bone metabolism. For female subjects, the biopsy was obtained at the same stage of the period. Informed consent was obtained from all volunteers and the study was approved by the Institutional Review Board (IRB) of the Second Affiliated Hospital, Wenzhou Medical University (IRB No. 2018-14). hBMSCs were isolated as we previously described (24). Cells from each donor were cultured independently, and experiments were repeated at least 3 times. hBMSCs were cultured in Dulbecco’s modified Eagle media (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 1% penicillin and streptomycin (Gibco) at 37°C in a 5% CO2 atmosphere. Cells grown to 80% to 90% confluency were subcultured by using 0.25% trypsin-EDTA (Gibco), and the third passage of cells was used in the following experiments. To determine the effects of Y1 receptor deficiency on osteogenic differentiation, hBMSCs were cultured in osteogenic media (growth culture media supplemented with 10−8 M dexamethasone (Sigma), 50 μg/ml ascorbic acid (Gibco) and 5 mM β-glycerol phosphate (Sigma) for 5 to 10 days. The media were refreshed twice a week during osteogenic differentiation.
Cell Proliferation Assay
Cell proliferation rates were evaluated by using a cell counting kit (CCK-8, Dojindo), according to the manufacturer’s instructions. Briefly, cells were seeded in 96-well plates and each well was treated with 10 μl CCK-8 reagent for 4 h. Absorbance at 570 nm was read on a microplate reader (Bio-Rad). Cell proliferation rates were expressed as fold changes relative to the con groups.
Alkaline Phosphatase/Alizarin Red S Staining and Assay
Alkaline phosphatase (ALP) staining was performed at day 5 in BMSCs during osteogenic differentiation. Cells rinsed with phosphate-buffered saline (PBS) were fixed with 4% paraformaldehyde, and then ALP staining was performed using an ALP substrate mixture (ALP staining kit, Sigma) for 30 min in darkness. ALP activity was colorimetrically determined at 405 nm using a Sigma kit with p-nitrophenyl phosphate (pNPP) as the substrate. The protein contents were measured using a protein assay kit (Bio-Rad) based on the bicinchoninic acid (BCA) method, and the overall enzyme activity was presented as units/mg protein.
Alizarin red S (ARS) staining was performed at day 10 during osteogenic differentiation. Cells were fixed in ice-cold 70% ethanol and then stained with 3% alizarin red S solution (Sigma) for 30 min. The bound dye was then colorimetrically detected at 595 nm after de-staining with 10% cetylpyridinium chloride monohydrate (Sigma) for 20 min. The quantification of ARS staining was conducted by drawing an Alizarin Red standard curve.
Real-time Polymerase Chain Reaction (PCR) Assay
Real-time PCR was performed as previously reported (8), to detect the expression of several osteogenic differentiation related marker genes (RUNX2, COL-1A1, ALP, OCN, OPN, BSP, SOX9 and PPAR-γ) at day 3. Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions. Total RNA was reverse-transcribed into cDNAs using an Omniscript kit (Qiagen). mRNA expression was measured by using DyNamo SYBR1 Green qPCR kit (Takara). Primer sequences were designed as follows: RUNX2, forward primer 5′-GCGTCAACACCATCATTCTG-3′ and reverse primer 5′-CAGACCAGCAGCACTCCATC-3′; COL-1A1, forward primer 5′-GAGAGCATGACCGAT GGAT-3′ and reverse primer 5′- ATGTTTTGGTGGTTCAGGAGG-3′; ALP, forward primer 5′-GAGAGC ATGACCGATGGAT-3′ and reverse primer 5′-ATGTTTTGGTGGTTCAGGAGG-3′; OCN, forward primer 5′-AGAGCCCCAGTCCCCTACCC-3′ and reverse primer 5′-AGGCCTCCTGAAAGCCGATG-3′; OPN, forward primer 5′-CCGTTGCCCAGGACCTGAA-3′ and reverse primer 5′-TGTGGCTGTGGGTTTCAGCA-3′; BSP, forward primer 5′- TGCCTTGAGCCTGCCTGCTTCC-3′ and reverse primer 5′- CAAAATTAAAGCAGTCTTCATTTTG-3′; Y1 receptor, forward primer CTCGCTGGTTCTCATCGCTGTGGAACGG and reverse primer GCGAATGTATATCTTGAAGTAG; SOX9, forward primer 5′- TGGGCAAGCTCTGGAGACTTC-3′ and reverse primer 5′- ATCCGGGTGGTCCTTCTTGTG-3′; PPAR-γ, forward primer 5′- GCCGAGAAGGAGAAGC-3′ and reverse primer 5′-TGGTCAGCGGGAAGG-3′; PKA, forward primer 5′-AAAGAATGGGCAACCAGTG′ and reverse primer 5′-GCTGACCCCTAAAATAATG-3′; GAPDH, forward primer 5′-ACCACAGTCCATGCCATCAC-3′ and reverse primer 5′-TCCAC- CACCCTGTTGCTGTA-3′. Gene expression was calculated by normalizing its level to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH, an internal control) using the 2−ΔΔCT methods.
Western Blot
Total protein was extracted with a passive lysis kit (Promega) and quantified. Protein resolved by SDS-PAGE (Sigma) was then transferred to PVDF membranes. Membranes were incubated with blocking buffer (5% bovine serum albumin (BSA, Sigma)) for 2 h, and then incubated at 4°C overnight with antibodies against Y1 receptor (Abcam, No. 35336), RUNX2 (Millipore, No. 05-1478), p-PKA (Cell Signaling, No.9621), total PKA (Cell Signaling, No.4782), p-CREB (Cell Signaling, No.9198), total CREB (Cell Signaling, No.9193), p-caspase3 (Cell Signaling, No.9664), p-JNK (Santa Cruz, No. sc-6254), p-ERK (Cell Signaling, No.9101), p-P38 (Cell Signaling, No. 9211), Lamin B (Santa Cruz, No. sc-374015), LDH (Santa Cruz, No. sc-133123). These antibodies were diluted in TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% (v/v) Tween 20). After washing, protein expression was detected by using a Western Chemiluminescent HRP Substrate Kit (Millipore), with GAPDH (Cell Signaling, No.8884) as the endogenous control. Quantitative densitometric values of the detected bands were quantified using the NIH Image J Software. We performed all Western blot assays at least three times.
Nuclear and Cytosolic Fractionation
Nuclear and cytoplasmic fractionation was conducted by using the Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Briefly, cells collected by trypsinization were washed with ice-cold PBS and lysed with ice-cold 0.1% NP-40 (Sigma). The lysates were collected as the “whole cell lysate.” After centrifugation, the supernatants were collected as the “cytosolic fraction.” The pelleted “nuclear fraction” was further lysed in RIPA solution (Beyotime) with protease inhibitor cocktail tablets (Roche) and then collected. Protein in these fractions was analyzed by Western blot analysis.
Small Hairpin RNA (shRNA) Plasmid Transduction
Methods for shRNA plasmid transduction and infection were previously described (8). In brief, hBMSCs were transfected with Y1 receptor shRNA plasmid (36098-SH, Santa Cruz), PKA shRNA plasmid (sc-39162-SH, Santa Cruz) and shCon by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. 48 h post-transfection, the cells were cultured in selective media including 4 μg/mL puromycin (Sigma). After the media being replaced every 3 days, the third passage of stable clones was collected in the following experiments. Resistant clones were pooled to generate polyclonal cells, and silence of targeted genes was verified by real-time PCR and Western blot analysis.
Luciferase Reporter Assay
To assess luciferase activity, hBMSCs were transfected with pCRE-luciferase reporter plasmid (Agilent Technologies) and the internal control pRL-SV40 (Promega) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were cultured in osteogenic media for 3 days with shRNA or H-89 treatment. The target luciferase activity was analyzed using the Dual-Luciferase1 Reporter Assay System (Promega) by normalizing to the Renilla luciferase activity.
Intracellular Cyclic Adenosine Monophosphate (cAMP) and PDE Activity Assay
Intracellular cAMP was quantitated using a direct cAMP ELISA kit (Enzo Life Sciences) according to the manufacturer’s instructions. Briefly, cells seeded into plates were scraped and lysed with 0.1 M HCl. Intracellular cAMP levels in the cell lysates were colorimetrically detected at 405 nm using the cAMP ELISA Kit. The final cAMP concentrations were calculated after being normalized by total protein content. For PDE activity assay, cell lysates were centrifuged and the supernatants were collected for further analysis. After purification by gel filtration, the supernatants were detected at 620 nm using a PDE Activity Assay Kit (Abcam) according to the manufacturer’s instructions. IBMX (a potent PDE inhibitor, 10 μM) was applied as the negative control. Quantification of the PDE activity was conducted with drawing a standard curve.
Statistical Analysis
Experiments in this study were repeated in triplicate in each of the 5 donors (N=5), and the data were presented as mean ± standard deviation (SD). Statistical analyses were determined by one-way ANOVA followed by a post hoc Dunnett’s test or Student’s t-test. The SPSS 18.0 software was used for data management. Values of P < 0.05 were considered statistically significant.
Results
Effects of Y1 Receptor Regulation on the Viability and Differentiation of BMSCs
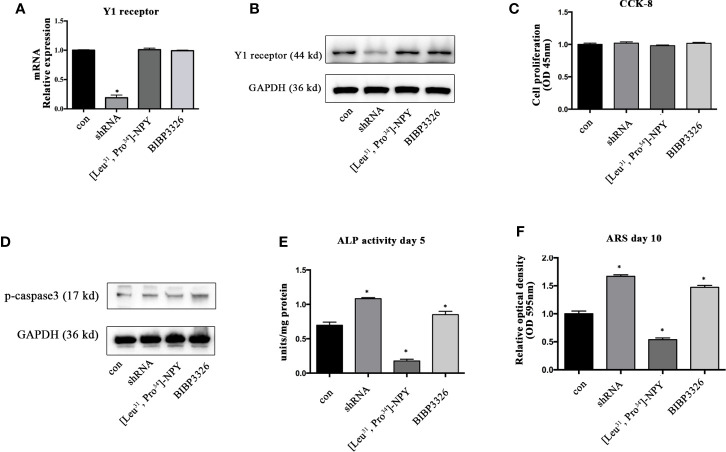
To regulate the expression levels of Y1 receptor in BMSCs, we used shRNA interference, pharmacological agonist [Leu31, Pro34]-NPY (10−7 M) and antagonist BIBP3226 (10−7 M). The level of Y1 receptor mRNA was significantly decreased after shRNA treatment, suggesting a high efficiency of shRNA interference. However, neither agonist nor antagonist of Y1 receptor influenced the mRNA expression of Y1 receptor ( Figure 1A ). The results of Western blot showed the similar trends in protein levels ( Figure 1B ). We then explored the effects of Y1 receptor regulation on cell proliferation and apoptosis. The results of CCK-8 demonstrated that gene and pharmacological regulation of Y1 receptor did not affect cell proliferation ( Figure 1C ). Since caspase-3 has been widely known as the key factor regulating cell apoptosis (25), we assessed cell apoptosis by detecting the phosphorylation of caspase-3 and found Y1 receptor did not affect cell apoptosis ( Figure 1D ). We then investigated the effects of Y1 receptor regulation on osteogenic differentiation in BMSCs by two measurements: ALP activity and calcium nodule formation. Inhibition of Y1 receptor signaling by shRNA or antagonist promoted osteogenic differentiation in BMSCs, as shown by an increase of alkaline phosphatase (ALP) activity and calcium nodules formation. In contrast, treatment with Y1 receptor agonist inhibited osteoblast differentiation of BMSCs with a low amount of ALP and calcium nodules ( Figures 1E, F ). These results suggested that inhibition of Y1 receptor signaling enhances osteoblast differentiation of BMSCs, which was consistent with the findings as we reported in the osteoblastic MC3T3 cells (8). Particularly, we found the pro-osteogenic effects of Y1 receptor deficiency by shRNA was relatively significant than that by pharmaceutical treatment, thus we used shRNA transfection to silenced Y1 receptor in the following study.
Figure 1.
The effects of Y1 receptor signaling regulation on the viability and osteogenic differentiation of BMSCs. (A, B) BMSCs were treated with Y1 receptor shRNA, Y1 receptor agonist [Leu33, Pro34]-NPY and antagonist BIBP3326. Levels of Y1 receptor mRNA were detected by real-time PCR at day 3, with GAPDH as the internal control; protein expression of Y1 receptor were assessed by Western blot at day 3. (C) Cell proliferation was measured using a CCK-8 kit at day 3. (D) Cell apoptosis was measured by expression levels of phosphorylated casepase-3 (p-casepase3) by Western blot at day 3. (E) BMSCs were cultured in osteogenic media for 5 days. ALP activity was colorimetrically measured at 405 nm to determine the extent of osteoblast differentiation. (F) Alizarin Red S staining (ARS) was performed in BMSCs cultured in the osteogenic medium for 10 days. Quantification of Alizarin Red S staining was colorimetrically detected at 595 nm to determine the calcium nodule formation (N = 5). Data are presented as mean ± SD; *p < 0.05 compared to control. con: cells cultured in a normal or osteogenic medium; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; OD, optical density (data presented in A, C, E, and F is pooled; data presented in B, D is pooled).
Inhibition of Y1 Receptor Signaling Promoted RUNX2 Expression Through p-PKA Activation During Osteogenic Differentiation
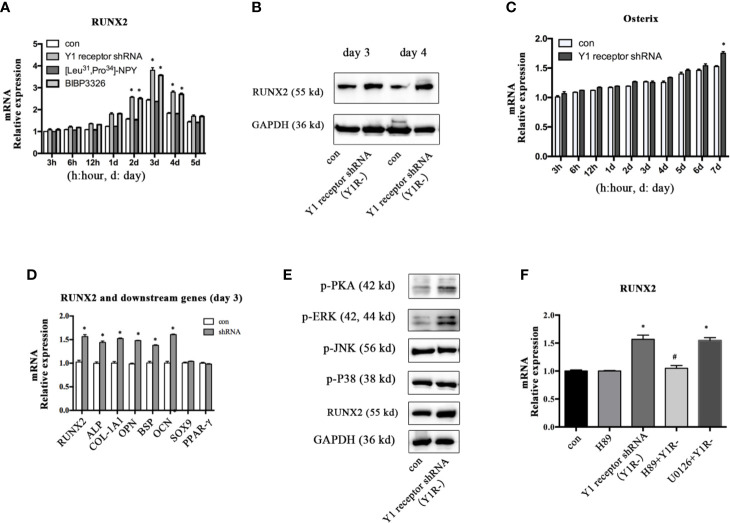
To explore the molecular mechanisms mediating the osteogenic effects of Y1 receptor deficiency, we analyzed the expression levels of the transcriptional factor RUNX2 and Osterix, which were known to play critical roles during osteogenic differentiation. The expression levels of RUNX2 mRNA peaked on day 3 and gradually decreased thereafter during BMSCs osteogenesis. Inhibition of Y1 receptor signaling enhanced the levels of RUNX2 mRNA during BMSCs osteogenesis ( Figure 2A ). Consistently, RUNX2 protein expressions on day 3 and day 4 were increased by Y1 receptor deficiency ( Figure 2B ). The expression levels of Osterix gradually increased during differentiation, and there was a significant difference in the expression levels of Osterix with Y1 receptor shRNA treatment for 7 days ( Figure 2C ). Since RUNX2 acted as the upstream of Osterix (12), and the most significant changes in RUNX2 expression occurred on day 3 during differentiation, we thus explored the levels of target genes of RUNX2 on day 3. The target osteogenic genes of RUNX2, such as ALP, COL-1A1, OPN, BSP, and OCN, were also increased by Y1 receptor deficiency on day 3 ( Figure 2D ). It has been reported that RUNX2 activity was modulated by several upstream signalings including PKA (26, 27), MAPK components (ERK, P38 and JNK pathways) (27, 28). We further investigated how Y1 receptor deficiency promoted RUNX2 expression by detecting these signaling pathways. The levels of p-P38 and p-JNK signaling proteins were not affected by Y1 receptor knockdown. While the levels of p-PKA and p-ERK signaling proteins were increased by Y1 receptor knockdown ( Figure 2E ), thus PKA and ERK signaling pathways were activated by Y1 receptor deficiency. Moreover, H-89 (a PKA inhibitor, 10−5 M) but not U0126 (a ERK inhibitor, 10−5 M) reversed RUNX2 induction by Y1 receptor deficiency ( Figure 2F ). These results demonstrated Y1 receptor deficiency enhanced RUNX2 expression through PKA pathway, which might be the main pathway mediating the osteogenic effects of Y1 receptor deficiency.
Figure 2.
Inhibition of Y1 receptor signaling promoted RUNX2 expression and p-PKA during osteogenic differentiation. (A) Expression of RUNX2 mRNA in BMSCs during osteogenic differentiation were measured by real-time PCR. (B) BMSCs were incubated in osteogenic media for 3 and 4 days, and RUNX2 protein expression were measured by Western blot. (C) Expression of Osterix mRNA in BMSCs during osteogenic differentiation were measured by real-time PCR. (D) The levels of osteogenic genes were assessed by real-time PCR at day 3 during differentiation. (E) The levels of p-PKA, p-ERK, p-JNK, p-P38, and RUNX2 were measured by Western blot at day 3. (F) Expression of RUNX2 mRNA in BMSCs treated with Y1 deficiency, H-89 (a PKA inhibitor) and U0126 (a ERK inhibitor) were measured by real-time PCR at day 3. Real-time PCR was conducted using GAPDH as the internal control (N = 5). con: cells cultured in osteogenic media. Data are presented as mean ± SD; *p < 0.05 compared to control. GAPDH: glyceraldehyde 3-phosphate dehydrogenase (data presented in A, B, D, and F are pooled; data presented in C, E are representative). #p < 0.05 compared to the group Y1 receptor shRNA.
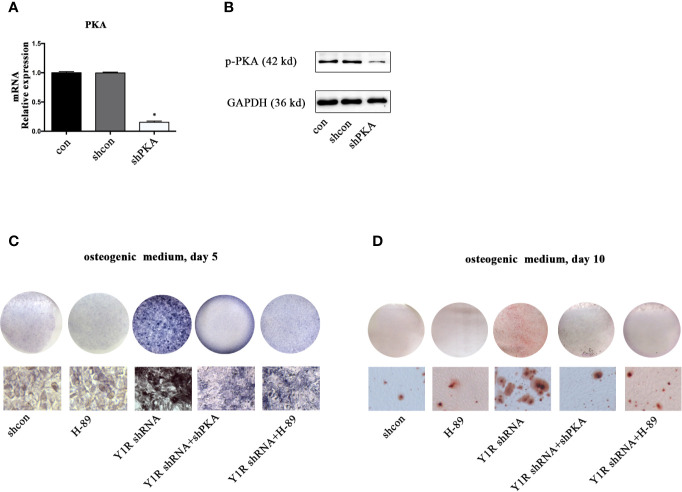
We then silenced PKA by using shRNA to verify this hypothesis. The results of real-time PCR and Western blot showed a high efficiency of shPKA ( Figures 3A, B ). Both genetic and pharmacological blockade of PKA signaling reduced ALP activity and mineralization of BMSCs ( Figures 3C, D ), indicating that Y1 receptor deficiency triggers osteogenic differentiation through activation of the PKA pathway.
Figure 3.
PKA inhibition abolished osteogenic effects of Y1 receptor deficiency in BMSCs. BMSCs were cultured in osteogenic media, with or without Y1 shRNA, shPKA and H-89 (0.1μM). ALP staining was performed on day 5 and Alizarin Red S staining was performed on day 10. (A, B) PKA expressions in mRNA and protein levels were assessed by real-time PCR and Western blot at day 3. (C) BMSCs treated with shcon, Y1 receptor shRNA, shPKA, and H-89 were cultured in osteogenic media; ALP staining was performed on day 5. (D) BMSCs treated with shcon, Y1 receptor shRNA, shPKA, and H-89 were cultured in osteogenic media; ARS staining was performed on day 10. Real-time PCR was conducted using GAPDH as the internal control (N = 5). con: cells cultured in osteogenic media (data presented in a is pooled; data presented in B–D are representative). *p < 0.05 compared to control.
PKA Inhibitor Abolished Phosphorylation of PKA Downstream Signaling Induced by Y1 Receptor Deficiency
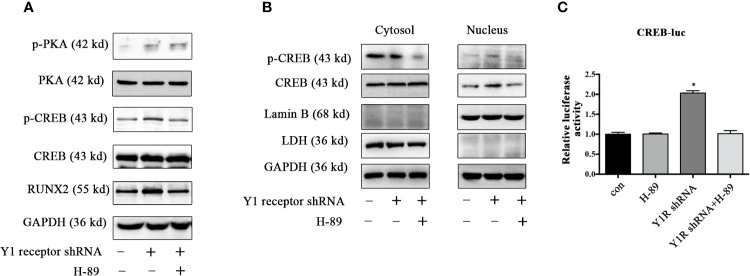
Since CREB was the main downstream signaling of PKA, we assessed the expression of p-CREB and found Y1 receptor knockdown upregulated the levels of p-PKA and p-CREB in BMSCs during osteogenic differentiation for 5 days ( Figure 4A ). Moreover, H-89 (a potent PKA inhibitor) suppressed RUNX2 and p-CREB expression induced by Y1 receptor deficiency ( Figure 4A ), indicating that PKA signaling mediated RUNX2 and p-CREB expression induced by Y1 receptor deficiency. We then assessed the cellular location of p-CREB, which was associated with nuclear import (29). The results showed that the p-CREB nuclear translocation was activated by Y1 receptor knockdown and was reversed by PKA inhibition during BMSCs differentiation ( Figure 4B ). To further investigate whether CREB transcription levels were regulated by CREB nuclear translocation induced by Y1 receptor deficiency, we conducted luciferase reporter assay thereafter. The results demonstrated that Y1 receptor deficiency increased transcription activity of CREB, while PKA inhibitor blocked this effect ( Figure 4C ). Altogether, our data indicated that PKA was the signaling pathway through which Y1 deficiency enhanced osteogenic differentiation.
Figure 4.
PKA inhibitor abolished phosphorylation of PKA downstream signaling induced by Y1 receptor deficiency. BMSCs were incubated in osteogenic media for 5 days. (A) Expression levels of PKA, CREB, and RUNX2 were quantified by Western blot analysis. (B) Cell lysates were collected and fractionated into nuclear and cytosolic extracts. Expression levels of p-CREB and CREB were analyzed by Western blot analysis. LDH was used as a control for cytosol protein samples, and Lamin B was used as a control for nuclear protein samples. (C) CREB transcriptional activity was assessed by luciferase reporter assay in BMSCs treated with Y1 shRNA or H-89 for 5 days during osteoblast differentiation. The luciferase activity of the cell lysates was measured using Dual-luciferase reporter assay system with normalization to Renilla luminescence (N = 5). Data are presented as mean ± SD; *p < 0.05 compared to control. GAPDH: glyceraldehyde 3-phosphate dehydrogenase. con: cells cultured in osteogenic media. CREB-luc: cAMP-response element binding protein (CREB) luciferase reporter plasmid (data presented in A, B are representative; data presented in C are pooled).
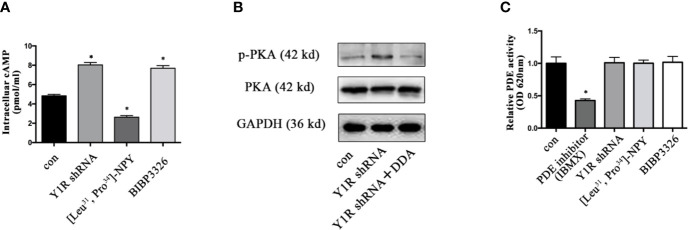
Upregulation of p-PKA Depended on the Induction of Intracellular cAMP
It was well established that cAMP induction enhanced gene expression of bone specific growth factors (15, 30), and PKA phosphorylation depended on cellular cAMP (31), we thus determined whether PKA signaling activation induced by Y1 deficiency was through cellular cAMP during BMSCs differentiation. The intracellular cAMP levels were increased in BMSCs with Y1 receptor knockdown ( Figure 5A ). Blockade of intracellular cAMP by adenylyl cyclase (AC) inhibitor 2′, 3′-dideoxyadenosine (DDA, 10−5 M, Sigma) (32, 33) significantly abolished p-PKA activation by Y1 receptor deficiency ( Figure 5B ). It has been reported phosphodiesterases (PDEs) was a hydrolase targeting cAMP (34); we thus evaluated its expression levels in BMSCs differentiation. The results showed no significant change of PDEs expression in BMSCs treated with Y1 receptor knockdown ( Figure 5C ). In conclusion, activation of the p-PKA by Y1 receptor deficiency was dependent on intracellular cAMP induction, without dependency on PDEs regulation.
Figure 5.
Y1 receptor deficiency increased the level of intracellular cAMP. (A) BMSCs were treated with Y1 receptor shRNA, [Leu31,Pro34]-NPY and BIBP3226 for 15 min. Intracellular cAMP production was measured by ELISA. (B): The expression levels of p-PKA were detected in BMSCs treated with Y1 receptor shRNA and DDA (a cAMP inhibitor, 10−5 M) by Western blot. (C) PDE activity was measured in BMSCs treated with IBMX (isobutylmethylxanthine, a PDE inhibitor, 10 µM), Y1 receptor shRNA, [Leu31, Pro34]-NPY and BIBP3226. N=5; Data are presented as mean ± SD; *p < 0.05 compared to control. con: cells cultured in a normal medium. PDE: phosphodiesterases, OD: optical density (data presented in A, C are pooled; data presented in B are representative).
Discussion
By using genetic and pharmacological approaches, our results reveal that Y1 receptor inhibition enhances osteogenesis of BMSCs through PKA signaling. This effect is mediated by nucleus translocation of p-CREB, and thus promotes RUNX2 expression. Particularly, Y1 receptor inhibition likewise increases the levels of intracellular cAMP, suggesting that intracellular cAMP mediates the activation of PKA signaling. In conclusion, we propose that inhibition of Y1 receptor promotes osteogenesis in BMSCs via cAMP/PKA/CREB pathway.
Neuropeptide Y (NPY) and its receptors are not only neuronal regulators of energy homeostasis (35), but are also known to be involved in bone homeostasis (36). As the only Y receptor expressed by BMSCs and mature osteoblast in situ (7), Y1 receptor has a negative effect on osteoblastic differentiation and bone formation. Germ-line Y1 receptor deletion mice results in a high bone mass phenotype with increased mineral node apposition (5). Furthermore, osteoblast specific knockout of Y1 receptor also enhances the differentiation potential of BMSCs and results in increased bone formation in mice, suggesting a critical role of Y1 receptor in osteoblast (7). Practically, RUNX2, a key transcription factor for osteogenesis, is upregulated by Y1 receptor silence in both the bones tissues (7) and MC3T3−E1 osteoblast cells (37). RUNX2 is critical for early-stage differentiation of BMSCs, which drives the expression of many osteoblast-specific genes (38, 39). RUNX2 knockout in mice results in lack of mature osteoblasts and decreased bone formation (11). Thus RUNX2 is one of the most important regulators in osteogenic differentiation, and we detected its activity in hBMSCs during differentiation. This study shows that RUNX2 expression and osteoblastic differentiation of hBMSCs are enhanced by Y1 receptor shRNA, which is consistent with previous reports.
To further determine the signaling pathways through which Y1 deficiency enhances osteogenic differentiation, we silence Y1 receptor by shRNA in BMSCs. The proliferation and apoptosis of BMSCs were not affected by Y1 receptor regulation, implying that Y1 receptor did not impact the viability of BMSCs. Thereafter, the osteogenic effect of Y1 receptor on BMSCs was independent of enhanced self-renewal capacity. Given the critical role of RUNX2 and Osterix in osteogenic differentiation, we then scanned their expression levels in BMSCs. Y1 receptor deficiency significantly increased RUNX2 expression at the early stage of osteogenic differentiation, which was consistent with previous reports (7, 37). In contrast, Y1 receptor deficiency enhanced Osterix expression at the relative late stage of osteogenic differentiation. Since RUNX2 is thought to be the upstream of Osterix (12), this study focuses on the expression level of RUNX2. We then hypothesize that the osteogenic effect of Y1 receptor deficiency is through upregulation of RUNX2 in BMSCs, and further explore the underlying mechanisms.
Y1 receptor, interacts with Gi/o G protein, has been reported to play an inhibitory role in adenylate cyclase (AC) activity and cAMP production (1, 19), while cAMP promotes osteoblast differentiation via PKA and CREB activation (15, 40). Moreover, cAMP production induced by Y1 receptor deficiency activates protein kinases A (PKA) and CREB phosphorylation in islets β-cells (21). It has been shown that RUNX2 activity was regulated by cAMP/PKA pathways and MAPK components (ERK, P38 and JNK pathways) (26–28). We next detected the expression levels of cAMP/PKA pathways as well as p38, JNK, and the ERK signaling in BMSCs treated with Y1 receptor silence during osteogenic differentiation. The result showed the upregulated expression of RUNX2 induced by Y1 receptor deficiency was through cAMP/PKA pathways, independent of MAPK components.
In this study, we find that cAMP production is increased by Y1 receptor deficiency in BMSCs. Since the expression of intracellular cAMP is regulated by adenylate cyclase (AC) and phosphodiesterases (PDE) activity (41), we detected the PDE activity and found it was not affected by Y1 receptor regulation. Therefore, the increased intracellular cAMP levels are rather dependent on regulation of AC than PDE activity. Y1 receptor deficiency enhances osteogenic differentiation via direct effects on cAMP production. Further support for this concept comes from studies where we could show that protein levels of p-CREB are significantly upregulated in Y1 receptor silence BMSCs.
Taken together, these data demonstrate that reduced Y1 receptor signaling exerts osteogenic effects in BMSCs by activation of RUNX2 via the cAMP/PKA/CREB pathway. Regulation of Y1 receptor signaling might offer promise as a potential therapy for osteoporosis and other bone loss conditions.
There are some limitations in this study. Primary human cells used in this study are known to produce high standard deviations. Moreover, this study focuses on the expression of RUNX2 in the early stage of differentiation, and hypothesizes that changes in RUNX2 as the cause of osteogenesis regulation by Y1 receptor. Since osteoblast differentiation takes place over a long timeframe, and transcription factor such as Osterix may also play an important role in the later stage of differentiation. This will be an issue we need to further study.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board (IRB) of the Second Affiliated Hospital, Wenzhou Medical University (IRB No. 2018-14). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
WY, S-DJ, and X-YP conceived and designed the experiments. WY, W-NX, and F-CC performed the experiments. S-LD and P-BC acquired and analyzed the data. WY drafted the manuscript. LY and X-YP helped perform the analysis with constructive discussions and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (Grant No.: 81672206); Zhejiang Province Technology Project (grant number 2015C33209); and Wenzhou Technology Project (grant number Y20150243).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all the staff of the Laboratory of the Orthopaedic Research Institute and the Scientific Research Center of the Second Affiliated Hospital of Wenzhou Medical University.
References
- 1. Zhang L, Bijker MS, Herzog H. The neuropeptide Y system: pathophysiological and therapeutic implications in obesity and cancer. Pharmacol Ther (2011) 131(1):91–113. 10.1016/j.pharmthera.2011.03.011 [DOI] [PubMed] [Google Scholar]
- 2. Czarnecka M, Lu C, Pons J, Maheswaran I, Ciborowski P, Zhang L, et al. Neuropeptide Y receptor interactions regulate its mitogenic activity. Neuropeptides (2019) 73:11–24. 10.1016/j.npep.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yulyaningsih E, Zhang L, Herzog H, Sainsbury A. NPY receptors as potential targets for anti-obesity drug development. Br J Pharmacol (2011) 163(6):1170–202. 10.1111/j.1476-5381.2011.01363.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sousa DM, Herzog H, Lamghari M. NPY signalling pathway in bone homeostasis: Y1 receptor as a potential drug target. Curr Drug Targets (2009) 10(1):9–19. 10.2174/138945009787122888 [DOI] [PubMed] [Google Scholar]
- 5. Baldock PA, Allison SJ, Lundberg P, Lee NJ, Slack K, Lin EJ, et al. Novel role of Y1 receptors in the coordinated regulation of bone and energy homeostasis. J Biol Chem (2007) 282(26):19092–102. 10.1074/jbc.M700644200 [DOI] [PubMed] [Google Scholar]
- 6. Sousa DM, Baldock PA, Enriquez RF, Zhang L, Sainsbury A, Lamghari M, et al. Neuropeptide Y Y1 receptor antagonism increases bone mass in mice. Bone (2012) 51(1):8–16. 10.1016/j.bone.2012.03.020 [DOI] [PubMed] [Google Scholar]
- 7. Lee NJ, Doyle KL, Sainsbury A, Enriquez RF, Hort YJ, Riepler SJ, et al. Critical role for Y1 receptors in mesenchymal progenitor cell differentiation and osteoblast activity. J Bone Mineral Res (2010) 25(8):1736–47. 10.1002/jbmr.61 [DOI] [PubMed] [Google Scholar]
- 8. Yu W, Zhu C, Xu W, Jiang L, Jiang S. Neuropeptide Y1 Receptor Regulates Glucocorticoid-Induced Inhibition of Osteoblast Differentiation in Murine MC3T3-E1 Cells via ERK Signaling. Int J Mol Sci (2016) 17(12). 10.3390/ijms17122150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Sci (N Y NY) (1999) 284(5411):143–7. 10.1126/science.284.5411.143 [DOI] [PubMed] [Google Scholar]
- 10. Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol (2011) 13(1):27–38. 10.1038/nrm3254 [DOI] [PubMed] [Google Scholar]
- 11. Adhami MD, Rashid H, Chen H, Clarke JC, Yang Y, Javed A. Loss of Runx2 in committed osteoblasts impairs postnatal skeletogenesis. J Bone Mineral Res (2015) 30(1):71–82. 10.1002/jbmr.2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell (2002) 108(1):17–29. 10.1016/s0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- 13. Cheng A, Genever PG. SOX9 determines RUNX2 transactivity by directing intracellular degradation. J Bone Mineral Res (2010) 25(12):2680–9. 10.1002/jbmr.174 [DOI] [PubMed] [Google Scholar]
- 14. Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, et al. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene (2004) 23(24):4315–29. 10.1038/sj.onc.1207676 [DOI] [PubMed] [Google Scholar]
- 15. Siddappa R, Martens A, Doorn J, Leusink A, Olivo C, Licht R, et al. cAMP/PKA pathway activation in human mesenchymal stem cells in vitro results in robust bone formation in vivo. Proc Natl Acad Sci U States America (2008) 105(20):7281–6. 10.1073/pnas.0711190105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang DC, Tsay HJ, Lin SY, Chiou SH, Li MJ, Chang TJ, et al. cAMP/PKA regulates osteogenesis, adipogenesis and ratio of RANKL/OPG mRNA expression in mesenchymal stem cells by suppressing leptin. PloS One (2008) 3(2):e1540. 10.1371/journal.pone.0001540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park KH, Choi Y, Yoon DS, Lee KM, Kim D, Lee JW. Zinc Promotes Osteoblast Differentiation in Human Mesenchymal Stem Cells Via Activation of the cAMP-PKA-CREB Signaling Pathway. Stem Cells Dev (2018) 27(16):1125–35. 10.1089/scd.2018.0023 [DOI] [PubMed] [Google Scholar]
- 18. Zhang ZR, Leung WN, Li G, Kong SK, Lu X, Wong YM, et al. Osthole Enhances Osteogenesis in Osteoblasts by Elevating Transcription Factor Osterix via cAMP/CREB Signaling In Vitro and In Vivo. Nutrients (2017) 9(6). 10.3390/nu9060588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kassis S, Olasmaa M, Terenius L, Fishman PH. Neuropeptide Y inhibits cardiac adenylate cyclase through a pertussis toxin-sensitive G protein. J Biol Chem (1987) 262(8):3429–31. [PubMed] [Google Scholar]
- 20. Selbie LA, Schmitz-Peiffer C, Sheng Y, Biden TJ. Molecular cloning and characterization of PKC iota, an atypical isoform of protein kinase C derived from insulin-secreting cells. J Biol Chem (1993) 268(32):24296–302. [PubMed] [Google Scholar]
- 21. Loh K, Shi YC, Walters S, Bensellam M, Lee K, Dezaki K, et al. Inhibition of Y1 receptor signaling improves islet transplant outcome. Nat Commun (2017) 8(1):490. 10.1038/s41467-017-00624-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bjurholm A, Kreicbergs A, Schultzberg M, Lerner UH. Neuroendocrine regulation of cyclic AMP formation in osteoblastic cell lines (UMR-106-01, ROS 17/2.8, MC3T3-E1, and Saos-2) and primary bone cells. J Bone Mineral Res (1992) 7(9):1011–9. 10.1002/jbmr.5650070903 [DOI] [PubMed] [Google Scholar]
- 23. Igwe JC, Jiang X, Paic F, Ma L, Adams DJ, Baldock PA, et al. Neuropeptide Y is expressed by osteocytes and can inhibit osteoblastic activity. J Cell Biochem (2009) 108(3):621–30. 10.1002/jcb.22294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ying X, Chen X, Liu H, Nie P, Shui X, Shen Y, et al. Silibinin alleviates high glucose-suppressed osteogenic differentiation of human bone marrow stromal cells via antioxidant effect and PI3K/Akt signaling. Eur J Pharmacol (2015) 765:394–401. 10.1016/j.ejphar.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 25. Bai J, Meng Z. Expression of caspase and apoptotic signal pathway induced by sulfur dioxide. Environ Mol Mutagenesis (2010) 51(2):112–22. 10.1002/em.20517 [DOI] [PubMed] [Google Scholar]
- 26. Li TF, Dong Y, Ionescu AM, Rosier RN, Zuscik MJ, Schwarz EM, et al. Parathyroid hormone-related peptide (PTHrP) inhibits Runx2 expression through the PKA signaling pathway. Exp Cell Res (2004) 299(1):128–36. 10.1016/j.yexcr.2004.05.025 [DOI] [PubMed] [Google Scholar]
- 27. Franceschi RT, Xiao G. Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways. J Cell Biochem (2003) 88(3):446–54. 10.1002/jcb.10369 [DOI] [PubMed] [Google Scholar]
- 28. Harrell CR, Markovic BS, Fellabaum C, Arsenijevic A, Volarevic V. Mesenchymal stem cell-based therapy of osteoarthritis: Current knowledge and future perspectives. Biomedi Pharmacother Biomed Pharmacother (2019) 109:2318–26. 10.1016/j.biopha.2018.11.099 [DOI] [PubMed] [Google Scholar]
- 29. Stevenson AS, Cartin L, Wellman TL, Dick MH, Nelson MT, Lounsbury KM. Membrane depolarization mediates phosphorylation and nuclear translocation of CREB in vascular smooth muscle cells. Exp Cell Res (2001) 263(1):118–30. 10.1006/excr.2000.5107 [DOI] [PubMed] [Google Scholar]
- 30. Doorn J, van de Peppel J, van Leeuwen JP, Groen N, van Blitterswijk CA, de Boer J. Pro-osteogenic trophic effects by PKA activation in human mesenchymal stromal cells. Biomaterials (2011) 32(26):6089–98. 10.1016/j.biomaterials.2011.05.010 [DOI] [PubMed] [Google Scholar]
- 31. Delghandi MP, Johannessen M, Moens U. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signal (2005) 17(11):1343–51. 10.1016/j.cellsig.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 32. Shi W, Gao Y, Wang Y, Zhou J, Wei Z, Ma X, et al. The flavonol glycoside icariin promotes bone formation in growing rats by activating the cAMP signaling pathway in primary cilia of osteoblasts. J Biol Chem (2017) 292(51):20883–96. 10.1074/jbc.M117.809517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haslam RJ, Davidson MM, Desjardins JV. Inhibition of adenylate cyclase by adenosine analogues in preparations of broken and intact human platelets. Evidence for the unidirectional control of platelet function by cyclic AMP. Biochem J (1978) 176(1):83–95. 10.1042/bj1760083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. von Bulow V, Rink L, Haase H. Zinc-mediated inhibition of cyclic nucleotide phosphodiesterase activity and expression suppresses TNF-alpha and IL-1 beta production in monocytes by elevation of guanosine 3′,5′-cyclic monophosphate. J Immunol (Baltimore Md 1950) (2005) 175(7):4697–705. 10.4049/jimmunol.175.7.4697 [DOI] [PubMed] [Google Scholar]
- 35. Derghal A, Djelloul M, Trouslard J, Mounien L. The Role of MicroRNA in the Modulation of the Melanocortinergic System. Front Neurosci (2017) 11:181:181. 10.3389/fnins.2017.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pedrazzini T. Importance of NPY Y1 receptor-mediated pathways: assessment using NPY Y1 receptor knockouts. Neuropeptides (2004) 38(4):267–75. 10.1016/j.npep.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 37. Yahara M, Tei K, Tamura M. Inhibition of neuropeptide Y Y1 receptor induces osteoblast differentiation in MC3T3E1 cells. Mol Med Rep (2017) 16(3):2779–84. 10.3892/mmr.2017.6866 [DOI] [PubMed] [Google Scholar]
- 38. Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell (1997) 89(5):747–54. 10.1016/s0092-8674(00)80257-3 [DOI] [PubMed] [Google Scholar]
- 39. Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem (2006) 99(5):1233–9. 10.1002/jcb.20958 [DOI] [PubMed] [Google Scholar]
- 40. Kim JM, Choi JS, Kim YH, Jin SH, Lim S, Jang HJ, et al. An activator of the cAMP/PKA/CREB pathway promotes osteogenesis from human mesenchymal stem cells. J Cell Physiol (2013) 228(3):617–26. 10.1002/jcp.24171 [DOI] [PubMed] [Google Scholar]
- 41. Wellinghausen N, Driessen C, Rink L. Stimulation of human peripheral blood mononuclear cells by zinc and related cations. Cytokine (1996) 8(10):767–71. 10.1006/cyto.1996.0102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.