Abstract
Pharmacological interventions for hepatocellular carcinoma (HCC) are hindered by complex factors, and rational combination therapy may be developed to improve therapeutic outcomes. Very recently, we have identified a bioengineered microRNA let-7c-5p (or let-7c) agent as an effective inhibitor against HCC in vitro and in vivo. In this study, we sought to identify small-molecule drugs that may synergistically act with let-7c against HCC. Interestingly, we found that let-7c exhibited a strong synergism with 5-fluorouracil (5-FU) in the inhibition of HCC cell viability as manifested by average combination indices of 0.3 and 0.5 in Hep3B and Huh7 cells, respectively. By contrast, coadministration of let-7c with doxorubicin or sorafenib inhibited HCC cell viability with, rather surprisingly, no or minimal synergy. Further studies showed that protein levels of multidrug resistance–associated protein (MRP) ATP-binding cassette subfamily C member 5 (MRP5/ABCC5), a 5-FU efflux transporter, were reduced around 50% by let-7c in HCC cells. This led to a greater degree of intracellular accumulation of 5-FU in Huh7 cells as well as the second messenger cyclic adenosine monophosphate, an endogenous substrate of MRP5. Since 5-FU is an irreversible inhibitor of thymidylate synthetase (TS), we investigated the interactions of let-7c with 5-FU at pharmacodynamic level. Interestingly, our data revealed that let-7c significantly reduced TS protein levels in Huh7 cells, which was associated with the suppression of upstream transcriptional factors as well as other regulatory factors. Collectively, these results indicate that let-7c interacts with 5-FU at both pharmacokinetic and pharmacodynamic levels, and these findings shall offer insight into molecular mechanisms of synergistic drug combinations.
SIGNIFICANCE STATEMENT
Combination therapy is a common strategy that generally involves pharmacodynamic interactions. After identifying a strong synergism between let-7c-5p and 5-fluorouracil (5-FU) against hepatocellular carcinoma cell viability, we reveal the involvement of both pharmacokinetic and pharmacodynamic mechanisms. In particular, let-7c enhances 5-FU exposure (via suppressing ABCC5/MRP5 expression) and cotargets thymidylate synthase with 5-FU (let-7c reduces protein expression, whereas 5-FU irreversibly inactivates enzyme). These findings provide insight into developing rational combination therapies based on pharmacological mechanisms.
Introduction
Targeted therapies for hepatocellular carcinoma (HCC) have been of recent interest (Chen et al., 2020) given the steady increase in HCC incidence over the past few decades (Siegel et al., 2020). The multikinase inhibitors sorafenib (SFB) (Gusani et al., 2009) and its second line analog regorafenib (Rimassa et al., 2017) have been major medications approved by the Food and Drug Administration for the treatment of HCC that only offer moderate benefit to improve overall survival. A randomized clinical trial among patients with advanced HCC revealed an approximate 6-month increase in mean survival by SFB treatment (Llovet et al., 2008). In patients who progressed on SFB, the mean survival was extended for another 6 months by regorafenib treatment (Bruix et al., 2017). Finally, although very recent approval of immunotherapy drugs, such as nivolumab, ramucirumab, pembrolizumab, atezolizumab, and bevacizumab alone or in combination, for the treatment of HCC has brought new hopes, progression-free survival remains low at several months to 1 year (El-Khoueiry et al., 2017; Zhu et al., 2017; Feun et al., 2019; Finn et al., 2020). As such, continued investment in more effective targeted therapies are needed for patients with HCC.
Very recently, our group has identified and characterized a bioengineered microRNA (miRNA or miR) let-7c-5p (or let-7c) agent for HCC therapy (Jilek et al., 2019). Using a large collection of miRNA agents with a common expression scaffold, we found that let-7c effectively inhibited HCC cell viability and tumor sphere formation in vitro through selective suppression of targeted (proto-)oncogenes. Additionally, liposome-polyplexed let-7c greatly released orthotopic HCC tumor burden and significantly extended overall survival of tumor-bearing mice in vivo (Jilek et al., 2019). Nevertheless, this bioengineered let-7c molecule alone did not completely block or eradicate tumor progression, warranting further improvement to enhance therapeutic efficacy.
Combination therapy is a common strategy for improved treatment of advanced cancer diseases (Webster, 2016), and we have been exploring new rational combinations according to their mechanisms of actions, including pharmacokinetic interactions between particular miRNAs and small-molecule drugs (Yu et al., 2016). SFB, the multikinase inhibitor used in first-line treatment of unresectable HCC (Llovet et al., 2008), is the top candidate to investigate possible combination therapy with let-7c against HCC. The DNA-intercalating doxorubicin (DOX) being approved for the management of various kinds of cancer is a chemotherapeutics being combined with other medications toward possible improvement of therapeutic outcomes among patients with advanced HCC (Abou-Alfa et al., 2019). Although it is indicated primarily for colorectal cancer, the antimetabolite 5-fluorouracil (5-FU) has been investigated for combination therapy for patients with advanced HCC (Qin et al., 2013; Goyal et al., 2019). The most well understood mechanism of action of 5-FU is irreversible inactivation of thymidylate synthetase (TS) by covalent linkage to its active metabolite, 5-fluoro-2′-deoxyuridylate (FdUMP), leading to cell death by impaired thymidine biosynthesis (Longley et al., 2003). Meanwhile, incorporation of fluorinated metabolites of 5-FU into DNA and RNA may contribute to its pharmacological effects.
In this study, we first compared the effectiveness of combination treatments of bioengineered let-7c with SFB, DOX, and 5-FU in the inhibition of HCC cell viability. Surprisingly, let-7c plus 5-FU combination treatment exhibited a strong synergism, whereas none or minimal was exhibited for let-7c plus SFB or DOX. We then provided metabolic and biochemical evidence to elucidate the mechanisms underlying synergy between 5-FU and let-7c, which involved both pharmacokinetic and pharmacodynamic interactions.
Materials and Methods
Chemicals.
5-FU, chlorzoxazone, and cAMP were purchased from Sigma-Aldrich (St. Louis, MO). SFB and DOX were purchased from LC Laboratories (Woburn, MA). Methanol, water, acetonitrile, ethyl acetate, and formic acid were all analytical grade and purchased from Thermo Fisher Scientific (Waltham, MA).
Cell Culture.
Huh7 cells were purchased from Japanese Collection of Research Bioresources and grown in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY). Hep3B cells were bought from American Type Culture Collection (Manassas, VA) and cultured in Eagle’s Minimum Essential Medium (Corning, Glendale, AZ). Cell culture medium was supplemented with 10% fetal bovine serum (Gibco) and 1% antibiotic/antimycotic (Corning). Cells were cultured at 37°C in a humidified incubator with 5% CO2.
Production of Recombinant RNA Agents.
Bioengineered let-7c and truncated control [Sephadex aptamer–tagged methionyl-transfer RNA (MSA)] were cloned, expressed in Escherichia coli, purified by anion exchange or size-exclusion chromatography, and assayed for homogeneity as previously described (Ho et al., 2018; Jilek et al., 2019).
Cell Viability, Dose-Response Relationship, and Calculation of Combination Index.
HCC cells were seeded in 96-well plates at a density of 5000 cells/well prior to transfection with let-7c or control MSA by using LipofectAMINE 3000 reagent (Life Technologies, Grand Island, NY) per the manufacturer’s instructions. After 6 hours, transfection media were removed and replaced with media containing various concentrations of 5-FU diluted in dimethyl sulfoxide (final concentration 0.1%, v/v). Seventy-two hours after initial treatment, cell viability was measured by using the CellTitre Glo 3.0 luminometric ATP assay (Promega, Madison, WI) and following the manufacturer’s instructions.
The degree of inhibition of cell viability was normalized to corresponding vehicle control (0% inhibition by 0 nM drug). Data were fit to an inhibitory dose-response model with variable slope (Prism; GraphPad, San Diego, CA) with acceptable goodness of fit (R2 = 0.90–0.99). The equation is:
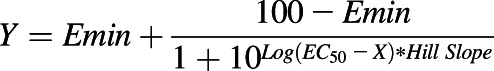 |
Drug combination index (CI) was calculated by using CompuSyn software. Briefly, percent inhibition was first converted to fraction affected (Fa) (i.e., 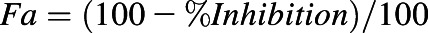 ). All combinations of RNA agents and small-molecule chemo-drugs with respective Fa values were analyzed by CompuSyn to generate respective CI values. The Chou-Talalay plots (Chou, 2010) were established by plotting log(CI) against Fa, in which log(CI) > 0 indicates antagonism, log(CI) = 0 indicates addition, and log(CI) < 0 indicates synergism.
). All combinations of RNA agents and small-molecule chemo-drugs with respective Fa values were analyzed by CompuSyn to generate respective CI values. The Chou-Talalay plots (Chou, 2010) were established by plotting log(CI) against Fa, in which log(CI) > 0 indicates antagonism, log(CI) = 0 indicates addition, and log(CI) < 0 indicates synergism.
Immunoblot Analysis.
Huh7 cells were seeded in six-well plates at a density of 300,000 cells/well and transfected with 15 nM of bioengineered let-7c or control MSA using LipofectAMINE 3000 reagent. Cells were harvested in ice-cold PBS at 72 hours post-transfection and lysed in radioimmunoprecipitation assay buffer consisting of complete proteinase inhibitors cocktail (Sigma). Protein concentrations were determined by bicinchoninic acid assay (BCA) assay (Pierce, Rockford, IL). Whole cell proteins (15–25 µg per sample) were denatured in Laemmli buffer, resolved on TGX Stain-Free polyacrylamide gel (Bio-Rad, Hercules, CA), immobilized on polyvinylidene difluoride (for transporter detection) or nitrocellulose (for all other protein detection), and imaged for total protein per the manufacturer’s instructions. Membranes were blocked in 5% nonfat milk in Tris-buffered saline, incubated overnight at 4°C in primary antibody diluted in 5% bovine serum albumin in Tris-buffered saline, and then incubated for 2 hours at room temperature in appropriate secondary antibodies. Relative protein expression was determined by enhanced chemiluminescence detection and imaging (Bio-Rad) and normalized to respective total protein levels. Antibodies used in this study were: anti-multidrug resistance–associated protein (MRP) 4 (ABCC4) rat monoclonal (3369; Abcam), anti-MRP5/ABCC5 rat polyclonal ( 180724; Abcam), anti–breast cancer resistance protein (BCRP) (ABCG2) rabbit monoclonal (42078; Cell Signaling), anti–pan-rat sarcoma (RAS) mouse monoclonal (Ras10; Thermo Fisher Scientific), anti-TS rabbit monoclonal (9045; Cell Signaling), anti–AT-rich interactive domain-containing protein 3B (ARID3B) rabbit polyclonal (92328, Abcam), anti–cellular myelocytomatosis oncogene (c-MYC) rabbit monoclonal (13987; Cell Signaling), anti–Lin-28 homolog B (LIN28B) rabbit monoclonal (11965; Cell Signaling), horseradish peroxidase (HRP)-conjugated anti-Rabbit IgG (111-035-003; Jackson ImmunoReseach), HRP-conjugated anti-Mouse IgG (7076; Cell Signaling), and HRP-conjugated anti-Rat IgG (7077; Cell Signaling).
Liquid Chromatography with Tandem Mass Spectrometry Quantification of 5-FU Uptake.
Huh7 cells were seeded in 24-well plates at a density of 50,000 cells/well and transfected with 5 nM of let-7c or MSA. After 72 hours, cells were washed twice with PBS and incubated with either 10 µM of 5-FU in Hank’s balanced salt solution (Gibco) for 30, 60, or 90 minutes. To terminate the incubation, 5-FU containing salt solution was removed, and cells were washed twice with ice-cold PBS and lysed directly on the plate with 500 µl of double distilled H2O containing 10 nM of chlorzoxazone as an internal standard (IS). The cell suspension was then subjected to a brief sonication, and an aliquot was used for the quantification of protein concentrations by using BCA assay (Pierce). Proteins were then precipitated by adding 1 ml ice-cold acetonitrile and removed by centrifugation at 13,000g for 10 minutes. Analytes were isolated by liquid-liquid extraction with 3 ml of ethyl acetate. Analytes were recovered by drying the upper (organic) phase over air and resuspending in 50 µl of water/methanol (1/1; v/v) consisting of 0.1% formic acid, and 5 µl was injected for liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis by using an AB Sciex 4000 QTRAP tandem mass spectrometry system (AB Sciex, Framingham, MA) equipped with a Shimadzu Prominence Ultra-Fast Liquid Chromatography system (Shimadzu Corporation, Kyoto, Japan).
5-FU and chlorzoxazone were separated over an Agilent Zorbax C18 column by isocratic elution with water/methanol (1/1; v/v) containing 0.1% formic acid at a flow rate of 0.6 ml/minute. Analytes were detected in negative electrospray ionization mode using the following source parameters: −4.5 kV ion spray voltage, 400°C source temperature, 20 psi curtain gas pressure, 25 psi gas 1 pressure, 75 psi gas 2 pressure, and −10 V entrance potential, with multiple reaction monitoring, mass to charge (m/z) 129.0→42.0 for 5-FU, and m/z 167.8→131.9 for the IS. Linear calibration range of 5-FU was 1−1000 nM. Accuracy was 98.4%–100%, and precision was <16%.
LC-MS/MS Quantification of Intracellular cAMP Levels.
Huh7 cells were seeded in six-well plates at a density of 500,000 cells/well and transfected with 10 nM of let-7c, 1 µM of 5-FU, or vehicle. After 72 hours, cells were harvested and washed twice with ice-cold PBS, scraped in ice-cold PBS, and pelleted at 700g for 5 minutes at 4°C. Cell pellets were resuspended in 100 µl of water and subjected to three consecutive freeze-thaw cycles (liquid nitrogen and 37°C water bath) to lyse cells, and an aliquot was removed for total protein quantification by BCA assay for normalization of intracellular nucleotide concentrations. Ice-cold acetonitrile (400 µl) containing 200 nM of chloro-phenylalanine (IS) was added to another aliquot of 80 µl cell lysate to precipitate proteins that were removed by centrifugation at 13,000g for 10 minutes at 4°C. The supernatant was dried over air, and the residue was reconstituted in 50 µl of water with 0.1% formic acid.
A Synergi Fusion-RP column (2.0 × 50 mm, 4 µm; Phenomenex) was used to separate cAMP and IS through gradient elution with solution A (water with 0.1% formic acid) and solvent B (methanol): 0% B (1 minute), 0%–20% B (5 minutes), 20%–100% B (0.5 minutes), 100% B (0.5 minutes), 100%–0% B (0.5 minutes), and 0% B (1 minute). Samples were analyzed in negative electrospray ionization mode using the following source parameters: −3.5 kV ion spray voltage, 600°C source temperature, 25 psi curtain gas pressure, 70 psi gas 1 pressure, 50 psi gas 2 pressure, and −10 V entrance potential, with multiple reaction monitoring, m/z 328.0→133.9 for cAMP, and m/z 197.8→136.9 for the IS. Linear calibration range ranging from 0.1 to 500 nM was established for cAMP. Accuracy was 92.6%–96.5%, and precision was within 10%.
Statistical Analysis.
Values are mean ± S.D.. Data were analyzed by one-way ANOVA with Bonferroni’s post-tests (GraphPad Prism). Difference was considered statistically significant when P < 0.05.
Results
Strong Synergism Is Identified for Let-7c and 5-FU Combination Treatment in the Inhibition of Human HCC Cell Viability.
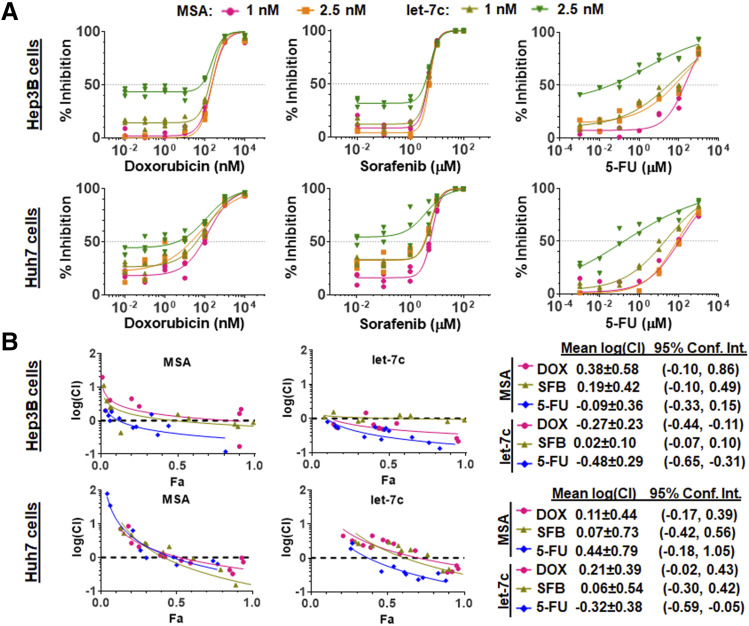
Three small-molecule drugs, SFB, DOX, and 5-FU, were chosen for the investigation of possible combination therapies with let-7c against HCC. Our data showed that all drugs exhibited sigmoidal dose-response curves in the presence of let-7c or control MSA against both Hep3B and Huh7 cell viability (Fig. 1A). All data fit well to the inhibitory dose-response model with variable slopes, and values of respective pharmacodynamic parameters, including EC50, Hill slope, and Emin, were estimated (Table 1 and 2). Consistent with our recent findings (Jilek et al., 2019), let-7c itself at a relatively higher concentration (2.5 nM) or in combination with lower concentrations of chemo-drugs (e.g., 0.02 nM DOX or 1 nM 5-FU) already showed an obvious reduction of HCC cell viability, which was also indicated by greater Emin values for let-7c combinations than for corresponding MSA control treatments (Table 1 and 2). The increase in Emin values for let-7c combinations relative to respective MSA control combinations was consistently shown for every small-molecule drug. Interestingly, coadministration of let-7c with 5-FU elicited significantly lower EC50 values than control MSA (Table 1 and 2) in both Hep3B and Huh7 cells, whereas minimal or insignificant changes were noted for let-7c combinations with DOX or SFB relevant to corresponding MSA controls.
Fig. 1.
Bioengineered let-7c exerts a strong synergism with 5-FU but none or minimal with DOX or SFB in the inhibition of HCC cell viability. (A) Huh7 and Hep3B cells transfected with 1 or 2.5 nM of bioengineered let-7c or control MSA were treated with various concentrations of 5-FU, DOX, or SFB. Inhibition of cell viability was measured by ATP luminometric assay, and the data were fit to an inhibitory dose-response model. Vehicle control was set as 0% inhibition. (B) The Chou-Talalay plots [log(CI) vs. Fa] revealed a strong synergism for let-7c plus 5-FU combination treatment.
TABLE 1.
Pharmacodynamic parameters estimated for DOX, SFB, and 5-FU as combined with let-7c or control MSA in the inhibition of Hep3B cell viability
| MSA | Let-7c | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.0 nM | 2.5 nM | 1.0 nM | 2.5 nM | ||||||||||
| EC50 | DOX (nM) | 230.3 | ± | 1.1 | 240.2 | ± | 1.1 | 205.9 | ± | 1.1 | 230.2 | ± | 1.2 |
| SFB (µM) | 4.8 | ± | 1.1 | 5.2 | ± | 1.0 | 4.9 | ± | 1.0 | 5.2 | ± | 1.0 | |
| 5-FU (µM) | 237.2 | ± | 1.3 | 132.6 | ± | 1.7 | 56.9 | ± | 1.9* | 2.4 | ± | 3.4* | |
| Emin | DOX (nM) | 1.7 | ± | 1.6 | >0.05 | 14.2 | ± | 1.1* | 43.2 | ± | 1.1* | ||
| SFB (µM) | 8.4 | ± | 1.5 | 3.8 | ± | 0.7 | 12.0 | ± | 1.0 | 31.7 | ± | 1.0* | |
| 5-FU (µM) | 7.2 | ± | 2.9 | 14.1 | ± | 4.3 | 9.9 | ± | 5.6 | 33.5 | ± | 8.0* | |
| Hill slope | DOX | 1.4 | ± | 0.2 | 1.6 | ± | 0.2 | 1.6 | ± | 0.2 | 1.6 | ± | 0.2 |
| SFB | 2.8 | ± | 0.4 | 2.9 | ± | 0.1 | 2.5 | ± | 0.2 | 2.4 | ± | 0.3 | |
| 5-FU | 0.8 | ± | 0.1 | 0.4 | ± | 0.1 | 0.3 | ± | 0.1 | 0.3 | ± | 0.1 | |
| R2 | DOX | 0.98 | 0.99 | 0.99 | 0.98 | ||||||||
| SFB | 0.99 | 0.99 | 0.99 | 0.99 | |||||||||
| 5-FU | 0.96 | 0.91 | 0.92 | 0.94 | |||||||||
P < 0.05 compared with corresponding MSA control treatment.
TABLE 2.
Pharmacodynamic parameters estimated for DOX, SFB, and 5-FU as combined with let-7c or control MSA in the inhibition of Huh7 cell viability
| MSA | Let-7c | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.0 nM | 2.5 nM | 1.0 nM | 2.5 nM | ||||||||||
| EC50 | DOX (nM) | 150.8 | ± | 1.2 | 69.9 | ± | 1.6 | 117.8 | ± | 1.2 | 120.9 | ± | 1.4 |
| SFB (µM) | 6.4 | ± | 1.0 | 5.4 | ± | 1.1 | 5.9 | ± | 1.1 | 3.9 | ± | 1.4 | |
| 5-FU (µM) | 113.9 | ± | 1.5 | 121.6 | ± | 1.4 | 23.8 | ± | 1.3* | 0.2 | ± | 14.9* | |
| Emin | DOX (nM) | 17.9 | ± | 1.9 | 21.7 | ± | 4.3 | 26.2 | ± | 2.0* | 44.1 | ± | 2.1* |
| SFB (µM) | 15.9 | ± | 1.4 | 32.3 | ± | 1.8 | 32.9 | ± | 1.3* | 54.1 | ± | 3.6* | |
| 5-FU (µM) | 1.3 | ± | 4.0 | 0.00 | ± | 4.47 | 3.2 | ± | 2.4 | 0.00 | ± | 31.7 | |
| Hill slope | DOX | 0.7 | ± | 0.1 | 0.5 | ± | 0.1 | 0.7 | ± | 0.1 | 0.6 | ± | 0.1 |
| SFB | 2.4 | ± | 0.3 | 2.3 | ± | 0.4 | 2.3 | ± | 0.3 | 1.1 | ± | 0.4 | |
| 5-FU | 0.5 | ± | 0.1 | 0.6 | ± | 0.1 | 0.4 | ± | 0.0 | 0.2 | ± | 0.1 | |
| R2 | DOX | 0.98 | 0.93 | 0.97 | 0.95 | ||||||||
| SFB | 0.99 | 0.97 | 0.99 | 0.86 | |||||||||
| 5-FU | 0.92 | 0.94 | 0.99 | 0.90 | |||||||||
P < 0.05 compared with corresponding MSA control treatment.
To define whether synergism occurs for specific combination treatment, we employed Chou-Talalay method (Chou, 2010) to calculate individual CI values and plotted them against the Fa values (Fig. 1B). When individual small-molecule drugs were combined with control MSA, all three combinations elicited either additive or slightly antagonistic effects because the average log(CI) values were not significantly less than zero (Fig. 1B). To our surprise, SFB plus let-7c combination did not produce any significant synergistic effects in either Hep3B or Huh7 cells, and DOX plus let-7c combination only showed a synergism in Hep3B cells. By contrast, combination treatment with 5-FU and let-7c consistently elicited a synergism for the inhibition of both Hep3B [mean log(CI) = −0.48 ± 0.29; 95% confidence interval −0.65 to −0.31] and Huh7 [mean log(CI) = −0.32 ± 0.38; 95% confidence interval −0.59 to −0.05] cells (Fig. 1B).
Transporter MRP5/ABCC5 Protein Levels Are Reduced by Let-7c in Huh7 Cells.
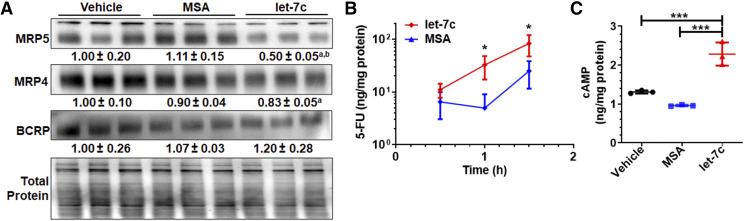
To determine potential pharmacokinetic mechanism underlying the synergism between let-7c and 5-FU against HCC cell viability, we evaluated the impact of let-7c on protein expression of MRP5 by immunoblot, as MRP5 confers resistance to 5-FU (Pratt et al., 2005). Our data showed that MRP5 protein levels were significantly decreased by let-7c for approximately 50% in Huh7 cells as compared with control MSA and vehicle treatments (Fig. 2A). Since resistance to 5-FU may be mediated by other efflux transporters (Yuan et al., 2009; Nambaru et al., 2011) overexpressed in Huh7 cells, the protein levels of MRP4/ABCC4 and BCRP/ABCG2 were also evaluated. A slight decrease of 17% in MRP4 expression was observed in cells treated with let-7c, but this is not statistically significant as compared with MSA treatment. In addition, BCRP protein levels were not altered by bioengineered let-7c in Huh7 cells (Fig. 2A). The results suggest potential involvement of let-7c–MRP5 pathway in the modulation of Huh7 cell sensitivity to 5-FU.
Fig. 2.
Let-7c reduces the expression of efflux transporters to improve intracellular 5-FU and cAMP accumulation in Huh7 cells. (A) Immunoblot analyses showed that MRP5 but not MRP4 or BCRP was the most significantly suppressed after let-7c treatment. aP < 0.05; bP < 0.05, as compared with the vehicle and MSA treatments, respectively. Cells were treated with let-7c or control RNA for 72 hours. Transporter levels were normalized to corresponding total protein levels, and vehicle control group was set as 1.00. This led to a greater intracellular accumulation of exogenous 5-FU (B) and retention of endogenous cAMP (C) compared with control MSA or vehicle treatment. Values are mean ± S.D. (N = 3/group). *P < 0.05; ***P < 0.001 (one-way ANOVA with Bonferroni’s post hoc test).
Intracellular Accumulation of Exogenous 5-FU and Endogenous cAMP Is Enhanced by Let-7c in Huh7 Cells.
We then directly examined the consequent effects on intracellular accumulation of 5-FU drug by using an accurate LC-MS/MS method. Our data showed that intracellular 5-FU concentrations were 6.6- and 3.3-fold higher in Huh7 cells treated with let-7c than those transfected with control MSA at 60 and 90 minutes postexposure to 10 µM of 5-FU, respectively (Fig. 2B). Furthermore, we established an LC-MS/MS method to examine the effects of let-7c on the intracellular levels of cAMP, an important “second messenger” and endogenous substrate of MRP4 and MRP5 (Wielinga et al., 2003). cAMP levels were revealed to be elevated approximately 2-fold in Huh7 cells at 72 hours post-treatment with 10 nM of let-7c compared with control MSA and vehicle treatments (Fig. 2C). Taken together, these data functionally corroborate the reduction of MRP5 expression by let-7c in Huh7 cells.
Let-7c Reduces the Expression of TS, the Direct Target of 5-FU Metabolite FdUMP.
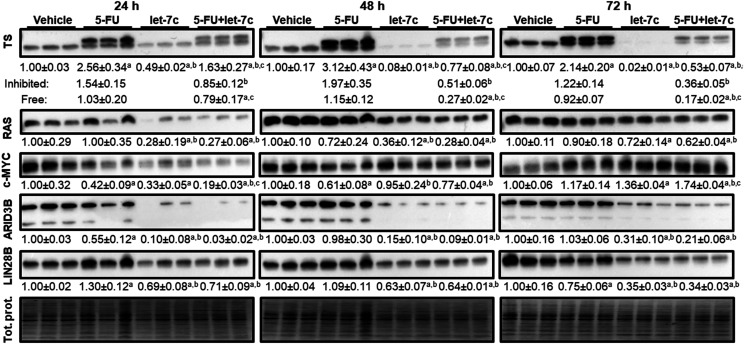
Given the modest perturbation in 5-FU uptake via let-7c–mediated suppression of MRP5 expression, we then evaluated the impact of let-7c on TS, the primary target of 5-FU. After the treatment with 5-FU, immunoblot analysis revealed two TS bands (Fig. 3), with one being FdUMP-bound or inactivated protein band (upper) and one being noninhibited or active protein band (lower), as reported (Drake et al., 1993). By contrast, only one active TS protein band was present in cells subjected to vehicle treatment. Interestingly, let-7c (10 nM) treatment alone sharply suppressed the protein levels of TS in Huh7 cells at 48 and 72 hours post-treatment (Fig. 3). Although this effect was marginally reversed in cells cotreated with let-7c and 5-FU (10 nM + 1 µM, respectively) relevant to let-7c alone, reduction of TS protein levels (unbound, FdUMP-bound, and total) by let-7c (let-7c plus 5-FU vs. 5-FU alone) was consistently demonstrated (Fig. 3).
Fig. 3.
Let-7c suppresses TS expression in Huh7 cells, whereas 5-FU inhibits 5-FU. Expression of TS and other related proteins in Huh7 cells was analyzed by immunoblot at 24, 48, and 72 hours post-treatment with 5-FU (1 µM) and let-7c (10 nM) alone or in combination. Cotargeting TS by 5-FU and let-7c is manifested by the appearance of 5-FU metabolite FdUMP-bound (inhibited, top band) TS in cells treated with 5-FU and significant reduction of unbound and total TS protein levels in cells treated with let-7c. Protein levels of known let-7c–targeted genes RAS, ARID3B, LIN28B, and cellular myelocytomatosis oncogene (c-MYC) were also significantly reduced by bioengineered let-7c. Protein expression level was normalized to corresponding total protein (Tol. prot.) level, and differences between each treatment groups were compared by one-way ANOVA with Bonferroni’s post hoc tests. Values are mean ± S.D. (N = 3/group). P < 0.05, compared with vehicle (a), 5-FU alone (b), or let-7c alone (c).
Since TS is not a direct target of let-7c, reduction of TS protein levels by let-7c may be attributable to the regulation of its upstream regulatory factors. Given the findings that let-7c directly represses the transcription factor c-MYC (Kim et al., 2009), which regulates TS (Mannava et al., 2008) as well as RAS (Johnson et al., 2005), that is able to control c-MYC expression (Kerkhoff et al., 1998; Sears et al., 2000), we further evaluated the expression of c-MYC and RAS. Our data showed that RAS protein levels were consistently reduced in Huh7 cells at 24–72 hours post-treatment with let-7c alone or combination with 5-FU compared with corresponding controls (Fig. 3). Furthermore, c-Myc protein levels were significantly lower in cells at 24 hours after let-7c treatments but recovered at 72 hours. In addition, protein levels of LIN28B and ARID3B, which are canonical targets of let-7c, were consistently suppressed in Huh7 cells by bioengineered let-7c treatments independent of 5-FU treatment. Together, these results demonstrate a cooperative targeting of TS by let-7c and 5-FU.
Discussion
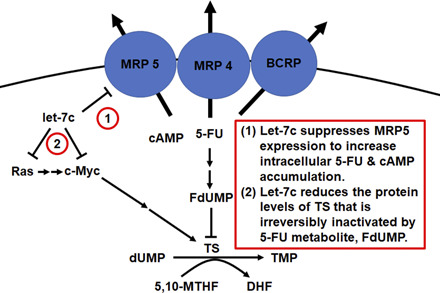
Combination therapy has posed a promising strategy in oncology (Webster, 2016) for the treatment of advanced, metastatic, and recurring cancer diseases when monotherapy is less or not effective. Significant challenges arise in experimental identification of effective drug combinations from large numbers of possible combinations, although bioinformatic methods (Preuer et al., 2018) might make some predictions. Furthermore, determination of synergistic, additive, or antagonistic effects requires valid experimental design and data analysis with correct algorisms (Chou, 2006, 2010; Tallarida, 2006), and the underlying mechanisms may involve complex interactions. Previously, RNA agents have been shown to modulate mainly the pharmacokinetics of small-molecule drugs coadministered or act on pharmacological targets different from chemotherapeutics, leading to the improvement of anticancer effects (Pan et al., 2009; Li et al., 2015, 2018; Zhao et al., 2015; Tu et al., 2019; Yi et al., 2020). In this study, we surprisingly identified that let-7c inhibits HCC cell viability synergistically with 5-FU but no or minimal with SFB or DOX, which was firmly established by using the Chou-Talalay method (Chou, 2006, 2010). Additionally, our new findings on the reduction of 5-FU transporter MRP5 and 5-FU–targeted TS by let-7c indicate the presence of both pharmacokinetic and pharmacodynamic mechanisms behind the synergism between let-7c and 5-FU.
DOX is a potent cytotoxic chemotherapeutic functioning primarily by DNA intercalation and suppression of topoisomerase II (Pommier et al., 2010). Since DOX is limited clinically by dose-limiting cumulative cardiotoxicity, dose reduction via combination therapy is a promising strategy. Additionally, high-dose DOX-mediated apoptosis is mitigated by B-cell lymphoma–extra large protein (Park et al., 2007), and let-7c–mediated suppression of B-cell lymphoma–extra large may potentially enhance DOX efficacy. However, we did not observe significant synergy between let-7c and DOX in HCC cells, which is different from our previous findings on synergism for rationally designed miR-34a plus DOX combination in osteosarcoma cells (Zhao et al., 2015; Jian et al., 2017). This observation may be in part a result of let-7 family–mediated suppression of caspase-3 (Tsang and Kwok, 2008) despite the general regard for let-7 as a tumor-suppressive miRNA. Furthermore, combination drug response is highly dependent on the experimental system. For example, the most potent target of multikinase inhibitor SFB is the vascular endothelial growth factor receptor (Wilhelm et al., 2004), which may likely be more relevant in suppression of angiogenesis in an in vivo system. Additionally, the absence of synergy between let-7c plus SFB combination against HCC cells may also be attributable to redundancies in targets because the mitogen-activated protein kinase pathway suppression via rapidly accelerated fibrosarcoma and RAS inhibition (by SFB and let-7c, respectively) may not elicit synergy in a cell-based system (Lyons et al., 2001).
Development of multidrug resistance is a common problem for antineoplastic drugs, and it may involve several mechanisms (Choi and Yu, 2014), including overexpression of efflux transporters, alteration of drug targets, or change of the tumor microenvironment. Common to many small-molecule chemotherapeutics, 5-FU efficacy is strongly hindered by overexpression of efflux transporters. Among them, MRP5 and MRP8 have been found to confer chemoresistance to 5-FU in multiple cancer types (Oguri et al., 2007; Nambaru et al., 2011), and respective strategies are under evaluation to reduce transporter expression or block transporter activity (Choi and Yu, 2014; Li et al., 2016). Alternatively, enhanced expression of TS, the primary pharmacological target of 5-FU, has been shown to confer chemoresistance in patients (Peters et al., 2002) and is induced after 5-FU treatment (Longley et al., 2002). Indeed, this study revealed a persistent expression of active TS in HCC cells after 5-FU treatment in addition to the inactivated or FdUMP-bound TS, suggesting the presence of compensatory mechanisms and likelihood of resistance. However, coadministered let-7c was able to reduce TS protein levels in HCC cells beyond the suppression of MRP5. As such, a strong synergy was observed for let-7c plus 5-FU combination treatment in the inhibition of HCC cell viability, offering insights into developing more effective therapeutic approaches.
In addition to pleiotropic regulation of target gene expression with contributions to oncogenesis, many miRNAs have been shown to contribute to drug metabolism and disposition via regulating the expression of enzymes, transporters, or their regulatory factors (Yu and Pan, 2012; Yu et al., 2016). Many MRP transporters were found to be dysregulated in patient tissues with HCC (Borel et al., 2012). Of these, MRP5 was significantly upregulated in HCC tissues and inversely correlated with let-7a levels. Since let-7 family miRNAs are predicted to act directly on the mRNA of MRP5 (Agarwal et al., 2015), further studies using luciferase reporter assay supported a direct regulation of MRP5 by let-7a (Borel et al., 2012). Meanwhile, we cannot exclude possible contributions from let-7–mediated regulation of any transcription factors upstream of MRP5, which is a common “indirect” mechanism underlying miRNA-controlled regulation of drug transporters and metabolic enzymes (Yu et al., 2016). Rather, the present study is the first to demonstrate that MRP5 protein expression was suppressed by a let-7 family miRNA in human carcinoma cells. In addition, we revealed that let-7c–mediated suppression of MRP5 subsequently enhanced intracellular accumulation of 5-FU levels. Functional consequence of let-7c–MRP5 signaling in Huh7 cells was also supported by a simultaneous increase of intracellular cAMP levels, an endogenous substrate of MRP5.
Notably, MRP5 expression was only reduced by 50% in Huh7 cells, whereas MRP4 was also overexpressed, suggesting that pharmacokinetic interactions may not exhaustively contribute to the strong synergism between let-7c and 5-FU observed in this study, which is evidenced by the CI values as well as an immense 100-fold reduction in 5-FU EC50 values after coadministration of let-7c. Since induction of TS is another mechanism of 5-FU resistance (Longley et al., 2002), additional pharmacodynamic interactions may explain the observed synergy between 5-FU and let-7c. In this study, we demonstrated for the first time that TS expression was remarkably suppressed by let-7c. Interestingly, the transcript of TS does not contain a target site for let-7 family miRNAs (Agarwal et al., 2015), suggesting that regulation of TS by let-7 may undergo an indirect path. Indeed, we confirmed the suppression of a number of signaling factors by let-7c in Huh7 cells, including c-MYC, RAS, LIN28, and ARID3B. Among them, c-MYC was known to regulate TS (Mannava et al., 2008), whereas c-MYC expression is regulated by RAS through various mechanisms (Kerkhoff et al., 1998; Sears et al., 2000), and LIN28 regulates both RAS and c-MYC signaling (Shyh-Chang and Daley, 2013). In addition, TS is regulated by E2F gene family transcription factors (DeGregori et al., 1995; Dong et al., 2000) that can be directly targeted by let-7 (Benhamed et al., 2012; Chafin et al., 2014) or modulated by let-7–targeted elements, such as RAS (Yoon et al., 2006). Although the synergism between let-7c and 5-FU in the inhibition of HCC cell viability was revealed in both Huh7 and Hep3B cells, the present study on the underlying pharmacokinetic and pharmacodynamic mechanisms is limited to the use of Huh7 cells. More extensive studies are warranted to define whether they are common mechanisms or whether there are any variations among various HCC cell lines and patient specimens.
In conclusion, our data support a strong synergism between let-7c and 5-FU in the inhibition of HCC cell viability. Furthermore, we have found that let-7c–dependent suppression of MRP5 enhances the intracellular 5-FU accumulation in HCC cells. Finally, let-7c sharply reduces TS protein expression levels, whereas 5-FU inactivates TS protein. Our results reveal the presence of both pharmacokinetic and pharmacodynamic mechanisms underlying synergistic let-7c–5-FU interactions. These findings shine light on rational design and development of combination therapies for the treatment of advanced cancer diseases.
Abbreviations
- ARID3B
AT-rich interactive domain-containing protein 3B
- ABC
ATP-binding cassette
- BCA
bicinchoninic acid assay
- BCRP
breast cancer resistance protein
- CI
combination index
- c-MYC
cellular myelocytomatosis oncogene
- DOX
doxorubicin
- Fa
fraction affected
- FdUMP
5-fluoro-2'-deoxyuridylate
- 5-FU
5-fluorouracil
- HCC
hepatocellular carcinoma
- HRP
horseradish peroxidase
- IS
internal standard
- LC-MS/MS
liquid chromatography with tandem mass spectrometry
- LIN28B
Lin-28 homolog B
- miRNA or miR
microRNA
- MRP
multidrug resistance–associated protein
- MSA
Sephadex aptamer–tagged methionyl-transfer RNA
- m/z
mass to charge
- RAS
rat sarcoma
- SFB
sorafenib
- TS
thymidylate synthetase
Authorship Contributions
Participated in research design: Jilek, Tu, Yu.
Conducted experiments: Jilek, Zhang.
Contributed new reagents or analytic tools: Jilek, Tu, Zhang, Yu.
Performed data analysis: Jilek.
Wrote or contributed to the writing of the manuscript: Jilek, Tu, Zhang, Yu.
Footnotes
This work was supported by National Institutes of Health National Institute of General Medical Sciences [Grant R01-GM113888] and National Cancer Institute [Grant R01-CA225958]. J.L.J. was supported by National Institutes of Health National Institutes of General Medical Sciences–funded Pharmacology Training Program [Grant T32-GM099608]. The authors also appreciate the access to the Molecular Pharmacology Shared Resources funded by the University of California, Davis Comprehensive Cancer Center Support Grant (CCSG) awarded by the National Cancer Institute [Grant P30-CA093373].
References
- Abou-Alfa GK, Shi Q, Knox JJ, Kaubisch A, Niedzwiecki D, Posey J, Tan BR, Jr, Kavan P, Goel R, Lammers PE, et al. (2019) Assessment of treatment with sorafenib plus doxorubicin vs sorafenib alone in patients with advanced hepatocellular carcinoma: phase 3 CALGB 80802 randomized clinical trial. JAMA Oncol 5:1582–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam JW, Bartel DP. (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. (2012) Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 14:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel F, Han R, Visser A, Petry H, van Deventer SJ, Jansen PL, Konstantinova P, Réseau Centre de Ressources Biologiques Foie (French Liver Biobanks Network), France (2012) Adenosine triphosphate-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular microRNAs. Hepatology 55:821–832. [DOI] [PubMed] [Google Scholar]
- Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al. RESORCE Investigators (2017) Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 389:56–66. [DOI] [PubMed] [Google Scholar]
- Chafin CB, Regna NL, Caudell DL, Reilly CM. (2014) MicroRNA-let-7a promotes E2F-mediated cell proliferation and NFκB activation in vitro. Cell Mol Immunol 11:79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Garmire L, Calvisi DF, Chua MS, Kelley RK, Chen X. (2020) Harnessing big ‘omics’ data and AI for drug discovery in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 17:238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, Yu AM. (2014) ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des 20:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC. (2006) Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 58:621–681. [DOI] [PubMed] [Google Scholar]
- Chou TC. (2010) Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70:440–446. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. (1995) Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol 15:4215–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Lester L, Johnson LF. (2000) Transcriptional control elements and complex initiation pattern of the TATA-less bidirectional human thymidylate synthase promoter. J Cell Biochem 77:50–64. [DOI] [PubMed] [Google Scholar]
- Drake JC, Allegra CJ, Johnston PG. (1993) Immunological quantitation of thymidylate synthase-FdUMP-5,10-methylenetetrahydrofolate ternary complex with the monoclonal antibody TS 106. Anticancer Drugs 4:431–435. [DOI] [PubMed] [Google Scholar]
- El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling THR, et al. (2017) Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389:2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feun LG, Li YY, Wu C, Wangpaichitr M, Jones PD, Richman SP, Madrazo B, Kwon D, Garcia-Buitrago M, Martin P, et al. (2019) Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer 125:3603–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, et al. IMbrave150 Investigators (2020) Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 382:1894–1905. [DOI] [PubMed] [Google Scholar]
- Goyal L, Zheng H, Abrams TA, Miksad R, Bullock AJ, Allen JN, Yurgelun MB, Clark JW, Kambadakone A, Muzikansky A, et al. (2019) A phase II and biomarker study of sorafenib combined with modified FOLFOX in patients with advanced hepatocellular carcinoma. Clin Cancer Res 25:80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusani NJ, Jiang Y, Kimchi ET, Staveley-O’Carroll KF, Cheng H, Ajani JA. (2009) New pharmacological developments in the treatment of hepatocellular cancer. Drugs 69:2533–2540. [DOI] [PubMed] [Google Scholar]
- Ho PY, Duan Z, Batra N, Jilek JL, Tu MJ, Qiu JX, Hu Z, Wun T, Lara PN, DeVere White RW, et al. (2018) Bioengineered noncoding RNAs selectively change cellular miRNome profiles for cancer therapy. J Pharmacol Exp Ther 365:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian C, Tu MJ, Ho PY, Duan Z, Zhang Q, Qiu JX, DeVere White RW, Wun T, Lara PN, Lam KS, et al. (2017) Co-targeting of DNA, RNA, and protein molecules provides optimal outcomes for treating osteosarcoma and pulmonary metastasis in spontaneous and experimental metastasis mouse models. Oncotarget 8:30742–30755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilek JL, Zhang Q-Y, Tu M-J, Ho PY, Duan Z, Qiu J-X, Yu A-M. (2019) Bioengineered let-7c inhibits orthotopic hepatocellular carcinoma and improves overall survival with minimal immunogenicity. Mol Ther Nucleic Acids 14:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. (2005) RAS is regulated by the let-7 microRNA family. Cell 120:635–647. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E, Houben R, Löffler S, Troppmair J, Lee JE, Rapp UR. (1998) Regulation of c-myc expression by Ras/Raf signalling. Oncogene 16:211–216. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23:1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Addepalli B, Tu MJ, Chen QX, Wang WP, Limbach PA, LaSalle JM, Zeng S, Huang M, Yu AM. (2015) Chimeric microRNA-1291 biosynthesized efficiently in escherichia coli is effective to reduce target gene expression in human carcinoma cells and improve chemosensitivity. Drug Metab Dispos 43:1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PC, Tu MJ, Ho PY, Jilek JL, Duan Z, Zhang QY, Yu AX, Yu AM. (2018) Bioengineered NRF2-siRNA is effective to interfere with NRF2 pathways and improve chemosensitivity of human cancer cells. Drug Metab Dispos 46:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang H, Assaraf YG, Zhao K, Xu X, Xie J, Yang DH, Chen ZS. (2016) Overcoming ABC transporter-mediated multidrug resistance: molecular mechanisms and novel therapeutic drug strategies. Drug Resist Updat 27:14–29. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. SHARP Investigators Study Group (2008) Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359:378–390. [DOI] [PubMed] [Google Scholar]
- Longley DB, Boyer J, Allen WL, Latif T, Ferguson PR, Maxwell PJ, McDermott U, Lynch M, Harkin DP, Johnston PG. (2002) The role of thymidylate synthase induction in modulating p53-regulated gene expression in response to 5-fluorouracil and antifolates. Cancer Res 62:2644–2649. [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. (2003) 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3:330–338. [DOI] [PubMed] [Google Scholar]
- Lyons JF, Wilhelm S, Hibner B, Bollag G. (2001) Discovery of a novel Raf kinase inhibitor. Endocr Relat Cancer 8:219–225. [DOI] [PubMed] [Google Scholar]
- Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA. (2008) Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 7:2392–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambaru PK, Hübner T, Köck K, Mews S, Grube M, Payen L, Guitton J, Sendler M, Jedlitschky G, Rimmbach C, et al. (2011) Drug efflux transporter multidrug resistance-associated protein 5 affects sensitivity of pancreatic cancer cell lines to the nucleoside anticancer drug 5-fluorouracil. Drug Metab Dispos 39:132–139. [DOI] [PubMed] [Google Scholar]
- Oguri T, Bessho Y, Achiwa H, Ozasa H, Maeno K, Maeda H, Sato S, Ueda R. (2007) MRP8/ABCC11 directly confers resistance to 5-fluorouracil. Mol Cancer Ther 6:122–127. [DOI] [PubMed] [Google Scholar]
- Pan YZ, Gao W, Yu AM. (2009) MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos 37:2112–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS, Kim MA, Eom YW, Choi KS. (2007) Bcl-xL blocks high dose doxorubicin-induced apoptosis but not low dose doxorubicin-induced cell death through mitotic catastrophe. Biochem Biophys Res Commun 363:1044–1049. [DOI] [PubMed] [Google Scholar]
- Peters GJ, Backus HH, Freemantle S, van Triest B, Codacci-Pisanelli G, van der Wilt CL, Smid K, Lunec J, Calvert AH, Marsh S, et al. (2002) Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta 1587:194–205. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Leo E, Zhang H, Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 17:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt S, Shepard RL, Kandasamy RA, Johnston PA, Perry W, III, Dantzig AH. (2005) The multidrug resistance protein 5 (ABCC5) confers resistance to 5-fluorouracil and transports its monophosphorylated metabolites. Mol Cancer Ther 4:855–863. [DOI] [PubMed] [Google Scholar]
- Preuer K, Lewis RPI, Hochreiter S, Bender A, Bulusu KC, Klambauer G. (2018) DeepSynergy: predicting anti-cancer drug synergy with Deep Learning. Bioinformatics 34:1538–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Bai Y, Lim HY, Thongprasert S, Chao Y, Fan J, Yang TS, Bhudhisawasdi V, Kang WK, Zhou Y, et al. (2013) Randomized, multicenter, open-label study of oxaliplatin plus fluorouracil/leucovorin versus doxorubicin as palliative chemotherapy in patients with advanced hepatocellular carcinoma from Asia. J Clin Oncol 31:3501–3508. [DOI] [PubMed] [Google Scholar]
- Rimassa L, Pressiani T, Personeni N, Santoro A. (2017) Regorafenib for the treatment of unresectable hepatocellular carcinoma. Expert Rev Anticancer Ther 17:567–576. [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. (2000) Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14:2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. (2013) Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. (2020) Cancer statistics, 2020. CA Cancer J Clin 70:7–30. [DOI] [PubMed] [Google Scholar]
- Tallarida RJ. (2006) An overview of drug combination analysis with isobolograms. J Pharmacol Exp Ther 319:1–7. [DOI] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT. (2008) Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis 13:1215–1222. [DOI] [PubMed] [Google Scholar]
- Tu MJ, Ho PY, Zhang QY, Jian C, Qiu JX, Kim EJ, Bold RJ, Gonzalez FJ, Bi H, Yu AM. (2019) Bioengineered miRNA-1291 prodrug therapy in pancreatic cancer cells and patient-derived xenograft mouse models. Cancer Lett 442:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RM. (2016) Combination therapies in oncology. Nat Rev Drug Discov 15:81–82. [DOI] [PubMed] [Google Scholar]
- Wielinga PR, van der Heijden I, Reid G, Beijnen JH, Wijnholds J, Borst P. (2003) Characterization of the MRP4- and MRP5-mediated transport of cyclic nucleotides from intact cells. J Biol Chem 278:17664–17671. [DOI] [PubMed] [Google Scholar]
- Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, et al. (2004) BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 64:7099–7109. [DOI] [PubMed] [Google Scholar]
- Yi W, Tu MJ, Liu Z, Zhang C, Batra N, Yu AX, Yu AM. (2020) Bioengineered miR-328-3p modulates GLUT1-mediated glucose uptake and metabolism to exert synergistic antiproliferative effects with chemotherapeutics. Acta Pharm Sin B 10:159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SO, Shin S, Mercurio AM. (2006) Ras stimulation of E2F activity and a consequent E2F regulation of integrin alpha6beta4 promote the invasion of breast carcinoma cells. Cancer Res 66:6288–6295. [DOI] [PubMed] [Google Scholar]
- Yu A-M, Pan Y-Z. (2012) Noncoding microRNAs: small RNAs play a big role in regulation of ADME? Acta Pharm Sin B 2:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A-M, Tian Y, Tu M-J, Ho PY, Jilek JL. (2016) MicroRNA pharmacoepigenetics: posttranscriptional regulation mechanisms behind variable drug disposition and strategy to develop more effective therapy. Drug Metab Dispos 44:308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Lv H, Peng B, Wang C, Yu Y, He Z. (2009) Role of BCRP as a biomarker for predicting resistance to 5-fluorouracil in breast cancer. Cancer Chemother Pharmacol 63:1103–1110. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tu M-J, Yu Y-F, Wang W-P, Chen Q-X, Qiu J-X, Yu A-X, Yu A-M. (2015) Combination therapy with bioengineered miR-34a prodrug and doxorubicin synergistically suppresses osteosarcoma growth. Biochem Pharmacol 98:602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AX, Baron AD, Malfertheiner P, Kudo M, Kawazoe S, Pezet D, Weissinger F, Brandi G, Barone CA, Okusaka T, et al. (2017) Ramucirumab as second-line treatment in patients with advanced hepatocellular carcinoma: analysis of REACH trial results by child-pugh score. JAMA Oncol 3:235–243. [DOI] [PubMed] [Google Scholar]