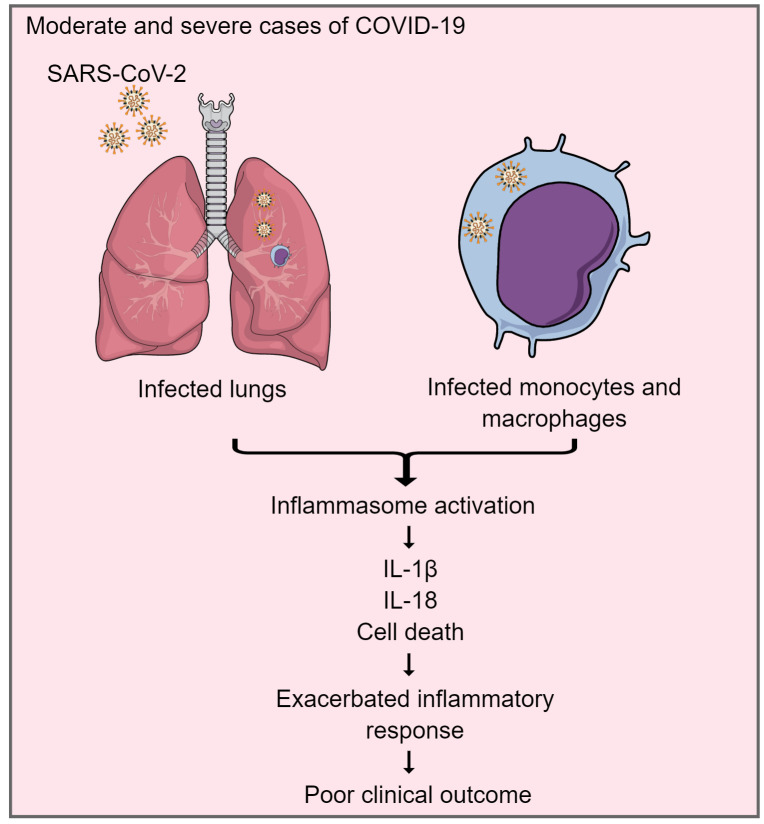
This work shows that inflammasomes are activated in response to SARS-CoV-2 infection in vitro and in COVID-19 patients. Activation of inflammasomes in moderate and severe cases of COVID-19 contributes to the exacerbated inflammatory response, impacting disease progression and clinical outcome.
Abstract
Severe cases of COVID-19 are characterized by a strong inflammatory process that may ultimately lead to organ failure and patient death. The NLRP3 inflammasome is a molecular platform that promotes inflammation via cleavage and activation of key inflammatory molecules including active caspase-1 (Casp1p20), IL-1β, and IL-18. Although participation of the inflammasome in COVID-19 has been highly speculated, the inflammasome activation and participation in the outcome of the disease are unknown. Here we demonstrate that the NLRP3 inflammasome is activated in response to SARS-CoV-2 infection and is active in COVID-19 patients. Studying moderate and severe COVID-19 patients, we found active NLRP3 inflammasome in PBMCs and tissues of postmortem patients upon autopsy. Inflammasome-derived products such as Casp1p20 and IL-18 in the sera correlated with the markers of COVID-19 severity, including IL-6 and LDH. Moreover, higher levels of IL-18 and Casp1p20 are associated with disease severity and poor clinical outcome. Our results suggest that inflammasomes participate in the pathophysiology of the disease, indicating that these platforms might be a marker of disease severity and a potential therapeutic target for COVID-19.
Graphical Abstract
Introduction
COVID-19 is an inflammatory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which can manifest as a broad spectrum of symptoms ranging from few or no symptoms to severe pneumonia that may evolve to acute respiratory distress syndrome and death (Merad and Martin, 2020). While the molecular mechanisms driving disease severity remain unclear, the clinical association of inflammatory mediators such as IL-6 and lactate dehydrogenase (LDH) with severe cases suggests that excessive inflammation is central to a poor clinical outcome (Chen et al., 2020; Han et al., 2020; Huang et al., 2020). The induction of inflammatory processes in the host cell often requires the engagement of inflammasomes, which are protein platforms that aggregate in the cytosol in response to different stimuli (Broz and Dixit, 2016). The NLRP3 inflammasome, the most studied one of such platforms, comprises the NLRP3 receptor, the adaptor molecule apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC), and caspase-1. Caspase-1 is activated by proteolytic cleavage and promotes the activation of substrates, including the inflammatory cytokines IL-1β and IL-18 and gasdermin-D, a pore-forming protein that induces an inflammatory form of cell death called pyroptosis (Broz and Dixit, 2016). NLRP3 activation in response to microbial infections, cell damage, or aggregates in the host cell cytoplasm promotes ASC polymerization, leading to the formation of a micron-sized structure called puncta (or speck) that is a hallmark of active inflammasomes in the cells (Hauenstein et al., 2015). The participation of inflammasomes in the pathogenesis of COVID-19 remains unknown. The presence of cell death and inflammasome-derived products such as IL-1β, IL-18, and LDH in patients’ sera suggests inflammasome engagement (Chen et al., 2020; Chi et al., 2020; Han et al., 2020; Huang et al., 2020; Lucas et al., 2020; Wen et al., 2020), but the definitive demonstration of inflammasome participation is still required because these products can be produced via inflammasome-independent pathways (Alfaidi et al., 2015; Guma et al., 2009; Joosten et al., 2009).
Here we enrolled 124 patients with severe and moderate COVID-19 and demonstrated the activation of the inflammasome during the disease and in postmortem tissues. We determined that the magnitude of the inflammasome activation is associated with the clinical outcome of the disease, supporting future studies to target inflammasomes as therapeutic strategies for COVID-19.
Results and discussion
SARS-CoV-2 infection triggers NLRP3 inflammasome in primary human monocytes
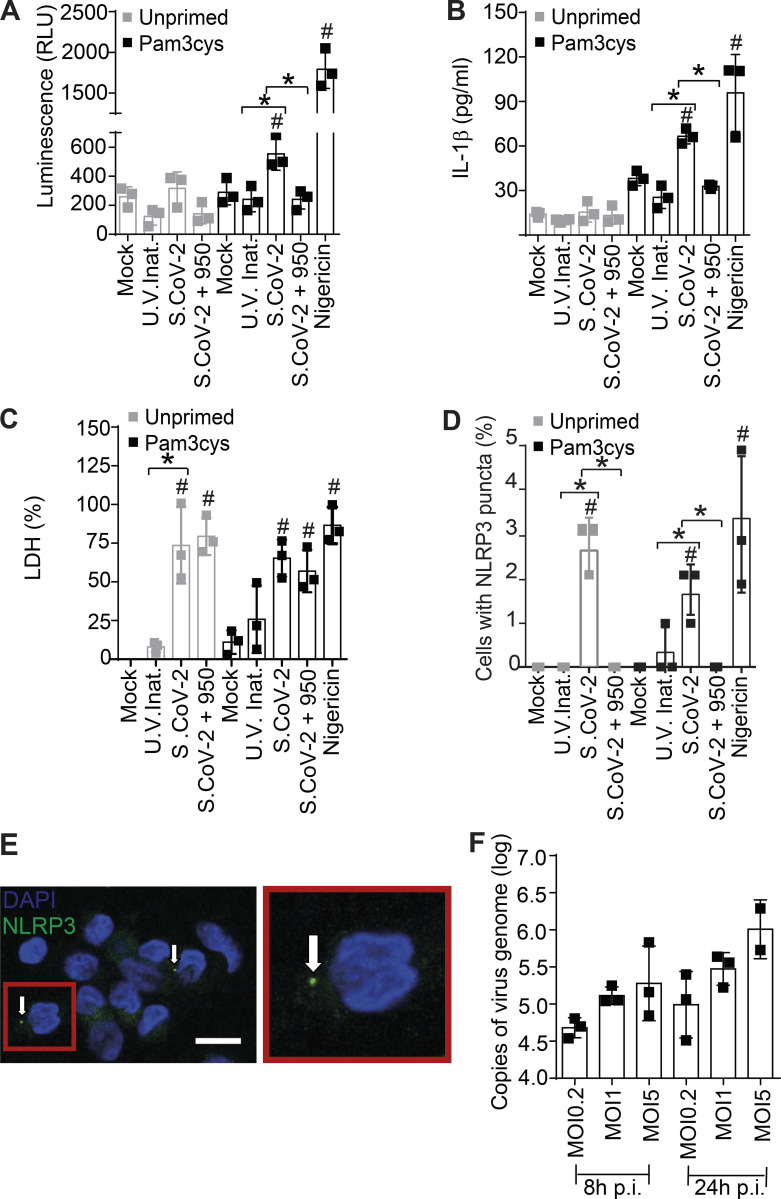
The pronounced inflammatory characteristics of COVID-19 and the correlation of disease severity with the pyroptosis marker LDH prompted us to investigate the activation of the inflammasome by SARS-CoV-2 and its role in disease development. Initially we infected primary human monocytes in vitro with SARS-CoV-2 and assessed inflammasome activation. Using monocytes from healthy donors, we found that SARS-CoV-2 infection triggers caspase-1 activation, as measured by the Caspase-Glo 1 luminescent assay that detects caspase-1 activity in the media (Fig. 1 A), and IL-1β production, quantified using ELISA (Fig. 1 B). Nigericin, a bacterial toxin known to trigger NLRP3 inflammasome, was used as a positive control. Caspase-1 activation and IL-1β production required viable SARS-CoV-2, as UV-inactivated virus did not induce caspase-1 activation and IL-1β production (Fig. 1, A and B). Importantly, SARS-CoV-2–induced caspase-1 activation and IL-1β production were inhibited by MCC950, a potent and selective inhibitor of NLRP3 (Coll et al., 2015). We also measured LDH release in response to infection and found that SARS-CoV-2 triggers LDH release in monocytes in a process that is independent of priming and requires viable SARS-CoV-2 (Fig. 1 C). MCC950 did not affect LDH release, suggesting that NLRP3-independent responses operate for induction of a lytic form of cell death induced by the virus (Fig. 1 C). Similar results were obtained when we simultaneously tested monocytes from three different donors (Fig. S1, A–C). Next, we measured the formation of NLRP3 and ASC puncta and found that SARS-CoV-2 triggers puncta formation in a process that requires viable virus (Fig. 1, D and E; and Fig. S1, D and E). As previously reported, priming is not required for NLRP3 activation in primary human monocytes (Gritsenko et al., 2020; Netea et al., 2009). Importantly, the formation of NLRP3 puncta was inhibited by MCC950, confirming the antibody specificity and assuring the engagement of NLRP3 inflammasome. MCC950 did not significantly inhibit the formation of ASC puncta, a feature that supports the hypothesis that additional ASC-dependent inflammasomes may be also activated by SARS-CoV-2 (Fig. S2 D). Finally, we measured the viral load using RT-PCR and confirmed that SARS-CoV-2 is able to infect human monocytes in vitro (Fig. 1 F). Collectively, these data indicate that SARS-CoV-2 infects human monocytes and triggers NLRP3 activation and a lytic form of cell death. Unlike caspase-1 activation, IL-1β production, and NLRP3 puncta formation, LDH release was not affected by MCC950 treatment, suggesting that additional pathways may operate for induction of lytic cell death in response to SARS-CoV-2. These questions and the determination of which specific viral proteins are involved in NLRP3 activation and induction of cell death shall be addressed in future studies. Regardless of the possible participation of additional inflammasomes, the experiments performed with MCC950, together with the antibody-staining data, caspase-1 activation, and IL-1β production, strongly support the engagement of the NLRP3 inflammasome in the innate immune responses to SARS-CoV-2.
Figure 1.
Infection of primary human monocytes with SARS-CoV-2 triggers NLRP3 inflammasome activation and cell death. Human CD14+ monocytes were primed or not with Pam3Cys (300 ng/ml) for 4 h and infected with SARS-CoV-2 at an MOI of 5 (or as indicated) for 24 h. Mock was used as a negative infection control and nigericin as a positive control for NLRP3 activation. The NLRP3 inhibitor MCC950 (10 µM) was added 1 h after viral infection and maintained (S.CoV-2 + 950). When indicated, the SARS-CoV-2 was inactivated by UV irradiation (U.V. Inat.). (A) Caspase-1 activity was measured in the tissue culture supernatants using the Caspase-Glo 1 assay. (B) IL-1β production in the tissue culture supernatants. (C) LDH release in the tissue culture supernatants. Triton X-100 (9%) was used to induce cell death and estimate 100% death. (D) Percentage of cells containing NLRP3 puncta was estimated by microscopy scoring. (E) A representative image of a monocyte containing NLRP3 puncta (green, indicated by arrows) is shown. Inset (red) shows a higher magnification of a cell containing NLRP3 puncta. DAPI stains cell nuclei (blue). Scale bar, 10 µm. (F) Viral loads in the cell culture supernatants were estimated by RT-PCR in monocytes infected for 8 and 24 h at the indicated MOI. #, P < 0.05 compared with mock treated cells; *, P < 0.05 comparing the indicated groups, as determined by Student’s t test. A, Pam3cys: Mock × S.CoV-2, P = 0.032; U.V. Inat. × S.Cov-2, P = 0.018; S.CoV-2 × S.Cov-2 + 950, P = 0.014; Mock × Nigericin, P = 0.0005. B, Pam3cys: Mock × S.CoV-2, P = 0.0022; U.V. Inat. × S.Cov-2, P = 0.0014; S.CoV-2 × S.Cov-2 + 950, P = 0.0004; Mock × Nigericin, P = 0.0194. C, unprimed: Mock × S.CoV-2, P = 0.0049; U.V. Inat. × S.Cov-2, P = 0.0104. C, Pam3cys: Mock × S.CoV-2, P = 0.0027; Mock × Nigericin, P = 0.0007. D, unprimed: Mock × S.CoV-2, P = 0.0013; U.V. Inat. × S.Cov-2, P = 0.0013; S.CoV-2 × S.Cov-2 + 950, P = 0.0075. D, Pam3cys: Mock × S.CoV-2, P = 0.0075; U.V. Inat. × S.Cov-2, P = 0.0474; S.CoV-2 × S.Cov-2 + 950, P = 0.0075; Mock × Nigericin, P = 0.0194. Box shows the average of triplicates ± SD of the values. Each technical triplicate is indicated as a square in the figure. Shown is one representative experiment out of three (A–F) experiments performed with similar results. p.i., postirradiation; RLU, relative light unit; S.CoV-2, SARS-CoV-2.
Figure S1.
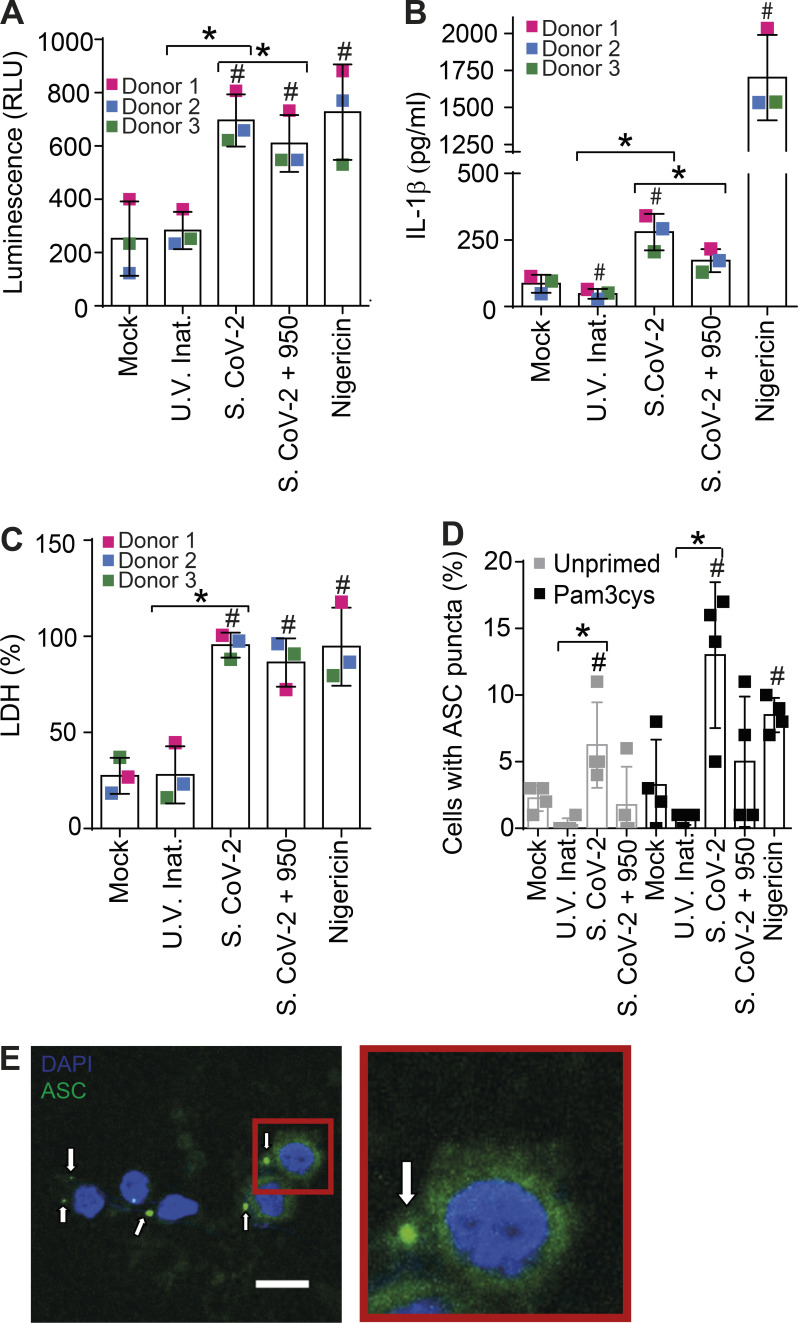
Infection of primary human monocytes with SARS-CoV-2 triggers NLRP3 inflammasome activation and cell death. (A–C) Human CD14+ monocytes obtained from three independent healthy donors (each labeled with a different color) were primed with Pam3Cys (300 ng/ml) for 4 h and infected with SARS-CoV-2 at an MOI of 5 for 24 h. Mock was used as a negative infection control and nigericin as a positive control for NLRP3 activation. The NLRP3 inhibitor MCC950 (10 µM) was added 1 h after viral infection and maintained (S.CoV-2 + 950). SARS-CoV-2 was inactivated by UV irradiation (U.V. Inat.). (A) Caspase-1 activity was measured in the tissue culture supernatants using the Caspase-Glo 1 assay. (B) IL-1β production in the tissue culture supernatants of monocytes. (C) LDH release measured in the tissue culture supernatants. Triton X-100 (9%) was used to induce cell death and estimate 100% death. (D) Human CD14+ monocytes obtained from one single donor were primed or not with Pam3Cys (300 ng/ml) for 4 h and infected with SARS-CoV-2 at a MOI of 5 for 24 h. The percentage of cell containing ASC puncta was estimated by microscopy scoring. (E) A representative image of monocytes containing ASC puncta (green, indicated by white arrows) is shown. Inset (red) shows a higher magnification of a cell containing ASC puncta. DAPI stains cell nuclei (blue). Scale bar 10 µm. #, P < 0.05 as compared with Mock treated cells; *, P < 0.05 comparing the two indicated groups, as determined by Student’s t test. Box shows the average ± SD of the values. (A–C) Each square represents data from a single donor (A–C) or data from four technical replicates performed using cells from a single donor (D). We show one representative experiment out of three independent experiments performed with similar results.
Figure S2.
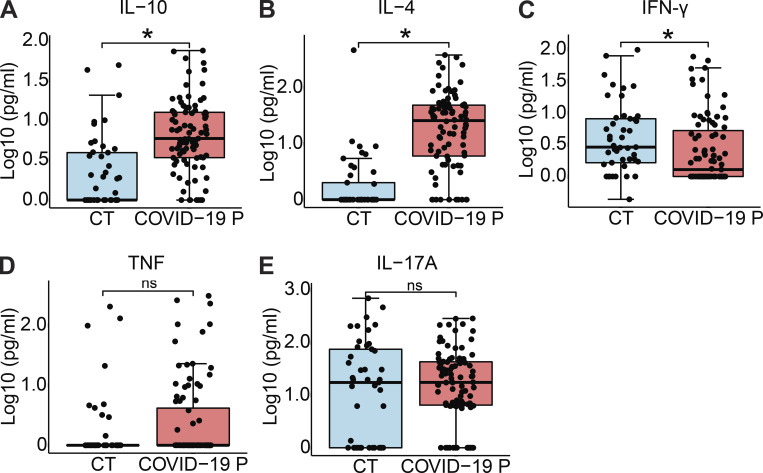
Cytokine production in COVID-19 patients. Cytokine concentration in the serum controls individuals (CT, n = 45) and COVID-19 patients (COVID-19 P, n = 92; all tested positive using RT-PCR). (A–E) IL-10 (A), IL-4 (B), IFN-γ (C), TNF-α (D), and IL-17A (E) were measured by CBA. Data are shown as Log10-transformed concentrations in picograms per milliliter. *, P < 0.05 as determined by Student’s t test (A, P = 0.001550; B, P = 0.000000394; C, P = 0.029000; D, P = 0.3; E, P = 0.49). Each dot represents the value from a single individual. Box shows average ± SD of the values. ns, not significant.
The NLRP3 inflammasome is active in COVID-19 patients
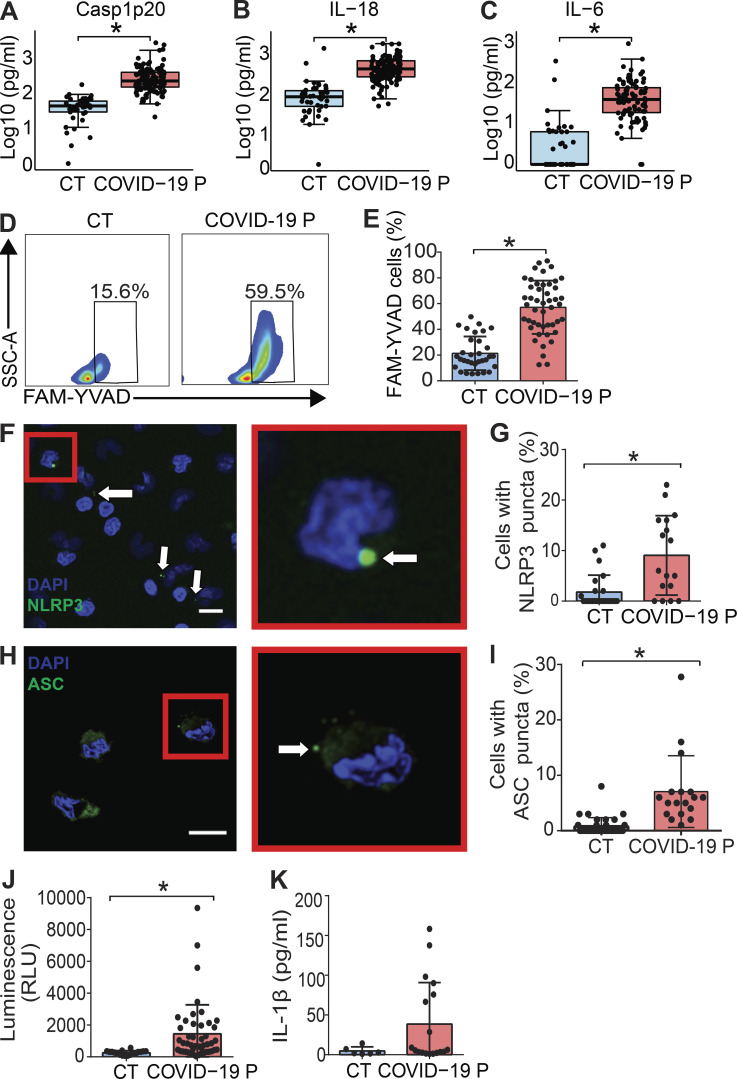
Next, we tested inflammasome activation in the sera of COVID-19 patients. It was previously shown that IL-1β is activated regardless of caspase-1 in vivo (Alfaidi et al., 2015; Guma et al., 2009; Joosten et al., 2009); therefore, we assessed active/cleaved caspase-1 (Casp1p20) and cleaved IL-18 as readouts for inflammasome activation in COVID-19 patients. We tested sera from 124 COVID-19 patients (Table S1) sampled on the day of hospitalization (all tested RT-PCR–positive for SARS-CoV-2) and compared them with sera of 42 controls that tested RT-PCR/serology–negative or that were collected before the COVID-19 pandemic. We found higher concentrations of Casp1p20 and IL-18 in the sera of patients (Fig. 2, A and B), suggesting active inflammasome in COVID-19 patients. We also found increased IL-6, IL-10, and IL-4 in patients when compared with controls, whereas we did not detect statistically significant differences for TNF-α and IL-17A (Fig. 2 C and Fig. S2, D and E). IFN-γ levels were slightly lower in COVID-19 patients compared with controls (Fig. S2 C). Lymphopenia is well reported in COVID-19 patients (Huang et al., 2020; Wang et al., 2020); thus, it is possible that the reduced numbers of circulating T cells in COVID-19 patients explain the reduced IFN-γ levels in patients’ sera (Fig. S2 C). We then measured inflammasome activation in peripheral blood mononuclear cells (PBMCs) from 47 patients and compared them with 32 healthy individuals. Using the FAM-YVAD assay that stains active intracellular caspase-1 (Zamboni et al., 2006), we found that on the day of hospitalization, the PBMCs from patients showed a higher percentage of FAM-YVAD+ cells compared with healthy controls (Fig. 2, D and E). Microscopy observation of these cells allows clear visualization of NLRP3 and ASC puncta in PBMCs, indicating active inflammasomes in cells from COVID-19 patients (Fig. 2, F–I). We further confirmed caspase-1 activation using a luminescent assay and found active caspase-1 in supernatants from 46 patients’ PBMCs cultures, as opposed to low caspase-1 activation in healthy donors (Fig. 2 J). We also detected IL-1β in the supernatants of PBMCs from some patients, but not from healthy donors (Fig. 2 K), further indicating inflammasome activation in PBMCs from COVID-19 patients.
Figure 2.
Inflammasome activation in COVID-19 patients. (A–C) Cytokine concentration in the serum control individuals (CT, n = 42 to ELISA and 45 to CBA) and COVID-19 patients (COVID-19 P, n = 124 to ELISA and 92 to CBA; all tested positive using RT-PCR). Active caspase-1 (Casp1p20; A) and IL-18 (B) were measured by ELISA, and IL-6 (C) was measured by CBA. Data shown as Log10-transformed concentrations in picograms per milliliter. (D–I) PBMCs were isolated from fresh blood of CT or COVID-19 P. (D and E) FAM-YVAD+ PBMCs were estimated by FACS using the FLICA Caspase-1 Assay Kit. (D) Representative histograms of one representative CT and one COVID-19 P indicate the gate for determination of the percentage of FAM-YVAD+ cells. (E) The percentage of FAM-YVAD+ cells for the 32 CT and 47 COVID-19 P. (F) PBMCs from COVID-19 P were stained with anti-NLRP3 (green) for determination of NLRP3 puncta (white arrows) and DAPI to stain the cell nuclei (blue). Inset (red) shows a higher magnification of a cell containing NLRP3 puncta. Scale bar, 10 µm. (G) The percentages of cells with NLRP3 puncta are shown for 24 CT and 17 COVID-19 P. (H) PBMCs from COVID-19 P were stained with anti-ASC (green) for determination of ASC puncta (white arrow). DAPI stains the cell nuclei (blue). Inset (red) shows a higher magnification of a cell containing ASC puncta. Scale bar, 10 µm. (I) The percentages of cells with ASC puncta are shown for 35 CT and 18 COVID-19 P. (J) PBMCs from 18 CT and 46 COVID-19 P were maintained in culture for 16 h, and the supernatants were assayed for caspase-1 activity using the Caspase-Glo 1 Assay. (K) PBMCs from 6 CT or 18 COVID-19 P were maintained in culture for 16 h and IL-1β production were estimated by ELISA. *, P < 0.05 as determined by Student’s t test (A, P = 0.00000025; B, P = 0.0000063; C, P = 0.00000025) or Mann–Whitney test (E, P = 0.00018; G, P = 0.00076; I, P = 0.0011; J, P = 0.0076; K, P = 0.1293). Each dot represents the value from a single individual. Box corresponds to average ± SD of the values. Data shown in E, G, and I–K result from multiple experiments that have been pooled to generate the figures. SSC-A, side scatter–A; RLU, relative light unit.
NLRP3 inflammasome is present in postmortem tissues of COVID-19 patients
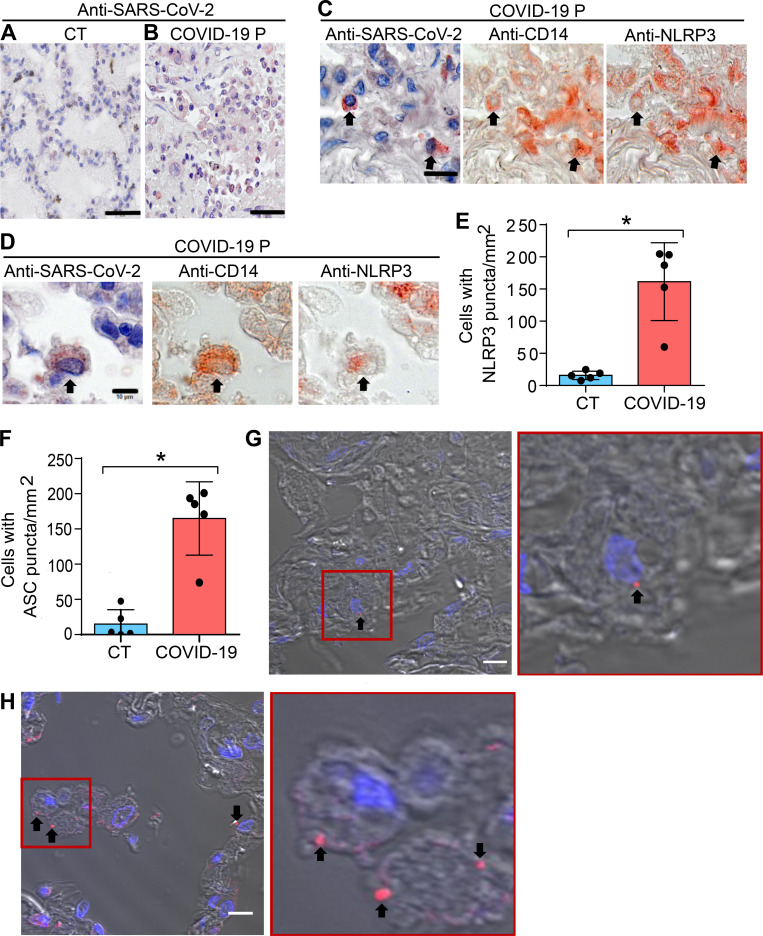
We further assessed inflammasome activation in lung tissues obtained from autopsies of deceased COVID-19 patients. Using an anti–SARS-CoV-2 antibody, we first confirmed virus presence by immunohistochemical staining of SARS-CoV-2 in postmortem lung tissues (Fig. 3, A and B). We then performed multiplex staining by sequential immunohistochemistry with SARS-CoV-2, anti-CD14, and anti-NLRP3 and identified infected CD14+ cells expressing NLRP3 in postmortem tissues (Fig. 3, C and D). Using multiphoton microscopy, we quantified the number of NLRP3 and ASC puncta in the lungs of five controls and five COVID-19 patients, and found that patients contain higher numbers of NLRP3 (Fig. 3 E) and ASC puncta (Fig. 3 F) compared with controls. Representative images of lung tissues stained with anti-NLRP3 (Fig. 3 G) or anti-ASC (Fig. 3 H) antibody illustrate the puncta formation, indicating active inflammasomes in lethal cases of COVID-19.
Figure 3.
Lung histopathological analysis and NLRP3 activation in fatal cases of COVID-19. Representative pulmonary histological findings in COVID-19 patient (COVID-19 P), autopsied by ultrasound-guided minimally invasive autopsy. (A and B) Representative immunohistochemical image of tissues from control (CT; A) or COVID-19 P (B) stained with anti–SARS-CoV-2. Scale bar, 50 µm. (C and D) Multiplex staining by sequential immunohistochemistry staining with anti–SARS-CoV-2, anti-CD14, and anti-NLRP3. Arrows indicate infected CD14+ cells expressing NLRP3. Scale bar, 20 µm (C) and 10 µm (D). (E and F) Quantification of NLRP3 (E) or ASC puncta (F) in pulmonary tissues of five CT and five COVID-19 P. Each dot in the figure represents the value obtained from each individual. Box corresponds to average ± SD of the values. *, P < 0.05, as determined by Student’s t test (E, P = 0.0079; F, P = 0.0079). (G and H) Multiphoton microscopy of pulmonary tissues stained with anti-NLRP3 (G) or anti-ASC (H) antibody showing inflammasome puncta (red, indicated by a black arrow). Insets (red) show a higher magnification of a cell containing NLRP3 (G) and ASC (H) puncta. DAPI stains cell nuclei (blue). Scale bar, 10 µm.
Inflammasome activation influences the clinical outcome of COVID-19
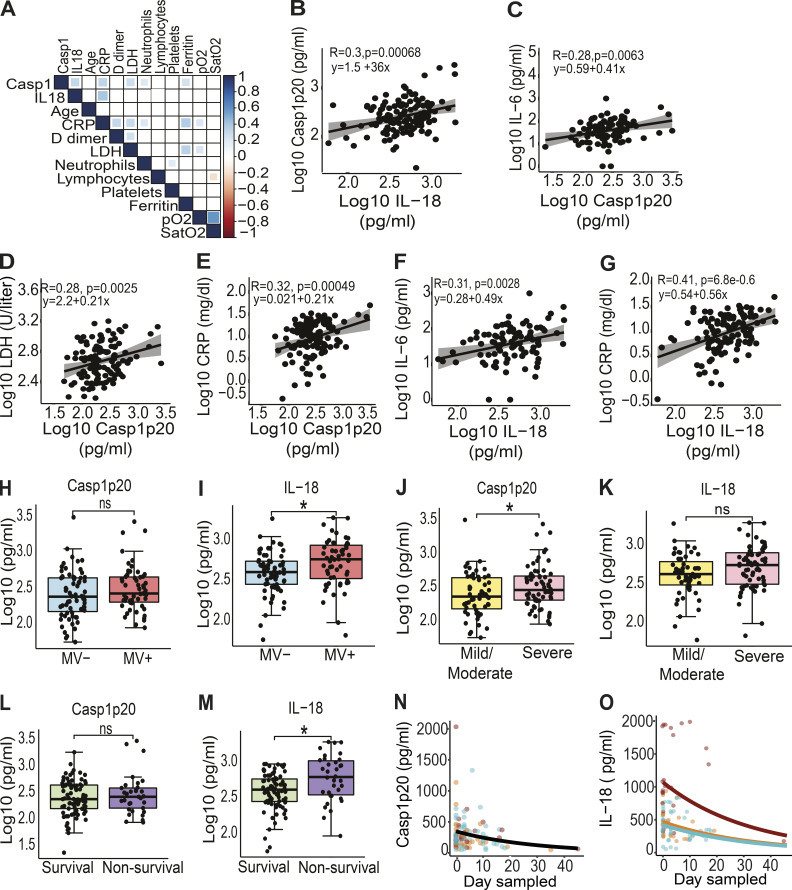
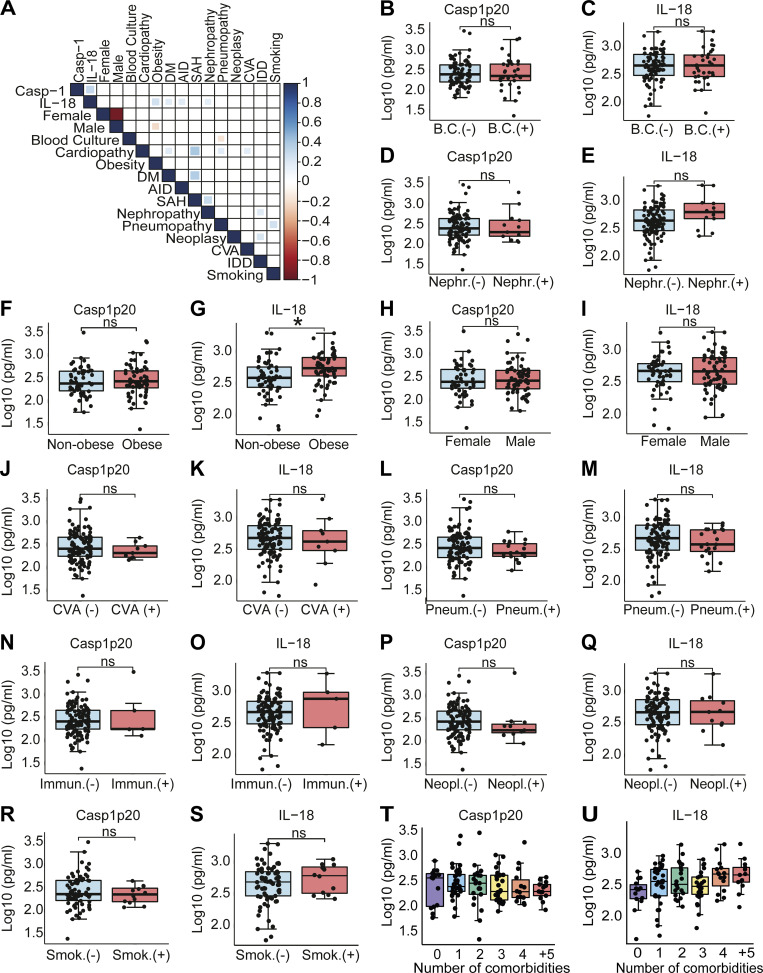
We next assessed the impact of the inflammasome activation in the clinical outcome of the disease. Initially, we performed analyses using the levels of Casp1p20 and IL-18 in 124 patients’ sera obtained on the day of hospitalization. We performed a Pearson correlation matrix to evaluate multiple parameters. These analyses were performed in R to generate the correlation matrix and the corrplot function to generate the graphical display of the correlation matrix. We report the correlations comprising P values < 0.05. We detected positive associations of Casp1p20 and/or IL-18 levels with inflammatory markers such as C-reactive protein (CRP), LDH, and ferritin (Fig. 4 A). As expected, we identified a positive correlation between Casp1p20 and IL-18 levels (Fig. 4 B). In addition, Casp1p20 positively correlated with IL-6, LDH, and CRP (Fig. 4, C–E). Furthermore, we found that IL-18 levels positively correlated with IL-6 and CRP levels (Fig. 4, F and G). We also evaluated whether the levels of Casp1p20 and IL-18 were affected by patients’ comorbidities and clinical parameters, including bacterial coinfections (cultivable bacteria in the blood), nephropathy, obesity, gender, cerebrovascular accident, pneumopathy, immunodeficiency, and neoplasia (Fig. S3). We only detected statistically significant differences when comparing obese with nonobese patients, as the levels of IL-18 were higher in patients with body mass index ≥30 (Fig. S3 G). We also analyzed patients according to the number of comorbidities and did not detect statistically significant differences in Casp1p20 and IL-18 levels according to the number of patients’ comorbidities (Fig. S3, T and U).
Figure 4.
Inflammasome activation influences the clinical outcome of COVID-19. (A) Correlation matrix of Casp1p20 and IL-18 levels in the serum of COVID-19 patients on the hospitalization day with patient characteristics and clinical parameters. (B–J) Correlations of Casp1p20 with IL-18 (B), Casp1p20 with IL-6 (C), Casp1p20 with LDH (D), Casp1p20 with CRP (E), IL-18 with IL-6 (F), and IL-18 with CRP (G). (H and I) Levels of Casp1p20 (H) and IL-18 (I) in patients who required MV (MV+, blue box) or not (MV−, red box). (J and K) Levels of Casp1p20 (J) and IL-18 (K) in patients with mild/moderate (yellow box) or severe COVID-19 (pink box). (L and M) Levels of Casp1p20 (L) and IL-18 (M) in survivors (green box) or nonsurvivors (purple box). The levels of Casp1p20 and IL-18 were measured by ELISA and are shown as Log10-transformed concentrations in picograms per milliliter. *, P < 0.05 as determined by Student’s t test (H, P = 0.160; I, P = 0.014; J, P = 0.036; K, P = 0.068; L, P = 0.430; M, P = 0.0044). Each dot represents value from a single individual. Box corresponds to average ± SD of the values. (N and O) Derived predictions from the best-fit models retained in Casp1p20 (N) and IL-18 (O) longitudinal analyses; IL-18 model (O) comprises variation in the intercept among patient groups: death (red), critical/recovery (orange), and mild/recovery (blue). ns, not significant.
Figure S3.
Association of inflammasome activation with clinical characteristics and comorbidities. (A) Matrix correlation of Casp1p20 and IL-18 levels in the serum of COVID-19 patients on the hospitalization day with clinical parameters and comorbidities. (B–S) Levels of Casp1p20 (B, D, F, H, J, L, N, P, and R) and IL-18 (C, E, G, I, K, M, O, Q, and S) in patients with such clinical parameters as cultivable bacteria in the blood (B and C), nephropathy (Nephr.; D and E), obesity (F and G), gender (H and I), cerebrovascular accident (CVA; J and K), pneumopathy (Pneum.; L and M), immunodeficiency (Immun.; N and O), neoplasia (Neopl.; P and Q), and smoking (Smok.; R and S). (T and U) Casp1p20 (T) and IL-18 (U) levels in the sera of COVID-19 patients associated with the number of patient comorbidities, ranging from no comorbidities (0) to more than five comorbidities (+5). The levels of Casp1p20 and IL-18 were measured by ELISA and are shown as Log10-transformed concentrations in picograms per milliliter. *, P < 0.05 as determined by Student’s t test (G, P = 0.0054). Each dot represents a value from a single individual. Box shows average ± SD of the values. B.C., blood culture; DM, diabetes mellitus; AID, autoimmune diseases; SAH, systemic arterial hypertension; CVA, cerebrovascular accident; IDD, immunodeficiency; ns, not significant.
Next we investigated whether Casp1p20 and IL-18 levels measured on the day of hospitalization correlated with the clinical outcome of the disease. Importantly, we found that IL-18 levels, but not Casp1p20, were higher in patients who required mechanical ventilation (MV) compared with patients who did not (Fig. 4, H and I). When we separated the patients according to the severity of the disease (mild/moderate versus severe), we found that the levels of Casp1p20 but not IL-18 were higher in patients with the severe form of COVID-19 (Fig. 4, J and K). We also observed that the levels of IL-18, but not Casp1p20, were higher in lethal cases of COVID-19 compared with survivors (Fig. 4, L and M). Finally, we performed longitudinal analyses of IL-18 and Casp1p20 production in 37 patients from the day of hospitalization (day zero) for up to 45 d after admission using generalized mixed models with “gamma” distribution and “log” link function as the best dataset prediction. For these analyses, we categorized the patients as death, mild–recovery (patients who were hospitalized but did not require MV and recovered), and critical–recovery (patients who required MV in the intensive care unit and recovered). For Casp1p20, the most parsimonious model indicated a significant effect on the day sampled, and the production level decreased with time regardless of sex or patient outcome (Fig. 4 N). For IL-18, the best fit model retained also described an overall reduction in IL-18 production along time (day sampled), which differed among patient groups. This model predicted a decrease in IL-18 at similar rates among the three groups, with patients who died presenting higher levels (intercept) that never reached those observed in mild–recovery and critical–recovery patients (Fig. 4 O), which supports our prediction that the magnitude of inflammasome activation impacts the disease outcome. For certain parameters, we detected statistically significant differences in IL-18, whereas in others, the differences were identified in Casp1p20. Caspase-1–independent activation of IL-18 has already been described (Sugawara et al., 2001; Tsuchiya et al., 2014; Tsutsui et al., 1999), and therefore one cannot exclude the possibility that an inflammasome-independent pathway may account for elevated IL-18 production in COVID-19 patients. However, this hypothesis is not supported by our data, indicating a significant correlation of IL-18 with Casp1p20 in COVID-19 sera (Fig. 4, A and B). In addition, our results demonstrating elevated levels of Casp1p20 in patients, together with the demonstration of NLRP3 and ASC specks in patients’ PBMCs and in postmortem tissues, strongly support our assertion that inflammasomes are active in COVID-19 patients and correlate with the outcome of COVID-19. In summary, our data demonstrate that the inflammasome is robustly active in COVID-19 patients who require hospitalization. Results also support that both the magnitude of inflammasome activation on the hospitalization day and the course of inflammasome activation during hospitalization influence the clinical outcome. In this study, we did not test asymptomatic patients for inflammasome activation, but one might expect that Casp1p20 and IL-18 levels would be intermediate between uninfected individuals and mild/moderate patients. It is possible that inflammasome activation in asymptomatic and mild patients is beneficial to the host and may account for protective host responses against the virus, as opposed to the detrimental effect of the inflammasome in generating exacerbated inflammation in patients with severe forms of COVID-19. Regardless of the possible protective role of the inflammasome in mild and asymptomatic patients, our observations suggest that the inflammasome is key for the induction of the massive inflammation observed in severe and fatal cases of COVID-19. Our study supports the use of inflammasome activation both as a marker of disease severity and prognostic and as a potential therapeutic target for COVID-19. It is worth mentioning that ongoing clinical trials aim the inhibition of inflammasome and IL-1 signaling using different drugs, including Anakinra (Cavalli et al., 2020; Huet et al., 2020). In agreement with our data, a recent study indicates that exacerbated IL-18 production is associated with disease severity (Lucas et al., 2020). Thus, a clinical approach that inhibits targets that are upstream of IL-1, such as inflammatory caspases and gasdermin-D, may be more promising than solely blocking IL-1 signaling. In this scenario, determination of specific mechanisms by which SARS-CoV-2 triggers the inflammasome activation, and understanding which specific inflammasome platforms are activated during the disease will generate important information for effective therapeutic strategies to target COVID-19.
Materials and methods
Patients
A total of 124 patients with COVID-19 who tested positive using RT-PCR as described previously (Corman et al., 2020; Nalla et al., 2020) were enrolled in this study. Patients were classified according to their clinical manifestations in (1) mild cases, in which the clinical symptoms are mild and no pneumonia manifestations can be found in imaging; (2) moderate cases, in which patients have symptoms such as fever and respiratory tract symptoms, and pneumonia manifestations identifiable in imaging; and (3) severe cases, meaning adults who met any of the following criteria: respiratory rate >30 breaths/min, oxygen saturations 93% at a rest state, and arterial partial pressure of oxygen (PaO2)/oxygen concentration (FiO2) <300 mm Hg (Wu and McGoogan, 2020). Patients were enrolled in Hospital das Clínicas, Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo from April 6 to July 2, 2020. Table S1 summarizes clinical, laboratory, and treatment records. We also collected samples from 73 age- and gender-matched healthy controls. Controls were collected before the COVID-19 pandemic or tested negative for COVID-19 using RT-PCR and/or serology (specific IgM and IgG antibodies; Asan Easy Test COVID-19 IgM/IgG kits; Asan Pharmaceutical Co.).
PBMC isolation
Whole blood was collected from healthy donors (Ethics Committee Protocol from the Clinical Hospital of Ribeirão Preto Universidade de São Paulo: Certificado de Apresentação para Apreciação Ética, no. 06825018.2.3001.5440) in tubes containing EDTA (BD Vacutainer CPT; BD Biosciences), according to the manufacturer’s instructions. The material was centrifuged at 400 ×g for 10 min at room temperature. Then the plasma was discarded, and the cell pellet was resuspended in PBS 1×, pH 7.4 (Gibco-BRL). The cells were applied to the Ficoll-Paque PLUS gradient column (GE Healthcare Biosciences AB). These were then centrifuged at 640 ×g for 30 min at room temperature to obtain the purified mononuclear fraction, which was carefully collected and transferred to a new tube. The cells were washed, and the pellet was resuspended in RPMI for subsequent analyses. For infection experiments in vitro, the PBMCs from healthy volunteers were quantified, and the monocytes (CD14+ cells) were purified using positive selection with magnetic nanoparticles (BD). Briefly, PBMCs were labeled with BD IMag Anti-human CD14 Magnetic Particles - DM. The cells were transferred to a 48-well culture plate and placed over a magnetic field of the cell separation. Labeled cells migrated toward the magnet (positive fraction), whereas unlabeled cells were drawn off (negative fraction). The plate was then removed from the magnetic field for resuspension of the positive fraction. The separation was repeated twice to increase the purity of the positive fraction, and the CD14+ monocytes were used for infection experiments.
Virus stock production
The SARS-CoV-2 used was the Brazil/SPBR-02/2020 strain, isolated from the first Brazilian case of COVID-19. Viral stock was propagated under BSL3 conditions in Vero American Type Culture Collection CCL81 cells, cultured in DMEM supplemented with heat-inactivated FBS (10%). For preparation of viral stocks, Vero cells were infected in DMEM in the presence of trypsin-TPCK (1 µg/µl) for 48 h at 37°C in a 5% CO2 atmosphere. When the virus induced a cytopathic effect, the cells were harvested with cell scrapers and centrifuged (10,000 ×g). Uninfected cultures of Vero cells were cultivated similarly in parallel to generate the mock controls to infection. The supernatant was stored at −80°C, and the virus titration was performed in Vero cells using standard limiting dilution to confirm the 50% tissue culture infectious dose.
In vitro infection of human monocytes
A total of 2 × 105 purified human monocytes were plated in 48-well plates and infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.2, 1, and/or 5. After 1 h of virus adsorption, fresh medium (RPMI 2% FBS without Phenol Red), with or without the NLRP3 inhibitor MCC950 (InvivoGen), 10 µM, was added. As a control for the virus activity, we inactivated SARS-CoV-2 using UV irradiation for 20 min and used them at the same MOI as indicated for the viable SARS-CoV-2. As a negative control, we used mock, which consisted of noninfected Vero cells medium. Nigericin (20 µM) was used as a positive control for inflammasome activation. Cells were incubated for 24 h at 37°C in the presence of a 5% CO2 atmosphere. After incubation, cells were processed for immunofluorescence assays, and the supernatant was collected for determination of caspase-1 activation, cytokine production, and LDH quantification.
For the purpose of virus genome measurements, the cells were washed 1 h after virus adsorption with PBS 1× to completely remove noninternalized viruses, and fresh medium (RPMI 2% FBS without Phenol Red) was added. The virus genome was measured in the supernatants at 8 and 24 h after infection.
RT-PCR for SARS-CoV-2
Detection of SARS-CoV-2 was performed with primer-probe sets for 2019-nCoV_N2 and gene E, according to US Centers for Disease Control and Prevention and Charité group protocols (Corman et al., 2020; Nalla et al., 2020). The genes evaluated (N2, E, and RNase-P housekeeping gene) were tested by one-step real-time RT-PCR using total nucleic acids extracted with TRIzol (Invitrogen) from 50 µl of cell supernatants in order to measure the genome viral load from the in vitro assays. All real-time PCR assays were done on a Step-One Plus real-time PCR thermocycler (Applied Biosystems). Briefly, RNA extraction was performed by TRIzol. A total of 100 ng of RNA was used for genome amplification, adding specific primers (20 µM), and probe (5 µM), and with TaqPath 1-Step quantitative RT-PCR Master Mix (Applied Biosystems), with the following parameters: 25°C for 2 min, 50°C for 15 min, and 95°C for 2 min, followed by 45 cycles of 94°C for 5 s and 60°C for 30 s. Primers used were the following: N2 forward: 5′-TTACAAACATTGGCCGCAAA-3′, N2 reverse: 5′-GCGCGACATTCCGAAGAA-3′; N2 probe: 5′-FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1-3′ (Nalla et al., 2020); E forward: 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′, E reverse: 5′-ATATTGCAGCAGTACGCACACA-3′; E probe: 5′-AM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ-1-3′ (Corman et al., 2020); RNase-P forward: 5′-AGATTTGGACCTGCGAGCG-3′, RNase-P reverse: 5′-GAGCGGCTGTCTCCACAAGT-3′; and RNase-P probe: 5′-FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ-1-3′ (Nalla et al., 2020).
Evaluation of active caspase-1 activity and LDH release in cultured cells
For caspase-1 and LDH determination, 2 × 105 human CD14+ monocytes were plated on 48-well plates in RPMI 10% FBS and incubated overnight. The following day, cells were infected with SARS-CoV-2 using MOI 5 in RPMI without Phenol Red (3.5 g/liter Hepes, 2 g/liter NaHCO3, 10.4 g/liter RPMI without Phenol Red, and 1% glutamine, pH 7.2). After 1 h of virus adsorption, fresh media (RPMI 2% FBS without Phenol Red) with or without MCC950 at 10 µM were added, and cells were incubated for 24 h. To measure caspase-1 activity, the supernatants were collected and incubated with the Luciferin WEHD-substrate provided by the Caspase-Glo 1 Assay (Promega). After 1 h of incubation at room temperature, luminescence was measured using the SpectraMax i3 system (Molecular Devices). LDH release was measured in the supernatants using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) following the manufacturer’s instructions.
Evaluation of caspase-1 activity in PBMCs from COVID-19 patients
To evaluate caspase-1 activation, 5 × 105 PBMCs from COVID-19 patients or healthy donors were centrifuged (400 ×g, 10 min), and cells were labeled for 30 min with the FLICA carboxyfluorescein reagent (FAM–YVAD–FMK; Immunochemistry Technologies), as recommended by the manufacturer. The cells were then washed two times with PBS 1× and fixed with fixative reagent provided by the manufacturer. Acquisition was performed in fixed cells in a flow cytometer (BD Accuri C6; BD Biosciences) and then analyzed using FlowJo (Tree Star) software. To evaluate caspase-1 activity in supernatants, 2 × 105 PBMCs from COVID-19 patients were plated in 96-well plates and incubated overnight. To measure caspase-1 activity, the supernatants were collected, and the Caspase-Glo 1 Assay (Promega) was performed following the manufacturer’s instructions.
Immunofluorescence staining of isolated cells
For staining PBMCs from COVID-19 patients, a total of 5 × 105 PBMCs were plated in 8-well chamber slides for 1 h in RPMI without FBS for monocyte adhesion before fixation. For staining cells infected in vitro, a total of 2 × 105 human monocytes were plated in 24-well plates containing coverslips and infected with SARS-CoV-2 at the indicated MOI for 16 h. For fixation of the samples, tissue culture supernatants were removed, and cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Paraformaldehyde was removed, cells were washed with PBS 1×, and the coverslips or chambers were processed for immunofluorescence as described. Cells were blocked and permeabilized using PBS 1× with goat serum and 0.05% saponin for 1 h at room temperature. Primary antibodies used were rabbit mAb anti-human NLRP3 (clone D2P5E, batch 2; 1:1,000; Cell Signaling) or rabbit polyclonal antibody anti-human ASC (1:2,000; Adipogen AL177). Antibodies were diluted in blocking solution and added to each chamber/coverslip. After 1 h of incubation, the samples were washed with PBS 1×, and secondary antibodies were added and incubated for 1 h at room temperature. Secondary antibodies used were goat anti-rabbit 488 (1:3,000; Invitrogen) and goat anti-rabbit 594 (1:3,000; Life Technologies). Slides were washed and mounted using DAPI (1 mM) and ProLong (Invitrogen).
Lung samples from autopsies and immunofluorescence and imaging
We performed adapted minimally invasive autopsies in COVID-19 patients (Avrahami et al., 1995; Damore et al., 2000; Vejrosta and Bilder, 1975). Briefly, a mini-thoracotomy (3 cm) was done under the main area of lung injury identified by prior x ray or computed tomography. The lung parenchyma was clamped by Collins Forceps, cut, and fixed in 10% buffered formalin. Pulmonary tissue samples were stained with H&E and immunostaining as reported (Fabro et al., 2015, 2019). The slides were incubated with the primary antibodies, rabbit anti-CD14 mAb (1:200; Abcam), rabbit anti-human NLRP3 mAb (clone D2P5E; 1:3,000; Cell Signaling), and rabbit anti-human ASC polyclonal antibody (1:2,000; Adipogen AL177) for 2 h at room temperature or overnight at 4°C. Goat anti-mouse Alexa Fluor 647 (Invitrogen) or goat anti-rabbit Alexa Fluor 594 (Invitrogen) was used as a secondary antibody. Images were acquired by an Axio Observer combined with an LSM 780 confocal device with 630× magnification (Carl Zeiss). Minimally invasive autopsies were approved by the Faculdade de Medicina de Ribeirão Preto/Universidade de São Paulo Ethical Committee (protocol no. 4.089.567).
Sequential immunoperoxidase labeling and erasing
Tissue sections from paraffin-embedded lung fragments obtained from COVID-19 fatal cases were tested by immunohistochemistry using anti–SARS-CoV-2 polyclonal antibody for in situ detection of SARS-CoV-2. Sequential immunoperoxidase labeling and erasing was then performed to determine additional markers after SARS-CoV-2 immune stain, using antibodies to CD14 (1:100 dilution; Abcam) and NLRP3 (1:100 dilution; Cell Signaling). After the incubation with primary antibody, the slides were incubated with an immunoperoxidase polymer anti-mouse visualization system (SPD-125; Spring Bioscience, Biogen) and then with the chromogen substrate 3-amino-9-ethylcarbazole (AEC) peroxidase system kit (SK-4200; Vector Laboratories). Microphotographs after immunostaining of tissue slides were scanned on a VS120 Olympus microscope. After high-resolution scanning, slides with coverslips were removed in PBS and dehydrated through an ethanol gradient to 95% ethanol. Slides were incubated in ethanol series until AEC color reaction was erased. Following rehydration, antibodies were eluted by incubating sections in 0.15 mM KMnO4/0.01 M H2SO4 solution for 2 min, followed immediately by a distilled water wash. Tissue was then restained as indicated in the blocking step.
Cytokine quantification in sera
Active caspase-1 (Casp1p20) and IL-18 levels were evaluated by ELISA assay (R&D Systems) in the serum from patients with COVID-19 or healthy donors following the manufacturer’s instructions. TNF-α, IL-4, IL-6, IL-10, IFN-γ, and IL-17 were quantified in the serum from patients with COVID-19 or health donors using a human cytometric bead array (CBA) cytokine kit (Th1/Th2/Th17 Cytokine Kit; BD Biosciences) following the manufacturer’s instructions. IL-1β in the tissue culture supernatants of human monocytes infected with SARS-CoV-2 was quantified by ELISA (R&D Systems) or by the CBA Human IL-1β flex set (BD Biosciences) following the manufacturer’s instructions.
Statistics
Statistical significance for the linear analysis was determined by two-tailed paired or unpaired Student t test for data that reached normal distribution, and the Mann–Whitney test was used for not normally distributed data. These statistical procedures and graph plots were performed with GraphPad Prism 8.4.2 software. In addition, longitudinal analyses were implemented to describe variation in IL-18 and Casp1p20 production over time, considering patients’ outcomes and sex as fixed factors. These analyses were performed in R (version 4.0.2) using RStudio (version 1.3.1056). The patients’ outcomes were divided into three categories (37 patients in total; 16 women and 21 men): death (n = 10 individuals, a total of 25 samples), mild–recovery (patients who were hospitalized but did not require MV; n = 17 individuals, a total of 53 samples), and critical–recovery (patients who required MV in the intensive care unit and recovered; n = 10 individuals, a total of 27 samples). Activation of IL-18 and Casp1 were evaluated separately using full models that considered the interaction of time (“Day.sampled”) with patients’ outcomes (“Outcome”) or sex (“Sex”) and included individuals (“Patient.ID”) as a random factor to control for repeated measures and individual effects. Normality and homoscedasticity of the dataset were verified and refuted for a time series dataset, and analyses were implemented using the glmmTMB package (Brooks et al., 2017) in a generalized mixed model approach with “gamma” distribution and “log” link function. An Akaike information criterion for finite samples was used to select the best models from the full finite samples using MuMIn (Barton, 2020). Models having Akaike information criterion for finite sample values within 2 U of the best-fit model were considered to have substantial support (Burnham and Anderson, 2002); adequate residuals distributions were confirmed, and representative graphics and tables were constructed using the packages DHARMa (Hartig, 2020), ggeffects (Lüdecke, 2018), and broom.mixed (Bolker and Robinson, 2020).
Study approval
The procedures followed in the study were approved by the National Ethics Committee, Brazil (Comissão Nacional de Ética em Pesquisa, Certificado de apresentação para Apreciação Ética: 30248420.9.0000.5440). Written informed consent was obtained from recruited patients.
Online supplemental material
Fig. S1 shows inflammasome activation and induction of cell death in human monocytes infected in vitro with SARS-CoV-2. Fig. S2 shows additional cytokines in the sera of COVID-19 patients and controls. Fig. S3 shows associations of inflammasome activation with clinical characteristics and comorbidities of COVID-19 patients. Table S1 shows clinical characteristics of COVID-19 patients enrolled in this study.
Supplementary Material
shows clinical characteristics of COVID-19 patients enrolled in this study.
Acknowledgments
We are grateful to Maira Nakamura, Bárbara Moreira de Carvalho e Silva, and Laura Khouri for technical assistance.
Funding was provided by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grants 2013/08216-2, 2019/11342-6, and 2020/04964-8), Center for Research in Inflammatory Disease (CRID/FAPESP grant 2013/08216-2), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq grant 304775/2016-9), Instituto Nacional de Ciência e Tecnologia de Vacinas (INCTV/CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES grant 88887.507253/2020-00).
Author contributions: T.S. Rodrigues and D.S. Zamboni conceived the study. T.S. Rodrigues, K.S.G. de Sá, A.Y. Ishimoto, A. Becerra, S. Oliveira, L. Almeida, A.V. Gonçalves, D.B. Perucello, and W.A. Andrade designed the experiments, defined parameters, collected and processed PBMC and sera samples, and analyzed data. R. Castro, J.E. Toller-Kawahisa, D.C. Nascimento, M.H.F. de Lima, C.M.S. Silva, and D.B. Caetite processed PBMC samples. L.D. Cunha supervised the collection of PBMC samples from patients. T.S. Rodrigues, R.B. Martins, I.A. Castro, and M.C. Pontelli performed the experiments with viral infections. A.T. Fabro, S.S. Batah, L. Siyuan, and M.N. Benatti performed minimally invasive autopsies. K.S.G. de Sá and F.P. Veras stained and analyzed autopsy tissues. A.Y. Ishimoto, F.C. de Barros, and T. Kohlsdorf performed bioinformatic analyses and designed and conducted statistical analyses of the data. N.B. do Amaral, M.C. Giannini, L.P. Bonjorno, M.I.F. Lopes, M.N. Benatti, R.C. Santana, F.C. Vilar, M. Auxiliadora-Martins, R. Luppino-Assad, S.C.L. de Almeida, F.R. de Oliveira, R.D.R. de Oliveira, and P. Louzada-Junior assisted in patient recruitment and collected patient specimens and the epidemiological and clinical data. P. Louzada-Junior supervised and R.D.R. de Oliveira helped with clinical data management. T.M. Cunha, J.C. Alves-Filho, F.Q. Cunha, L.D. Cunha, F.G. Frantz, T. Kohlsdorf, A.T. Fabro, E. Arruda, R.D.R. de Oliveira, and P. Louzada-Junior helped with interpretations of the data. T.S. Rodrigues and D.S. Zamboni drafted the manuscript. All authors helped in editing the manuscript. D.S. Zamboni secured funds and supervised the project.
References
- Alfaidi, M., Wilson H., Daigneault M., Burnett A., Ridger V., Chamberlain J., and Francis S.. 2015. Neutrophil elastase promotes interleukin-1β secretion from human coronary endothelium. J. Biol. Chem. 290:24067–24078. 10.1074/jbc.M115.659029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami, R., Watemberg S., Hiss Y., and Deutsch A.A.. 1995. Laparoscopic vs conventional autopsy. A promising perspective. Arch. Surg. 130:407–409. 10.1001/archsurg.1995.01430040069014 [DOI] [PubMed] [Google Scholar]
- Barton, K. 2020. MuMIn: Multi-Model Inference. R package version 1.43.17. https://CRAN.R-project.org/package=MuMIn (accessed April 14, 2020)
- Bolker, B., and Robinson D.. 2020. broom.mixed: Tidying Methods for Mixed Models. R package version 0.2.6. https://CRAN.R-project.org/package=broom.mixed (accessed May 17, 2020)
- Brooks, M.E., Kristensen K., van Benthem K.J., Magnusson A., Berg C.W., Nielsen A., Skaug H.J., Maechler M., and Bolker B.M.. 2017. glmmTMB Balances Speed and Flexibility Among Packages for Zero-inflated Generalized Linear Mixed Modeling. R J. 9:378–400. 10.32614/RJ-2017-066 [DOI] [Google Scholar]
- Broz, P., and Dixit V.M.. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16:407–420. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Burnham, K.P., and Anderson D.R.. 2002. Model Selection and Multimodel Inference: a practical information-theoretic approach. Springer-Verlag New York, New York. [Google Scholar]
- Cavalli, G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., et al. 2020. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2:e325–e331. 10.1016/S2665-9913(20)30127-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. 2020. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 130:2620–2629. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y., Ge Y., Wu B., Zhang W., Wu T., Wen T., Liu J., Guo X., Huang C., Jiao Y., et al. 2020. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 in China. J. Infect. Dis. 222:746–754. 10.1093/infdis/jiaa363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, R.C., Robertson A.A., Chae J.J., Higgins S.C., Muñoz-Planillo R., Inserra M.C., Vetter I., Dungan L.S., Monks B.G., Stutz A., et al. 2015. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21:248–255. 10.1038/nm.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damore, L.J. II, Barth R.F., Morrison C.D., Frankel W.L., and Melvin W.S.. 2000. Laparoscopic postmortem examination: a minimally invasive approach to the autopsy. Ann. Diagn. Pathol. 4:95–98. 10.1016/S1092-9134(00)90018-2 [DOI] [PubMed] [Google Scholar]
- Fabro, A.T., da Silva P.H., Zocolaro W.S., de Almeida M.S., Rangel M.P., de Oliveira C.C., Minatel I.O., Prando E.D., Rainho C.A., Teodoro W.R., et al. 2015. The Th17 pathway in the peripheral lung microenvironment interacts with expression of collagen V in the late state of experimental pulmonary fibrosis. Immunobiology. 220:124–135. 10.1016/j.imbio.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Fabro, A.T., Engelman G.G., Ferreira N.N., Velloni J.M.F., Espósito D.L.A., da Fonseca B.A.L., and Brunaldi M.O.. 2019. Yellow Fever-induced Acute Lung Injury. Am. J. Respir. Crit. Care Med. 200:250–252. 10.1164/rccm.201711-2267IM [DOI] [PubMed] [Google Scholar]
- Gritsenko, A., Yu S., Martin-Sanchez F., Diaz-Del-Olmo I., Nichols E.M., Davis D.M., Brough D., and Lopez-Castejon G.. 2020. Priming Is Dispensable for NLRP3 Inflammasome Activation in Human Monocytes In Vitro. Front. Immunol. 11:565924. 10.3389/fimmu.2020.565924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guma, M., Ronacher L., Liu-Bryan R., Takai S., Karin M., and Corr M.. 2009. Caspase 1-independent activation of interleukin-1beta in neutrophil-predominant inflammation. Arthritis Rheum. 60:3642–3650. 10.1002/art.24959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y., Zhang H., Mu S., Wei W., Jin C., Tong C., Song Z., Zha Y., Xue Y., and Gu G.. 2020. Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: a retrospective and observational study. Aging (Albany NY). 12:11245–11258. 10.18632/aging.103372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig, F. 2020. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level / Mixed) Regression Models. R package version 0.3.2.0. https://CRAN.R-project.org/package=DHARMa (accessed September 8, 2020)
- Hauenstein, A.V., Zhang L., and Wu H.. 2015. The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines. Curr. Opin. Struct. Biol. 31:75–83. 10.1016/j.sbi.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet, T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., Sacco E., Naccache J.M., Bézie Y., Laplanche S., et al. 2020. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2:e393–e400. 10.1016/S2665-9913(20)30164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, L.A., Netea M.G., Fantuzzi G., Koenders M.I., Helsen M.M., Sparrer H., Pham C.T., van der Meer J.W., Dinarello C.A., and van den Berg W.B.. 2009. Inflammatory arthritis in caspase 1 gene-deficient mice: contribution of proteinase 3 to caspase 1-independent production of bioactive interleukin-1beta. Arthritis Rheum. 60:3651–3662. 10.1002/art.25006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team . 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 584:463–469. 10.1038/s41586-020-2588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüdecke, D. 2018. ggeffects: Tidy Data Frames of Marginal Effects from Regression Models. J. Open Source Softw. 3:772. 10.21105/joss.00772 [DOI] [Google Scholar]
- Merad, M., and Martin J.C.. 2020. Author Correction: Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 20:448. 10.1038/s41577-020-0353-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla, A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., and Greninger A.L.. 2020. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 58:e00557–e20. 10.1128/JCM.00557-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea, M.G., Nold-Petry C.A., Nold M.F., Joosten L.A.B., Opitz B., van der Meer J.H.M., van de Veerdonk F.L., Ferwerda G., Heinhuis B., Devesa I., et al. 2009. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood. 113:2324–2335. 10.1182/blood-2008-03-146720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, S., Uehara A., Nochi T., Yamaguchi T., Ueda H., Sugiyama A., Hanzawa K., Kumagai K., Okamura H., and Takada H.. 2001. Neutrophil proteinase 3-mediated induction of bioactive IL-18 secretion by human oral epithelial cells. J. Immunol. 167:6568–6575. 10.4049/jimmunol.167.11.6568 [DOI] [PubMed] [Google Scholar]
- Tsuchiya, K., Hara H., Fang R., Hernandez-Cuellar E., Sakai S., Daim S., Chen X., Dewamitta S.R., Qu H., Mitsuyama M., and Kawamura I.. 2014. The adaptor ASC exacerbates lethal Listeria monocytogenes infection by mediating IL-18 production in an inflammasome-dependent and -independent manner. Eur. J. Immunol. 44:3696–3707. 10.1002/eji.201444673 [DOI] [PubMed] [Google Scholar]
- Tsutsui, H., Kayagaki N., Kuida K., Nakano H., Hayashi N., Takeda K., Matsui K., Kashiwamura S., Hada T., Akira S., et al. 1999. Caspase-1-independent, Fas/Fas ligand-mediated IL-18 secretion from macrophages causes acute liver injury in mice. Immunity. 11:359–367. 10.1016/S1074-7613(00)80111-9 [DOI] [PubMed] [Google Scholar]
- Vejrosta, Z., and Bilder J.. 1975. [Pathogenesis of excessive mobility of the temporomandibular joint]. Cesk. Stomatol. 75:119–124. [PubMed] [Google Scholar]
- Wang, D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. 2020. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 323:1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., et al. 2020. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 6:31. 10.1038/s41421-020-0168-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., and McGoogan J.M.. 2020. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 323:1239–1242. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- Zamboni, D.S., Kobayashi K.S., Kohlsdorf T., Ogura Y., Long E.M., Vance R.E., Kuida K., Mariathasan S., Dixit V.M., Flavell R.A., et al. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7:318–325. 10.1038/ni1305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows clinical characteristics of COVID-19 patients enrolled in this study.