Abstract
Pedobacter cryoconitis strain UP508 was isolated from a soil sample using a mixture of ampicillin, kanamycin, and nalidixic acid for selection. UP508 was found to produce >30 unknown antibacterial peptides, of which eight, isopedopeptins A–H (1–8), were isolated by bioassay-guided fractionation and characterized with respect to structures and biological properties. Compounds 1–8 were all composed of nine amino acid residues and one 3-hydroxy fatty acid residue, and the structures were ring-closed via an ester bond from the C-terminal aspartic acid to the 3-hydroxy fatty acid. The differences between the peptides were the size and branching of the 3-hydroxy fatty acid and the presence of a valine or a 3-hydroxyvaline residue. The isopedopeptins mainly had activity against Gram-negative bacteria, and isopedopeptin B (2), which had the best combination of antibacterial activity, in vitro cytotoxicity, and hemolytic properties, was selected for further studies against a larger panel of Gram-negative bacteria. Isopedopeptin B was found to have good activity against strains of WHO top-priority Gram-negative bacteria, i.e., carbapenem-resistant Acinetobacter baumannii, Escherichia coli, and Pseudomonas aeruginosa, with minimal inhibitory concentrations (MIC) down to 1, 2, and 4 μg/mL, respectively. Furthermore, compound 2 had activity against colistin-resistant strains of A. baumannii, E. coli, and Klebsiella pneumoniae, with a MIC down to 8, 2, and 4 μg/mL, respectively. Compound 6 was tested in an E. coli liposome system where it induced significant leakage, indicating membrane disruption as one mechanism involved in isopedopeptin antibacterial activity. Isopedopeptin B stands out as a promising candidate for further studies with the goal to develop a new antibiotic drug.
The global spread of antibiotic resistance threatens many aspects of modern healthcare, and in the future even the simplest infection or any routine surgery may be dangerous without access to efficient and safe antibiotics. In Europe, infections caused by antibiotic-resistant bacteria are responsible for the deaths of more than 30 000 individuals per year,1 and estimations in the U.S. show similar figures.2 The situation is expected to escalate in the years to come. To manage this serious problem, humanity needs access to new antibiotic drugs but also regulations to limit the spread of resistance against present and future antibiotic drugs.
A large proportion of the antibacterial drugs used today, and in the past, originate from nature—either as molecules isolated from microorganisms or developed from such molecules. Fungi and bacteria produce compounds with antimicrobial properties, with the assumed function to mediate interactions with competing microorganisms, to bring benefits to the producers. One important group of natural compounds used as antibiotic drugs are peptides produced by fungi or bacteria, and vancomycin, penicillins, and cephalosporins are all classical examples of such peptides which influence cell-wall synthesis in the target bacteria. Over the years, these peptides have served as models for the development of related improved antibiotic drugs, such as extended-spectrum penicillins, fourth-generation cephalosporins, and the recent semisynthetic vancomycin derivative telavancin. Another important group of peptide antibiotics are the cyclic lipopeptides, which have a cyclic peptide core carrying a fatty acid tail, exemplified by the cationic peptide colistin and the anionic peptide daptomycin. Colistin was introduced more than 60 years ago for the treatment of Gram-negative infections, but due to toxicity issues, its use decreased in the 1980s, and now it is used mainly as a last-resort antibiotic against multidrug-resistant infections. Daptomycin was approved by the FDA in 2003, for treatment of, e.g., complicated skin and skin structure infections caused by Gram-positive bacteria. Cationic peptides typically exert their antibacterial activity by interaction with negatively charged components of the bacterial membrane and disrupting its cohesion, and in the case of colistin, this includes specific interaction with bacterial lipopolysaccharides (LPS).3,4 The mechanism of action of the anionic daptomycin involves calcium dependent oligomerization and membrane insertion, causing membrane leakage.5 Antibacterial peptides, produced by microorganisms, are commonly produced by nonribosomal peptide synthases (NRPS). Nonribosomal peptides often contain nonproteinogenic amino acid residues, such as d-amino acids and a great variety of modified amino acid residues. Examples of such modified residues are the oxidized and chlorinated tyrosine residues and cross-linked phenylglycin residues in vancomycin and the β-lactam-thiazolidine structure of penicillins, that is formed by modification of an NRPS produced tripeptide precursor.
Despite the impressive track record of antibiotics discovery in nature, it was out of fashion for a couple of decades to screen microorganisms for the production of new antibiotics candidates, but in recent years, fungi and bacteria are again considered as promising sources for new chemical entities for development into antibiotic drugs.6,7 One major challenge when searching for new antibiotic compounds in microorganisms is to find novel drug-producing microbial strains to investigate. The recently described drug candidate teixobactin8 was discovered in a bacterial strain isolated using methods targeting previously uncultured bacteria,9 which are estimated to be well over 90% of all bacterial species in environmental samples. This methodology is expected to open up an exciting untapped source of bacterial strains for the production of novel and interesting bacterial compounds. Another recent approach to obtain previously unexploited drug-producing microbial strains is to use antibiotic resistance-based isolation of microorganisms.10 This technique is based on the inherent need for antibiotic producing microorganisms to carry genes for “self-resistance,” which makes resistant microorganisms likely candidates to produce antibiotic compounds.
The present paper describes the isolation and characterization of a family of antibacterial cyclic lipodepsipeptides, isopedopeptin A–H (1–8) from the bacterium Pedobacter cryoconitis strain UP508, which was isolated from a soil sample using a resistance-based approach. Several of these peptides display activity against many important clinical strains of Gram-negative bacteria, including strains of all the WHO top-priority human pathogens: carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae, as well as colistin-resistant strains of A. baumannii, Escherichia coli, and Klebsiella pneumoniae.
Results and Discussion
Isolation of Compounds
Culture extracts of P. cryoconitis UP508 were analyzed by UHPLC-MS, followed by database and literature comparison. The observation of the following doubly charged ions ([M + 2H]2+), m/z 566.3251 (1), 559.3170 (2), 572.3243 (3), 551.3214 (4), 558.3278 (5), 580.3398 (6), 571.3356 (7), and 571.3352 (8), suggested the presence of several unknown compounds in the extracts. Most of the compounds also had [M + 3H]3+ and [M + H]+ ions, and the presence of several different charge states suggested the compounds to be peptides. Subsequent bioassay-guided fractionation of these P. cryoconitis UP508 culture extracts resulted in many fractions with activity against both Gram-positive and Gram-negative bacteria, including the bacterial pathogens E. coli, K. pneumoniae, Enterobacter cloacae, P. aeruginosa, and A. baumannii. Antibacterial fractions were analyzed by UHPLC-MS, followed by database mining, which suggested the presence of a large number (>30) of unknown antibacterial compounds with molecular weights in the range 1100–1160 Da, and all the above suggested peptides (1–8) were present in antibacterial fractions. Subsequently, small amounts of compounds 1 and 2, along with several of the other compounds above, were obtained in semipure form after another round of preparative HPLC. MIC determination gave preliminary values <10 μg/mL against strains of E. coli, A. baumannii, E. cloacae, and K. pneumoniae for several of the compounds, indicating them to be interesting for further studies. Subsequently, the cultivation of the UP508 isolate was optimized along with the method for isolation of the compounds, which allowed the isolation of compounds 1–8 (between 0.66 mg and 7.95 mg each), for further characterization with respect to structure and biological properties.
Structure Determination
Compound 1 was found to have an [M + 2H]2+ ion of m/z 566.3243, corresponding to m/z 1131.6413 for the [M + H]+ ion. NMR in DMSO-d6 (Table 1) gave spin-systems in accord with the presence of the standard amino acid residues Phe, Thr, Leu, and Asp. In addition, several spin-systems suggested several nonproteinogenic amino acids to be present in 1, along with a 3-hydroxy fatty acid residue (3OHFA), as outlined below. Compound 1 was thus concluded to be a peptide. MSMS on compound 1 gave poor yields of fragment ions, which indicated a cyclic structure for 1.
Table 1. 1H and 13C NMR Data (600 and 150 MHz, resp) for Compound 1 (DMSO-d6, 30°C).
| pos. | 13C | 1H | mult (J, Hz) | pos. | 13C | 1H | mult (J, Hz) |
|---|---|---|---|---|---|---|---|
| 3OHFA | Thr | ||||||
| 1 | 167.6 | NH | 8.42 | d (7.6) | |||
| 2 | 39.4 | 2.69 | dd (13.8, 3.5) | 1 | 169.4 | ||
| 2.17 | dd (13.8, 4.1) | 2 | 59.5 | 4.20 | dd (7.8, 2.3) | ||
| 3 | 70.7 | 4.97 | m | 3 | 65.0 | 4.35 | dq (2.5, 6.5) |
| 4 | 31.2 | 1.49 | m | 4 | 20.4 | 1.22 | d (6.5) |
| 5 | 26.1 | 1.20 | obsc | DAPA2 | |||
| 6 | 28.6 | 1.21 | obsc | NH | 8.18 | d (8.7) | |
| 7 | 25.0 | 1.20 | obsc | 1 | 169.1 | ||
| 8 | 38.0 | 1.12 | obsc | 2 | 53.2 | 4.22 | ddd (8.7, 8.7, 5.7) |
| 9 | 27.0 | 1.49 | obsc | 3 | 40.6 | 3.04 | dd (13.1, 5.7) |
| 10 | 22.1 | 0.84 | d (6.6) | 2.87 | obsc | ||
| 11 | 22.1 | 0.84 | d (6.6) | Phe | |||
| DAPA1 | NH | 7.26 | d (9.0) | ||||
| NH | 8.57 | d (9.1) | 1 | 171.5 | |||
| 1 | 170.8 | 2 | 53.5 | 4.76 | obsc | ||
| 2 | 50.6 | 4.73 | obsc | 3 | 37.6 | 2.89 | obsc |
| 3 | 43.9 | 3.12 | dd (13.1, 5.4) | 2.72 | obsc | ||
| 2.97 | dd (13.1, 4.1) | 4 | 137.2 | ||||
| Leu | 5/9 | 129.2 | 7.34 | m | |||
| NH | 8.89 | br s | 6/8 | 127.6 | 7.21 | m | |
| 1 | 173.1 | 7 | 125.8 | 7.15 | m | ||
| 2 | 52.8 | 4.33 | obsc | 3OHVal | |||
| 3 | 39.1 | 1.65 | m | NH | 8.59 | d (9.6) | |
| 1.46 | m | 1 | 169.4 | ||||
| 4 | 24.0 | 1.65 | obsc | 2 | 58.0 | 4.72 | obsc |
| 5 | 22.2 | 0.88 | d (6.2) | 3 | 70.6 | ||
| 6 | 22.2 | 0.84 | d (6.1) | 4 | 26.4 | 1.05 | s |
| DABA | 5 | 26.5 | 0.92 | s | |||
| NH | 8.65 | d (9.3) | Asp | ||||
| 1 | 168.0 | NH | 8.52 | d (9.6) | |||
| 2 | 49.5 | 4.64 | ddd (9.3, 7.0, 7.0) | 1 | 173.9 | ||
| 3 | 29.2 | 1.82 | m | 2 | 48.4 | 4.74 | obsc |
| 4 | 35.4 | 2.75 | m | 3 | 40.5 | 2.84 | dd (16.1, 4.5) |
| 2.61 | m | 2.23 | d (16.1, 3.8) | ||||
| ABA | 4 | 174.4 | |||||
| NH | 10.22 | br s | |||||
| 1 | 165.4 | ||||||
| 2 | 132.6 | ||||||
| 3 | 114.1 | 5.57 | q (7.3) | ||||
| 4 | 12.5 | 1.77 | d (7.3) |
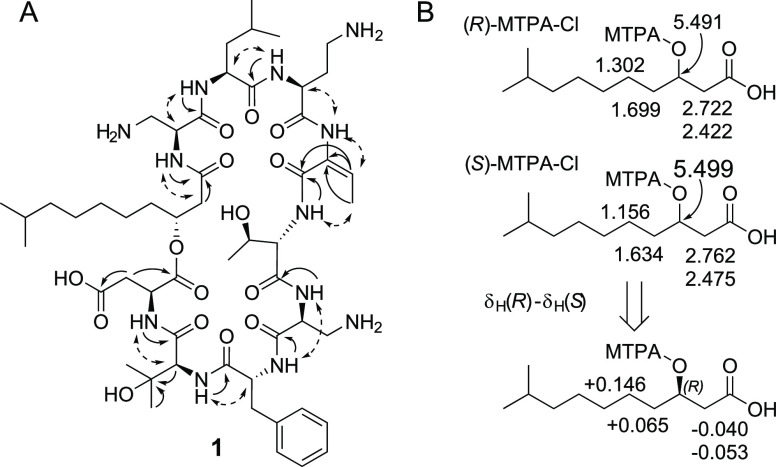
Two spin systems, NH–CH–CH2, with chemical shifts for the CH2 groups in agreement with linkages to amino groups (δH 3.12/2.91, δC 43.9, and δH 3.04/2.87, δH 40.6, respectively), suggested the presence of two 2,3-diaminopropanoic acid residues (DAPA) in compound 1. A similar spin-system, but extended by a CH2 group, i.e., NH–CH–CH2–CH2, suggested a 2,4-diaminobutanoic acid residue (DABA) to be present in 1. Three basic amino acids in 1, i.e., two DAPA residues and one DABA residue, were in agreement with the presence of an intense [M + 3H]3+ ion for the peptide in MS analysis. One further spin system was NH–CH, and by HMBC correlations (Figure 1A) the methine group was determined to be linked to a quarternary carbon (δC 70.6) which was linked to two methyl groups and, as judged by the chemical shift, to a hydroxy group. This suggested the presence of a 3-hydroxyvaline residue (3OHVal) in compound 1. The final proposed nonproteinogenic amino acid residue in 1 had a methyl group (δH 1.77, δC 12.5) which was linked to an sp2 CH (δH 5.57, δC 114.1). By HMBC (Figure 1A), this sp2 carbon was determined to be linked to another sp2 carbon (δC 132.6) and subsequently to a carbonyl (δC 165.4). If the carbon at δC 132.6 was linked to an amino function, this would make up a 2-amino-2-butenoic acid residue (ABA), described to be present in many microbial nonribosomal peptides, with Z-geometry11 or with E-geometry.12 In the 1D 1H NMR spectrum of 1, there were nine signals from putative amide protons, and eight of these signals were already assigned to the amino acid residues Phe, Thr, Leu, Asp, 2 × DAPA, DABA, and OHVal, using data from COSY, TOCSY, and HSQC-TOCSY. The remaining putative amide signal (broad signal at δH 10.22) was thus assigned as the tentative ABA amide proton. Subsequently, ROESY experiments gave a cross-peak between this signal and the ABA olefinic signal (Figure 1A), which supported the assignment of the signal at δH 10.22 as the ABA amide proton, but also suggested the ABA residue to be in the E-configuration in 1. The proposed 3OHFA, with the spin-system CH2–CH–CH2–CH2–(CH2)n–CH2–CH(CH3)–CH3, was identified by combination of COSY, TOCSY, HSQC, and HMBC NMR data. The terminal CH2 had chemical shifts and a coupling pattern in agreement with the linkage to a carbonyl (δH 2.69/2.17, 2J 13.8 Hz, δC 39.4) and chemical shifts for the adjacent CH in accord with linkage to an esterified hydroxy function (δH 4.97, δC 70.7). HMBC data verified the linkage of the terminal CH2 to a carbonyl group (δC 167.6).
Figure 1.
(A) Key HMBC (solid single-headed arrows) and ROESY (dashed double-headed arrows) correlations for structure determination of compound 1. (B) Determination of absolute configuration of the 3-hydroxydecanoic acid originating from compound 1, by NMR analysis of Mosher esters.13 Selected 1H NMR chemical shifts obtained in CHCl3-d are displayed on top, and the calculated chemical shifts differences (R-S) below, along with the indicated absolute configuration.
By comparison of the m/z for the [M + H]+ ion of 1 (1131.6413), with the sum of the individual masses of the different amino acid residues (Phe, Thr, Leu, Asp, 2 × DAPA, DABA, 3OHVal and ABA), a nice fit was obtained when adding the mass for a 3-hydroxy-9-methylhydroxydecanoic acid residue (theoretical m/z 1131.6408 for the [M + H]+ ion, difference 0.4 ppm). Thus, compound 1 was suggested to be a cyclic lipodepsipeptide containing a 3-hydroxy-9-methyldecanoic acid together with nine amino acid residues.
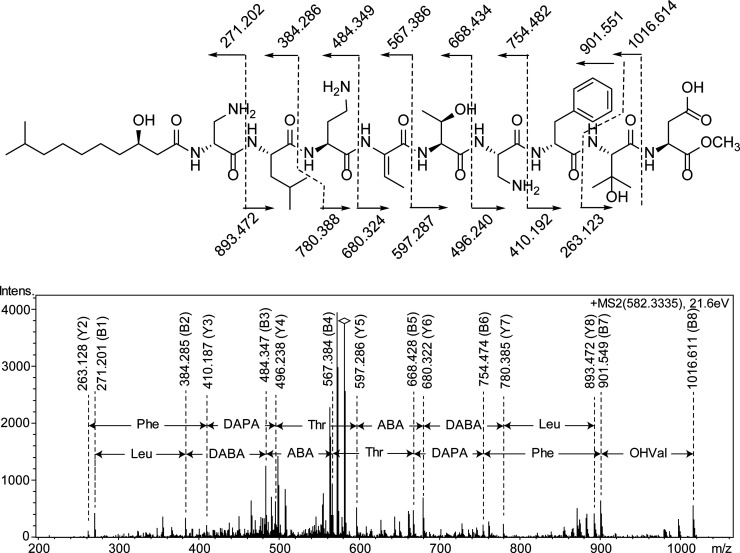
As mentioned above, MSMS on the native peptide gave poor yields of sequence fragment ions, in accord with 1 being a cyclic lipodepsipeptide. Thus, the peptide was treated with dilute NaOMe in MeOH to open the presumed lactone. Analysis by MS showed that MeOH indeed had been added to the structure ([M + 2H]2+m/z 582.334), and subsequent MSMS analysis gave informative fragment ions which were used for the determination of the amino acid sequence of 1 (Figure 2). The B-fragment ion series B1–B8 was in good agreement with the sequence (3-hydroxy-9-methyldecanoyl-DAPA)-Leu-DABA-ABA-Thr-DAPA-Phe-3OHVal-Asp-OMe, and this sequence was corroborated by the Y-fragment ion series Y2–Y8. The 3-hydroxy-9-methyldecanoyl CH group had chemical shifts (δH 4.97, δC 70.7) in accord with linkage to an esterified hydroxy function, and furthermore, if the lactone would have been closed to the hydroxy function of either the Thr or 3OHVal residue, informative N-terminal sequence fragment ions would have been expected in MSMS analysis of the native peptide. Thus, compound 1 was proposed to have the structure cyclo(3-hydroxy-9-methyldecanoyl-DAPA1-Leu-DABA-ABA-Thr-DAPA2-Phe-OHVal-Asp) (Figure 3).
Figure 2.
MSMS spectrum of compound 1 after ring-opening with NaOMe in MeOH. The m/z for sequence ions B1–B8 and Y2–Y8 are shown in the spectrum, along with the corresponding amino acid sequences. Theoretical m/z values for the observed B- and Y-series ions are shown in the structure on top.
Figure 3.
Structures of isopedopeptins A–H (1–8).
The MS–MS based amino acid sequence was verified by ROESY and HMBC experiments (Figure 1A). The following ROESY cross-peaks were observed: 3OHFA H-2/DAPA1 NH, DAPA1 H-2/Leu NH, Leu H-2/DABA NH, DABA H-2/ABA NH, ABA–CH3/Thr NH, Thr NH/DAPA2 NH, DAPA2 NH/Phe NH, Phe H-2/3OHVal NH, 3OHVal H-2/Asp NH (Figure 1), along with the following HMBC cross-peaks: 3OHFA CO/DAPA1 NH, Leu CO/DABA NH, ABA CO/Thr NH, Thr CO/DAPA2 NH, DAPA2 CO/Phe NH, Phe CO/3OHVal NH, and 3OHVal CO/Asp NH (Figure 1A).
The configuration of the amino acids was determined using the advanced Marfey’s method,14,15 by acidic hydrolysis and derivatization with 1-fluoro-2,4-dinitrophenyl-5-l-leucinamide (FDLA), followed by UHPLC-MS analysis. Comparison with commercial reference amino acids enabled identification of the amino acids as 2 × L-DAPA, l-Leu, L-DABA, l-Thr, d-Phe, l-OHVal, and l-Asp. d-DAPA was also detected in peptide hydrolysates, but hydrolysis of peptides in H2O/D2O mixtures demonstrated that D-DAPA was formed by epimerization, as previously observed,16 and was thus not present in the peptide. Similarly, Phe was also shown to epimerize during the acidic hydrolysis, but to a lesser extent than DAPA.
The configuration of the 3-hydroxy-9-methyldecanoyl residue in 1 was determined by Mosher’s method.13 After release of the fatty acid by acidic hydrolysis of the peptide, the fatty acid was derivatized with (R)- and (S)-2-methoxy-2-trifluoromethyl-2-phenylacetyl (MTPA) chloride. The crude products were analyzed by NMR in pyridine-d5 and chloroform-d. Comparison of the proton chemical shifts of H-2, H-4, and H-5, between the products obtained with R- and S-reagents (i.e., (R)-MTPA–Cl product – (S)-MTPA–Cl product) gave differences in line with R-configuration of the 3-hydroxydecanoyl residue (Figure 1B). This was valid for both NMR solvents. Thus, analysis by NMR and MS, along with chemical degradations and derivatizations, resulted in the full structure of compound 1 (Figure 3).
Compound 2 was found to have an [M + 2H]2+ ion of m/z 559.3169, corresponding to m/z 1117.6265 for the [M + H]+ ion, which suggested a difference compared to 1 by a −CH2– group. NMR in DMSO-d6 (Supplementary Table 2) indicated the presence of the same amino acids as in 1, along with an unbranched 3-hydroxy fatty acid residue. By comparison of the m/z for the [M + H]+ ion of 2 (1117.6265), with the added individual masses of the different amino acid residues, a nice fit was obtained when adding the mass for a 3-hydroxydecanoic acid residue (theoretical m/z 1117.6252 for the [M + H]+ ion, difference 1.2 ppm). Thus, compound 2 was suggested to be a cyclic lipodepsipeptide containing a 3-hydroxydecanoic acid together with nine amino acid residues. Subsequent NaOMe/MeOH lactone opening and MSMS analysis gave the sequence (3-hydroxydecanoyl-DAPA)-Leu-DABA-ABA-Thr-DAPA-Phe-OHVal-Asp-OMe, and this sequence was corroborated by ROESY and HMBC correlations. The configuration of the amino acids was determined by acidic hydrolysis and derivatization with FDLA, followed by UHPLC-MS analysis, to be the same as for 1. Assuming the same configuration of the 3-hydroxydecanoyl residue in 2 as for the (R)-3-hydroxy-9-methyldecanoyl residue of 1 resulted in the full structure of 2 (Figure 3).
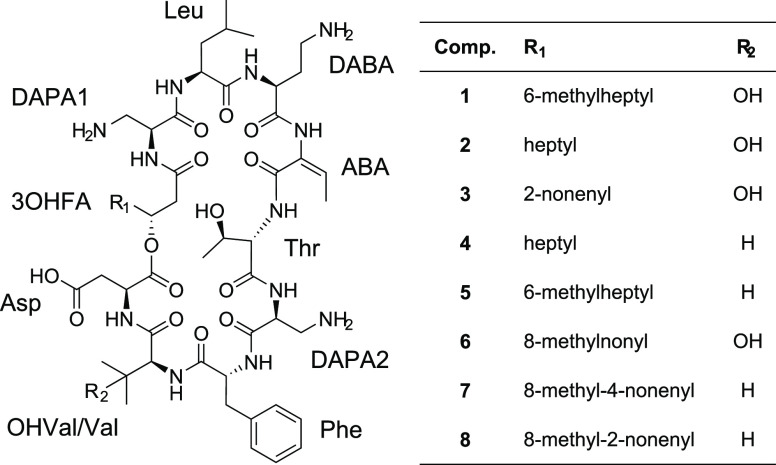
Subsequently, compounds 3–8 were analyzed in analogy with compounds 1 and 2, and their structures were found to be very similar to 1 and 2, including the configuration of all amino acid residues (d-Phe and l-configuration for all other residues). The R-configuration of the 3OHFA residues was only determined on 1 and was assumed to be the same on compounds 2–8. The main differences between compounds 3–8 and compounds 1 and 2 are described below.
MS analysis of compound 3 resulted in m/z 572.3246 for the [M+2H]2+ ion (m/z 1143.6419 for the [M + H]+ ion). Compared to compounds 1 and 2, MSMS showed the structure difference to be in the 3OHFA part of the structure. The 3OHFA of 3 was found to have 12 carbon atoms, and additionally, one unsaturation or ring in the 3OHFA. NMR analysis (Supplementary Table 3) showed the 3OHFA to be unbranched and to contain one double bond (two olefinic protons at δH 5.27 and δH 5.40, respectively). By COSY experiments, it was concluded that the double bond was located at C-5/C-6 of the 3OHFA. The H-5/H-6 3J was 11 Hz, which strongly suggested the 3OHFA in 3 to be a cis-3-hydroxy-5-dodecenoyl residue (Figure 3).
Compound 4 had a [M + 2H]2+ ion at m/z 551.3194 and a [M + H]+ ion at m/z 1101.6315, suggesting a difference corresponding to one oxygen atom compared to compound 2. MSMS analysis showed the 3OHVal residue to be absent in 4, and instead the data suggested a Val residue to be present. This difference was later confirmed by NMR analysis (Supplementary Table 4), which also showed the presence of a 3-hydroxydecanoyl residue in 4, just as in 2 (Figure 3).
The [M + 2H]2+ ion of compound 5 had m/z 558.3275, corresponding to m/z 1115.6477 for the [M + H]+ ion. Just as for 4, compound 5 was found by MSMS to have a Val residue instead of a 3OHVal residue, and the difference compared to 4 was located in the 3OHFA part of the structure, which was found to have 11 carbon atoms. NMR analysis (Supplementary Table 5) subsequently showed this 3OHFA was to be a 3-hydroxy-9-methyldecanoyl residue, just as in 1 (Figure 3).
Compound 6 was by MS ([M+2H]2+m/z 580.3401, [M + H]+m/z 1159.6729), MSMS, and NMR (Supplementary Table 6) found to be similar to compound 1, the only difference being the number of carbons in the 3OHFA. Compound 6 had a 3-hydroxy-11-methyldodecanoyl residue compared to a 3-hydroxy-9-methyldecanoyl residue in 1 (Figure 3).
Analysis by MS, MSMS, and NMR (Supplementary Tables 7 and 8) showed compounds 7 and 8 to be very similar to each other. They both contained Val instead of 3OHVal residues, and they both contained cis-3-hydroxy-11-methyldodecenoyl residues, but the location of the double bond was at C-7/C-8 in 7 and at C-5/C-6 in 8 (Figure 3).
Similar peptides, pedopeptin A–C and B12489A–C, have previously been isolated from Pedobacter sp. SANK 72003.17,18 These peptides are all cyclic lipodepsipeptides based on one 3-hydroxy fatty acid residue and nine amino acid residues, including one E-ABA residue. The main difference between pedopeptin A–C/B12489A–C and compounds 1–8 is that Leu and Phe have the opposite positions in the pedopeptins/B12489A–C compared to those in 1–8, and the Thr of 1–8 is exchanged to Leu in pedopeptin A–C/B12489A–C. Additionally, the structures of the 3-hydroxy fatty acid residues are different in the pedopeptins/B12489A–C compared to 1–8, with 3-hydroxyoctanoyl or 3-hydroxy-7-methyloctanoyl residues in the pedopeptins/B12489A–C compared to 3-hydroxydecanoyl and larger residues in 1–8. On the basis of the similarities and differences with the pedopeptins/B12489A–C, compounds 1–8 were named isopedopeptin A–H.
Biological Properties
Compounds 1–8 were tested against a panel of Gram-negative and Gram-positive bacteria, as well as against two fungi (Table 2). In general, the activity against Gram-negative bacteria was better than against Gram-positive bacteria, and the activity against fungi was poor (Table 2). Compound 1 displayed the highest antibacterial activities, with MIC values down to 4 μg/mL against the β-lactamase producing E. coli LMG15862 and A. baumannii LMG1041T, whereas compounds 2 and 7 had the second-best antibacterial activities with MIC 8 μg/mL against these pathogens. Compounds 1, 2, and 7 also had promising MICs against P. aeruginosa LMG6395T and ESBL K. pneumoniae LMG20218, in the range 8–16 μg/mL (Table 2). The IC50 values against HepG2 cells varied substantially between the different compounds (Table 2). Compound 2 had the highest IC50 value (56 μg/mL) and 2 also had an acceptable hemolysis rate (<0.8%), whereas compounds 1 and 7 had lower IC50 values and substantially higher hemolysis rates (Table 2).
Table 2. MIC Values (μg/mL) for Compounds 1–8 against the Primary Panel of Bacteria and Fungi, along with IC50 Values (μg/mL) against HepG2 Cells and Haemolysis Frequencies (ESBL, extended-spectrum β-lactamase; MRSA, methicillin-resistant S. aureus).
| compound |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pathogen | strain | type | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| E. coli | LMG15862 | β-lactam. | 4 | 8 | 16 | 32 | >32 | 16 | 8 | 16 |
| A. baumannii | LMG1041T | WT | 4 | 8 | 16 | 16 | 32 | 16 | 8 | 16 |
| E. cloacae | LMG2783T | WT | 16 | 32 | 32 | 32 | >32 | 32 | 16 | 16 |
| K. pneumoniae | LMG20218 | ESBL | 8 | 16 | 32 | >32 | 32 | 32 | 16 | 16 |
| P. aeruginosa | LMG6395T | WT | 16 | 16 | 32 | >32 | 32 | 32 | 8 | 16 |
| S. aureus | LMG15975 | MRSA | >32 | 32 | >32 | >32 | >32 | 32 | 16 | 16 |
| B. cereus | CCUG7414T | >32 | >32 | 32 | 32 | 16 | 32 | 16 | 32 | |
| A. fumigatus | J7 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | |
| C. albicans | H-29 | WT | 32 | 32 | 32 | >32 | >32 | >32 | >32 | >32 |
| IC50(μM) | HepG2 | liver | 29 | 50 | 29 | 9 | 13 | 9 | 11 | 14 |
| IC50(μg/mL) | HepG2 | liver | 33 | 56 | 33 | 10 | 14 | 10 | 13 | 16 |
| Hemolysis (%) | 4.2 | 0.8 | 3.0 | 9.1 | 5.2 | 27.5 | 14.1 | 52.3 | ||
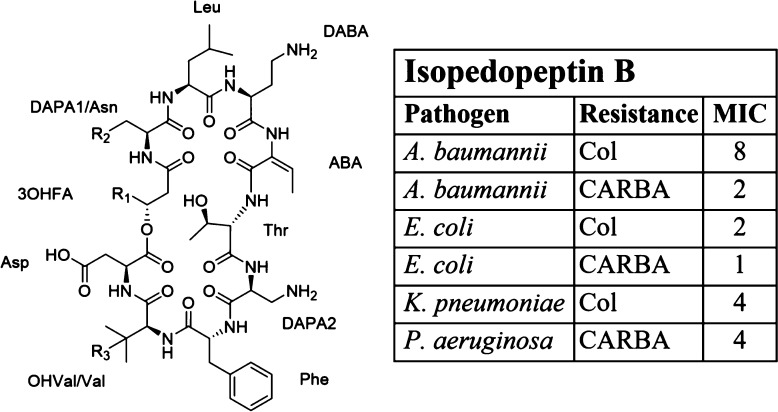
Given the interesting pattern of antibacterial activities of 2, along with the favorable IC50 value and hemolysis rate, compound 2 was selected for further studies of biological properties, and the compound was thus tested against a wide range of clinically relevant bacterial isolates (Table 3). The compound had good activity against the WHO top-priority pathogens carbapenem-resistant A. baumannii, E. coli, and P. aeruginosa, with MICs ranging from 1 to 4 μg/mL. Compound 2 was also active against colistin-resistant strains of A. baumannii, E. coli, and K. pneumoniae, with MICs of 8 μg/mL, 2 μg/mL, and 4 μg/mL, respectively, as well as against the clinical multidrug-resistant K. pneumoniae CH3498, MIC 8 μg/mL (Table 3). The activity of 2 against these important pathogens makes this compound very interesting for further studies, with the ultimate aim to develop a new antibiotic drug.
Table 3. Selected MIC Values (μg/mL) for Compound 2 after Testing against an Extended Panel Bacterial Isolates (WT, wild type; MDR, multidrug-resistant; Col, Colistin; CARBA, Carbapenem; ESC, Extended-spectrum cephalosporin; CIP, Ciprofloxacin; GEN, Gentamicin; SXT, Trimethoprim-sulfamethoxazole; mcr-1, Mobilized colistin resistance gene; AMK, Amikacin; TET, Tetracycline).
| pathogen | strain ID | type/resistance pattern | MIC |
|---|---|---|---|
| A. baumannii | ATCC19606 | WT | 8 |
| A. baumannii | BM4454 | MDR clinical | 8 |
| A. baumannii | BM4652 | efflux-defective derivative of BM4454 | 8 |
| A. baumannii | EN0287 | Col | 8 |
| A. baumannii | A219 | CARBA | 2a |
| A. baumannii | A250 | CARBA | 2a |
| E. coli | ATCC25922 | WT | 8 |
| E. coli | CH3130 | ΔtolC mutant isogenic to ATCC25922 | 8 |
| E. coli | D22 | lpxC mutant, hypersensitive | 4 |
| E. coli | CH3491 | ESC, CIP, GEN, SXT | 8 |
| E. coli | CH9623 | mcr-1 | 2 |
| E. coli | CH9624 | mcr-1 | 2 |
| E. coli | ATCC25922 | WT | 1a |
| E. coli | EC4129 | CARBA | 2a |
| E. coli | EC4163 | CARBA | 1a |
| K. pneumoniae | ATCC13833 | WT | 4 |
| K. pneumoniae | 1161486 | WT | 8 |
| K. pneumoniae | 1161486a | ΔtolC mutant isogenic to 1161486 | 8 |
| K. pneumoniae | CH3498 | ESC, CARBA, CIP, GEN, AMK, SXT, TET SXT, TET | 8 |
| K. pneumoniae | CH9625 | Col | 4 |
| K. pneumoniae | CH9626 | Col | 4 |
| P. aeruginosa | PAO1 | WT | 32 |
| P. aeruginosa | PAO750 | efflux-defective isogenic to PAO1 | 32 |
| P. aeruginosa | ATCC27853 | WT | 4a |
| P. aeruginosa | PS826 | CARBA | 4a |
| P. aeruginosa | PS992 | CARBA | 4a |
Determined at the Public Health Agency of Sweden.
The previously described pedopeptins17 were shown to have antibacterial activity against E. coli, S. aureus, and S. epidermis, with MICs down to 2 μg/mL against the two tested E. coli strains and down to 4 μg/mL against the two tested Staphylococcii.19 The MICs for pedopeptins against two different E. coli strains are similar to MICs determined for compound 2 against several E. coli strains, but the activity against S. aureus is strikingly different for 2 compared to the pedopeptins. Possibly, this may be due to differences between the bacterial strains used in the two studies, but it may also depend on the structural differences between 2 and the pedopeptins.
To investigate the mechanism of action of 2 and to measure the frequency of resistance (FoR), two E. coli strains were incubated with compound 2 at 4 × MIC and 8 × MIC. In these experiments, however, no resistant mutants were observed (FoR < 2 × 10–9 and <3 × 10–9, respectively), and thus, no information on the mechanism of action could be obtained by sequencing of resistant mutants. The pedopeptins, similar peptides from Pedobacter sp. SANK 72003,17 have been shown to inhibit the binding of bacterial LPS to cellular receptors at very low concentrations (IC50 11–47 nM).19 Given the similarities between the pedopeptins and the isopedopeptins (1–8), it is possible that also 1–8 may interact with cellular LPS receptors. In the same study, Kozuma et al. found that also polymyxin B inhibits the binding of LPS to cellular receptors at even lower concentrations (3.6 nM) than the pedopeptins.19 The antibiotic compound colistin, which is a polymyxin B analogue, has been described to have specific interactions with bacterial LPS but also general detergent type membrane effects.3 Colistin and polymyxin B are both polycationic due to the presence of the amino functions of the five DABA side-chains in their structures. Compounds 1–8 and the pedopeptins, with one DABA and two DAPA residues, are also cationic at physiological pH. Cationic peptides are well-documented to interact with negative structure elements of membrane components,5 and likely, 1–8 exert their activity by interaction with bacterial membranes. Potentially, 1–8 and the pedopeptins have a similar mechanism of action to that of colistin, i.e., binding to LPS and a general detergent effect on bacterial cytoplasmic membranes. However, in the present study, compound 2 was found to have potent activity also against colistin-resistant strains of A. baumannii, E. coli, and K. pneumoniae (Table 3). The E. coli strains carrying the mobilized colistin resistance gene (mcr-1, Table 3) have an LPS with phosphoetanolamine residues bound to the lipid A part, which decreases the affinity for colistin.20 Thus, if there is an interaction of 2 with LPS, this must be different compared to the interaction of colistin and polymyxin B with LPS.
In addition to colistin type peptides, also other antibacterial lipopeptides have been reported to interact with bacterial membranes, e.g., the antibiotic drug daptomycin, which has been shown to have a calcium-dependent aggregation in bacterial cell membranes, which creates leakage.5 To investigate if permeabilization of bacterial membranes could be responsible for the activity of the cationic cyclic isopedopeptins, compound 6 was tested in an in vitro bacterial membrane system using large unilamellar liposomes made from an E. coli phospholipid extract. Indeed, compound 6 was found to cause membrane leakage, and the half maximal effective concentration (EC50) was 2.0 μM (Figure 4). In comparison, the cyclic lipopeptide polymyxin B and the human antimicrobial peptide LL-37, two highly potent membrane disrupting antibacterial agents, gave EC50’s of 1 μM and 0.6 μM, respectively (Figure 4). Bacterial liposome permeabilization has a strong correlation with antibacterial activity and indicates that compound 6 exerts antibacterial activity by membrane disruption. Due to the structural similarities between all the isopedopeptins (1–8), it is reasonable to suggest that this could be a shared mechanism of action that at least in part explains their antibacterial activity.
Figure 4.
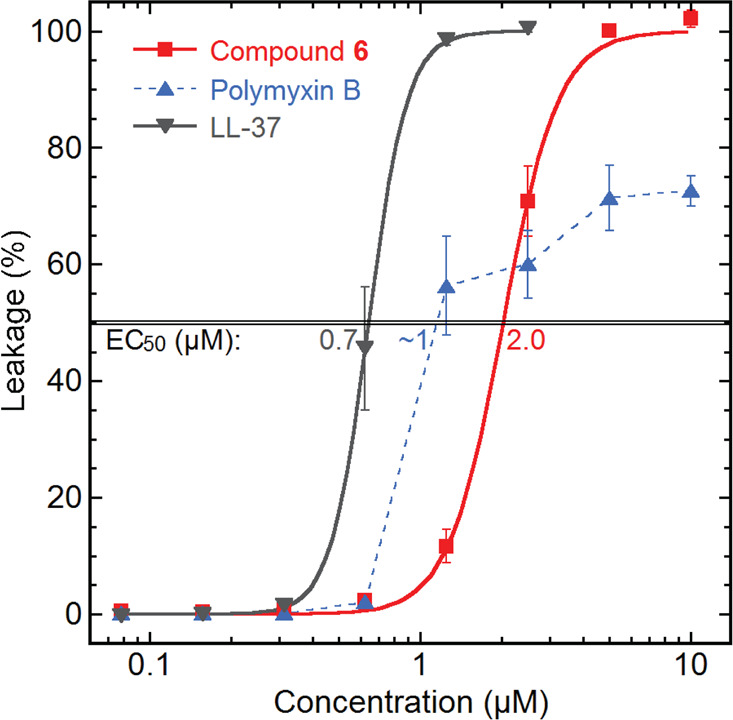
Compound 6 tested for leakage generation on E. coli liposomes, along with the known membrane disrupting lipopeptide polymyxin B and the human antimicrobial peptide LL-37.
Conclusions
The cyclic lipodepsipeptides isopedopeptin A–H (1–8) described in the present paper have interesting activity against WHO top-prioritized Gram-negative bacteria. In particular, isopedopeptin B (2), with potent antibacterial activity combined with acceptable cytotoxicity and hemolysis rates, along with a low FoR, is a promising candidate for further studies and optimization with the goal to develop a new antibiotic drug. The isopedopeptins form the basis of a recently published Patent Cooperation Treaty (PCT) application.21
Acknowledgments
Funding from Ultupharma AB to C.N., J.B., J.J.L., B.G., and A.B. is gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.0c00568.
Methods, tabulated NMR data for compounds 1–8, one-dimensional 1H NMR spectra for 1–8, one-dimensional 13C NMR spectra for 1 and 2, 1H–1H COSY spectra for 1–8, 1H–13C HSQC spectra for 1–8, 1H–13C HMBC spectra for 1–8, ROESY spectra for 1–8, 1H–1H TOCSY spectra for 1–2, HR-MS spectra for 1–8, MSMS spectra for 1–8, and retention times for 1–8 from advanced Marfey’s analysis (PDF)
Author Contributions
C.N., J.B., J.J.L., B.G., B.Ö., and A.B. designed the study. C.N., J.B., J.J.L., and A.B. did the laboratory work, except for the membrane permeabilization assay (A.A.S.), the cytotoxicity assays (R.L.), the extended MIC determination (D.H and S.C.), the FoR determination (D.H and S.C.), and hemolysis assay (D.H. and S.C.). A.B. wrote the first manuscript draft, supported by all authors, and all authors contributed to the subsequent editing of the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): C.N., J.B., J.J.L., B.G., B.O., and A.B. are shareholders of Ultupharma AB, which owns the IP rights to the published findings.
Supplementary Material
References
- Cassini A.; Hogberg L. D.; Plachouras D.; Quattrocchi A.; Hoxha A.; Simonsen G. S.; Colomb-Cotinat M.; Kretzschmar M. E; Devleesschauwer B.; Cecchini M.; Ouakrim D. A.; Oliveira T. C.; Struelens M. J; Suetens C.; Monnet D. L; Strauss R.; Mertens K.; Struyf T.; Catry B.; Latour K.; Ivanov I. N; Dobreva E. G; Tambic Andrasevic A.; Soprek S.; Budimir A.; Paphitou N.; Zemlickova H.; Schytte Olsen S.; Wolff Sonksen U.; Martin P.; Ivanova M.; Lyytikainen O.; Jalava J.; Coignard B.; Eckmanns T.; Abu Sin M.; Haller S.; Daikos G. L; Gikas A.; Tsiodras S.; Kontopidou F.; Toth A.; Hajdu A.; Guolaugsson O.; Kristinsson K. G; Murchan S.; Burns K.; Pezzotti P.; Gagliotti C.; Dumpis U.; Liuimiene A.; Perrin M.; Borg M. A; de Greeff S. C; Monen J. C.; Koek M. B.; Elstrøm P.; Zabicka D.; Deptula A.; Hryniewicz W.; Canica M.; Nogueira P. J.; Fernandes P. A.; Manageiro V.; Popescu G. A; Serban R. I; Schreterova E.; Litvova S.; Stefkovicova M.; Kolman J.; Klavs I.; Korosec A.; Aracil B.; Asensio A.; Perez-Vazquez M.; Billstrom H.; Larsson S.; Reilly J. S; Johnson A.; Hopkins S. (2019) Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Antibiotic Resistance Threats in the United States, 2019. U.S. Department of Health and Human Services, CDC: Atlanta, GA, 2019. [Google Scholar]
- Trimble M. J.; Mlynárčik P.; Kolář M.; Hancock R. E. W. (2016) Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harbor Perspect. Med. 6, a025288. 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Qin W.; Lin J.; Fang S.; Qiu J. (2015) Antibacterial Mechanisms of Polymyxin and Bacterial Resistance. BioMed Res. Int. 2015, 1–11. 10.1155/2015/679109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus K. S.; Hancock R. E. W. (2006) Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta, Biomembr. 1758, 1215–1223. 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Harvey A. L.; Edrada-Ebel R.; Quinn R. J. (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discovery 14, 111–129. 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- Genilloud O. (2014) The re-emerging role of microbial natural products in antibiotic discovery. Antonie van Leeuwenhoek 106, 173–188. 10.1007/s10482-014-0204-6. [DOI] [PubMed] [Google Scholar]
- Ling L. L.; Schneider T.; Peoples A. J.; Spoering A. L.; Engels I.; Conlon B. P.; Mueller A.; Schäberle T. F.; Hughes D. E.; Epstein S.; Jones M.; Lazarides L.; Steadman V. A.; Cohen D. R.; Felix C. R.; Fetterman K. A.; Millett W. P.; Nitti A. G.; Zullo A. M.; Chen C.; Lewis K. (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols D.; Cahoon N.; Trakhtenberg E. M.; Pham L.; Mehta A.; Belanger A.; Kanigan T.; Lewis K.; Epstein S. S. (2010) Use of Ichip for High-Throughput In Situ Cultivation of “Uncultivable” Microbial Species. Appl. Environ. Microbiol. 76, 2445–2450. 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker M. N.; Wang W.; Spanogiannopoulos P.; Waglechner N.; King A. M.; Medina R.; Wright G. D. (2013) Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 31, 922–929. 10.1038/nbt.2685. [DOI] [PubMed] [Google Scholar]
- Namikoshi M.; Choi B. W.; Sakai R.; Sun F.; Rinehart K. L.; Carmichael W. W.; Evans W. R.; Cruz P.; Munro M. H. G.; Blunt J. W. (1994) New Nodularins: A General Method for Structure Assignment. J. Org. Chem. 59, 2349–2357. 10.1021/jo00088a014. [DOI] [Google Scholar]
- Sano T.; Kaya K. (1998) Two New (E)-2-Amino-2-Butenoic Acid (Dhb)-Containing Microcystins Isolated from Oscillatoria agardhii. Tetrahedron 54, 463–470. 10.1016/S0040-4020(97)10291-5. [DOI] [Google Scholar]
- Dale J. A.; Mosher H. S. (1973) Nuclear magnetic resonance enantiomer reagents. Configurational correlation via nuclear magnetic resonance chemical shifts of diastereomericmandelate, O-methylmandelate, and α-methoxy-α-trifluorophenylacetate (MTPA) esters. J. Am. Chem. Soc. 95, 512–519. 10.1021/ja00783a034. [DOI] [Google Scholar]
- Harada K.; Fujii K.; Mayumi T.; Hibino Y.; Suzuki M.; Ikai Y.; Oka H. (1995) Constituent Amino Acids in Peptide --- Advanced Marfey’s Method. Tetrahedron Lett. 36, 1515–1518. 10.1016/0040-4039(95)00078-Q. [DOI] [Google Scholar]
- Fujii K.; Ikai Y.; Mayumi T.; Oka H.; Suzuki M.; Harada K. (1997) A Nonempirical Method Using LC/MS for Determination of the Absolute Configuration of Constituent Amino Acids in a Peptide: Elucidation of Limitations of Marfey’s Method and of Its Separation Mechanism. Anal. Chem. 69, 3346–3352. 10.1021/ac9701795. [DOI] [Google Scholar]
- Kjær A.; Larsen P. O.; Sillen L. G.; Andersson G.; Stenhagen E.; Palmstierna H. (1959) Amino Acid Studies. Part II. Structure and Synthesis of Albizziine (L-2-Amino-3-ureidopropionic Acid), and Amino Acid from Higher Plants. Acta Chem. Scand. 13, 1565–1574. 10.3891/acta.chem.scand.13-1565. [DOI] [Google Scholar]
- Hirota-Takahata Y.; Kozuma S.; Kuraya N.; Fukuda D.; Nakajima M.; Ando O. (2014) Pedopeptins, novel inhibitors of LPS: Taxonomy of producing organism, fermentation, isolation, physicochemical properties and structural elucidation. J. Antibiot. 67, 243–251. 10.1038/ja.2013.122. [DOI] [PubMed] [Google Scholar]
- Mutsuo N., Yuki H., Osamu A., Nahojiyu K., Shiho K., and Daisuke F.. B-12489 Substance and Method for Producing the Same. JP2005200324(A), July 28th, 2005.
- Kozuma S.; Hirota-Takahata Y.; Fukuda D.; Kuraya N.; Nakajima M.; Ando O. (2014) Screening and biological activities of pedopeptins, novel inhibitors of LPS produced by soil bacteria. J. Antibiot. 67, 237–242. 10.1038/ja.2013.121. [DOI] [PubMed] [Google Scholar]
- Liu Y.-Y.; Wang Y.; Walsh T. R.; Yi L.-X.; Zhang R.; Spencer J.; Doi Y.; Tian G.; Dong B.; Huang X.; Yu L.-F.; Gu D.; Ren H.; Chen X.; Lv L.; He D.; Zhou H.; Liang Z.; Liu J.-H.; Shen J. (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- Öberg B., Broberg A., Guss B., Levenfors J., Bjerketorp J., and Nord C.. Peptide compounds. WO 2020/046190A1, March 5th, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.