Abstract
Background
Epigenome-wide association studies (EWAS) have been widely applied to identify methylation CpG sites associated with human disease. To date, the Infinium MethylationEPIC array (EPIC) is commonly used for high-throughput DNA methylation profiling. However, the EPIC array covers only 30% of the human methylome. Methylation Capture bisulfite sequencing (MC-seq) captures target regions of methylome and has advantages of extensive coverage in the methylome at an affordable price.
Methods
Epigenome-wide DNA methylation in four peripheral blood mononuclear cell samples was profiled by using SureSelectXT Methyl-Seq for MC-seq and EPIC platforms separately. CpG site-based reproducibility of MC-seq was assessed with DNA sample inputs ranging in quantity of high (> 1000 ng), medium (300–1000 ng), and low (150 ng–300 ng). To compare the performance of MC-seq and the EPIC arrays, we conducted a Pearson correlation and methylation value difference at each CpG site that was detected by both MC-seq and EPIC. We compared the percentage and counts in each CpG island and gene annotation between MC-seq and the EPIC array.
Results
After quality control, an average of 3,708,550 CpG sites per sample were detected by MC-seq with DNA quantity > 1000 ng. Reproducibility of DNA methylation in MC-seq-detected CpG sites was high among samples with high, medium, and low DNA inputs (r > 0.96). The EPIC array captured an average of 846,464 CpG sites per sample. Compared with the EPIC array, MC-seq detected more CpGs in coding regions and CpG islands. Among the 472,540 CpG sites captured by both platforms, methylation of a majority of CpG sites was highly correlated in the same sample (r: 0.98–0.99). However, methylation for a small proportion of CpGs (N = 235) differed significantly between the two platforms, with differences in beta values of greater than 0.5.
Conclusions
Our results show that MC-seq is an efficient and reliable platform for methylome profiling with a broader coverage of the methylome than the array-based platform. Although methylation measurements in majority of CpGs are highly correlated, a number of CpG sites show large discrepancy between the two platforms, which warrants further investigation and needs cautious interpretation.
Keywords: Methylation capture sequencing, EPIC, DNA methylation, Peripheral blood mononuclear cells
Introduction
The rapid increase in the number of epigenome-wide association studies (EWAS) have successfully identified differentially methylated CpG sites that are associated with environmental exposures and diseases [1–6]. Such DNA methylation marks have been used as biomarkers for diagnosing, subtyping, and monitoring disease progression [7–11]. The most popular and affordable methods to profile epigenome-wide DNA methylation are array-based platforms, primarily the Illumina Human Methylation 450 K (450 K) and Infinium MethylationEPIC (EPIC) BeadChips (Illumina Inc, San Diego, CA). These arrays utilize Illumina’s beadchip technology that does not require polymerase chain reaction (PCR), but is subject to dye intensity biases between the two platforms [12]. These arrays have limited coverage of the methylome and can only detect up to 870,000 CpGs across the epigenome, leaving a large proportion of CpG sites unmeasured. Moreover, the EPIC array offers improved but still suboptimal coverage of regulatory elements [13]. Whole-genome bisulfite sequencing (WGBS) is able to capture more than 28 million CpGs, but the feasibility remains low for the population-based EWAS due to high cost and large genomic DNA input requirements to compensate for degradation during DNA bisulfite treatment. Alternatively, Methylation Capture Sequencing (MC-seq) is able to detect DNA methylation at single-nucleotide resolution utilizing a targeted next-generation sequencing approach [14]. It permits profiling of significantly more CpG sites than the EPIC array, requires less genomic DNA input than WGBS, and less expensive than WGBS, but can be susceptible to bias due to the presence of PCR duplicates. Feature-to-cost comparisons among different platforms can help understand the utilities of each platform and provide guidance for investigators in choosing a methylation profiling platform.
A few studies have compared the CpG coverage, reproducibility, and performance of array-based and MC-seq platforms [15–17]. Teh et al. compared MC-seq and the 450 K array in seven DNA samples extracted from saliva [15]. A recent study compared the EPIC array and TruSeq targeted bisulfite sequencing in four cord blood DNA samples [17]. However, no comparisons of MC-seq and array-based methylome profiling of peripheral blood mononuclear cells (PBMCs) has been reported. Here, we profiled the DNA methylome in PBMCs using the Agilent SureSelect Methyl-Seq platform and compared the results to the EPIC array in DNA samples extracted from PBMCs.
Methods
Methylation capture sequencing (MC-seq)
DNA samples description
DNA was extracted from de-identified PBMCs collected from four individuals. Genomic DNA quality was determined by estimating the A260/A280 and A260/A230 ratios by spectrophotometry and concentration by fluorometry. DNA integrity and fragment size were confirmed using a microfluidic chip run on an Agilent Bioanalyzer. To assess the reproducibility of MC-seq by DNA quantity, DNA samples from each participant were profiled in triplicate times with high (> 1000 ng), medium (300–1000 ng), and low (150–300 ng) DNA input. In total, 12 DNA samples were measured by MC-seq. Bisulfate conversion was conducted for each DNA sample as described below.
Methyl-seq target enrichment library prep
Indexed paired-end whole-genome sequencing libraries were prepared using the SureSelect XT Methyl-Seq kit (Agilent, part#G9651B). Genomic DNA was sheared to a fragment length of 150–200 bp using focused acoustic energy delivered by the Covaris E220 system (Covaris, part#500003). Fragmented sample size distribution was determined using the Caliper LabChip GX system (PerkinElmer, Part#122000). Fragmented DNA ends were repaired with T4 DNA Polymerase and Polynucleotide Kinase and “A” base was added using Klenow fragment in a single reaction followed by AMPure XP bead-based purification (Beckman Coulter, part#A63882). The methylated adapters were ligated using T4 DNA ligase followed by AMPure XP bead purification. Quality and quantity of adapter-ligated DNA were assessed using the Caliper LabChip GX system. Samples yielding > 350 ng were enriched for targeted methylation sites by using the custom SureSelect Methyl-Seq Capture Library. Hybridization was performed at 65 °C for 16 h using a C1000 Thermal Cycler (BIO-RAD, part# 1851197). Once the enrichment was completed, the samples were mixed with streptavidin-coated beads (Thermo Fisher Scientific, part#65602) and washed with a series of buffers to remove non-specific bound DNA fragments. DNA fragments were eluted from beads with 0.1 M NaOH. Unmethylated C residues of enriched DNA were modified by bisulfite conversion using the EZ DNA Methylation-Gold Kit (Zymo Research, part#D5005). The SureSelect enriched, bisulfite-converted libraries were PCR amplified using custom-made indexed primers (IDT, Coralville, Iowa). Dual-indexed libraries were quantified by quantitative polymerase chain reaction (qPCR) using the Library Quantification Kit (KAPA Biosystems, Part#KK4854) and inserts size distribution was assessed using the Caliper LabChip GX system. Samples with a yield of ≥ 2 ng/μl were proceeded to sequencing.
Flow cell preparation and sequencing
Sample concentrations were normalized to 10 nM and loaded onto an Illumina NovaSeq flow cell at a concentration that yields 40 million passing filter clusters per sample. Samples were sequenced using 100 bp paired-end sequencing on an Illumina HiSeq NovaSeq according to Illumina standard protocol. The 10 bp dual index was read during additional sequencing reads that automatically follows the completion of the first read. Data generated during sequencing runs were simultaneously transferred to the Yale Center for Genome Analysis high-performance computing cluster. A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time.
Preprocessing and quality control
Signal intensities were converted to individual base calls during a run using the system’s Real Time Analysis (RTA) software. Sample de-multiplexing and alignment to the human genome was performed using Illumina’s CASAVA 1.8.2 software suite. The sample error rate was required to be less than 1% and the distribution of reads per sample in a lane was required to be within reasonable tolerance.
Quality control (QC) on MC-seq was conducted following standard procedure as previously described [18]. Quality of sequence data was examined by using FastQC (ver. 0.11.8). Adapter sequences and fragments at 5′ and 3′ (phred score < 20) with poor quality were removed by Trim_galore (ver. 0.6.3_dev). We used Bismark pipelines (ver. v0.22.1_dev) to align the reads to the bisulfite human genome (hg19) with default parameters [19]. Quality-trimmed paired-end reads were transformed into a bisulfite converted forward strand version (C → T conversion) or into a bisulfite-treated reverse strand (G → A conversion of the forward strand). Duplicated reads were removed from the Bismark mapping output by deduplicate_bismark and CpG, CHG, and CHH (where H = A, T, or C) were extracted by bismark_methylation_extractor.
All CpG sites were grouped by sequencing coverage, also known as read depth. The groups with coverage of 1× to 100× were used to test the relationship between coverage and number of CpG sites. Only the CpG sites with coverage > 10× depth were used for final comparisons to ensure MC-seq data quality. Genes were annotated using Homer annotatePeaks.pl, including intergenic, 5′UTR, promoter, exon, intron, 3′UTR, transcription start site (TTS), and non-coding categories. CpG island, shore, shelf, and open sea annotation were defined by locally developed bash and R scripts based on genomic coordinates (hg19) of CpG islands from the UCSC genome browser. CpG shores was defined as up to 2 kb from CpG islands and CpG shelf was defined as up to 2 kb from a CpG shore.
Assessment of reproducibility
We assessed CpG- and participant-based reproducibility for MC-seq among 12 samples with DNA quantity of high, medium, and low input in two ways. First, CpG-based reproducibility was assessed by calculating Pearson correlations using the CpG sites in common of the samples from the same participant with different input DNA quantities. Scatterplots were rendered showing 10,000 randomly selected common CpG sites comparing samples with high and medium, high and low, and medium and low DNA inputs. Second, participant-based reproducibility was assessed by comparing methylation profiles among pairs of participants using the samples with high DNA inputs, by calculating Pearson correlations of common CpG sites.
EPIC array data preprocessing
The Infinium MethylationEPIC array (Illumina, San Diego, CA, USA) was used to measure PBMC DNA methylation profiles from the same four participants. These four samples with DNA input of 1000 ng were preprocessed using standard procedures as previously described [20]. Briefly, the predicted sex based on methylome was consistent with self-reported sex for all samples. All samples had a call rate greater than 0.15. A total of 19,090 CpG sites on X chromosomes and 537 CpG sites on Y chromosomes were filtered. A total of 846,464 CpG sites passed quality control.
Comparison of methylation at each CpG site between MC-seq and EPIC array
The overall distribution of gene annotation in relation to CpG island and genetic region between MC-seq and EPIC array data from the four participants was compared. Common CpG sites between MC-seq and EPIC array assays were defined according to genomic coordinates. Pearson correlation and the absolute beta-difference value (Δβ) were calculated among common CpG sites between MC-seq methylation percentage values and EPIC methylation beta values by using R (ver. 3.5.1). If median Δβ of the common CpG site between two platforms was > 0.1, it was defined as a discordant CpG pair; otherwise, the CpG site was defined as a concordant CpG pair. The density plot of Δβ and a Manhattan plot showing the distribution of Δβ across epigenome were illustrated. Scatterplots were rendered showing the correlation of β values from 10,000 randomly selected CpG sites measured by both MC-seq and EPIC array.
Results
MC-seq overview and reproducibility
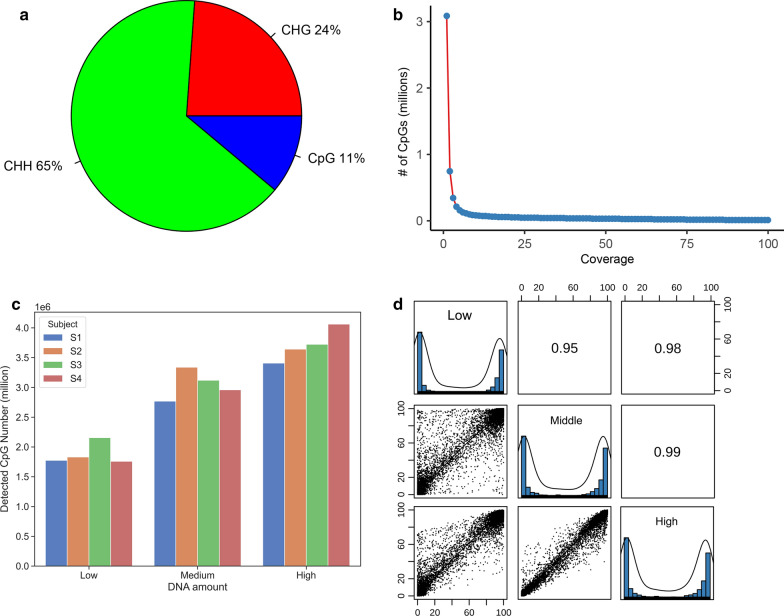
In MC-seq, all sequences were efficiently mapped to the reference genome with greater than 89% mapping efficiency. Interestingly, the number of non-CpG sites was significantly greater than the number of CpG sites. Among all detected methylation sites by MC-seq, 11% were CpG sites, 65% were CHH sites, and 24% were CHG sites (Fig. 1a).
Fig. 1.
Methylation Capture Sequencing (MC-seq). a Distribution of methylation sequence context (CpG, CHH, CHG); b Coverage depth versus a number of detected CpG sites; c Detected CpG sites in low, medium, and high DNA inputs for four participants using MC-seq with minimum coverage ≥ 10×; d Scatterplots comparing 10,000 randomly selected common CpG sites among samples with high, medium, and low DNA input quantities and their Pearson correlations
Figure 1b shows the relationship of the number of detected CpG sites and depth of sequence coverage by MC-seq in one sample. The depth of read at which the majority of sites were sequenced was estimated to be approximately 10× coverage, observed as the inflection point of the distribution of Fig. 1b. An increase of depth only slightly increased the capture of CpG sites and the inflection point is on 10× coverage, consistent with previous literature [15, 17]. Thus, the number of CpG sites with coverage ≥ 10× from MC-seq was used in subsequent analyses.
After quality control filtering, MC-seq captured an average of 2,878,207 methylation CpG sites with coverage ≥ 10× among the 12 DNA samples, with an average of 3,708,550 CpG sites among samples with high DNA input (> 1000 ng), an average of 3,046,172 CpG sites among samples with medium DNA input (300–1000 ng), and an average of 1,879,898 CpG sites among samples with low DNA input (150–300 ng) (Fig. 1c and Table 1). Despite the fact that the detected number of CpG sites varied depending on DNA input quantity, CpG-based correlation among the common CpG sites between samples with high and medium, high and low DNA input quantities exceeded r > 0.95. Correlations of common CpG sites between medium and low DNA inputs were also high with r in 0.92–0.94 (Table 2). Figure 1d shows the scatterplot of 10,000 randomly selected common CpGs between samples with high and medium, high and low, and medium and low DNA input quantities. Pair-wise participant-based correlations were high as r > 0.98 among common CpG sites (Table 3). Overall, MC-seq exhibited good reproducibility. The methylation profile generating in high DNA input from each participant was used for subsequent analyses.
Table 1.
Detected CpG number by DNA amount in MC-seq with coverage ≥ 10×
| DNA amount | Participant ID | CpG number | Average CpG number |
|---|---|---|---|
| Low | S1 | 1,774,940 | 1,879,898 |
| S2 | 1,831,086 | ||
| S3 | 2,154,732 | ||
| S4 | 1,758,834 | ||
| Medium | S1 | 2,768,456 | 3,046,172 |
| S2 | 3,338,200 | ||
| S3 | 3,119,259 | ||
| S4 | 2,958,772 | ||
| High | S1 | 3,406,879 | 3,708,550 |
| S2 | 3,642,776 | ||
| S3 | 3,722,552 | ||
| S4 | 4,061,994 | ||
| Total average | 2,878,207 |
Table 2.
Comparison of MC-seq between samples with high, medium, and low DNA input amount
| Participant ID | DNA amount | |||
|---|---|---|---|---|
| High | Medium | Common CpG | Pearson correlation | |
| S1 | 3,406,879 | 2,768,456 | 2,747,844 | 0.984 |
| S2 | 3,642,776 | 3,338,200 | 3,283,296 | 0.984 |
| S3 | 3,722,552 | 3,119,259 | 3,101,938 | 0.977 |
| S4 | 4,061,994 | 2,958,772 | 2,957,239 | 0.979 |
| DNA amount | ||||
|---|---|---|---|---|
| High | Low | Common CpG | Pearson correlation | |
| S1 | 3,406,879 | 1,774,940 | 1,771,936 | 0.960 |
| S2 | 3,642,776 | 1,831,086 | 1,829,919 | 0.966 |
| S3 | 3,722,552 | 2,154,732 | 2,153,175 | 0.974 |
| S4 | 4,061,994 | 1,758,834 | 1,758,622 | 0.963 |
| DNA amount | ||||
|---|---|---|---|---|
| Medium | Low | Common CpG | Pearson correlation | |
| S1 | 2,768,456 | 1,774,940 | 1,745,241 | 0.942 |
| S2 | 3,338,200 | 1,831,086 | 1,827,536 | 0.943 |
| S3 | 3,119,259 | 2,154,732 | 2,135,980 | 0.939 |
| S4 | 2,958,772 | 1,758,834 | 1,744,416 | 0.928 |
Table 3.
Overlap of detected CpG across samples with high DNA input amount by MC-seq
| Participant ID 1 | Participant ID 2 | Common CpG | Pearson R |
|---|---|---|---|
| S1 | S2 | 3,336,037 | 0.980 |
| S1 | S3 | 3,350,314 | 0.976 |
| S1 | S4 | 3,394,970 | 0.982 |
| S2 | S3 | 3,519,772 | 0.978 |
| S2 | S4 | 3,613,753 | 0.982 |
| S3 | S4 | 3,676,406 | 0.978 |
Distribution of methylome regions by MC-seq and EPIC
We compared genome-wide DNA methylation captured by MC-seq and by EPIC array in the four high DNA input samples. An average of 3,708,550 CpG sites were detected by MC-seq and 846,464 CpG sites by EPIC array. Overall, MC-seq detected 11.5 times more CpG sites in exons and 10.2 times more CpG sites in 5′ UTR region compared to the EPIC array, and 4.8 to 8.9 times more CpG site in other categories of genomic regions by MC-seq compared to EPIC array. However, the proportion of CpGs out of all CpGs successfully measured that map to gene regions in MC-seq as compared to the EPIC array did not significantly differ between these two platforms. For example, the proportion of CpG sites in transcription termination site (TTS) regions was similar between two platforms. MC-seq showed slightly greater proportions of CpG sites in 5′UTR and exon regions, while the EPIC array detected a greater proportion of CpG sites in promoter regions (Fig. 2a). In terms of CpG sites in relation to CpG islands including open seas, shelves, and shores, MC-seq detected 10.9 times more CpG sites located on CpG islands and 5.4–6.2 times more on other regions compared with the EPIC array. The proportion of CpG islands detected by MC-seq was greater than by the EPIC array (42% versus 29%), while the EPIC array detected a modestly higher percentage of CpG sites located in open seas than the MC-seq (39% versus 31%) (Fig. 2b).
Fig. 2.
Comparison of CpG proportion in epigenomic regions between MC-seq and EPIC. a Distribution of genomic regions (intergenic, promoter, 5′UTR, exon, intron, non-coding, 3′UTR, transcription termination site (TTS), and non-coding). b Distribution of CpG position relative to CpG islands (CpG island, shore, shelf, and open sea)
Comparison of Common CpG sites Measured by MC-seq and EPIC
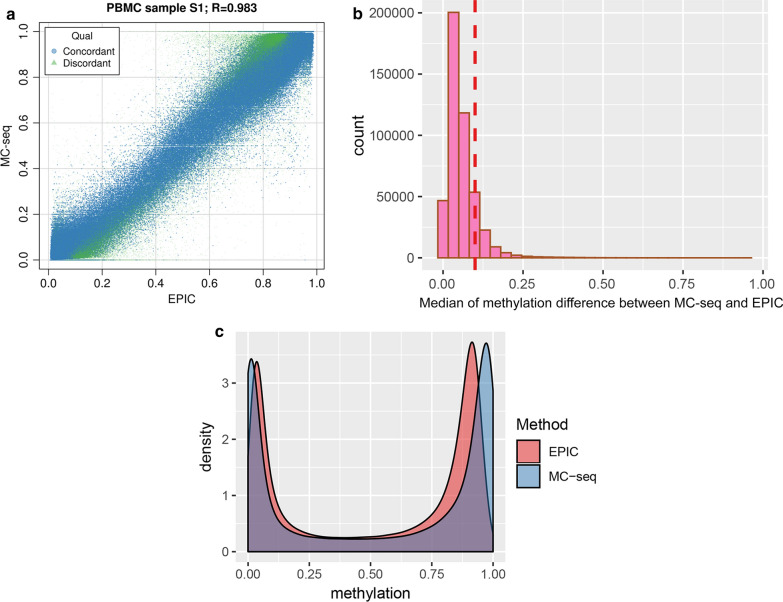
A total of 472,540 CpG sites were measured by both platforms. Overall, the correlations of these shared CpG sites was high, ranging from r = 0.983 to 0.985 across the four samples (Fig. 3a). Figure 3b presents the distribution of the absolute difference of methylation β values between MC-seq and EPIC. A small proportion of CpG sites (1.4%) were discordant (i.e., Δβ > 0.1), while 98.6% of CpG sites were concordant (i.e., Δβ < 0.1). Figure 3a presents the concordant (blue) and discordant CpG sites (green) between MC-seq and EPIC for participant S1 (Fig. 3a). The 60,753 discordant CpG sites appeared to be randomly distributed across the epigenome (Additional file 1: Figure S1). Among the discordant CpG sites, we identified 239 CpG sites with highly discrepant methylation (i.e., Δβ > 0.5) (Table 4). Addition file 2: Table S1 presents top 100 discordant CpG sites with medium discrepant methylation (Dβ = 0.1 ~ 0.4)Additional file 3: Figure S2 shows that participants S2, S3, and S4 have similar distribution of concordant and discordant plots as participant S1.
Fig. 3.
Comparing methylation values among common CpG sites between MC-seq and EPIC. a Correlation of methylation values measured by MC-seq and EPIC array among common CpG sites in participant S1. Blue dots represent concordant CpGs with Δβ < 0.1 between the two platforms and green dots represent discordant quality with Δβ ≥ 0.1; b The distribution of median Δβ in common CpG sites between MC-seq and EPIC array. The red dotted line represents Δβ = 0.1 as a cutoff for concordant CpG site between two platforms. c The density plot of methylation values among common CpG sites profiled by MC-seq and EPIC array in participant S1
Table 4.
Discordant CpG sites between MC-seq and EPIC (difference>0.5)
| Probe | Chr | Position | Gene | MC-seq median | EPIC median | Median Difference between MC-seq and EPIC | Refgene group | Relation to CpG island |
|---|---|---|---|---|---|---|---|---|
| cg09156519 | 9 | 103361572 | 0.009 | 0.960 | 0.95 | S_Shore | ||
| cg18176117 | 9 | 96097296 | C9orf129 | 0.000 | 0.932 | 0.93 | Body | N_Shore |
| cg14268958 | 10 | 133453066 | 0.000 | 0.899 | 0.89 | S_Shelf | ||
| cg10576280 | 10 | 124133822 | PLEKHA1 | 0.072 | 0.948 | 0.88 | TSS1500 | N_Shore |
| cg01005486 | 3 | 13246006 | 0.047 | 0.886 | 0.84 | Island | ||
| cg23433318 | 19 | 667542 | 0.007 | 0.866 | 0.84 | Island | ||
| cg10766172 | 7 | 27498479 | 0.989 | 0.147 | 0.82 | |||
| cg11812439 | 4 | 68928706 | LOC550113;SYT14P1;TMPRSS11F | 0.973 | 0.155 | 0.82 | Body; Body; Body | |
| cg23950473 | 5 | 154393265 | KIF4B; KIF4B | 0.992 | 0.174 | 0.82 | 1stExon; 5’UTR | |
| cg00259849 | 8 | 4183880 | CSMD1 | 0.000 | 0.821 | 0.82 | Body | |
| cg23981150 | 1 | 161111090 | 1.000 | 0.217 | 0.78 | Island | ||
| cg09698465 | 12 | 133000178 | 0.906 | 0.080 | 0.78 | Island | ||
| cg20450977 | 11 | 10529463 | MTRNR2L8; MTRNR2L8 | 0.964 | 0.189 | 0.77 | 3’UTR; 1stExon | |
| cg01053463 | 1 | 26186087 | C1orf135 | 0.021 | 0.757 | 0.76 | TSS1500 | Island |
| cg12499827 | 2 | 202004893 | CFLAR; CFLAR; CFLAR; CFLAR; CFLAR; CFLAR; CFLAR; CFLAR; CFLAR | 0.971 | 0.215 | 0.76 | TSS200; Body; Body; Body; Body; Body; Body; Body; Body | |
| cg03133777 | 2 | 170361364 | BBS5 | 1.000 | 0.244 | 0.75 | 3’UTR | |
| cg19040702 | 17 | 22023833 | 0.969 | 0.230 | 0.75 | |||
| cg21675871 | 11 | 69813397 | 0.217 | 0.960 | 0.74 | Island | ||
| cg04240493 | 3 | 148414664 | AGTR1; AGTR1; AGTR1; AGTR1 | 0.979 | 0.258 | 0.72 | TSS1500; TSS1500; TSS1500; TSS1500 | N_Shore |
| cg16889427 | 10 | 127584375 | FANK1 | 0.036 | 0.759 | 0.71 | TSS1500 | Island |
| cg25916505 | 18 | 32820654 | ZNF397; ZNF397 | 0.000 | 0.711 | 0.71 | TSS1500; TSS1500 | N_Shore |
| cg13525026 | 17 | 18061071 | MYO15A | 1.000 | 0.290 | 0.71 | Body | |
| cg07825433 | 4 | 1215099 | CTBP1; CTBP1 | 0.000 | 0.716 | 0.71 | Body; Body | N_Shelf |
| cg03846641 | 2 | 109746751 | LOC100287216; SH3RF3 | 0.239 | 0.952 | 0.71 | TSS200; Body | Island |
| cg19188207 | 2 | 10340837 | C2orf48 | 1.000 | 0.290 | 0.71 | Body | |
| cg11495544 | 17 | 73402155 | GRB2; GRB2 | 0.753 | 0.049 | 0.70 | TSS1500; TSS1500 | S_Shore |
| cg06931905 | 8 | 42036940 | PLAT; PLAT | 0.896 | 0.197 | 0.70 | Body; Body | |
| cg03348902 | 1 | 569603 | 0.869 | 0.168 | 0.70 | |||
| cg27120934 | 6 | 129480619 | LAMA2; LAMA2 | 0.979 | 0.297 | 0.69 | Body; Body | |
| cg07576219 | 1 | 55012408 | ACOT11; ACOT11 | 0.927 | 0.250 | 0.69 | TSS1500; TSS1500 | S_Shelf |
| cg08400246 | 5 | 156570642 | MED7; MED7 | 0.153 | 0.870 | 0.68 | TSS1500; TSS1500 | S_Shore |
| cg27626141 | 8 | 103876469 | AZIN1; AZIN1 | 0.000 | 0.682 | 0.68 | TSS200; TSS200 | Island |
| cg26688472 | 2 | 203638928 | ICA1L | 0.984 | 0.303 | 0.68 | 3’UTR | Island |
| cg26101183 | 10 | 65930786 | 0.957 | 0.279 | 0.68 | Island | ||
| cg27090007 | 13 | 28519388 | ATP5EP2 | 0.985 | 0.321 | 0.67 | Body | |
| cg11896012 | 19 | 53696753 | ZNF665 | 0.048 | 0.700 | 0.67 | TSS200 | S_Shore |
| cg02606018 | 12 | 10658281 | 0.979 | 0.323 | 0.66 | |||
| cg00438164 | 4 | 100870480 | H2AFZ; LOC256880 | 0.004 | 0.650 | 0.65 | Body; TSS1500 | Island |
| cg21164300 | 9 | 136098495 | 0.000 | 0.644 | 0.64 | N_Shelf | ||
| cg15891076 | 10 | 65930618 | 0.971 | 0.328 | 0.64 | Island | ||
| cg05948389 | 5 | 1641924 | 0.014 | 0.660 | 0.64 | N_Shelf | ||
| cg10507965 | 10 | 102107251 | SCD; SCD | 0.011 | 0.642 | 0.64 | 5’UTR; 1stExon | Island |
| cg21662326 | 11 | 14521493 | COPB1; COPB1; COPB1 | 0.643 | 0.012 | 0.63 | TSS200; TSS200; TSS200 | |
| cg09646578 | 8 | 5019363 | 0.310 | 0.934 | 0.63 | |||
| cg24717964 | 20 | 61477008 | DPH3B; DPH3B; TCFL5 | 0.986 | 0.356 | 0.63 | 1stExon; 5’UTR; Body | |
| cg07437919 | 8 | 142234483 | SLC45A4 | 0.957 | 0.313 | 0.63 | Body | N_Shore |
| cg01105403 | 2 | 240723304 | 0.050 | 0.890 | 0.63 | |||
| cg20482143 | 7 | 64340804 | 0.982 | 0.346 | 0.63 | |||
| cg11187452 | 22 | 49698612 | 0.017 | 0.653 | 0.63 | Island | ||
| cg24504954 | 3 | 61237217 | FHIT; FHIT | 0.017 | 0.649 | 0.62 | TSS200; TSS200 | Island |
| cg27434351 | 11 | 14521491 | COPB1; COPB1; COPB1 | 0.639 | 0.016 | 0.62 | TSS200; TSS200; TSS200 | |
| cg15864074 | 2 | 120974042 | 0.976 | 0.354 | 0.62 | |||
| cg00913521 | 12 | 89893799 | WDR51B | 0.977 | 0.339 | 0.62 | Body | |
| cg27534567 | 1 | 568536 | 0.834 | 0.262 | 0.62 | |||
| cg24515136 | 17 | 49024834 | 0.949 | 0.328 | 0.62 | S_Shelf | ||
| cg01417615 | 1 | 52456419 | RAB3B | 0.629 | 0.015 | 0.61 | TSS200 | Island |
| cg00236302 | 12 | 69004867 | RAP1B; RAP1B | 0.000 | 0.612 | 0.61 | 5’UTR; 5’UTR | Island |
| cg10747603 | 22 | 29197018 | XBP1; XBP1 | 0.022 | 0.627 | 0.61 | TSS1500; TSS1500 | S_Shore |
| cg03594447 | 1 | 20359744 | 1.000 | 0.358 | 0.61 | |||
| cg23045277 | 4 | 190587808 | 0.299 | 0.910 | 0.61 | |||
| cg02218809 | 16 | 29973300 | TMEM219; TMEM219 | 0.020 | 0.612 | 0.61 | TSS200; TSS200 | Island |
| cg05646491 | 10 | 135379754 | SYCE1; SYCE1; SYCE1 | 0.988 | 0.382 | 0.60 | TSS1500; 5’UTR; TSS1500 | Island |
| cg07596174 | 20 | 55926107 | RAE1; RAE1 | 0.014 | 0.613 | 0.60 | TSS1500; TSS200 | N_Shore |
| cg03543448 | 16 | 4384967 | GLIS2 | 0.927 | 0.315 | 0.60 | Body | |
| cg25793197 | 5 | 31923469 | PDZD2 | 0.976 | 0.379 | 0.60 | Body | |
| cg21392229 | 2 | 161223778 | RBMS1; RBMS1 | 1.000 | 0.384 | 0.60 | Body; Body | |
| cg05607320 | 12 | 53342553 | KRT18; KRT18 | 0.064 | 0.651 | 0.60 | TSS200; TSS1500 | N_Shore |
| cg13896861 | 9 | 94878241 | SPTLC1; SPTLC1 | 0.117 | 0.711 | 0.60 | TSS1500; TSS1500 | S_Shore |
| cg03064900 | 4 | 190566141 | 0.323 | 0.921 | 0.60 | N_Shore | ||
| cg16199859 | 3 | 75263685 | 0.861 | 0.276 | 0.60 | |||
| cg15006843 | 1 | 205720633 | NUCKS1 | 0.880 | 0.260 | 0.60 | TSS1500 | S_Shore |
| cg02498218 | 4 | 26361371 | RBPJ; RBPJ; RBPJ; RBPJ | 0.979 | 0.388 | 0.59 | Body; Body; 5’UTR; Body | Island |
| cg07116712 | 15 | 96887959 | 0.091 | 0.681 | 0.59 | Island | ||
| cg11643306 | 20 | 34204831 | SPAG4 | 0.038 | 0.630 | 0.59 | Body | S_Shore |
| cg08568561 | 7 | 42834498 | 0.981 | 0.392 | 0.59 | |||
| cg06669598 | 6 | 127622363 | ECHDC1; ECHDC1; ECHDC1; ECHDC1; ECHDC1 | 0.984 | 0.351 | 0.59 | 3’UTR; 3’UTR; Body; Body; Body | |
| cg22805431 | 3 | 113955600 | ZNF80 | 0.983 | 0.409 | 0.59 | 1stExon | |
| cg24636332 | 17 | 4437925 | SPNS2 | 0.301 | 0.939 | 0.59 | Body | N_Shore |
| cg05924191 | 15 | 35279830 | ZNF770 | 0.008 | 0.605 | 0.59 | 5’UTR | N_Shore |
| cg14402194 | 14 | 23398944 | PRMT5; PRMT5; PRMT5; PRMT5; PRMT5; PRMT5; LOC101926933 | 0.028 | 0.592 | 0.58 | TSS200; TSS200; TSS200; TSS200; TSS200; TSS200; Body | S_Shore |
| cg01737532 | 4 | 190862170 | FRG1 | 0.000 | 0.584 | 0.58 | 1stExon | Island |
| cg25744017 | 15 | 52819324 | MYO5A; MYO5A | 0.957 | 0.379 | 0.58 | Body;Body | N_Shore |
| cg03432151 | 15 | 89745000 | ABHD2; ABHD2 | 0.948 | 0.360 | 0.58 | 3’UTR; 3’UTR | |
| cg27196695 | 10 | 134571377 | INPP5A | 1.000 | 0.420 | 0.58 | Body | |
| cg27571351 | 10 | 17619364 | 0.986 | 0.407 | 0.58 | |||
| cg02775804 | 2 | 120974080 | 0.977 | 0.401 | 0.58 | |||
| cg16461530 | 10 | 134798264 | 0.664 | 0.106 | 0.58 | |||
| cg12654770 | 10 | 52487693 | 0.962 | 0.385 | 0.58 | |||
| cg16112880 | 1 | 201123745 | TMEM9 | 0.003 | 0.579 | 0.58 | TSS200 | Island |
| cg20641423 | 8 | 125315065 | 0.911 | 0.338 | 0.57 | S_Shore | ||
| cg23248615 | 10 | 2005709 | 0.905 | 0.313 | 0.57 | |||
| cg25550279 | 7 | 53254983 | 0.965 | 0.381 | 0.57 | Island | ||
| cg01070250 | 1 | 569687 | 0.843 | 0.271 | 0.57 | |||
| cg06977575 | 4 | 139481990 | 0.953 | 0.376 | 0.57 | Island | ||
| cg14511644 | 9 | 15055021 | 0.977 | 0.399 | 0.57 | |||
| cg08947542 | 8 | 35383200 | UNC5D | 0.879 | 0.309 | 0.57 | Body | |
| cg10258063 | 2 | 217363243 | RPL37A | 0.043 | 0.613 | 0.57 | TSS1500 | N_Shore |
| cg24209723 | 18 | 12913133 | 0.973 | 0.399 | 0.57 | S_Shore | ||
| cg02265379 | 5 | 87898506 | LOC645323 | 0.971 | 0.384 | 0.57 | Body | Island |
| cg18925601 | 7 | 158752715 | 0.006 | 0.574 | 0.57 | Island | ||
| cg01406075 | 11 | 58731104 | 0.885 | 0.309 | 0.57 | N_Shore | ||
| cg13545297 | 12 | 54404315 | HOXC8 | 0.229 | 0.791 | 0.57 | Body | S_Shore |
| cg09036531 | 10 | 96991505 | 0.968 | 0.402 | 0.57 | |||
| cg25649283 | 9 | 140714075 | EHMT1 | 0.382 | 0.950 | 0.57 | Body | Island |
| cg06204030 | 17 | 7792051 | CHD3; CHD3; CHD3 | 0.761 | 0.141 | 0.57 | TSS200; TSS200; Body | S_Shelf |
| cg18627328 | 19 | 621561 | POLRMT | 0.980 | 0.411 | 0.56 | Body | Island |
| cg13085681 | 8 | 48920761 | UBE2V2 | 0.009 | 0.576 | 0.56 | TSS1500 | N_Shore |
| cg00999469 | 6 | 25107287 | CMAHP | 0.043 | 0.931 | 0.56 | Body | |
| cg20960039 | 9 | 130213605 | LRSAM1; RPL12; LRSAM1; LRSAM1; LRSAM1; RPL12 | 0.020 | 0.582 | 0.56 | TSS1500; 1stExon; TSS200; TSS200; TSS200; 5’UTR | Island |
| cg04400841 | 2 | 208988863 | CRYGD | 0.215 | 0.764 | 0.56 | Body | Island |
| cg12476298 | 19 | 58426697 | ZNF417 | 0.977 | 0.407 | 0.56 | Body | N_Shore |
| cg23997402 | 19 | 14275669 | LPHN1; LPHN1 | 0.972 | 0.442 | 0.56 | Body; Body | S_Shore |
| cg16935370 | 5 | 154393281 | KIF4B; KIF4B | 0.981 | 0.414 | 0.56 | 1stExon; 5′UTR | |
| cg04222159 | 1 | 204981786 | NFASC; NFASC; NFASC; NFASC | 0.630 | 0.073 | 0.56 | Body; Body; Body; Body | |
| cg06396237 | 8 | 120779442 | TAF2 | 0.985 | 0.430 | 0.56 | Body | |
| cg06599543 | 6 | 165749446 | PDE10A;PDE10A | 0.857 | 0.332 | 0.56 | Body;Body | S_Shore |
| cg11566832 | 10 | 88659593 | BMPR1A | 0.145 | 0.693 | 0.55 | Body | |
| cg20334010 | 15 | 41047916 | RMDN3; RMDN3 | 0.020 | 0.570 | 0.55 | TSS1500; TSS1500 | S_Shore |
| cg18245781 | 5 | 3659697 | 0.283 | 0.852 | 0.55 | |||
| cg03761810 | 2 | 10264850 | RRM2; RRM2 | 0.018 | 0.567 | 0.55 | Body; Body | S_Shore |
| cg02122372 | 3 | 149657597 | RNF13; RNF13 | 1.000 | 0.439 | 0.55 | Body; Body | |
| cg06753227 | 18 | 9475508 | RALBP1 | 0.000 | 0.553 | 0.55 | TSS200 | Island |
| cg14131834 | 13 | 45914250 | LOC100190939; TPT1 | 0.040 | 0.594 | 0.55 | TSS1500; Body | N_Shore |
| cg25583180 | 5 | 177614382 | GMCL1L | 1.000 | 0.451 | 0.55 | Body | Island |
| cg09112623 | 6 | 33756905 | LEMD2; LEMD2 | 0.568 | 0.019 | 0.55 | 5’UTR; 1stExon | Island |
| cg11759477 | 4 | 190861959 | FRG1 | 0.000 | 0.548 | 0.55 | TSS200 | Island |
| cg12796755 | 14 | 51132292 | SAV1 | 0.932 | 0.383 | 0.55 | Body | N_Shelf |
| cg19693446 | 14 | 102144192 | 0.958 | 0.407 | 0.55 | |||
| cg25187648 | 3 | 49395165 | GPX1; GPX1; GPX1 | 0.018 | 0.568 | 0.55 | Body; 3’UTR; 1stExon | Island |
| cg20391833 | 6 | 167116208 | RPS6KA2 | 0.964 | 0.431 | 0.55 | Body | |
| cg09705232 | 6 | 97611802 | MIR548H3; C6orf167 | 0.974 | 0.428 | 0.55 | Body; Body | |
| cg04643437 | 12 | 14518655 | ATF7IP; ATF7IP | 0.000 | 0.561 | 0.55 | 1stExon; 5’UTR | Island |
| cg17558062 | 13 | 45965415 | LOC100190939 | 1.000 | 0.456 | 0.54 | Body | Island |
| cg26825848 | 4 | 190566175 | 0.350 | 0.899 | 0.54 | N_Shore | ||
| cg13943141 | 9 | 93205862 | 0.846 | 0.296 | 0.54 | |||
| cg26951705 | 19 | 56612697 | ZNF787 | 0.000 | 0.542 | 0.54 | Body | Island |
| cg24654094 | 1 | 160340832 | NHLH1 | 0.964 | 0.433 | 0.54 | Body | Island |
| cg02996355 | 14 | 81879375 | 0.909 | 0.364 | 0.54 | |||
| cg11914812 | 12 | 56904792 | 1.000 | 0.459 | 0.54 | |||
| cg24895977 | 19 | 35861796 | 0.990 | 0.450 | 0.54 | |||
| cg11637682 | 6 | 147124984 | LOC729176; C6orf103 | 0.867 | 0.300 | 0.54 | TSS200; Body | |
| cg07089633 | 14 | 73396378 | DCAF4; DCAF4; DCAF4; DCAF4; DCAF4 | 1.000 | 0.450 | 0.54 | 5’UTR; 5’UTR; 5’UTR; 5’UTR; 5’UTR | S_Shelf |
| cg20360416 | 4 | 7246127 | SORCS2 | 0.018 | 0.659 | 0.54 | Body | |
| cg25627920 | 17 | 39992620 | NT5C3B; NT5C3B; NT5C3B; KLHL10 | 0.016 | 0.552 | 0.54 | TSS200; TSS200; TSS200; TSS1500 | Island |
| cg24000259 | 5 | 55488291 | ANKRD55 | 0.961 | 0.411 | 0.54 | Body | |
| cg09138437 | 11 | 64527189 | PYGM; PYGM | 0.993 | 0.445 | 0.54 | 1stExon; 1stExon | |
| cg02673636 | 1 | 109647056 | 0.976 | 0.437 | 0.54 | S_Shelf | ||
| cg18740872 | 5 | 39220260 | FYB; FYB | 1.000 | 0.466 | 0.53 | TSS1500; TSS1500 | |
| cg14354292 | 7 | 63353606 | 0.960 | 0.426 | 0.53 | |||
| cg17704839 | 19 | 9939038 | UBL5; UBL5 | 0.014 | 0.542 | 0.53 | Body; Body | S_Shore |
| cg05971373 | 7 | 157498604 | PTPRN2; PTPRN2; PTPRN2 | 1.000 | 0.467 | 0.53 | Body; Body; Body | S_Shelf |
| cg05291429 | 17 | 1494566 | SLC43A2 | 0.402 | 0.969 | 0.53 | Body | S_Shelf |
| cg08841342 | 3 | 156528470 | PA2G4P4 | 0.976 | 0.444 | 0.53 | Body | |
| cg04096697 | 6 | 37012867 | 0.983 | 0.451 | 0.53 | Island | ||
| cg26878995 | 1 | 168106731 | GPR161 | 0.052 | 0.570 | 0.53 | TSS1500 | S_Shore |
| cg24031524 | 20 | 19804606 | 0.990 | 0.468 | 0.53 | |||
| cg19311470 | 4 | 39460490 | RPL9; RPL9; LIAS; LIAS | 0.004 | 0.529 | 0.53 | TSS1500; 5’UTR; TSS200; TSS200 | Island |
| cg02181482 | 5 | 178942685 | 0.956 | 0.449 | 0.53 | |||
| cg05346902 | 19 | 47910374 | MEIS3; MEIS3 | 0.068 | 0.593 | 0.53 | Body; Body | Island |
| cg16470772 | 10 | 8203304 | 0.971 | 0.445 | 0.53 | |||
| cg10115022 | 1 | 27527942 | 0.974 | 0.464 | 0.53 | Island | ||
| cg27231717 | 6 | 26319377 | 0.905 | 0.386 | 0.53 | |||
| cg09451549 | 19 | 8386408 | RPS28; NDUFA7; RPS28 | 0.000 | 0.527 | 0.53 | 5’UTR; TSS200; 1stExon | Island |
| cg07628841 | 2 | 27851430 | GPN1; CCDC121; GPN1; GPN1; CCDC121; CCDC121; GPN1; GPN1 | 0.010 | 0.536 | 0.53 | TSS200; 1stExon; TSS200; TSS1500; 5’UTR; 1stExon; TSS200; TSS1500 | |
| cg03816081 | 10 | 29577743 | LYZL1 | 0.863 | 0.301 | 0.52 | TSS1500 | |
| cg00762003 | 21 | 45393541 | AGPAT3; AGPAT3 | 0.383 | 0.892 | 0.52 | Body; Body | Island |
| cg19466922 | 7 | 130138026 | MEST; MEST; MEST; MEST; MEST; MEST | 1.000 | 0.476 | 0.52 | Body; Body; Body; Body; Body; Body | |
| cg07712165 | 17 | 80899280 | TBCD | 0.440 | 0.959 | 0.52 | Body | Island |
| cg01199952 | 13 | 25591486 | 0.984 | 0.456 | 0.52 | N_Shore | ||
| cg11374834 | 3 | 75263691 | 0.950 | 0.428 | 0.52 | |||
| cg02974491 | 1 | 1162280 | SDF4; SDF4 | 0.403 | 0.964 | 0.52 | Body; Body | Island |
| cg10555853 | 1 | 33516627 | 0.929 | 0.407 | 0.52 | Island | ||
| cg21216606 | 2 | 207275704 | 0.985 | 0.464 | 0.52 | |||
| cg17711541 | 6 | 26124704 | HIST1H2AC; HIST1H2BC | 0.007 | 0.529 | 0.52 | 1stExon; TSS1500 | Island |
| cg06412823 | 7 | 22541074 | STEAP1B; STEAP1B | 0.196 | 0.739 | 0.52 | TSS1500; TSS1500 | S_Shore |
| cg03054343 | 11 | 50238214 | 1.000 | 0.463 | 0.52 | Island | ||
| cg05766605 | 1 | 19384827 | 0.423 | 0.938 | 0.52 | |||
| cg07684215 | 10 | 132976057 | TCERG1L | 0.181 | 0.923 | 0.52 | Body | |
| cg27193858 | 6 | 41169120 | TREML2 | 0.181 | 0.691 | 0.52 | TSS200 | |
| cg00964321 | 16 | 15083956 | PDXDC1 | 0.906 | 0.386 | 0.52 | Body | Island |
| cg25394572 | 11 | 56457777 | OR8U8 | 0.949 | 0.429 | 0.52 | Body | |
| cg10667969 | 3 | 149181941 | 0.967 | 0.458 | 0.52 | |||
| cg18394854 | 5 | 8457818 | 0.212 | 0.732 | 0.52 | Island | ||
| cg05741225 | 10 | 133917303 | JAKMIP3 | 0.906 | 0.370 | 0.52 | TSS1500 | |
| cg06026769 | 12 | 20704492 | PDE3A | 0.993 | 0.473 | 0.52 | Body | N_Shore |
| cg09032630 | 6 | 27831956 | HIST1H2AL | 0.802 | 0.311 | 0.52 | TSS1500 | N_Shore |
| cg16626480 | 22 | 25575426 | KIAA1671 | 0.950 | 0.422 | 0.52 | Body | Island |
| cg24534731 | 17 | 36888147 | CISD3 | 0.969 | 0.457 | 0.52 | Body | S_Shore |
| cg16202259 | 14 | 104625420 | KIF26A | 0.058 | 0.962 | 0.52 | Body | Island |
| cg25325592 | 8 | 1439535 | 0.408 | 0.923 | 0.52 | N_Shore | ||
| cg00391025 | 3 | 100427239 | TFG; TFG | 1.000 | 0.477 | 0.52 | TSS1500; TSS1500 | N_Shore |
| cg25149037 | 17 | 39736213 | 0.820 | 0.454 | 0.52 | |||
| cg19120749 | 11 | 1431650 | BRSK2; BRSK2; BRSK2; BRSK2; BRSK2; BRSK2 | 0.468 | 0.983 | 0.52 | TSS1500; TSS200; Body; Body; Body; Body | Island |
| cg24270624 | 10 | 95721318 | PIPSL | 0.947 | 0.421 | 0.52 | Body | |
| cg16346588 | 10 | 242978 | ZMYND11; ZMYND11; ZMYND11 | 0.970 | 0.459 | 0.51 | Body; Body; Body | |
| cg02750322 | 15 | 83673816 | C15orf40; C15orf40; C15orf40; C15orf40; C15orf40 | 0.966 | 0.451 | 0.51 | Body; 3’UTR; Body; Body; Body | |
| cg04363536 | 3 | 49466872 | NICN1 | 0.000 | 0.515 | 0.51 | TSS200 | S_Shore |
| cg17883371 | 1 | 91359225 | 1.000 | 0.480 | 0.51 | Island | ||
| cg25018832 | 1 | 564471 | LOC101928626 | 0.602 | 0.088 | 0.51 | TSS200 | |
| cg16838729 | 4 | 43901032 | 0.903 | 0.394 | 0.51 | |||
| cg23222247 | 17 | 47302219 | PHOSPHO1; PHOSPHO1 | 0.009 | 0.531 | 0.51 | Body;Body | Island |
| cg19496566 | 19 | 48249018 | GLTSCR2 | 0.009 | 0.535 | 0.51 | 1stExon | Island |
| cg03165426 | 7 | 30726958 | CRHR2; CRHR2 | 0.429 | 0.942 | 0.51 | Body; 5’UTR | |
| cg19600494 | 2 | 106959525 | 0.968 | 0.455 | 0.51 | Island | ||
| cg10854807 | 17 | 79479308 | ACTG1 | 0.004 | 0.522 | 0.51 | Body | Island |
| cg20699097 | 11 | 111957680 | TIMM8B; TIMM8B; SDHD | 0.009 | 0.524 | 0.51 | TSS200; TSS200; 1stExon | Island |
| cg22819767 | 10 | 11866910 | C10orf47 | 0.958 | 0.440 | 0.51 | 5’UTR | S_Shore |
| cg20254251 | 8 | 144557206 | ZC3H3 | 0.993 | 0.450 | 0.51 | Body | |
| cg00590830 | 1 | 32385224 | PTP4A2; PTP4A2; PTP4A2; PTP4A2; PTP4A2 | 0.971 | 0.437 | 0.51 | 1stExon; 1stExon; 5’UTR; 5’UTR; 5’UTR | |
| cg03877767 | 2 | 11680057 | GREB1; GREB1 | 0.172 | 0.683 | 0.51 | 5’UTR; TSS200 | |
| cg00487526 | 15 | 90818384 | 0.956 | 0.444 | 0.51 | Island | ||
| cg17501384 | 2 | 217364031 | RPL37A | 0.017 | 0.521 | 0.51 | Body | S_Shore |
| cg17646418 | 6 | 166911767 | RPS6KA2; RPS6KA2 | 0.987 | 0.468 | 0.51 | Body; Body | |
| cg06757405 | 5 | 140789450 | PCDHGA4; PCDHGA9; PCDHGA1; PCDHGB1; PCDHGB6; PCDHGB6; PCDHGB3; PCDHGA6; PCDHGA8; PCDHGA5; PCDHGB4; PCDHGA3; PCDHGA2; PCDHGB2; PCDHGA7; PCDHGB5 | 0.108 | 0.607 | 0.51 | Body; Body; Body; Body; 1stExon; 1stExon; Body; Body; Body; Body; Body; Body; Body; Body; Body; Body | Island |
| cg13448596 | 8 | 2031599 | MYOM2 | 0.381 | 0.887 | 0.51 | Body | |
| cg16711165 | 11 | 111957658 | TIMM8B; TIMM8B; SDHD | 0.017 | 0.522 | 0.51 | TSS200; TSS200; 1stExon | Island |
| cg16786640 | 4 | 3485263 | DOK7; DOK7 | 0.447 | 0.954 | 0.51 | Body; Body | N_Shore |
| cg07638938 | 10 | 131348599 | MGMT | 0.986 | 0.492 | 0.51 | Body | |
| cg05407710 | 8 | 143329409 | TSNARE1 | 0.960 | 0.451 | 0.51 | Body | N_Shelf |
| cg07869994 | 3 | 174095190 | 0.988 | 0.482 | 0.51 | Island | ||
| cg07455406 | 14 | 21077527 | 0.017 | 0.523 | 0.51 | N_Shore | ||
| cg04576847 | 17 | 12623611 | MYOCD; MYOCD; MYOCD; MYOCD | 0.958 | 0.447 | 0.51 | Body; 5’UTR; Body; 1stExon | |
| cg15699853 | 18 | 57684747 | 0.976 | 0.467 | 0.51 | |||
| cg11231240 | 8 | 82434638 | 1.000 | 0.482 | 0.51 | Island | ||
| cg06157924 | 4 | 942005 | TMEM175 | 0.451 | 0.957 | 0.51 | Body | S_Shore |
| cg23679141 | 4 | 165118930 | MARCH1; ANP32C | 0.946 | 0.445 | 0.51 | 5’UTR; TSS200 | |
| cg03053358 | 17 | 1029917 | ABR; ABR | 0.446 | 0.964 | 0.51 | Body; 5’UTR | S_Shore |
| cg13705894 | 9 | 138305338 | 0.978 | 0.495 | 0.51 | S_Shore | ||
| cg18512780 | 17 | 76117734 | TMC6; TMC6 | 0.033 | 0.529 | 0.50 | Body; Body | |
| cg06307940 | 16 | 46660818 | 0.986 | 0.487 | 0.50 | |||
| cg15541008 | 5 | 95297508 | ELL2; ELL2 | 0.000 | 0.516 | 0.50 | 1stExon; 5’UTR | S_Shore |
| cg19969624 | 13 | 95954210 | ABCC4; ABCC4; ABCC4; ABCC4 | 0.011 | 0.507 | 0.50 | TSS1500; TSS1500; TSS1500; TSS1500 | Island |
| cg01758870 | 7 | 23719630 | C7orf46; C7orf46; C7orf46 | 0.028 | 0.545 | 0.50 | TSS200;TSS200;TSS200 | Island |
| cg08323201 | 15 | 101835348 | SNRPA1 | 0.000 | 0.503 | 0.50 | 1stExon | Island |
| cg21863998 | 11 | 19770288 | NAV2; NAV2; NAV2 | 0.439 | 0.905 | 0.50 | Body; Body; Body | |
| cg11471802 | 8 | 47529015 | 0.348 | 0.882 | 0.50 | Island | ||
| cg22281935 | 2 | 162934111 | 0.982 | 0.481 | 0.50 | S_Shelf | ||
| cg06032540 | 15 | 43941563 | CATSPER2; CATSPER2; CATSPER2 | 1.000 | 0.500 | 0.50 | TSS1500; TSS1500; TSS1500 | Island |
| cg18761878 | 1 | 568475 | 0.893 | 0.401 | 0.50 |
Density plots of methylation β showed bimodal distribution using both the MC-seq and the EPIC array platforms (Fig. 3c). Density of methylated CpG sites was slightly higher than the density of unmethylated CpG sites on both platforms. However, the two peaks in the EPIC array density plot were closer than the two peaks in the MC-seq density plot (Fig. 3c), indicating that MC-seq captures a higher dynamic range (i.e., more methylated and unmethylated) of CpG sites than the EPIC array. Additional file 4: Figure S3 shows that participants S2, S3, and S4 have similar density plots.
Discussion
We profiled the same PBMC samples using the MC-seq and EPIC array platforms and compared their performance. Our results show that the Agilent SureSelect Methyl-Seq targeted enrichment platform produced high-quality DNA methylation sequencing data at single base-pair resolution. MC-seq can reliably detect CpG sites with DNA input quantities as low as 300 ng. Overall, MC-seq detected 3–4 times more CpG sites than the EPIC array; however, the proportion of CpG sites mapped on functional genomic regions was similar between the two platforms. Methylation at a majority of CpG sites between the two platforms was highly correlated, while methylation at a low percentage of CpG sites differed significantly between the two platforms. Specifically, we found that methylation at 239 CpG sites differed significantly between the two platforms with absolute Δβ values greater than 0.5, which suggests that these CpG sites should be interpreted with caution in EWAS studies.
Our results show that MC-seq produces highly reliable CpG site methylation estimates across the genome. The observed CpG-based reproducibility is high, suggesting that technical variation on CpG calls is low. Inter-personal methylation variation is important for EWAS analysis. We found that our participant-based methylation on common CpG sites across four participants is also highly correlated, which further demonstrates the high reproducibility of this platform.
One disadvantage of sequencing-based approaches is the requirement for a larger quantity of input DNA than array-based approaches for methylation profiling. The recommended input DNA for Agilent SureSelect platform is 1ug, while input DNA quantity for EPIC array can be as low as 250 ng. Input DNA quantity is one important consideration influencing study design and methylation assay platform selection for population-based EWAS. Agilent has reported that DNA quantity can be as low as 250 ng for SureSelect sequencing [14]. To examine whether DNA quantity impacts the performance of MC-seq and to test whether low input DNA quantity also produces reliable CpG detection, we compared the capacity of CpG site detection across three different DNA input quantities. We found that medium DNA input quantity (i.e., 300 ng to 1000 ng) reliably detected CpG sites is comparable to the number of CpG sites captured by high DNA input quantity (i.e., greater than 1000 ng). Low DNA input quantity (i.e., less than 300 ng) detected the lowest number of CpG sites compared with high and medium DNA input quantity. For samples with low DNA input quantity, additional PCR cycles are needed to ensure post-capture library yield that results in extensive duplicate reads. In the four low DNA input samples, the duplicate rate exceeds 80%. Thus, removing duplicate reads is an important step in the QC process for MC-seq. We found that the number of CpG sites in low DNA input samples without duplicated reads still is significantly higher than the number of CpG sites detected by the EPIC array.
Consistent with previous reports, we found that methylation at the majority of CpG sites measured by both approaches (> 98%) is highly consistent between MC-seq and array-based methods. However, we identified 1.4% of CpG sites with discrepancies in CpG methylation that exceeds 10%. More importantly, 239 out of 60,753 discordant CpG sites had methylation differences exceeding 50%. These CpG sites are located on 159 gene regions (Table 4). Some of these genes have been previously reported to be associated with diseases. For example, SLC45A4 was reported to harbor an epigenetic marker for adiposity [21]. The methylation β differs on the CpG site of this gene by as much as 0.63 between the two platforms. We have also identified those CpG sites that showed less but still apparent discrepancy between the two assay platforms (i.e., absolute difference of beta values between 0.1 and 0.5). The top 100 CpG sites discrepant in a range of 0.1–0.4 between two platforms are presented in Table S2 to allow investigators to consider this potential source of bias in EWAS findings. The discrepancy might be due to bias in the performance of the beadchip assay at these positions, sequence context-dependent impacts on the performance of sequencing, batch effects, or a combination of these possibilities. This large discrepancy warrants further investigation and interpretation of findings at these CpG sites must be interpreted with caution.
One of the limitations of this study is the small number of participants used to estimate inter-sample variability. A previous study used a benchmark approach to evaluate performance of different platforms [17] and concluded that the EPIC array performed better than the MC-seq platform. However, the study did not remove duplicate reads as part of their data processing, which may have compromised the QC for MC-seq data processing as discussed above. Future studies, including benchmarking using a larger sample size, could further improve the analysis of platform performance. Of note, MC-seq detected high percentages of CHG and CHH sites across four methylome, which is consistent with previous reports [15]. The significances of those methylation sites warrant further investigation.
New approaches to measurement of DNA methylation continue to emerge that may warrant similar investigation in an ongoing effort to provide users with empiric comparisons to inform decisions about platform selection. One recent approach is enzymatic methyl-sequencing (EM-seq) (e.g., NEBNext EM-seq by New England Biolabs, Ipswich, MA) [22]. The input genomic DNA requirement is low 10–200 ng and EM-seq has comparable performance to WGBS [22], but its performance in relation to array- or capture sequencing-based approaches has not been reported. Should EM-seq gain popularity, it would be important to directly compare the performance of MC-seq and EM-seq to provide empiric evidence to users to inform platform selection.
Nevertheless, we have demonstrated that MC-seq is an efficient, reliable, and affordable platform that allows medium input quantity of DNA input (i.e., > 300 ng), which is equivalent to DNA input required for EPIC array. MC-seq has the advantage of capturing significantly more CpG sites than the EPIC array. Although methylation measurements between the two platforms are highly consistent, we have identified a small number of CpG sites that must be interpreted with caution if they are associated with a trait of interest because they showed significant discrepancies between the two platforms.
Conclusions
Our results show that MC-seq is an efficient and reliable platform for methylome profiling with a broader coverage of the methylome than the array-based platform. Although methylation measurements in majority of CpGs are highly correlated, a number of CpG sites show large discrepancy between the two platforms, which warrants further investigation and needs cautious interpretation.
Supplementary information
Additional file 1: Figure S1. A Manhattan plot showing the distribution of Δβ between MC-seq and EPIC array in PBMC by chromosome positions. Blue line represents Δβ = 0.1 and red line represents Δβ = 0.5.
Additional file 2: Table S1. Top 100 discordant CpG sites between MC-seq and EPIC array (Δβ = 0.1 ~ 0.4).
Additional file 3: Figure S2. Comparison of methylation values measured by MC-seq and EPIC array among common CpG sites in participant S2, S3, and S4. Blue dots represent concordant CpGs with Δβ < 0.1 between the two platforms and green dots represent discordant quality with Δβ ≥ 0.1
Additional file 4: Figure S3. The density plot of methylation values among CpG sites assayed in common by MC-seq and EPIC array in participant S2, S3, and S4
Acknowledgements
The project was supported by the National Institute on Drug Abuse (R03DA039745, R01DA038632, R01DA047063, R01DA047820). The authors appreciate the support of the Yale Center of Genomic Analysis and Women’s Interagency HIV Study.
Abbreviations
- EM-seq
Enzymatic methyl-seq
- EPIC
Illumina Infinium MethylationEPIC Beadchip
- EWAS
Epigenome-wide association study
- MC-seq
Methylation capture sequencing
- PBMC
Peripheral blood mononuclear cell
- PCR
Polymerase chain reaction
- QC
Quality control
- RTA
Real-time analysis
- TTS
Transcription termination site
- WGBS
Whole-genome bisulfite sequencing
Authors’ contributions
CS contributed to data analysis and the first draft of manuscript. XZ contributed to data processing, quality control, analysis, and manuscript preparation. BA was involved in manuscript preparation and provided peripheral blood monocyte cells. KX contributed to study design, analytical strategies, and manuscript preparation. All the authors read and approved the final manuscript.
Data availability
All methylation data from MC-seq and EPIC platforms are deposited in GEO (GSE152922).
Ethics approval and consent to participate
The study was approved by the committee of the Human Research Subject Protection at Yale University and the Institutional Research Board Committee of the Connecticut Veteran Healthcare System. De-identifiable samples were from Women’s Interagency HIV Study cohort. All participants provided written consents.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13072-020-00372-6.
References
- 1.Bakusic J, Schaufeli W, Claes S, Godderis L. Stress, burnout and depression: a systematic review on DNA methylation mechanisms. J Psychosom Res. 2017;92:34–44. doi: 10.1016/j.jpsychores.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Kraiczy J, Nayak KM, Howell KJ, Ross A, Forbester J, Salvestrini C, et al. DNA methylation defines regional identity of human intestinal epithelial organoids and undergoes dynamic changes during development. Gut. 2019;68(1):49–61. doi: 10.1136/gutjnl-2017-314817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam K, Pan K, Linnekamp JF, Medema JP, Kandimalla R. DNA methylation based biomarkers in colorectal cancer: a systematic review. Biochim Biophys Acta Rev Cancer. 2016;1866(1):106–120. doi: 10.1016/j.bbcan.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Li M, D’Arcy C, Li X, Zhang T, Joober R, Meng X. What do DNA methylation studies tell us about depression? A systematic review. Transl Psychiatry. 2019;9(1):68. doi: 10.1038/s41398-019-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nano J, Ghanbari M, Wang W, de Vries PS, Dhana K, Muka T, et al. Epigenome-wide association study identifies methylation sites associated with liver enzymes and hepatic steatosis. Gastroenterology. 2017;153(4):1096–1106. doi: 10.1053/j.gastro.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Teroganova N, Girshkin L, Suter CM, Green MJ. DNA methylation in peripheral tissue of schizophrenia and bipolar disorder: a systematic review. BMC Genet. 2016;17(1):27. doi: 10.1186/s12863-016-0332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delpu Y, Cordelier P, Cho W, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14(7):15029–15058. doi: 10.3390/ijms140715029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jönsson G, Olsson H, et al. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010;12(3):R36. doi: 10.1186/bcr2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet. 2019;20(2):109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 11.Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med. 2019;25(3):403–418. doi: 10.1038/s41591-019-0376-8. [DOI] [PubMed] [Google Scholar]
- 12.Dedeurwaerder S, Defrance M, Bizet M, Calonne E, Bontempi G, Fuks F. A comprehensive overview of Infinium HumanMethylation450 data processing. Brief Bioinform. 2013;15(6):929–941. doi: 10.1093/bib/bbt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):1–17. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JZ, Pabon C, Napier M. Agilent SureSelectXT Methyl-Seq applications with low-input DNA and smaller capture libraries. 2017. https://www.agilent.com/cs/library/applications/5991-7838EN.pdf.
- 15.Teh AL, Pan H, Lin X, Lim YI, Patro CP, Cheong CY, et al. Comparison of methyl-capture sequencing vs. Infinium 450 K methylation array for methylome analysis in clinical samples. Epigenetics. 2016;11(1):36–48. doi: 10.1080/15592294.2015.1132136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Z, Cunningham J, Slager S, Kocher JP. Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis. Epigenomics. 2015;7(5):813–828. doi: 10.2217/epi.15.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiss JA, Brennan KJ, Baccarelli AA, Tellez-Rojo MM, Estrada-Gutierrez G, Wright RO, et al. Battle of epigenetic proportions: comparing Illumina’s EPIC methylation microarrays and TruSeq targeted bisulfite sequencing. Epigenetics. 2020;15(1–2):174–182. doi: 10.1080/15592294.2019.1656159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wreczycka K, Gosdschan A, Yusuf D, Gruning B, Assenov Y, Akalin A. Strategies for analyzing bisulfite sequencing data. J Biotechnol. 2017;261:105–115. doi: 10.1016/j.jbiotec.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Hu Y, Justice AC, Li B, Wang Z, Zhao H, et al. DNA methylation signatures of illicit drug injection and hepatitis C are associated with HIV frailty. Nat Commun. 2017;8(1):2243. doi: 10.1038/s41467-017-02326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillycrop KA, Garratt ES, Titcombe P, Melton PE, Murray RJ, Barton SJ, et al. Differential SLC6A4 methylation: a predictive epigenetic marker of adiposity from birth to adulthood. Int J Obes. 2019;43(5):974–988. doi: 10.1038/s41366-018-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams L, Bei Y, Church HE, Dai N, Dimalanta ET, Ettwiller LM, et al. Enzymatic Methyl-seq: the next generation of methylome analysis. NEB Expressions. 2019;.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. A Manhattan plot showing the distribution of Δβ between MC-seq and EPIC array in PBMC by chromosome positions. Blue line represents Δβ = 0.1 and red line represents Δβ = 0.5.
Additional file 2: Table S1. Top 100 discordant CpG sites between MC-seq and EPIC array (Δβ = 0.1 ~ 0.4).
Additional file 3: Figure S2. Comparison of methylation values measured by MC-seq and EPIC array among common CpG sites in participant S2, S3, and S4. Blue dots represent concordant CpGs with Δβ < 0.1 between the two platforms and green dots represent discordant quality with Δβ ≥ 0.1
Additional file 4: Figure S3. The density plot of methylation values among CpG sites assayed in common by MC-seq and EPIC array in participant S2, S3, and S4
Data Availability Statement
All methylation data from MC-seq and EPIC platforms are deposited in GEO (GSE152922).