Abstract
COVID-19 respiratory disease caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has rapidly become a global health issue since it emerged in December 2019. While great global efforts are underway to develop vaccines and to discover or repurpose therapeutic agents for this disease, as of this writing only the nucleoside drug remdesivir has been approved under Emergency Use Authorization to treat COVID-19. The RNA-dependent RNA polymerase (RdRP), a viral enzyme for viral RNA replication in host cells, is one of the most intriguing and promising drug targets for SARS-CoV-2 drug development. Because RdRP is a viral enzyme with no host cell homologs, selective SARS-CoV-2 RdRP inhibitors can be developed that have improved potency and fewer off-target effects against human host proteins and thus are safer and more effective therapeutics for treating COVID-19. This review focuses on biochemical enzyme and cell-based assays for RdRPs that could be used in high-throughput screening to discover new and repurposed drugs against SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, coronavirus infection, RNA-dependent RNA polymerase, RdRP assays, RdRP inhibitors
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified to cause coronavirus disease-2019 (COVID-19) shortly after its emergence in late 2019. Its name was officially announced by the World Health Organization (WHO) based on its phylogeny, taxonomy, and established practice subsequent to the previous SARS outbreak in 2002 and 2003.1 An exponential growth in the number of SARS-CoV-2 infections in the beginning of 2020 was observed in many countries, and the WHO declared COVID-19 a global pandemic on March 11, 2020. COVID-19 vaccines are actively under development, with several already in Phase 1 clinical trials. Specific and effective small-molecule drugs for COVID-19 are not yet available, however, making the development of targeted drugs for treating COVID-19 an urgent unmet translational and medical need.
Viruses are simple but diverse organisms that can be categorized based on their differences in genetic material (DNA or RNA), configuration (double- or single-stranded), and the orientation of the encoded genes.2 Most DNA viruses have double-stranded DNA (dsDNA) and rely on DNA-dependent DNA polymerase for replication, similar to mammalian cells. RNA viruses are grouped into single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) viruses. ssRNA viruses can be either positive- or negative-sense. Positive-sense ssRNA is functionally like viral “mRNA” (messenger RNA) that can be directly translated into viral proteins in the host cell.3 Negative-sense ssRNA viruses carry complementary RNA that needs to be reverse-transcribed into positive-sense RNA through the enzymatic action of viral-specific RNA-dependent RNA polymerase (RdRP) before it can be translated to protein.3 RdRP function is also needed for positive-sense viruses during viral genome replication.
Coronaviruses are large, enveloped, positive-sense RNA viruses containing an ssRNA genome with a 5′-terminal cap structure and 3′-polyadenylation that can be as large as 32 kb.4 , 5 Previously, the most pathogenic human coronaviruses were SARS-CoV (8000 confirmed cases) and Middle East respiratory syndrome coronavirus (MERS-CoV) (2500 confirmed cases), with mortality rates of approximately 10% and 36%, respectively. The clinical manifestations of both diseases included fever, cough, fatigue, shortness of breath, and occasionally watery diarrhea.6 Some patients developed pneumonia, severe acute respiratory syndrome, and kidney failure that eventually led to death.7 Compared to SARS-CoV and MERS-CoV, SARS-CoV-2 appears to be more infectious, with more efficient human-to-human transmission that quickly resulted in a worldwide public health emergency.8 , 9
SARS-CoV-2 has four structural proteins—spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins—wrapped with an RNA genome to form the viral particles ( Fig. 1A and 1B). Its genome is approximately 30 kb in size and can be transcribed in 14 open reading frames (ORFs). The 5′ ORF1a/1b represents approximately 67% of the entire genome, and encodes pp1a and pp1ab precursor polyproteins (by a –1 ribosomal frameshift) that are hydrolyzed to 16 nonstructural proteins (nsp1 to nsp16) through proteolytic processing.10 These 16 nsps form the replicase–transcriptase complex (RTC) that consists of multiple enzymes, including papain-like protease (nsp3), chymotrypsin-like main protease (3CL protease, nsp5), primase complex (nsp7–nsp8), RdRP (nsp12), helicase (nsp13), and exoribonuclease (nsp14).11 As many as 13 of the remaining ORFs at the 3′ end encode the four structural proteins and nine putative accessory factors.11
Figure 1.
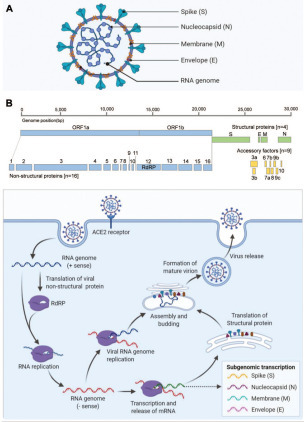
(A) Illustration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that consists of four structural proteins and RNA genome. (B) Schematic illustration of SARS-CoV-2 RNA genome11 and virus life cycle in host cell. The SARS-CoV-2 RNA genome encodes 16 nsps, 4 structural proteins, and 9 putative accessory factors. In the virus life circle, SARS-CoV-2 binds to angiotensin-converting enzyme-2 (ACE2) receptor and then releases RNA genome to cytosol to initiate the RNA replication and the formation of new virions. Created with Biorender.com.
Multiple viral proteins play critical roles for viral entry into the host cell and for intracellular replication. They are attractive targets for antiviral therapeutic development due to the lack of closely related host cell counterparts. For example, the S glycoprotein of SARS-CoV-2 binds to the host cell angiotensin-converting enzyme-2 (ACE2) receptor to initiate viral entry. Therefore, molecules that block spike protein binding to ACE2 are a burgeoning area of therapeutic development. Nonstructural proteins that function as viral enzymes, such as RdRP, 3CL protease, papain-like protease, and helicase, are also potential targets for drug development against SARS-CoV-2. In this review, we discuss drug development efforts for RdRPs and provide a perspective for assay strategies.
RdRP Structure and Function
All RNA viruses and some DNA viruses encode RdRPs that facilitate viral gene transcription and replication in concert with other viral and host factors.12 RdRPs for all RNA viral classes, positive-sense RNA, negative-sense RNA, and dsRNA share multiple sequence motifs and tertiary structures. The core structure of RdRP resembles the shape of a right hand, complete with palm, thumb, and finger domains. Five of the seven classical RdRP catalytic motifs (A–E) are located within the most conserved palm domain, while the other two (F and G) are within the finger domains.12, 13, 14 The structurally conserved RdRP core and the related motifs are essential for viral RdRP catalytic function and thereby represent potential targets for drug intervention.15
Although substrate requirements differ, all characterized RdRPs share the same catalytic mechanism. On infection of the host cell, viral RdRP participates in the formation of the genome replication machinery by complexing with other factors.16 RdRP initiates and governs the elongation of the RNA strand that includes the addition of hundreds to thousands of nucleotides ( Fig. 2A). Nucleotide analogs can stop the RNA elongation catalyzed by RdRP once they are inserted into the newly synthesized RNA chain (Fig. 2B). Viral RNA synthesis by RdRP from the 3′ end of the RNA template can be either primer-dependent or de novo (primer-independent). De novo RNA synthesis by RdRP is capable of using nucleotides to begin RNA synthesis in the absence of primer.17 In contrast, RdRP primer-dependent RNA synthesis relies on a short oligonucleotide or a protein covalently linked to nucleotides serving as primer;17 the primer anneals to the template RNA to provide a starting point for RNA synthesis.
Figure 2.
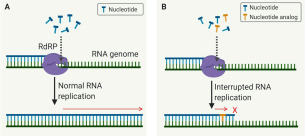
(A) The catalytic mechanism of RNA-dependent RNA polymerase (RdRP) in RNA replication. (B) The intervention of nucleotide analog as an inhibitor (an insertion of the nucleotide analog stops the RNA elongation after a few nucleotides that is catalyzed by RdRP).
Drug Development Targeting RdRP
RdRP plays essential roles in the RNA virus life cycle and has no host cell homolog. This opens the door for antiviral drug development and reduces the risk that a protein in human cells will be affected. Generally, viral RdRPs are regarded as low-fidelity enzymes largely due to lack of proofreading functions.18 Thus, a wide array of chain terminators or mutagenic nucleoside analog inhibitors targeting RdRP have been explored.18 It was found that nucleoside analogs in the form of adenine or guanine derivatives block viral RNA synthesis for a broad spectrum of RNA viruses, including human coronaviruses.19 , 20 Two such nucleoside analogs, the influenza drug favipiravir and experimental Ebola virus drug remdesivir (approved on May 1, 2020, for emergency use for the treatment of hospitalized COVID-19 patients), are currently being evaluated in clinical trials for the treatment of COVID-19. Table 1 shows a summary of US Food and Drug Administration (FDA)-approved antiviral drugs and clinical-stage investigational drugs that target viral RdRPs.
Table 1.
RdRP Inhibitors for Treatments of Viral Infections.
| Antiviral Agents | Approved Clinical Use | Reported Mechanism of Action | Approval Date or Clinical Status | Ref. |
|---|---|---|---|---|
| Favipiravir | Influenza viruses A, B, and C | Binds to catalytic domain of RdRP and prevents the inclusion of nucleotides for viral RNA replication | • Approved in March 2014 for influenza in Japan • Randomized trial for COVID-19 |
21, 22 |
| Ribavirin | HCV, RSV, and viral hemorrhagic fever | Inhibits viral RNA synthesis and mRNA capping | • Approved in December 1985 • Randomized trial for COVID-19 |
23 |
| Sofosbuvir | HCV genotype 2 or 3 | Binds to Mg2+ ions in RdRP of HCV, and inhibits HCV replication | Approved in December 2013 | 24 |
| Baloxavir | Influenza viruses A and B | Inhibits cap-dependent endonuclease in PA unit of influenza virus RdRP | Approved in 2018 | 25 |
| Dasabuvir | HCV genotype 1 | Binds to the palm-1 (P1) site of the influenza virus RdRP to stop virus replication | Approved in 2014 for use in combination with ombitasvir/paritaprevir/ritonavir | 26 |
| Remdesivir | Emergency Use Authorization for COVID-19 | Compete with ATA and terminates the nucleotide incorporation | Emergency Use Authorization for COVID-19 in May 2020 | 27 |
| Galidesivir | Inhibits viral RdRP function by terminating nonobligate RNA chain | Phase 1 for yellow fever, Marburg virus, and COVID-19 | 28 | |
| Pimodivir | Inhibits the PB2 cap-binding subunit of influenza A viruses RdRP | Phase 3 for influenza virus A | 29 | |
| Beclabuvir | Binds the NS5B (HCV RdRP) thumb pocket-1 allosteric site to inhibit RNA replication | Phase 2 for HCV and HIV/HCV co-infection in combination with asunaprevir and daclatasvir | 30 |
COVID-19, Coronavirus disease-2019; HCV, hepatitis C virus; mRNA, messenger RNA; PA, polymerase acidic; PB2, polymerase basic protein-2; RdRP, RNA-dependent RNA polymerase; RSV, respiratory syncytial virus.
Favipiravir
Favipiravir is a guanine analog antiviral drug targeting RdRP of RNA viruses. It was discovered by Toyama Chemical Co. (Tokyo, Japan) through a chemical library screen against influenza RdRP.22 Favipiravir has been approved in Japan and China, and it is being evaluated in the United States in a Phase 3 clinical trial for treatment of influenza.22 Favipiravir is effective against all subtypes and strains of influenza viruses, including those that are sensitive and resistant to other marketed drugs.22 Favipiravir is a prodrug that undergoes phosphoribosylation and phosphorylation to its active form, favipiravir-ribofuranosyl-5′-triphosphate (F-RTP), in human cells.31 F-RTP is recognized as a purine nucleotide by RdRP, but it inhibits RdRP enzyme activity and, thus, blocks viral RNA synthesis.22 In competition experiments with purine nucleosides or purine bases, the antiviral activity of favipiravir was attenuated, which confirmed its mechanism of action.21 Although the precise interaction of F-RTP with RdRP has not yet been elucidated, it is hypothesized that F-RTP may be misincorporated in a nascent viral RNA or bind to the catalytic domain of RdRP, thus preventing further addition of nucleotides in the viral RNA replication process.21 The catalytic domain of RdRP is conserved in various types of RNA viruses, underpinning a potential broad antiviral spectrum for favipiravir. Of particular interest is the observation that favipiravir has no effects on human DNA polymerases and RNA transcriptase up to 100 µg/mL, indicating a high selectivity index.21 Several clinical trials have been launched to evaluate the safety and efficacy of favipiravir for COVID-19 patents.32 In a clinical trial comparing favipiravir with umifenovir (Arbidol, an influenza virus entry inhibitor), favipiravir showed an improved clinical recovery rate on day 7 (71.43% vs. 55.86%), and reduced fever and cough duration compared to the umifenovir group for moderate patients with COVID-19, but there was no statistical difference regarding auxiliary oxygen therapy or the noninvasive mechanical ventilation rate.33 Finally, in targeting the viral RdRP, favipiravir could be promising in combination with other antiviral agents for treatment of COVID-19. As such, a clinical trial for COVID-19 treatment currently underway combines favipiravir with tocilizumab [an anti-interleukin-6 (IL6) humanized monoclonal antibody] (NCT04310228). It is the goal that guanine analogs such as favipiravir prove effective in double-blind clinical trials against SARS-CoV-2 so that clinicians may have another tool to treat patients in the emergency room.
Remdesivir
Remdesivir is a monophosphoramidate prodrug of an adenosine analog (GS-441524 is the active form) that has broad-spectrum antiviral activity against several viral families, including filoviruses, paramyxoviruses, and coronaviruses.34, 35, 36, 37 Remdesivir was originally developed as a treatment for Ebola virus disease. Remdesivir inhibited Ebola virus replication in multiple human cell lines in vitro with EC50 in the submicromolar range, and it showed positive therapeutic effects in nonhuman primates.38 A pharmacokinetics study showed remdesivir had high first-pass hepatic extraction due to its phosphoramidate prodrug features; therefore, oral administration was not explored.39 Moreover, the infection of Ebola virus also affects gastrointestinal systems, which may impair the absorption of orally administered drugs to reach effective doses.39 Thus, remdesivir was developed as an agent for intravenous injection in clinical development. Remdesivir has two hydroxyl groups, which may become orally active through chemical modification by masking one of them as an ester, like the modification on another anti-coronavirus drug, N-hydroxycytidine.40 Although an oral dosage form is under development, the route of administration of remdesivir remains intravenous.
On cell entry, remdesivir converts to a nucleoside triphosphate (NTP), suggesting that the corresponding triphosphate (TP) is the active form.38 Studies have confirmed that remdesivir-TP competes with adenosine triphosphate (ATP), the natural nucleotide, for incorporation into the nascent RNA strand41 , 42 and acts as an alternative substrate for the purified Ebola virus RdRP complex.34 Following the incorporation of remdesivir-TP, the growth of the RNA strand is halted. Because such inhibition does not occur immediately after the incorporation of inhibitor, the mechanism of its inhibitory effect is categorized as delayed RNA chain termination. For example, in the case of Ebola virus, the incorporated inhibitor at position i did not interrupt incorporation of the nucleotide at position i+1, but rather at position i+5.34 In a clinical trial for Ebola, remdesivir showed less efficacy compared to monoclonal antibody treatments such as mAb114 and REGN-EB3, but its safety profile was established.43
Remdesivir has been confirmed to inhibit coronaviruses, including SARS-CoV and MERS-CoV, in cell culture and animal models.44 Compared to Ebola virus RdRP, in which remdesivir causes delayed chain termination at position i+5, RNA synthesis was arrested at position i+3 for MERS-CoV.41 Coronaviruses usually have some proofreading ability to detect and correct the incorporation of incorrect nucleoside analogs, but remdesivir has been shown to outpace this protective barrier to maintain its antiviral activity.42 The existing reports on these anti-coronavirus effects inspired researchers to test remdesivir in COVID-19 clinical trials. Several compassionate and multisite clinical trials for COVID-19 have been reported or are currently underway.37 A recent report observed that the severe COVID-19 patients treated with compassionate-use remdesivir exhibited a clinical improvement of 68% (36 of 53).45 The interpretation of these results is limited, however, because the size of the patient cohort was small, the follow-up duration was relatively short, and there was no randomized control group.45 An improved SARS-CoV-2-specific RdRP inhibitor with better potency is still needed.
The strong scientific rationale, discussed above for RdRP as an opportune target for the development of novel or drug-repurposing therapeutics for treating COVID-19, stresses the need for rapid development of robust drug discovery assays focused on SARS-CoV-2 RdRP. In many cases, such new assays can be adapted from state-of-the-art high-throughput assay detection technologies that were developed previously for RdRPs from other viruses. Several such assay approaches are reviewed and discussed in the following sections.
Biochemical RdRP Enzyme Activity Assays
Polymerase Elongation Template Element (PETE) Assay for RdRP
Because RdRP catalyzes the incorporation of NTPs during RNA elongation, a PETE assay can be developed to detect the elongation activity of RdRP.46 In this assay approach, an oligonucleotide at the 5′ end of an RNA template is labeled with a fluorescent probe for fluorescence polarization (FP) measurements. The polarization signal from the fluorescent probe increases as its mobility becomes low following the elongation of the newly synthesized complementary RNA chain by RdRP.18 Inhibition of RdRP activity by a compound reduces the FP signal as the elongation of the complementary RNA chain stops. A schematic view of this assay is given in Figure 3A. This assay is suitable and has been used for high-throughput screening (HTS) of a compound library for lead discovery.18
Figure 3.
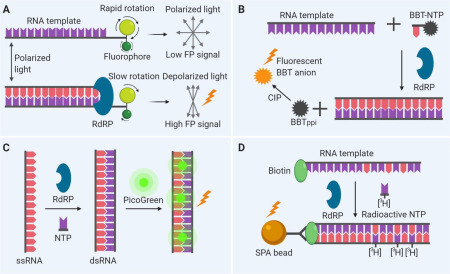
Schematic of biomedical assay designs. (A) Polymerase elongation template element (PETE) assay. An RNA oligonucleotide template is labeled with a fluorophore that can undergo rapid molecular rotation. The addition of nucleoside triphosphates (NTPs) to the RNA template mediated by RNA-dependent RNA polymerase (RdRP) sterically hinders the rotation of the fluorophore, resulting in an increase of the fluorescence polarization (FP) signal. (B) Fluorescence-based alkaline phosphatase–coupled polymerase assay (FAPA). The incorporation of (2-[2-benzothiazoyl]-6-hydroxybenzothiazole) conjugated adenosine triphosphate (BBT-ATP) into the RNA strand by RdRP results in the generation of BBT pyrophosphate (BBTPPi) that can be further catalyzed by reacting with the calf intestinal alkaline phosphatase (CIP) to produce fluorescent BBT anion. (C) Fluorometric RdRP activity assay. Using a single-stranded RNA (ssRNA) template, RdRP catalyzes the formation of double-stranded RNA (dsRNA) that can be detected by PicoGreen. (D) Scintillation proximity assay (SPA). SPA beads bind to the synthesized RNA containing [3H]-GTPs (guanosine triphosphates), leading to the close proximity that generates the SPA signal.
Fluorescence-Based Alkaline Phosphatase–Coupled Polymerase Assay (FAPA)
The FAPA approach includes a modified nucleotide analog in the substrate system during RNA synthesis by RdRP. As the polymerase reaction proceeds, incorporation of modified nucleotide analog results in the release of the fluorophore, allowing detection. For example, a modified nucleotide analog (2-[2-benzothiazoyl]-6-hydroxybenzothiazole) conjugated adenosine triphosphate (BBT-ATP) incorporated into the growing RNA chain was catalyzed by RdRP, resulting in a by-product of BBT, pyrophosphate (PPi).47 The BBTPPi subsequently was reacted with alkaline phosphatase to produce a highly fluorescent BBT anion.47 Figure 3B shows the principle of this assay. A screening of 40,572 compounds using this assay identified RdRP inhibitors of dengue virus.47 Besides BBT-ATP, the other nucleotide analogs can be also modified in different ways.48 , 49 Instead of applying alkaline phosphatase to produce a fluorophore, the intrinsic fluorescence of other by-products can also be used as reporters with modifications.50
Fluorometric RdRP Activity Assay
Fluorophores have been extensively used for the detection of RNA and DNA. In this fluorometric RdRP activity assay, fluorophores are used to detect dsRNA formation from the ssRNA template (Fig. 3C). One application of this assay was to screen the inhibitors of hepatitis C virus (HCV) RdRP.51 By using a poly(C) RNA template, HCV RdRP catalyzed the primer-independent synthesis of dsRNA that was detected by fluorescent dye PicoGreen.51 PicoGreen was originally developed to quantify dsDNA, but it was subsequently found to also preferentially bind dsRNA instead of ssRNA.51 This assay can be easily adapted to compound screening for RdRP inhibitors for many types of viruses. In addition to PicoGreen, other fluorophores have also been used to distinguish dsRNA from ssRNA, and they are useful for this type of RdRP assay.52 , 53
Scintillation Proximity Assay (SPA)
SPA has also been used in RdRP enzyme assays for HTS.54 This assay relies on the incorporation of radioactive nucleotides to the newly synthesized RNA chain catalyzed by RdRP using a biotinylated primer-template in the presence of 3H-GTP (Fig. 3D).55 Application of streptavidin-coupled SPA detection beads in this radioactive enzyme assay enables homogeneous assay detection that avoids the labor-intensive filtration and washing steps from the original radioactive NTP incorporation assay. Because they are radioactive assays, however, specific safety precautions and waste handling are required that may be inconvenient and require enhanced safety protocols. Therefore, most radioactive assays have been replaced by fluorometric assays in recent years.47
Cell-Based Enzyme Activity Assays for RdRP
While cell-free enzyme assays have the advantage of high-throughput capabilities for primary compound screening, several disadvantages of biochemical enzyme assays have been recognized by the field. First, enzyme assays do not require a compound to pass through cell membranes. If the inhibitors identified from enzyme assays cannot enter cells, they will not be active in subsequent cell-based assays and in animal models. This characteristic would not be determined in a biochemical assay. Second, cellular metabolic effects on compounds are not measured. Thus, biochemical enzyme assays cannot be used to determine the activities of prodrugs that need to be hydrolyzed by cellular enzymes to be converted to active components. In addition, some active compounds identified in cell-free enzyme assays may not be active in cell-based enzyme assays because they may be inactivated by intracellular enzymes. Third, the recombinant enzyme proteins may not be folded properly or may be missing natural subunits or cofactors compared to the RdRP expressed in cells. This can cause discrepancies of inhibitor potencies between biochemical enzyme assays and cell-based enzyme assays.
To complement biochemical RdRP enzyme assays, cell-based viral RdRP assays have been developed.56 , 57 In a cell-based RdRP assay, cells are transfected to express the viral RdRP or RdRP complex together with luciferase RNA in a negative-sense orientation that is then transcribed to the positive-sense luciferase RNA by RdRP for reporter protein synthesis.56 As a result, the intensity of the measured luciferase signal is proportional to the level of intracellular RdRP activity. Enzymatic activity of RdRP can be achieved by solely expressing one RdRP protein, such as the NS5B for HCV.57 But for some viruses, a complex of RdRP with a few other viral proteins is needed for the functional enzymatic activity. For instance, the functional RdRP complex for influenza virus requires the co-expression of influenza nucleoprotein (NP), polymerase acidic (PA), polymerase basic protein-1 (PB1), and PB2.56 Similarly, for Lassa virus, the minimal viral trans-acting factors required for genome replication are the L protein, which contains the RdRP activity, and the NP.58 These proteins were stably expressed in a cell line along with Gaussia luciferase and green fluorescent protein (GFP) reporters.58
Validation of Cell-Based RdRP Assays
Once developed, the cell-based assays must be rigorously validated to ensure that the reporter specifically reflects RdRP activity. To this end, control compounds, which can inhibit the activity of viral RdRP, are usually used. Ideal control compounds would be the well-defined RdRP inhibitors for the viruses studied. If available, it is encouraged to include different types of compounds that inhibit RdRP activities through various mechanisms. In the case of the RdRP cell-based assay for HCV, two types of three well-characterized NS5B inhibitors were used to examine the dependency of luciferase expression on the RdRP activity. They include one nucleotide, 2′-C-methylcytidine, and two non-nucleotides, cyclosporine A and wedelolactone.57 Among them, 2′-C-methylcytidine is a premature chain terminator of RdRP in the process of viral genome synthesis,59 cyclosporine A inhibits the functional association of cellular cyclophilin B with RdRP,60 and wedelolactone interferes with HCV NS5B–RNA binary complex formation.61 Their EC50 values acquired in the cell-based assay were compared with other previously reported assay systems to determine the assay sensitivity and accuracy for high-throughput operations in drug discovery.
Previous RdRP Screens for SARS-CoV and MERS-CoV
Molecular modeling was first applied to predict potential inhibitors that can be developed prior to the determination of SARS-CoV and MERS-CoV crystal structures. Xu et al. built a three-dimensional model of the catalytic domain based on the RdRP crystal structures of five other known RNA viruses.62 Using this model, they identified nucleoside analogs with specific features targeting RdRP. In a recent study, Elfiky et al. identified seven nucleotide inhibitors, including four novel compounds, against MERS-CoV RdRP using molecular modeling and docking simulation strategies.63 The EC50 values of these nucleotide analogs, however, were usually determined by viral yield or viral plaque assays that do not directly measure RdRP activity and are not suitable for the biosafety level 2 (BSL-2) laboratory setting.64, 65, 66
To develop HTS assays that specifically target RdRP of a coronavirus, a functional RdRP complex must first be achieved. Based on the knowledge acquired from study of SARS and MERS, RdRP (nsp12) protein alone exhibits limited activity without its complex partners.67 For SARS-CoV, the RNA replication machinery needs the participation of nsp7 and nsp8, which greatly improved the RdRP activity.67 The exact contribution of nsp7 and nsp8 in this machinery is still unclear. Kirchdoerfer et al. hypothesized that the nsp7–nsp8 heterodimer stabilizes the polymerase domain to permit template recognition through binding to the index-finger loop in RdRP.67 An alternative hypothesis is that nsp8, serving as a primase, synthesizes short oligonucleotide primers for the subsequent extension by the nsp12 “canonical” RdRP.68 Different from SARS-CoV, an active RdRP complex for MERS-CoV was obtained with only nsp8 and nsp12.41 In this report, a DNA plasmid encoding for a portion of the MERS-CoV 1ab polyprotein was used for protein expression in insect cells.41 This polyprotein contains only nsp5, nsp7, nsp8, and nsp12. After purification, the polyprotein was posttranslationally cleaved to yield a complex composed of nsp8 and nsp12 only. The activity of this MERS-CoV complex was confirmed by a radioactive NTP incorporation assay in the presence of a short primer and an RNA template. The IC50 of remdesivir–TP against this MERS-CoV RdRP complex was 0.032 µM at a fixed concentration of NTPs (0.02 µM).
Of note, besides nsp7, nsp8, and nsp12, it has been reported that nsp14 also plays significant roles in RNA replication, contributing to proofreading functions to safeguard coronavirus replication fidelity.4 Compared to other RNA viruses, the incorporation of coronavirus nsp14 conferred up to a 20-fold increase in replication fidelity and was responsible for the high resistance of coronaviruses to many nucleoside analogs.66
Such studies have demonstrated that the RNA-synthesizing machinery in coronaviruses requires incorporation of RdRP together with other key nsps to form a fully functional polymerase complex. In this context, cell-based RdRP assays offer the advantage that several nsps can be co-expressed and can self-assemble into functionally representative RNA replication machinery supporting drug development strategies targeting SARS-CoV-2 RdRP. At this point, however, the exact composition of this complex is still unclear.
Perspectives on SARS-CoV-2 RdRP Inhibitor Discovery
The rapid global spread of the COVID-19 pandemic has emphasized the urgent need to develop new therapeutics. Approved small-molecule drugs that target viral proteins critical to virus replication such as RdRP would be useful agents to fight SARS-CoV-2 infection.69 A key advantage of targeting the SARS-CoV-2 RdRP for drug development is that there has been rapid progress in developing drug-like polymerase inhibitors for other viruses in the past few years, although SARS-CoV-2 RdRP assays for HTS are yet to be developed and validated. Advances in the structural analysis of SARS-CoV-2 RdRP will also be critical in aiding the design of new drug candidates. In fact, a recent study has elucidated the cryo-structure of the SARS-CoV-2 full-length nsp12 monomer, and of its complex with nsp7 and nsp8.13 It was found that SARS-CoV-2 nsp12 has a newly identified β-hairpin domain at its N-terminus in addition to the conserved viral family polymerase architecture composed of fingers, palm, and thumb subdomains. The catalytic metal ions are not observed in the absence of primer-template RNA and NTPs, despite these having being present in several structures of viral polymerases that synthesize RNA.13 This structural information could be used to explain the inhibition mechanism of remdesivir and favipiravir,13 and could contribute to the discovery of new drugs. Recently, an open-label, Phase 3 clinical trial for remdesivir revealed encouraging results that showed patients who received remdesivir had a 31% faster time to recovery than those who received placebo (p < 0.001); it also suggested a survival benefit, with a mortality rate of 8.0% for the group receiving remdesivir versus 11.6% for the placebo group (p = 0.059).70 Following this result, the FDA granted remdesivir Emergency Use Authorization on May 1, 2020.27 Although the treatment efficacy is limited, it validated the utility of RdRP inhibitor for treatment of severe COVID-19 patients.
Considering the similarity of the key drug-binding pockets between SARS-CoV-2, SARS-CoV, and MERS-CoV RdRPs,13 repurposing known SARS-CoV and MERS-CoV inhibitors for SARS-CoV-2 remains a promising strategy. In addition, combination therapy of RdRP inhibitors with approved or clinical-stage drug candidates targeting other viral proteins may provide better therapeutic efficacy for treating COVID-19.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
The authors appreciate valuable and constructive suggestions from anonymous reviewers.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health. The authors received no other financial supports for the research, authorship, and/or publication of this article.
ORCID iD
Wei Zheng  https://orcid.org/0000-0003-1034-0757
https://orcid.org/0000-0003-1034-0757
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming It SARS-CoV-2. Nature Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmonds P., Aiewsakun P. Virus Classification: Where Do You Draw the Line? Arch. Virol. 2018;163:2037–2046. doi: 10.1007/s00705-018-3938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durmuş S., Ülgen K.Ö. Comparative Interactomics for Virus–Human Protein–Protein Interactions: DNA Viruses versus RNA Viruses. FEBS Open Bio. 2017;7:96–107. doi: 10.1002/2211-5463.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subissi L., Posthuma C.C., Collet A., et al. One Severe Acute Respiratory Syndrome Coronavirus Protein Complex Integrates Processive RNA Polymerase and Exonuclease Activities. Proc. Natl. Acad. Sci. 2014;111:E3900. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo P.C.Y., Lau S.K.P., Lam C.S.F., et al. Discovery of a Novel Bottlenose Dolphin Coronavirus Reveals a Distinct Species of Marine Mammal Coronavirus in Gammacoronavirus. J. Virol. 2014;88:1318. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.de Wit E., van Doremalen N., Falzarano D., et al. SARS and MERS: Recent Insights into Emerging Coronaviruses. Nature Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paules C.I., Marston H.D., Fauci A.S. Coronavirus Infections—More Than Just the Common Cold. JAMA. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 8.Chan J.F.-W., Yuan S., Kok K.-H., et al. A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. The Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen N., Zhou M., Dong X., et al. Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. The Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F., Zhao S., Yu B., et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon D. E., Jang G. M., Bouhaddou M.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature. 2020. doi:10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed]
- 12.Gorbalenya A.E., Pringle F.M., Zeddam J.-L., et al. The Palm Subdomain-Based Active Site Is Internally Permuted in Viral RNA-Dependent RNA Polymerases of an Ancient Lineage. J. Molec. Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y., Yan L., Huang Y., et al. Structure of the RNA-Dependent RNA Polymerase from COVID-19 Virus. Science. 2020:eabb7498. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataraman S., Prasad B.V.L.S., Selvarajan R. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu B., Gong P. Structural Basis of Viral RNA-Dependent RNA Polymerase Catalysis and Translocation. Proc. Natl. Acad. Sci. 2016:201602591. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahlquist P. RNA-Dependent RNA Polymerases, Viruses, and RNA Silencing. Science. 2002;296:1270. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 17.Ranjith-Kumar C.T., Gutshall L., Kim M.-J., et al. Requirements for De Novo Initiation of RNA Synthesis by Recombinant Flaviviral RNA-Dependent RNA Polymerases. J. Virol. 2002;76:12526. doi: 10.1128/JVI.76.24.12526-12536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campagnola G., Gong P., Peersen O.B. High-Throughput Screening Identification of Poliovirus RNA-Dependent RNA Polymerase Inhibitors. Antiviral Res. 2011;91:241–251. doi: 10.1016/j.antiviral.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Clercq E. New Nucleoside Analogues for the Treatment of Hemorrhagic Fever Virus Infections. Chem. Asian J. 2019;14:3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G., De Clercq E. Therapeutic Options for the 2019 Novel Coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 21.Furuta Y., Takahashi K., Kuno-Maekawa M., et al. Mechanism of Action of T-705 against Influenza Virus. Antimicrob. Agents Chemo. 2005;49:981. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a Broad Spectrum Inhibitor of Viral RNA Polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graci J.D., Cameron C.E. Mechanisms of Action of Ribavirin against Distinct Viruses. Rev. Med. Virol. 2006;16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Clercq E., Li G. Approved Antiviral Drugs over the Past 50 Years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noshi T., Kitano M., Taniguchi K., et al. In Vitro Characterization of Baloxavir Acid, a First-in-Class Cap-Dependent Endonuclease Inhibitor of the Influenza Virus Polymerase PA Subunit. Antiviral Res. 2018;160:109–117. doi: 10.1016/j.antiviral.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Eltahla A.A., Luciani F., White P.A., et al. Inhibitors of the Hepatitis C Virus Polymerase; Mode of Action and Resistance. Viruses. 2015;7:5206–5224. doi: 10.3390/v7102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.https://www.fda.gov/media/137564/download.
- 28.Warren T.K., Wells J., Panchal R.G., et al. Protection against Filovirus Diseases by a Novel Broad-Spectrum Nucleoside Analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark M.P., Ledeboer M.W., Davies I., et al. Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2. J. Med. Chem. 2014;57:6668–6678. doi: 10.1021/jm5007275. [DOI] [PubMed] [Google Scholar]
- 30.Lemm J.A., Liu M., Gentles R.G., et al. Preclinical Characterization of BMS-791325, an Allosteric Inhibitor of Hepatitis C Virus NS5B Polymerase. Antimicrob. Agents Chemo. 2014;58:3485. doi: 10.1128/AAC.02495-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiso M., Takahashi K., Sakai-Tagawa Y., et al. T-705 (Favipiravir) Activity against Lethal H5N1 Influenza A Viruses. Proc. Natl. Acad. Sci. 2010;107:882. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Y.-X., Chen X.-P. Favipiravir: Pharmacokinetics and Concerns about Clinical Trials for 2019-nCoV Infection. Clin. Pharma. Ther.2020. doi:10.1002/cpt.1844. [DOI] [PubMed]
- 33.Chen C., Zhang Y., Huang J.; et al. Favipiravir versus Arbidol for COVID-19: A Randomized Clinical Trial. medRxiv. 2020, 2020.03.17.20037432.
- 34.Tchesnokov E.P., Feng J.Y., Porter D.P., et al. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy B.G., Perron M., Murakami E., et al. The Nucleoside Analog GS-441524 Strongly Inhibits Feline Infectious Peritonitis (FIP) Virus in Tissue Culture and Experimental Cat Infection Studies. Vet. Microbiol. 2018;219:226–233. doi: 10.1016/j.vetmic.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choy K.-T., Wong A.Y.-L., Kaewpreedee P., et al. Remdesivir, Lopinavir, Emetine, and Homoharringtonine Inhibit SARS-CoV-2 Replication In Vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eastman R.T., Roth J.S., Brimacombe K.R., et al. Remdesivir: A Review of Its Discovery and Development Leading to Human Clinical Trials for Treatment of COVID-19. Preprints. 2020:2020040299. doi: 10.1021/acscentsci.0c00747. doi:10.20944/preprints202004.0299.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren T.K., Jordan R., Lo M.K., et al. Therapeutic Efficacy of the Small Molecule GS-5734 against Ebola Virus in Rhesus Monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel D., Hui H.C., Doerffler E., et al. Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J. Med. Chem. 2017;60:1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 40.Toots M., Yoon J., Cox R.M., et al. Characterization of Orally Efficacious Influenza Drug with High Resistance Barrier in Ferrets and Human Airway Epithelia. Sci. Transl. Med. 2019;11:eaax5866. doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon C.J., Tchesnokov E.P., Feng J.Y., et al. The Antiviral Compound Remdesivir Potently Inhibits RNA-Dependent RNA Polymerase from Middle East Respiratory Syndrome Coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amirian E.S., Levy J.K. Current Knowledge about the Antivirals Remdesivir (GS-5734) and GS-441524 as Therapeutic Options for Coronaviruses. One Health. 2020;9:100128. doi: 10.1016/j.onehlt.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulangu S., Dodd L.E., Davey R.T., et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. New Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheahan T.P., Sims A.C., Graham R.L., et al. Broad-Spectrum Antiviral GS-5734 Inhibits Both Epidemic and Zoonotic Coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grein J., Ohmagari N., Shin D.; et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. New Engl. J. Med.2020. doi:10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed]
- 46.Mestas S.P., Sholders A.J., Peersen O.B. A Fluorescence Polarization-Based Screening Assay for Nucleic Acid Polymerase Elongation Activity. Anal. Biochem. 2007;365:194–200. doi: 10.1016/j.ab.2007.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niyomrattanakit P., Abas S.N., Lim C.C., et al. A Fluorescence-Based Alkaline Phosphatase–Coupled Polymerase Assay for Identification of Inhibitors of Dengue Virus RNA-Dependent RNA Polymerase. J. Biomol. Screen. 2011;16:201–210. doi: 10.1177/1087057110389323. [DOI] [PubMed] [Google Scholar]
- 48.Kozlov M., Bergendahl V., Burgess R., et al. Homogeneous Fluorescent Assay for RNA Polymerase. Anal. Biochem. 2005;342:206–213. doi: 10.1016/j.ab.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 49.Smith T.M., Lim S.P., Yue K., et al. Identifying Initiation and Elongation Inhibitors of Dengue Virus RNA Polymerase in a High-Throughput Lead-Finding Campaign. J. Biomol. Screen. 2014;20:153–163. doi: 10.1177/1087057114551141. [DOI] [PubMed] [Google Scholar]
- 50.Bhat J., Rane R., Solapure S.M., et al. High-Throughput Screening of RNA Polymerase Inhibitors Using a Fluorescent UTP Analog. J. Biomol. Screen. 2006;11:968–976. doi: 10.1177/1087057106291978. [DOI] [PubMed] [Google Scholar]
- 51.Eltahla A.A., Lackovic K., Marquis C., et al. A Fluorescence-Based High-Throughput Screen to Identify Small Compound Inhibitors of the Genotype 3a Hepatitis C Virus RNA Polymerase. J. Biomol. Screen. 2013;18:1027–1034. doi: 10.1177/1087057113489883. [DOI] [PubMed] [Google Scholar]
- 52.Kocabas F., Turan R.D., Aslan G.S. Fluorometric RdRp Assay with Self-Priming RNA. Virus Genes. 2015;50:498–504. doi: 10.1007/s11262-015-1187-8. [DOI] [PubMed] [Google Scholar]
- 53.Sáez-Álvarez Y., Arias A., Del Águila C., et al. Development of a Fluorescence-Based Method for the Rapid Determination of Zika Virus Polymerase Activity and the Screening of Antiviral Drugs. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-41998-1. 5397–5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong E.Y., Kenens H., Ivens T., et al. In: Antiviral Methods and Protocols. Gong E.Y., editor. Humana Press; Totowa, NJ: 2013. Expression and Purification of Dengue Virus NS5 Polymerase and Development of a High-Throughput Enzymatic Assay for Screening Inhibitors of Dengue Polymerase; pp. 237–247. [DOI] [PubMed] [Google Scholar]
- 55.Nasiri A.H., Nasiri H.R. Polymerase Assays for Lead Discovery: An Overall Review of Methodologies and Approaches. Anal. Biochem. 2018;563:40–50. doi: 10.1016/j.ab.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 56.Su C.-Y., Cheng T.-J.R., Lin M.-I., et al. High-Throughput Identification of Compounds Targeting Influenza RNA-Dependent RNA Polymerase Activity. Proc. Natl. Acad. Sci. 2010;107:19151. doi: 10.1073/pnas.1013592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J.-C., Tseng C.-K., Chen K.-J., et al. A Cell-Based Reporter Assay for Inhibitor Screening of Hepatitis C Virus RNA-Dependent RNA Polymerase. Anal. Biochem. 2010;403:52–62. doi: 10.1016/j.ab.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 58.Cubitt B., Ortiz-Riano E., Cheng B.Y., et al. A Cell-Based, Infectious-Free, Platform to Identify Inhibitors of Lassa Virus Ribonucleoprotein (vRNP) Activity. Antiviral Res. 2020;173 doi: 10.1016/j.antiviral.2019.104667. 104667–104667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clark J.L., Hollecker L., Mason J.C., et al. Design, Synthesis, and Antiviral Activity of 2′-Deoxy-2′-Fluoro-2′-C-methylcytidine, a Potent Inhibitor of Hepatitis C Virus Replication. J. Med. Chem. 2005;48:5504–5508. doi: 10.1021/jm0502788. [DOI] [PubMed] [Google Scholar]
- 60.Qing M., Yang F., Zhang B., et al. Cyclosporine Inhibits Flavivirus Replication through Blocking the Interaction between Host Cyclophilins and Viral NS5 Protein. Antimicrob. Agents Chemo. 2009;53:3226–3235. doi: 10.1128/AAC.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaushik-Basu N., Bopda-Waffo A., Talele T.T., et al. Identification and Characterization of Coumestans as Novel HCV NS5B Polymerase Inhibitors. Nucl. Acids Res. 2008;36:1482–1496. doi: 10.1093/nar/gkm1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu X., Liu Y., Weiss S., et al. Molecular Model of SARS Coronavirus Polymerase: Implications for Biochemical Functions and Drug Design. Nucl. Acids Res. 2003;31:7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elfiky A. A., Azzam E. B. Novel Guanosine Derivatives against MERS CoV Polymerase: An In Silico Perspective. J. Biomol. Struct. Dynam.2020, doi:10.1080/07391102.2020.1758789. [DOI] [PMC free article] [PubMed]
- 64.Barnard D.L., Hubbard V.D., Burton J., et al. Inhibition of Severe Acute Respiratory Syndrome-Associated Coronavirus (SARSCoV) by Calpain Inhibitors and β-D-N4-Hydroxycytidine. Antiviral Chem Chemo. 2004;15:15–22. doi: 10.1177/095632020401500102. [DOI] [PubMed] [Google Scholar]
- 65.Chan J.F.W., Chan K.-H., Kao R.Y.T., et al. Broad-Spectrum Antivirals for the Emerging Middle East Respiratory Syndrome Coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pruijssers A.J., Denison M.R. Nucleoside Analogues for the Treatment of Coronavirus Infections. Curr. Opin. Virol. 2019;35:57–62. doi: 10.1016/j.coviro.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 Polymerase Bound to nsp7 and nsp8 Co-Factors. Nature Comm. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Imbert I., Guillemot J.-C., Bourhis J.-M., et al. A Second, Non-Canonical RNA-Dependent RNA Polymerase in SARS Coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kruse R.L. Therapeutic Strategies in an Outbreak Scenario to Treat the Novel Coronavirus Originating in Wuhan, China. F1000Res. 2020;9 doi: 10.12688/f1000research.22211.2. 72–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.https://www.nih.gov/news-events/news-releases/nih-clinical-trial-shows-remdesivir-accelerates-recovery-advanced-covid-19.


