Abstract
Background
In 2016 the first-in-human phase I study of a miRNA-based cancer therapy with a liposomal mimic of microRNA-34a-5p (miR-34a-5p) was closed due to five immune related serious adverse events (SAEs) resulting in four patient deaths. For future applications of miRNA mimics in cancer therapy it is mandatory to unravel the miRNA effects both on the tumor tissue and on immune cells. Here, we set out to analyze the impact of miR-34a-5p over-expression on the CXCL10/CXCL11/CXCR3 axis, which is central for the development of an effective cancer control.
Methods
We performed a whole genome expression analysis of miR-34a-5p transfected M1 macrophages followed by an over-representation and a protein–protein network analysis. In-silico miRNA target prediction and dual luciferase assays were used for target identification and verification. Target genes involved in chemokine signaling were functionally analyzed in M1 macrophages, CD4+ and CD8+ T cells.
Results
A whole genome expression analysis of M1 macrophages with induced miR-34a-5p over-expression revealed an interaction network of downregulated target mRNAs including CXCL10 and CXCL11. In-silico target prediction in combination with dual luciferase assays identified direct binding of miR-34a-5p to the 3′UTRs of CXCL10 and CXCL11. Decreased CXCL10 and CXCL11 secretion was shown on the endogenous protein level and in the supernatant of miR-34a-5p transfected and activated M1 macrophages. To complete the analysis of the CXCL10/CXCL11/CXCR3 axis, we activated miR-34a-5p transfected CD4+ and CD8+ T cells by PMA/Ionomycin and found reduced levels of endogenous CXCR3 and CXCR3 on the cell surface.
Conclusions
MiR-34a-5p mimic administered by intravenous administration will likely not only be up-taken by the tumor cells but also by the immune cells. Our results indicate that miR-34a-5p over-expression leads in M1 macrophages to a reduced secretion of CXCL10 and CXCL11 chemokines and in CD4+ and CD8+ T cells to a reduced expression of CXCR3. As a result, less immune cells will be attracted to the tumor site. Furthermore, high levels of miR-34a-5p in naive CD4+ T cells can in turn hinder Th1 cell polarization through the downregulation of CXCR3 leading to a less pronounced activation of cytotoxic T lymphocytes, natural killer, and natural killer T cells and possibly contributing to lymphocytopenia.
Keywords: CD4-Positive T-Lymphocytes, cytokines, gene expression profiling, immunotherapy, macrophages
Background
Micro(mi)RNAs are small non-coding RNA molecules of ~21–24 nucleotides (nt) in length and function as post-transcriptional regulators of gene expression.1 The specific binding of miRNAs to target sites, mostly in 3′ untranslated regions (3′UTRs) of their target mRNAs, leads to an inhibition of the synthesis of the target proteins.2 In few cases miRNAs can bind within 5′ untranslated regions or open reading frames.3 MiRNAs play a pivotal role in carcinogenesis as shown for a variety of cancer types.4 Among the most prominent tumor related miRNAs, tumor suppressor miR-34a-5p is lost or downregulated in broad range of cancer types.5–7 In 2013 a clinical trial (NCT01829971) with a liposomal miR-34a-5p mimic (MRX34) has been started for patients with unresectable primary liver cancer, advanced liver cancer, or metastatic liver cancer. In 2016 the phase I of this study was stopped due to immune-mediated serious adverse events (SAEs) leading to death of four patients.8 At the same time, there was first evidence for increased levels of miR-34a-5p in the serum or whole blood of cancer patients.9 10 A later study demonstrated an upregulation of miR-34a-5p in different blood cells including monocytes, natural killer cells, B cells and CD3+ T cells of patients with lung cancer.11 We recently provided evidence for a role of miR-34a-5p in T cell signaling and in the formation of the immunological synapse.12 A network-based approach revealed a functional role of miR-34a-5p in intracellular calcium signaling and in NF-κB signaling as part of T cell regulation networks.13–15 Here, we set out to investigate the role of miR-34a-5p in chemokine signaling, which is central for orchestrating a directed antitumor-response.
Methods
Cell lines, tissue culture
Human HEK 293T cells were purchased from the German collection of micro-organisms and cell cultures (DSMZ) and authenticated using STR DNA typing. HEK 293T cells were cultured as described previously.12 The cell line was cultured for less than 6 months after receipt.
Isolation and differentiation of macrophages
M1 macrophages were differentiated from freshly obtained PBMC using M1 macrophage Generation Medium DXF according to the manufacturer’s protocol (PromoCell GmbH, Heidelberg, Germany). In brief, freshly isolated PBMCs were seeded out in an appropriate amount of Monocyte Attachment Medium (C-28051, PromoCell GmbH) in a density of 1 million/cm2 and incubated for 1–1.5 hours at 5% CO2, 37°C. Subsequently, the non-adherent cells were removed by three washing steps with warm monocyte attachment medium, the M1 macrophage Generation Medium DXF (C-28055, PromoCell GmbH) was added to the remaining adherent monocytes and the cells were incubated with refreshment of the M1 macrophage Generation Medium DXF every 3 days for 9 days at 5% CO2 and 37°C. At day 10, the differentiated M1-Macrophages were checked by flow cytometry for correct differentiation, transfected as described below and used for the mRNA microarrays and ELISAs.
mRNA microarray
For detection of mRNA expression changes in activated and miR-34a-5p transfected M1 macrophages the Agilent Low Input, one-color, Quick Amp Labeling Kit and SurePrint G3 Human Gene Expression 8×60Kv3 Microarray (Cat. No. G4851C, Agilent Technologies, Santa Clara, California, USA) was used corresponding to the manufacturer’s protocols. In brief, 100 ng total RNA was reversed transcribed at 40°C for 2 hours using T7 Primer. Subsequently, the cRNA was labeled with Cyanine3-pCp using the T7 RNA polymerase for 2 hours at 40°C. The purification of the labeled cDNA was performed with RNeasy Mini kit (Qiagen, Hilden, Germany) according to Agilent’s One-Color Gene Expression Microarray Protocol followed by the quantification using a NanoDrop2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The hybridization of 600 ng labeled cRNA to the microarray slide was carried out at 65°C, 17 hours, 10 rpm in the SureHyb chambers (Agilent Technologies). After two washing steps the microarray slide was scanned on the Agilent Microarray Scanner G2565BA (Agilent Technologies). The raw fluorescence signals were analyzed with the Agilent AGW Feature Extraction software (V.10.7.1.1, Agilent Technologies). Normalization of the background corrected values was done with the Biological Significance analysis using GeneSpring (V.14.9, Agilent Technologies). In our analysis only mRNAs with RefSeqAccession number were included that were detected in at least four of the tested samples. The fold change of the mRNAs from miR-34a-5p transfected M1-macrophages was calculated by normalization of the expression values to the mean expression values of the control samples.
Expression and reporter vectors
The pSG5-mir-34a expression plasmid was synthesized and cloned by Eurofins Genomics and harbors the nucleotides 9151617-9151816 of chromosome 1 (Eurofins Genomics). The 3′UTRs of CXCR1, CXCR2, CXCR3, CXCL1, CXCL2, CXCL5, CXCL10, CXCL11, CXCL12, CXCL14, and CXCL16 were PCR amplified using specific primers and ligated via SpeI, SacI, or NaeI restriction sites into the pMIR-RNL-TK vector, which was described in Beitzinger et al.16 The target sites of positively tested target genes were mutated using specific primers by site-directed mutagenesis with the QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies). The sequences of all specific cloning and mutagenesis primers are given in online supplemental tables 1 and 2.
jitc-2020-001617supp001.pdf (26.8KB, pdf)
jitc-2020-001617supp002.pdf (24.8KB, pdf)
Dual luciferase reporter assays
The dual luciferase assays were performed as described previously.17 In brief, HEK 293T cells were seeded out and transfected after 24 hours with the respective combinations of reporter and expression vectors. PolyFect transfection reagent (Qiagen) was used for the transient transfections and the Dual-Luciferase Reporter Assay System Kit (Promega, Mannheim, Germany) for conducting the dual luciferase assays. All dual luciferase assays were conducted in duplicates and have been repeated four times.
CD4+ and CD8+ T cell isolation, transfection, stimulation, flow cytometry, and western blot analysis of CD4+ and CD8+ T cells
CD4+ or CD8+ T cells were isolated, purity was confirmed, and the cells were cultured as described previously.15 17 The gating strategy for the CD4+ and CD8+ T cells is shown in online supplemental figure 1. CD4+ and CD8+ T cells were transfected with hsa-miR-34a-5p miScript miRNA Mimic (MIMAT0000255: 5′UGGCAGUGUCUUAGCUGGUUGU) or the allstars negative control (ANC) using HiPerFect transfection reagent (Qiagen) as mentioned earlier.15 Twenty-four hours post transfection CD4+ and CD8+ T cells were stimulated with PMA/Ionomycin (5 ng/500 ng) for 24 hours. After stimulation, 1×105 of the transfected CD4+ and CD8+ T cells were stained with anti-CD4-FITC (RPA-T4, BD), anti-CD8-FITC (RPA-T8, BD) and anti-CXCR3-APC (1C6/CXCR3 (RUO), BD) or respective conjugated isotype control antibodies, fixed in 1% paraformaldehyde and analyzed by flow cytometry (FACS Canto II, BD Biosciences). The remaining transfected CD4+ and CD8+ T cells were used for western blot analysis, performed as described previously.15 CXCR3 was stained with a monoclonal rabbit anti human CXCR3 antibody (6H1L8, Thermo Fisher Scientific). α-tubulin served as loading control and was stained with a monoclonal rabbit anti human α-tubulin antibody (11H10, Cell Signaling Technology). The secondary antirabbit antibody was purchased from Sigma Aldrich (A0545, Sigma Aldrich).
jitc-2020-001617supp003.pdf (332.1KB, pdf)
Quantification of CXCL10 and CXCL11 secretion by ELISA and endogenous levels of CXCL10 and CXCL11 by western blot analysis
1.5×106 differentiated macrophages of two donors/mL/12well were transfected with 150 ng hsa-miScript miRNA Mimic miR-34a-5p (MIMAT0000255: 5′UGGCAGUGUCU UAGCUGGUUGU) or the ANC using HiPerFect transfection reagent (Qiagen). The transfections were carried out in three independent experiments from two donors. After 48 hours of transfection the M1 macrophages were activated using IFN-γ (50 ng/mL, Miltenyi Biotec) and LPS (10 ng/mL, Sigma Aldrich). Four hours after activation the supernatants of the transfected M1 macrophages were collected and CXCL10 as well as the CXCL11 secretion was quantified by the human CXCL10 or CXCL11 DuoSet ELISA Kit (R&D Systems) according to manufacturer’s protocol. The remaining M1 macrophages were stored in 700 µL Qiazol (Qiagen) at −80°C for subsequent RNA isolation.
After 48 hours following transfection, the M1 macrophages were activated using IFN-γ (50 ng/mL, Miltenyi Biotec) and LPS (10 ng/mL, Sigma Aldrich) for 20 hours. After stimulation, 1.5×106 of the transfected M1 macrophages were used for western blot analysis, performed as described previously.15 CXCL10 was stained with a monoclonal rabbit anti human CXCL10 antibody (D5L5L, Cell Signaling Technology). CXCL11 was stained with a monoclonal mouse anti human CXCL11 antibody (10C6, Thermo Fisher Scientific). α-tubulin served as loading control and was stained with a monoclonal rabbit anti human α-tubulin antibody (11H10, Cell Signaling Technology). All secondary antibodies were purchased from Sigma Aldrich.
RNA-isolation, quantitative real time PCR
48 hours post transfection of ANC or miR-34a-5p into 1.5×106 macrophages of two donors RNA isolation was conducted using the miRNeasy Mini Kit according to the manufacturer’s protocol (Qiagen). The levels of hsa-miR-34a-5p in the transfected M1 macrophages was determined by quantitative real time PCR (qRT-PCR) using the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, USA) and the miScript PCR System (Qiagen) corresponding to the manufacturer’s protocols. In brief 200 ng total RNA was reverse transcribed into cDNA using the miScript RT II Kit with the miScript HiSpec Buffer (Qiagen). RNU6 served as endogenous control for miRNA expression. Ectopic expression of miR-34a-5p in M1 macrophages of the two donors is depicted in online supplemental figure 2.
jitc-2020-001617supp004.pdf (128.6KB, pdf)
Data analysis and web tools
Statistical analysis of the luciferase assays, the western blots, the FACS analysis and ELISA was performed with SigmaPlot 10 (Systat, Chicago, USA) applying Student’s t-test. Quantification of the western blots was carried out with Image Lab Software V.5.2.1 (Bio-Rad Laboratories, Hercules, California, USA). The asterisks in the figures correspond) to the statistical significance as calculated by Student’s t-test: *=0.01< p≤0.05; **=0.001<p≤0.01; ***=p≤0.001. For the pathway analysis of the deregulated mRNAs and predicted target genes by an over-representation analysis (ORA), we used GeneTrail3 using default settings (Null hypothesis (for p value computation): two-sided; method to adjust p values: Benjamini-Yekutieli; significance level: 0.05; reference set: all protein coding genes) (http://genetrail.bioinf.uni-sb.de/).18 For the protein–protein interaction network analysis we used the STRING database V.11 (https://string-db.org/) with default settings including the 20 most downregulated and upregulated mRNAs on miR-34a-5p over-expression to obtain a concise network.19 The in-silico target prediction was carried out using miRWalk 2.0 that uses 10 algorithms including DIANAmT3.0, miRanda (2010), miRDB (2009), miRWalk, RNAhybrid (V.2.1), PICTAR4 (2006), PICTAR5 (2007), PITA (2008), RNA22 (2008), and TargetScan5.1.20
Results
Analysis of differentially expressed mRNAs in M1 macrophages upon miR-34a-5p over-expression
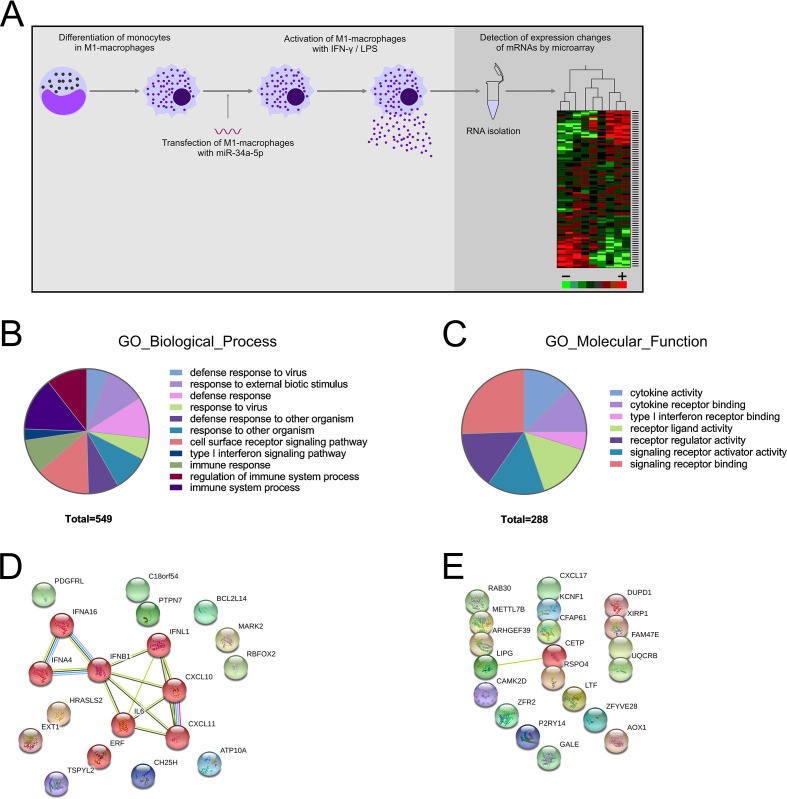
In a former study we found miR-34a-5p over-expressed in CD56+ (natural killer cells), CD19+ (B cells), CD3+ (T cells) as well as CD14+ (monocytes) cells of patients with lung cancer.11 Subsequently, we identified miR-34a-5p as modulator of intracellular calcium signaling, NF-κB signaling in CD4+/CD8+ T cells and as major hub of T cell regulation networks.13 15 17 Here we set out to investigate the impact of miR-34a-5p over-expression on chemokine signaling by analyzing the mRNA expression changes in the M1 macrophages transfected with miR-34a-5p or ANC from two different donors of two independent transfection experiments (figure 1A). Out of 58 000 mRNAs contained in the Agilent microarray, 27 285 transcripts were detected in at least four out of the eight samples. In total, 480 protein-coding transcripts showed an at least 1.5-fold change in the miR-34a-5p transfected cells as compared with the control cells. Out of the 480 mRNAs, 184 were upregulated and 296 downregulated. An ORA of 480 deregulated mRNAs by GeneTrail3 identified 140 significant pathways in Gene Ontology (GO) category “biological process” and 12 significant pathways in the GO category “molecular function” for the downregulated mRNAs. Figure 1B, C displays a representative selection of the significant pathways of these two GO categories. For example, the five most significant pathways in the GO category “molecular function” were “cytokine activity” (p value ≤0.001), “cytokine receptor binding” (p value ≤0.001), “type I interferon receptor binding” (p value ≤0.001), “receptor ligand activity” (p value ≤0.001), and “signaling receptor activator activity” (p value ≤0.001). For the upregulated mRNAs only one significant pathway, GO molecular function: “nucleic acids binding” was detected. Online supplemental tables 3 and 4 summarize the complete results of the ORA analysis for the downregulated and upregulated mRNAs. To identify protein–protein interaction networks we conducted a protein–protein association analysis using the STRING database with the 20 most downregulated and upregulated mRNAs on miR-34a-5p over-expression applying an interaction score of ≥0.4. This analysis highlighted a central interaction network for the downregulated mRNAs consisting of IFNA16, IFNA4, IFNB1, INFL1, IL6, CXCL10, and CXCL11 (figure 1D). For the upregulated mRNAs no interaction network could be established (figure 1E).
Figure 1.
Gene expression analysis of the effects of miR-34a-5p over-expression in M1 macrophages. (A) Experimental workflow. (B-C) Over-representation analysis of 296 mRNAs, which were at least 1.5-fold downregulated on miR-34a-5p over-expression by GeneTrail3. (B) Representative significant enriched pathways in Gene Ontology (GO) category “biological process.” (C) Representative significant enriched pathways in the GO category “molecular function.” (D) Protein–protein interaction networks of the 20 most downregulated mRNAs using the STRING database V.11 (https://string-db.org/). (E) Protein–protein interaction networks of the 20 most upregulated mRNAs using the STRING database V.11.
jitc-2020-001617supp005.pdf (39.6KB, pdf)
jitc-2020-001617supp006.pdf (17.5KB, pdf)
C-X-C motif ligands and C-X-C motif receptors are predicted target genes of miR-34a-5p
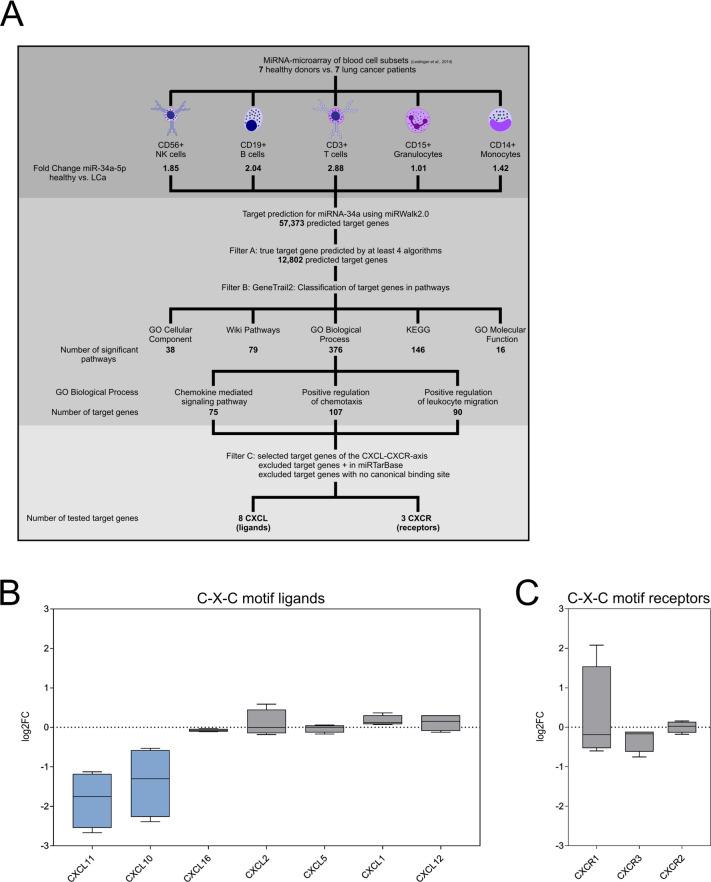
To explore the function of miR-34a-5p in the regulation of the immune response via cytokines, we performed a combination of in-silico target prediction with downstream pathway analysis to identify potential miR-34a-5p target genes. First, we conducted an in-silico target prediction by miRWalk V.2.0. In total, we identified 57 373 potential target gene 3′UTRs of miR-34a-5p (figure 2A). We subsequently reduced the number of putative target genes to 12 802 by considering only target gene 3′UTRs, which were predicted by at least four different algorithms. To identify potential miR-34a-5p target genes related to chemokine signaling, we mapped the predicted target genes to the according pathways by GeneTrail3 and identified 655 significant pathways. Out of these pathways, 38 significant pathways belonged to GO Cellular Component, 16 to GO Molecular Function, 79 to Wiki Pathways, 146 to KEGG (Kyoto Encyclopedia of Genes and Genomes), and 375 to GO biological processes. The GO biological process category included the three significant pathways, “chemokine mediated signaling pathway” with 75 potential miR-34a-5p target genes, “positive regulation of chemotaxis” with 107 and “positive regulation of leukocyte migration” with 90 potential target genes. We subsequently deleted target genes, which have been validated by others according to miRTarBase21 and genes without canonical binding site for miR-34a-5p. Following these selection steps, we identified eight C-X-C motif ligands and three C-X-C motif receptors with miR-34a-5p binding sites in their 3′UTRs in the three pathways that is, “chemokine mediated signaling pathway,” “positive regulation of chemotaxis,” and “positive regulation of leukocyte migration.” The C-X-C motif ligands included CXCL1, CXCL2, CXCL5, CXCL10, CXCL11, CXCL12, CXCL14, CXCL16 and the C-X-C motif receptors included CXCR1, CXCR2, CXCR3.
Figure 2.
(A) Workflow for the prediction and pathway analysis of miR-34a-5p target genes associated with chemokine signaling. (B) mRNA expression changes of C-X-C motif ligands on miR-34a-5p over-expression. Blue color represents a log2 fold change ≤−0.2. (C) mRNA expression changes of C-X-C motif receptors on miR-34a-5p over-expression.
Regulation of C-X-C motif ligands and C-X-C motif receptors mRNAs upon miR-34a-5p over-expression in M1 macrophages
Table 1 displays the 20 most upregulated or downregulated mRNAs in the miR-34a-5p transfected M1 macrophages including CXCL10 and CXCL11. Among the mRNAs encoding CXCLs and CXCRs with miR-34a-5p binding sites in their 3′UTRs, the mRNAs for CXCL10, CXCL11, and CXCR3 showed the strongest decrease in mRNA expression on miR-34a-5p over-expression. In detail, CXCL11 mRNA showed a median log2 fold change of −1.75, CXCL10 mRNA a change of −1.3, and CXCR3 a change of −0.295 (figure 2B, C, online supplemental table 5).
Table 1.
20 most upregulated and downregulated mRNAs in miR-34a-5p transfected M1 macrophages. Upper panel displays the 20 most downregulated mRNAs and the lower panel the 20 most upregulated mRNAs
| Downregulated mRNAs | ||
| GeneSymbol | RefSeqAccession | Fold change: miR-34a-5p vs ANC |
| ERF | NM_006494 | −8.11 |
| RBFOX2 | NM_001031695 | −4.27 |
| ATP10A | NM_024490 | −3.71 |
| CXCL11 | NM_005409 | −3.54 |
| MARK2 | NM_001039469 | −3.52 |
| IFNL1 | NM_172140 | −3.21 |
| HRASLS2 | NM_017878 | −2.89 |
| CH25H | NM_003956 | −2.79 |
| IFNB1 | NM_002176 | −2.77 |
| CXCL10 | NM_001565 | −2.71 |
| BCL2L14 | NM_138722 | −2.58 |
| PTPN7 | NM_080588 | −2.57 |
| PDGFRL | NM_006207 | −2.54 |
| IFNA16 | NM_002173 | −2.51 |
| CXCL10 | NM_001565 | −2.51 |
| IFNA4 | NM_021068 | −2.49 |
| EXT1 | NM_000127 | −2.47 |
| C18orf54 | NM_173529 | −2.47 |
| TSPYL2 | NM_022117 | −2.45 |
| IL6 | NM_000600 | −2.41 |
| Upregulated mRNAs | ||
| GeneSymbol | RefSeqAccession | Fold change: miR-34a-5p vs ANC |
| GALE | NM_000403 | 6.40 |
| CAMK2D | NM_001221 | 5.08 |
| P2RY14 | NM_014879 | 3.37 |
| ZFYVE28 | NM_001172657 | 3.25 |
| KCNF1 | NM_002236 | 2.83 |
| FAM47E | NM_001242936 | 2.66 |
| XIRP1 | NM_194293 | 2.60 |
| ZFR2 | NM_015174 | 2.50 |
| LIPG | NM_006033 | 2.45 |
| DUPD1 | NM_001003892 | 2.42 |
| CFAP61 | NM_015585 | 2.41 |
| METTL7B | NM_152637 | 2.33 |
| CXCL17 | NM_198477 | 2.24 |
| RAB30 | NM_014488 | 2.23 |
| ARHGEF39 | NM_032818 | 2.22 |
| UQCRB | NM_001254752 | 2.21 |
| CETP | NM_000078 | 2.15 |
| LTF | NM_002343 | 2.06 |
| AOX1 | NM_001159 | 2.05 |
| RSPO4 | NM_001029871 | 2.05 |
ANC, allstars negative control.
jitc-2020-001617supp007.pdf (9.1KB, pdf)
C-X-C motif ligands and C-X-C motif receptors as miR-34a-5p targets identified by dual luciferase assays
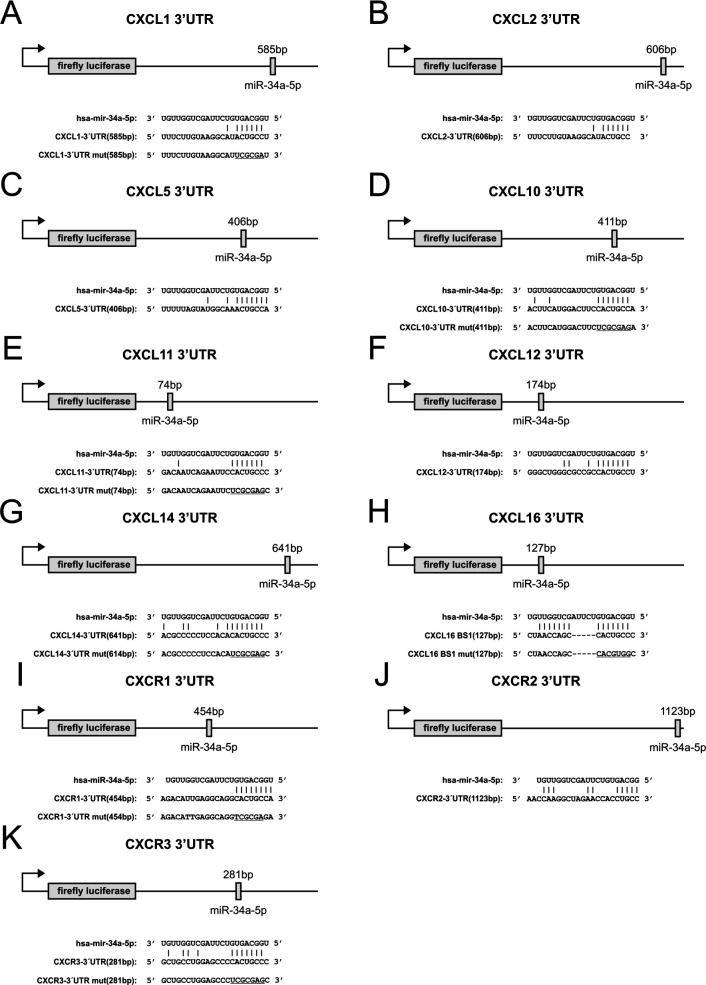
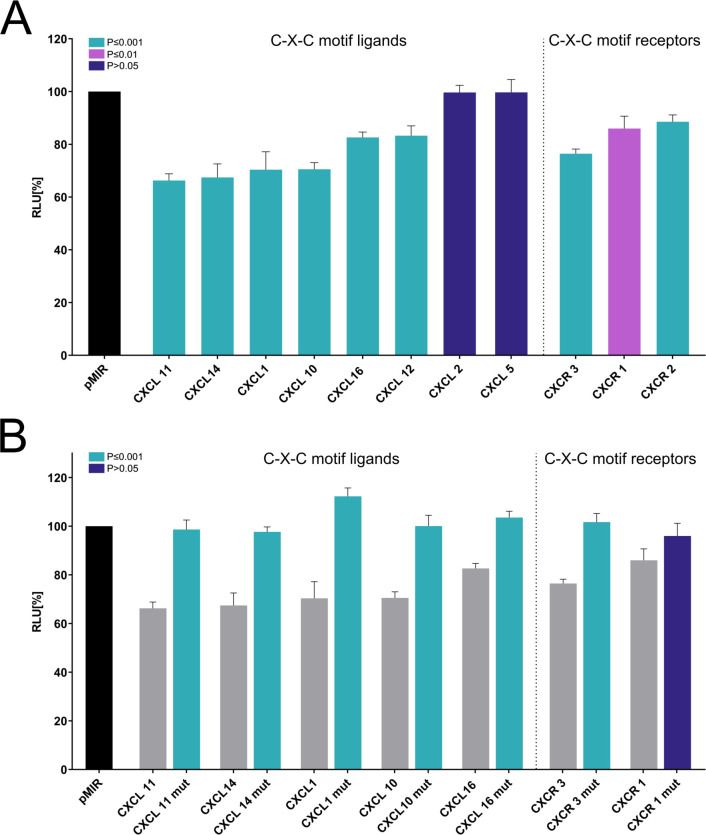
Our in-silico target prediction identified eight C-X-C motif ligands and three C-X-C motif receptors with miR-34a-5p binding sites in their 3′UTRs including CXCL1, CXCL2, CXCL5, CXCL10, CXCL11, CXCL12, CXCL14, CXCL16 as well as CXCR1, CXCR2, CXCR3 (figure 3). We cloned the respective 3′UTR sequences into the pMIR-RNL-TK reporter vector and cotransfected these recombinants together with a miR-34a-5p expression vector in HEK 293T cells. Dual luciferase assays were conducted in duplicates and repeated four times. The luciferase activities (RLU (relative light units)) of the wild-type reporters were normalized against the RLU of the empty reporter vector. Among the C-X-C motif ligands, we found the strongest reduction of the RLU for CXCL11, which was reduced to 66.3% (p value ≤0.001). Except for the ligands CXCL2 and CXCL5, all other remaining C-X-C ligands were also significantly repressed by miR-34a-5p including CXCL14 (67.4%), CXCL1 (70.4%), CXCL10 (70.5%), CXCL16 (82.6%), and CXCL12 (83.3%) (figure 4A, left panel). Among the C-X-C motif receptors, we observed the strongest reduction of the RLU for CXCR3, which was reduced to 76.5% (p value ≤0.001). The remaining C-X-R receptors were also significantly repressed by miR-34a-5p including CXCR1 (86%) and CXCR2 (88.5%) (figure 4A, right panel). To validate the direct binding of miR-34a-5p to its target sites, we mutated the binding sites in the 3′UTRs of CXCL ligands CXCL1, CXCL10, CXCL11, CXCL14, CXCL16 and in the 3′UTRs of CXCR receptors CXCR1 and CXCR3. Except for CXCR1, all mutated reporter vectors showed a significant increase of the RLU when cotransfected with miR-34a-5p compared with the respective non-mutated recombinants indicating the direct binding of miR-34a-5p to its binding sites in the respective 3′UTRs (figure 4B).
Figure 3.
Schematic diagram of reporter gene plasmids. The position of the predicted miR-34a-5p binding sites in the respective 3′UTR reporter plasmids and their corresponding sequences as well as the sequences of the mutated binding sites (underlined) are shown. (A) CXCL1-3′UTR reporter vector, (B) CXCL2-3′UTR reporter vector, (C) CXCL5-3′UTR reporter vector, (D) CXCL10-3′UTR reporter vector, (E) CXCL11-3′UTR reporter vector, (F) CXCL12-3′UTR reporter vector, (G) CXCL14-3′UTR reporter vector, (H) CXCL16-3′UTR reporter vector, (I) CXCR1-3′UTR reporter vector, (J) CXCR2-3′UTR reporter vector, (K) CXCR3-3′UTR reporter vector.
Figure 4.
(A) Dual luciferase reporter gene assays. Left panel: dual luciferase reporter gene assays of C-X-C motif ligands, right panel: dual luciferase reporter gene assays of C-X-C motif receptors. HEK 293T cells were cotransfected with miR-34a-5p and reporter vectors of the target genes as indicated. The luciferase activities were normalized with respect to the luciferase activity measured with empty reporter construct. The results represent the mean of four independent experiments carried out in duplicates. Columns colored in turquois represent a significant reduction of the luciferase activity with a p value ≤0.001. Columns colored in magenta represent a significant reduction of the luciferase activity with a p value ≤0.01 and ≥0.001. Columns colored in dark blue represent a non-significant reduction of the luciferase activity with a p value ≥0.05. Data are represented as mean±SEM, (B) dual luciferase reporter gene assays with mutated reporter constructs, left panel: dual luciferase reporter gene assays of mutated C-X-C motif ligand constructs, right panel: dual luciferase reporter gene assays of mutated C-X-C motif receptor constructs. HEK 293T cells were cotransfected with the miR-34a-5p and the wild type reporter plasmids of the respective target genes or mutated reporter plasmids (mut) of the respective target genes as indicated in the diagram. The luciferase activities were normalized with respect to the luciferase activity measured with empty reporter construct. Columns colored in turquois represent a significant induction of the luciferase activity of the respective mutated reporter vector with a p value ≤0.001. Columns colored in dark blue represent a non-significant induction of the luciferase activity of the respective mutated reporter vector with a p value ≥0.05. Data are represented as mean±SEM. RLU, relative light units.
MiR-34a-5p over-expression decreased CXCR3 levels in CD4+ and CD8+ T cells as well as CXCL10 and CXCL11 levels in M1 macrophages
To analyze the downstream effects of miR-34a-5p over-expression in immune cells, we focused on the CXCL10/CXCL1/CXCR3 axis, which is central for the development of an effective cancer control, the differentiation of naive T cells to T helper 1 (Th1) cells, and the direction of immune cells to their functional sites by a chemotactic gradient of the chemokines CXCL9, CXCL10, CXCL11 binding to CXCR3.22–24
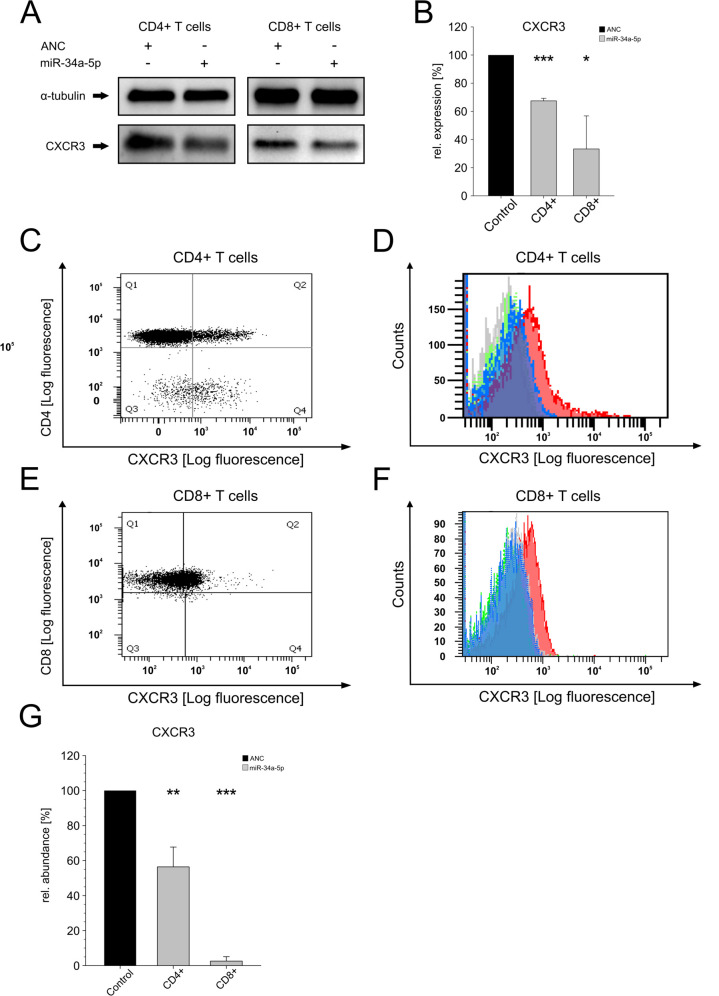
We transfected CD4+ and CD8+ T cells with miR-34a-5p mimic or with “ANC” and activated the transfected T cells for 4 hours with PMA/Ionomycin 48 hours after transfection. Western blotting with an antibody directed against CXCR3 showed a reduction of endogenous CXCR3 protein to 67.4% in the miR-34a-5p transfected CD4+ T cells (p value ≤0.001) and to 33.2% in the miR-34a-5p transfected CD8+ T cells (0.01<p≤0.05) (figure 5A, B). Flow cytometry was used to analyze the effect of miR-34a-5p over-expression on CXCR3 cell surface expression. Following transfection and activation, we found a downregulation of CXCR3 cell surface expression to 56.4% in miR-34a-5p transfected CD4+ T cells (p value 0.001<p≤0.01) and to 2.5% in miR-34a-5p transfected CD8+ T cells compared with control transfected cells (p value ≤0.001) (figure 5C–G).
Figure 5.
Effect of miR-34a-5p over-expression on CXCR3 of CD4+ and CD8+ T cells. (A) Western blot analysis of CXCR3 in miR-34a-5p transfected CD4+ (left panel) and CD8+ T cells (right panel). The cells were transfected either with allstars negative control (ANC) or miR-34a-5p mimic. 48 hours after transfection the cells were activated using PMA/ionomycin and the endogenous protein level of CXCR3 was analyzed by western blotting using specific antibodies against CXCR3. α-tubulin served as loading control. One representative western blot is displayed. (B) Quantification of endogenous CXCR3 protein levels in miR-34a-5p transfected CD4+ (left panel) and CD8+ T cells (right panel). Three independent western blots were quantified by densitometry using image lab software. The protein expression of CXCR3 in CD4+ and CD8+ T cells was normalized with respect to the corresponding α-tubulin signals of the appropriate samples. The three asterisks correspond to a p value ≤0.001 and one asterisk to a p value 0.01<p≤0.05. (C) CD4+ T cells were stained for CD4 and costained for CXCR3 for 30 min at 4°C. Cells were analyzed by flow cytometry. gated CD4+ T cells were analyzed for CXCR3 expression. (D) Mean fluorescence intensities of CXCR3 of CD4+ T cells. ANC-transfected (red) or miR-34a-5p mimic-transfected (blue) CD4+ T cells or respective isotype controls (green and gray) were analyzed by flow cytometry. (E) CD8+ T cells were stained for CD8 and costained for CXCR3 for 30 min at 4°C. Cells were analyzed by flow cytometry. Gated CD8+T cells were analyzed for CXCR3 expression. (F) Mean fluorescence intensities of CXCR3 of CD8+ T cells. ANC-transfected (red) or miR-34a-5p mimic-transfected (blue) CD8+ T cells or respective isotype controls (green and gray) were analyzed by flow cytometry. (G) Quantification of cell surface levels of CXCR3 on miR-34a-5p transfection on CD4+ and CD8+ T cells. FACS data from four independent experiments from two different donors performed in duplicates were summarized. ANC transfected control is displayed in black and miR-34a-5p transfected T cells in gray. Two asterisks correspond to a p value ≤0.01 and ≥0.001 and three asterisks to a p value ≤0.001. Data are represented as mean±SEM.
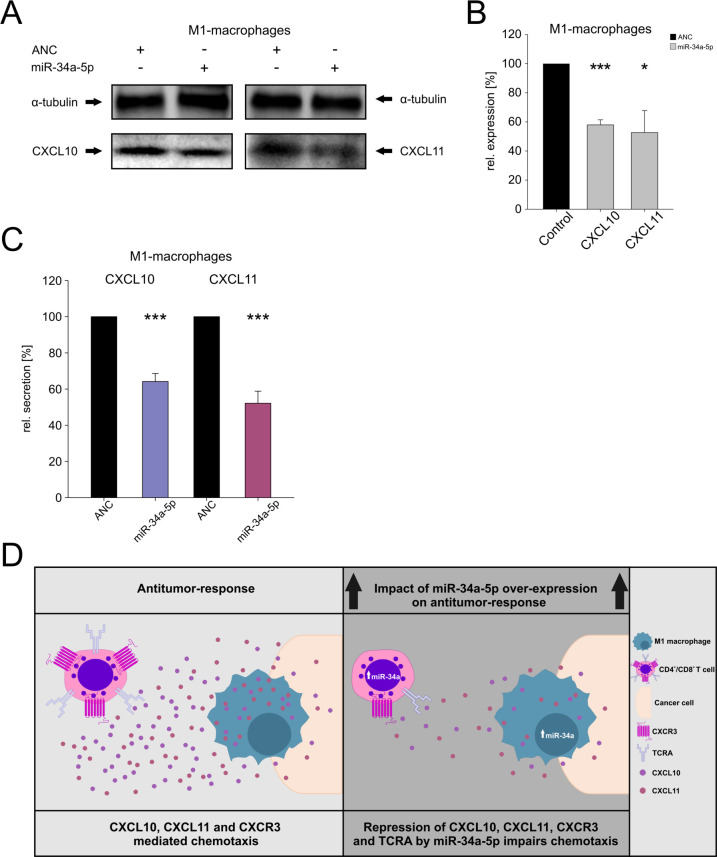
We next investigated the effect of miR-34a-5p on the CXCL10, CXCL11 secretion and the endogenous levels in M1-macrophages, which were recently identified to predominantly express CXCL10, and CXCL11 after immune checkpoint blockade.25 We differentiated monocytes in M1 macrophages, transfected the cells with miR-34a-5p mimics or with ANC, and activated the transfected cells with IFN-γ and LPS. Western blot analysis of three independent experiments from two different donors with antibodies against CXCL10 and CXCL11 showed a decrease of endogenous CXCL10 protein to 57.9% (p value ≤0.001) on miR-34a-5p over-expression and of endogenous CXCL11 protein to 52.6% (0.01<p≤0.05) (figure 6A, B). Using ELISA we found in three independent experiments from two different donors a reduced CXCL10 secretion to 69.3% (p value ≤0.001) and a reduced CXCL11 secretion to 59.2% (p value ≤0.001) in the supernatants of miR-34a-5p transfected M1 macrophages (figure 6C).
Figure 6.
(A–C) Effects of miR-34a-5p over-expression on the endogenous levels and secretion of CXCL10 and CXCL11 in M1 macrophages. Monocytes were differentiated in M1 macrophages, transfected with ANC, or miR-34a-5p mimic, and activated with IFN-γ and LPS. Subsequently, the CXCL10 and CXCL11 endogenous levels were measured by western blots (A, one representative western blot is displayed) and quantified from three independent experiments from two different donors (B) Three asterisks correspond to p value ≤0.001 and one asterisk to a p value 0.01<p≤0.05. The secretion was measured by ELISA (C). ELISA data were quantified from three independent experiments from two different donors. Three asterisks correspond to p value ≤0.001. Data are represented as mean±SEM. (D) Hypothesis of the impact of miR-34a-5p over-expression on antitumor-response.
Discussion
We determined the role of miR-34a-5p in chemokine signaling and identified miR-34a-5p as key modulator of the CXCL10/CXCL11/CXCR3 axis. This axis is central for the development of an effective cancer control as recently summarized in a review by Tokunaga et al.22 A single cell RNA-seq analysis of tumor-infiltrating immune cells after immune checkpoint blockade showed that CXCL10 is predominantly expressed by macrophages.25 CXCL10 is induced by IFN-γ, IFN-α/β, and regulated by NF-kB.26 27 In response to IFN-γ, CXCL10 is secreted by monocytes, endothelial cells, fibroblasts, and cancer cells.28 First evidence for a direct impact of the interplay between miR-34a-5p and CXCL10 on the tumor development stems from a study showing that the inhibition of CXCL10 by miR-34a-5p in breast cancer cells leads to a suppression of invasion and migration.29 Further evidence for a central role of CXCL10 in tumor development stems from studies on murine malignant pleural effusion (MPE) models. The intrapleural injection of CXCL10-deficient tumor cells resulted in a decrease of Th1 and Th17 cells in MPE, an increase of MPE volume, and a reduction of survival.30 Our analysis shows that the CXCL10 secretion can be reduced by miR-34a-5p over-expression in M1 macrophages suggesting differential roles of miR-34a-5p in cancer and immune cells.
The second ligand CXCL11, which is regulated by miR-34a-5p can also be induced by IFN-γ and IFN-β.31 CXCL11 has the highest affinity to the CXCR3 chemokine receptor of the three selective ligands followed by CXCL10 and CXCL9.32 33 Like CXCL10, CXCL11 is secreted in response to IFN-γ by monocytes, endothelial cells, fibroblasts, and cancer cells.28 Cancer cell secreted CXCL11 can promote CD8+ T cells infiltration in non-small lung cancer by docetaxel treatment.34 CXCL11 as genetic adjuvant increase vaccine antigen-specific CD8+ T cells to a greater degree than the other two CXCR3 ligands.35
Like the ligands, the miR-34a-5p regulated receptor CXCR3 plays a pivotal role in tumor development. CXCR3 is strongly expressed on activated Th1 cells, cytotoxic T lymphocytes (CTLs), natural killer (NK) cells, and natural killer T (NKT) cells.36 On naive T cells CXCR3 is downregulated but rapidly induced by antigen-presenting dendritic cells causing Th1 polarization following Th1 dependent activation of CTLs, NK, and NKT cells by INF-γ.37 In humans three isoforms of CXCR3 CXCR3-A, CXCR3-B, and CXCR3-Alt were identified exhibiting differences in ligand-binding properties, signaling pathways, and cellular responses.38 In a murine lung cancer model IL-7 induces antitumor reactivity of T cells in a CXCR3 ligand dependent manner.39
To fully appreciate the role of miR-34a-5p in tumor development, it is necessary to consider its impact on the entire CXCL10/CXCL11/CXCR3 axis. The axis primarily mediates migration, differentiation, and activation of immune cells. The paracrine CXCL10/CXCL11/CXCR3 signaling in tumor models displays antitumor activity mediated by Th1, CTLs, NK, and NKT cells.40 The autocrine CXCL10/CXCL11/CXCR3 axis in cancer cells increases proliferation, angiogenesis, and metastasis. Former studies showed that CXCR3+ cancer cells have a disposition to migrate to ligand rich metastatic sites.41 42 Specific aspects of this axis in cancer are controversially discussed as for example the association between CXCL10 and poor prognosis.43 44 Nevertheless, the autocrine and the paracrine signaling offers the possibility for cancer treatment by activating the paracrine axis and inhibiting the autocrine signaling.22 An immunotherapy may combine ectopic expression of CXCL10/CXCL11 in the tumor, for example, by intratumor injections of CXCL1045 and the inhibition of CXCR3 on cancer cells, for example, by CXCR3 antagonists (like AMG487).42 With regard to immune cells, our results indicate that over-expression of miR-34a-5p in CD4+, CD8+ T cells, and M1 macrophages can impact the antitumor-response. Due to miR-34a-5p over-expression, M1 macrophages synthesize and secret less CXCL10 and CXCL11 chemokines and CD4+ and CD8+ T cells synthesize and express less CXCR3 on their surface. Hence, it can be assumed that far fewer immune cells will be attracted to the tumor site (figure 6D). Taken together with the results of our former studies that showed a downregulation of VAMP2 expression and PRF1 secretion together with a reduced killing efficiency of miR-34a-5p over-expressing CD8+ T cells, our data provide strong evidence that miR-34a-5p over-expression will not solely impact the chemotaxis but also their effector functions at the tumor site.14 15
Our data on the effect of miR-34a-5p in immune cells may also help to explain the immune related SAEs, especially the lymphocytopenia observed with patients that have undergone the immunotherapy with MRX34. It is very likely that the miR-34a-5p mimic, which was administered by intravenous administration was not only up-taken by the tumor cells but also by the immune cells. High levels of miR-34a-5p in naive CD4+ T cells can in turn hinder Th1 polarization through the downregulation of CXCR3 leading to a less pronounced activation of CTLs, NK and NKT cells. In addition, elevated levels of miR-34a-5p in CD4+ and CD8+ T cells also impact central components of the intracellular calcium signaling and the NF-κB signaling and are associated with a decreased CD8+ mediated killing efficiency in the tumor microenvironment.13 14 Consequently, intravenously administered miR-34a-5p mimic may cause immune related SAEs including lymphocytopenia by several cellular routes.
To limit off target effects in immune cells, it is necessary to allow a directed and specific delivery of miR-34a-5p to the tumor cells. As shown for multiple myeloma the miR-125b-dependent stimulation of miR-34a-5p expression via the IL-6R/STAT3/miR-34a feedback loop offers a possibility to increase miR-34a-5p-levels in tumor cells while avoiding off target effects in immune cells.46 Another possibility for a directed delivery is to encapsulate the miR-34a-5-p mimic in nanocarriers. Former studies used stable nucleic acid lipid vesicles (SNALPs) encapsulating miR-34a-5p for the treatment of multiple myeloma cells.47 These SNALPs were conjugated with transferrin to target multiple myeloma cells that overexpress transferrin receptors. This ensured an efficient delivery to these cells in an in vivo mouse model avoiding evident toxicity and prolonging the survival of the miR-34a-5p-SNALP treated mice.48
Conclusions
First, miR-34a-5p over-expression in immune cells impacts the antitumor-response of M1-macrophages, CD4+ and CD8+ T cells by downregulation of CXCR3 and its ligands CXCL10 and CXCL11. As a result, less immune cells will be attracted to the tumor site. Second, miR-34a-5p over-expression in M1-macrophages, CD4+ and CD8+T cells may partly explain the immune-mediated SAEs including lymphocytopenia that have been observed in the phase I study of the miRNA-34a-5p-based cancer therapy. Our results favor the idea of intratumor injections of MRX34 favorably encapsulated in nanocarriers to allow specific downregulation of CXCR3 and other oncogenic miR-34a-5p target genes in the cancer cells while avoiding systemic SAEs caused by the intravenous administration of the miR-34a-5p mimic.
Footnotes
MH and LN contributed equally.
Contributors: MH, BW-R, AK, EM conceived and designed the experiments. MH, LN, BW-R, LK, SR, CD, TT, TK performed the experiments. MH, LN, LK, BW-R, TK analyzed the data. MH, BW, MS, HPL, AK, EM contributed to the writing of the manuscript.
Funding: This work was supported by the European Union’s Seventh Framework Program for Research, Technological Development and Demonstration [grant number: 600841] and by the Michael J. Fox Foundation [grant number: 14446].
Competing interests: No, there are no competing interests.
Patient consent for publication: Not required.
Ethics approval: The study was carried out according to the Declaration of Helsinki and was approved by the local ethics committee (Ref.-No. 213/08). All study participants gave written informed consent to participate in this study.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Ambros V, Bartel B, Bartel DP, et al. . A uniform system for microRNA annotation. RNA 2003;9:277–9. 10.1261/rna.2183803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene 2006;25:6163–9. 10.1038/sj.onc.1209909 [DOI] [PubMed] [Google Scholar]
- 3.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5' untranslated region or the open reading frame. RNA 2010;16:2493–502. 10.1261/rna.2384610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garzon R, Calin GA, Croce CM. Micrornas in cancer. Annu Rev Med 2009;60:167–79. 10.1146/annurev.med.59.053006.104707 [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Guessous F, Zhang Y, et al. . Microrna-34A inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 2009;69:7569–76. 10.1158/0008-5472.CAN-09-0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig N, Kim Y-J, Mueller SC, et al. . Posttranscriptional deregulation of signaling pathways in meningioma subtypes by differential expression of miRNAs. Neuro Oncol 2015;17:1250–60. 10.1093/neuonc/nov014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bu P, Chen K-Y, Chen JH, et al. . A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell 2013;12:602–15. 10.1016/j.stem.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong DS, Kang Y-K, Borad M, et al. . Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer 2020;122:1630–7. 10.1038/s41416-020-0802-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel D, Boufraqech M, Jain M, et al. . Mir-34A and miR-483-5p are candidate serum biomarkers for adrenocortical tumors. Surgery 2013;154:1224–9. discussion 29. 10.1016/j.surg.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichelser C, Flesch-Janys D, Chang-Claude J, et al. . Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem 2013;59:1489–96. 10.1373/clinchem.2013.205161 [DOI] [PubMed] [Google Scholar]
- 11.Leidinger P, Backes C, Dahmke IN, et al. . What makes a blood cell based miRNA expression pattern disease specific?--a miRNome analysis of blood cell subsets in lung cancer patients and healthy controls. Oncotarget 2014;5:9484–97. 10.18632/oncotarget.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart M, Rheinheimer S, Leidinger P, et al. . Identification of miR-34a-target interactions by a combined network based and experimental approach. Oncotarget 2016;7:34288–99. 10.18632/oncotarget.9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diener C, Hart M, Alansary D, et al. . Modulation of intracellular calcium signaling by microRNA-34a-5p. Cell Death Dis 2018;9:1008. 10.1038/s41419-018-1050-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart M, Walch-Rückheim B, Friedmann KS, et al. . miR-34A: a new player in the regulation of T cell function by modulation of NF-κB signaling. Cell Death Dis 2019;10:46 10.1038/s41419-018-1295-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart M, Walch-Rückheim B, Krammes L, et al. . miR-34A as hub of T cell regulation networks. J Immunother Cancer 2019;7:187. 10.1186/s40425-019-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beitzinger M, Peters L, Zhu JY, et al. . Identification of human microRNA targets from isolated Argonaute protein complexes. RNA Biol 2007;4:76–84. 10.4161/rna.4.2.4640 [DOI] [PubMed] [Google Scholar]
- 17.Hart M, Walch-Rückheim B, Friedmann KS, et al. . miR-34A: a new player in the regulation of T cell function by modulation of NF-κB signaling. Cell Death Dis 2019;10:46. 10.1038/s41419-018-1295-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerstner N, Kehl T, Lenhof K, et al. . GeneTrail 3: advanced high-throughput enrichment analysis. Nucleic Acids Res 2020;48:W515–20. 10.1093/nar/gkaa306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Gable AL, Lyon D, et al. . String v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. 10.1038/nmeth.3485 [DOI] [PubMed] [Google Scholar]
- 21.Chou C-H, Shrestha S, Yang C-D, et al. . miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46:D296–302. 10.1093/nar/gkx1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokunaga R, Zhang W, Naseem M, et al. . CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat Rev 2018;63:40–7. 10.1016/j.ctrv.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannenbaum CS, Tubbs R, Armstrong D, et al. . The CXC chemokines IP-10 and mig are necessary for IL-12-mediated regression of the mouse RENCA tumor. J Immunol 1998;161:927–32. [PubMed] [Google Scholar]
- 24.Tensen CP, Flier J, Van Der Raaij-Helmer EM, et al. . Human IP-9: a keratinocyte-derived high affinity CXC-chemokine ligand for the IP-10/Mig receptor (CXCR3). J Invest Dermatol 1999;112:716–22. 10.1046/j.1523-1747.1999.00581.x [DOI] [PubMed] [Google Scholar]
- 25.House IG, Savas P, Lai J, et al. . Macrophage-Derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res 2020;26:487–504. 10.1158/1078-0432.CCR-19-1868 [DOI] [PubMed] [Google Scholar]
- 26.Qian C, An H, Yu Y, et al. . Tlr agonists induce regulatory dendritic cells to recruit Th1 cells via preferential IP-10 secretion and inhibit Th1 proliferation. Blood 2007;109:3308–15. 10.1182/blood-2006-08-040337 [DOI] [PubMed] [Google Scholar]
- 27.Schmid H, Boucherot A, Yasuda Y, et al. . Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 2006;55:2993–3003. 10.2337/db06-0477 [DOI] [PubMed] [Google Scholar]
- 28.Ohmori Y, Schreiber RD, Hamilton TA. Synergy between interferon-gamma and tumor necrosis factor-alpha in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor kappaB. J Biol Chem 1997;272:14899–907. 10.1074/jbc.272.23.14899 [DOI] [PubMed] [Google Scholar]
- 29.Xu M, Li D, Yang C, et al. . Microrna-34A inhibition of the TLR signaling pathway via CXCL10 suppresses breast cancer cell invasion and migration. Cell Physiol Biochem 2018;46:1286–304. 10.1159/000489111 [DOI] [PubMed] [Google Scholar]
- 30.Wu X-Z, Zhai K, Yi F-S, et al. . IL-10 promotes malignant pleural effusion in mice by regulating TH 1- and TH 17-cell differentiation and migration. Eur J Immunol 2019;49:653–65. 10.1002/eji.201847685 [DOI] [PubMed] [Google Scholar]
- 31.Rani MR, Foster GR, Leung S, et al. . Characterization of beta-R1, a gene that is selectively induced by interferon beta (IFN-beta) compared with IFN-alpha. J Biol Chem 1996;271:22878–84. 10.1074/jbc.271.37.22878 [DOI] [PubMed] [Google Scholar]
- 32.Weng Y, Siciliano SJ, Waldburger KE, et al. . Binding and functional properties of recombinant and endogenous CXCR3 chemokine receptors. J Biol Chem 1998;273:18288–91. 10.1074/jbc.273.29.18288 [DOI] [PubMed] [Google Scholar]
- 33.Cole KE, Strick CA, Paradis TJ, et al. . Interferon-Inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med 1998;187:2009–21. 10.1084/jem.187.12.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Q, Wang S, Chen X, et al. . Cancer-cell-secreted CXCL11 promoted CD8+ T cells infiltration through docetaxel-induced-release of HMGB1 in NSCLC. J Immunother Cancer 2019;7:42. 10.1186/s40425-019-0511-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namkoong H, Song M-Y, Seo YB, et al. . Enhancement of antigen-specific CD8 T cell responses by co-delivery of Fc-fused CXCL11. Vaccine 2014;32:1205–12. 10.1016/j.vaccine.2013.07.066 [DOI] [PubMed] [Google Scholar]
- 36.Qin S, Rottman JB, Myers P, et al. . The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest 1998;101:746–54. 10.1172/JCI1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim CH, Rott L, Kunkel EJ, et al. . Rules of chemokine receptor association with T cell polarization in vivo. J Clin Invest 2001;108:1331–9. 10.1172/JCI13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berchiche YA, Sakmar TP. Cxc chemokine receptor 3 alternative splice variants selectively activate different signaling pathways. Mol Pharmacol 2016;90:483–95. 10.1124/mol.116.105502 [DOI] [PubMed] [Google Scholar]
- 39.Andersson A, Yang S-C, Huang M, et al. . Il-7 promotes CXCR3 ligand-dependent T cell antitumor reactivity in lung cancer. J Immunol 2009;182:6951–8. 10.4049/jimmunol.0803340 [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Chu Y, Wang Y, et al. . Targeted in vivo expression of IFN-gamma-inducible protein 10 induces specific antitumor activity. J Leukoc Biol 2006;80:1434–44. 10.1189/jlb.0306212 [DOI] [PubMed] [Google Scholar]
- 41.Cambien B, Karimdjee BF, Richard-Fiardo P, et al. . Organ-Specific inhibition of metastatic colon carcinoma by CXCR3 antagonism. Br J Cancer 2009;100:1755–64. 10.1038/sj.bjc.6605078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu G, Yan HH, Pang Y, et al. . Cxcr3 as a molecular target in breast cancer metastasis: inhibition of tumor cell migration and promotion of host anti-tumor immunity. Oncotarget 2015;6:43408–19. 10.18632/oncotarget.6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wightman SC, Uppal A, Pitroda SP, et al. . Oncogenic CXCL10 signalling drives metastasis development and poor clinical outcome. Br J Cancer 2015;113:327–35. 10.1038/bjc.2015.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato Y, Motoyama S, Nanjo H, et al. . Cxcl10 expression status is prognostic in patients with advanced thoracic esophageal squamous cell carcinoma. Ann Surg Oncol 2016;23:936–42. 10.1245/s10434-015-4909-1 [DOI] [PubMed] [Google Scholar]
- 45.Arenberg DA, White ES, Burdick MD, et al. . Improved survival in tumor-bearing SCID mice treated with interferon-gamma-inducible protein 10 (IP-10/CXCL10). Cancer Immunol Immunother 2001;50:533–8. 10.1007/s00262-001-0231-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Misso G, Zarone MR, Lombardi A, et al. . Mir-125B upregulates miR-34a and sequentially activates stress adaption and cell death mechanisms in multiple myeloma. Mol Ther Nucleic Acids 2019;16:391–406. 10.1016/j.omtn.2019.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scognamiglio I, Di Martino MT, Campani V, et al. . Transferrin-conjugated SNALPs encapsulating 2'-O-methylated miR-34a for the treatment of multiple myeloma. Biomed Res Int 2014;2014:1–7. 10.1155/2014/217365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Martino MT, Campani V, Misso G, et al. . In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS One 2014;9:e90005. 10.1371/journal.pone.0090005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001617supp001.pdf (26.8KB, pdf)
jitc-2020-001617supp002.pdf (24.8KB, pdf)
jitc-2020-001617supp003.pdf (332.1KB, pdf)
jitc-2020-001617supp004.pdf (128.6KB, pdf)
jitc-2020-001617supp005.pdf (39.6KB, pdf)
jitc-2020-001617supp006.pdf (17.5KB, pdf)
jitc-2020-001617supp007.pdf (9.1KB, pdf)