Abstract
Background
We aimed to assess the impact of genomic human leukocyte antigen (HLA)-I/II homozygosity on the survival benefit of patients with unresectable locally advanced, metastatic non-small lung cancer treated by single-agent programmed cell death protein-1/programmed death ligand 1 (PD1/PDL1) inhibitors.
Methods
We collected blood from 170 patients with advanced lung cancer treated with immunotherapy at two major oncology centers in Western Australia. Genomic DNA was extracted from white blood cells and used for HLA-I/II high-resolution typing. HLA-I/II homozygosity was tested for association with survival outcomes. Univariable and multivariable Cox regression models were constructed to determine whether HLA homozygosity was an independent prognostic factor affecting Overall Survival (OS) and Progression Free Survival (PFS). We also investigated the association between individual HLA-A and -B supertypes with OS.
Results
Homozygosity at HLA-I loci, but not HLA-II, was significantly associated with shorter OS (HR=2.17, 95% CI 1.13 to 4.17, p=0.02) in both univariable and multivariable analysis. The effect of HLA-I homozygosity in OS was particularly relevant for patients with tumors expressing PDL1 ≥50% (HR=3.93, 95% CI 1.30 to 11.85, p<0.001). The adverse effect of HLA-I homozygosity on PFS was only apparent after controlling for interactions between PDL1 status and HLA-I genotype (HR=2.21, 95% CI 1.04 to 4.70, p=0.038). The presence of HLA-A02 supertype was the only HLA-I supertype to be associated with improved OS (HR=0.56, 95% CI 0.34 to 0.93, p=0.023).
Conclusion
Our results suggest that homozygosity at ≥1 HLA-I loci is associated with short OS and PFS in patients with advanced non-small cell lung cancer with PDL1 ≥50% treated with single-agent immunotherapy. Carriers of HLA-A02 supertype reported better survival outcomes in this cohort of patients.
Keywords: immunotherapy, lung neoplasms, biomarkers, tumor
Background
Lung cancer is the second most common cancer worldwide in 2019 and the leading cancer-related cause of death among both men and women.1 While the introduction of immunotherapy into the treatment of advanced and metastatic non-small cell lung cancer (NSCLC) has improved survival when compared with chemotherapy, the 5 years survival rate continues to be as low as 23.2% and 15.5% in the first-line and second-line setting, respectively.2 While programmed death ligand (PDL1) expression on tumor cells is to date the only FDA (Food and Drug Administration) approved biomarker of response to anti-programmed cell death protein-1/programmed death ligand 1 (PD1/PDL1) therapies, the 3-year and 5-year survival remains suboptimal even in patients with PDL1 expression on more than 50% of the viable tumor cells, that is, their tumor proportion score (TPS) is ≥50%.2 3 Therefore, tumor PDL1 expression cannot be used as the only biomarker to predict response and survival in patients treated with anti-PD1/PDL1 therapy. There is ongoing research to identify affordable and easily obtained biomarkers that can be used as adjunct to tumor PDL1 expression for precision medicine.
Human leukocyte antigen (HLA) is expressed on somatic as well as immune cells. HLA molecules introduce internally processed cellular antigens to T lymphocytes to trigger a series of signals to activate or inhibit T cell proliferation and differentiation.4 5 HLA molecules are divided into two types, HLA-I and HLA-II. HLA-I is expressed on all nucleated cells, including somatic, cancer and immune cells and presents cell peptides to activate CD8+T cells. HLA-II, on the other hand, is expressed on antigen-presenting cells and mediates the activation of CD4+ helper or regulatory T cells. HLA molecules are encoded by the HLA gene complex located on the short arm of chromosome 6 at locus 21 (6p21).6 7 The HLA locus is not only polygenic but also highly polymorphic with each locus having multiple alleles.8 9 HLA alleles can be grouped based on their peptide-binding repertoire into supertypes.10 Only HLA-A and HLA-B supertypes are well-characterized, with Sidney et al (2008) describing nine supertypes for HLA-A and HLA-B alleles based on their specificity for the anchor amino acids.10
HLA molecule downregulation in cancer tissue is a well-established mechanism of cancer evading the immune system.11–15 The low expression or low diversity of HLA on the cell surface is directly related to the range of antigens that can be presented to T cells. Many studies have looked at the correlation between expressed HLA-I on tumor cells and its effect on survival and response to immunotherapy.16–22 However, the effect of low HLA genetic diversity (or homozygosity) on response to immunotherapy has not been explored, with the exception of two recent studies.23 24 Chowell et al investigated the relationship between genetic HLA homozygosity and survival in 100 NSCLC and 269 patients with melanoma treated with immunotherapy, either with anti-PD1/PDL1 or anti-CTLA4 or both. Reduced survival was correlated with homozygosity in at least one HLA-I locus. Therefore, it was postulated that maximal heterozygosity of HLAs might be associated with higher diversity of antigens presented to the T cells, compared with an individual with homozygosity at one or more HLA loci.23 Furthermore, they observed that HLA-B62 supertype was strongly associated with reduced survival, while the HLA-B44 supertype associated with longer survival.23 Hence genomic HLA can be explored as an economical and easily obtained non-invasive biomarker that can help guide treatment choice among patients with NSCLC patients treated with immunotherapy.
Here, we aim to assess whether homozygosity in HLA-I or HLA-II is associated with survival outcomes among patients with advanced NSCLC treated with single agent anti-programmed cell death protein-1 (PD1) or anti-PDL1 therapy in the first-line or second-line setting.
Methods
Patients
Patients were recruited from two major teaching hospitals in Western Australia. Recruited patients were 18 years or older, diagnosed with unresectable locally advanced NSCLC and treated with single agent pembrolizumab, atezolizumab or nivolumab in the first-line or the second-line setting. Given that genomic HLA would not be altered by infection, treatment or any other environmental factors, patients were recruited both prospectively and retrospectively. Patient demographics and clinicopathological features that may affect outcome were collected from the clinical records and included: age, Eastern Cooperative Oncology Group (ECOG) performance status and smoking status. Pre-treatment neutrophil/lymphocyte ratio (NLR) was calculated using the pre-treatment full blood count (FBC) results. Tumor characteristics included: histopathology, PDL1 expression and genetic alterations (epidermal growth factor receptor (EGFR) mutations, echinoderm microtubule-associated protein like-4-anaplastic lymphoma kinase (EML4/ALK) fusion, c-ros oncogene 1 (ROS1) fusions and Kirsten RAt Sarcoma (KRAS) GTPase mutations). The latter was only performed in NSCLC with non-squamous cell carcinoma histology.
DNA extraction and genomic HLA-typing
Blood was collected in EDTA blood tubes, and either stored as whole blood or white blood cell pellet in a freezer at −80°C. QIAamp DNA Blood Maxi Kit was used for DNA extraction. Extracted DNA was used for high-resolution HLA typing at the Institute of Immunology and Infectious disease (IIID) at Murdoch University.
HLA testing at IIID has been accredited by the American Society for Histocompatibility and Immunogenetics (ASHI) and the National Association of Testing Authorities (NATA). Specific HLA loci on the extracted DNA are PCR amplified using sample-specific molecular indexed primers (MID-tagged) that amplify polymorphic exons from Class I (A, B, C Exons 2 and 3) and Class II (DQ, Exons 2 and 3; DRB and DPB1, Exon 1) HLA genes. MID-tagged primers have been optimized to minimize allele dropouts and primer bias. Amplified DNA products from unique MID-tagged products are pooled in equimolar ratios and subjected to library preparation. Post QC and quantitation the normalized libraries are then sequenced on the Illumina MiSeq platform using the MiSeq V3 600-cycle kit (2×300 bp reads). Sequences are separated by MID tags and alleles called using an in-house accredited HLA allele caller software pipeline that minimizes the influence of sequencing errors. Alleles are called using the latest IMGT HLA allele database as the allele reference library. Sample to report integrity was tracked and checked using proprietary and accredited Laboratory Information and Management System and HLA analyze reporting software that performs comprehensive allele balance and contamination checks on the final data set.25 Genomic HLA-A and HLA-B alleles were classified into supertypes using the method described by Sidney et al.10
PDL1 analysis
Immunohistochemistry, using the Dako 22C3 clone for all patients, for PDL1 testing was performed by PathWest Pathology Services as per standard diagnostic services and reported as TPS.
Statistical analysis
Progression free survival (PFS) was defined as the time interval between the start of therapy and the date of first progression/death. Progression was determined by clinician assessment based on both radiological and clinical presentation of the patient. Overall survival (OS) was defined as the time between the start of immunotherapy and death. The minimum follow-up time was 6 months. Those who did not experience disease progression or were still alive at the time of analysis (January 2020) were censored.
Patients were dichotomised based on HLA homozygosity at one or more loci and heterozygosity; or based on the presence of specific HLA-A and HLA-B supertypes. PFS and OS were compared between groups using log-rank (Mantel-Cox) test and Kaplan-Meier plots using GraphPad Prism V.8 (GraphPad Software, Inc, San Diego, California).
A power calculation was carried out assuming a frequency of homozygosity in one HLA loci in the general population of around 20%, with a median follow-up period of 36 months from the time of starting immunotherapy, and a median survival for homozygous patients of approximately 12 months (based on Chowell et al 2018 cohort 1).23 If the true HR is 2 for overall survival of homozygotes relative heterozygotes, 161 patients will be sufficient to reject the null hypothesis with probability 0.86 and a Type I error probability of 0.05.
Cox regression analyzes were used to compare the effect of HLA-I homozygosity on OS outcomes in subgroups of patients stratified according to age, sex, PDL1 tumor proportion score, pre-treatment NLR, ECOG performance scores, smoking status, histopathology, the presence of KRAS mutations (only among non-squamous cell carcinoma (non-SCC) cases), therapy modality and line of treatment.
We conducted univariable and multivariable analyzes using Cox regression models in SPSS V.25 (IBM, Armonk, New York) correlating HLA homozygosity with PFS and OS and controlling for tumor PDL1, pre-treatment NLR, age, ECOG status and therapy type (anti-PD1 and anti-PDL1). As the expression of PDL1 protein on cancer cells might be dependent on the first signal of interaction between HLA-I molecule on tumor cells and the T-cell receptor (TCR), we investigated the interactions between HLA homozygosity and tumor PDL1 expression and to reduce the effect of different confounders on the studied variable.
Results
Patients characteristics
A total of 170 patients were recruited between May 2018 and June 2019. Nine patients were excluded: six patients were excluded as they were receiving targeted therapy or immunotherapy in combination with another anticancer agent, one patient died from another cause before reaching the response assessment period, and two patients were lost to follow-up. A total of 161 patients treated with pembrolizumab (n=37), nivolumab (n=72) and atezolizumab (n=52) were included in the study. Pembrolizumab was administered as first-line therapy, while atezolizumab or nivolumab were used as second-line or subsequent lines of treatment. Patients’ characteristics and cancer histopathology in the studied cohort were similar to the general NSCLC population, with most of the patients being smokers (81%), men (56%), more than 65 years old (57%) and had good performance status (ECOG 0 to 1) (table 1). Adenocarcinoma was the predominant lung histology (66%). KRAS mutation was the predominant molecular alteration in the non-SCC lung cancer, especially in the PDL1 known group. PDL1 status was available for 114 patients, of whom 61 patients (54%) expressed PDL1 in more than 50% of tumor cells. Standard of care at the time required that treatment with first-line pembrolizumab was only allowed for patients with a PDL1 TPS score of ≥50%. Hence, all patients in the PDL1 known group treated with pembrolizumab as first-line therapy had their PDL1 expression tested and demonstrated ≥50% PDL1 expression in tumor cells. Those treated with nivolumab or atezolizumab in the second-line setting had TPS scores <50% or were not tested for PDL1 expression. This explains the higher proportion of first-line therapy patients in the PDL1 known group. Overall, both groups had very similar characteristics (table 1).
Table 1.
Demographics and disease characteristics of patients at baseline
| Full cohort N (%) |
PDL1 known group N (%) |
|
| Age | ||
| ≥65 | 92 (57) | 70 (61) |
| <65 | 69 (43) | 44 (39) |
| Sex | ||
| M | 90 (56) | 66 (58) |
| F | 71 (44) | 48 (42) |
| ECOG | ||
| ≤1 | 137 (85) | 96 (84) |
| >1 | 23 (14) | 17 (15) |
| Unknown | 2 (1) | 1 (1) |
| Smoking | ||
| Yes | 131 (81) | 91 (80) |
| No | 19 (12) | 14 (12) |
| Unknown | 11 (7) | 9 (8) |
| Histopathology | ||
| Adenocarcinoma | 106 (66) | 74 (65) |
| SCC | 45 (28) | 32 (28) |
| Others | 10 (6) | 8 (7) |
| Molecular status* | ||
| KRAS mutant | 55 (47) | 41 (50) |
| KRAS wild type | 46 (40) | 36 (44) |
| KRAS unknown | 10 (9) | 2 (2) |
| EGFR, ALK or ROS1 mutant | 5 (4) | 3 (4) |
| Line of treatment | ||
| First line | 37 (23) | 37 (32) |
| Second or more | 124 (77) | 77 (68) |
| Total | 161 | 114 |
*Molecular status was only examined in NSCLC with non-squamous cell carcinoma histology (116 patients in full cohort and 82 in the PDL1 known group).
ALK, echinoderm microtubule-associated protein like-4-anaplastic lymphoma kinase (EML4/ALK) fusion; ECOG, Eastern Cooperative Oncology Group; EGFR, epidermal growth factor receptor; KRAS, Kirsten Rat Sarcoma GTPase; NSCLC, non-small cell lung cancer; PDL1, programmed death ligand; ROS1, echinoderm c-ros oncogene 1 fusion; SCC, squamous cell carcinoma.
Genomic HLA-I homozygosity at one or more loci prevalence was 20%, with a similar proportion (18%) observed in the PDL1 group. The prevalence of genomic HLA-II homozygosity at one or more loci was moderately higher in the PDL1 known group, being 35% compared with 33% in the full cohort (online supplemental table 1). Comparison of clinical characteristics between patients with HLA homozygosity and those with maximal heterozygosity did not reveal unevenness between groups (online supplemental table 2).
jitc-2020-001620supp001.pdf (611.2KB, pdf)
Prognostic value of HLA homozygosity
Survival analysis showed that NSCLC patients who were treated with single-agent immunotherapy and are homozygous at one or more HLA-I loci had reduced overall survival, compared with those who are heterozygous at all HLA-I loci (HR=1.96, 95% CI 1.02 to 3.78, p=0.04) (figure 1A). The effect of homozygosity at one or more HLA-I loci trended towards improved PFS but it was not statistically significant (figure 1B). However, we did not find a statistically significant effect on OS or PFS, when HLA zygosity was evaluated for HLA-A, HLA-B or HLA-C separately (data not shown). Moreover, no statistically significant differences were observed in OS or PFS between patients that were homozygous at one or more HLA-II loci and those that were heterozygous at all HLA-II loci (figure 1C, D).
Figure 1.
Effect of HLA-I/II homozygosity on OS and PFS in patients with NSCLC treated with anti-PD1/PD-L1 immunotherapy. A and B, comparison of patients with homozygosity at one or more HLA-I loci on OS and PFS, respectively. C and D, comparison of patients with homozygosity at one or more HLA-II loci on OS and PFS, respectively. Kaplan-Meier curves were compared using log-rank analysis. HLA, human leukocyte antigen; NSCLC, non-small cell lung cancer; OS, overallsurvival; PD1/PDL1, programmed cell death protein-1/programmed death ligand 1; PFS, progression free survival.
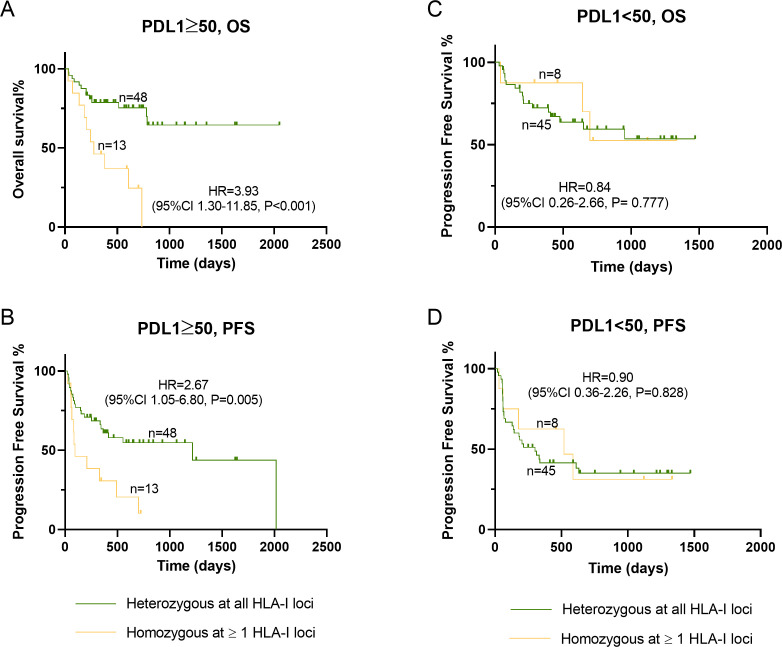
Subgroup analysis suggested that homozygosity at one or more HLA-I loci was associated with shorter survival especially in patients older than 65, those who have high ECOG status (≥2) and those treated with anti-PD1 rather than anti-PDL1 immunotherapy (figure 2). In particular, HLA homozygosity seemed to significantly affect the survival of patients expressing PDL1 in more than 50% of their tumor tissue (p<0.001). Kaplan-Meier curves of PDL1 status-based subgroups, revealed that the negative effect of HLA homozygosity on OS and PFS was only statistically significant in the subgroup with more than 50% PDL1-expressing cells, and separation of the curves was evident (HR=3.93, 95% CI 1.30 to 11.85, p<0.001) (figure 3). The effect of HLA-I zygosity on OS among patients with PDL1 ≥50% was driven by homozygosity in HLA-A (HR=6.46, 95% CI 1.41 to 29.2, p=0.015, (online supplemental figure 1). A very limited number of cases were found homozygous for HLA-B or HLA-C (n=3 and 4, respectively), and therefore not suitable for statistical analysis. No differences in OS or PFS were observed in patients with PDL1 less than 50%, based on their HLA genotype.
Figure 2.
Association between HLA-I homozygosity and OS. Subgroups were stratified according to age, sex, PDL1 tumor proportion score, pre-treatment neutrophil-lymphocyte ratio (NLR), Eastern Cooperative Oncology Group (ECOG) performance scores, smoking status, histopathology, the presence of KRAS mutations (only among non-SCC cases), therapy modality and line of treatment and analyzed using of Cox regression. F, female; HLA, human leukocyte antigen; M, male; OS, overall survival; PD1, programmed cell death protein-1; PDL1, programmed death ligand; SCC, squamous cell carcinoma.
Figure 3.
Effect of HLA-I homozygosity on survival and progression free survival in patients with NSCLC treated with anti-PD1/PDL1 immunotherapy based on their PDL1 status (≥50% vs <50%). A and B, association between homozygosity at one or more HLA-I loci on OS and PFS, respectively, among patients whom their cancer express PDL1 in ≥50% of the cancer tissue. C and D, association between homozygosity at one or more HLA-II loci on OS and PFS, respectively, among patients whom their cancer express PDL1 in <50% of the cancer tissue. HLA, human leukocyte antigen; NSCLC, non-small cell lungcancer; OS, overall survival; PD1/PDL1, programmed cell death protein-1/programmeddeath ligand 1; PFS, progression free survival.
We carried out univariable and multivariable Cox regression analyzes including other variables that may affect survival in these patients (age, sex, PDL1 expression, ECOG, NLR and therapy type (anti-PD1 and anti-PDL1)) (table 2). Homozygosity at one or more HLA-I loci and type of checkpoint inhibitor used (anti-PD1 vs anti-PDL1) were the only markers associated with OS in the univariable analysis (table 2). In the multivariable analysis, NLR also emerged as a prognostic marker of OS together with treatment type and HLA-I genotype. Notably, after controlling for interaction between PDL1 and HLA-I in the model, HLA-I homozygosity emerged as having a highly significant association with OS (HR=3.70, 95% CI 1.53 to 8.95, p=0.004). The adverse effect of homozygosity at one or more HLA-I loci on PFS was only apparent after controlling for interactions between PDL1 status and HLA-I genotype (HR=2.18, 95% CI 1.02 to 4.66, p=0.043) (table 2). No significant interactions were found between HLA-I and the other variables (data not shown).
Table 2.
Univariable and multivariable Cox regression analysis (n=114)
| OS | Univariable | Multivariable | Multivariable +interaction | |||||||||
| P value | HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | ||||
| Lower | Upper | Lower | Upper | Lower | Upper | |||||||
| HLA-I zygosity (hetero vs homo) | 0.020 | 2.17 | 1.13 | 4.17 | 0.023 | 2.19 | 1.12 | 4.30 | 0.004 | 3.70 | 1.53 | 8.95 |
| Age (<65 vs ≥65) | 0.534 | 0.83 | 0.45 | 1.51 | 0.229 | 0.67 | 0.34 | 1.29 | 0.400 | 0.75 | 0.38 | 1.47 |
| Sex (M vs F) | 0.598 | 1.14 | 0.70 | 1.88 | 0.094 | 1.70 | 0.91 | 3.16 | 0.254 | 1.44 | 0.77 | 2.71 |
| PDL1 (≥50% vs <50%) | 0.916 | 0.97 | 0.53 | 1.77 | 0.498 | 0.81 | 0.43 | 1.50 | 0.755 | 1.12 | 0.54 | 2.35 |
| ECOG (≤1 vs ≥2) | 0.216 | 1.63 | 0.72 | 3.51 | 0.141 | 1.81 | 0.82 | 3.97 | 0.113 | 1.89 | 0.86 | 4.17 |
| NLR (≤5 vs >5) | 0.059 | 1.78 | 0.98 | 3.24 | 0.002 | 2.95 | 1.51 | 5.75 | 0.001 | 2.97 | 1.53 | 5.76 |
| Therapy type (αPD1 vs αPDL1) | <0.001 | 3.16 | 1.66 | 5.99 | <0.001 | 4.36 | 2.18 | 8.75 | <0.001 | 4.26 | 2.12 | 8.59 |
| HLA-I*PDL1 | 0.094 | 0.26 | 0.05 | 1.26 | ||||||||
| PFS | ||||||||||||
| HLA-I zygosity (hetero vs homo) | 0.093 | 1.62 | 0.92 | 2.86 | 0.059 | 1.77 | 0.98 | 3.20 | 0.043 | 2.18 | 1.02 | 4.66 |
| Age (<65 vs ≥65) | 0.352 | 0.79 | 0.48 | 1.30 | 0.262 | 0.74 | 0.43 | 1.26 | 0.369 | 0.78 | 0.45 | 1.34 |
| Sex (M vs F) | 0.780 | 1.06 | 0.71 | 1.59 | 0.111 | 1.51 | 0.91 | 2.49 | 0.176 | 1.43 | 0.85 | 2.38 |
| PDL1 (≥50% vs <50%) | 0.347 | 1.26 | 0.78 | 2.06 | 0.721 | 1.10 | 0.66 | 1.82 | 0.470 | 1.24 | 0.69 | 2.23 |
| ECOG (≤1 vs ≥2) | 0.426 | 1.32 | 0.67 | 2.58 | 0.401 | 1.35 | 0.67 | 2.73 | 0.395 | 1.36 | 0.67 | 2.75 |
| NLR (≤5 vs >5) | 0.626 | 1.13 | 0.67 | 1.86 | 0.113 | 1.56 | 0.90 | 2.70 | 0.119 | 1.54 | 0.89 | 2.67 |
| Therapy type (αPD1 vs αPDL1) | <0.001 | 2.93 | 1.75 | 4.88 | <0.001 | 3.18 | 1.84 | 5.47 | <0.001 | 3.07 | 1.77 | 5.32 |
| HLA-I*PDL1 | 0.404 | 0.58 | 0.16 | 2.10 | ||||||||
ECOG, Eastern Cooperative Oncology Group; F, female; HLA, human leukocyte antigen; M, male; NLR, neutrophil/lymphocyte ratio; NSCLC, non-small cell lung cancer; OS, overall survival; PD1/PDL1, programmed cell death protein-1/programmed death ligand 1; PFS, progression free survival.
Prognostic value of HLA-I supertypes
We also investigated the clinical relevance of HLA-I supertypes on patient survival after treatment with anti-PD1/PDL1 antibodies. Among the eight supertypes evaluated, only HLA-A02 had a statistically significant association with improved survival (HR=0.56, 95% CI 0.34 to 0.93, p=0.023) (table 3, online supplemental figure 1). The effect of homozygosity at one or more HLA-I loci among PDL1 ≥50% group was driven by the presence of HLA-A02 (HR=0.36, 95% CI 0.15 to 0.89, p=0.042) (online supplemental figure 2). However, the multivariable analysis indicated that HLA-A02 status was not an independent prognostic variable (online supplemental table 3).
Table 3.
Association of HLA-I supertypes with overall survival of NSCLC patients treated with anti-PD1/PDL1
| HLA-I supertype | Frequency N (%) | HR (95% CI) | P value |
| A24 | 38 (23.6) | 1.34 (0.74 to 2.41) | 0.295 |
| A01 | 71 (44.1) | 0.95 (0.57 to 1.57) | 0.834 |
| A02 | 80 (49.7) | 0.56 (0.34 to 0.93) | 0.023 |
| A03 | 84 (52.2) | 1.27 (0.77 to 2.08) | 0.356 |
| B58 | 13 (8.1) | 0.98 (0.40 to 2.43) | 0.966 |
| B62 | 30 (18.6) | 1.55 (0.80 to 3.00) | 0.137 |
| B27 | 32 (19.9) | 0.91 (0.55 to 1.49) | 0.075 |
| B44 | 90 (55.9) | 0.83 (0.50 to 1.38) | 0.470 |
| B07 | 75 (46.6) | 0.91 (0.55 to 1.49) | 0.705 |
| B08 | 37 (23) | 1.11 (0.60 to 2.07) | 0.724 |
| A01/A24 | 13 (8.1) | 1.29 (0.51 to 3.30) | 0.548 |
HLA, human leukocyte antigen;NSCLC, non-small cell lung cancer; PD1/PDL1, programmed cell death protein-1/programmed death ligand 1.
Discussion
Tumor specific markers such as PDL1 expression, tumor mutational burden and mismatch repair deficiency have been described as predictors of long-term benefit to immune checkpoint inhibitor blockade.26 However, variations in outcomes can also be influenced by host-specific factors. Chowell et al 2018 showed in a cohort of melanoma and patients with NSCLC treated with single-agent anti-PD1 or in combination with anti-CTLA-4, that HLA-I homozygosity at one or more loci was associated with shorter survival comparing to those who were heterozygous at all HLA-I loci (HR=1.4). However, the study did not demonstrate the effect of HLA homozygosity in patients with NSCLC specifically. The results of our study indicate that homozygosity at one or more HLA-I loci is associated with poor outcomes in patients with NSCLC treated with single-agent anti-PD1/PDL1 in the first-line or the second-line setting.
HLA-I expression in cancer tissue was found to be a key factor determining increased immune cell infiltration into NSCLC tissue, and independent of PDL1 status.27 While there is enough evidence to suggest that downregulation of HLA-I molecule is one of the mechanisms used by the tumor to escape the immune system,13 14 a coordinated balance needs to be achieved to avoid destruction by natural killer cells (NK cells)28 and this might reduce the impact of HLA-I downregulation as a mechanism to evade the immune system. On the other hand, loss of heterozygosity in cancer tissue was found to be associated with poor treatment outcomes.29 30 However, loss of genomic heterozygosity in tumor occurs only in 11% of cases.23 As shown in our study, germline homozygosity of HLA-I at one or more loci also has a clear impact on survival, underscoring the role that HLA-I molecules play on the immune response against NSCLC.
The effect of HLA-I homozygosity on survival was more pronounced in patients with strong PDL1 expression (≥50%). The expression of PDL1 on tumor tissue, as an immune regulatory mechanism has to be preceded by an interaction between peptide/HLA-I molecule and T cell receptors. This implies that the expressed HLA-I molecule is not downregulated in tumor cells and PDL1 expression is the primary mechanism of immune evasion in these cases.27 Therefore, the HLA-I homozygosity may have a profound impact on survival among patients with PDL1 ≥50%, when treated with PD1/PDL1 inhibitors. This also implies that patients with PDL1 <50% utilize other immune evasion mechanisms, including downregulation of HLA in cancer cells.
Our results are in contrast with the report by Negrao et al,24 which did not find a significant correlation between HLA zygosity and survival in an NSCLC cohort treated with PD-1/PDL1 at the M. D. Anderson Cancer Center. Notably, a large proportion (25%) of their patient cohort received anti-PD-1/PDL1 as third-line therapy, while all our study participants were treated either in the first-line or second-line setting. Moreover, their dichotomisation for PDL1 expression was carried out based on 1% TPS, while our subgroup analysis was based on a 50% PDL1 expression cut-off.
Chowell et al showed that the presence of HLA-B62 and HLA-B44 in patients with melanoma treated with immune checkpoint inhibitors are associated with worse (HR=2.29) and better prognosis (HR 0.61), respectively. However, they did not evaluate the role of HLA supertypes in their cohort of patients with NSCLC. In our cohort of patients with NSCLC treated with anti-PD1/PDL1 therapy, the presence of HLA-A02 supertype appears to positively affect survival. This may reflect differences in cancer biology between melanoma and NSCLC, which is reflected by the degree of response to immunotherapy and in particular to anti-CTLA4 agents. The differential effect of HLA supertypes in melanoma and NSCLC, may also reflect the differences in the type of mutations and therefore neoepitopes between these two cancers.31 HLA-A02 molecule binds small aliphatic and small hydrophobic/aliphatic amino acids in the B and F pockets,10 for example, KRAS G12V neoantigen represent the tumor specific antigen bound to HLA-A*02:01.32 This might help predict the type of cancer neoantigens that are associated with better survival among patients with NSCLC treated with anti-PD1/PDL-1. HLA-B is usually expressed more frequently on the cell surface compared with HLA-A.33 Nevertheless, the beneficial effects in survival outcomes observed for carriers of the HLA-A02 supertype suggest that NSCLC might escape this downregulation mechanism.
Homozygosity at one or more genomic HLA-II loci in patients with NSCLC treated with single agent anti-PD1/PDL1 treatment did not demonstrate a direct influence on survival and may require larger numbers to show its impact. HLA-II molecule is rarely expressed on cancer tissue and more likely expressed on antigen-presenting cells.34 Its expression on tumor infiltrating lymphocytes rather than NSCLC tissue is associated with better PFS and OS.35 HLA-II is responsible for the activation of CD4+ helper T-lymphocytes which4 5 then help to activate B-cells and CD8+T cells, as well as promote the induction of memory CD8+T cell.36 37 The role of CD4+T cells is much less understood than CD8+T cells in cancer biology.34 All these factors potentially contributed to our observation of a lack of statistically significant effect of homozygosity at one or more HLA-II molecule on OS and PFS of patients with NSCLC treated with single agent anti-PD1/PDL1.
A secondary observation from our analysis was that anti-PD1 therapy had longer OS and PFS when compared with anti-PDL1 immunotherapy. This observation is consistent with the meta-analysis conducted by Tartarone et al 2019.38 However, the latter study only carried out an indirect comparison and analyzed PFS only. We also considered that the results might be largely biased by our retrospective cohort, which included a large number of long-term survivors who received nivolumab (anti-PD1). However, in the absence of an interaction between the type of therapy and genomic HLA-I, we believe that this finding is unrelated to the main theme of this research project.
Our study has some limitations that require discussion. We did not incorporate the role of tumor mutation burden (TMB) in this analysis, which is thought to play a role in response to immunotherapy.39 However, Chowell et al 201823 showed that low TMB among patients with melanoma did not predict but that rather had an additive effect in the prognostic value of genomic HLA-I in survival. In addition, we had limited access to cancer tissue and were unable to assess somatic loss of HLA-I heterozygosity or HLA expression, which have been correlated individually with shorter OS and PFS.23 40 For some of our patients, a fine needle aspiration (FNA) was utilized for assessment of PDL1 status and other molecular testing. Limited tissue availability after PDL1 analysis and other molecular testing is a significant roadblock in the field. In fact, Hurkmans et al40 reported of having to exclude 70% of the patients recruited due to tissue inadequacy to perform HLA immunohistochemistry. Moreover, our study is unable to assess the role of homozygosity at one or more HLA-I loci as a predictive marker for response to anti-PD1/PDL1 among patients with NSCLC. Finally, while our subgroup analysis showed a strong association between HLA and survival in patients with PDL1 ≥50% TPS, the small number of homozygous events resulted in large CIs. Thus, more extensive studies are needed to validate the clinical prognosis value of HLA zygosity in NSCLC treated with anti-PD1/PDL1 therapy.
Our study provides enough evidence that genomic homozygosity is associated with a worse prognosis in patients with NSCLC treated with single-agent immunotherapy. This is of special importance in NSCLC which often relies on FNA for diagnosis, and therefore tumor material can be exhausted after performing mutational profiling and PDL1 staining. We propose that, in addition to PDL1 TPS, HLA-I typing should be considered as a non-invasive and cost-effective biomarker to guide treatment personalisation. Given that patients with HLA-I homozygosity are less likely to derive clinical benefit from single-agent anti-PD1/PDL1 therapy, consideration of a more aggressive treatment combinations may be needed to improve their prognosis, especially among those with PDL1 TPS of 50% or more.
Acknowledgments
We would like to acknowledge the contribution of patients and their families without whom it would not be possible to perform the study. We would also like to thank the members of the Edith Cowan University Research Group for receiving and processing patient samples, in particular Michael Morici and Emmanuel Acheampong.
Footnotes
Twitter: @ElinSGrayPhD
Contributors: AA and MAK formulated the hypothesis; AA, MM and ESG designed the study; AA, SC, SB and MAK recruited patients; AC and MW oversaw experiments; AA, LC, JL and ESG performed analyses; AA and ESG wrote the original manuscript article; MM and ESG supervise the study. All authors reviewed and approved the final version of the manuscript.
Funding: This work was supported by the clinical trial unit at Fiona Stanley Hospital, Murdoch, Western Australia; a research grant from the Lung Foundation Australia – Ellen Yates Memorial Grant in Aid for Lung Cancer Research; a fellowship to AA from the Western Australia Cancer Council Palliative Care Network; and a fellowship to EG from the Cancer Research Trust and Cancer Council WA.
Competing interests: MM sits on advisory boards for Merck Sharp and Dohme (MSD), Bristol-Myers Squibb (BMS) and AstraZeneca (AZ). MAK sits on advisory boards of MSD, BMS and Merck Serono. ESG has received travel support from MSD.
Patient consent for publication: Not required.
Ethics approval: Procedures have been approved by the Human Research Ethics Committees (HREC) at Edith Cowan University (ECU) (No. 19857), Sir Charles Gairdner Hospital (SCGH) (No. 2013-246) and Fiona Stanley Hospital (FSH) (2014-144). All participants signed a consent form which is saved at ECU research database.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data is available upon reasonable request.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.AIHW Cancer data in Australia, 2019. [Google Scholar]
- 2.Garon EB, Hellmann MD, Rizvi NA, et al. . Five-Year overall survival for patients with advanced Non‒Small-Cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study. J Clin Oncol 2019;37:2518–27. 10.1200/JCO.19.00934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rodríguez-Abreu D, Robinson AG, et al. . OA14.01 KEYNOTE-024 3-year survival update: pembrolizumab vs platinum-based chemotherapy for advanced Non–Small-Cell lung cancer. J Thorac Oncol 2019;14:S243 10.1016/j.jtho.2019.08.483 [DOI] [PubMed] [Google Scholar]
- 4.Arasanz H, Gato-Cañas M, Zuazo M, et al. . Pd1 signal transduction pathways in T cells. Oncotarget 2017;8:51936–45. 10.18632/oncotarget.17232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corthay A, Lorvik KB, Bogen B. Is secretion of tumour-specific antigen important for cancer eradication by CD4(+) T cells?--Implications for cancer immunotherapy by adoptive T cell transfer. Scand J Immunol 2011;73:527–30. 10.1111/j.1365-3083.2011.02558.x [DOI] [PubMed] [Google Scholar]
- 6.Shiina T, Hosomichi K, Inoko H, et al. . The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet 2009;54:15–39. 10.1038/jhg.2008.5 [DOI] [PubMed] [Google Scholar]
- 7.Schwab M. Encyclopedia of cancer. Berlin, Heidelberg: Springer Berlin Heidelberg, 2009: 1393–93. [Google Scholar]
- 8.Janeway CA Jr TP, Walport M, et al. . Immunobiology: the immune system in health and disease. New York: Garland Science, 2001. [Google Scholar]
- 9.Hla alleles numbers: Anthony Nolan research, 2019. Available: http://hla.alleles.org/nomenclature/stats.html [Accessed 29 Feb 2020].
- 10.Sidney J, Peters B, Frahm N, et al. . Hla class I supertypes: a revised and updated classification. BMC Immunol 2008;9:1. 10.1186/1471-2172-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Algarra I, Collado A, Garrido F. Altered MHC class I antigens in tumors. Int J Clin Lab Res 1997;27:95–102. 10.1007/BF02912442 [DOI] [PubMed] [Google Scholar]
- 12.Chew G-L, Campbell AE, De Neef E, et al. . Dux4 suppresses MHC class I to promote cancer immune evasion and resistance to checkpoint blockade. Dev Cell 2019;50:658–71. 10.1016/j.devcel.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bubeník J. Tumour MHC class I downregulation and immunotherapy (review). Oncol Rep 2003;10:2005–8. [PubMed] [Google Scholar]
- 14.Leclerc M, Mezquita L, Guillebot De Nerville G, et al. . Recent advances in lung cancer immunotherapy: input of T-cell epitopes associated with impaired peptide processing. Front Immunol 2019;10:1505–05. 10.3389/fimmu.2019.01505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park IA, Hwang S-H, Song IH, et al. . Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLoS One 2017;12:e0182786. 10.1371/journal.pone.0182786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champine PJ, Michaelson J, Weimer BC, et al. . Microarray analysis reveals potential mechanisms of BRMS1-mediated metastasis suppression. Clin Exp Metastasis 2007;24:551–65. 10.1007/s10585-007-9092-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dave SS, Fu K, Wright GW, et al. . Molecular diagnosis of Burkitt's lymphoma. N Engl J Med 2006;354:2431–42. 10.1056/NEJMoa055759 [DOI] [PubMed] [Google Scholar]
- 18.Rimsza LM, Roberts RA, Campo E, et al. . Loss of major histocompatibility class II expression in non-immune-privileged site diffuse large B-cell lymphoma is highly coordinated and not due to chromosomal deletions. Blood 2006;107:1101–7. 10.1182/blood-2005-04-1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlecht NF, Burk RD, Adrien L, et al. . Gene expression profiles in HPV-infected head and neck cancer. J Pathol 2007;213:283–93. 10.1002/path.2227 [DOI] [PubMed] [Google Scholar]
- 20.Perea F, Sánchez-Palencia A, Gómez-Morales M, et al. . Hla class I loss and PD-L1 expression in lung cancer: impact on T-cell infiltration and immune escape. Oncotarget 2018;9:4120–33. 10.18632/oncotarget.23469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai W, Zhou D, Wu W, et al. . Mhc class II restricted neoantigen peptides predicted by clonal mutation analysis in lung adenocarcinoma patients: implications on prognostic immunological biomarker and vaccine design. BMC Genomics 2018;19:582. 10.1186/s12864-018-4958-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thibodeau J, Bourgeois-Daigneault M-C, Lapointe R. Targeting the MHC class II antigen presentation pathway in cancer immunotherapy. Oncoimmunology 2012;1:908–16. 10.4161/onci.21205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowell D, Morris LGT, Grigg CM, et al. . Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582–7. 10.1126/science.aao4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Negrao MV, Lam VK, Reuben A, et al. . Pd-L1 expression, tumor mutational burden, and cancer gene mutations are stronger predictors of benefit from immune checkpoint blockade than HLA class I genotype in non-small cell lung cancer. J Thorac Oncol 2019;14:1021–31. 10.1016/j.jtho.2019.02.008 [DOI] [PubMed] [Google Scholar]
- 25.Currenti J, Chopra A, John M, et al. . Deep sequence analysis of HIV adaptation following vertical transmission reveals the impact of immune pressure on the evolution of HIV. PLoS Pathog 2019;15:e1008177. 10.1371/journal.ppat.1008177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burdett N, Desai J. New biomarkers for checkpoint inhibitor therapy. ESMO Open 2020;5. 10.1136/esmoopen-2019-000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perea F, Sánchez-Palencia A, Gómez-Morales M, et al. . Hla class I loss and PD-L1 expression in lung cancer: impact on T-cell infiltration and immune escape. Oncotarget 2018;9:4120-4133. 10.18632/oncotarget.23469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulendran B, Katsikis PD, Schoenberger SP. Natural killer cell licensing during viral infection. crossroads between innate and adaptive immunity III. New York, NY: Springer New York, 2011. [Google Scholar]
- 29.McGranahan N, Rosenthal R, Hiley CT, et al. . Allele-Specific HLA loss and immune escape in lung cancer evolution. Cell 2017;171:e11:1259–71. 10.1016/j.cell.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shim JH, Kim HS, Cha H, et al. . HLA-corrected tumor mutation burden and homologous recombination deficiency for the prediction of response to PD-(L)1 blockade in advanced non-small-cell lung cancer patients. Ann Oncol 2020;31:902–11. 10.1016/j.annonc.2020.04.004 [DOI] [PubMed] [Google Scholar]
- 31.Alexandrov LB, Kim J, Haradhvala NJ, et al. . The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishto M, Mansurkhodzhaev A, Ying G, et al. . An in silico-in vitro Pipeline Identifying an HLA-A*02:01+ KRAS G12V+ Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients. Front Immunol 2019;10:2572. 10.3389/fimmu.2019.02572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crux NB, Elahi S. Human leukocyte antigen (HLA) and immune regulation: how do classical and non-classical HLA alleles modulate immune response to human immunodeficiency virus and hepatitis C virus infections? Front Immunol 2017;8:832. 10.3389/fimmu.2017.00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostroumov D, Fekete-Drimusz N, Saborowski M, et al. . Cd4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell Mol Life Sci 2018;75:689–713. 10.1007/s00018-017-2686-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y, Rozeboom L, Rivard CJ, et al. . Mhc class II expression in lung cancer. Lung Cancer 2017;112:75–80. 10.1016/j.lungcan.2017.07.030 [DOI] [PubMed] [Google Scholar]
- 36.Koretzky GA. Multiple roles of CD4 and CD8 in T cell activation. J Immunol 2010;185:2643. 10.4049/jimmunol.1090076 [DOI] [PubMed] [Google Scholar]
- 37.Laidlaw BJ, Craft JE, Kaech SM. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol 2016;16:102–11. 10.1038/nri.2015.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tartarone A, Roviello G, Lerose R, et al. . Anti-Pd-1 versus anti-PD-L1 therapy in patients with pretreated advanced non-small-cell lung cancer: a meta-analysis. Future Oncol 2019;15:2423–33. 10.2217/fon-2018-0868 [DOI] [PubMed] [Google Scholar]
- 39.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133–50. 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurkmans DP, Kuipers ME, Smit J, et al. . Tumor mutational load, CD8+ T cells, expression of PD-L1 and HLA class I to guide immunotherapy decisions in NSCLC patients. Cancer Immunol Immunother 2020;69:771–7. 10.1007/s00262-020-02506-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001620supp001.pdf (611.2KB, pdf)