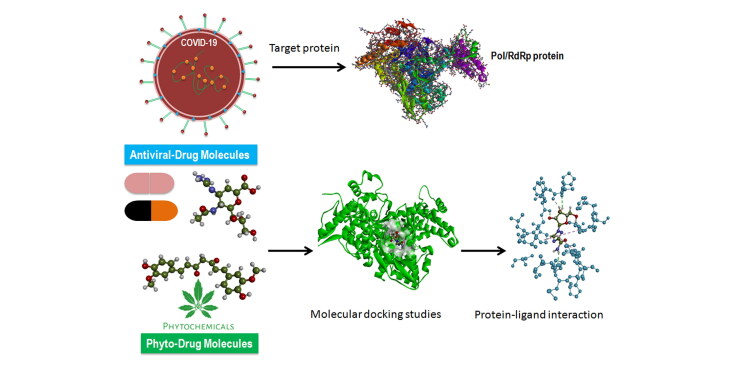
Abstract
Since the SARS/MERS epidemic, scientists across the world have been racing to identify the novel-CoVs as it has been predicted that next epidemic can very well be a result from a new mutation of CoV, for which hundred mutations have already been discovered, and the same fear has come true with world facing a raging pandemic due to COVID-19, an infectious disease caused by a newly discovered coronavirus. COVID-19 or Severe acute respiratory syndrome coronavirus2 (SARS-CoV-2), is a single stranded RNA virus. COVID −19 is highly contagious and has resulted in current global pandemic with almost no country of the world unaffected by this virus. Owing to the lack of effective therapeutics or vaccines, the best measures to control human coronaviruses remain a strong public health surveillance system coupled with rapid diagnostic testing and quarantine/social; distancing/lockdowns as and when necessary. In the present study, we have used the insilico approach for the prediction of novel drug molecules from available antiviral drugs and also from natural compounds that can be best target against RNA-dependent RNA-polymerase (Pol/RdRp) protein of SARS-CoV-2 which can be suitable drugs for the treatment of COVID-19 virus. From the current study we observed that three antiviral and three phyto-chemicals have minimum binding energy with the target protein which were further evaluated in molecular dynamics studies and could specifically bind to RdRp protein of COVID-19. Based on results we suggest that these drugs may act as best or novel inhibitor that may be used for the treatment of SARS-CoV-2.
Communicated by Ramaswamy H. Sarma
Keywords: Phytochemicals, antiviral drugs, pharmacokinetics, binding pocket, molecular docking, structural activity relationship
Graphical Abstract
1. Introduction
An RNA-dependent RNA polymerase (RdRp) is the central catalytic subunit of the RNA-synthesizing machinery of all positive-strand RNA viruses. It is presumed that replication and transcription of the ∼30-kb severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) RNA genome are catalyzed by an RdRp domain in the C-terminal part of nonstructural protein 12 (nsp12), one of 16 replicase subunits which is responsible for the production of negative strand RNA (–RNA), new genome molecules and in many virus groups also subgenomic (sg) messenger RNAs (mRNAs) (Ahlquist et al., 2003; Miller & Koev, 2000). Coronaviruses (CoVs) are a family of + strand single stranded RNA (+ssRNA) viruses that have large RNA genome and cause gamut of diseases in mammals and births to respiratory infections in humans which can be potentially lethal (Fehr & Perlman, 2015). SARS-CoV-2 is genetically similar to bat coronaviruses, giving strong credence to the theory that bats were the primary sources for this virus, also virus found in bats have the same receptor angiotensin converting enzyme 2 (ACE2) as in humans (Benvenuto et al., 2020; Perlman, 2020; Zhou et al., 2020). An intermediate animal reservoir such as a pangolin is also suspected to have been the primary source of transmission of this virus to humans. SARS-CoV-2 virions are typically 50–200 nanometers in diameter (Chen et al., 2020). All coronaviruses including SARS-CoV-2 has four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; with N protein holding the RNA genome, and the S, E, and M proteins together create the viral envelope (Hasan et al., 2020). The spike protein with millennia of evolution is perfectly shaped to attach to the membrane of a host cell in stealth mode (Chatterjee, 2020; Lai & Cavanagh, 1997; Woo et al., 2010). Once the CoV has attached to the host cell it gets a clear path to infiltrate the host cells even before the immune system has chance to know that an external virus has attacked the host organism. Protein modeling experiments on the spike protein of the virus soon suggested that SARS-CoV-2 has sufficient affinity to the angiotensin converting enzyme 2 (ACE2) receptors of human cells to use them as a mechanism of cell entry (Batlle et al., 2020; Chen & Hao, 2020; Walls et al., 2020). After attaching the host cell SARS-CoV-2, the virion then releases RNA into the cell, forcing the cell to produce copies of the virus that further infect more CoV which in turn infect even more host cells.This virus is assumed to have originated in infamous wet markets of China from where it spread to humans (Andersen et al., 2020; Boopathi et al., 2020; Cyranoski, 2020). SARS-CoV primarily infects epithelial cells within the lung. The virus is capable of entering macrophages and dendritic cells but only leads to an abortive infection (Chu et al., 2020; Conti et al., 2020).
In the current COVID-19 outbreak and the previous SARS outbreak it is critical to identify an etiological agent which helps in diagnosing cases in locations where a severe CoV outbreak is occurring. The identification of cases will guide the development, of public health measures to control outbreaks. To date we have not been able to develop anti-viral therapeutics that specifically target human coronaviruses, so treatments are only supportive. The SARS and MERS outbreaks in the past and COVID-19 have stimulated research on these viruses and the ongoing research have identified a large number of suitable anti-viral targets, such as viral proteases, polymerases, and entry proteins. Significant work remains, however, to develop drugs that target these processes and have the ability to inhibit viral replication. In the present study, we have used insilico approach for the prediction of novel drug molecules from FDA approved antiviral drugs including Adefovir, Amantadine, Oseltamivir, Ribavirin and Zanamivir and seven phytochemical compounds from natural plants Curcumin, Demethoxycurcumin, Flavonoid, Isoflavonee, Terpinen-4-ol, Nimbin and Piperine that can be best targeted against RNA-dependent RNA polymerase (Pol/RdRp) protein of severe acute respiratory syndrome coronavirus 2 which is responsible for replication and transcription of the viral RNA genome. Our study selected five antiviral drugs which have been already approved by FDA hence providing a faster access to patients suffering from COVID-19 if there is any positive correlation is found any of drug and its impact in either clearing the viral load or reducing the severity of the COVID-19 infection. Also as these drugs already have the FDA approval hence there is already sufficient evidence of safety of administering these drugs to humans. The seven natural compounds from different plants have been reported to have many properties like curcumin found to possess antiviral, antifungal, antibacterial and antimicrobial activities (Moghadamtousi et al., 2014). Demethoxycurcumin has antiviral, antitumor, antifungal, antibacterial, antidiabetic and antimicrobial properties (Agrawal & Goel, 2016). Flavonoid possesses antiviral effect on human viruses and protective effect on viral Infections (Kaul et al., 1985; Zakaryan et al., 2017). Isoflavonee is shown to inhibit infectivity of enveloped or non-enveloped viruses, as well as single-stranded or double-stranded RNA or DNA viruses (Andres et al., 2009). Terpinen-4-ol is reported to have an inhibitory effect on influenza virus replication (Garozzo et al., 2011). Nimbin is a known anti-viral with beneficial effect in viral infections (Badam et al., 1999). Piperine have many health benefits and disease preventing properties like e.g. anti-inflammatory, antiviral, antipyretic, immune and bioavailability enhancing qualities (Mair et al., 2016). Based on the above properties we recommend in vitro testing and clinical trials of these highly beneficial compounds for their impact on recovery of patients suffering from Covid-19, potential reduction in severity and duration of the infections. These compounds can potentially help in slowing down the replication of CoV in host providing effective therapeutic strategy against this dreaded virus.
2. Methods
2.1. Sequence retrieval and conserved region identification
The protein sequences of 4 RNA-dependent RNA polymerase (RdRp) from different variants of severe acute respiratory syndrome coronavirus (SARS-CoV) including SARS‐CoV-2 (NCBI ID: YP_009725307), SARS‐CoV (NCBI Reference Sequence: NP-828869.1), MERS‐CoV (NCBI Reference Sequence: YP_009047223.1) and Bat-CoV (NCBI reference sequence: AID16545.1) were retrieved in FASTA format from National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) database. Jalview v2 (Thompson et al., 1994) was used for the identification of the conserved region among the sequences through multiple-sequence alignment (MSA) with Clustal Omega (Hall, 1999). BLOSUM62 distance matrix method through Jalview software was used to construct phylograms from the MSA, and to scrutinise divergence among different strains of the CoV (Waterhouse et al., 2009).
2.2. Molecular modelling and structure assessment
The selected target protein RdRp structure was modelled through homology methods. The template selection and validation of the target protein (RdRp) was done using BLASTP against PDB database to find the best template (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). PDB ID: 6NUR, protein name Chain A, NSP12 (Severe acute respiratory syndrome-related coronavirus) was found as a best template. After the selection of template, a 3 D protein model of target protein was generated using SWISS MODEL method (https://swissmodel.expasy.org/.) (Waterhouse et al., 2018). Prediction of secondary structure of RdRp model was predicted through PSIPRED 4.0 (http://bioinf.cs.ucl.ac.uk/psipred/) (Jones, 1999). This algorithm assigns secondary structural elements (α-helices, β sheets and turns).
2.3. Predictive accuracy, model optimization and superimposition
The accuracy of predicted 3 D structure of target protein and its stereo-chemical properties were analysed using SWISS-MODEL online tool (https://swissmodel.expasy.org/) (Waterhouse et al., 2018). SWISS MODEL structure assessment, MolProbity reports (http://molprobity.biochem.duke.edu/), QMEAN (Quantitative Model Energy Analysis), Z-score, Clash score, Ramachandran plot, Ramachandran outliers, Ramachandran favoured, Rotamer outliers, C-beta deviations, bad bonds and bad angles were used to determine stereo-chemical properties of target protein. The model structure was optimized by energy minimization through YASARA server (http://www.yasara.org/minimizationserver.htm) (Krieger et al., 2009), which performs an energy minimization using the YASARA force field. Superimposition of RdRp model and template was done using FATCAT online web tool (http://fatcat.godziklab.org/) (Yuzhen & Godzik, 2004) which calculates Root Mean Square Deviation (RMSD) values, p-value, differential distance matrix decomposition and Eigen value distribution.
2.4. Binding site prediction
Protein–ligand binding sites are the active sites on protein surface that perform protein functions. Thus, the identification of these binding sites is often the first step to study protein functions and structure-based drug design. Active sites (ligand binding sites) of the receptor protein were analyzed by Prank Web online tool (http://prankweb.cz/). Binding sites are the distribution of surrounding residues in the active sites and act as the catalytic residues (Jendele et al., 2019; Lukas et al., 2019).
2.5. Ligand preparation
FDA approved antiviral drugs: Adefovir, Amantadine, Oseltamivir, Ribavirin and Zanamivir and seven phyto-chemical compounds from natural plants Curcumin, Demethoxycurcumin, Flavonoid, Isoflavonee, Terpinen-4-ol, Nimbin and Piperine were selected as ligands for binding with target protein (Pol/RdRp) of COVID-19. The structure of these drug compounds were downloaded from PubChem data base (http://pubchem.ncbi.nlm.nih.gov/). The drug files was downloaded in SDF file format and were converted into PDB files using Discovery Studio Visualizer which includes 2 D to 3 D conversion, verification and optimization of structures. The selected compounds were subjected to Lipinski rules of five. We analysed the rules of five by using online insilico program Lipinski Rule of Five which is available on the Web link (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) (LipinskiCA, 2004).
2.6. Molecular docking
Molecular docking study of selected five FDA approved antiviral drug molecule structures and seven molecule structures from natural plants was performed against the binding site of target protein i.e. RdRp protein of COVID-19. AutoDock vina PyRx software was used to dock protein and ligand molecules (Trott & Olson, 2010). The target receptor file and all ligand files were prepared to AutoDock Vina for the docking with individual drug molecules. A grid map was set grid center X: 141.1104, Y: 142.3415, Z: 147.8521 and number of points X: 93, Y: 70, Z: 70, spacing was 0.3750 Angstrom. After docking process was complete the complex structure results were analysed. Fifteen different compounds were tested against COVID-19 RdRp, including 5 FDA approved antiviral drugs, 7 Natural compounds and 3 nucleotides (ATP,GTP and UTP) (Lam et al., 2012). The nucleotides were used as control in the docking process.
2.7. Pharmacokinetic properties
FAFDrugs4 (http://fafdrugs4.mti.univ-paris-diderot.fr/) (David et al., 2008) and AdmetSAR (http://lmmd.ecust.edu.cn/admetsar) (Cheng et al., 2012) server was used to analyze the absorption, distribution, metabolism, and excretion properties of all ligand compounds. ADMET properties of all the ligand molecules were checked in optimal descriptors (hydrogen bonds, charge) at pH = 7.4. The oral toxicity properties and LD50 was also analysed by using PROTOX web server (http://tox.charite.de/protox_II/) (Drwal et al., 2014).
2.8. Molecular dynamics simulation
The Desmond module was used to carry out MD simulations for this study. Firstly, water model were developed and also add sodium and chloride ions to neutralize the drug and target complex by using ‘system builder’ tool of Desmond, Schrödinger. Liquid simulations optimization was minimized at OPLS3 force field. After creating frames for 100ps interval, the complexes were submitted for 50 ns simulations. We performed 50 ns molecular dynamics studies of all the ligand-target complexes. We performed molecular dynamic simulation experiment at 50 ns of all the selected 6 (3 were FDA approved and 3 were phyto-drug molecules) ligands which showed good docking score in virtual screening.
3. Results
3.1. Protein sequence retrieval and multiple sequence alignment
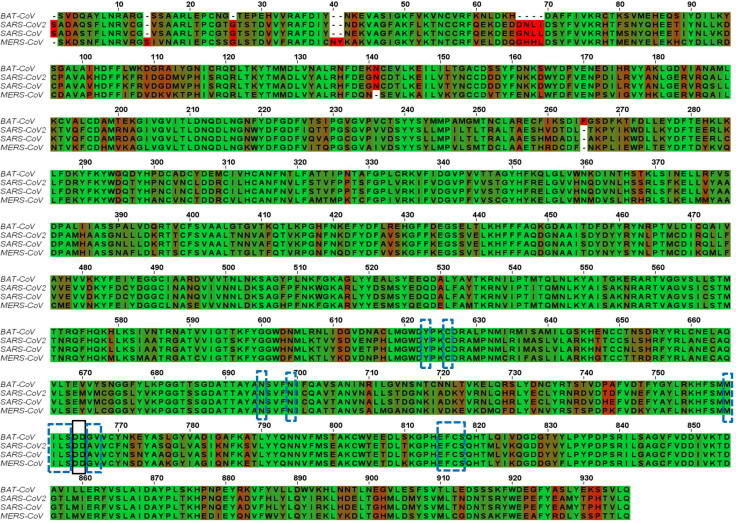
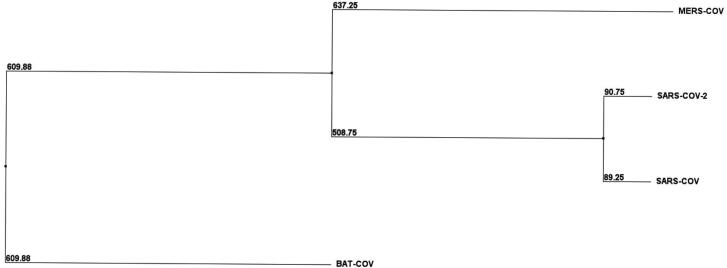
A total of 4 RNA-dependent RNA polymerase (RdRp) protein sequences from different variants of severe acute respiratory syndrome coronavirus (SARS-CoV) were retrieved in FASTA format from National Centre for Biotechnology Information database. The multiple sequence alignment (MSA) of proteins SARS‐CoV 2 (NCBI ID: YP_009725307), SARS‐CoV (NCBI Reference Sequence: NP_828869.1), MERS‐CoV (NCBI Reference Sequence: YP_009047223.1) and Bat-CoV RdRp sequence (NCBI reference sequence: AID16545.1) was performed using Clustal omega software (https://www.ebi.ac.uk/Tools/msa/clustalo/). In the multiple sequence alignment results the black-dashed rectangle marks active site residues (successive aspartate residues D764 and D765). The active site aspartates are projecting from the beta-turn joining the β15 and β16. As implied from the MSA, the active site is highly conserved (highlighted in green colour). Also, the 5 Å region surrounding the D764 and D765 are highly conserved in all CoVs strains as shown by the blue-dashed rectangles. This region includes Y-619, C-622, N-691, N-695, M-756, I-757, L-758, S-759, A-762, V-763, E-811, F-812, C-813, S-814. The active site residues and most of the 5 Å region surrounding it are surface accessible, though can bind to the free nucleotides (ATP, GTP and UTP) (Elfiky et al., 2020). The selected conserved region from the MSA is represented in the Figure 1. BLOSUM62 distance matrix method through Jalview software was used to construct phylograms from the MSA, with the aim to examine the divergence among the retrieved sequences (Thompson et al., 1994). Phylogram of RNA-dependent RNA polymerase is shown in Figure 2.
Figure 1.
Multiple sequence alignment of 4 strains of CoV (Bat-CoV, SARS-Cov-2, SARS-CoV and MERS-CoV) RdRp Sequences. Green colour highlighted indicated identical residues while brown and red colour highlighted residues are less conserved. The black dashed rectangles mark active site aspartates while blue rectangles mark the residues lying in the 5 Å region surrounding the active aspartates. The alignment was made using the Clustal omega web server and Jalview software.
Figure 2.
Phylogenetic tree displaying the evolutionary divergence among the different RNA-dependent RNA polymerase proteins, all selected 4 strains of CoV (Bat-CoV, SARS-Cov-2, SARS-CoV and MERS-CoV). BLOSUM62 distance matrix method through Jalview software was used for the construction of phylogenetic tree.
3.2. Template selection and structural modelling
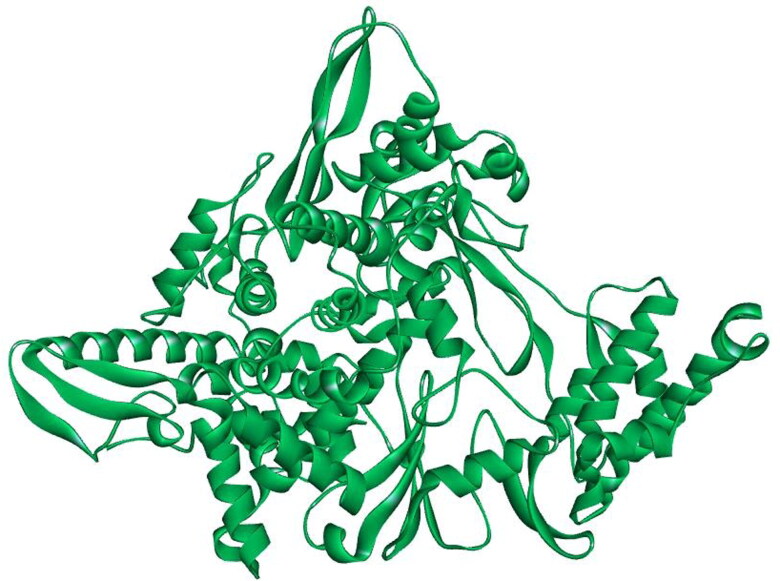
Next with the aim to determine the best ligands against target protein RdRp of COVID-19 (SARS-CoV-2), the selected target protein RdRp of COVID-19 structure was modelled through homology modelling methods using the Swiss Model web server. The template selection and validation of the target protein (RdRp) was done using BLAST-P against PDB database to find the best template.The PDB ID: 6NUR, protein name Chain A, NSP12 of COVID-19 was employed as a best template. On analysis we found that ‘template’ protein model built in our study was the best template with 100% score (maximum score 1889 from a total score 1889), query cover 99%, E-value was 0.0 and sequence identity was 96.35% suggesting it to be best template for the further study. The MMDB ID of the selected template was 175857 with electron microscope as experimental method and the resolution of selected template was 3.1 Å (Kirchdoerfer & Ward, 2019). In addition homology modelling was also done by SWISS-MODEL. On analysis from the Swiss Model database we found large number of models with different sequence identity, the model which exhibited a very high sequence identity (96.35%) to the template sequence of PDB ID: 6NUR of NSP12 of COVID-19 was selected. All other models with sequence identity that scored less than 96.35% were eliminated from the study. The predicted 3 D structure of the target protein is represented in Figure 3. The structure assessment was performed by analysing the secondary structure of RdRp 3 D protein using PSIPRED 4.0 online web tool (Predict Secondary Structure) (Jones, 1999). The results predicted large number of beta sheets, alpha helix and turns further suggesting that modelled structure were accurate (Figure 4).
Figure 3.
Representative image indicating 3 D modelled structure of target protein (POL/RdRp) generated by Swiss-Model Database.
Figure 4.
Representative secondary structure of target protein RdRp displaying helix strand, coils and membrane interactions. Yellow colour showing beta strands, pink colour showing helix and gray colour showing coils.
3.3. Assessment of model quality and analysis of stereo-chemical properties
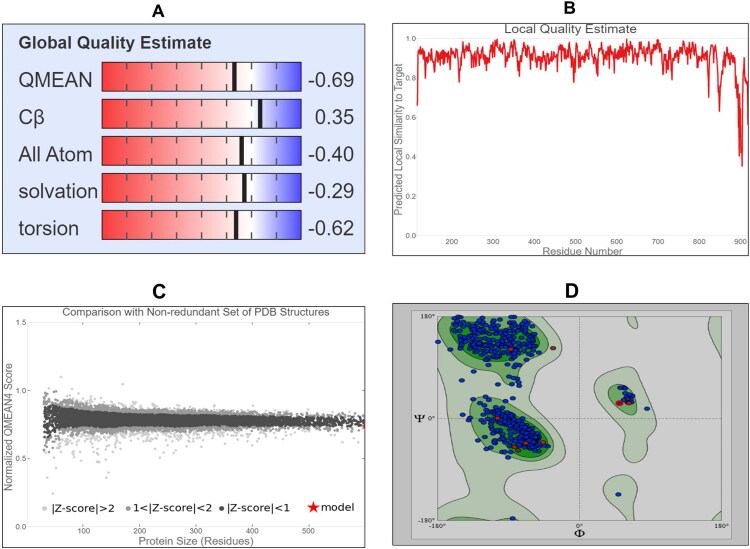
After the generation of 3 D structure of target protein, next we tested the quality of the model of the target protein and its stereo-chemical properties using SWISS MODEL online program. We observed that target model protein with clash score 0.24, good Global Model Quality Estimation (GMQE) score 0.83 and Qualitative Model Energy Analysis (QMEAN) score was −0.69 which guarantees the best model quality and also Z score was estimated (Figure 5A–C). To know the active binding site(s) of ‘target’ protein structure, we first did the validation of the 3 D structure by Ramachandran plot and we found 97.50% residues were in the favoured region, MolProbity score was 0.69 (Figure 5D). In addition we found that there were no observations of any Bad bonds, Cβ deviations were 11, and BAD angles observed were 62 out of 8994 angles, and thus validates the quality of the 3 D structure of the target protein (Table 1).
Figure 5.
Representative image indicating (A) Global Quality Estimation (B) Local Quality Estimation (C) Z-score of 3 D structure of RdRp (D) Ramachandran plot retrieved from SWISS-MODEL database of target protein showing 97.50% of the residues in the favoured region with good validation score, proving the validity of the model.
Table 1.
Table indicating stereo-chemical properties of modelled 3 D structure of RdRp of COVID-19.
| Properties | RdRp from COVID-19 |
|---|---|
| Residues score | |
| MolProbity score | 0.69 |
| Clash score | 0.24 |
| Ramachandran favoured | 97.50% |
| No of residues in favoured region | 786 |
| No of residues in allowed region | 15 |
| Ramachandran outliers | 0.0% |
| Rotamer outliers | 0.28% |
| C-Beta deviations | 11 |
| Bad bonds | 0/6624 |
| Bad angles | 62/8994 |
| Cis prolines | 1/26 |
| QMEAN | −0.69 |
| GMQUE | 0.83 |
| Sequence identity | 96.35% |
| Template name | 6nur |
| E value | 0.0 |
3.4. Model Optimization and superimposition
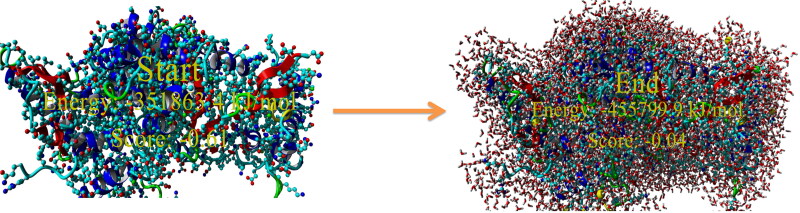
The model structure was then optimized by energy minimization through YASARA Energy Minimization Server (Krieger et al., 2009). The structure validation Z-scores and force field energies before and after the minimization were calculated through this tool. The energies of the structure before and after optimization were −351863.4 and −455799.9 kJ/mol respectively. This specifies that the protein structure was stabilized by an amount of −103,936.5 kJ/mol. Moreover, the structure validation Z-scores before and after optimization was −0.61 and −0.04 indicating good optimization of the target protein. The structure of RdRp after energy minimization is presented in Figure 6. Superimposition of RdRp model and template was done using FATCAT online web tool. p-value is another estimator utilized to assess structural similarity in the FATCAT-flexible method. According to the FATCAT-flexible method, a P-value < 0.05 means that the two structures compared are significantly similar. These two structures shown in Figure 6 are significantly similar with p-value of 0.00e + 00 (raw FATCAT score was 2346.50). The Root Mean Square Deviation (RMSD) is the measure of the average distance between two aligned proteins (Kufareva & Abagyan, 2012). In general, smaller RMSD values are accompanied with protein structure pairs with greater similarity (Carugo & Pongor, 2001). These two structures have 793 equivalent positions with an RMSD of 0.07Å without twists. Differential distance matrix decomposition distance range from −0.5 to 0.6 Å and Eigen value distribution range from −26.2 to 19.6 (Figure 7A–C). These algorithms further confirmed that the target model that we generated is the best quality model.
Figure 6.
Representative figure displaying energy minimization of modelled structure through YASARA web server.
Figure 7.
Superimposition of RdRp target protein and template (A) Representative comparative FATCAT image between RdRp protein and template. Horizontal axis represent target protein PDB and vertical axis indicate template PDB (B) Image displaying differential distance matrix decomposition distance range for both template and target protein with values ranging from −0.5–0.6 Å (C) Eigen value distribution curve for both template and target protein ranging from −26.2–19.6.
3.5. Reliable active sites prediction
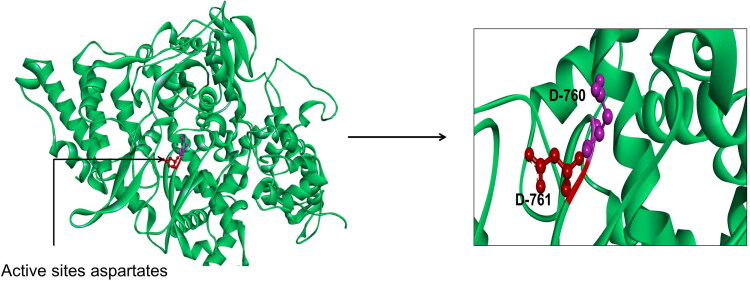
After the validation of this predicted 3 D structure of target protein, next we were concerned in finding the ligand binding site residues in Pol/RdRp protein using PrankWeb online tool (Jendele et al., 2019). On analysis of the 3 D structure of RdRp target protein of COVID-19, we found there were top three active sites (meta-pockets) considered as ligand binding sites in this protein. We observed aspartate active site residues in all the Metapockets (Table 2). Since these three meta-pocket(s) were found to contain aspartate active site residues, they were then studied for further analysis. The aspartate active site residues and most of the 5 Å region surrounding it are surface accessible, though can bind to the free nucleotides (ATP, GTP, and UTP). The active site aspartates are protruding from the beta-turn joining the β15 and β16. The active site is surface accessible as we can see from the surface representation allowing the interaction with the free nucleotides passing through the nucleotide channel of the RdRp (Elfiky, 2020). The 3 D structure showing the position of aspartate active sites (D-760 and D-761) is represented in Figure 8.
Table 2.
Table indicating amino acid positions of active sites within these meta-pockets. Blue colour residues indicate aspartate active site residues which are surface accessible. Metapockets were analysed using Prank Web online tool (http://prankweb.cz/).
| Metapockets region | Amino acids name and position |
|---|---|
| Metapocket region −1 | L-270, L-271, K-272, Y-273, V-315, T-319, P-323, T-324, S-325, F-326, G-327, P-328, L-329,V-330, Y-346, R-349, E-350, D-378, P-379, A-382, A-383, A-394, C-395, F-396, V-398,Y-456, R-457, N-459, L-460, P-461, D-618, Y-619, C-622, N-628, S-664, M-666, V-675, K-676, P-677, N-691, N-695, M-756, I-757, L-758, S-759, D-760, D-761, A-762, V-763, E-811, F-812, C-813, S-814 |
| Metapocket region −2 | D-452, Y-455, Y-456, Q-541, M-542, K-551, R-553, A-554, R-555, T-556, A-558, D-618, Y-619, P-620, K-621, C-622, D-623, R-624, E-665, V-666, K-676, T-680, S-681, S-682, L-758, S-759, D-761, K-798 |
| Metapocket region −3 | A-125, V-128, Y-129, R-132, H-133, V-204, T-206, L-207, D-208, S-236, L-240, D-465, Q-468, V-675, N-691, N-705, L-708, S-709, Y-728, Y-732, C-813 |
Figure 8.
Representative figure displaying of newly emerged COVID-19 RdRp model built by Swiss Model in the green cartoon representation. The active site aspartates are represented in red and purple sticks for clarification (see the enlarged panel).
3.6. Ligand molecules preparation and lipinski rule of five properties
After finalizing the meta-pocket sites on the basis of important amino acid residues, a total of 12 compounds were selected, out of which five were FDA approved antiviral drugs: Adefovir, Amantadine, Oseltamivir, Ribavirin and Zanamivir and seven phyto-chemical compounds from natural plants were Curcumin, Demethoxycurcumin, Flavonoid, Isoflavone, Terpinen-4-ol, Nimbin and Piperine. Table 3 summarizes 7 major phyto-compounds with their pharmacological properties.
Table 3.
List of seven major phyto-compounds with pharmacological properties.
| Compound name | Activities | References | Plant source |
|---|---|---|---|
| Curcumin | Antiviral, Antifungal, Antibacterial, Antimicrobial |
Moghadamtousi et al. (2014) | Turmeric (Curcuma longa) |
| Demethoxycurcumin | Antiviral, Antitumor, Antifungal, Antibacterial, Antidiabetic, Antimicrobial |
Agrawal & Goel (2016) | Turmeric (Curcuma longa) |
| Flavonoid | Antiviral Effect of Flavonoids on Human Viruses Flavonoids: Promising Natural Compounds Against Viral Infections |
Kaul et al. (1985), Zakaryan et al. (2017) |
Parsley (Petroselinum crispum) |
| Isoflavone | Shown to inhibit the infectivity of enveloped or non-enveloped viruses, as well as single-stranded or double-stranded RNA or DNA viruses | Andres et al. (2009) | Red clover (Trifoliumpratense L.) |
| Terpinen-4-ol | Have an inhibitory effect on influenza virus replication. | Garozzo et al. (2011) | Tea tree (Melaleucaalternifolia ) |
| Nimbin | The antiviral and virucidal effect of methanolic extract fraction of leaves of neem | Badam et al. (1999) | Neem (Azadirachtaindica) |
| Piperine | Health beneficial and disease preventing properties, like anti-inflammatory, antiviral, antipyretic, immune and bioavailability enhancing qualities |
Mair et al. (2016) | Black Pepper (Piper nigrum) |
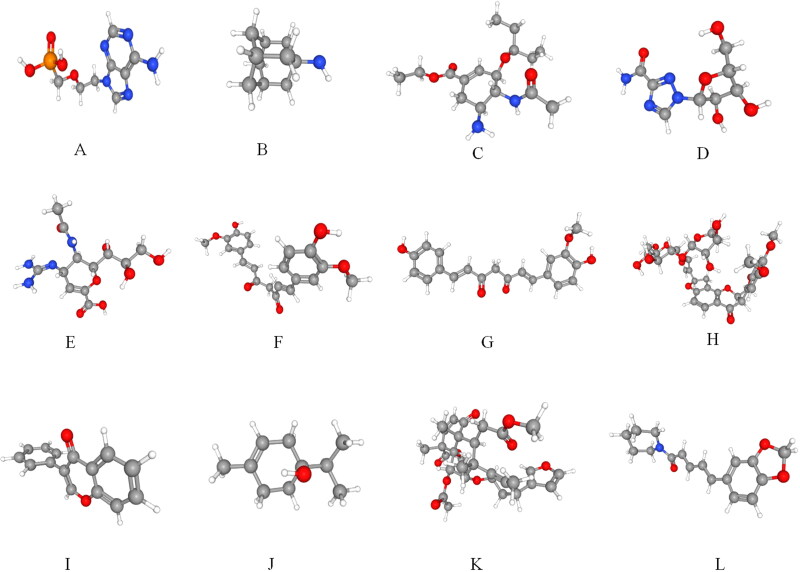
All these compounds were used as ligands for docking with the selected COVID-19 target protein. The 3 D structures of all the 12 compounds were downloaded from the NCBI PubChem Database (Figure 9A,L). These 12 compounds were then subjected to Lipinski rule of five to evaluate drug-likeness (Lipinski, 2004) and we found that all the 12 selected compounds followed the Lipinski rule of five suggesting their role as a drug compounds (Table 4).
Figure 9.
A–L: Structural and chemical properties of antiviral and Phyto Drugs: Representative 3 D structure of FDA approved antiviral drugs A) Adefovir B) Amantadine C) Oseltamivir D) Ribavirin E) Zanamivir. 3 D structure of natural drug molecules F) Curcumin G) Piperine H) Terpinen-4-ol I) Nimbin J) Isoflavone K) Demethoxycurcumin L) Flavonoid. SDF structure retrieved from PUBCHEM database was converted to PDB structure using Discovery-studio visualizer free software.
Table 4.
Table specifying selected compounds follow Lipinski rule of five. Chemical properties were predicted by online web tool (www.scfbio-iitd.res.in) Lipinski Rule of five.
| Sr. no. | Compound CID | Compound name | Molecular mass | LOgP | Hydrogen bond donors | Hydrogen bond acceptors | Molar refractivity |
|---|---|---|---|---|---|---|---|
| Natural drugs | |||||||
| 1 | 969516 | Curcumin | 368 | 3.36 | 2 | 6 | 102.0165 |
| 2 | 5469424 | Demethoxycurcumin | 338 | 3.361 | 2 | 5 | 95.4645 |
| 3 | 122792 | Flavonoid | 360 | 2.6254 | 3 | 8 | 90.376 |
| 4 | 5281797 | Isoflavone | 208 | 3.6195 | 0 | 1 | 66.2499 |
| 5 | 11230 | Terpinen-4-ol | 154 | 2.5 | 1 | 1 | 47.39 |
| 6 | 108058 | Nimbin | 540 | 3.92 | 0 | 9 | 137 |
| 7 | 638024 | Piperine | 285 | 2.99 | 0 | 4 | 81.1 |
| FDA approved drugs | |||||||
| 1 | 60172 | Adefovir | 273 | −0.43 | 4 | 8 | 62.5 |
| 2 | 2130 | Amantadine | 151 | 1.91 | 2 | 1 | 45.09 |
| 3 | 65028 | Oseltamivir | 312 | 1.28 | 3 | 6 | 84.15 |
| 4 | 37542 | Ribavirin | 244 | −3.0 | 5 | 8 | 51.5 |
| 5 | 60855 | Zanamavir | 332 | −3.78 | 9 | 11 | 76.89 |
3.7. Molecular docking studies
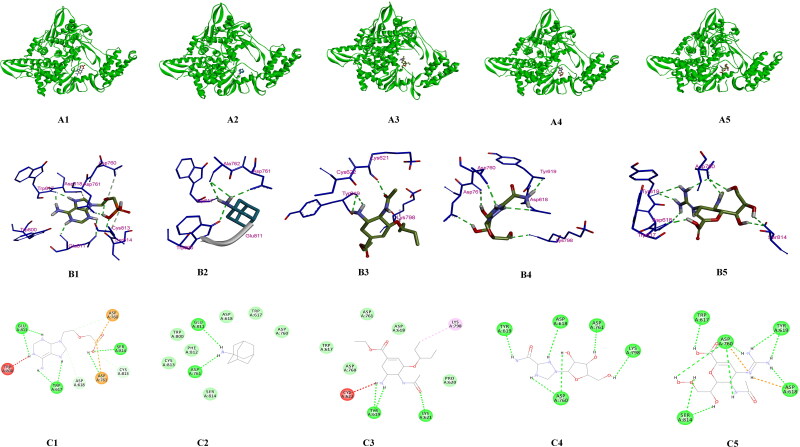
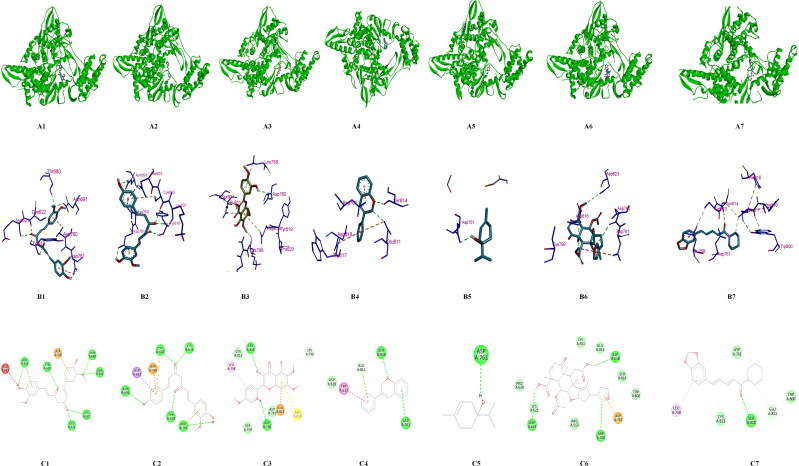
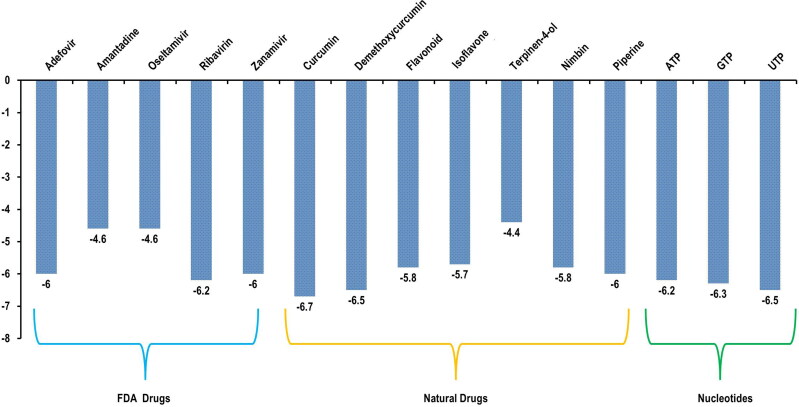
These 12 drug compounds and three control nucleotides (ATP, GTP and UTP) were then checked for their binding with target protein and docking score was evaluated using AutoDock vina PyRx software (Trott & Olson, 2010). The target receptor file and all ligand files were prepared to AutoDock Vina for the docking with individual drug molecules. A grid map was set grid center X: 141.1104, Y: 142.3415, Z: 147.8521 and number of points X: 93, Y: 70, Z: 70, spacing was 0.3750 Angstrom. For each compound, many docking poses were obtained with the target protein. The binding interaction(s) of all the docked 3 D and 2 D poses of receptor target protein of COVID-19 with various antiviral drugs is represented in Figure 10A–C. Similarly interaction of phytochemicals is represented in 3 D and 2 D poses in Figure 11A–C. After docking process was complete the results were further analysed for binding energy (ΔG), interacting residues and number of H-bonds (Table 5). The complex structure of antiviral and phyto drugs with target protein as a 3 D cartoon is represented in Figure 10A1–A5 and Figure 11A1–A7 respectively. Various hydrogen bond interactions in 3 D form shown by the antiviral drug and phto drug molecules are represented in Figure 10B1–B5 and Figure 11B1–B7 respectively. Similarly number of residue interactions and different bond interactions of all antiviral drug molecules and Phyto-drug molecules in 2 D form is shown in Figure 10C1–C5 and Figure 11C1 to C7 respectively. Out of many docking poses, only those were selected which had the minimum binding energy, good hydrogen bond interaction and which interacted with aspartate active sites. From our results we found that three FDA approved antiviral drugs including Zanamivir, Ribavirin and Adefovir molecules and three Phyto-drug molecule comprising Curcumin, Piperine and Demethoxycurcumin showed minimum binding energy, maximum hydrogen bond interactions and binding with aspartate active residues with the target protein (RdRp target protein of COVID-19) which is shown in Table 5. As aspartate active site residues are surface accessible residue which allows the interaction with free nucleotides passing through the nucleotide channel of RdRp protein. We have also docked these three nucleotides (ATP, GTP and UTP) as a control drug which also showed the minimum binding energy. Graphical representation and comparison of all drugs and control molecules are shown in Figure 12.
Figure 10.
A-C: Molecular Modelling and docking studies of antiviral with pol/RdRp protein of COVID-19: Representative 3 D structure of complex docked target protein molecule (POL/RdRp) with antiviral drugs A1) Protein + Adefovir, A2) Protein + Amantadine, A3) Protein + Oseltamivir, A4) Protein + Ribavirin, A5) Protein + Zanamivir (A1-A5). Representative bonding interaction(s) between drug molecules and target protein receptor B1) Protein + Adefovir, B2) Protein + Amantadine, B3) Protein + Oseltamivir, B4) Protein + Ribavirin, B5) Protein + Zanamivir (B1-B5). Representation of 2 D complex structure of antiviral drugs and target protein presenting different types of bonds interacting with number of amino acid residues C1) Protein + Adefovir, C2) Protein + Amantadine, C3) Protein + Oseltamivir, C4) Protein + Ribavirin, C5) Protein + Zanamivir (C1-C5).
Figure 11.
A–C: Molecular Modelling and docking studies of antiviral with pol/RdRp protein of COVID-19: Representative 3 D structure of complex docked target protein molecules with Phyto Drugs A1) Protein + Curcumin, A2) Protein + Demethoxycurcumin, A3) Protein + Flavonoid, A4) Protein + Isoflavone, A5) Protein + Terpinen-4-ol A6) Protein + Nimbin, A7) Protein + Piperine (B1-B2). Representative bonding interaction(s) between Phyto-Drug molecules and target protein receptor B1) Protein + Curcumin, B2) Protein + Demethoxycurcumin B3) Protein + Flavonoid, B4) Protein + Isoflavone, B5) Protein + Terpinen-4-ol, B6) Protein + Nimbin, B7) Protein + Piperine (C1-C2). Representation of 2 D complex structure of Phyto-Drugs and target protein presenting different types of bonds interacting with number of amino acid residues C1) Protein + Curcumin, C2) Protein + Demethoxycurcumin, C3) Protein + Flavonoid, C4) Protein + Isoflavone, C5) Protein + Terpinen-4-ol, F6) Protein + Nimbin, C7) Protein + Piperine. All the 2 D and 3Dstructures were generated from the Discovery studio visualize software and PYMOL visualize.
Table 5.
Interacting amino acids and docking score of antiviral drugs and Phyto-Drugs molecules using molecular docking software (AutoDock vina).
| Name of compound | Binding energy Kcal/mol (ΔG) |
Interacting residues | No of H bonds |
|---|---|---|---|
| FDA approved drugs | |||
| Adefovir | −6.0 | ASP760, ASP761, SER814, CYS813, ASP618, TRP617, GLU811, TRP800 | 9 |
| Amantadine | −4.6 | ASP761, GLU811, TRP800, ALA762 | 4 |
| Oseltamivir | −4.6 | TYR619, LYS621, CYS622, LYS798 | 3 |
| Ribavirin | −6.2 | ASP760, ASP761, ASP618, TYR619, LYS798 | 6 |
| Zanamivir | −6.0 | SER814, ASP760, TRP617, TYR619, ASP618 | 9 |
| Natural drugs | |||
| Curcumin | −6.7 | ASP761, TYR619, GLU811, ASP760, THR680, ASN691, LYS621, CYS622 | 6 |
| Demethoxycurcumin | −6.5 | ASN691, ASP623, ASP760, CYS622, LYS621, TYR619, ASP761 | 5 |
| Flavonoid | −5.8 | ASP760, SER814, LYS798, GLU811, ASP618, LEU758 | 4 |
| Isoflavone | −5.7 | ASP761, SER814, GLU811, TRP617 | 3 |
| Terpinen-4-ol | −4.4 | ASP761 | 1 |
| Nimbin | −5.8 | ASP623, ASP760, ASP761, ASP618 | 3 |
| Piperine | −6.0 | LEU758, SER814, GLU811, CYS813 | 2 |
Figure 12.
Graphical representation of binding energy calculated by AutoDock vina for FDA approved antiviral drugs, phyto-drug molecules and nucleotides (ATP, GTP and UTP) from minimum to maximum energy score in kcal/mol.
3.8. Pharmacokinetics properties predictions
After the confirmation of the interaction of these drugs with target protein RdRp of COVID-19, next the aim of the study was to scrutinize the pharmacokinetics for absorption, distribution, metabolism, and excretion (ADME) properties of all these drug compounds using FAF Drugs3 (Lagorce et al., 2015) and admetSAR (Cheng et al., 2012) server. ADMET stands for Absorption, Distribution, Metabolism, Excretion and Toxicity. The prediction of the ADMET properties plays an important role in the drug design process because these properties account for the failure of about 60% of all drugs in the clinical phases. We also analysed these drugs for oral toxicity (LD50) properties by using PROTOX web server (Drwal et al., 2014). We found that Adefovir, Amantadine, Oseltamivir, Ribavirin and Zanamivir have LD-50 values 13 mg/kg, 157 mg/kg, 260 mg/kg, 2700 mg/kg and 5000 mg/kg respectively and the phyto-drugs including Curcumin, Demethoxycurcumin, Isoflavone, Flavonoid, Terpinen-4-ol, Nimbin and Piperine have LD-50 values 2000 mg/kg, 2000 mg/kg, 500 mg/kg, 5000 mg/kg, 1016 mg/kg, 1000 mg/kg and 350 mg/kg. ADMET properties and toxicity calculation of drug molecules which interacted with the target protein of COVID-19 are shown in Tables 6 and 7. Our results showed that all the antiviral drug molecules and phyto-compounds we selected have promising and good ADMET properties and also showed high quality QSAR models.
Table 6.
Pharmacokinetic ADMET properties and oral toxicity studies of FDA approved antiviral drug molecules.
| Compound name/ Properties |
Adefovir | Amantadine | Oseltamivir | Ribavirin | Zanamivir |
|---|---|---|---|---|---|
| LD50 | 13 mg/kg | 157 mg/kg | 260 mg/kg | 2700 mg/kg | 5000 mg/kg |
| Human ether-a-go-go-related gene | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| Inhibition | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| AMES toxicity | Non AMES toxic | Non AMES toxic | AMES toxic | Non AMES toxic | Non AMES toxic |
| Carcinogens | Non-carcinogens | Non- carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens |
| Fish toxicity | High FHMT | High FHMT | High FHMT | High FHMT | Low FHMT |
| TetrahymenaPyriformis toxicity | High TPT | High TPT | High TPT | High TPT | High TPT |
| Honey bee toxicity | High HBT | High HBT | High HBT | High HBT | High HBT |
| Biodegradation | Not ready biodegradable | Not ready biodegradable | Ready biodegradable | Not ready biodegradable | ready biodegradable |
| Acute oral toxicity | III | III | III | III | III |
Table 7.
Pharmacokinetic ADMET properties and oral toxicity studies of phyto-drugs.
| Compound name/ Properties |
Curcumin | Demethoxycur-cumin | Isoflavone | Flavonoid | Terpinen-4-ol | Nimbin | Piperine |
|---|---|---|---|---|---|---|---|
| LD50 | 2000 mg/kg | 2000 mg/kg | 500 mg/kg | 5000 mg/kg | 1016 mg/kg | 1000 mg/kg | 350 mg/kg |
| Human ether-a-go-go- related gene | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| Inhibition | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| AMES toxicity | Non AMES toxic | Non AMES toxic | AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic |
| Carcinogens | Non-carcinogens | Non- carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens |
| Fish toxicity | High FHMT | High FHMT | High FHMT | High FHMT | High FHMT | Low FHMT | High FHMT |
| TetrahymenaPyriformis toxicity | High TPT | High TPT | High TPT | High TPT | High TPT | High TPT | High TPT |
| Honey bee toxicity | High HBT | High HBT | High HBT | High HBT | High HBT | High HBT | High HBT |
| Bio-degradation | Not ready bio-degradable | Not ready bio-degradable | Ready bio-degradable | Not ready bio-degradable | Not ready bio-degradable | Not ready bio-degradable | Ready bio-degradable |
| Acute oral toxicity | III | III | III | III | III | III | III |
3.9. Molecular dynamics studies
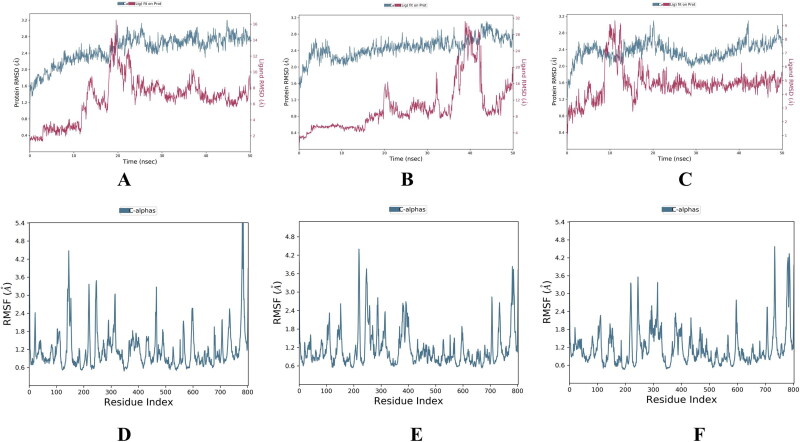
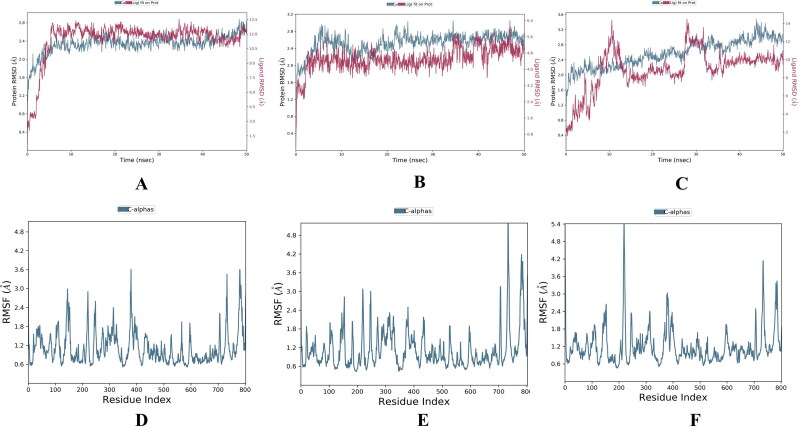
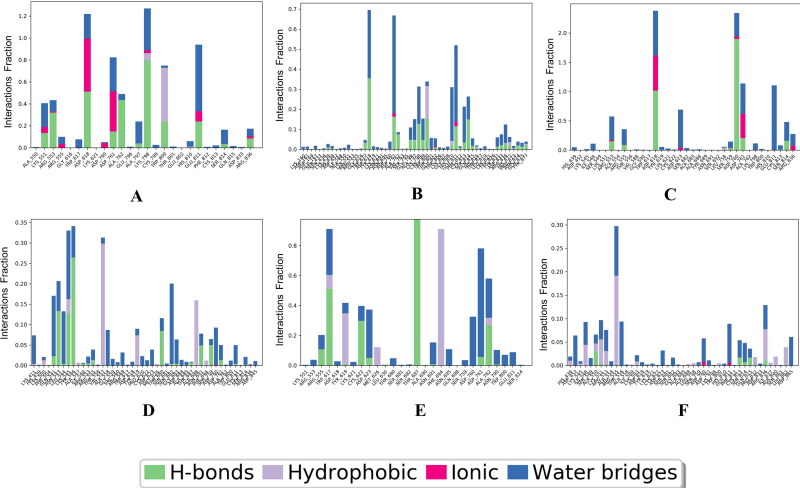
We performed molecular dynamic simulation experiment at 50 ns of all the selected 6 (3 were FDA approved & 3 were phyto-drug molecules) ligand which showed good docking score in virtual screening. In our study of 50 ns MD simulation study we found that the average RMSD and RMSF value of all top drug molecules Adefovir, Ribavirin, Zanamivir, Curcumin, Demethoxycurcumin and Piperine was found to be (RMSD) 3.2 Å, 3.0 Å, 3.1 Å, 2.8 Å, 3.0 Å, 3.2 Å and (RMSF) 0.8 Å, 0.6 Å, 0.8 Å, 0.6 Å, 0.8 Å, 0.8 Å. (Figures 13–14 and Table 8). When comparing the RMSF value of the all the drug molecules we could find that higher fluctuations were noted and details of interactions of these ligands with the target protein are showed in Figure 15.
Figure 13.
RMSD and RMSF of FDA approved molecules bound forms of selected molecules. Adefovir, Ribavirin, Zanamivir; (A-C) RMSD value and (D-F) RMSF values of these molecules.
Figure 14.
RMSD and RMSF of Phyto-drug molecules bound forms of selected molecules. Curcumin, Demethoxycurcumin and Piperine; (A-C) RMSD value and (D-F) RMSF values of these molecules.
Table 8.
Molecular dynamics studies of all selected molecules showing RMSD, RMSF and interacting residues.
| Ligands | Simulation times (ns) | RMSD | RMSF | Residues interaction |
|---|---|---|---|---|
| Adefovir | 50 ns | 3.2 Å | 0.8 Å | Arg-553,Asp-618,Asp-761,Ala-762,Lys-798,Glu-811,Arg-836 |
| Ribavirin | 50 ns | 3.0 Å | 0.6 Å | Asp-618,Asp-761,lys-798,Glu-811,Ser-814 |
| Zanamivir | 50 ns | 3.1 Å | 0.8 Å | Asp-618,Asp-760,Asp-761,Ser-814 |
| Curcumin | 50 ns | 2.8 Å | 0.6 Å | Asn-543,Tyr-546,Thr-680,As-691,Asp-761 |
| Demethoxycurcumin | 50 ns | 3.0 Å | 0.8 Å | Arg-555,Cys-622,Thr-687,Asp-761,Ala-762 |
| Piperine | 50 ns | 3.2 Å | 0.8 Å | Ala-550,Asp-760,Cys-813,Ser-814,Gln-815 |
Figure 15.
Details of protein Ligand contact profile of selected ligands A) Adefovir, B) Ribavirin, C) Zanamivir, and D) Curcumin, E) Demethoxycurcumin, F) Piperine.
4. Discussion
Molecular docking studies explored the binding of FDA approved antiviral drugs: Adefovir, Amantadine, Oseltamivir, Ribavirin and Zanamivir and seven phyto-chemical compounds from natural plants Curcumin, Demethoxycurcumin, Flavonoid, Isoflavone, Terpinen-4-ol, Nimbin and Piperine and three control molecules (ATP, GTP and UTP) for interaction with target protein (Pol/RdRp) of SARS coronavirus 2 (COVID-19). The molecular docking study showed that selected three out of five FDA approved drug molecules (Zanamivir, Ribavirin and Adefovir) and three out of seven different phyto-compounds (Curcumin, Piperine and Demethoxycurcumin) and control molecules specifically bind to RdRp protein of COVID-19 showing minimum binding energy which attests to the good binding affinity of these drugs to the target protein RdRp of COVID-19. Three FDA approved antiviral drugs including Zanamivir, Ribavirin and Adefovir molecules showed the minimum binding energy (ΔG/kcal/mol: −6.0, −6.2, −6.0 respectively) and three Phyto-drug molecule comprising of Curcumin, Demethoxycurcumin and Piperine have the minimum binding energy (ΔG/kcal/mol = −6.7, −6.5, −6.0 respectively) (Table 5). In addition, all these shortlisted drugs have also been found to interact with aspartate active site residues. Similar to our study, Elfiky et al. (2020) showed the five approved drugs i.e. Galidesivir, Remdesivir, Tenofovir, Sofosbuvir, and Ribavirin also have the ability to bind the SARS-CoV-2 RdRp, with binding energies of −7.0, −7.6, −6.9, −7.5, and −7.8 kcal/mol, respectively. Huang et al., 2020 also showed many herbal compounds used in traditional Chinese medicine therapy have been screened and found to have free binding affinity against RdRp protein of SARSCoV-2. In another study Pandit & Latha, 2020 reported the binding affinity of phyto-compounds derived from Silybum marianum (Silybin), Withania somnifera (Withaferin A), Tinospora cordifolia (Cordioside) and Aloe barbadensis (Catechin and Quercetin) with SARS-CoV-2 target. The binding energy of phyto-compounds was more than the widely used for hydroxychloroquine and other repurposed drugs used for treatment of COVID–19 infection. Tatar and Turhan (2020) reported the binding affinity of nafamostat, rapamycin, saracatinib, Imatinib and Camostat with binding energy −10.24, −9.28, −9.66, −9.23 and 9.07 respectively with N-Terminal domain of SARS-CoV-2 Nucleocapsid. Gupta et al. (2020) and Muralidharan et al. (2020) have also done similar studies. Ubani et al. (2020) reported best binding affinity of scopodulcic acid and 249 Dammarenolic acid with the Spike glycoprotein (6VSB) and the Mpro 250 (6FLU7) respectively.
It is fascinating to find that three FDA approved antiviral drugs including Zanamivir, Ribavirin and Adefovir molecules and three Phyto-drug molecule comprising Piperin, Curcumin and Demethoxycurcumin showed minimum binding energy and hydrogen bond interactions alongwith interaction with aspartate active site with the target protein (RdRp target protein of COVID-19 (Table 5). Interestingly, we observed that LD50 of Zanamivir and Ribavirin was 5000 mg/kg and 2700 mg/kg respectively signifying that these compounds are less toxic. Likewise Curcumin and Demethoxycurcumin compound are also found to be non-toxic with high LD50 value (LD50 > 1000 indicates that drug molecules are non-toxic while LD50 < 1000 suggests drugs are highly toxic). Furthermore, all the three FDA approved antiviral drugs and three Phyto-drug molecules fulfill all the criteria of Lipinski’s rule of five and ADME/T. Similar to our study, Ahmed and Shohael, (2019) reported that the chrysophanol, aloe-emodin, rhein and emodin all fulfil the criteria of Lipinski’s rule of five and ADME/T. Collectively the data and our study suggests that these selected compounds i.e. Zanamivir, Ribavirin, Adefovir and Curcumin, demehoxycurcumin, Piperine with minimum binding energy with the target protein may act as potential candidates for the design of novel drugs for the treatment of COVID-19.
5. Conclusion
In this study we studied twelve drugs like compounds five from available antiviral drugs and seven from natural chemical compounds. The three best FDA approved antiviral drugs including Zanamivir, Ribavirin and Adefovir used for other viral diseases and three phyto-drug like compounds like Curcumin, Demethoxycurcumin and Piperine could specifically bind to RdRp protein of COVID-19 showing minimum binding energy which attests to the good binding affinity of these drugs to the target protein. On the basis of molecular docking energy score of all selected three FDA approved drug molecules and three phyto-drug molecules from different plants suggest that it may act as best or novel inhibitor that may be used for the treatment of severe acute respiratory syndrome coronavirus 2. We need in-vitro proof of concept studies of these identified agents to find out a potential inhibitor of SARS-2. Hence this information may be useful for the designing of the novel drug molecules against COVID-19.
Funding Statement
No funding received as it is an insilico study.
Disclosure statement
There is no conflict of interest.
References
- Agrawal, S., & Goel, R. K. (2016). Curcumin and its protective and therapeutic uses. National Journal of Physiology, Pharmacy and Pharmacology, 6(1), 1–8. [Google Scholar]
- Ahlquist, P., Noueiry, A. O., Lee, W. M., Kushner, D. B., & Dye, B. T. (2003). Host factors in positive-strand RNA virus genome replication. Journal of Virology, 77(15), 8181–8186. 10.1128/jvi.77.15.8181-8186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S., & Shohael, A. M. (2019). In silico studies of four anthraquinones of Sennaalata l. As potential antifungal compounds. Pharmacology Online, 2, 259–268. [Google Scholar]
- Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., & Garry, R. F. (2020). Correspondence: The proximal origin of SARS-CoV-2. Nature Medicine, 26 (4), 450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres, A., Donovan, S. M., & Kuhlenschmidt, M. S. (2009). Soy isoflavones and virus infections. The Journal of Nutritional Biochemistry, 20(8), 563–569. 10.1016/j.jnutbio.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badam, L., Joshi, S. P., & Bedekar, S. S. (1999). In vitro' antiviral activity of neem (Azadirachtaindica. A. Juss) leaf extract against group B coxsackieviruses. The Journal of Communicable Diseases, 31 (2), 79–90. [PubMed] [Google Scholar]
- Batlle, D., Wysocki, J., & Satchell, K. (2020). Soluble angiotensin-converting enzyme 2: A potential approach for coronavirus infection therapy. Clinical Science, 134(5), 543–545. 10.1042/CS20200163 [DOI] [PubMed] [Google Scholar]
- Benvenuto, D., Giovanetti, M., Ciccozzi, A., Spoto, S., Angeletti, S., & Ciccozzi, M. (2020). The 2019-new coronavirus epidemic: Evidence for virus evolution. Journal of Medical Virology, 92 (4), 455–459. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo, O., & Pongor, S. (2001). A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Science, 10(7), 1470–1473. 10.1110/ps.690101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. (2020). Understanding the nature of variations in structural sequences coding for coronavirus spike, envelope, membrane and nucleocapsid proteins of SARS-CoV-2. Envelope, membrane and nucleocapsid proteins of SARS-CoV-2. https://ssrn.com/abstract=3562504
- Chen, L., & Hao, G. (2020). The role of angiotensin converting enzyme 2 in coronaviruses/influenza viruses and cardiovascular disease. Cardiovasc Research, 116(12), 1932-1936. 10.1093/cvr/cvaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., Yang, Q., Wang, J., Liu, Y., Wei, Y., Xia, J., Yu, T., Zhang, X., & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395 (10223), 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, F., Li, W., Zhou, Y., Shen, J., Wu, Z., Liu, G., Lee, P. W., & Tang, Y. (2012). admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. Journal of Chemical Information and Modeling, 52(11), 3099–3105. 10.1021/ci300367a [DOI] [PubMed] [Google Scholar]
- Chu, H., Chan, J. F. W., Wang, Y., Yuen, T. T. T., Chai, Y., Hou, Y., Shuai, H., Yang, D., Hu, B., Huang, X., Zhang, X., Cai, J. P., Zhou, J., Yuan, S., Kok, K. H., Wang, K. K., Chan, I. H. E., Yee, Zhang, A. J., … Yuen, K. Y. (2020). Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: An ex vivo study with implications for the pathogenesis of COVID-19. Clinical Infectious Diseases, 71(6), 1400–1409. 10.1093/cid/ciaa410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, P., Ronconi, G., Caraffa, A. L., Gallenga, C. E., Ross, R., Frydas, I., & Kritas, S. K. (2020). Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. Journal of Biological Regulators and Homeostatic Agents, 34, 1. [DOI] [PubMed] [Google Scholar]
- Cyranoski, D. (2020). Mystery deepens over animal source of coronavirus. Nature, 579 (7797), 18–19. 10.1038/d41586-020-00548-w [DOI] [PubMed] [Google Scholar]
- David, L., Olivier, S., Hervé, G., Maria, A. M., & Bruno, O. V. (2008). FAF-Drugs2: Free ADME/tox filtering tool to assist drug discovery and chemical biology projects. BMC Bioinformatics, 9, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drwal, M. N., Banerjee, P., Dunkel, M., Wettig, M. R., & Preissner, R. (2014). ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Research, 42(Web Server issue), W53–8. 10.1093/nar/gku401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9.doi: 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses, 1282, 1–23. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garozzo, A., Timpanaro, R., Stivala, A., Bisignano, G., & Castro, A. (2011). Activity of Melaleuca alternifolia (tea tree) oil on Influenza virus A/PR/8: study on the mechanism of action. Antiviral Research, 89(1), 83–88. 10.1016/j.antiviral.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 41, 95–98. [Google Scholar]
- Hasan, A., Paray, B. A., Hussain, A., Qadir, F. A., Attar, F., Aziz, F. M., Sharifi, M., Derakhshankhah, H., Rasti, B., & Mehrabi, M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 22, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F., Li, Y., Leung, E. L.-H., Liu, X., Liu, K., Wang, Q., Lan, Y., Li, X., Yu, H., Cui, L., Luo, H., & Luo, L. (2020). A review of therapeutic agents and Chinese herbal medicines against SARS-COV-2 (COVID-19). Pharmacological Research, 158, 104929. 10.1016/j.phrs.2020.104929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendele, L., Krivak, R., Skoda, P., Novotny, M., & Hoksza, D. (2019). PrankWeb: A web server for ligand binding site prediction and visualization. Nucleic Acids Research, 47(W1), W345–W349. 10.1093/nar/gkz424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology, 292(2), 195–202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- Kaul, T. N., Middleton, E., & Ogra, P. L. (1985). Antiviral effect of flavonoids on human viruses. Journal of Medical Virology, 15(1), 71–79. 10.1002/jmv.1890150110 [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer, R. N., & Ward, A. B. (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nature Communications, 10(1), 2342. 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, E., Joo, K., Lee, J., Lee, J., Raman, S., Thompson, J., Tyka, M., Baker, D., & Karplus, K. (2009). Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins: Structure, Function, and Bioinformatics, 77 (S9), 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva, I., & Abagyan, R. (2012). Methods of protein structure comparison. Methods in Molecular Biology, 857, 231–257. 10.1007/978-1-61779-588-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M. M., & Cavanagh, D. (1997). The molecular biology of coronaviruses. Advances in Virus Research, 48, 1–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagorce, D., Sperandio, O., Baell, J. B., Miteva, M. A., & Villoutreix, B. O. (2015). FAF-Drugs3: A web server for compound property calculation and chemical library design. Nucleic Acids Research, 43(W1), W200–W207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, A. M., Espiritu, C., Bansal, S., Steuer, H. M. M., Niu, C., Zennou, V., Keilman, M., Zhu, Y., Lan, S., Otto, M. J., & Furman, P. A. (2012). Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrobial Agents and Chemotherapy, 56(6), 3359–3368. 10.1128/AAC.00054-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today. Technologies, 1(4), 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Lukas, J., Radoslav, K., Petr, S., Marian, N., & David, H. (2019). Prank Web: A web server for ligand binding site prediction and visualization. Nucleic Acids Research, 47(W1), W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, C. E., Liu, R., Atanasov, A. G., Schmidtke, M., Dirsch, V. M., & Rollinger, J. M. (2016). Antiviral and anti-proliferative in vitro activities of piperamides from black pepper. Planta Medica, 81(S 01), S1–S381. [Google Scholar]
- Miller, W. A., & Koev, G. (2000). Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology, 273(1), 1–8. 10.1006/viro.2000.0421 [DOI] [PubMed] [Google Scholar]
- Moghadamtousi, S. Z., Kadir, H. A., Hassandarvish, P., Tajik, H., Abubakar, S., & Zandi, K. (2014). A Review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed Research International, 2014, 186864. 10.1155/2014/186864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Pandit, M., & Latha, N. (2020). In silico studies reveal potential antiviral activity of phytochemicals from medicinal plants for the treatment of COVID-19 infection. Structural Biology Bioinformatics. 10.21203/rs.3.rs-22687/v1 [DOI] [Google Scholar]
- Perlman, S. (2020). Another decade, another coronavirus. The New England Journal of Medicine, 382(8), 760–762. 10.1056/NEJMe2001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar, G., & Turhan, K. (2020). Investigation of N terminal domain of SARS CoV 2 nucleocapsid protein with antiviral compounds based on molecular modeling approach. ScienceOpen. 10.14293/S2199-1006.1.SOR-.PPPT99I.v1 [DOI] [Google Scholar]
- Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res, 22(22), 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubani, A., Agwom, F., Shehu, N. Y., Luka, P., Umera, A., Umar, U., Omale, S. N. E., & Aguiyi, J. C. (2020). Molecular docking analysis of some phytochemicals on two SARS-COV-2 targets. bioRxiv doi.org/https://doi.org/ 10.1101/2020.03.31.017657. [DOI]
- Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., & Barton, G. J. (2009). Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford, England)), 25(9), 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C., Huang, Y., Lau, S. K., & Yuen, K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses, 2(8), 1804–1820. 10.3390/v2081803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhen, Y., & Godzik, A. (2004). FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Research, 32(Web Server issue), W582–W585. 10.1093/nar/gkh430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan, H., Arabyan, E., Oo, A., & Zandi, K. (2017). Flavonoids: Promising natural compounds against viral infections. Archives of Virology, 162(9), 2539–2551. 10.1007/s00705-017-3417-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., Si, H., Zhu, Y., Li, B., Huang, C. L., Chen, H. D., Chen, J., Luo, Y., Guo, H., Jiang, R. D., Liu, M. Q., Chen, Y., Shen, X. R., Wang, … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]