Abstract
Background
Xenorhabdus and Photorhabdus are entomopathogenic bacteria that cause septicemia and toxemia in insects. They produce secondary metabolites to induce host immunosuppression. Their metabolite compositions vary among bacterial species. Little is known about the relationship between metabolite compositions and the bacterial pathogenicity. The objective of this study was to compare pathogenicity and production of secondary metabolites of 14 bacterial isolates (species or strains) of Xenorhabdus and Photorhabdus.
Results
All bacterial isolates exhibited insecticidal activities after hemocoelic injection to Spodoptera exigua (a lepidopteran insect) larvae, with median lethal doses ranging from 168.8 to 641.3 CFU per larva. Bacterial infection also led to immunosuppression by inhibiting eicosanoid biosynthesis. Bacterial culture broth was fractionated into four different organic extracts. All four organic extracts of each bacterial species exhibited insecticidal activities and resulted in immunosuppression. These organic extracts were subjected to GC-MS analysis which predicted 182 compounds, showing differential compositions for 14 bacteria isolates. There were positive correlations between total number of secondary metabolites produced by each bacterial culture broth and its bacterial pathogenicity based on immunosuppression and insecticidal activity. From these correlation results, 70 virulent compounds were selected from secondary metabolites of high virulent bacterial isolates by deducting those of low virulent bacterial isolates. These selected virulent compounds exhibited significant immunosuppressive activities by inhibiting eicosanoid biosynthesis. They also exhibited relatively high insecticidal activities.
Conclusion
Virulence variation between Xenorhabdus and Photorhabdus is determined by their different compositions of secondary metabolites, of which PLA2 inhibitors play a crucial role.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-020-02042-9.
Keywords: Xenorhabdus, Photorhabdus, Secondary metabolite, Pathogenicity, Eicosanoid, Immunity
Background
Xenorhabdus and Photorhabdus are entomopathogenic bacteria that exhibit mutualistic symbiosis with entomopathogenic nematodes Steinernema and Heterorhabditis, respectively [1]. These nematodes can carry and release these bacteria into insect hemocoel by infecting susceptible insect larvae, in which these bacteria can kill insects and convert the cadaver into a food source suitable for nematode growth and development [2]. When nematode population increases to a certain carrying level in a specific insect host, these nematodes are re-associated with specific symbiotic bacteria before emerging from the insect cadaver to search for a new host [3]. There are complex chemical communications in tripartite interactions of bacteria-nematode for symbiosis, bacteria-insect for pathogenicity, and nematode-insect for host recognition. Pathogenic interactions between bacteria and susceptible insects have been relatively well studied regarding the production of specialized metabolites derived from non-ribosomal peptide synthetase (NRPS) or polyketide synthase (PKS) [4]. However, there are secondary compounds other than NRPS-PKS produced from synthetic machineries of bacteria [5].
Virulence of entomopathogenic bacteria exhibits variations among species and strains [6, 7]. Bacteria can secrete several virulence factors in insect hemocoel to suppress insect immune responses and cause fatal septicemia [8, 9]. To induce immunosuppression, both bacterial genera commonly inhibit phospholipase A2 (PLA2) activity of insects [10]. PLA2 is known to catalyze the release of arachidonic acid from phospholipids, which is a committed step to produce various eicosanoids [11]. Eicosanoids mediate cellular and humoral immune responses against various microbial pathogens in insects [12]. Indeed, X. nematophila can secrete at least eight secondary metabolites to suppress insect immunity by inhibiting PLA2 [13]. Inter-specific variations in Xenorhabdus bacterial virulence have been explained by variations in their inhibitory activities against PLA2 activities [14]. Intra-specific variations in bacterial virulence of X. nematophila have also been reported and explained by differences in immunosuppression due to differential inhibitory effects on PLA2 catalytic activity [15]. Park et al. [16] have explained that different virulence due to phase variation is associated with the expression of a specific outer membrane protein. This discovery on virulence gene is further extended by the finding that the expression level of leucine-responsive protein (Lrp), a global transcriptional factor, can modulate bacterial pathogenicity [17]. Expression levels of Lrp and other transcriptional factors can modulate secondary metabolite production [18], suggesting that the production of secondary metabolites might be positively correlated with bacterial pathogenicity.
Secondary metabolites produced by NRPS and PKS of different species of Xenorhabdus and Photorhabdus are different in their compositions [19]. Different secondary metabolites can inhibit diverse physiological molecules of susceptible insects to induce immunosuppression. For example, rhabducin, an isocyanide-containing compound produced from biosynthetic gene cluster, can inhibit the activity of phenoloxidase (PO) in Galleria mellonella [20]. More than 70 kinds of rhabdopeptide/xenortide peptides derived from NRPS are structurally similar to protease inhibitors. They might degrade various proteins associated with immunity [21, 22]. Phurealipids produced from NRPS/PKS can prevent the expression of antimicrobial peptide genes [23]. Thus, diverse secondary metabolites produced by entomopathogenic bacteria might effectively suppress insect immune responses to induce septicemia.
The aim of this study was to determine virulent secondary metabolites produced by Xenorhabdus or Photorhabdus based on their inhibitory activities against insect PLA2. To this end, this study compared virulence and secondary metabolites of 14 different bacterial isolates of Xenorhabdus and Photorhabdus.
Results
Variations in bacterial virulence
Insecticidal activities of 14 bacterial isolates (species or strains) of Xenorhabdus and Photorhabdus were assessed by hemocoelic injection of freshly grown live bacteria into L5 larvae of S. exigua (Table 1). All bacterial treatments exhibited insecticidal activities. However, their insecticidal activities were different, with LD50 ranging from 168.8 (P. temperata temperata: ‘Ptt’) to 641.3 (X. ehlersii: ‘Xe’) CFU/larva.
Table 1.
Insecticidal activities of 14 entomopathogenic bacteria (EPB) of Xenorhabdus and Photorhabdus against L5 larvae of S. exigua. Larvae were hemocoelically injected with different doses of freshly cultured bacteria. Before injection, larvae were surface sterilized with 70% ethanol. For each test dose, 10 larvae were used with three replications
| EPBa | LD50 (cfu/larva) (95% CI) |
Slope ± SE | df | χ2 |
|---|---|---|---|---|
| Ptt | 168.8 (110.4–318.3) | 0.87 ± 0.27 | 1 | 0.817 |
| Xh | 179.2 (156.9–345.6) | 0.77 ± 0.29 | 2 | 0.953 |
| Xn F | 216.9 (114.6–406.8) | 0.82 ± 0.27 | 2 | 0.906 |
| Xb | 222.4 (120.3–432.7) | 0.77 ± 0.28 | 2 | 0.835 |
| Xn M | 244.3 (147.5–480.4) | 0.87 ± 0.26 | 2 | 0.959 |
| Pl 193 | 244.3 (128.6–487.3) | 0.87 ± 0.26 | 2 | 0.959 |
| Pl laum | 274.8 (150.3–512.6) | 0.74 ± 0.29 | 2 | 0.985 |
| Xn SK2 | 274.9 (151.4–523.2) | 0.74 ± 0.29 | 2 | 0.985 |
| Pl thra | 282.87 (148.8–551.4) | 0.97 ± 0.24 | 2 | 0.911 |
| Xn 12,145 | 360.7 (201.7–680.4) | 0.85 ± 0.26 | 2 | 0.849 |
| Xn K1 | 550.9 (296.8–1079.8) | 0.64 ± 0.32 | 1 | 0.879 |
| Xn SK1 | 552.3 (294.2–1089.5) | 0.64 ± 0.32 | 2 | 0.987 |
| Xp | 556.9 (289.5–1069.5) | 0.64 ± 0.32 | 1 | 0.784 |
| Xe | 641.3 (346.4–1192.7) | 0.66 ± 0.31 | 1 | 0.964 |
aEPBs include Photorhabdus temperata Ssp. temperata ANU101 (‘Ptt’), Xenorhabdus hominickii ANU101 (‘Xh’), X. nematophila France (‘XnF’), X. bovienii (‘Xb’), X. nematophila Mexico (‘XnM’), Photorhabdus luminescens KACC12123 (‘Pl 193’), P. luminescens subsp. laumondii KACC12283 (‘Pl laum’), X. nematophila SK2 (‘XnSK2’), P. luminescens subsp. thracensis KACC12284 (‘Pl thra’), X. nematophila KACC12145 (‘Xn12145’), Xenorhabdus nematophila K1 (‘XnK1’), X. poinarii (‘Xp’), and X. ehlersii KSY (‘Xe’)
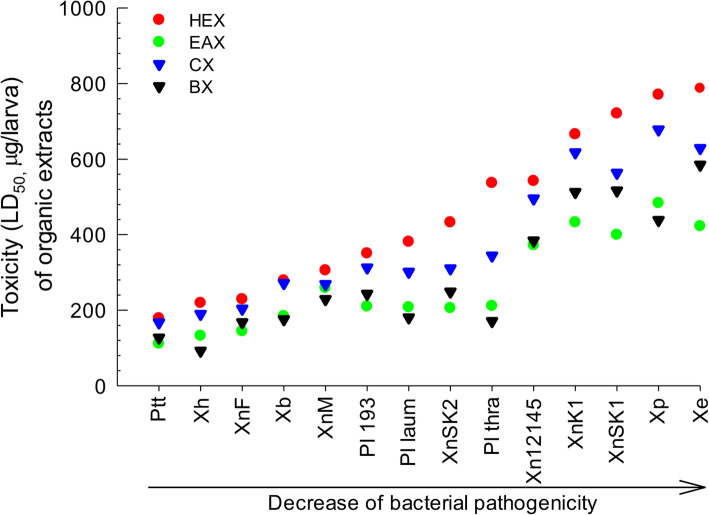
To analyze variations in bacterial insecticidal activities, their secondary metabolites were extracted from bacterial culture broth with four different organic solvents: hexane (‘HEX’), ethyl acetate (‘EAX’), chloroform (‘CX’), and butanol (‘BX’) extracts. These organic extracts showed variations in insecticidal activities, with LD50 values ranging from 92.4 (BX from X. hominickii: ‘Xh’) to 787.6 (HEX from ‘Xe’) μg/larva (Table S1). Insecticidal activities of bacteria and their extracts exhibited high positive correlations (Fig. 1): bacterial toxicity with HEX (r = 0.954; P < 0.0001), EAX (r = 0.938; P < 0.0001), CX (r = 0.967; P < 0.0001), and BX (r = 0.967; P < 0.0001).
Fig. 1.
Correlations between insecticidal activities of bacteria and their organic extracts. Bacteria used in this assay were: Photorhabdus temperata Subsp. temperata ANU101 (‘Ptt’), Xenorhabdus hominickii ANU101 (‘Xh’), X. nematophila K1 (‘XnK1’), X. ehlersii KSY (‘Xe’),, X. nematophila SK1 (‘XnSK1’), X. nematophila SK2 (‘XnSK2’), Photorhabdus luminescens KACC12123 (‘Pl 193’), P. luminescens subsp. laumondii KACC12283 (‘Pl laum’), P. luminescens subsp. thracensis KACC12284 (‘Pl thra’), X. nematophila KACC12145 (‘Xn12145’), X. nematophila Mexico (‘XnM’), X. nematophila France (‘XnF’), X. bovienii (‘Xb’), and X. poinarii (‘Xp’). Bacterial pathogenicity is presented in Table 1. Their cultured broths were fractionated with four different organic solvents: hexane (‘HEX’), ethyl acetate (‘EAX’), chloroform (‘CX’), and butanol (‘BX’). For each treatment, three replications were performed using 10 larvae per replication
Comparative analysis of bacterial extracts for insect immunosuppression
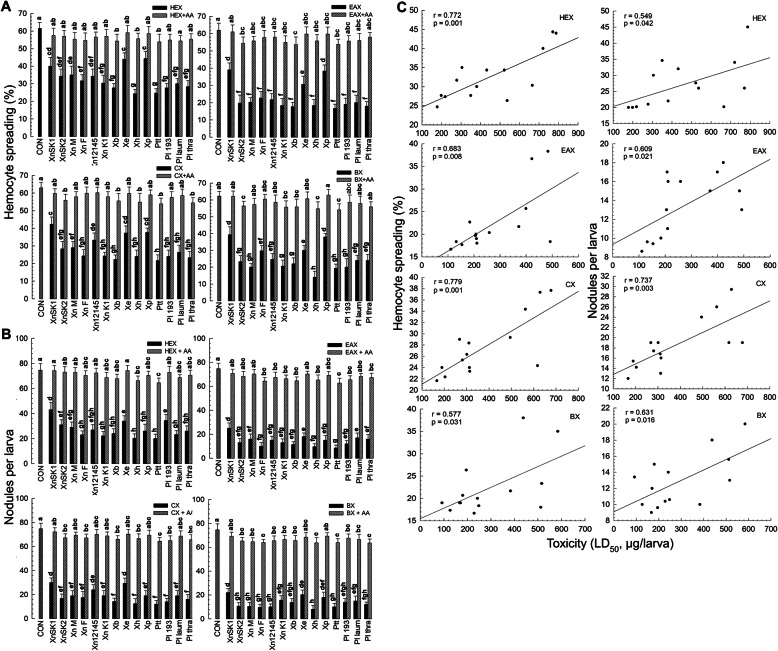
Variations in insecticidal activities among entomopathogenic bacteria might be caused by their differential immunosuppressive capabilities against target insects. To test this hypothesis, effects of bacterial extracts on cellular immune responses were assessed using hemocyte-spreading behaviour and nodulation assays after bacterial infection. All extracts significantly (p < 0.05) inhibited the hemocyte-spreading behavior (Fig. 2a) and nodulation (Fig. 2b). Immunosuppression was significantly (p < 0.05) rescued by the addition of arachidonic acid (a catalytic product of PLA2), suggesting that the immunosuppression was caused by the inhibition of PLA2. However, inhibitory activities of these bacterial extracts were different among bacterial species. When bacterial insecticidal activities were compared with immunosuppressive activities, these two parameters showed high positive correlations for all extracts (Fig. 2c).
Fig. 2.
Suppression of cellular immune responses by four organic extracts of 14 bacterial isolates and its correlation with bacterial pathogenicity. Bacteria used in this assay were: Photorhabdus temperata Subsp. temperata ANU101 (‘Ptt’), Xenorhabdus hominickii ANU101 (‘Xh’), X. nematophila K1 (‘XnK1’), X. ehlersii KSY (‘Xe’),, X. nematophila SK1 (‘XnSK1’), X. nematophila SK2 (‘XnSK2’), Photorhabdus luminescens KACC12123 (‘Pl 193’), P. luminescens subsp. laumondii KACC12283 (‘Pl laum’), P. luminescens subsp. thracensis KACC12284 (‘Pl thra’), X. nematophila KACC12145 (‘Xn12145’), X. nematophila Mexico (‘XnM’), X. nematophila France (‘XnF’), X. bovienii (‘Xb’), and X. poinarii (‘Xp’). Their cultured broths were extracted with four different organic solvents: hexane (‘HEX’), ethyl acetate (‘EAX’), chloroform (‘CX’), and butanol (‘BX’). a Effects of organic extracts on hemocyte-spreading behavior. For each treatment, three independently prepared hemocyte mixtures were used. To determine the spreading behavior, 100 hemocytes were randomly chosen. b Effects of organic extracts on hemocyte nodulation in response to bacterial challenge. Each L5 larva of S. exigua was injected with bacterial extract (10 μg/larva) along with heat-killed E. coli (4 × 104 cells). For each treatment, three replications were used with five larvae per replication. Arachidonic acid (AA, a catalytic product of PLA2) was used to rescue the inhibition. Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 (LSD test). c Correlations (r) between insecticidal activities of bacterial extracts (Fig. 1) and their immunosuppressive activities. Lines represent the best-fit regression
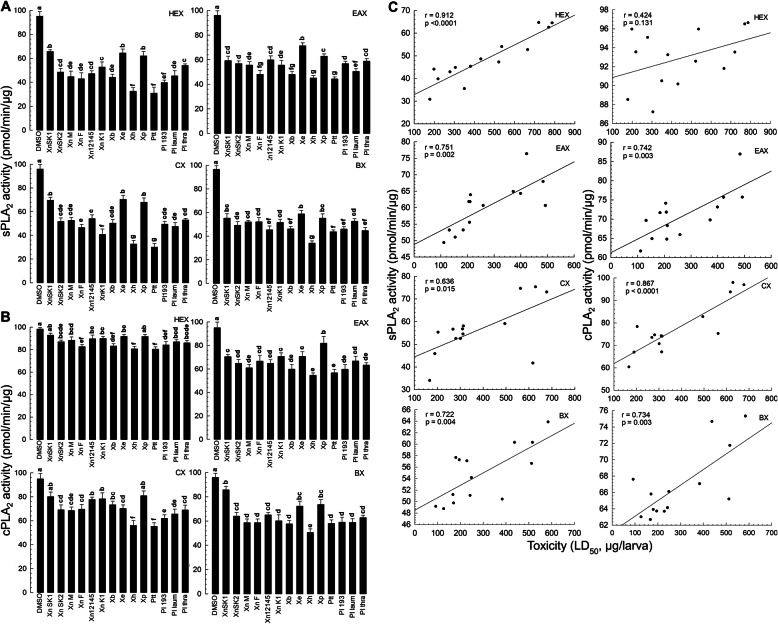
To determine whether immunosuppressive activities of these bacterial extracts were caused by inhibition of insect PLA2, these extracts were incubated with PLA2 extract obtained from S. exigua hemocytes. All bacterial extracts significantly inhibited sPLA2 activity, although they showed variations in inhibiting sPLA2 (Fig. 3a). For cPLA2, some extracts failed to inhibit its enzyme activity (Fig. 3b). Compared to HEX and CX, EAX and BX appeared to show higher inhibitory activities. Ptt and Xh extracts showed significantly higher inhibitory activities than the other bacterial extracts. There were positive correlations between bacterial insecticidal activities and PLA2 inhibition except HEX against cPLA2 (Fig. 3c).
Fig. 3.
Inhibitory activities of four organic extracts of 14 bacterial isolates against PLA2 enzyme and their correlation with bacterial pathogenicity. Bacteria used in this assay included 14 isolates: Photorhabdus temperata Subsp. temperata ANU101 (‘Ptt’), Xenorhabdus hominickii ANU101 (‘Xh’), X. nematophila K1 (‘XnK1’), X. ehlersii KSY (‘Xe’),, X. nematophila SK1 (‘XnSK1’), X. nematophila SK2 (‘XnSK2’), Photorhabdus luminescens KACC12123 (‘Pl 193’), P. luminescens subsp. laumondii KACC12283 (‘Pl laum’), P. luminescens subsp. thracensis KACC12284 (‘Pl thra’), X. nematophila KACC12145 (‘Xn12145’), X. nematophila Mexico (‘XnM’), X. nematophila France (‘XnF’), X. bovienii (‘Xb’), and X. poinarii (‘Xp’). Their cultured broths were extracted with four different organic solvents: hexane (‘HEX’), ethyl acetate (‘EAX’), chloroform (‘CX’), and butanol (‘BX’). a Effects of organic extracts on sPLA2. b Effects of organic extracts on cPLA2. DMSO was used as control. Each treatment was replicated three times using independent samples. Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 (LSD test). c Correlations (r) between insecticidal activities of bacterial extracts (Fig. 1) and their inhibitory activities against PLA2 enzyme. Lines represent the best-fit regression
Prediction of virulent secondary metabolites produced by Xenorhabdus and Photorhabdus
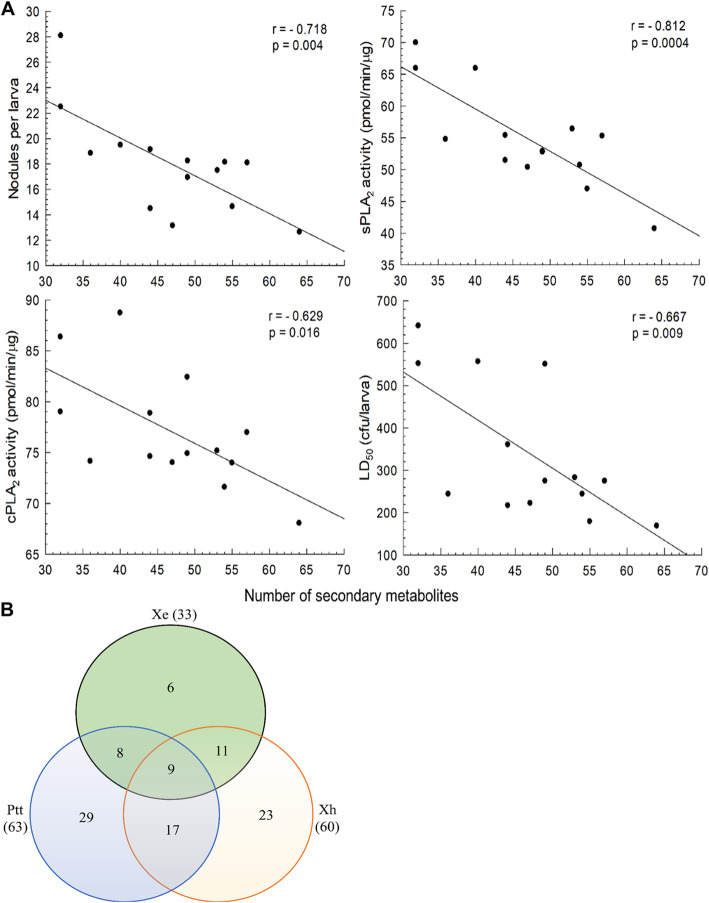
Immunosuppressive activities of bacterial extracts suggested that their secondary metabolites contained inhibitory compounds. To predict functional secondary metabolites, all extracts were assessed by TLC to confirm the presence of compounds. These compounds were then analyzed by GC-MS (Figure S1). Predicted compounds of four extracts for each bacterial species were combined and resulting 182 compounds were compared between bacterial isolates (Table S2). In addition to different compositions among bacterial isolates, the total number of secondary metabolites varied from 32 compounds of X. nematophila SK1 to 63 compounds of Ptt. Interestingly, there was a significant (p < 0.05) negative correlation between the total number of secondary metabolites and target insect immune responses measured by nodulation, sPLA2 activity, or cPLA2 (Fig. 4a). In addition, if bacteria had more secondary metabolites, they exhibited higher insecticidal activities.
Fig. 4.
Prediction of virulent secondary metabolites produced by Xenorhabdus and Photorhabdus. a Correlations (r) between total number of secondary metabolites and virulent parameters of hemocyte nodulation suppression, inhibition of sPLA2 or cPLA2, and insecticidal activities (LD50) of 14 bacterial isolates (b) Venn diagram analysis of secondary metabolites produced by Photorhabdus temperata Subsp. temperata (‘Ptt’), Xenorhabdus hominickii (‘Xh’), and X. ehlersii (‘Xe’). Figures in parentheses indicate total numbers of bacterial secondary metabolites predicted by GC-MS
To select virulent bacterial compounds, two most potent bacteria, Ptt and Xh, were compared with the least potent Xe for their secondary metabolites (Fig. 4b). Ptt and Xh shared over 44% metabolites. Almost 82% metabolites of Xe were detected in the culture broth of Ptt or Xh. Thus, Ptt- (29 compounds) or Xh- (23 compounds) specific compounds and overlapping (17 compounds) compounds were chosen as possible virulent compounds (Table 2).
Table 2.
Prediction of 70 virulent secondary metabolites derived from X. hominickii (‘Xh’) and P. temperata temperata (‘Ptt’). These metabolites were predicted from GC-MS analysis of extracts of bacterial culture broths
| Groupa | No | Compounds |
|---|---|---|
| Ptt + Xh | 1 | 1-Butanamine, N-butyl- |
| 2 | Benzyl alcohol | |
| 3 | 5-Thiazoleethanol, 4-methyl- | |
| 4 | Indole | |
| 5 | Benzeneethanol, 4-hydroxy- | |
| 6 | Phthalimide | |
| 7 | o-Cyanobenzoic acid | |
| 8 | Acetamide, N-(2-phenylethyl)- | |
| 9 | Indole-3-pyruvic acid | |
| 10 | Tryptophol | |
| 11 | 1H-Indole-3-acetic acid, hydrazide | |
| 12 | 2-Mercaptophenol | |
| 13 | Pyrrolo[1,2-a] pyrazine-1,4-dione, hexahydro- | |
| 14 | 1H-Indole-3-ethanol, acetate (ester) | |
| 15 | 2-Dodecen-1-yl (−) succinic anhydride | |
| 16 | Pyrrolo[1,2-a] pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl)- | |
| 17 | 2,5-Piperazinedione, 3-(phenylmethyl)- | |
| Xh | 1 | 1-Hexanol, 2-ethyl- |
| 2 | Pyrazine, 3-ethyl-2,5-dimethyl- | |
| 3 | Benzeneethanamine | |
| 4 | Hexanoic acid, 5-oxo-, ethyl ester | |
| 5 | 1,1-Diisobutoxy-isobutane | |
| 6 | Formamide, N, N-dibutyl- | |
| 7 | Cyclohexasiloxane, dodecamethyl- | |
| 8 | Butanoic acid, butyl ester | |
| 9 | Propanoic acid, 2-methyl-, butyl ester | |
| 10 | 2-Tetradecene, (E)- | |
| 11 | 3-Ethoxy-4-Methoxyphenol | |
| 12 | 7,9-Dimethyl-1,4-dioxa-7,9-diazacycloundecane-8-thione | |
| 13 | Heptadecane, 2,6-dimethyl- | |
| 14 | 1H-Indole-3-acetic acid, methyl ester | |
| 15 | 1H-Indene, 2-butyl-5-hexyloctahydro- | |
| 16 | L-Proline, N-valeryl-, decyl ester | |
| 17 | Stannane, tetraethyl- | |
| 18 | Fluorene, 4-[1,2-dihydroxyethyl]- | |
| 19 | 1-Eicosene | |
| 20 | Nonadecanenitrile | |
| 21 | Heptadecanoic acid, 14-methyl-, methyl ester | |
| 22 | E-8-Methyl-9-tetradecen-1-ol acetate | |
| 23 | Pentanamide, N-[2-(indol-3-yl)] ethyl- | |
| Ptt | 1 | 4-Ethylamino-n-butylamine |
| 2 | 2,5-Dimethyl-4-hydroxy-3(2H)-furanone | |
| 3 | Octanoic acid | |
| 4 | Benzothiazole | |
| 5 | 1,2-Ethanediol, 1-phenyl- | |
| 6 | n-Decanoic acid | |
| 7 | Phenol, 2,6-bis(1,1-dimethylethyl)- | |
| 8 | Dodecanoic acid | |
| 9 | 1-Pentadecene | |
| 10 | 1H-Benzimidazole, 2-(methylthio)- | |
| 11 | Propanamide, 2-amino-3-(3-indolyl)- | |
| 12 | Hexadecane, 7,9-dimethyl- | |
| 13 | 1-Hexadecanol, 2-methyl- | |
| 14 | Dicyclohexyldisulphide | |
| 15 | 1-Nonadecene | |
| 16 | Propanamide, 2,2,3,3,3-pentafluoro-N-(2-phenylethyl)- | |
| 17 | 1,13-Tetradecadien-3-one | |
| 18 | Hexadecanoic acid, methyl ester | |
| 19 | Diethyl Phthalate | |
| 20 | E-15-Heptadecenal | |
| 21 | 3-Phenylbicyclo (3.2.2) nona-3,6-dien-2-one | |
| 22 | E-11-Methyl-12-tetradecen-1-ol acetate | |
| 23 | 2-Methyl-E-7-octadecene | |
| 24 | Octadecanenitrile | |
| 25 | Pyrene, 4-methyl- | |
| 26 | 9-Octadecenamide, (Z)- | |
| 27 | Di-n-octyl phthalate | |
| 28 | Zinc, bis (dimethylcarbamodithioato-S, S′)-, (T-4)- | |
| 29 | Zinc dibutyldithiocarbamate |
aGroups are classified by bacterial metabolites from both Ptt and Xh (‘Ptt + Xh’), only Xh (‘Xh’), and only Ptt (‘Ptt’)
Validation of virulent secondary metabolites for their immunosuppression and insecticidal activities
Twelve compounds were selected from 70 virulent candidates produced by Ptt or Xh (Table S3). As a reference, one compound (2-mercaptobenzothiazole: ‘MT’) was randomly selected from metabolites produced by Xe as well as Ptt and Xh. All 12 virulent compounds highly suppressed hemocytic nodulation in response to bacterial infection (Fig. 5a). MT also suppressed immune responses, although it exhibited much lower inhibitory activity than those 12 virulent compounds. Compounds that showed the highest inhibitory activities were indole (‘IND’), ethoxymethoxyphenol (‘EMP’), and dimethylhydroxyfuranone (‘DHF’). These 12 virulent compounds significantly (p < 0.05) inhibited sPLA2 activity (Fig. 5b) and cPLA2 activity (Fig. 5c) whereas MT did not. IND, EMP, and DHF also showed high inhibitory activities against PLA2. These 12 virulent compounds also showed high insecticidal activities (Table 3). In contrast, MT had relatively low toxicities to the test insect species. However, IND, EMP, and DHF exhibited high insecticidal activities, with LD50 ranging from 4.29 to 5.12 μg/larva.
Fig. 5.
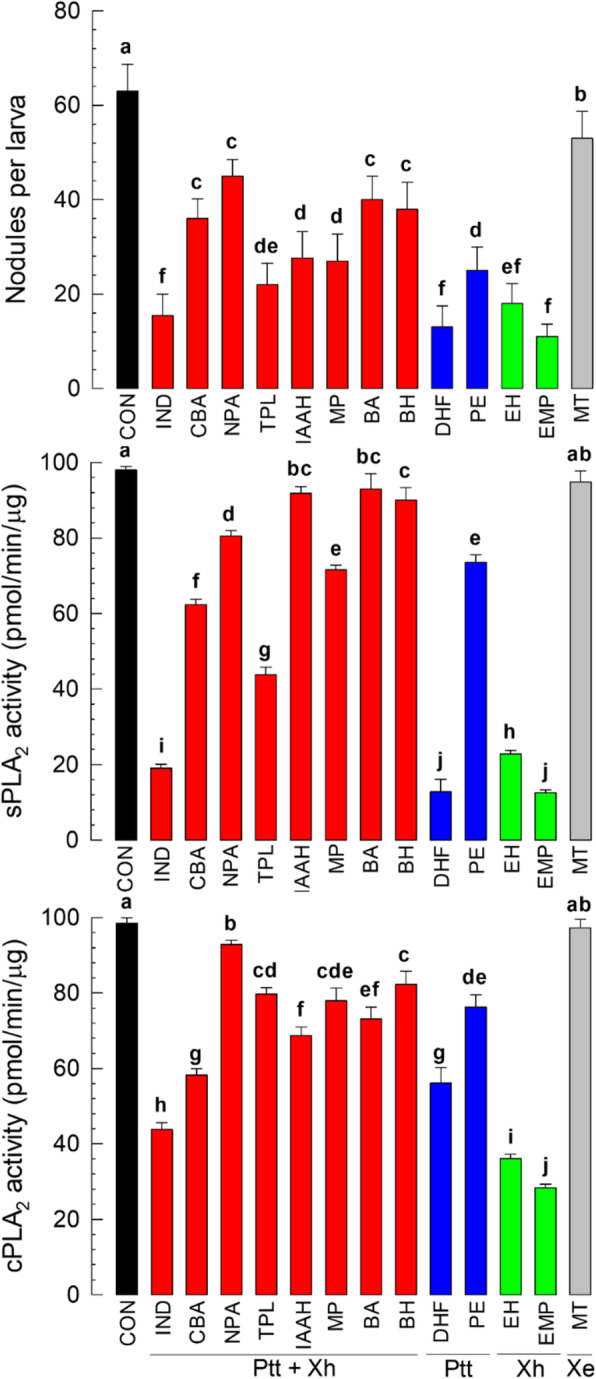
Validation of virulence activities of 12 virulent candidate secondary metabolites produced by two highly potent bacterial isolates of Photorhabdus temperata Subsp. temperata (‘Ptt’) and Xenorhabdus hominickii (‘Xh’). A reference metabolite was selected from compounds produced by a low virulent isolate of X. ehlersii (‘Xe’). For virulence tests, suppression of hemocyte nodulation, inhibition of sPLA2, and inhibition of cPLA2 were determined. To induce hemocyte nodulation, 4 × 104 cfu/larva of E. coli was injected to L5 larvae of S. exigua. Bacterial metabolites (10 μg/larvae) were injected to inhibit nodule formation. For each treatment, 15 larvae were used. To measure sPLA2 and cPLA2 enzyme activities, a commercial assay kit was used with PLA2-specific substrate as described in Materials and methods. As control (‘CON’), DMSO was used. Each treatment was independently replicated three times. Different letters above standard error bars indicates significant differences among means at Type I Error = 0.05 (LSD test). Bacterial metabolites used in this assay included benzyl alcohol (‘BA’), benzeneethanol-4-hydroxy (‘BH’), o-cyanobenzoic acid (‘CBA’), 2,5-dimethyl-4-hydroxy-3(2H)-furanone (‘DHF’), 2-ethyl-1-hexanol (‘EH’), 3-ethoxy-4-methoxyphenol (‘EMP’), indole-3-aceticacid hydrazide (‘IAAH’), indole (‘IND’), 2-mercaptophenol (‘MP’), 2-mercaptobenzothiazole (‘MT’), N-(2-phenylethyl) acetamide (‘NPA’), 1-phenyl-1,2-ethanediol (‘PE’), and tryptophol (‘TPL’). These compounds are classified as bacterial metabolites synthesized by both Ptt and Xh (‘Ptt + Xh’), only Xh (‘Xh’), only Ptt (‘Ptt’), and common to Ptt, Xh, and Xe (‘Xe’)
Table 3.
Insecticidal activities of virulent secondary metabolites of P. temperata temperata (Ptt) and X. hominickii (Xh) against S. exigua. Virulent metabolites were compared with the reference compound produced by a less virulent X. ehlersii (Xe). Bioassays were conducted by hemocoelic injection to L5 larvae. For each treatment dose, 10 larvae were used
| Groupa | Compoundb | LD50 (μg/larva) (95% CI) |
Slope ± SE | df | χ2 |
|---|---|---|---|---|---|
| Ptt + Xh | IND | 5.12 (2.51–10.2) | 0.50 ± 0.41 | 3 | 0.813 |
| CBA | 8.52 (4.2–16.4) | 0.47 ± 0.44 | 3 | 0.774 | |
| NPA | 12.79 (6.1–24.5) | 0.32 ± 0.59 | 3 | 0.953 | |
| TPL | 7.44 (8.8–34.9) | 0.44 ± 0.47 | 3 | 0.905 | |
| IAAH | 14.14 (7.6–27.6) | 0.31 ± 0.61 | 3 | 0.944 | |
| MP | 15.15 (7.9–30.3) | 0.29 ± 0.66 | 3 | 0.993 | |
| BA | 17.93 (9.3–34.7) | 0.31 ± 0.63 | 3 | 0.940 | |
| BH | 22.47 (17.4–62.9) | 0.35 ± 0.58 | 3 | 0.701 | |
| Ptt | DHF | 4.94 (3.2–8.7) | 0.49 ± 0.42 | 3 | 0.718 |
| PE | 24.28 (12.6–47.5) | 0.42 ± 0.49 | 3 | 0.819 | |
| Xh | EH | 8.93 (4.8–17.5) | 0.43 ± 0.47 | 3 | 0.492 |
| EMP | 4.29 (2.1–8.4) | 0.36 ± 0.53 | 3 | 0.926 | |
| Xe | MT | 38.75 (20.0–75.6) | 0.38 ± 0.54 | 3 | 0.691 |
aGroup is classified by bacterial compounds specific to (1) both Ptt and Xh (‘Ptt + Xh’), (2) only Ptt (‘Ptt’), (3) only Xh (‘Xh’), and (4) common to Ptt, Xh, and Xe (‘Xe’)
bCompound acronyms are: benzyl alcohol (‘BA’), benzeneethanol-4-hydroxy (‘BH’), o-cyanobenzoic acid (‘CBA’), 2,5-dimethyl-4-hydroxy-3(2H)-furanone (‘DHF’), 2-ethyl-1-hexanol (‘EH’), 3-ethoxy-4-methoxyphenol (‘EMP’), indole-3-aceticacid hydrazide (‘IAAH’), indole (‘IND’), 2-mercaptophenol (‘MP’), 2-mercaptobenzothiazole (‘MT’), N-(2-phenylethyl) acetamide (‘NPA’), 1-phenyl-1,2-ethanediol (‘PE’), and tryptophol (‘TPL’)
Discussion
Bacterial species in genera Xenorhabdus and Photorhabdus can produce various secondary metabolites to maintain their mutualistic symbiosis with host entomopathogenic nematodes [24]. Some of metabolites are required for their colonization to specific nematode hosts while others are crucial for pathogenesis specifically targeting insect’s immune system [3]. This study assessed all metabolites extracted with four organic solvents from bacterial culture broth of Xenorhabdus and Photorhabdus to identify their virulent secondary metabolites against insect immune responses.
Organic extracts of bacterial culture broth of seven different species classified in Xenorhabdus and Photorhabdus showed immunosuppressive and insecticidal activities. However, pathogenic activities of these extracts were different among 14 bacterial isolates and highly correlated with their bacterial insecticidal activities. These results suggest that these bacteria can induce immunosuppression of target insects via their secondary metabolites, supporting the hypothesis that immunosuppression is required for bacterial pathogenicity. Protein toxins that also induce the immunosuppression and finally kill target insects are known in Xenorhabdus and Photorhabdus [25]. However, acute attack by a target insect’s immune system should be avoided and actively suppressed by the secondary metabolites [26].
Immunosuppression of target insects by Xenorhabdus and Photorhabdus infection is caused by inhibition of PLA2 by bacterial secondary metabolites [10]. The first identified PLA2 inhibitor produced by bacteria was benzylideneacetone (BZA) produced by X. nematophila [27]. Subsequently, eight different PLA2 inhibitors were identified from bacterial culture broths of X. nematophila and P. temperata temperata [13]. On the other hand, these PLA2 inhibitors induced apoptosis-associated cytotoxicity and insecticidal activities [28]. This explains why bacterial metabolites containing PLA2 inhibitors in our current study showed insecticidal activities. Indeed, the degree of PLA2 inhibition by bacteria has been found to be highly correlated with bacterial pathogenicity [15]. These findings suggested that bacterial metabolites that inhibited PLA2 activities in this current study contained virulent secondary metabolites.
Virulent secondary metabolites produced by Xenorhabdus and Photorhabdus were chosen based on comparative analysis of secondary metabolites among bacterial isolates. GC-MS analyses for bacterial culture broths of 14 isolates predicted 182 compounds. Most of them were phenol derivatives, tryptophan derivatives, peptides, and fatty acid derivatives including secondary metabolites synthesized by NRPS-PKS [4]. In general, predicted and identified compounds from NRPS-PKS of Xenorhabdus and Photorhabdus genomes are associated with bacterial pathogenicity [19]. This is supported by the high correlation between the number of predicted compounds in each bacterial isolate and bacterial pathogenicity found in the current study. This also suggests that these compounds predicted by GC-MS might contain virulent secondary metabolites. To select virulent compounds, predicted metabolites of highly pathogenic bacterial isolates (Ptt and Xh) were compared with those of a low pathogenic isolate (Xe). The comparison for 103 total compounds resulted in the selection of 70 compounds, including Ptt-specific, Xh-specific, and Ptt + Xh common metabolites. Among 70 virulent candidates, 12 randomly chosen compounds showed high immunosuppressive and insecticidal activities, validating the prediction of these 70 virulent secondary metabolites of Xenorhabdus and Photorhabdus. This analysis did not include 80 compounds (182 compounds from 14 total isolates – 103 compounds of 3 isolates used for the comparison) because other bacterial isolates were less virulent than Ptt and Xh. Among 12 virulent compounds, three (IND, EMP, and DHF) were highly potent in inhibiting insect immune responses, exhibiting high insecticidal activities. These compounds are not likely to be the secondary metabolites produced from NRPS-PKS. Although secondary metabolites derived from NRPS-PKS contribute to induce the bacterial pathogenicity as mentioned above, non-NRPS-PKS metabolites may also play crucial role in expressing insecticidal activity of the bacteria. This is supported by the fact that a mutant X. szentirmaii in a specific phosphopantethienyl transferase, which activates the carrier protein domain of PKS, NRPS, and fatty acid synthase [29], loses the production of NRPS-PKS metabolites but exhibits insecticidal virulence [30]. IND has been found to be a PLA2 inhibitor from entomopathogenic bacteria [13]. Unlike tryptophan derivatives including indole, EMP is a phenol derivative that shares structural similarities with other sPLA2 inhibitors such as 4-bromophenacyl bromide and BZA. DHF is a furanone and a relatively new chemical compound that can inhibit PLA2 among bacterial metabolites. Derivatives containing these three different skeletons need to be compared for their activities to inhibit insect PLA2 to develop potent immunosuppressants. In summary, this study newly proposes other PLA2 inhibitors including EMP and DHF.
Conclusions
Bacterial isolates of Xenorhabdus and Photorhabdus were pathogenic by suppressing insect immune responses via PLA2 inhibition using their secondary metabolites. Such immunosuppression was highly correlated with insecticidal activity of bacteria. Based on this correlation, 70 virulent compounds were predicted by comparative analysis of secondary metabolites between high and low pathogenic bacteria.
Methods
Insect and bacteria
Larvae of Spodoptera exigua were reared at temperature of 25 ± 2 °C and relative humidity of 65 ± 5% under 16 h light and 8 h dark condition with an artificial diet [31]. Under these conditions, larvae underwent five instars (L1-L5). Adults were provided with 10% sucrose solution. For bioassays to determine bacterial pathogenicity, larvae were taken from a cohort.
Ten strains of Xenorhabdus and four strains of Photorhabdus were collected from previous stocks: [Xenorhabdus nematophila K1 (‘XnK1’) [32], X. hominickii ANU101 (‘Xh’) [16], X. ehlersii KSY (‘Xe’) [33], Photorhabdus temperata Ssp. temperata ANU101 (‘Ptt’) [34], X. nematophila SK1 (‘XnSK1’) and X. nematophila SK2 (‘XnSK2’) [15], Korean Agricultural Culture Collection (KACC, RDA, Jeonju, Korea) [Photorhabdus luminescens KACC12123 (‘Pl 193’), P. luminescens subsp. laumondii KACC12283 (‘Pl laum’), P. luminescens subsp. thracensis KACC12284 (‘Pl thra’), X. nematophila KACC12145 (‘Xn12145’), X. nematophila Mexico (‘XnM’), X. nematophila France (‘XnF’), X. bovienii (‘Xb’), and X. poinarii (‘Xp’)]. These bacteria were grown in tryptic soy broth (TSB: Difco, Sparks, MD, USA) for 48 h at 28 °C with shaking at 180 rpm [16, 32]. Escherichia coli Top 10 was purchased from Invitrogen (Carlsbad, CA, USA) and cultured overnight in Luria-Bertani (LB) medium at 37 °C with shaking at 180 rpm. For immune challenge experiment, heat killed (80 °C for 10 min) E. coli were used. Their cell number was counted using a hemocytometer (Neubauer improved bright line, Cat. No. 0640010, Superior Marienfeld, Lauda-Konigshofen, Germany) under a phase contrast microscope (BX41, Olympus, Tokyo, Japan). Different bacterial concentrations were prepared through serial dilution in sterilized deionized distilled water.
Chemicals
Arachidonic acid (5,8,11,14-eicosatetraenoic acid; AA), tryptophol (TPL), indole (IND), indole-3-aceticacid hydrazide (IAAH), o-cyanobenzoic acid (CBA), 2-ethyl-1-hexanol (EH), 2-mercaptophenol (MP), and 2-mercaptobenzothiazole (MT) were purchased from Sigma-Aldrich Korea (Seoul, Korea) and dissolved in dimethyl sulfoxide (DMSO). 1,2-bis (heptanoylthio) phosphatidylcholine (PC) was used as secretory PLA2 (sPLA2) substrate while arachidonoyl thio-PC was used as cellular PLA2 (cPLA2) substrate. These substrates were purchased from Cayman Chemical (Ann Arbor, MI, USA). Benzyl alcohol (BA) and benzeneethanol-4-hydroxy (BH) were purchased from Alfa Aesar China (Shanghai, China). 3-ethoxy-4-methoxyphenol (EMP) was obtained from BOC Sciences (Shirley, NY, USA). 1-phenyl-1,2-ethanediol (PE) was bought from Acros Organics (NJ, USA). 2,5-dimethyl-4-hydroxy-3(2H)-furanone (DHF) was purchased from Tokyo Chemical Industry (Tokyo, Japan). N-(2-phenylethyl) acetamide (NPA) was kindly provided by Professor Helge Bode (Goethe University, Frankfurt, Germany). Phosphate-buffered saline (PBS, pH 7.4) was prepared with 100 mM of phosphate and 0.7% NaCl. Anticoagulant buffer (ACB, pH 4.5) was prepared with NaCl (186 mM), Na2EDTA (17 mM) and citric acid (41 mM).
Fractionation of bacterial culture broth and TLC analysis
All bacterial isolates were cultured individually in TSB (1 L) at 28 °C with shaking at 180 rpm. After 48 h, bacterial pellets were separated from supernatant after centrifuging cultured broth at 10,000×g for 20 min at 4 °C. The resulting supernatant was subjected to fractionation of secondary metabolites by successively adding four different organic solvents (hexane, ethyl acetate, chloroform, and butanol) as described previously by Mollah et al. [35]. Resulting extracts were resuspended in methanol (0.4 mg/mL) and further diluted with DMSO or methanol to desired concentrations based on experimental purposes. During each fractionation step, the metabolite extraction was subjected to thin layer chromatography (TLC) by spotting it on a silica gel plate (20 × 20 cm: Merck, Darmstadt, Germany). A mixture of chloroform, methanol, and acetic acid (7:2.5:0.5, v/v) was used as an eluent. Spots in the silica gel plate were visualized and marked under a fluorescence analysis cabinet (Spectroline, CM-10, Westbury, NY, USA).
Gas chromatography coupled with mass spectrophotometer (GC-MS) analysis
GC (7890B, Agilent Technologies, Santa Clara, CA, USA) equipped with MS (5977A Network, Agilent Technologies) was used for the prediction of the bacterial extracts in the methanol resuspension. It was performed as described by Mollah et al. [35]. An HP 5 MS column (non-polar column, Agilent Technologies) was used for GC. Helium was used as carrier gas at a flow rate of 1 mL/min. Split mode of injection with split ratio 10:1 was followed at 200 °C. The initial temperature was 100 °C for 3 min. It was raised to 300 °C at 5 °C/min. The oven temperature was maintained at 300 °C for 10 min. For recording mass spectra, EI mode at 70 eV with a scanning range of 33–550 m/z was used. Mass spectra were used for identification of purified samples using NIST11 database (U.S. Department of Commerce, Gaithersburg, MD, USA) and literature data (http://nistmassspeclibrary.com).
Hemocyte-spreading behavior assay
Hemolymph (~ 250 μL) was collected from L5 larvae of S. exigua by cutting prolegs. It was mixed with 750 μL ACB. The hemolymph suspension was centrifuged at 800×g for 5 min and 800 μL supernatant was mixed with filter-sterilized 200 μL TC100 insect culture medium (Hyclone, Daegu, Korea). This hemocyte suspension (9 μL) and each bacterial metabolite (1 μL) were mounted on a glass slide to assess hemocyte-spreading behavior. For rescue experiment with AA (10 μg/μL in DMSO), 8 μL of cell suspension, 1 μL of bacterial metabolite, and 1 μL of AA were placed on a glass slide. After incubating the mixture at room temperature under darkness for 40 min, hemocytes were observed under a phase contrast microscope (Olympus) at 800 × magnification. As control, the dilution solvent (DMSO) of metabolites was used. Spread hemocytes were recognized based on cytoplasmic extension out of cell boundary. Each treatment was replicated three times with separately prepared suspension mixture. In each replication, 100 hemocytes were randomly chosen to assess hemocyte-spreading behavior.
Nodulation assay
Hemocyte nodule formation was observed using 3 days old L5 larvae of S. exigua after injecting 3 μL of overnight grown E. coli Top 10 (104 cells/larva) and 2 μL of DMSO to the hemocoel through prolegs using a microsyringe (Hamilton, Reno, NV, USA). To assess any inhibitory effect of bacterial extracts or secondary metabolites, 1 μL (1 μg/μL in DMSO) of test sample was co-injected with 3 μL of E. coli and 1 μL of DMSO. For AA rescue experiment, 1 μL of AA (10 μg/μL in DMSO) was co-injected with 1 μL of bacterial metabolite and 3 μL of E. coli. After incubation at 25 °C for 8 h, injected larvae were dissected to count melanized nodules under a stereomicroscope (Stemi SV11, Zeiss, Jena, Germany) at 50 × magnification. Each treatment was replicated three times using five larvae per replication.
PLA2 enzyme assay
PLA2 activity was assessed using processes previously described by Vatanparast et al. [36]. Briefly, enzymes were extracted after grinding hemocytes of L5 larvae of S. exigua with PBS followed by centrifugation at 12,500×g for 5 min. The resulting supernatant containing the cytosol and membrane mixture was used as enzyme source. The reaction mixture (225 μL) contained 10 μL of PLA2 enzyme, 10 μL of Ellman’s reagent (5,5-di-thio-bis-(2-nitrobenzoicacid)), 5 μL of test inhibitor, and 200 μL of sPLA2 or cPLA2 substrate. DMSO was used instead of test inhibitor as control. Change in absorbance of reaction product was measured at wavelength of 405 nm using a microplate reader (Victor, PerkinElmer, Waltham, MA, USA). Each treatment was replicated three times with biologically independent samples.
Bacterial pathogenicity test
To determine insecticidal activities of 14 bacterial isolates, bacterial cells after 24 h of culture were centrifuged at 10,000×g for 2 min at 4 °C. Cell pellet was re-suspended in PBS for injection. Bacterial suspension was diluted with PBS to obtain different concentrations (0, 101, 102, 103, 104, 105 and 106 colony-forming unit (cfu)/larva) and injected to hemocoel of L5 S. exigua larvae using a microsyringe. Before injection, larvae were surface sterilized with 70% ethanol. Bacterial dose was measured after plating the suspension onto LB agar medium followed by culturing at 28 °C for 48 h. Mortality was counted at 24 h after bacterial injection. As control bacterial treatment, E. coli was used. Injected larvae were incubated at room temperature with sufficient diet. Larvae were considered to be dead if there was no movement upon touching. Each treatment consisted of three replications using 10 larvae for each replication.
Toxicity tests of bacterial metabolites and synthetic compounds
To determine toxicities of bacterial extracts and predicted metabolites, hemocoelic injection was performed. Dried bacterial extracts and metabolites were weighed and dissolved in DMSO to prepare different concentrations (0.001, 0.01, 0.1, 1 and 10 μg/μL) for toxicity assays. A 10 μL microsyringe was used for hemocoelic injection. Each test sample (3 μL) was injected to L5 larvae of S. exigua. DMSO was injected as control. Treated larvae were placed in 9 cm diameter dishes and incubated at room temperature. Mortality was determined every 24 h for 96 h after treatment. Larvae were considered as dead if there was no movement upon touching. Each test concentration was replicated three times using 10 larvae per replication.
Statistical analysis
All assay data for continuous variables were subjected to one-way analysis of variance (ANOVA) using PROG GLM in SAS program [37]. For ANOVA, mortality data were subjected to arcsine transformation. Means were compared with the least significant difference (LSD) test at 0.05 level of Type I error. Median lethal dose (LD50) was calculated using EPA Probit Analysis Program, ver. 1.5 (U.S. Environmental Protection Agency, Washington, D.C., USA).
Supplementary Information
Additional file 1: Table S1. Insecticidal activities of four organic extracts of culture broth of 14 entomopathogenic bacteria (EPB) of Xenorhabdus and Photorhabdus against L5 larvae of S. exigua. The bioassay was carried out by injecting 3 μL of bacterial culture broth extracts from different concentrations into the hemocoel of L5 larva. Each treatment dose used 10 larvae and each treatment replicated three times. Table S2. GC-MS prediction of secondary metabolites in organic extracts of bacterial culture broth from 14 species of Xenorhabdus and Photorhabdus. Table S3. Secondary metabolites predicted from Photorhabdus temperata temperata (Ptt), Xenorhabdus hominickii (Xh) and Xenorhabdus ehlersii (Xe) which were used for biological activity analysis. Figure S1. Chromatograms from the GC-MS analysis of the organic extracts of 14 bacterial culture broths for the prediction of bacterial metabolites. Four organic solvents such as hexane, ethyl acetate, chloroform and butanol were used to get organic extracts ‘HEX’, ‘EAX’, ‘CX’ and ‘BX’ from the bacteria, Photorhabdus temperata Subsp. temperata ANU101 (‘Ptt’), Xenorhabdus hominickii ANU101 (‘Xh’), X. nematophila K1 (‘XnK1’), X. ehlersii KSY (‘Xe’),, X. nematophila SK1 (‘XnSK1’), X. nematophila SK2 (‘XnSK2’), Photorhabdus luminescens KACC12123 (‘Pl 193’), P. luminescens subsp. laumondii KACC12283 (‘Pl laum’), P. luminescens subsp. 2 thracensis KACC12284 (‘Pl thra’), X. nematophila KACC12145 (‘Xn12145’), X. nematophila Mexico (‘XnM’), X. nematophila France (‘XnF’), X. bovienii (‘Xb’), and X. poinarii (‘Xp’). The extracts were dried using rotary evaporator and the resulting product was dissolved in methanol. After filtration, the sample was used for GC analysis and MS recording. NIST 11 database were used to predict compounds based on mass spectra. X axis indicates the retention time in min.
Acknowledgments
The authors thank Youngim Song for supplying all materials and Miltan Chandra Roy for rearing test insects.
Abbreviations
- AA
Arachidonic acid
- ACB
Anticoagulant buffer
- ANOVA
Analysis of variance
- BA
Benzyl alcohol
- BZA
Benzylideneacetone
- BH
Benzeneethanol-4-hydroxy
- CBA
Cyanobenzoic acid
- CFU
Colony-forming unit
- DHF
Dimethylhydroxyfuranone
- DMSO
Dimethyl sulfoxide
- EH
Ethyl-1-hexanol
- EMP
Ethoxymethoxyphenol
- IAAH
Indole-3-aceticacid hydrazide
- IND
Indole
- LD50
Median lethal dose
- LSD
Least significant difference
- MP
Mercaptophenol
- MT
Mercaptobenzothiazole
- NRPS
Non-ribosomal peptide synthetase
- PC
Phosphatidylcholine
- PKS
Polyketide synthase
- PLA2
Phospholipase A2
- Lrp
Leucine-responsive protein
- NPA
N-(2-phenylethyl) acetamide
- PE
Phenyl-1,2-ethanediol
- PO
Phenoloxidase
- Pl
Photorhabdus luminescens
- Ptt
Photorhabdus temperata temperata
- TPL
Tryptophol
- TSB
Tryptic soy broth
- Xb
Xenorhabdus bovienii
- Xe
Xenorhabdus ehlersii
- Xh
Xenorhabdus hominickii
- Xn
Xenorhabdus nematophila
- Xp
Xenorhabdus poinarii
Authors’ contributions
YK designed experiments and revised the manuscript draft. MIM performed all experiments and data analyses. MIM prepared the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by a grant (No. 2017R1A2133009815) of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning, Republic of Korea.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information.
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Akhurst RJ. Neoaplectana species: specificity of association with bacteria of the genus Xenorhabdus. Exp Parasitol. 1983;55:258–263. doi: 10.1016/0014-4894(83)90020-6. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro-Ilan DI, Han R, Dolinksi C. Entomopathogenic nematode production and application technology. J Nematol. 2012;44:206–217. [PMC free article] [PubMed] [Google Scholar]
- 3.Goodrich-Blair H, Clarke DJ. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- 4.Tobias NJ, Wolff H, Djahanschiri B, Grundmann F, Kronenwerth M, Shi YM, et al. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat Microbiol. 2017;2:1676–1685. doi: 10.1038/s41564-017-0039-9. [DOI] [PubMed] [Google Scholar]
- 5.Mollah MI, Roy MC, Choi D, Hasan MA, Al Baki MA, Yeom HS, et al. Variations of indole metabolites and NRPS-PKS loci in two different virulent strains of Xenorhabdus hominickii. Front Microbiol. 2020;00:000. doi: 10.3389/fmicb.2020.583594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sajjadian SM, Kim Y. Dual oxidase-derived reactive oxygen species against Bacillus thuringiensis and its suppression by eicosanoid biosynthesis inhibitors. Front Microbiol. 2020;11:528. doi: 10.3389/fmicb.2020.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park Y, Herbert EE, Cowles CE, Cowles KN, Menard ML, Orchard SS, et al. Clonal variation in Xenorhabdus nematophila virulence and suppression of Manduca sexta immunity. Cell Microbiol. 2007;9:645–656. doi: 10.1111/j.1462-5822.2006.00815.x. [DOI] [PubMed] [Google Scholar]
- 8.Sergeant M, Baxter L, Jarrett P, Shaw E, Ousley M, Winstanley C, et al. Identification, typing, and insecticidal activity of Xenorhabdus isolates from entomopathogenic nematodes in United Kingdom soil and characterization of the xpt toxin loci. Appl Environ Microbiol. 2006;72:5895–5907. doi: 10.1128/AEM.00217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrestha S, Kim Y. An entomopathogenic bacterium, Xenorhabdus nematophila, inhibits hemocyte phagocytosis of Spodoptera exigua by inhibiting phospholipase A2. J Invertebr Pathol. 2007;96:64–70. doi: 10.1016/j.jip.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Cho JD, Park Y. Two groups of entomopathogenic bacteria, Photorhabdus and Xenorhabdus, share an inhibitory action against phospholipase A2 to induce host immunodepression. J Invertebr Pathol. 2005;89:258–264. doi: 10.1016/j.jip.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Mouchlis VD, Dennis EA. Phospholipase A2 catalysis and lipid mediator lipidomics. Biochim Biophys Acta. 2019;1864:766–771. doi: 10.1016/j.bbalip.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y, Ahmed S, Stanley D, An C. Eicosanoid-mediated immunity in insects. Dev Comp Immunol. 2018;83:130–143. doi: 10.1016/j.dci.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Seo S, Lee S, Hong Y, Kim Y. Phospholipase A2 inhibitors synthesized by two entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus temperata subsp. temperata. Appl Environ Microbiol. 2012;78:3816–3823. doi: 10.1128/AEM.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed S, Kim Y. Differential immunosuppression by inhibiting PLA2 affects virulence of Xenorhabdus hominickii and Photorhabdus temperata temperata. J Invertebr Pathol. 2018;157:136–146. doi: 10.1016/j.jip.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Hasan MA, Ahmed S, Mollah MI, Lee D, Kim Y. Variation in pathogenicity of different strains of Xenorhabdus nematophila: differential immunosuppressive activities and secondary metabolite production. J Invertebr Pathol. 2019;166:107221. doi: 10.1016/j.jip.2019.107221. [DOI] [PubMed] [Google Scholar]
- 16.Park Y, Kang S, Sadekuzzaman M, Kim H, Jung JK, Kim Y. Identification and bacterial characteristics of Xenorhabdus hominickii ANU101 from an entomopathogenic nematode, Steinernema monticolum. J Invertebr Pathol. 2017;144:74–87. doi: 10.1016/j.jip.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Casanova-Torres AM, Shokal U, Morag N, Eleftherianos I, Goodrich-Blair H. The global transcription factor Lrp is both essential for and inhibitory to Xenorhabdus nematophila insecticidal activity. Appl Environ Microbiol. 2017;83:e00185–e00117. doi: 10.1128/AEM.00185-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel Y, Windhorst C, Lu X, Goodrich-Blair H, Bode HB. The global regulators Lrp, LeuO, and HexA control secondary metabolism in entomopathogenic bacteria. Front Microbiol. 2017;8:209. doi: 10.3389/fmicb.2017.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi YM, Bode HB. Chemical language and warfare of bacterial natural products in bacteria-nematode-insect interactions. Nat Prod Rep. 2018;35:309–335. doi: 10.1039/C7NP00054E. [DOI] [PubMed] [Google Scholar]
- 20.Crawford JM, Portmann C, Zhang X, Roeffaers MB, Clardy J. Small molecule perimeter defense in entomopathogenic bacteria. Proc Natl Acad Sci U S A. 2012;109:10821–10826. doi: 10.1073/pnas.1201160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai X, Nowak S, Wesche F, Bischoff I, Kaiser M, Furst R, et al. Entomopathogenic bacteria use multiple mechanisms for bioactive peptide library design. Nat Chem. 2016;9:379–386. doi: 10.1038/nchem.2671. [DOI] [PubMed] [Google Scholar]
- 22.Sussmuth RD, Mainz A. Nonribosomal peptide synthesis-principles and prospects. Angew Chem Int Ed Eng. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 23.Nollmann FI, Heinrich AK, Brachmann AO, Morrisseau C, Mukherjee K, Casanova-Torres AM, et al. A Photorhabdus natural product inhibits insect juvenile hormone epoxide hydrolase. Chembiochem. 2015;16:766–771. doi: 10.1002/cbic.201402650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobias NJ, Shi YM, Bode HB. Refining the natural product repertoire in entomopathogenic bacteria. Trends Microbiol. 2018;26:833–840. doi: 10.1016/j.tim.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Yang Q, Zhang J, Li T, Liu S, Song P, Nangong Z, et al. PirAB protein from Xenorhabdus nematophila HB310 exhibits a binary toxin with insecticidal activity and cytotoxicity in Galleria mellonella. J Invertebr Pathol. 2017;148:43–50. doi: 10.1016/j.jip.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Choi D, Jung J, Kim Y. Eicosanoid mediation of immune responses at early bacterial infection stage and its inhibition by Photorhabdus temperata subsp. temperata, an entomopathogenic bacterium. Arch Insect Biochem Physiol. 2018;99:e21502. doi: 10.1002/arch.21502. [DOI] [PubMed] [Google Scholar]
- 27.Ji D, Yi Y, Kim GH, Choi YH, Kim P, Baek NI, et al. Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophila against major plant-pathogenic bacteria. FEMS Microbiol Lett. 2004;239:241–248. doi: 10.1016/j.femsle.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 28.Mollah MI, Yeasman F, Kim Y. Benzylideneacetone and other phenylethylamide bacterial metabolites induce apoptosis to kill insects. J Asia Pac Entomol. 2020;23:449–457. doi: 10.1016/j.aspen.2020.03.008. [DOI] [Google Scholar]
- 29.Bunet R, Riclea R, Laureti L, Hôtel L, Paris C, Girardet JM, Spiteller D, Dickschat JS, Leblond P, Aigle B. A single Sfp-type phosphopantetheinyl transferase plays a major role in the biosynthesis of PKS and NRPS derived metabolites in Streptomyces ambofaciens ATCC23877. PLoS One. 2014;9:e87607. doi: 10.1371/journal.pone.0087607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciezki K, Wesener S, Jaber D, Mirza S, Forst S. ngrA-dependent natural products are required for interspecies competition and virulence in the insect pathogenic bacterium Xenorhabdus szentirmaii. Microbiology. 2019;165:538–553. doi: 10.1099/mic.0.000793. [DOI] [PubMed] [Google Scholar]
- 31.Goh HG, Lee SG, Lee BP, Choi KM, Kim JH. Simple mass-rearing of beet armyworm, Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae), on an artificial diet. Korean J Appl Entomol. 1990;29:180–183. [Google Scholar]
- 32.Park Y, Kim Y, Yi Y. Identification and characterization of a symbiotic bacterium associated with Steinernema carpocapsae in Korea. J Asia Pac Entomol. 1999;2:105–111. doi: 10.1016/S1226-8615(08)60038-2. [DOI] [Google Scholar]
- 33.Kim H, Keum S, Hasan A, Kim H, Jung Y, Lee D, et al. Identification of an entomopathogenic bacterium, Xenorhabdus ehlersii KSY, from Steinernema longicaudum GNUS101 and its immunosuppressive activity against insect host by inhibiting eicosanoid biosynthesis. J Invertebr Pathol. 2018;159:6–17. doi: 10.1016/j.jip.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Kang S, Han S, Kim Y. Identification of an entomopathogenic bacterium, Photorhabdus temperata subsp. temperata, in Korea. J Asia Pac Entomol. 2004;7:331–337. doi: 10.1016/S1226-8615(08)60235-6. [DOI] [Google Scholar]
- 35.Mollah MI, Dekebo A, Kim Y. Immunosuppressive activities of novel PLA2 inhibitors from Xenorhabdus hominickii, an entomopathogenic bacterium. Insects. 2020;11:505. doi: 10.3390/insects11080505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vatanparast M, Ahmed S, Herrero S, Kim Y. A non-venomous sPLA2 of a lepidopteran insect: its physiological functions in development and immunity. Dev Comp Immunol. 2018;89:83–92. doi: 10.1016/j.dci.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 37.SAS Institute Inc. SAS/STAT user’s guide, Release 6.03, Ed. Cary, NC. 1989.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Insecticidal activities of four organic extracts of culture broth of 14 entomopathogenic bacteria (EPB) of Xenorhabdus and Photorhabdus against L5 larvae of S. exigua. The bioassay was carried out by injecting 3 μL of bacterial culture broth extracts from different concentrations into the hemocoel of L5 larva. Each treatment dose used 10 larvae and each treatment replicated three times. Table S2. GC-MS prediction of secondary metabolites in organic extracts of bacterial culture broth from 14 species of Xenorhabdus and Photorhabdus. Table S3. Secondary metabolites predicted from Photorhabdus temperata temperata (Ptt), Xenorhabdus hominickii (Xh) and Xenorhabdus ehlersii (Xe) which were used for biological activity analysis. Figure S1. Chromatograms from the GC-MS analysis of the organic extracts of 14 bacterial culture broths for the prediction of bacterial metabolites. Four organic solvents such as hexane, ethyl acetate, chloroform and butanol were used to get organic extracts ‘HEX’, ‘EAX’, ‘CX’ and ‘BX’ from the bacteria, Photorhabdus temperata Subsp. temperata ANU101 (‘Ptt’), Xenorhabdus hominickii ANU101 (‘Xh’), X. nematophila K1 (‘XnK1’), X. ehlersii KSY (‘Xe’),, X. nematophila SK1 (‘XnSK1’), X. nematophila SK2 (‘XnSK2’), Photorhabdus luminescens KACC12123 (‘Pl 193’), P. luminescens subsp. laumondii KACC12283 (‘Pl laum’), P. luminescens subsp. 2 thracensis KACC12284 (‘Pl thra’), X. nematophila KACC12145 (‘Xn12145’), X. nematophila Mexico (‘XnM’), X. nematophila France (‘XnF’), X. bovienii (‘Xb’), and X. poinarii (‘Xp’). The extracts were dried using rotary evaporator and the resulting product was dissolved in methanol. After filtration, the sample was used for GC analysis and MS recording. NIST 11 database were used to predict compounds based on mass spectra. X axis indicates the retention time in min.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information.