Abstract
Background
Taxonomic profiles of vaginal microbial communities can be sorted into a discrete number of categories termed community state types (CSTs). This approach is advantageous because collapsing a hyper-dimensional taxonomic profile into a single categorical variable enables efforts such as data exploration, epidemiological studies, and statistical modeling. Vaginal communities are typically assigned to CSTs based on the results of hierarchical clustering of the pairwise distances between samples. However, this approach is problematic because it complicates between-study comparisons and because the results are entirely dependent on the particular set of samples that were analyzed. We sought to standardize and advance the assignment of samples to CSTs.
Results
We developed VALENCIA (VAginaL community state typE Nearest CentroId clAssifier), a nearest centroid-based tool which classifies samples based on their similarity to a set of reference centroids. The references were defined using a comprehensive set of 13,160 taxonomic profiles from 1975 women in the USA. This large dataset allowed us to comprehensively identify, define, and characterize vaginal CSTs common to reproductive age women and expand upon the CSTs that had been defined in previous studies. We validated the broad applicability of VALENCIA for the classification of vaginal microbial communities by using it to classify three test datasets which included reproductive age eastern and southern African women, adolescent girls, and a racially/ethnically and geographically diverse sample of postmenopausal women. VALENCIA performed well on all three datasets despite the substantial variations in sequencing strategies and bioinformatics pipelines, indicating its broad application to vaginal microbiota. We further describe the relationships between community characteristics (vaginal pH, Nugent score) and participant demographics (race, age) and the CSTs defined by VALENCIA.
Conclusion
VALENCIA provides a much-needed solution for the robust and reproducible assignment of vaginal community state types. This will allow unbiased analysis of both small and large vaginal microbiota datasets, comparisons between datasets and meta-analyses that combine multiple datasets.
Video abstract.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40168-020-00934-6.
Introduction
It is human nature to group objects and observations into categories based on their commonalities [1]. Doing so allows us to identify similarities and provides a unified framework for thought. This approach is particularly useful when the underlying set of objects or observations are multidimensional and difficult to grasp through intuition. When confronted with the diversity present in the microbial communities that inhabit the human body, microbiome scientists have often turned toward categorization [2]. These communities routinely include hundreds of species with a long tail of taxonomic diversity [3]. Variation in the community composition between specific body sites and individuals can be high, leading many to suggest that each person has their own “microbial fingerprint” [4]. Yet commonalities exist in the taxonomic compositions of these diverse communities and have enabled their categorization into types. This approach has been widely applied to human enteric [5, 6], vaginal [7, 8], skin [9], lung [10], and oral microbial communities [11] and has provided critical insights into the structure, function, and epidemiology of the human microbiome.
Hierarchical clustering (HC) is perhaps the most common approach used to categorize microbiota based on their composition and is performed on a matrix containing the distance between all pairwise combinations of samples. This approach is problematic for at least two reasons. First, the samples are typically clustered only within the study, complicating the interpretation of the findings in the context of other studies. This is especially problematic given the increasing volume of studies published on the human microbiome [12]. Second, because HC relies on pairwise distances between samples, the categories provided are entirely dependent on the particular set of samples included in the clustering. This means that assignments derived from HC can be unstable [13]. Removing a single sample from the dataset has a cascading effect on the assignment of the remaining samples. To overcome these limitations, we developed a nearest centroid classification algorithm to reproducibly place microbial communities into categories based on their composition and structure. This approach leverages a training dataset to define the centroid of each category and then places new data into categories based on the centroid to which they bear the highest similarity. It has been used previously to categorize proteins based on mass spectrometry data [14] and tumor subtypes based on gene expression patterns [15]. The resulting assignments are not dependent on within-study comparisons and therefore do not suffer from the same limitations as those provided by HC. Nearest centroid classification assignments are generally robust and can be compared across studies.
To demonstrate the utility of the nearest centroid classification, we implemented it for the assignment of vaginal microbial community profiles to community state types (CSTs). The concept of CSTs was introduced in 2011 by Ravel et al. to categorize vaginal microbial communities routinely observed among reproductive age women [7], and built upon prior methods to categorize these communities [16]. That study, and many subsequent studies [7, 17–21], have indicated that there are at least five vaginal CSTs, four of which are each dominated by different Lactobacillus spp. and another characterized by a more even community of facultative and obligate anaerobic bacteria (some studies have also distinguished subtypes within this CST [22]). Additional longitudinal studies have demonstrated that there can be a high degree of variation in community composition within a reproductive age woman over time [22–24], making it more appropriate to think of CSTs as a snapshot of the community at the time of sampling (i.e. state type) rather than a “type” which implies it is static over time. The dimensionality reduction provided by the CST approach has allowed epidemiologists to link variation in the vaginal microbiota with vaginal inflammation [25], STI occurrence [21, 26, 27], Candida detection [28], signs of vaginal atrophy [29], and increased risk of preterm birth [30].
To develop our nearest centroid classifier, we leveraged a large dataset of vaginal bacterial community profiles as defined by 16S rRNA gene amplicon sequencing (> 13,000 samples from > 1900 women). This dataset allowed us to comprehensively identify, define, and characterize vaginal CSTs common to North American reproductive age women using HC. In doing so, we recapitulated and expanded upon the CSTs that had been defined in previous studies. We then constructed reference centroids and applied the nearest centroid classification algorithm for the assignment of vaginal microbiota profiles to CSTs. We demonstrated the utility and robustness of the resulting tool, VALENCIA (VAginaL community state typE Nearest CentroId clAssifier), using several publicly available test datasets that contained vaginal samples from adolescent girls [31], postmenopausal women (from the Study of Women’s Health Across the Nation (SWAN)), and eastern and southern African women [32]. These test datasets were also derived from the sequencing of different 16S rRNA variable regions and different bioinformatics pipelines. Finally, we examine relationships between vaginal CSTs, as defined by VALENCIA, and host (race, age) and community (pH, Nugent score) characteristics. We anticipate that VALENCIA will prove a critical tool for vaginal microbiota research and potential future clinical application by providing robust and reproducible assignments of samples to CSTs.
Results
Assemblage of the largest dataset of human vaginal microbiota profiles
We compiled a dataset of vaginal community compositions from 13,160 vaginal swab or lavage specimens that had been collected by our research group from three locations around the USA: Baltimore, MD; Birmingham, AL; and Atlanta, GA. The samples originated from 1975 North American women and included participants who self-identified as Black (n = 1343, 68%), White (n = 403, 20.4%), Hispanic (n = 110, 5.6%), Asian (n = 95, 4.8%), as well as 17 women who identified as a different race and 7 women who did not self-identify. Many of the women had participated in longitudinal studies (n = 916) and therefore contributed more than one sample to the compiled dataset; the median number of samples contributed per participant was three and ranged from one to seventy. All of the women included in this study were of reproductive age as defined by recent menstruation and were not pregnant at the time of sampling. The participant age range was 13–53 with a median participant age of 25. Eleven of the women were younger than 15 and two were older than 49. The composition of their vaginal microbiota was established by deep sequencing of the V3–V4 region of the 16S rRNA gene with an average of 54,898 reads per sample (range 1005–411,805).
Construction of community state type reference centroids
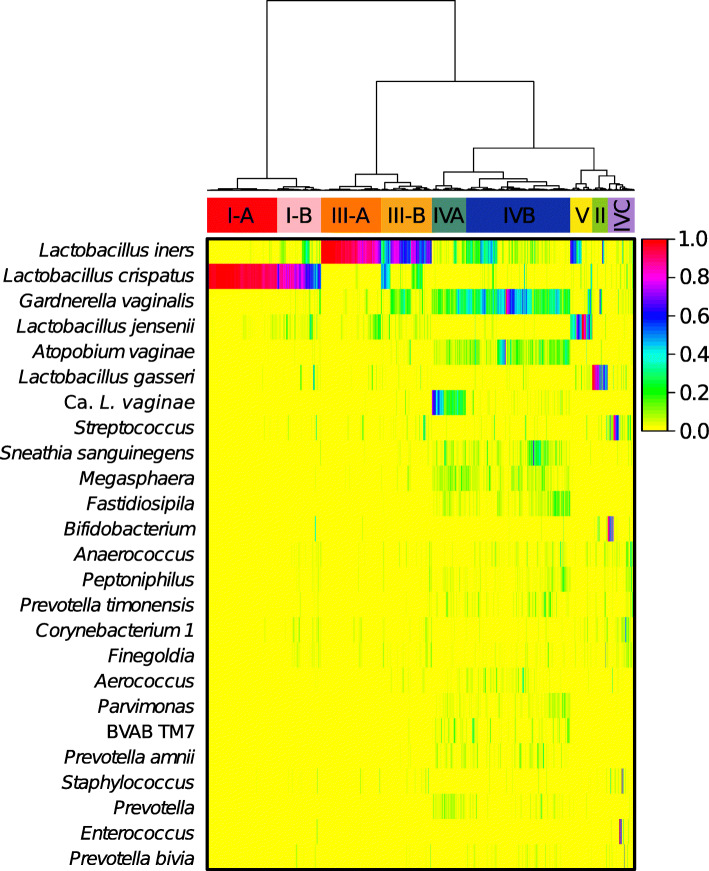
The compiled dataset of 13,160 vaginal microbiota profiles includes representations of all previously identified CSTs and was used as a comprehensive training dataset. We first defined the CSTs in the training dataset using hierarchical clustering of the pairwise Bray-Curtis distances between samples with Ward linkage (Fig. 1). We then identified seven CSTs, four of which had a high relative abundance of Lactobacillus species. These seven CSTs could be further broken down into thirteen sub-CSTs. To conform with previous studies, we name these as follows: CST I—L. crispatus dominated, CST II—L. gasseri dominated, CST III—L. iners dominated, and CST V—L. jensenii dominated. CSTs I and III were more common in this dataset than CSTs II and V and were each split into two sub-CSTs denoted with A and B. The “A” version represents samples that had a higher relative abundance of the focal species, with the “B” version representing samples with a somewhat lower relative abundance of that species. We also identified three CSTs which did not have a high relative abundance of lactobacilli which we term CST IV-A, IV-B, and IV-C. CST IV-A had a high relative abundance of Candidatus Lachnocurva vaginae (formerly known as BVAB1 [33]) and a moderate relative abundance of G. vaginalis, while IV-B had a high relative abundance of G. vaginalis and low relative abundance of Ca. L. vaginae. Both IV-A and IV-B had moderate relative abundances of Atopobium vaginae. Samples assigned to CST IV-C had a low relative abundance of Lactobacillus spp., G. vaginalis, A. vaginae, and Ca. L. vaginae and were instead characterized by the abundance of a diverse array of facultative and strictly anaerobic bacteria. We thus further split CST IV-C into 5 sub-CSTs as follows: CST IV-C0—an even community with moderate amount of Prevotella, CST IV-C1—Streptococcus dominated, CST IV-C2—Enterococcus dominated, CST IV-C3—Bifidobacterium dominated, and CST IV-C4—Staphylococcus dominated. Samples assigned to CST IV-C represented 6% (n = 802) of the training dataset.
Fig. 1.
Heatmap displaying the taxonomic composition of 13,160 vaginal microbial communities using the 25 most abundant phylotypes across all samples. Hierarchical clustering was performed using Bray-Curtis dissimilarity with Ward linkage. Seven community state types were defined, four of which were dominated by a single species of Lactobacillus and three which were not. This dataset was used to train VALENCIA
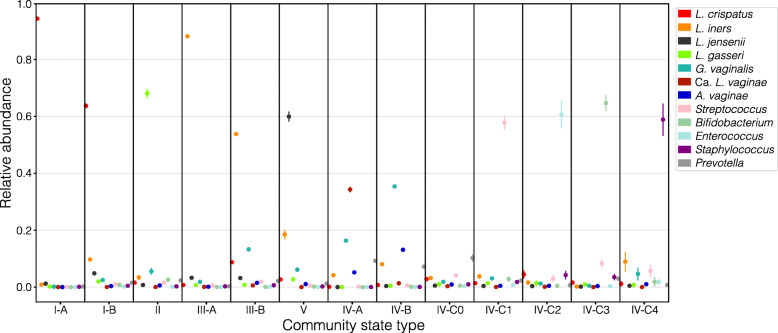
We next constructed a reference centroid for each of the thirteen sub-CSTs identified in the training dataset by averaging the relative abundances of each taxa across the samples assigned to the sub-CST. These centroids represent the average community composition of each sub-CST, as defined by the training dataset, and can be used as a stable reference for the assignment of vaginal microbiota profiles to CSTs (Fig. 2).
Fig. 2.
Average relative abundance of twelve key taxa across all of the samples used to define each of the thirteen sub-CSTs. Error bars represent the standard error of the mean as defined using 100 bootstraps of ten percent of the training dataset. These “average” communities define the reference centroids used by VALENCIA to assign new samples to sub-CSTs. Sub-CST IV-C0 is not dominated by any one species. CST V has 20% relative abundance of L. iners in addition to L. jensenii, indicating these two species can co-occur. This relationship is maintained over extended periods of time in some longitudinal profiles
VALENCIA: a novel method for assigning samples to community state types
We implemented a nearest centroid classification algorithm, which we term VALENCIA, to leverage the training dataset for reproducible and robust assignment of vaginal microbiota to community state types. The similarity of a vaginal microbiota profile to each of the thirteen reference centroids is evaluated using Yue and Clayton’s θ [34], a similarity measure based on species proportions. This yields an array of thirteen similarity scores, ranging from 0.0 (no shared taxa) to 1.0 (all taxa shared and at the same relative abundance) for each sample. The reference centroid to which the sample bears the highest similarity provides an optimal assignment to a sub-CST. These similarity scores can also be used to gauge confidence in the assignment and are particularly useful for handling cases where the sample either does not match any of the centroids or has close matches to multiple centroids. For these cases, VALENCIA yields a low confidence score to indicate the degree of ambiguity in the CST assignment.
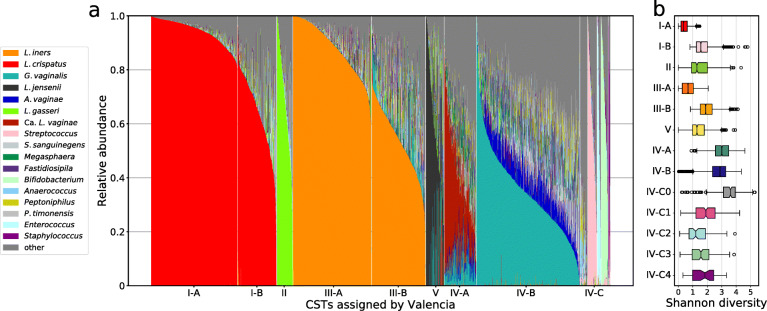
We first used VALENCIA to reassign CSTs to our training dataset. The taxonomic composition of samples assigned to each CST generally matched that of the associated reference centroid (Fig. 3a). As expected, differences in Shannon diversity (H) were observed among the sub-CSTs (Fig. 3b). Samples assigned to sub-CSTs defined by the dominance of a single phylotype were found to have lower values of H. A comparison between the initial hierarchal clustering and the assignments provided by VALENCIA revealed 1454 disagreements (11% of the samples). Discordant samples largely originated from communities which bore some degree of similarity to multiple CSTs. These communities exist in the grey areas between two or more community state types and are therefore difficult to classify. Based on the taxonomic profiles for these discordant samples, we have more confidence in the assignment provided by VALENCIA than that provided by hierarchical clustering (Additional file 1). For example, a number of samples were assigned to CST V by the hierarchical clustering but only contained ~ 20% relative abundance of L. jensenii and were instead majority L. iners. VALENCIA assigned these samples to CST III-B, which we find more agreeable. A similar pattern can be seen for discordant sample hierarchical clustering assigned to CST II which, based on the reference centroid for this community state type, should have a majority of L. gasseri. In this case, the discordant CST II samples had either a majority L. iners, which VALENCIA assigned to CST III-B or a majority G. vaginalis, which VALENCIA assigned to CST IV-B.
Fig. 3.
Taxonomic composition of all samples (n = 13,160) in the training data set categorized by sub-CST assignment according to VALENCIA (a). Distribution of Shannon diversity index values by sub-CST assignment (b). Shannon diversity was calculated using the log base 2
Validation of VALENCIA against outside datasets
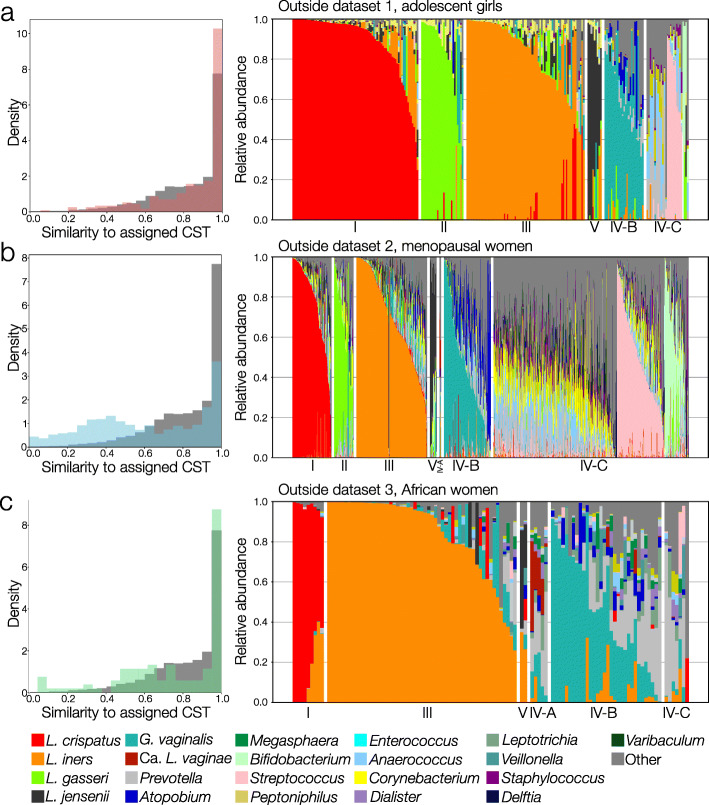
To further demonstrate the broad applicability of VALENCIA, we applied it to a number of test datasets that had not been included in the training dataset. These test datasets are from studies which had sampled women outside the age range of the training dataset, from non-North American populations, or had sequenced different 16S rRNA gene variable regions. Here we present our analysis of three such datasets: Test dataset 1 contained publicly available microbiota profiles from adolescent girls (n = 245, aged 10 to 15) derived from sequencing the 16S rRNA gene V1–V3 region [31]. Test dataset 2 was generated from SWAN analyzed in-house and contained microbiota profiles from postmenopausal women (n = 1380, aged 60 to 72) derived from sequencing the 16S rRNA gene V3–V4 region. Finally, test dataset 3 contained publicly available microbiota profiles from eastern and southern African reproductive age women (n = 110) derived from sequencing the 16S rRNA gene V4 region [32]. For test dataset 2, we applied our own taxonomic annotation pipeline to the data while for test datasets 1 and 3 we relied on the taxonomic annotations provided in the original studies. We assigned all of the samples in each of these three datasets to community state types using VALENCIA. In general, we find the CST assignments made by VALENCIA to be acceptable, although there is no “ground truth” from which to benchmark (Fig. 4). The distributions of similarity scores between the samples and their matching reference centroid for outside datasets 1 and 3 did not substantially differ from the same distribution provided by the reclassification of the training dataset (Fig. 4). Similarity scores for test dataset 2 were typically lower than those for the training dataset, although this is primarily driven by the high prevalence of CST IV-C and low prevalence of Lactobacillus spp.-dominant CSTs in postmenopausal women. For test dataset 1, the authors made their CST assignments (which were derived from hierarchical clustering) publicly available [31]. We find 17 instances of discordance between our and the original assignments (6.7%). As was the case for the reclassification of the training dataset, these discordant samples occupy the grey space between CSTs. For example, six instances of discordance come from communities which were assigned to CST I by Hickey et al. but had more L. iners than L. crispatus—VALENCIA assigned these samples to CST III-B.
Fig. 4.
Validation of VALENCIA using three test datasets of vaginal taxonomic profiles derived from sequencing of the 16S rRNA gene. For each dataset, the similarity of each sample to its assigned sub-CST is plotted as a normalized histogram (left, a red, b blue, c green) versus that for the training dataset (dark grey). The taxonomic composition of each sample in the dataset is also provided (right). Test dataset 1 (a) was published by Hickey et al., contained 245 samples, was derived from sequencing of the V1V3 region, and contained samples from adolescent girls. Test dataset 2 (b) contained 1380 samples from menopausal women and was derived from sequencing of the V3V4 region. Test dataset 3 (c) was published by McClelland et al., contained 110 samples from eastern and southern African women, and was derived from sequencing of the V4 region
Relationships between VALENCIA-defined community state types, Nugent score, and vaginal pH
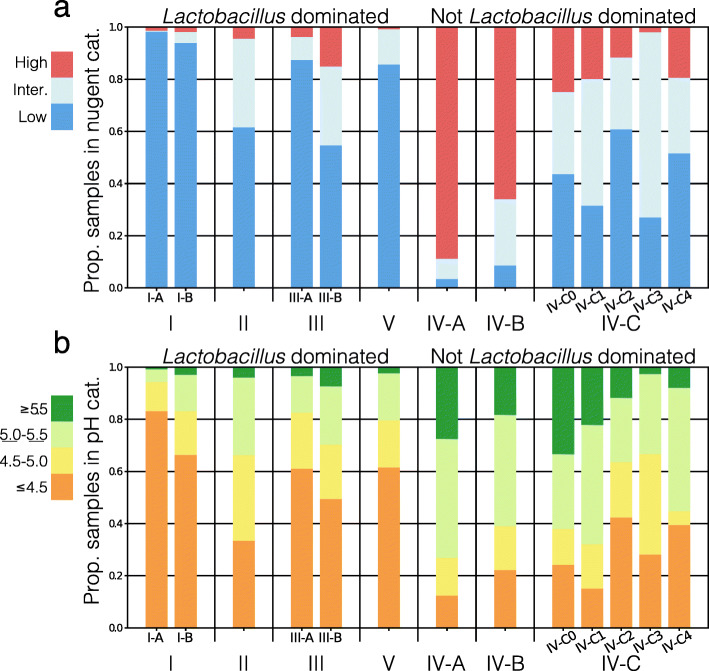
Prior to the application of amplicon sequencing to define microbiota composition, and still today, researchers and clinicians used the Nugent scoring system to evaluate and categorize vaginal microbial communities for the diagnosis of bacterial vaginosis (BV), a common vaginal condition [35]. The Nugent score is primarily based on the morphology of Gram-stained bacterial cells viewed under light microscopy. An abundance of Lactobacillus morphotypes, large Gram-positive rods, yields a low Nugent score while the presence of Gram-variable small (G. vaginalis) and/or curved rods (Ca. L. vaginae [36], Mobiluncus spp.) yields a high Nugent score, indicating Nugent-BV. We examined the relationship between VALENCIA-defined CSTs and Nugent score categories and observed high concordance (Fig. 5a). Lactobacillus-dominant CSTs typically have low Nugent scores, while communities assigned to other CSTs have higher Nugent scores. CST IV-A VALENCIA-assigned communities, which are enriched of Ca. L. vaginae, had the highest Nugent scores, consistent with the Nugent scoring system [37]. However, we were interested to see how the Nugent score system evaluated communities VALENCIA assigned to CST IV-C. These communities do not have a high relative abundance of Lactobacillus spp., G. vaginalis, or Ca. L. vaginae and instead have a high relative abundance of Streptococcus, Enterococcus, Staphylococcus, Prevotella, and Bifidobacterium. We find that the Nugent scoring system does not reliably assign these communities to a single category and instead gives mixed results. Of the five subtypes of CST IV-C, CST IV-C2 (Enterococcus), and IV-C4 (Staphylococcus) are most often assigned low Nugent scores although these communities are not dominated by Lactobacillus spp. Besides the ambiguities observed for these CST IV-C communities, VALENCIA-defined CSTs and the Nugent scoring system are largely in accordance with the definition of the Nugent score.
Fig. 5.
The relationship between each VALENCIA-assigned sub-CST and Nugent score (a) and vaginal pH (b). Nugent score was separated into high (score 8–10), intermediate (score 4–7), and low (score 0–3) categories. Vaginal pH was split into four categories: less than or equal to 4.5, between 4.5 and 5.0, between 5.0 and 5.5, and greater than or equal to 5.5
The microbiota is thought to be the primary driver of vaginal pH through the release of acidic fermentation end products. A low vaginal pH (≤ 4.5) has been associated with decreased risk of adverse health outcomes and is usually achieved via the production of lactic acid by lactobacilli [38–40]. Not surprisingly then, we found that the communities VALENCIA assigned to CSTs which are dominated by Lactobacillus spp. were associated with lower vaginal pH than those VALENCIA assigned to other CSTs (Fig. 5b, see supplemental figure 2 for odds of vaginal pH > 4.5 for each sub-CST). Communities which were dominated by L. crispatus (CST I) had the lowest pH (78% of samples had a pH ≤ 4.5) with those dominated by L. iners (CST III) and L. jensenii (CST V) following close behind. L. gasseri-dominated communities (CST II) were associated with the highest vaginal pH among the Lactobacillus spp.-dominant CSTs. Also as expected, communities which were deficient in Lactobacillus spp. typically had a higher vaginal pH. Communities VALENCIA assigned to CST IV-A had the highest pH followed closely by those assigned to CST IV-B. Due to the large number of samples included in this dataset, we were also able to examine pH for samples assigned to the less common CSTs. Among the subtypes of CST IV-C, we find that communities with Enterococcus, Staphylococcus, or Bifidobacterium were associated with lower vaginal pH while the majority of Streptococcus communities typically had a higher pH. None of these CST IV-C sub-CSTs (percent of samples with pH ≤ 4.5: IV-C0 24%; IV-C1 15%; IV-C2 42%; IV-C3 28%; IV-C4 39%) consistently reach the low vaginal pH achieved by CSTs I, III, or V (percent of samples with pH ≤ 4.5: I-A 83%; III-A 61%; V 61%, Fig. 5b).
Associations between community state types and participant’s race and age
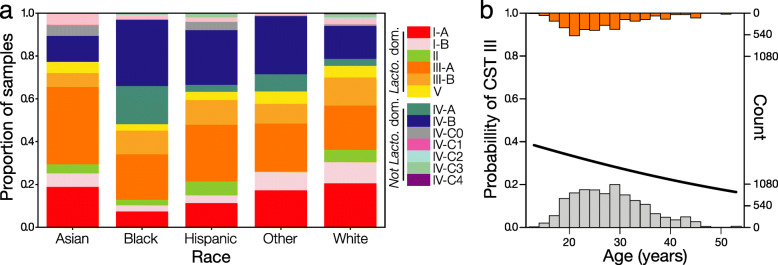
Statistical associations between CSTs and demographic, medical, and/or behavior data have been used to identify factors that influence the composition of vaginal microbiota. Our training dataset contained microbiota compositions from over 1900 allowing us to re-examine some previously observed associations with more statistical power. We sought to determine whether a participant’s race or age influenced their likelihood of having particular community state types. For each community state type with sufficient sample size (I, II, III, IV-A, IV-B, IV-C), we modeled their presence or absence as a binomial response variable with race and age as fixed predictor variables and the subject as a random variable (Fig. 6a). We found that women who self-identify as Black or African American were less likely to have CST I than women who identify as White or Asian (z = 4.6, p < 0.001; z = 2.9, p = 0.0235). Black women were also more likely to have CST IV-A than White women (z = 4.9, p < 0.001) and CST IV-B than women who identified as White or Asian (odds ratios 6.91 versus 0.96 and 0.05; p < 0.001, p < 0.001). None of the Asian women included in this study were found to have CST IV-A. CST IV-B was also more common for Hispanic women than White women (z = 3.3, p = 0.006). Finally, we found that Asian women were more likely to have CST III than either Black or White women, although this association was weaker (odds ratios 1.72 versus 0.34 and 0.36; p = 0.025, p = 0.050). No significant associations with race were found for CSTs II, V, or IV-C, which may be due to sample size limitations as these three CSTs are less prevalent than the others. Our results agree with previous studies [7, 41] and show a more refined association between raise and the prevalence of CSTs.
Fig. 6.
The relationship between the prevalence of each VALENCIA-assigned sub-CST and a woman’s self-identified race (a). Each bar represents the proportion of samples assigned to each CST in women whose race is Asian (n = 95), Black (n = 1,343), Hispanic (n = 110), White (n = 403), or Other (n = 17). For subjects who contributed multiple samples, the within subject relative prevalence of each CST was used in the calculation instead of their individual CST counts. We also examined relationships between the prevalence of each CST and a woman’s age (b). Only the prevalence of CST III was found to have a relationship with age among reproductive-age women. Bars represent the age distribution of subjects whose samples were (orange) or were not (grey) assigned to CST III. Older reproductive age women were less likely to have communities assigned to CST III than younger reproductive age women
We were also able to examine associations between a participant’s age and their likelihood of having each CST using the same logistic regression models. Our analyses indicate that the representation of CST III varies with age, after adjusting for race (Fig. 6b). Among reproductive age women, we observed that older women were less likely to have L. iners-dominated CSTs than younger women (z = − 3.589, p < 0.001). The probability of having CST III ranged from 40% for the youngest women included in the study, down to 20% for the oldest women. No significant associations with age were observed for the other CSTs. However, in our validation of VALENCIA, we analyzed the SWAN dataset of postmenopausal women between the ages of 60 and 72 (Fig. 4b). As expected, the representation of CSTs clearly differs between this dataset of postmenopausal women and the training dataset of reproductive age women.
Discussion
The classification of vaginal microbial communities into CSTs has proven to be highly valuable. Since their inception in 2011 [7], studies have shown associations between vaginal CSTs and host immune profiles [25] and have further linked particular CSTs to sexually transmitted infections [18, 21, 26, 27] and experiencing spontaneous preterm birth [8, 42, 43]. From these studies, it is clear that the CST classification system captures meaningful information about these communities, despite its apparent simplicity. One of the advantages of the CST classification approach is that it enables the usage of standard and vetted statistical models to demonstrate these associations. On the other hand, a common criticism of the CST approach is that it simplifies the communities by distilling their composition down to a single categorical variable. One alternative has been to instead model the abundance of each taxon individually. However, these analyses are complicated by spurious correlations introduced because each taxa’s abundance is expressed relative to the others when assessed via amplicon sequencing [44]. Others have used species-specific quantitative PCR assays to determine their absolute abundances. Yet this method is expensive, requires the development and implementation of many individual qPCR assays, and only quantifies the targeted species. New statistical and methodological approaches are needed that can facilitate such taxa-level association studies which take into account the specific statistical challenges of compositional data [45]. Even then, there appears to be a place for “CST-level” analyses in vaginal microbiome research as the approach provides a higher-level overview and epidemiological characterization of the vaginal microbial community across populations and serves as a guide for more in-depth analyses.
In order for a classification scheme to be useful, it must be reproducible. It immediately becomes difficult to compare the results from multiple studies if the samples were not assigned to categories in the same manner. Prior work on assignment of vaginal microbial communities to CSTs has primarily been accomplished through within-study hierarchical clustering of the pairwise distances between samples, which does not yield reproducible assignments. Other studies have used taxa-specific relative abundance thresholds to assign CSTs (e.g., communities with > 30% relative abundance of L. crispatus are assigned to CST I [19, 46]). However, this approach does not consider the entire microbial community and may offer a limited view of the vaginal microbiota when only a few taxa are considered. VALENCIA provides a unified method to accomplish this task which is based on the overall structure and composition of the community. The CST assignments provided by VALENCIA are robust and reproducible across studies and will enable researchers to leverage the numerous existing vaginal microbiota datasets for use in large-scale meta-analyses. On the other end of the scale, we also expect that VALENCIA will assist the analysis of small datasets, which are often plagued by poor CST assignments. Because VALENCIA is reference-based, every sample is treated independently, making it scalable to large or small datasets—VALENCIA can even be used to assign a single sample to a CST. The structured nature of VALENCIA CSTs also allows the researcher to tune the number of CSTs considered to the size of the study (e.g., five, seven, nine or thirteen). Unlike previous classification methods, VALENCIA also provides an estimate of confidence derived from similarity of each sample to each reference centroid. These values can be used in a resampling scheme to investigate the effect of shuffling communities which bear similarity to multiple CSTs.
Overall, there is an astonishing concordance in the makeup of the vaginal microbiota. Most women were found to have communities which fit neatly within the CSTs that we have defined, driving the broad applicability of VALENCIA for the classification of vaginal microbiota. Despite this, we and others have shown that there are differences in the prevalence of particular CSTs that are associated with a woman’s race [7, 41]. Care should be taken in conveying precisely what these differences are and what they are not. In particular, we have found that women of African descent are less likely to have a L. crispatus-dominated community and more likely to have CST IV-B than women of European or Asian descent. It is important to note that these results are not consistent with there being distinct or systematic differences in the taxonomic composition of the communities. In this study, we identified every CST in women from each self-identified racial category with the exception of CST IV-A in Asian women. It is merely that some CSTs are more prevalent in women of a certain race while other CSTs are less prevalent. The factors that drive these differences in the representation of vaginal CSTs have yet to be determined, are likely to be multifaceted, and could depend on host and/or microbial factors. For example, our recent study indicated that there may be racial differences in the interplay between vaginal microbial communities and the host immune system [43].
The number of samples included in our training dataset allowed us to define and investigate some of the less prevalent types of Lactobacillus-deficient vaginal communities. We placed these communities into CST IV-C, and further defined five subtypes. Of these, CST IV-C1 (Streptococcus-dominated) and CST IV-C3 (Bifidobacterium-dominated) were the most common among reproductive age women. The Nugent scoring system was not designed with either of these taxa in mind [35] and it is not clear how they relate to vaginal health [47]. While group B Streptococcus is a known neonatal pathogen, it is not known whether its pathogenic potential is at all realized in the vaginal environment [48]. Like the lactobacilli, many Streptococcus spp. can produce lactic acid as a fermentation end product [49] and therefore might be able to lower vaginal pH to a similar degree. Alas, we found these communities to instead be associated with high vaginal pH, indicating that vaginal Streptococcus spp. might produce other fermentation end products in the vagina. Bifidobacterium, on the other hand, has a reputation for being a “healthy” microbe based on its activity in the gut environment [50]. Due to its rarity, associations between Bifidobacterium-dominated communities and vaginal health have yet to be assessed. Although they are generally capable of producing L-lactic acid [51], we found that only about a quarter of women with Bifidobacterium-dominated communities had a vaginal pH ≤ 4.5, indicating it likely does not provide the same level of pH-mediated protection as a Lactobacillus-dominated community. Going forward, we hope that VALENCIA will enable association studies between these two uncommon CSTs and vaginal health. However, both the Streptococcus and Bifidobacterium genera include a diverse array of described species. Species- and strain-level characterization using metagenomic approaches may be necessary to understand their ecology in the vaginal environment.
As other reference-based approaches, one potential limitation of VALENCIA is that it is not able to classify communities which were not included in the training dataset. Though VALENCIA was demonstrated to be applicable on different populations, age ranges, and 16S rRNA regions, misclassification could happen for samples with novel bacteria or different community structures. The only potential issues we observed were related to the presence of community profiles which did not completely match those in the reference. For example, CST IV communities from some African populations tend to have a higher relative abundance of Prevotella spp. than CST IV communities from North American women [21, 25, 32]. We have shown, in our analysis of test dataset 3, that VALENCIA assigns these communities to one of the subtypes of CST IV depending on the presence and abundance of other taxa. However, an argument could be made for the addition of a novel CST and reference centroid which is Prevotella-dominated as defined by samples from African women. VALENCIA can be expanded by the addition of novel CSTs not found in the training dataset, when appropriate. This could be achieved either by adding data defining these novel CSTs to the existing training dataset or by constructing new training datasets from samples originating from a specific population of interest. As we collect more data from women across the globe, we plan to expand, update, and maintain VALENCIA.
We used the nearest centroid categorization approach to assign vaginal microbiota profiles to community state types. Our training and test datasets were all derived from 16S rRNA gene amplicon sequencing, but VALENCIA could also be used to categorize vaginal communities based on composition as established by shotgun metagenome data. This would likely not require additional changes to the tool or training dataset. In addition to this natural extension, a similar nearest centroid approach could be applied to the classification of other microbial communities into types. In many ways, vaginal microbiota are perhaps the easiest to categorize based on taxonomic composition because of their tendency to be dominated by a single species, which results in fairly distinct lines between community state types. The microbial communities which inhabit other body sites [52] or other environments (e.g., soil [53], ocean water [54]) tend to have communities which are more even and species rich. This is likely to blur the lines between community types and may complicate the use of the nearest centroid approach for their categorization. Microbial communities could also be categorized based on their functional and metabolomic composition. Classification of these multidimensional microbiome datasets would likely make their analysis and interpretation less complicated. A reference-based approach like the nearest centroid classification used here would provide robust and reproducible assignments.
Conclusion
We used a large dataset of over 13,160 vaginal microbiota profiles to train a nearest centroid classifier (VALENCIA) to infer community state types. The large training dataset allowed us to define CSTs which represent more uncommon vaginal microbiota compositions (e.g., those dominated by Bifidobacterium spp.). Our validation efforts demonstrated that VALENCIA provides robust and reproducible assignments of vaginal microbiota profiles to CSTs that are insensitive to a women’s age or geographic location. VALENCIA assignments are also largely unaffected by which variable region of the 16S rRNA gene was sequenced or which bioinformatics pipeline was used to taxonomically identify the resulting sequences. We expect that VALENCIA will enable epidemiological investigations into the factors that drive changes in the vaginal microbiota and associations between these communities and vaginal health. The reproducibility of this approach will allow for much-needed meta-analyses that combine the results of the myriad of existing studies on the vaginal microbiota.
Methods
Participants and sampling procedures
The training dataset of 13,160 vaginal microbiota profiles originated from several different studies, all of which have been published previously [7, 22, 55–59]. A detailed explanation of the sample procedures and study populations can be found in the original publications. Samples were either self-collected or physician-collected by swabbing the mid-vagina (n = 11,387) or physician collected via a vaginal lavage with 3 mL of sterile deionized water (n = 1844). Vaginal swabs and vaginal lavage samples were frozen at − 80 °C. Participants also provided behavior and lifestyle information. Nugent scoring was performed as previously described [7, 35]. Vaginal pH was established using the VpH glove (Inverness Medical) and binned into categories (≤ 4.5, greater than 4.5 but less than 5.0, between 5.0 and 5.5 inclusive, and ≥ 5.5). All studies were performed under Institutional Review Board-approved protocols, and samples were collected after obtaining written informed consent from all the participants.
DNA extraction
DNA was extracted from the samples using a combination of enzymatic digestion and mechanical disruption as described in Holm et al. [60]. Briefly, vaginal swab or lavage specimens were resuspended in phosphate buffer saline solution. A 0.5 mL aliquot of the cell suspension was incubated at 37 °C for 30 min following the addition of an enzymatic digestion cocktail (contents: 5 μl of lysozyme at 10 mg/ml, EMD Chemicals, Gibbstown, NJ; 13 μl of mutanolysin at 11,700 U/ml; Sigma-Aldrich, St. Louis, MO; and 3.2 μl of lysostaphin a 1 mg/ml; Ambi Products, LLC, Lawrence, NY). This was followed by the addition of 10 μl of Proteinase K (20 mg/ml; Invitrogen), 50 μl of 10% SDS (Sigma-Aldrich, St. Louis, MO), and 2 μl of RNase A (10 mg/ml; Invitrogen, Carlsbad, CA) and a further 45 min of incubation at 55 °C. Mechanical disruption was then performed using a FastPrep homogenizer at 6 m/s for 40 s. DNA was then purified from the crude lysates using the QS DSP virus/pathogen midi kit on the QIAsymphony robotic platform (Qiagen, Hilden, Germany) according to the manufacturer’s specifications.
16S rRNA gene amplification, sequencing, and analysis
PCR amplification of the V3V4 region of the 16S rRNA gene was conducted using either the “one-step” or the “two-step” amplification protocols described and validated in Holm et al. [60]. Primer sequences can be found in supplemental table 1. For the one-step protocol, extracted DNA (0.5 μl) was added to Phusion Taq master mix (ThermoFisher, Waltham) with 3% dimethylsulfoxide (DMSO) and each primer (final concentration 0.4 μM). Initial denaturation was performed at 98 °C for 30 s, followed by 30 cycles of denaturation (98 °C, 15 s), annealing (58 °C, 15 s), and extension (72 °C, 15 s). Final extension was conducted at 72 °C for 60 s. The two-step protocol utilized two rounds of PCR amplification: the first amplifies the V3V4 region of the 16S rRNA gene and adds Illumina sequencing primers, and the second adds 8-bp barcode sequences. Both two-step reactions utilized the same Phusion Taq master mix with 3% DMSO and primers at 0.4 μM. The first round also included 0.5 μl of extracted DNA. Initial denaturization was conducted at 94 °C for 3 min, followed by 20 cycles of denaturation (94 °C, 30 s), annealing (58 °C, 30 s), extension (72 °C, 1 min), and a final extension step at 72 °C for 7 min. The second round of amplification used 1 μl of 1:20 diluted PCR product from the first round as input. Initial denaturation was conducted at 94 °C for 30 s, followed by 10 cycles of denaturation (94 °C, 30 s), annealing (58 °C,30 s), extension (72 °C, 60 s), and then a final extension (72 °C, 5 min). Deep sequencing was accomplished on either an Illumina MiSeq or an Illumina HiSeq 2500 [60]. The resulting paired end sequences were processed using DADA2 [61] to identify amplicon sequence variants (ASVs) and remove chimeric sequences following general practices (https://benjjneb.github.io/dada2/bigdata.html). Following processing by DADA2, each sample had an average of 54,898 reads (range 1005–411,805). Taxonomy was assigned to each ASV using the RDP Naïve Bayesian Classifier [62] trained with the SILVA 16S rRNA gene database [63]. For several key genera (e.g., Lactobacillus, Prevotella, Sneathia, Mobiluncus), the ASVs were further classified to the species level using speciateIT (version 1.0, http://ravel-lab.org/speciateIT). Read counts for ASVs that were assigned to the same phylotype were combined. The final dataset contained 199 taxa following removal of those we identified as contaminants as well as those taxa present at a frequency of less than 10−5 study wide. The same bioinformatics procedures were used to analyze the in-house test dataset 2.
Construction of the reference centroids
Hierarchical clustering of the 13,160 taxonomic profiles using Bray-Curtis distances and ward linkage was first employed to define the vaginal CSTs (Fig. 1). This analysis recovered the canonical five CSTs as described in Ravel et al. [7] but went further in delineating subtypes among the five CSTs. Cluster selection was made using the cutree function from the R stats package (version 3.6.0) on the dendrogram produced by hierarchical clustering. Cluster numbers from 2 to 20 were produced and then evaluated using the Davies Bouldin score. Support was found for nine clusters (Supplemental Figure 3c). For the L. crispatus-dominated CSTs, we were able to distinguish between communities which had mostly just L. crispatus (CST I-A) and those that had a lower, moderate relative abundance of the species (CST I-B). The same paradigm was observed for L. iners-dominated communities. Communities dominated by L. gasseri and L. jensenii more uncommon and were therefore not split into sub-CSTs. We were also able to distinguish three non-CSTs with a paucity of lactobacilli: CST IV-A, which contained Ca. L. vaginae, G. vaginalis, A. vaginae, and Prevotella; CST IV-B which contained G. vaginalis, A. vaginae, and Prevotella; and CST IV-C which was characterized by a paucity of Lactobacillus spp., G. vaginalis, Ca. L. vaginae, and A. vaginae. A second round of hierarchical clustering was performed (Bray-Curtis distances, ward linkage) on just the CST IV-C communities to further split this diverse collection of communities into additional sub-CSTs. Cutree was used on the resulting dendrogram to split IV-C into five subtypes, four of which had a characteristic phylotype and one which had a more even taxonomic composition. This decision improved assignments to IV-C, enhanced the interpretation of these communities, and resulted in a better Davies Bouldin score (Supplemental Figure 3). Reference centroids were constructed by averaging the relative abundances of each phylotype across the samples in training dataset which were included in each of the 13 sub-CSTs. Shannon diversity of samples assigned to each sub-CST was calculated using the log base 2.
Implementation of the nearest centroid classification
VALENCIA uses the nearest centroid approach to classification to assign new samples to sub-CSTs based on their taxonomic composition and was implemented in python (version 3.6) and has the pandas module as a dependency [64]. The similarity of each sample to each reference centroid is assessed using Yue-Clayton’s θ [34], which considers the number and proportion of shared and unique phylotypes in its measure of similarity. Compared to Bray-Curtis or Jensen-Shannon, the Yue-Clayton θ measure depends more on the high relative abundance phylotypes than those that are at lower relative abundances. Samples are assigned to the sub-CST to which they bear the highest similarity and the degree of similarity to that sub-CST can be taken as a measure of confidence in the assignment. VALENCIA reports which sub-CST a sample was assigned to, as well as the set of similarity scores to each of the thirteen sub-CSTs. Also included in the output is a higher-order CST assignment which does not differentiate between the subtypes of I, III, or IV.
Running VALENCIA
The expected input of VALENCIA is a table of taxa read counts in each sample with the phylotypes as columns and the samples as rows. The first column should contain a unique identifier for the sample with the column heading “sampleID”. The second column should contain the total read count for the sample with the column name “read_count”. The remaining columns should contain the read count for each phylotype in the dataset. It is imperative that phylotype column headings match those used by the VALENCIA reference centroids which generally take the form of “phylotype rank underscore phylotype name” (e.g., g_Bifidobacterium). All phylotypes should be summarized to the genus rank or higher except for the following: Lactobacillus spp., Gardnerella spp., Prevotella spp., Atopobium spp., Sneathia spp, Mobiluncus spp. These key phylotypes appear as “Genus underscore species” (e.g., Lactobacillus_crispatus, Gardnerella_vaginalis). The other required input is a provided file which contains the reference centroids. The expected output is a modified version of the input data table with added columns indicating the CST and sub-CST designations, the similarity of the sample to the assigned CST, and the array of similarities scores to all of the reference centroids. The tool also generates a figure illustrating the performance of VALENCIA on the dataset which displays the distribution of similarity scores for samples assigned to each sub-CST, compared to the median similarity score for the sub-CST in the training dataset. Substantial differences may indicate either an incongruence in taxonomic names or the need for an additional sub-CST and associated centroid. VALENCIA, the reference centroids, and the training dataset are all available at github.com/ravel-lab/VALENCIA.
Validation efforts
Although there is no “gold-standard” to benchmark VALENCIA against, we did perform a number of tests to validate its use for the classification of vaginal microbial communities. First, we reclassified the training dataset using VALENCIA and compared the new assignments to those provided by the initial HC. We also tested the use of VALENCIA on other populations and on taxonomic compositions that had been generated by the sequencing of other 16S rRNA variable regions and other bioinformatics pipelines. Three datasets were used—two were published and made available by other groups and one which had been generated in-house. Test dataset 1 was published by Hickey et al in 2015 [31] and contained samples from adolescent girls aged 12–15. Test dataset 2 was generated in-house and contained samples from menopausal women above the age of 60. These data are available at github.com/ravel-lab/VALENCIA. Test dataset 3 was published by McClelland et al. [32] and contained samples from reproductive age eastern and southern African women. Test dataset 1 was derived from sequencing the V1V3 region of the 16S rRNA gene, test dataset 2 from the V3V4 region, and test dataset 3 from the V4 region. For test datasets 1 and 3, the published taxonomic assignments were used with adjustments to match the phylotype naming scheme used by VALENCIA. The in-house data included in test dataset 2 was generated via the same methods used in the generation of the training dataset. All three test datasets were classified using VALENCIA, the results of which are shown in Fig. 4. Although many factors can introduce bias in the assessment of taxonomic composition (e.g., DNA extraction, PCR primer selection [65, 66]), we did not find substantial irregularities in the assignment of CSTs by VALENCIA on these test datasets.
Statistical analysis
Associations between the representation of each CST (I, II, III, IV-A, IV-B, IV-C, V) and a participant’s race and age were tested using separate generalized logistic regression models. The presence or absence of each CST was used as a response variable and the participant’s race and age were included as categorical and continuous predictor variables, respectively. Because many of the participants had included multiple samples, participant was also included as a random effect. The glmer function from the lme4 package [67] (version 1.1-21) for R [68] (version 3.6.0) was used with the bobyqa optimization function and 105 iterations. Effect sizes were exponentiated using the R package broom.mixed [69] (version 0.2.4). All scripts used in the statistical analysis are available at github.com/ravel-lab/VALENCIA. The same approach was used to model the association between vaginal pH and each sub-CST. A single generalized logistic regression model was used to model vaginal pH as a binary variable with categories: ≤ 4.5 or > 4.5. The VALENCIA-defined sub-CST was used as a fixed predictor variable along with the subject as a random effect.
Supplementary information
Additional file 1: Supplemental Figure 1. Illustration of cluster selection for construction of VALENCIA sub-CST centroids. Stretched version of dendrogram from Figure 1 with horizontal line indicating chosen cluster threshold (a). Subset heatmap and dendrogram of samples in CST IV-C (b). Hierarchical clustering was performed on this subset using Bray-Curtis dissimilarity with Ward linkage and the horizontal line indicating cluster threshold. Silhouette and Davies-Bouldin scores for a range of number of clusters (c). Horizontal lines indicate the values for the final 13 sub-CSTs, after sub-clustering the CST IV-C samples.
Additional file 2: Supplemental Figure 2. Odds of women with each sub-CST having a vaginal pH >4.5.
Additional file 3. Supplemental Figure 3.
Acknowledgements
The authors acknowledge Courtney Robinson for assistance in compiling the data and insightful discussions. Data from the Study of Women’s Health Across the Nation (SWAN) were used in this study; we would like to acknowledge the critical contribution of SWAN. SWAN has grant support from the National Institutes of Health (NIH), DHHS, through the National Institute on Aging (NIA), the National Institute of Nursing Research (NINR) and the NIH Office of Research on Women’s Health (ORWH) (Grants U01NR004061; U01AG012505, U01AG012535, U01AG012531, U01AG012539, U01AG012546, U01AG012553, U01AG012554, U01AG012495). Clinical Centers: University of Michigan, Ann Arbor—Siobán Harlow, PI 2011–present, MaryFran Sowers, PI 1994–2011; Massachusetts General Hospital, Boston, MA—Joel Finkelstein, PI 1999–present; Robert Neer, PI 1994–1999; Rush University, Rush University Medical Center, Chicago, IL—Howard Kravitz, PI 2009–present; Lynda Powell, PI 1994–2009; University of California, Davis/Kaiser—Ellen Gold, PI; University of California, Los Angeles—Gail Greendale, PI; Albert Einstein College of Medicine, Bronx, NY—Carol Derby, PI 2011–present, Rachel Wildman, PI 2010–2011; Nanette Santoro, PI 2004–2010; University of Medicine and Dentistry—New Jersey Medical School, Newark—Gerson Weiss, PI 1994–2004; and the University of Pittsburgh, Pittsburgh, PA—Karen Matthews, PI. NIH Program Office: National Institute on Aging, Bethesda, MD—Chhanda Dutta 2016–present; Winifred Rossi 2012–2016; Sherry Sherman 1994–2012; Marcia Ory 1994–2001; National Institute of Nursing Research, Bethesda, MD—Program Officers. Central Laboratory: University of Michigan, Ann Arbor—Daniel McConnell (Central Ligand Assay Satellite Services). Coordinating Center: University of Pittsburgh, Pittsburgh, PA—Maria Mori Brooks, PI 2012–present; Kim Sutton-Tyrrell, PI 2001–2012; New England Research Institutes, Watertown, MA—Sonja McKinlay, PI 1995–2001. Steering Committee: Susan Johnson, Current Chair; Chris Gallagher, Former Chair. We thank the study staff at each site and all the women who participated in SWAN.
Abbreviations
- CST
Community state type
- HC
Hierarchical clustering
Authors’ contributions
MF, JR, and RB devised the study. MF designed and wrote VALENCIA. MF, BM, and PG performed the data analysis. SB and JH compiled the data and metadata. MH generated the data. MF, BM, and JR wrote the manuscript with edits from RB, SB, and JH. All authors read and approved the final manuscript.
Funding
The research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers U19AI084044, UH2AI083264, R01AI119012, and R01NR015495. The content of this article is the sole responsibility of the authors and does not necessarily represent the official views of the NIAID, NIA, NINR, ORWH, or the NIH.
Availability of data and materials
VALENCIA, the training dataset, test dataset 2, and all scripts used to analyze and display the data are available at https://github.com/ravel-lab/VALENCIA. Information on how to obtain test datasets 1 and 3 can be found in their respective papers.
Ethics approval and consent to participate
The internal data used to build and test VALENCIA were derived from archived and de-identified cervicovaginal lavages and swabs. The samples were originally collected after obtaining informed consent by all participants, who also provided consent for storage of the samples and used in future research studies related to women’s health. The original study was approved by the University of Maryland School of Medicine Institutional Review Board.
Consent for publication
Not applicable
Competing interests
JR is co-founder of LUCA Biologics, a biotechnology company focusing on translating microbiome research into live biotherapeutics drugs for women’s health. All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Plato. 1925. Statesman. Philebus. Ion.trans. Harold N. Fowler, W. R. M. Lamb.
- 2.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(649-662):e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meadow JF, Altrichter AE, Bateman AC, Stenson J, Brown GZ, Green JL, et al. Humans differ in their personal microbial cloud. PeerJ. 2015;3:e1258. doi: 10.7717/peerj.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costea PI, Hildebrand F, Arumugam M, Backhed F, Blaser MJ, Bushman FD, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2018;3:8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, Mcculle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women., vol 108, p 4680-4687. [DOI] [PMC free article] [PubMed]
- 8.Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The vaginal microbiome and preterm birth. Nat Med. 2019;25:1012–1021. doi: 10.1038/s41591-019-0450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal LN, Clemente JC, Tsay J-CJ, Koralov SB, Keller BC, Wu BG, Li Y, Shen N, Ghedin E, Morris A, Diaz P, Huang L, Wikoff WR, Ubeda C, Artacho A, Rom WN, Sterman DH, Collman RG, Blaser MJ, Weiden MD. 2016. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nature Microbiology 1. [DOI] [PMC free article] [PubMed]
- 11.Belstrom D, Holmstrup P, Bardow A, Kokaras A, Fiehn NE, Paster BJ. Temporal stability of the salivary microbiota in oral health. PLoS One. 2016;11:e0147472. doi: 10.1371/journal.pone.0147472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Team NIHHMPA A review of 10 years of human microbiome research activities at the US National Institutes of Health, fiscal years 2007-2016. Microbiome. 2019;7:31. doi: 10.1186/s40168-019-0620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Hur A, Guyon I. Detecting stable clusters using principal component analysis, p 159-182. Totowa, NJ: Humana Press Inc.; 2003. [DOI] [PubMed] [Google Scholar]
- 14.Levner I. Feature selection and nearest centroid classification for protein mass spectrometry. BMC Bioinformatics. 2005;6:68. doi: 10.1186/1471-2105-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabney AR. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–4154. doi: 10.1093/bioinformatics/bti681. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, Brown C, Abdo Z, Davis C, Hansmann M, Joyce P, et al. Disparity in the vaginal microbial community composition of healthy Caucasian and black women. ISME J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 17.Muzny CA, Blanchard E, Taylor CM, Aaron KJ, Talluri R, Griswold ME, et al. Identification of key bacteria involved in the induction of incident bacterial vaginosis: a prospective study. J Infect Dis. 2018;218:966–978. doi: 10.1093/infdis/jiy243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamarelle J, de Barbeyrac B, Le Hen I, Thiebaut A, Bebear C, Ravel J, et al. Vaginal microbiota composition and association with prevalent chlamydia trachomatis infection: a cross-sectional study of young women attending a STI clinic in France. Sex Transm Infect. 2018;94:616–618. doi: 10.1136/sextrans-2017-053346. [DOI] [PubMed] [Google Scholar]
- 19.Serrano MG, Parikh HI, Brooks JP, Edwards DJ, Arodz TJ, Edupuganti L, Huang B, Girerd PH, Bokhari YA, Bradley SP, Brooks JL, Dickinson MR, Drake JI, Duckworth RA, 3rd, Fong SS, Glascock AL, Jean S, Jimenez NR, Khoury J, Koparde VN, Lara AM, Lee V, Matveyev AV, Milton SH, Mistry SD, Rozycki SK, Sheth NU, Smirnova E, Vivadelli SC, Wijesooriya NR, Xu J, Xu P, Chaffin DO, Sexton AL, Gravett MG, Rubens CE, Hendricks-Munoz KD, Jefferson KK, Strauss JF, 3rd, Fettweis JM, Buck GA. 2019. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat Med 25:1001-1011. [DOI] [PMC free article] [PubMed]
- 20.MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulos N, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:8988. doi: 10.1038/srep08988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosmann C, Anahtar MN, Handley SA, Huttenhower C, Farcasanu M, Abu-Ali G, Bowen BP, Padavattan N, Desai C, Droit L, Moodley A, Dong M, Chen Y, Ismail N, Ndung'u T, Ghebremichael MS, Wesemann DR, Mitchell CM, Dong KL, Huttenhower C, Walker BD, Virgin HW, Kwon DS. 2017. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women, vol 46, p 29-37. [DOI] [PMC free article] [PubMed]
- 22.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UME, Zhong X, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brotman RM, Ravel J, Cone RA, Zenilman JM. Rapid fluctuation of the vaginal microbiota measured by gram stain analysis. Sex Transm Infect. 2010;86:297–302. doi: 10.1136/sti.2009.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan S, Liu C, Mitchell CM, Fiedler TL, Thomas KK, Agnew KJ, et al. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS One. 2010;5:e10197. doi: 10.1371/journal.pone.0010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brotman RM, Bradford LL, Conrad M, Gajer P, Ault K, Peralta L, et al. Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sex Transm Dis. 2012;39:807–812. doi: 10.1097/OLQ.0b013e3182631c79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Houdt R, Ma B, Bruisten SM, Speksnijder A, Ravel J, de Vries HJC. Lactobacillus iners-dominated vaginal microbiota is associated with increased susceptibility to chlamydia trachomatis infection in Dutch women: a case-control study. Sex Transm Infect. 2018;94:117–123. doi: 10.1136/sextrans-2017-053133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown SE, Schwartz J, Robinson C, O'Hanlon ED, Bradford LL, Xin H, et al. The vaginal microbiota and behavioral factors associated with genital Candida albicans detection in reproductive-age women. Sex Transm Dis. 2019;46:753–758. doi: 10.1097/OLQ.0000000000001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brotman RM, Shardell MD, Gajer P, Fadrosh D, Chang K, Silver MI, et al. Association between the vaginal microbiota, menopause status, and signs of vulvovaginal atrophy. Menopause. 2014;21:450–458. doi: 10.1097/GME.0b013e3182a4690b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard DX, Brown G, Julian DXX, Lee S, Ann DXX, Denise DX, Holly DXX, Lindsay DXX, Tom DXX, Phillip DX, X DX. 2019. Establishment of vaginal microbiota composition in early pregnancy and its association with subsequent preterm prelabor rupture of the fetal membranes. Translational Research doi:10.1016/j.trsl.2018.12.005:in press. [DOI] [PMC free article] [PubMed]
- 31.Hickey RJ, Zhou X, Settles ML, Erb J, Malone K, Hansmann MA, Shew ML, Pol BVD, Fortenberry JD, Forney LJ, Forney J. 2015. Vaginal microbiota of adolescent girls prior to the onset of menarche resemble those of reproductive-age women. mBio 6:e00097-15. [DOI] [PMC free article] [PubMed]
- 32.McClelland RS, Lingappa JR, Srinivasan S, Kinuthia J, John-Stewart GC, Jaoko W, et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect Dis. 2018;18:554–564. doi: 10.1016/S1473-3099(18)30058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm JB, France MT, Ma B, McComb E, Robinson CK, Mehta A, et al. Comparative metagenome-assembled genome analysis of "Candidatus Lachnocurva vaginae", formerly known as bacterial vaginosis-associated Bacterium-1 (BVAB1) Front Cell Infect Microbiol. 2020;10:117. doi: 10.3389/fcimb.2020.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue JC, Clayton MK. A similarity measure based on species proportions. Communications in Statistics - Theory and Methods. 2005;34:2123–2131. doi: 10.1080/STA-200066418. [DOI] [Google Scholar]
- 35.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by standardized method of gram stain interpretation. J Clin Microbiol. 1991;29:297–301. doi: 10.1128/JCM.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srinivasan S, Morgan MT, Liu C, Matsen FA, Hoffman NG, Fiedler TL, et al. More than meets the eye: associations of vaginal bacteria with gram stain morphotypes using molecular phylogenetic analysis. PLoS One. 2013;8:e78633. doi: 10.1371/journal.pone.0078633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muzny C, Sunesara IR, Griswold ME, Kumar R, Lefkowitz EJ, Mena LA, et al. Association between BVAB1 and high Nugent scores among women with bacterial vaginosis. Diag Microbiol Infect Dis. 2014;80:321–323. doi: 10.1016/j.diagmicrobio.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith SB, Ravel J. The vaginal microbiota, host defence and reproductive physiology. J Physiol. 2017;595:451–463. doi: 10.1113/JP271694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Hanlon DE, Moench TR, Cone RA. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS One. 2013;8:e80074. doi: 10.1371/journal.pone.0080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boskey ER, Ra C, Whaley KJ, Moench TR. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Human reproduction (Oxford, England) 16:1809-1813. 2001. [DOI] [PubMed]
- 41.Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, et al. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology. 2014;160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett PR, Lee YS, Holmes E, Teoh TG, Kindinger LM, Marchesi JR, et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome. 2017;5. [DOI] [PMC free article] [PubMed]
- 43.Elovitz MA, Gajer P, Riis V, Brown AG, Humphrys MS, Holm JB, et al. Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat Commun. 2019;10:1305. doi: 10.1038/s41467-019-09285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:1–6. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandal S, Van Treuren W, White RA, Eggesbo M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callahan BJ, DiGiulio DB, Aliaga Goltsman DS, Sun CL, Costello EK, Jeganathan P, Biggio JR, Wong RJ, Druzin ML, Shaw GM, Stevenson DK, Holmes SP, Relman DA, Contributions A, designed research D, performed research D, contributed new reagents D, tools a, analyzed data D. 2017. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. 114. [DOI] [PMC free article] [PubMed]
- 47.van de Wijgert J, Verwijs MC, Gill AC, Borgdorff H, van der Veer C, Mayaud P. Pathobionts in the vaginal microbiota: individual participant data meta-analysis of three sequencing studies. Front Cell Infect Microbiol. 2020;10:129. doi: 10.3389/fcimb.2020.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nizet V. 2018. Group B Streptococcal maternal colonization and neonatal disease: molecular mechanisms and preventative approaches. 6:1-17. [DOI] [PMC free article] [PubMed]
- 49.Smith PA, Sherman JM. The lactic acid fermentation of streptococci. J Bacteriol. 1941;43:725–731. doi: 10.1128/JB.43.6.725-731.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callaghan AO, Sinderen DV. 2016. Bifidobacteria and their role as members of the human gut microbiota. 7. [DOI] [PMC free article] [PubMed]
- 51.Freitas AC, Hill JE. Quantification, isolation and characterization of Bifidobacterium from the vaginal microbiomes of reproductive aged women. Anaerobe. 2017;47:145–156. doi: 10.1016/j.anaerobe.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 54.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, Cornejo-castillo FM, Costea PI, Cruaud C, Ovidio F, Engelen S, Ferrera I, Gasol JM, Guidi L, Hildebrand F, Kokoszka F, Lepoivre C. 2015. Structure and function of the global ocean microbiome. 348:1-10. [DOI] [PubMed]
- 55.Brotman RM, Klebanoff MA, Nansel TR, Yu KF, Andrews WW, Zhang J, et al. Bacterial vaginosis assessed by gram stain and diminished colonization resistance to incident gonococcal, chlamydial, and trichomonal genital infection. J Infect Dis. 2010;202:1907–1915. doi: 10.1086/657320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ravel J, Brotman RM, Gajer P, Ma B, Nandy M, Fadrosh DW, Sakamoto J, Koenig SSK, Fu L, Zhou X, Hickey RJ, Schwebke JR, Forney LJ. 2013. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis.1-6. [DOI] [PMC free article] [PubMed]
- 57.Tamarelle J, Ma B, Gajer P, Humphrys MS, Terplan M, Mark KS, et al. Nonoptimal vaginal microbiota after azithromycin treatment for chlamydia trachomatis infection. J Infect Dis. 2020;221:627–635. doi: 10.1093/infdis/jiz499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tuddenham S, Ghanem KG, Caulfield LE, Rovner AJ, Robinson C, Shivakoti R, et al. Associations between dietary micronutrient intake and molecular-bacterial vaginosis. Reprod Health. 2019;16:151. doi: 10.1186/s12978-019-0814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brotman RM, Klebanoff MA, Nansel TR, Andrews WW, Schwebke JR, Zhang J, et al. A longitudinal study of vaginal douching and bacterial vaginosis--a marginal structural modeling analysis. Am J Epidemiol. 2008;168:188–196. doi: 10.1093/aje/kwn103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holm JB, Humphrys MS, Robinson CK, Settles ML, Ott S, Fu L, Yang H, Gajer P, He X, McComb E, Gravitt PE, Ghanem KG, Brotman RM, Ravel J. 2019. Ultrahigh-throughput multiplexing and sequencing of >500-base-pair amplicon regions on the Illumina HiSeq 2500 platform. mSystems 4. [DOI] [PMC free article] [PubMed]
- 61.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKinney W. Data structures for statistical computing in python. 2010. [Google Scholar]
- 65.Greathouse KL, Sinha R, Vogtmann E. DNA extraction for human microbiome studies: the issue of standardization. Genome Biol. 2019;20:212. doi: 10.1186/s13059-019-1843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLaren MR, Willis AD, Callahan BJ. Consistent and correctable bias in metagenomic sequencing experiments. Elife. 2019;8. [DOI] [PMC free article] [PubMed]
- 67.Bates D, Maechler M. Package 'lme4'. 2010. [Google Scholar]
- 68.Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 69.Bolker B. 2019. Package “broom.mixed”.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Figure 1. Illustration of cluster selection for construction of VALENCIA sub-CST centroids. Stretched version of dendrogram from Figure 1 with horizontal line indicating chosen cluster threshold (a). Subset heatmap and dendrogram of samples in CST IV-C (b). Hierarchical clustering was performed on this subset using Bray-Curtis dissimilarity with Ward linkage and the horizontal line indicating cluster threshold. Silhouette and Davies-Bouldin scores for a range of number of clusters (c). Horizontal lines indicate the values for the final 13 sub-CSTs, after sub-clustering the CST IV-C samples.
Additional file 2: Supplemental Figure 2. Odds of women with each sub-CST having a vaginal pH >4.5.
Additional file 3. Supplemental Figure 3.
Data Availability Statement
VALENCIA, the training dataset, test dataset 2, and all scripts used to analyze and display the data are available at https://github.com/ravel-lab/VALENCIA. Information on how to obtain test datasets 1 and 3 can be found in their respective papers.