Abstract
Interleukin-10 (IL-10) is a dimeric cytokine with both immunosuppressive and immunostimulatory activities; however, IL-10-based therapies have shown only marginal clinical benefits. Here, we explored whether the stability of the IL-10-receptor complex contributes to the immunomodulatory potency of IL-10. We generated an IL-10 mutant with enhanced affinity for its IL-10Rβ receptor using yeast surface display. Compared to the wild-type cytokine, the affinity-enhanced IL-10 variants recruited IL-10Rβ more efficiently into active cell surface signalling complexes and triggered greater STAT1 and STAT3 activation in human monocytes and CD8+ T cells. These effects in turn led to more robust induction of IL-10-mediated gene expression programs at low ligand concentrations in both human cell subsets. IL-10-regulated genes are involved in monocyte energy homeostasis, migration and trafficking, and in CD8+ T cell exhaustion. At non-saturating doses, IL-10 did not induce key components of its gene expression program, which may explain its lack of efficacy in clinical settings. Our engineered IL-10 variant showed a more robust bioactivity profile than that of wild-type IL-10 at low doses in monocytes and CD8+ T cells. Moreover, CAR-modified T cells expanded with the engineered IL-10 variant displayed superior cytolytic activity than those expanded with wild-type IL-10. Our study provides insights into how IL-10-receptor complex stability fine-tunes IL-10 biology and opens new opportunities to revitalise failed IL-10 therapies.
INTRODUCTION:
Interleukin-10 (IL-10) is a hallmark cytokine for immune regulation that elicits potent anti-inflammatory responses. IL-10 regulates the adaptive arm of the immune response by reducing the antigen presentation potential of innate cells by decreasing their surface major histocompatibility complex (MHC) levels and costimulatory molecules (1, 2). In addition, IL-10 potently suppresses the production of proinflammatory cytokines from various cell types including monocytes, macrophages and T cells (3, 4), further contributing to an anti-inflammatory environment. IL-10’s critical contribution to a healthy immune response is further highlighted by the finding that IL-10 deficient humans develop severe autoimmune diseases such as Crohn’s disease and colitis (5, 6). Despite IL-10’s relevancy for human health, the molecular bases allowing IL-10 to elicit its broad spectrum of anti-inflammatory activities are poorly understood.
Because of its potent anti-inflammatory properties, recombinant IL-10 therapy was regarded as an attractive biological approach to treat autoimmune disorders. However, despite efficacy in mouse studies (7, 8), IL-10 therapies fail to elicit beneficial results in the clinic, with several clinical trials showing only mild efficacy and biased responses in patients (9, 10). A leading hypothesis to explain the poor clinical efficacy of IL-10 against bowel diseases such as IBD and colitis is that during IL-10 therapies, low levels of this cytokine reach the gastrointestinal tract, thus failing to produce an effective response (11). To date we have a poor understanding of how IL-10 doses influence its immunomodulatory potential. Supporting this model, the development of strategies for a more targeted IL-10 delivery show enhanced clinical efficacy, although these studies are still at an early stage (8, 12–14). An IL-10 variant with the ability to elicit robust responses at therapeutically relevant doses would be highly desirable.
In addition to its anti-inflammatory activities, IL-10 can increase the cytotoxic function of CD8+ T cells, augmenting their ability to target tumours and boosting the anti-cancer response (15). This property seems paradoxical because IL-10 in the tumour microenvironment is linked to tumour evasion of the immune response, most likely due to IL-10’s inhibitory effects on antigen presentation (16, 17). Despite this paradox, several studies have elegantly demonstrated that IL-10 can improve production of the CD8+ effector molecules granzyme B and interferon-γ (IFN- γ) both in vitro and in vivo (18–20). Currently, several clinical trials are testing the antitumour properties of IL-10 with already initial promising results (21). In these trials, high doses of PEGylated IL-10 (Pegilodekakin) were used, which resulted in prolonged IL-10 retention in the circulation to ensure efficacy, again highlighting that effective IL-10 in vivo responses need high concentrations and sustained levels of IL-10.
IL-10 is a dimeric cytokine which exerts its effects by binding to a surface receptor comprised of two IL-10Rα and two IL-10Rβ receptor subunits, which triggers the activation of the JAK1 (Janus kinase 1)/TYK2 (tyrosine kinase 2)/STAT3 (signal transducer and activator of transcription 3)/ STAT1 signalling pathway and the induction of specific gene expression programs. IL-10 binds with high affinity to IL-10Rα and with much lower affinity to IL-10Rβ, in the order of high micromolar/low millimolar range (22). Submicromolar affinities are required to ensure efficient cytokine receptor dimerisation (23, 24). Thus, assembly of active IL-10 receptor signalling complexes in the plasma membrane is probably limited by recruitment of IL-10Rβ, making this system sensitive to changes in either ligand concentrations and/or receptor densities in the plasma membrane. We therefore hypothesise that IL-10’s poor clinical activities result from its weak affinity for the IL-10Rβ subunit. An IL-10 variant that binds to IL-10Rβ with enhanced affinity could improve the efficacy of IL-10 based therapies. To test this hypothesis, we used the yeast surface display engineering platform to generate a new IL-10 variant which binds IL-10Rβ with much higher affinity than wild-type (WT) IL-10. Due to the dimeric nature of WT IL-10, which makes its manipulation challenging, we have used the previously described monomeric IL-10 variant as an engineering scaffold (25). Initially, we generated a high affinity monomeric IL-10 variant, which we then translated into its natural dimeric conformation, thus obtaining four ligands to test for activity: WT dimer (WTD), WT monomer (WTM), high affinity dimer (R5A11D) and high affinity monomer (R5A11M). These molecules provided us with the opportunity to assess the contributions of IL-10 receptor binding affinity as well as IL-10 receptor complex stoichiometry to IL-10 biology. Our data showed that increasing the affinity of IL-10 for IL-10Rβ enhanced IL-10’s known properties at both the molecular and cellular level. Quantitative imaging studies revealed that our high affinity variants, either in monomeric or dimeric forms, produced enhanced receptor heterodimerisation, which in turn, resulted in more potent activation of STAT1 and STAT3. In agreement with their improved signalling profiles, the affinity enhanced IL-10 variants induced more robust gene expression programs at low doses than the WT ligands in monocytes and CD8+ T cells and stronger cellular responses. Accordingly, chimeric antigen receptor (CAR)-modified T cells cultured with R5A11D displayed robust cytolytic activities in vitro against a target leukemic cell line. Overall, our study provides insights into how IL-10 doses regulate its immunomodulatory activities and show that our engineered IL-10 variants confer an advantage over WT IL-10 by eliciting more robust bioactivities at a wider range of doses.
RESULTS:
Engineering IL-10 variants with enhanced affinity towards IL-10Rβ
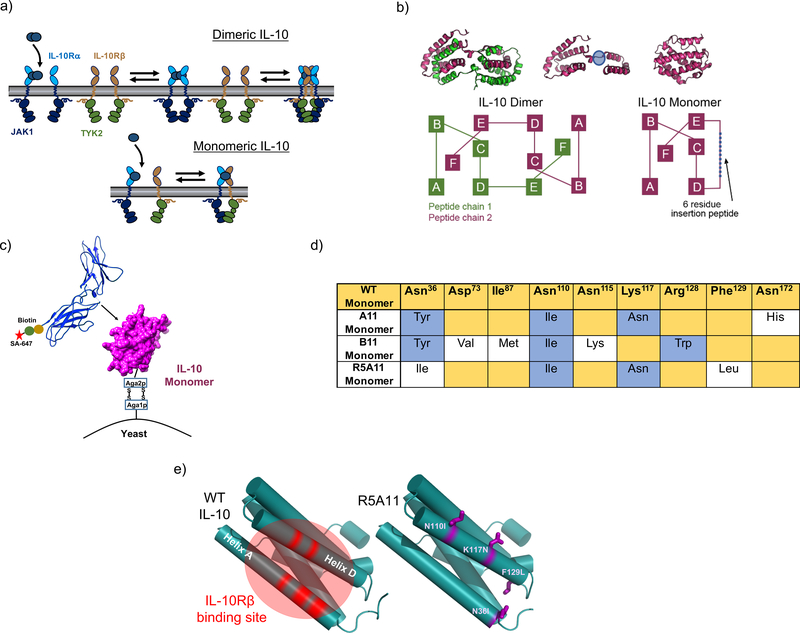
IL-10 probably engages its tetrameric receptor complex in a multi-step binding process. IL-10 is recruited to the cell surface by the fast and high affinity interaction with IL-10Rα. Subsequently, a second IL-10Rα subunit and two IL-10Rβ subunits, which only recognize the IL-10/IL-10Rα complex (26), can be recruited to initiate signalling (Fig. 1a, top panel). Although the exact stoichiometry of the signalling complex in the plasma membrane is still unclear, signal activation strictly requires the formation of IL-10Rα/IL-10Rβ heterodimers. However, IL-10 has poor binding affinity for IL-10Rβ (~mM range), which we hypothesised acts as a rate-limiting step in IL-10’s biological activities. Thus, we asked whether an IL-10Rβ affinity-enhanced IL-10 variant would overcome this in vivo rate-limiting-step by inducing robust responses at a wide range of ligand concentrations. To address this question, we used yeast surface display to increase the binding affinity of IL-10 for IL-10Rβ and study the signalling and activity profiles induced by these new affinity-enhanced IL-10 variants. A caveat to engineering IL-10 is its dimeric nature, which makes the correct display of this cytokine on the yeast surface challenging. We used a previously described monomeric IL-10 variant (25) as an engineering scaffold to overcome this limitation. The monomeric IL-10 was generated by extending the connecting linker between helices D and E in IL-10 by 6 peptides, consequently allowing helices E and F to fold into its own hydrophobic core to form an IL-10 monomer (Fig. 1b). Monomeric IL-10 recruits one molecule each of IL-10Rα and IL-10Rβ to form an active signalling trimeric complex (Fig. 1a, bottom panel). Although monomeric IL-10 can trigger IL-10-mediated responses, it does so with lower potency than its dimeric counterpart (22, 25).
Figure 1. Generation of high affinity IL-10 variants by yeast surface display.
(a). Schematic of IL-10 stepwise receptor assembly for IL-10 dimer and IL-10 monomer. (b). Schematic of IL-10 dimer and IL-10 monomer secondary structure organisation, adapted from (25, 26). The extended linker region is highlighted in blue. (c). Representation of IL-10 displayed on yeast cell surface used in screen with fluorescently labelled recombinant IL-10Rβ. (d). Table for amino acid changes found in high affinity mutants. WT sequence is shown in yellow and is based on the amino acid sequence reported for IL-10 including leader sequence on UniProt database (P22301) with the addition of the 6 amino acid linker at position 134 to allow the formation of the monomer. Conserved changes between mutants are shown in blue. Individual mutations are shown in white. (e). Left: WT IL-10 structure with helices A and D emphasised in red as the area predicted to be the IL-10Rβ binding site (28). Right: Cartoon representation of the IL-10 monomer with the positions of the mutations found in the high affinity variant R5A11 highlighted in purple.
First, we transfected yeast with the monomeric IL-10 construct to test whether binding to IL-10Rα and IL-10Rβ receptor subunits was preserved in the context of the yeast surface. We used biotinylated ectodomains of IL-10Rα and IL-10Rβ receptors in combination with AlexaFluor647 fluorescently labelled streptavidin to measure receptor binding by flow cytometry (Fig. 1c). Monomeric IL-10 retained binding to IL-10Rα, confirming that it was correctly displayed on the surface of the yeast (fig. S1a). We could not detect binding of monomeric IL-10 to IL-10Rβ in the presence or absence of IL-10Rα, confirming its weak binding to this receptor subunit (fig. S1a). Without a crystal structure of IL-10 bound to IL-10Rβ to guide us in the design of a site-directed mutant library, we undertook an unbiased error-prone approach to generate IL-10 mutants with enhanced affinity for IL-10Rβ. The gene encoding the monomeric IL-10 variant was subject to error-prone PCR and the amplified PCR product subsequently electroporated into the S. cerevisiae strain EBY100 following previously described protocols (27, 28). Eight rounds of selection were performed in which the concentration of IL-10Rβ was gradually decreased to isolate variants of IL-10 that bound to IL-10Rβ with enhanced affinity (fig. S1b). Initial rounds of selection were performed with high concentrations (1 μM) of biotinylated IL-10Rβ in the presence of 100nM non-biotinylated IL-10Rα to stabilize the surface complex and recover low affinity binding variants. After round 6 of selection, the library was comprised of variants that bound to IL-10Rβ even in the absence of IL-10Rα and by round, 8 the library contained variants that bound to concentrations of IL-10Rβ in the low nanomolar range (fig. S1c and S1d). At this point, we picked individual yeast colonies and isolated several clones (A11, B11, R5A11) that bound to IL-10Rβ with enhanced affinity when compared to WT IL-10 (fig. S1d). In the IL-10 structure, these mutations localised to the region along helices A and D that was previously predicted to bind IL-10Rβ (28), thereby validating our selection process (Fig. 1d and 1e).
Engineered IL-10 variants bind to IL-10Rβ with nanomolar affinities
Next, we characterized the biophysical properties of recombinantly expressed IL-10 variants. As expected, the IL-10 variants behaved as monomers when run in a gel filtration column (fig. S1e and S1f). To validate the apparent binding affinities seen in the on-yeast binding titration experiments, we carried out surface plasmon resonance (SPR) studies in which the IL-10 variants A11, B11 and R5A11 were flowed across biotinylated IL-10Rβ immobilised onto the chip surface (fig. S2a). We could not detect substantial binding of the WT monomeric IL-10 (WTM) at the micromolar range of doses used in this study, confirming the low binding affinity exhibited by WTM for IL-10Rβ (fig S2b and S2c). The IL-10 variants bound to IL-10Rβ with KD values in the low micromolar range (fig S2b and S2c), thus confirming their improved binding affinities.
IL-10 displays cooperative binding to its receptor subunits whereby its affinity for IL-10Rβ is enhanced once pre-bound to IL-10Rα (26). Thus, we investigated whether our mutants preserved this property by performing SPR measurements using the high affinity IL-10 variants pre-bound to soluble IL-10Rα (fig. S2d). We could not detect substantial binding of the WTM/IL-10Rα complex to IL-10Rβ, highlighting again its poor binding affinity towards IL-10Rβ (fig. S2e and S2f). All IL-10 variants exhibited enhanced binding to IL-10Rβ when complexed to IL-10Rα (in the nanomolar range), confirming their cooperative binding and suggesting that the canonical IL-10 receptor complex binding topology had not been perturbed by the mutations introduced in our new variants (fig. S2e and S2f). These results confirm successful engineering of new IL-10 variants which engage IL-10Rβ with higher binding affinity than their WT counterpart.
Enhanced IL-10Rβ binding affinity increases receptor heterodimerization.
Thus far, we had carried out the biophysical characterisation of our high affinity IL-10 variants with the monomeric form of the cytokine because it was necessary for the protein engineering methodologies used. To study the native IL-10/IL-10 receptor complex stoichiometry, we recombinantly expressed our high affinity IL-10 mutant, R5A11, in dimeric form (R5A11D) in addition to the monomeric form (R5A11M) (fig. S1a and S1b). We selected this mutant based on its higher expression yields compared to other isolated variants. Comparisons between this variant, WTD, and WTM allowed us to examine how increased binding affinity and stoichiometry contributed to IL-10’s molecular and cellular activities.
To test how increasing the binding affinity to IL-10Rβ supported receptor assembly at the plasma membrane of live cells, we probed diffusion and interaction of both receptor chains by dual colour total internal reflection fluorescence (TIRF) microscopy. We expressed IL-10Rα and IL-10Rβ, both carrying N-terminally tagged variants of non-fluorescent (Y67F) mEGFP, in HeLa cells. The tags were designed to specifically recognise either one of two different anti-GFP nanobodies (29). These nanobodies were conjugated to photostable organic fluorophores ATTO Rho11 and ATTO 643, which are suitable for simultaneous dual-colour single molecule tracking of IL-10Rα and IL-10Rβ in the plasma membrane of live cells as shown for other cytokine receptor systems (30–32) (Fig. 2a and fig. S3a).
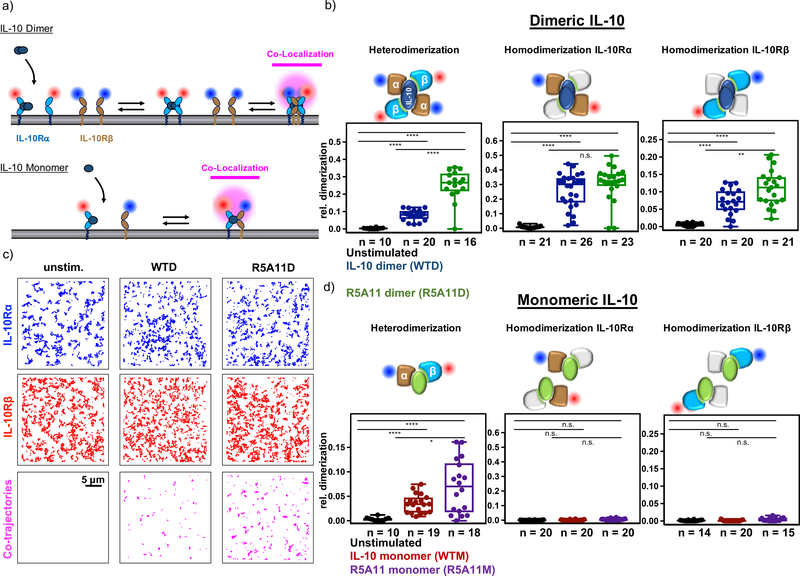
Figure 2. Increased IL-10Rβ binding affinity enhances IL-10 receptor dimerization.
(a). Quantifying receptor dimerization in the plasma membrane by dual-color single molecule colocalization and co-tracking. IL-10Rα and IL-10Rβ with N-terminally fused variants of monomeric ECFP and EGFP, respectively, were labelled with nanobodies ENAT643 and MIRho11, respectively. (b). Heterodimerization of IL-10Rα and IL-10Rβ (left), and homodimerization of IL-10Rα (center) and IL-10Rβ (right) induced by dimeric IL-10 variants as quantified by co-locomotion analysis. Each data point represents a cell with the number of cells of each experiment indicated under the box plot. c). Trajectories of IL-10Rα (blue), IL-10Rβ (red) and co-localized IL-10Rα:IL-10Rβ (magenta) in the absence of IL-10 (left column) and in the presence of WTD (middle column) and R5A11D (right column). Scale bar represents 5 μm. (d). Homo- and heterodimerization of IL-10Rα and IL-10Rβ induced by monomeric IL-10 variants quantified by co-locomotion analysis. Each data point represents a cell with the number of cells of each experiments indicated under the box plot. P value calculated by two-tailed student’s T-test.
After cell surface labelling, we found that both receptor subunits randomly diffused in the plasma membrane (Fig. 2b). Dimerization of receptor subunits was quantified by co-tracking analysis. Receptors were considered to be dimerized if two individual particles in both spectral channels persistently colocalized for ≥10 consecutive steps (~320 ms) in a proximity of 150 nm. These colocalization/cotracking thresholds allowed reliable elimination of density-dependent random colocalizations (31). In the absence of IL-10, we did not observe heterodimerization of IL-10Rα and IL-10Rβ (Fig. 2b and 2c). Stimulation with saturating concentrations of IL-10 WTD significantly dimerized IL-10Rα and IL-10Rβ. IL-10 R5A11D induced a substantially higher level of receptor heterodimers (Fig. 2b and 2c). This finding was also confirmed for the monomeric versions of both WT and high affinity IL-10 variants although at lower levels than seen for the dimeric versions (Fig. 2d). This observation is in line with the 50% reduced probability to observe heterodimers expected for the monomeric compared to dimeric ligand. Ligand stimulation led to a significant decrease of diffusion mobility, particularly for IL-10Rα (fig. S3b), which agrees with previous reports on cytokine receptor dimerization (23, 24, 33).
We also probed homodimerization of IL-10Rα and IL-10Rβ, respectively. We stochastically labelled either of the receptor chains with both dyes (fig. S3a), taking into account that only half of the dimers would be labelled with both dye species and thus would be picked up by co-tracking analysis. Stimulation with the dimeric IL-10 induced strong homodimerization of IL-10Rα with no difference between both cytokine variants because the IL-10Rα binding interface was unaltered in R5A11 (Fig. 2b). Instead, homodimerization of IL-10Rβ was significantly increased for the engineered variant R5A11D compared to WTD. For the monomeric IL-10 variants, all homodimerization experiments failed to induce receptor homodimers, in agreement with the monomeric nature of the ligands (Fig. 2d) (25). Together, these results confirmed that, compared to WT IL-10, the engineered R5A11 variants increased recruitment of IL-10Rβ into the signalling complex at the plasma membrane. An interesting point to consider is whether the IL-10Rβ homodimers induced by R5A11D can signal in the absence of IL-10Rα. However, based on previous work, this possibility seems unlikely. IL-10Rβ knockout cells do not respond to IL-10 treatment (34), and neutralising antibodies against IL-10Rα inhibit STAT3 activation in response to IL-10 (35).
IL-10 variants exhibit enhanced signalling activities in human primary monocytes
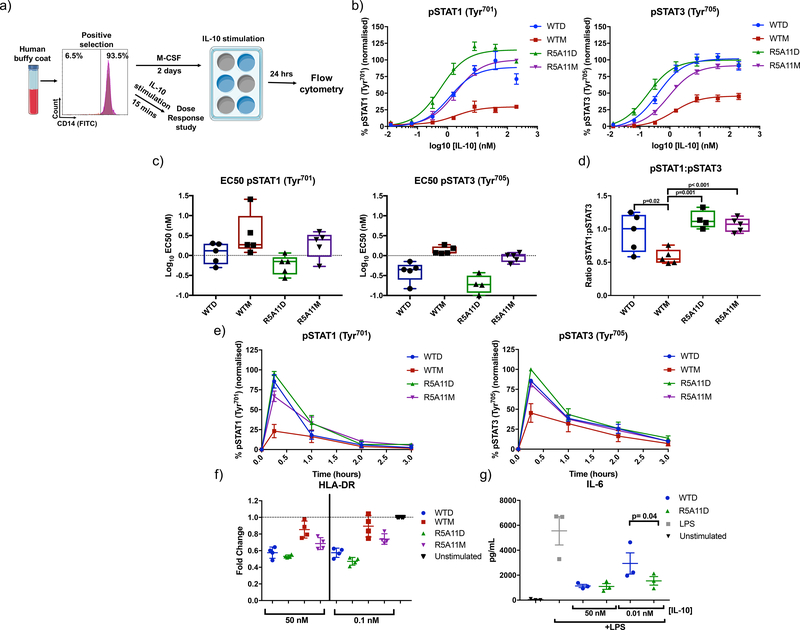
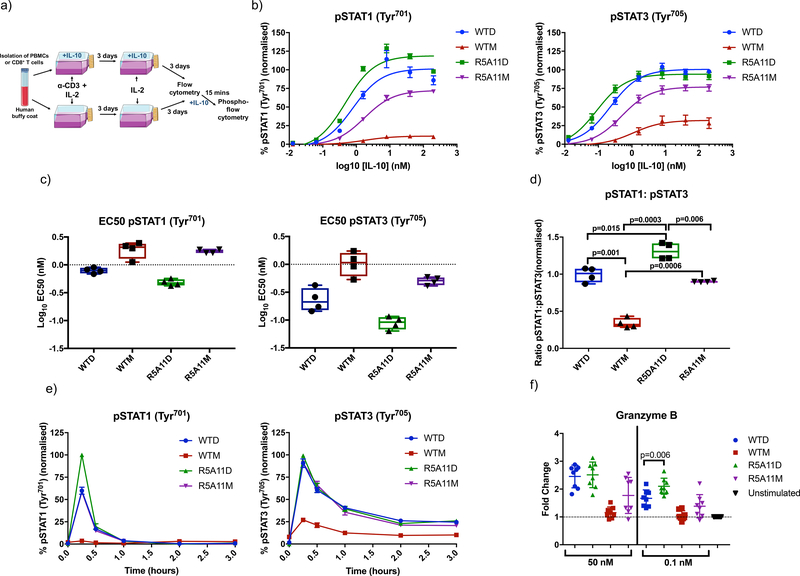
IL-10 inhibits inflammatory processes by modulating the activities of different innate cells including monocytes. We next performed signalling and activity assays in human monocytes to investigate the anti-inflammatory potential of our engineered variants. Monocytes (CD14+ cells) were isolated from human buffy coats and stimulated with WT IL-10 and high affinity monomer and dimers for 15 minutes for measurement of STAT1 and STAT3 phosphorylation or rested for two days before a 24 hour stimulation for measurement of HLA-DR levels (Fig. 3a). Levels of STAT1 and STAT3 phosphorylation upon ligand stimulation were measured by flow cytometry because these two transcription factors represent the major signalling pathway engaged by IL-10 (36, 37). At saturating concentrations, R5A11D and WTD activated STAT1 and STAT3 to a comparable extent (Fig. 3b). However, R5A11D induced enhanced phosphorylation of both STAT1 and STAT3 at subsaturating concentrations, which translated into a decrease in EC50 values compared to WTD (Fig. 3b and 3c). WTM showed poor activation of STAT1 and STAT3 with amplitudes of activation reaching less than fifty percent of those elicited by WTD (Fig. 3b). Although WTD, R5A11D and R5A11M showed similar pSTAT1:pSTAT3 ratios, the bias of WTM in inducing more pSTAT3 than pSTAT1 (Fig. 3d) agrees with previous observations from our laboratory describing biased signalling by short-lived cytokine-receptor complexes (30). R5A11M induced the activation of both STAT1 and STAT3 to levels comparable to those induced by the dimeric cytokines at saturating doses, suggesting that the defective signalling elicited by WTM results from its weak IL-10Rβ binding affinity (Fig. 3b and 3c). Time course analyses showed that the signalling profiles of the variants were not caused by differences in signalling kinetics. The four IL-10 ligands triggered comparable signalling kinetics in human monocytes (Fig. 3e and fig. S4), confirming that their different signalling profiles result from their different binding affinities for IL-10Rβ.
Figure 3. High affinity variants improve signalling capabilities of IL-10 in monocytes.
(a). Monocytes were isolated from human buffy coat samples by selecting for CD14-positive MACS. Cells were stimulated with WT IL-10 and IL-10 variants for 15 mins for phospho-flow cytometry analysis. Cells were rested in monocyte colony stimulating factor (M-CSF)-containing media for 2 days and then stimulated for 24 hours for analysis of HLA-DR levels. (b). Dose response curves for phosphorylated STAT1 (pSTAT1) and pSTAT3 in IL-10 treated monocytes. Cells were stimulated with WT and high affinity variants of IL-10 for 15 minutes. Activation of STAT1 and STAT3 was analysed by phospho-flow cytometry. Sigmoidal curves were fitted with GraphPad Prism software. Data shown are the mean of five biological replicates with error bars depicting the standard error of the mean (SEM). Each biological replicate was normalised by assigning the highest MFI value of the top concentration as 100% and the lowest MFI value of an untreated control as 0%. The rest of the samples and conditions were normalized accordingly (c). Log10 EC50 values for pSTAT1 and pSTAT3 from dose response curves in (b). Each data point represents a biological replicate (n=5) with a line indicating the mean and error bars showing the minimum and maximum values. (d). Ratio of pSTAT1 to pSTAT3 in IL-10-stimulated monocytes was calculated by taking the percentage activation of pSTAT3 and pSTAT1 at 40 nM and dividing pSTAT1 by pSTAT3 values. Each data point represents a biological replicate (n=5) with a line indicating the mean and error bars showing the minimum and maximum values. P value calculated by two-tailed paired t-test. (e). Kinetics of pSTAT3 and pSTAT1 induced by IL-10. Monocytes were stimulated with IL-10 for the indicated time periods before fixation. Data shown are the mean of four biological replicates with error bars depicting SEM. Each biological replicate was normalised by assigning the highest MFI value at 15 mins as 100% and the lowest MFI value of an untreated control as 0%. (f). Measurement of HLA-DR cell surface expression in monocytes after 24 hours IL-10 treatment. Each data point represents a biological replicate (n=4) and error bars indicate the standard deviation. Fold change is calculated for each biological replicate by dividing the MFI of the treated samples by a non-IL-10 treated control (unstimulated). (g). Measurement of IL-6 secretion by monocytes stimulated with LPS for 8 hours in the presence of IL-10. Each data point represents a biological replicate (n=3). P value calculated by two-tailed ratio paired t-test.
IL-10 exerts its anti-inflammatory properties by inhibiting antigen presentation in innate cells such as monocytes and dendritic cells (38). Thus, we next studied whether IL-10 binding affinity to IL-10Rβ influenced its ability to decrease human leukocyte antigen DR (HLA-DR) expression in human primary monocytes. WTD and R5A11D reduced the levels of HLA-DR surface levels to a similar extent (50%) at saturating doses, in agreement with their comparable signalling profiles (Fig. 3f). At subsaturating doses, however, R5A11D induced greater inhibition of HLA-DR expression (Fig. 3f). WTM induced a mild reduction of HLA-DR surface levels (20%) which paralleled its poor signalling potency (Fig. 3f). R5A11M induced only a 30% reduction of the surface HLA-DR levels, despite activating STAT1/STAT3 to a similar extent as the dimeric ligands (Fig. 3f), suggesting an additional dimer-dependent mechanism by which IL-10 reduces HLA-DR expression. We next investigated how IL-10Rβ binding affinity correlated with IL-10’s ability to inhibit proinflammatory cytokine production by monocytes. We measured IL-6 secretion from monocytes upon LPS stimulation in the presence of various doses of WTD and R5A11D (Fig. 3g). At saturating concentrations, WTD and R5A11D effectively inhibited IL-6 secretion to a similar extent (Fig. 3g). However, at subsaturating doses, R5A11D retained the ability to inhibit IL-6 secretion, unlike WTD (Fig. 3g). Together, our data highlight that IL-10 variants exhibiting enhanced binding towards IL-10Rβ gain a functional advantage at subsaturating doses, such as those that would be attained during therapeutic interventions.
Increased IL-10Rβ affinity enhances the transcriptional effects of IL-10 in monocytes
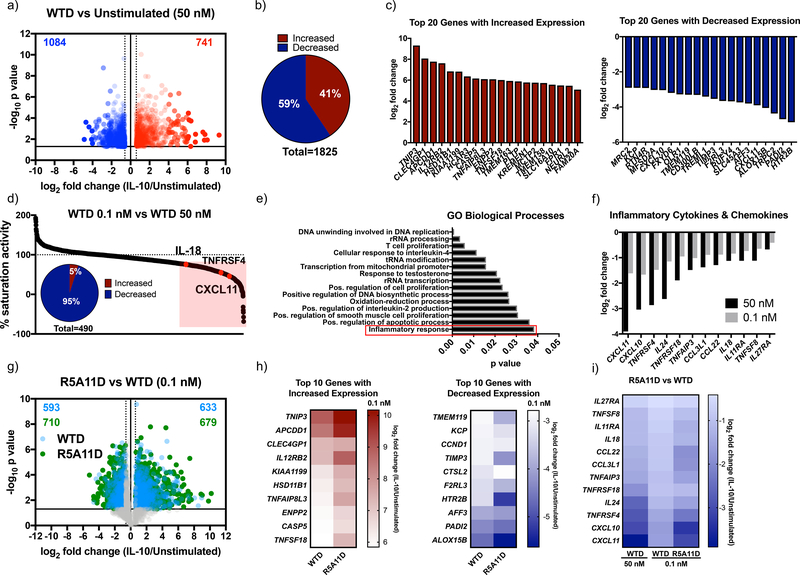
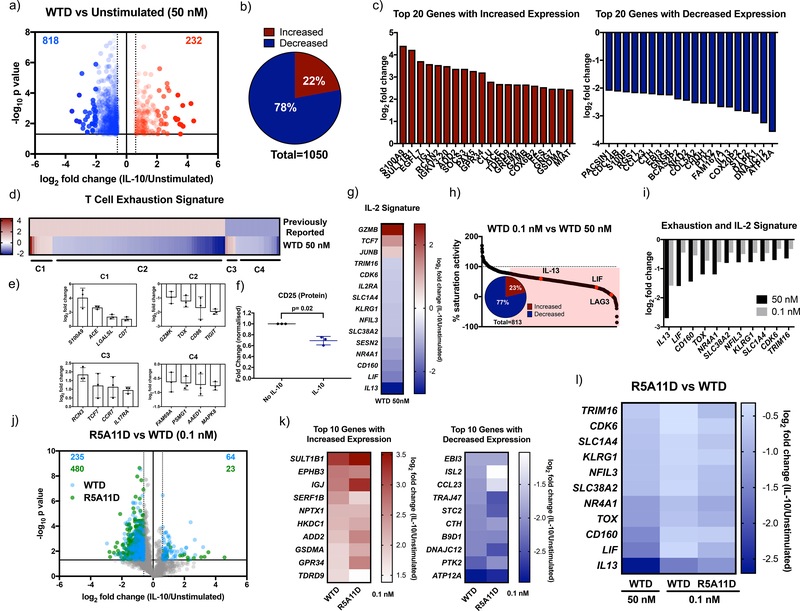
Our initial studies in monocytes were focused on two classical markers attenuated by IL-10, HLA-DR levels and IL-6 expression. To gain a broader understanding of how our variants affect human monocyte activities, we performed a detailed transcriptional analysis of human monocytes stimulated with the different IL-10 ligands for 24 hrs (fig. S5a). WTD treatment elicited strong transcriptional changes in human monocytes. We observed increased expression of 741 genes and decreased expression of 1084 genes (Fig. 4a and 4b, data file S1). Genes showing a large increase or decrease in expression after IL-10 treatment are shown (Fig. 4c). KEGG pathway analysis showed that IL-10 treatment affected the expression of a number of genes involved in pyruvate metabolism (fig. S5b) and the expression of other key metabolic genes (fig. S5c). WTD treatment affected expression of genes encoding hexokinase-2 and hexokinase-3, key enzymes in glycolysis. Expression of genes associated with acyl-CoA synthesis (ACSS2, ACSL4, ACSL1) were significantly increased by IL-10 treatment, highlighting a potential regulation of lipid biosynthesis by this cytokine (fig. S5c). In addition to metabolism-related genes, WTD treatment induced changes in the expression of genes encoding cytokines, chemokines and their receptors (fig. S5c). For instance, the expression of cytokine receptor-encoding genes such as IL-12Rβ2, IL-21R and IL-4R were increased, whereas those encoding cytokines such as IL-8, IL-18 and IL-24 were decreased (fig. S5c). The expression of the chemokine-encoding genes CXCL1, CCL22, CCL24, CCL18, CXCL10 and CXCL11 were also modulated by IL-10 treatment, which would be expected to contribute to the production of an anti-inflammatory environment. Futhermore, changes were observed in expression of various genes encoding cell surface proteins by WTD treatment, including CD93 – which encodes a receptor critical for monocyte phagocytosis and CD44 and CD9 – which encode proteins involved in cell surface adhesion (Fig. S5c); and an inhibition of the type I IFN gene signature, in agreement with previous studies (39, 40) (fig. S5c). Overall, our transcriptome study revealed a broad regulation of monocyte biology by IL-10, that included fine-tuning energy homeostasis, migration and trafficking. 73% of genes whose expression was changed by WTD at saturating doses were induced to a similar extent when sub-saturating doses of WTD were used, highlighting the robustness of the IL-10 response (Fig. 4d). However, of the 27% of genes differentially expressed by WTD treatment at the two doses investigated (0.1 nM and 50 nM), 95% of those corresponded to genes attenuated by IL-10 treatment and include those encoding critical pro-inflammatory chemokines and cytokines (Fig. 4d and 4e). A list of some differentially expressed genes is provided (Fig. 4f). Our data show that at low doses, IL-10 loses the ability to reduce the expression of genes encoding key cytokines and chemokines that critically contribute to enhance the inflammatory response.
Figure 4. Characterisation of transcriptional activity induced by IL-10 and high affinity variants in human monocytes.
(a). Volcano plot of monocyte genes whose expression was significantly increased by IL-10 wild type dimer ≧ 0.6 log2 fold change (red) and significantly decreased ≦ −0.6 log2 fold change compared to non-IL-10 stimulated cells. Fold change was calculated by dividing 50 nM WTD by unstimulated values for each donor. The average fold change was calculated and the log2 of this value was plotted. P values were calculated by two sample equal variance two-tailed t-test of the log2 fold change of 50 nM WTD/unstimulated genes for each donor. Genes that were not significantly changed or were ≦ 0.6 ≧ −0.6 log2 fold change were excluded. (b). Proportion of monocyte genes whose expression was significantly increased or decreased by 50 nM WTD ≧ 0.6 or ≦ −0.6 log2 fold change compared to unstimulated cells. (c). Log2 fold change for the top 20 protein coding genes whose expression was significantly increased (red) and decreased (blue) by 50 nM WTD in monocytes. (d). Percentage activity of low dose (0.1 nM) compared to high dose (50 nM) WTD. The log2 fold change of 0.1 nM WTD/unstimulated was divided by the log2 fold change of 50 nM WTD/unstimulated and multiplied by 100. Genes which showed ≦ 75% of high dose activity (490 genes) are highlighted in red. Insert shows the percentage of these genes whose expression was increased or decreased by IL-10. (e). Gene ontology biological processes analysis for the 490 genes that showed corresponding changes in expression in response to changes in WTD concentration. (f). Log2 fold change for a sample of inflammatory cytokine and chemokine genes for 50 nM and 0.1 nM WTD. (g). Volcano plot of gene expression changed by WTD (blue) and R5A11D (green) at 0.1 nM concentration. Only genes whose expression had already been shown to be significantly changed by 50 nM WTD were plotted. (h). Heatmap of the log2 fold change of the top 10 genes whose expression was increased or decreased by 0.1 nM WTD compared to 0.1 nM R5A11D. (i). Heatmap of inflammatory cytokine and chemokine genes whose expression was inhibited by WTD at 50 nM and 0.1 nM and R5A11D at 0.1 nM. Monocytes from 3 donors were analysed in (a) to (i).
Next, we studied how the engineered IL-10 variants affected gene expression programs in monocytes. WTM induced a poor transcriptional response, in line with its weak signal activation profile (fig. S5d and S5e). R5A11M triggered a more potent transcriptional response when compared to WTM but failed to reach the same potency induced by the dimeric ligands (fig. S5d and S5e). 40% of genes whose expression was altered by WTD showed reduced regulation in response to R5A11M (fig. S5f). This effect contrasted with its ability to activate STAT1 and STAT3 to levels comparable to those induced by the dimeric ligands, suggesting that STAT activation does not directly correlate with transcriptional activity in response to IL-10. In agreement with our signalling studies, R5A11D induced a more robust gene expression profile at subsaturating doses when compared to WTD (fig. S5d). At these doses, R5A11D treatment enhanced the expression of 19% of genes compared to WTD treatment, with only 6% of genes showing increased expression upon WTD treatment compared to R5A11D treatment (fig. S5d and S5g, Fig. 4g). Of the top 10 genes with increased or decreased expression by IL-10, most displayed more pronounced changes in response to R5A11D treatment (Fig. 4h). This pattern held true when genes were grouped by families, such as those encoding cytokines, chemokines, and CD markers and genes involved in MAPK signalling (fig. S5e). Genes encoding key proinflammatory cytokines were inhibited to a greater extent by R5A11D compared to WTD at the same subsaturating dose (Fig. 4i). Overall, our transcriptional data shows that IL-10 controls monocyte biology at different levels and that by exhibiting enhanced affinity towards IL-10Rβ, R5A11D elicits more robust responses at low ligand concentrations, thus suggesting that it may make IL-10 based therapies more clinically effective at targeting inflammatory disorders.
IL-10 variants exhibit enhanced signalling activities in human primary CD8+ T cells
In addition to its potent anti-inflammatory effects, IL-10 also stimulates cytotoxic CD8+ T cells under specific circumstances, enhancing the production of effector molecules and increasing their cytotoxic activity (41). We next investigated whether the enhanced activities exhibited by our affinity-matured variants in monocytes would translate into CD8+ T cells. Human primary CD8+ T cells were grown and activated (Fig. 5a) and the activation of STAT1 and STAT3 in response to the indicated concentrations of IL-10 variants was measured by flow cytometry (Fig. 5b). WTD and R5A11D induced similar STAT phosphorylation levels at saturating doses, but R5A11D showed a decreased EC50 value and stronger signalling at subsaturating doses (Fig. 5b–d), agreeing with our results in monocytes. R5A11D induced a more potent activation of STAT1 over STAT3 which we did not observe in monocytes, suggesting that long-lived IL-10 receptor complexes are more effective at activating STAT1 in CD8+ T cells. WTM produced weak activation of STAT1 and STAT3, inducing less than 25% of the activation amplitudes elicited by the dimeric molecules, and exhibited a bias towards STAT3 activation (Fig. 5b–d). In contrast to our observations in monocytes, R5A11M also elicited a STAT3-biased response, activating STAT3 to 80% of the levels induced by the dimeric molecules and STAT1 to 60% of the levels induced by the dimeric molecules (Fig. 5b–d), suggesting that signalling downstream of the IL-10 receptor complex differs between monocytes and CD8+ T cells. As with monocytes, the observed differences in signalling output by the different IL-10 ligands were not a result of altered signalling activation kinetics in CD8+ T cells (Fig. 5e).
Figure 5. High affinity variants of IL-10 show increased signalling capabilities in human CD8+ T cells.
(a). PBMCs were isolated from human buffy coat samples and/or CD8+ cells were purified by CD8 positive MACS selection. PBMCs and/or purified CD8+ cells were activated for three days using soluble anti-CD3 (100 ng/mL) (PBMCs) or anti-CD3/anti-CD28 beads (CD8+ cells) with IL-2 (20 ng/mL) in the presence or absence of IL-10. On day 3, activation media was removed and the cell populations were placed in media containing IL-2 ± IL-10 for a further 2–3 days before analysis. (b). Dose response curves for pSTAT3 and pSTAT1 in activated CD8+ cells in a PBMC population (activated in the absence of IL-10). Cells were placed in media lacking IL-2 overnight before being stimulated with WT and mutant IL-10 for 15 minutes. Data shown are the mean of four biological replicates with error bars depicting standard error of the mean. Each biological replicate was normalised by assigning the highest MFI value of the top concentration as 100% and the lowest MFI value of an untreated control as 0%. The rest of the samples and conditions were normalized accordingly. (c). Log10 EC50 values for pSTAT3 and pSTAT1 from dose response curves in (b). Each data point represents a biological replicate with a line indicating the mean and error bars showing the minimum and maximum values. Activated CD8+ cells in a PBMC population from 4 donors were analyzed. (d). Ratio of pSTAT1 to pSTAT3 in IL-10-stimulated CD8+ cells in a PBMC population. Ratios were calculated by taking the percentage activation of pSTAT3 and pSTAT1 at 40 nM for four biological replicates and dividing pSTAT1 values by pSTAT3 values. Each data point represents a biological replicate with a line indicating the mean and error bars showing the minimum and maximum values. P value calculated by paired t tests. Activated CD8+ cells in a PBMC population from 4 donors were analyzed. (e). Kinetics of pSTAT3 and pSTAT1 induced by IL-10. Non-activated CD8+ cells in a PBMC population were stimulated with IL-10 for the indicated time periods before fixation. Data shown are the mean of three biological replicates with error bars depicting SEM. Each biological replicate is normalised by assigning the highest MFI value at 15 mins as 100% and the lowest MFI value of an untreated control as 0%. (f). Measurement of granzyme B protein abundance in activated CD8+ T cells (grown and stimulated as in (a)) in the presence of IL-10). Granzyme B protein was quantified by flow cytometry of fixed and permeabilized cells. Fold change was calculated by normalising to a non-IL-10 treated control for each donor. Each data point represents a biological replicate (n=8) and error bars indicate the standard deviation. P value calculated by paired t-test.
Granzyme B is a potent cytotoxic effector molecule that is increased in CD8+ T cells upon IL-10 stimulation (42). As previously reported, IL-10 stimulation did not greatly affect the abundance of the classical early (CD69) and late (CD71) activation markers or the inhibitory receptors LAG-3 and PD-1 or affect CD8+ cell proliferation (fig. S6a and S6b). On the other hand, IL-10 stimulation led to increased granzyme B in CD8+ T cells both at the mRNA and protein levels, independently of whether CD8+ T cells were activated in the context of a PBMC population or a purified CD+8 T cell population (fig. S6c). At saturating concentrations, WTD and R5A11D enhanced granzyme B production to a similar extent, 2.5-fold higher than granzyme B levels induced by TCR stimulation alone (Fig. 5f). WTM poorly induced granzyme B production, consistent with its weak STAT activation. At a subsaturating concentration, we again observed a stronger increase in granzyme B levels induced by R5A11D. R5A11M stimulation resulted in two major populations, with half of the cells expressing granzyme B at levels similar to those in WTM-treated cells and the other half showing increased granzyme B to levels comparable to those induced by the dimeric molecules. Overall, our results show that enhanced affinity for IL-10Rβ bestows IL-10 with robust activities over a wide range of ligand doses and immune cell subsets.
Increased IL-10Rβ affinity enhances the transcriptional effects of IL-10 in CD8+ T cells
To obtain a greater understanding of how IL-10 influences CD8+ T cell responses, we next performed transcriptional studies on CD8+ T cells treated with the different IL-10 ligands. Human CD8+ T cells were purified by positive selection and activated in the presence of WTD and our variants over 6 days (fig. S7a). Fewer transcriptional changes were induced by WTD in CD8+ T cells than in monocytes. 1050 genes were significantly changed, with 78% of those genes’ expression being decreased by IL-10 treatment (Fig. 6a and 6b, data file S2). The top 20 genes with increased or decreased expression by WTD are shown in Figure 6c. KEGG pathway analysis showed that IL-10 affected genes are involved in cytokine-cytokine receptor interaction (fig. S7b). We observed that IL-10 induced a decrease in expression of genes classically associated with CD8+ T cell exhaustion (Fig. 6d). Comparison to a previously published list of exhaustion-specific CD8+ T cell genes (43) identified four clusters of exhaustion genes whose expression was altered by IL-10. Cluster 1 comprised genes with increased expression in both exhausted T cells and in T cells treated with IL-10. Cluster 2, the largest cluster, consisted of genes with increased expression in exhausted T cells but decreased expression by IL-10 treatment. Cluster 3 contained genes with decreased expression in exhausted T cells but increased expression by IL-10 treatment and cluster 4 comprised genes with decreased expression in both exhausted T cells and IL-10 treated T cells. A representative sample of genes from each cluster is shown (Fig. 6e). These results suggest that IL-10 may enhance CD8+ T cell activities by preventing their exhaustion. We also observed a significant decrease of IL-2Rα by IL-10 treatment both at the mRNA (fig. S7c) and protein level (Fig. 6f), which was associated with a reduction of expression of IL-2 dependent genes, such as IL-13, LIF, SLC1A4 and NFIL3 (Fig. 6g) (44). Our results suggest that IL-10 increases the cytotoxic activity of CD8+ T cells by limiting their sensitivity to IL-2. As with monocytes, subsaturating doses of WTD differentially affected a subset of genes whose expression was changed by IL-10, with the expression of most of those genes being decreased by IL-10 treatment (Fig. 6h). At subsaturating doses, WTD failed to decrease the expression of classical IL-2 dependent genes like IL-13 and LIF to the same extent as the saturating dose, suggesting that inhibition of IL-2 activities by IL-10 requires high IL-10 doses (Fig. 6i).
Figure 6. Characterisation of transcriptional activity induced by IL-10 and high affinity variants in human CD8+ T cells.
(a). Volcano plot of CD8+ T cell genes significantly increased by WTD 50 nM ≧ 0.6 log2 fold change (red) and significantly decreased ≦ −0.6 log2 fold change compared to unstimulated cells. Fold change was calculated by dividing 50 nM WTD values by unstimulated values for each donor. The average fold change was calculated and the log2 of this value is plotted. P values were calculated by two sample equal variance two-tailed t-test of the log2 fold change of 50 nM WTD/unstimulated genes for each donor. Genes that were not significantly changed (p>0.05) or were ≦ 0.6 ≧ −0.6 log2 fold change were excluded. (b). Proportion of CD8+ T cell genes whose expression was significantly increased or decreased by 50 nM WTD ≧ 0.6 or ≦ −0.6 log2 fold change compared to unstimulated cells. (c). Log2 fold change for the top 20 protein-coding genes whose expression was significantly increased (red) and decreased (blue) by 50 nM WTD in CD8+ T cells. (d). Heat map comparing a list of previously reported exhaustion-associated genes (43) to genes whose expression was significantly changed by 50 nM WTD. Previously reported genes were given a nominal value of 1 for genes with increased expression and −1 for genes with decreased expression. Log2 fold change for 50 nM WTD was plotted. Cluster 1 (C1) represents genes with increased expression in exhausted cells and with increased expression induced by 50 nM WTD. C2 represents genes with increased expression in exhausted cells and decreased expression induced by 50 nM WTD. C3 represents genes with decreased expression in exhausted cells and with increased expression induced by 50 nM WTD. C4 represents genes with decreased expression in exhausted cells and with increased expression induced by 50 nM WTD. (e). The log2 fold change induced by 50 nM WTD for selected genes from each cluster is shown. Each point represents one biological replicate. (f). Measurement of CD25 protein levels by flow cytometry in activated CD8+ T cells in a PBMC population in the presence or absence of IL-10. Fold change was calculated by dividing CD25 levels of IL-10 stimulated by non-IL-10 stimulated control values for each donor. Each point represents one biological replicate. P value was calculated by paired t test. (g). Heatmap showing the log2 fold change induced by 50 nM WTD for genes whose protein expression was previously reported to be altered by IL-2 in CD8+ T cells (44). (h). Percentage gene expression activity of low dose WTD (0.1 nM) compared to high dose WTD (50 nM). The log2 fold change of 0.1 nM WTD/unstimulated was divided by that for 50 nM WTD/unstimulated and multiplied by 100. Genes that showed ≦ 75% of high dose activity (813 genes) are highlighted in red. Insert shows the percentage of these genes with increased or decreased expression by WTD. (i). Log2 fold change for genes associated with exhaustion or IL-2 stimulation and their expression after treatment with WTD at 50 nM or 0.1 nM. (j). Volcano plot of genes whose expression was changed by WTD (blue) or R5A11D (green) at 0.1 nM in CD8+ T cells. Only genes that had already been shown to be significantly changed by 50 nM WTD are plotted. (k). Heatmap of the top 10 CD8+ T cell genes with increased or decreased expression by 0.1 nM WTD compared to 0.1 nM R5A11D. (l). Heatmap of exhaustion or IL-2 associated genes changed by WTD at 50 nM and 0.1 nM and R5A11D at 0.1 nM. CD8+ T cells from 3 donors were analysed in (a) to (l).
As seen for monocytes, WTM showed poor induction of gene expression (fig. S7d and S7e), in line with its suboptimal STAT activation. R5A11M again enhanced the transcriptional response when compared to WTM but induced expression at levels below those induced by the dimeric ligands despite similar STAT signalling profiles (fig. S7d and S7e). Indeed, 58% of the genes that showed altered expression in response to WTD were changed to a lesser extent by the high affinity monomer R5A11M (fig. S7f). Similar to the results obtained with monocytes, R5A11D at 0.1 nM produced enhanced transcriptional responses compared to WTD at 0.1 nM, supporting its ability to act effectively at low concentrations (fig. S7d and S7e). 39% of genes showed enhanced expression in response to R5A11D compared to WTD (Fig. 6j and fig. S7g), as reflected in the comparison of the top 10 genes with the greatest increase or decrease in expression in response to WTD and R5A11D at 0.1 nM (Fig. 6k). Classical IL-2 dependent genes showed greater inhibition by R5A11D compared to WTD at this low concentration (Fig. 6l). Together, our data confirms that IL-10 variants with enhanced affinity for IL-10Rβ exhibit more robust activity at a wider range of ligand concentrations, which opens new avenues to boost IL-10-based anti-cancer immune-therapies.
R5A11D enhances the anti-tumor function of chimeric antigen receptor (CAR) T-cells
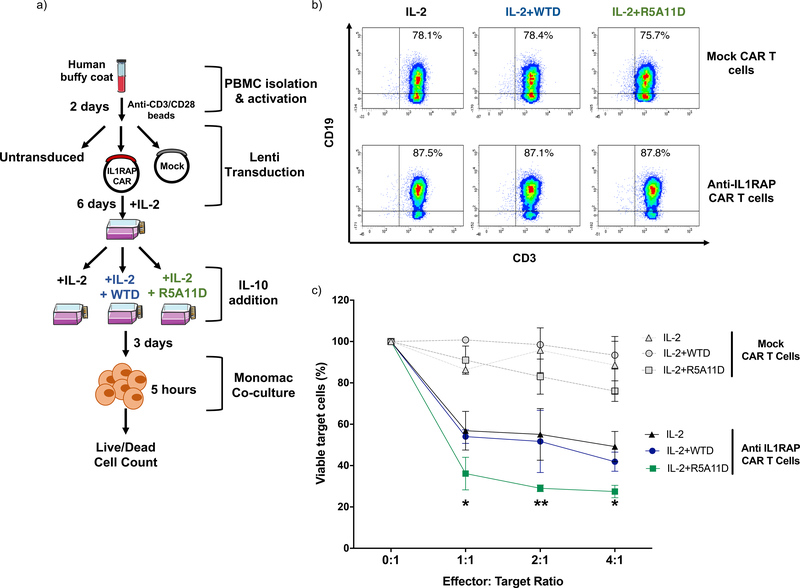
Chimeric antigen receptor (CAR) T cells have provided the basis for promising advances in cancer immune therapies. Thus, we determined whether the IL-10 variants would enhance the cytotoxic activities elicited by CAR T cells using a CAR T cell based model that targets acute myeloid leukemic cells (45). Human T cells were activated and transduced with anti-IL-1RAP-CD28−-41BB-CD3ζ CAR or mock CAR (without anti-IL-1RAP) and initially expanded for 6 days in IL-2-conditioned media. After that initial expansion, cells were cultured in IL-2-containing media containing either WTD or R5A11D for an additional 3 days before a cytotoxicity killing assay of target IL-1RAP-expressing Mono-Mac-6 cells was performed (Fig. 7a and 7b). IL-1RAP CAR T cells cultivated in the presence of IL-2 with or without the IL-10 variants showed specific cytotoxicity against Mono-Mac-6 cells in a ratio-dependent manner when compared to mock CAR-transfected T cells (Fig. 7c). At the different ratios tested, IL-1RAP CAR T cells cultured with IL-2 and R5A11D exhibited higher levels of cytotoxicity than IL-1RAP CAR T cells cultured with IL-2 and WTD or IL-2 alone. Only 36% of Mono-mac-6 cells remained viable after incubation with IL-1RAP CAR T cells treated with IL-2 and R5A11D, as compared with 54% and 56% of Mono-mac-6 cells remaining viable after incubation with IL-1RAP CAR T cells treated with IL-2 and WTD or IL-2, respectively. Collectively, our data demonstrate superior efficacy of R5A11D over WT IL-10 in boosting the cytotoxic activities of CAR T cells.
Figure 7. Evaluation of the effect of WTD and R5A11D IL-10 on CAR T-cell cytotoxicity.
(a) Schematic of cytotoxic assay for CAR T cells. Total T cells were isolated from PBMCs (n=3 donors), activated with anti-CD3/CD28 dynabeads for 2 days, and transduced with lentivirus. Effector T-cells (untransduced T-cells, mock T-cells or IL-1RAP (interleukin-1 receptor accessory protein) CAR T-cells) were cultivated in the presence of IL-2 with or without WTD (25 nM) or R5A11D (25 nM) for 3 days. The target cells, Mono-Mac-6 cells (IL-1RAP positive leukemic cell line), were co-cultured at the indicated effector:target (E:T) ratios. After 5 hours, the percentage of viable Mono-Mac-6 cells was measured by 7-AAD staining by flow cytometry. (b) Representative dot plots of expression profiles as analyzed by truncated CD19 marker expression on anti-IL-1RAP CAR T cells and mock T cells cultured in the presence of IL-2 with or without WTD or R5A11D for 3 days. PBMCs from n=3 healthy donors were analysed. (c) Target cell viability after 5 hours co-culture with effector cells. Data are presented as mean ±SEM (PBMCs from n=3 healthy donors). *p<0.001, **p<0.0001 (IL-2 alone or IL-2+WTD vs. IL-2+R5A11D).
DISCUSSION:
IL-10 is an important immunomodulatory cytokine that attenuates inflammatory responses and enhances the cytotoxic activities of CD8+ T cells (26, 41, 46). Despite its central role in preserving immune homeostasis, there is still a dearth of knowledge of the exact molecular mechanisms through which IL-10 carries out its functions. We postulated that the weak binding affinity that IL-10 exhibits for IL-10Rβ critically contributed to its functional fitness, by limiting the range of concentrations at which IL-10 elicits its full immunomodulatory potential. Here, we engineered variants of IL-10 with enhanced affinity for IL-10Rβ to investigate whether the stability of the IL-10 receptor complex determines the bioactivity potency of IL-10. A major finding of our study was that affinity-enhanced IL-10 variants triggered more robust responses at a wide range of ligand concentrations and in different immune cell subsets than did WT IL-10. The second major finding was that the stoichiometry of the IL-10-receptor complex contributed to the bioactivity potency of IL-10 beyond the regulation of STAT activation. More generally, this work outlines a strategy to improve the potency of cytokines with low binding affinities for their cognate receptors and presents new molecular and cellular data with the potential to revitalise failed IL-10 therapies.
IL-10 affected the monocytic transcriptional program in our studies, agreeing with previous work (46). IL-10 treatment inhibited antigen presentation by monocytes, limited their ability to recruit inflammatory immune cell subsets through inhibition of chemokines and chemokine receptor expression, and boosted their phagocytic activity through the upregulation of scavenger receptor-encoding genes such as CD93, CD47, CD163 and cytokine receptor-encoding genes such as IL-21R. In addition, IL-10 treatment modulated the metabolic activity of monocytes by altering their glycolytic and lipid biosynthesis potential, in line with prior data (47). IL-10 effects were slightly biased towards gene repression, with 59% of differentially expressed genes being inhibited by IL-10. Indeed, several studies have reported the ability of STAT3 to inhibit transcription induced by other STATs (48–50), suggesting that STAT3-activating cytokines may elicit their functions by disrupting transcriptional programs induced by other cytokines. In agreement with this model, we reported that IL-6, another STAT3 activating cytokine, promotes strong STAT3 binding to chromatin but poor gene expression (30). Our study was done in resting monocytes. However, in an inflammatory environment, activated monocytes secrete proinflammatory cytokines such as IFN-γ, which interfere with IL-10 activity (51). Thus, it would be of interest to investigate whether our IL-10 variants preserve their inhibitory potential in the presence of inflammatory cytokines.
Most reports in the literature that describe IL-10 activities have focused on myeloid cells and use a single dose of IL-10, often at saturation (3, 52, 53). However, we have a poor understanding of the range of IL-10 doses at which this cytokine elicits a full response in myeloid cells, a critical aspect when considering clinical translation of this cytokine. Here, we provided transcriptional data from monocytes stimulated with a saturating and a subsaturating dose of IL-10, with the latter more closely resembling the doses achieved during IL-10 therapies (42). The expression of 27% of genes differentially affected by IL-10 were sensitive to changes in IL-10 dose. The vast majority of affected genes (95%) were genes inhibited by IL-10 and those encoding proteins that critically contribute to establishing an inflammatory environment, such as the chemokines and cytokines IL-24, CXCL10, CXCL11, and CCL22. These data suggest that the anti-inflammatory activities of IL-10 specifically require high and sustained doses to reach their full effect, explaining in part the failure of IL-10 therapies. Our engineered IL-10 variant exhibited a more robust activity at subsaturating doses and potently inhibited the expression of genes encoding proinflammatory chemokines and cytokines, such as IL-24, CXCL10 and CCL22. It is thus tempting to speculate that our engineered variant could make IL-10 therapies therapeutically viable by promoting anti-inflammatory activities at low ligand doses.
The anti-inflammatory activities elicited by IL-10 and its effects on monocytes and macrophages are well documented. On the other hand, how IL-10 influences the activity of CD8+ T cells is less clear and more controversial (41). Although IL-10 has been reported to enhance the function of CD8+ T cells and their ability to kill tumour cells (18), others report that the presence of IL-10 in the tumour microenvironment predicts poor responses due to inhibition of T cell activation (54). Our results agree with a positive effect of IL-10 treatment on the cytotoxic activities of CD8+ T cells. CD8+ T cells stimulated in the presence of IL-10 exhibited enhanced levels of effector molecules such as granzyme B, agreeing with clinical trials that show an improvement in the tumour response of patients treated with PEGylated-IL-10 (21). In addition, our engineered IL-10 variant improved the killing activity of CAR-T cells in vitro. However, the molecular bases by which IL-10 boosts the anti-tumour CD8+ T cell response remains poorly defined. Our transcriptional studies highlighted that CD8+ T cells stimulated with IL-10 exhibited a reduction in an exhaustion gene signature and were more functionally fit. IL-10-treated CD8+ T cells also expressed lower levels of IL-2Rα, which correlated with a reduced IL-2 gene signature in these cells. Together, our data agree with a model in which IL-10 may reduce the sensitivity of CD8+ T cells to IL-2 and their transition towards an exhausted phenotype. Our engineered IL-10 variant outperformed WT IL-10 in every readout tested when subsaturating doses were used, reproducing our observations in monocytes and highlighting its potential to boost anti-tumour responses at therapeutic doses.
The importance of the dimeric IL-10 architecture for generating its biological responses is not yet fully understood. WTD binds IL-10Rα 60-fold more avidly than WTM, which contributes to its more efficient recruitment of IL-10Rβ to the signalling complex and its more potent activities (55). Paradoxically, R5A11M, which binds IL-10Rβ with higher affinity and elicits receptor assembly levels similar to WTD, triggers weaker transcriptional responses, despite activating STAT transcription factors to a similar extent as WTD. In addition, viral IL-10 (also a dimeric ligand) induces the same specific activity as WTD although it binds to IL-10Rα with lower affinity than WTM (56, 57). Overall these observations suggest that in addition to receptor binding affinity, the stoichiometry of the IL-10-receptor complex helps to fine-tune IL-10 bioactivity potencies. We showed that the number of phospho-tyrosines available in cytokine receptor intracellular domains critically contributes to defining signalling identity by cytokines (30). IL-6 variants that triggered partial phosphorylation of Tyr residues available in the gp130 intracellular domain exhibited a biased STAT3 versus STAT1 activation (30). A similar model could be invoked to explain functional differences between monomeric and dimeric IL-10 ligands. Different patterns of phospho-Tyr in IL-10Rα and IL-10Rβ could be induced in the hexameric receptor complex formed by the dimeric ligands, which could trigger the activation of additional signalling pathways and provide functional specificity. In agreement with this model, WTM and R5A11M elicited biased STAT3 activation in CD8+ T cells. Future studies will be needed to address whether the larger number of Tyr residues available in the hexameric complex engaged by WTD helps to define its signalling signature and biological identity.
MATERIALS AND METHODS:
Protein expression and purification
Monomeric wild type IL-10 (25), monomeric high affinity variants and IL-10Rα ectodomain (amino acids 22–235) were cloned and expressed as described in (30). Briefly, protein sequences were cloned into the pAcGP67-A vector (BD Biosciences) in frame with an N-terminal gp67 signal sequence, driving protein secretion, and a C-terminal hexahistidine tag. The baculovirus expression system was used for protein production as outlined in (58). Spodoptera frugiperda (SF9) cells, grown in SF900III media (Invitrogen), were transfected to produce P0 baculovirus stocks that were then expanded in SF9 cells to produce P1 virus stock. Protein expression was performed using Trichoplusiani ni (High Five) with cells grown in InsectXpress media (Lonza).
Purification was performed using a previously described method (59). Briefly, the cells were pelleted with centrifugation at 2000 rpm before precipitation through the addition of Tris pH 8.0, CaCl2 and NiCl2 to final concentrations of 200mM, 50mM and 1mM respectively. The precipitate formed was then removed through centrifugation at 6000 rpm. Nickel-NTA agarose beads (Qiagen) were added and the target proteins purified through batch binding followed by column elution in HBS, 200mM imidazole, pH 7.2. Target proteins were concentrated and further purified by size exclusion chromatography on an ENrich SEC 650 300 column (Biorad) (dimeric cytokines) and GE Healthcare Sephadex column (30/100GL) (monomeric cytokines); equilibrated in 10 mM HEPES (pH 7.2), 150 mM NaCl. IL-10Rα was biotinylated using EZ-Link NHS biotinylation kit (Thermo) according to the manufacturer’s protocols.
To express biotinylated IL-10Rβ, the ectodomain (amino acids 20–220) was cloned into the pAcGP67-A vector carrying a C-terminal biotin acceptor peptide (BAP)-LNDIFEAQKIEWHW followed by a hexahistidine tag. The purified protein was biotinylated with BirA ligase following a previously described protocol (59).
To express dimeric wild type IL-10 and dimeric high affinity variants (amino acids 26–178), synthesised gene blocks (IDT) were cloned into the pET21 vector in frame with an N-terminal hexahistidine tag and a lac promotor and transformed into E. Coli BL21 cells. Protein production was induced using a final concentration of 1 mM IPTG (Formedium) followed by incubation at 37⁰C for 3 to 5 hours. Cells were harvested by centrifugation at 6000 xg for 15 minutes. The cell pellets were resuspended in 50 mM Tris-HCl (pH 8.0), 25% (w/v) sucrose, 1 mM Na EDTA, 10 mM DTT, 0.2 mM PMSF per litre of original culture and frozen at −80⁰C overnight.
The recombinant protein was expressed in inclusion bodies. Cell pellets were lysed in 100 mM Tris-HCl (pH 8.0), 2% (v/v) TritonX-100, 200 mM NaCl, 2500 units Benzonase, 10 mM DTT, 5 mM MgCl2, 0.2 mM PMSF and incubated for 20 minutes with stirring at room temperature. 10 mM EDTA final concentration was added to the suspension and the cells were sonicated (8–10 cycles of 15 seconds on/off, 15 micron amplitude, Soniprep 150) in an ice bath. The solution was centrifuged at 7000 xg for 15 mins (4⁰C) and the pellet was resuspended in 50mM Tris-HCl pH 8.0, 0.5% Triton X-100, 100 mM NaCl, 1 mM Na EDTA, 1 mM DTT, 0.2 mM PMSF. This step was repeated for a total of at least three washes until the preparation appeared white in colour. The final pellet was then washed once in detergent free buffer (50 mM Tris-HCl pH 8.0, 1 mM Na EDTA, 1 mM DTT, 0.2 mM PMSF).
The purified inclusion bodies were solubilised in 10 mls of 6M GuHCl per litre of original culture, for 30 minutes at room temperature. The solution was clarified by a centrifugation at 7000 xg for 15 minutes and the solubilised protein was decanted. Refolding was performed through dropwise addition of the solubilised protein solution into refolding buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM EDTA, 2 mM reduced glutathione (GSH) and 0.2 mM oxidized glutathione (GSSG)) at a ratio of 1:20 solution:buffer at 4⁰C. This solution was incubated with gentle stirring overnight at 4⁰C. The solution was then filtered to remove any precipitant and dialysis performed against 10 mM HEPES (pH 7.2),150 mM NaCl, using dialysis membrane with a 14 kDa MW cut off. After dialysis protein was then further purified using Ni-NTA beads and by size exclusion on a Superdex75 increase 10/300 column (GE Healthcare). To remove endotoxin, 1 mL of Ni-NTA agarose was added to a polyprep column and equilibrated with 10 mls of HBS before protein was added. The column was washed with 50 column volumes of ice-cold HBS, 150 mM NaCl, 20 mM imidazole, 0.1% Triton-X114 (pH 7.2) to remove endotoxin. The column was then washed with a further 20 column volumes of HBS, 20 mM imidazole (pH 7.2). The now endotoxin-free protein was eluted using 4 column volumes of HBS, 200 mM imidiazole (pH 7.2). The protein was buffer exchanged into 10 mM HEPES, 150 mM NaCl (pH 7.2) using PD-10 columns (GE Healthcare). Endotoxin levels were measured using Pierce LAL Chromogenic Endotoxin Quantitation Kit (Thermo) following the manufacturer’s protocol. For all dimeric proteins endotoxin levels were below detection levels of the kit.
Surface Plasmon Resonance
Surface plasmon resonance was used to determine the binding affinity of the recombinantly produced monomeric WT IL-10 and variants to IL-10Rβ in the presence or absence of IL-10Rα. Biotinylated IL-10Rβ was immobilised onto the chip surface with streptavidin. Series S Sensor SA (GE Healthcare) chips were primed in 10 mM HEPES, 150 mM NaCl, 0.02% TWEEN-20, prior to immobilisation of the biotinylated receptor. Analysis runs were performed in 10 mM HEPES, 150 mM NaCl, 0.05% TWEEN-20 and 0.5% BSA. A Biacore T100 (T200 Sensitivity Enhanced) was used for measurement and Biacore T200 Evaluation Software 3.0 was used for data analysis.
Cell culture
Human buffy coats were obtained from the Scottish Blood Transfusion Service. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation following the manufacturer’s protocol (Lymphoprep, StemCell Technologies). PBMCs were grown in complete RPMI-1640 media (10% v/v FBS, 100 U/mL penicillin-streptomycin) (Gibco) and cytokines for proliferation and activation. For three days, media was supplemented with 100 ng/mL anti-CD3 (human UltraLEAF, Biolegend) and 20 ng/mL IL-2 (Proleukin, Novartis) in the absence or presence of the IL-10 variants. Cells were centrifuged and resuspended in media supplemented with 20 ng/mL of IL-2 in the absence or presence of IL-10 variants. Cell populations were allowed to expand for 2–3 days.
CD8+ T cells were isolated from PBMCs by magnetic separation (MACS Miltenyi) after staining with anti-CD8aFITC antibody (Biolegend #30906). Purified CD8+ T cells were activated with ImmunoCult Human CD3/CD28 T cell Activator (Stem Cell), used following the manufacturer’s protocol, and 20 ng/mL IL-2 in the presence or absence of IL-10 variants. Cells were activated for 3 days and then the media was replaced with complete RPMI media supplemented with 20 ng/mL IL-2 in the absence or presence of IL-10 variants for 2–3 days.
Monocytes were isolated from PBMC populations using CD14+ selection. An anti-CD14FITC antibody (Biolegend #367116) was used to stain cells and isolation was performed by magnetic separation following the manufacturer’s protocol (MACS Miltenyi). Monocytes were cultured in complete RPMI media (as above) supplemented with M-CSF (20 ng/mL, Biolegend) and stimulated with IL-10 variants.
CAR-T cytotoxicity assay
The Interleukin-1 Receptor Accessory Protein (IL-1RAP) (Interleukin-1 Receptor Accessory Protein) CAR lentiviral construct was generated as previously described (45). Briefly, this vector carries a 3rd generation CAR, an inducible Caspase 9 (iCASP9) (inducible Caspase 9) suicide gene and a delta CD19 surface gene. The mock control vector does not contain the CAR sequence.
PBMCs were isolated by Ficoll gradient density centrifugation using Ficoll-Paque (Velizy-Villacoublay, France) with anonymous blood samples collected from healthy donors at a French blood center (Besançon, France). Donors provided written informed consent. The study was conducted in accordance with the ethical guideline (declaration of Helsinki) and approved by the local ethical the CPP-Est committee (France).
The human tumor Mono-Mac-6 cell line was obtained from DSMZ German collection of microorganisms and cell culture GmbH and stored in a master cell bank. The cells were cultivated in RPMI-1640 media supplemented with 10% FBS penicillin-streptomycin.
CD3/CD28 activated cells were established from healthy donors and transduced with lentiviral supernatant encoding the IL-1RAP CAR or mock sequences. At day 6 post-transduction, cells were put in different culture conditions: IL-2 (500 UI/mL) with or without WTD or R5A11D at 25nM for 3 days.
To assess CAR T-cell cytotoxicity, effector cells (untransduced T-cells, Mock T-cells and IL-1RAP CAR T-cells) were labeled with e-FluorTM v450 (eBioscience, #65–0842-85) before co-culture to differenciate them from tumor cells. Effector cells were co-cultured with Mono-mac-6 cells at different effector:target (E:T) ratios (1:1; 2:1 and 4:1). After 5 hours, cells were stained with 7-amino-actinomycin D (7-AAD; BD Bioscience, #559925) and the percentage of alive Mono-Mac-6 cells was determined by flow cytometry using a FACSCanto II flow cytometer (BD Bioscience).
Flow cytometry staining and antibodies
For live cell surface staining of HLA-DRPE (Biolegend #307605) or HLA-DRAPC (Bioloegend #307610), cells were kept at 4⁰C or on ice during live cell surface marker staining and staining was done in 96-well v-bottom plates (Greiner) unless otherwise stated. Non-adherent monocytes were removed from culture by centrifugation and resuspension in cold PBS. Adherent monocytes were detached using Accutase (StemCell Technologies) at room temperature for 5 to 10 minutes. Non-adherent and detached cells were combined and resuspended in FcR blocking reagent (Miltenyi) for 10 minutes at 4⁰C in a volume of 50 μL per condition. Cells were washed in PBS/0.5% BSA and resuspend in 50 μL of antibody mixture diluted 1/100 in FcR blocking reagent. Antibody incubation was done for 30 to 60 minutes at 4⁰C in the dark. Cells were washed twice before being resuspended in 100 μL per well for analysis on the CytoFlex flow cytometer (Beckman Coulter). Mean fluorescence intensity (MFI) was quantified for all populations. Data was normalised within each donor by dividing the MFI of IL-10 treated cells by an untreated control sample from the same donor to calculate fold change.
For granzyme B intracellular staining, either PBMCs or CD8+ cells were fixed with 2% paraformaldehyde for 10 minutes at room temperature before washing in PBS. Cells were permeabilised in 0.1% Triton-X100/PBS for 10 minutes and washed in PBS/0.5% BSA. Cells were stained with anti-CD8aAlexaFluor700 (Biolegend #300920), anti-CD4PE (Biolegend #357404), anti-CD3BrilliantViolet510 (Biolegend #300448) and anti-granzyme BFITC (Biolegend #515403) at 1/100 dilution in PBS/0.5% BSA for one hour before washing. MFI was quantified for all populations and normalisation was performed as described above.
For phospho-flow analysis of STAT1 and STAT3, cells were plated at 50 μL of cell suspension per well at a density of 2 × 105 cells per well in 96-well V bottom plates. For dose-response studies, cells were left unstimulated or simulated with 7-fold serially diluted IL-10 variants (50 μL per well) for 15 minutes at 37°C before fixation with 2% paraformaldehyde for 10 minutes at room temperature. For kinetic studies, cells were stimulated with a saturating concentration of IL-10 variants (50 nM) at defined time points before fixation with 2% paraformaldehyde. Cells were washed in PBS and permeabilised in ice-cold 100% methanol and incubated on ice for a minimum of 30 minutes. Cells were fluorescently barcoded as previously described (30, 60). Briefly, a panel of 16 combinations of two NHS-dyes (Pacific Blue and DyLight800, Thermo) was used to stain individual wells on ice for 35 minutes before stopping the reaction by washing in PBS/0.5% BSA. Once barcoded, the 16 populations were pooled together for antibody staining. PBMCs, CD8+ cells and monocytes were stained with the cell surface markers described above as well as anti-pSTAT3Alexa488 (Biolegend #651006) and anti-pSTAT1Alexa647 (Cell Signalling Technologies #8009). During acquisition, individual populations were identified according to the barcoding pattern and pSTAT3Alexa488 and pSTAT1Alexa647 MFI was quantified for all populations. MFI was plotted and sigmoidal dose response curves were fitted using Prism software (Version 7, GraphPad). Data was normalised by assigning the highest MFI of the top concentration of all stimuli as 100% and the lowest MFI as 0% within each donor group.
Yeast display library
The yeast surface display protocol was adapted from previous protocols (30, 61). To create an IL-10 yeast display library, the monomeric IL-10 gene (25) was subject to error-prone PCR as previously described (28). This product was amplified and transformed with a linearized pCT302 vector into the Saccharomyces cerevisiae strain EBY100 and grown in selective dextrose casamino acids (SDCAA) media at 30°C for two days. Yeast cells were placed in selective galactose casamino acid media (SGCAA) at 20°C for two days to induce cell surface expression of IL-10 variants as previously described (27). Magnetic activated cell sorting (MACS, Miltenyi) was used to select for IL-10 variants with increased binding affinity for IL-10Rβ as described previously for other systems (62). Briefly, the first round of selection was performed using high concentrations of streptavidin beads to remove any yeast that displayed variants that bound streptavain. The second round of selection selected for yeast that displayed variants with the c-myc tag at their C-terminus, ensuring that displayed proteins were properly folded. The subsequent rounds of selection were carried out by incubating induced yeast with decreasing concentrations of recombinantly produced biotinylated IL-10Rβ for 2 hours then with fluorescently labelled streptavidin (AlexaFluor647) a 15 minute incubation. Magnetic activated cell sorting (MACS, Miltenyi) selected for yeast that displayed IL-10 variants that bound to IL-10Rβ. Once the concentration of IL-10Rβ required for binding had decreased sufficiently as compared to WT monomeric IL-10, the yeast were plated on SDCAA agar and single colonies were isolated for dose response studies to determine the EC50 values of the mutants.
Plasmoid preparations were then made from those yeast colonies displaying promising IL-10 variants using Zymoprep kits (ZymoResearch). The plasmids were transformed into competent DH5α E. coli for plasmid amplification prior to miniprep purification (Promega, Wizard Plus SV). DNA sequencing of each was then carried out to determine the sequence of the generated high-affinity monomeric IL-10 varients. These variant genes were then cloned into the baculovirus expression vector pACgp67BN and recombinantly expressed as described above.
RT-qPCR
Cells were subject to RNA isolation using the Qiagen RNeasy kit. 50ng RNA was reverse transcribed to cDNA using iScript cDNA synthesis kit (Biolegend), which was used as template for qPCR. SYBR premix Ex-Taq (Clontech) was used for the reaction with the following GZMB primers (used at 10 μM) (63): forward (5’−3’) tgggggacccagagattaaaa and reverse (5’−3’) tttcgtccataggagacaatgc. GAPDH and 18S were used as housekeeping genes for normalisation. Each sample was done in duplicate.
Measurement of IL-6 secretion
Monocytes were stimulated with LPS (100 ng/mL) (E. coli O26:B6, Sigma) plus IL-10 variants at various concentration for 8 hours. The supernatants were then removed and used for enzyme linked immunosorbent assays (ELISA) for IL-6 detection (Biolegend, #430501), following the manufacturer’s protocol. 96-well half-area plates (Sigma) were coated in capture antibody and incubated overnight at 4°C. Plates were washed in PBS/0.05% Tween-20 and blocked for 1 hour in assay diluent and then washed. The supernatants were diluted 1 to 10 in assay buffer before they were added to the plate. The plates were incubated at room temperature for two hours with gentle shaking. The plates were washed again and incubated for 1 hour with the detection antibody. After washing, avidin-HRP was added and incubated for 30 minutes, then the TMB substrate solution was incubated for 15 minutes. The reaction was stopped by addition of H2SO4 and absorbance was measured at 450 nm and 570 nm on a plate reader (CLARIOstar) with absorbance at 570 nm being subtracted from 450 nm. Standard curves were fitted and concentrations were calculated using the CLARIOstar software.
RNA Transcriptome Sequencing
Human primary monocytes and CD8+ T cells from three human donors each (isolated from PBMCs bought from StemCell Technologies) were stimulated as described above. Cells were washed in Hank’s balanced salt solution (HBSS, Gibco) and snap frozen for storage. RNA was isolated using the RNeasy Kit (Qiagen) according to the manufacturer’s protocol. All RNA 260/280 ratios were above 1.9. 1 μg of RNA was used per sample. Transcriptomic analysis was done by Novogene as follows. Sequencing libraries were generated using NEBNext® UltraTM RNALibrary Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First StrandSynthesis Reaction Buffer (5X). First strand cDNA was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends by exonuclease/polymerase activities. After adenylation of the 3’ ends of the DNA fragments, NEBNext Adaptors with a hairpin loop structure were ligated to prepare for hybridization. To select cDNA fragments of preferentially 150~200 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, USA). 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min then at 95 °C for 5 min before PCR. PCR was the performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. Finally, PCR products were purified (AMPure XP system) and the library quality was assessed using the Agilent Bioanalyzer 2100 system.
RNA Sequencing Data Analysis
Primary data analysis for quality control, mapping to reference genome, and quantification was conducted by Novogene as follows. In the quality control step, raw data (raw reads) of FASTQ format were firstly processed to obtain clean data (clean reads) by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. At the same time, Q20, Q30 and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Paired-end clean reads were mapped to the reference genome (GRCh37/hg19) directly downloaded from https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ using HISAT2 software. HISAT2 uses a large set of small Graph FM indexes that collectively cover the whole genome. These small indexes (called local indexes), combined with several alignment strategies, enable rapid and accurate alignment of sequencing reads. To quantify gene expression, HTSeq was used to count the read numbers mapped for each gene, including known and novel genes. The RPKM (reads per kilobase of exon model per million mapped reads) of each gene was calculated based on the length of the gene and reads count mapped to this gene. RPKM considers the effect of sequencing depth and gene length for the reads count at the same time and is currently the most commonly used method for estimating gene expression levels.
Statistical analysis of the Novagene output was performed as follows. The fold change was calculated by dividing the IL-10 stimulated expression levels by the unstimulated control expression levels within each donor. The average fold change was calculated for each stimulation across the three donors and the log2 of this average was calculated. To calculate significance, the log2 of the fold change between IL-10 stimulated and unstimulated expression levels of each donor was calculated separately and a two sample equal variance two tailed t test was used to generate the p value. The log10 of this p value was plotted against the previously calculated log2 average fold change. Genes with an RPKM of less than 1 in two or more donors in both WTD 50 nm and unstimulated conditions were excluded from analysis so as to remove genes whose expression was near the detection limit.Genes which were significantly (p≦ 0.05) changed greater than 0.6 or less than −0.6 log2 fold change in the WT IL-10 dimer (WTD) 50 nM condition were taken as a set list of genes against which all other IL-10 stimulations were compared, regardless of the level of significance in the other IL-10 stimulations. Genes with increased expression were denoted as genes ≧ 0.6 log2 fold change and genes with decreased expression were denoted as genes ≦ −0.6 log2 fold change. To compare WTD 50 nM stimulation to other IL-10 variant stimulations, the average log2 fold changes of the variants/unstimulated were divided by the average log2 fold change of WTD/unstimulated.
Functional annotation of genes (KEGG pathways, GO terms) was done using DAVID Bioinformatics Resource functional annotation tool (64, 65). Clustered heatmaps were generated using the R Pheatmap package from Kolde (2019). RNA sequencing data has been deposited in the GEO Database with the accession number GSE146438. This includes all RAW and normalized files.
Live-cell dual color single molecule imaging studies
Receptor homo- and heterodimerization was quantified by two-colour single-molecule cotracking as described previously (23, 31, 33). Receptor dimerization experiments were performed in HeLa cells transienty expressing IL-10Rα and IL-10Rβ with N-terminally fused variants of monomeric ECFP and EGFP, respectively. Cell surface Labelling was achieved using anti-GFP nanobodies Minimizer(MI) and Enhancer(EN), respectively, site-specifically conjugated with photostable fluorophores by an engineered cysteine residue. For quantification of receptor heterodimerization, IL-10Rα and IL-10Rβ were labelled with MIRho11 (ATTO Rho11, ATTO-TEC GmbH) and ENAT643 (ATTO 643, ATTO-TEC GmbH), respectively. To quantify homodimerization, IL-10Rα was labelled with MIRho11 and MIAT643, or IL-10Rβ was labelled with ENRho11 and ENAT643. Overexpression of the other receptor subunit was ensured by labelling with ENAT488 or MIAT488 (ATTO 488, ATTO-TEC GmbH), respectively. Time-lapse dual-color imaging of individual IL-10Rα and IL-10Rβ in the plasma membrane was carried out by total internal reflection fluorescence microscopy with excitation at 561 nm and 640 nm and detection with a single EMCCD camera (Andor iXon Ultra 897, Andor) using an image splitter (QuadView QV2, Photometrics). Molecules were localized using the multiple-target tracing (MTT) algorithm (66). Receptor dimers were identified as molecules that colocalized within a distance threshold of 150 nm for at least 10 consecutive frames as described in detail previously (23, 31, 33).
Supplementary Material
Supplementary Figure 1. Yeast surface display selection for high affinity monomeric IL-10 variants and recombinant expression of WT and high affinity IL-10 monomeric and dimeric variants.
Supplementary Figure 2. Biophysical characterization of the high affinity IL-10 variants
Supplementary Figure 3. Single molecule imaging of IL-10 receptors by TIRF microscopy.
Supplementary Figure 4. Extended kinetics of IL-10 and variants in human monocytes.
Supplementary Figure 5. Analysis of gene expression profiles induced by WT IL-10 and high affinity variants in human monocytes.
Supplementary Figure 6. Characterization of the IL-10-treated CD8+ T cell phenotype.
Supplementary Figure 7. Analysis of gene expression profiles induced by WT IL-10 and high affinity variants in human CD8+ T cells.
Data file S1. RNA sequencing data set for monocytes.
Data file S2. RNA sequencing data set for CD8+ T cells.
Acknowledgments:
We thank members of the Moraga, Mitra, Walter, Ferrand and Piehler laboratories for helpful advice and discussion. We thank Prof. Charles Taylor and Prof. Carmen Molina-Paris with their help with statistical analyses
Funding: This work was supported by the Wellcome Trust 203752/Z/16/Z (C.G.), by the Wellcome-Trust-202323/Z/16/Z and ERC-206-STG grant (I.M.), by EMBO (W.S. 454–2017) and by the DFG (SFB 944, P8/Z, J.P.) and NIH (R01 AI143554, M.R.W.). The Mitra Lab is supported by the Projects Fondation ARC 2019 and La Ligue Contre le Cancer grants.
Footnotes
Competing interests: C.G., S.M., and I.M. are inventors on patent application 2003428.6 submitted by University of Dundee and Institut National de la Santé et de al Recherche Médicale that covers the IL-10 mutants. The other authors declare that they have no competing interests.
Data materials and availability: The RNA sequencing data has been deposited in NCBI’s Gene Expression Omnibus with the accession number GSE146438, which includes all RAW and normalized files. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.de Waal Malefyt R et al. , Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J Exp Med 174, 915–924 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willems F et al. , Interleukin-10 inhibits B7 and intercellular adhesion molecule-1 expression on human monocytes. Eur J Immunol 24, 1007–1009 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A, IL-10 inhibits cytokine production by activated macrophages. J Immunol 147, 3815–3822 (1991). [PubMed] [Google Scholar]
- 4.Fiorentino DF et al. , IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J Immunol 146, 3444–3451 (1991). [PubMed] [Google Scholar]
- 5.Correa I et al. , Defective IL-10 production in severe phenotypes of Crohn’s disease. J Leukoc Biol 85, 896–903 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Zhu L et al. , IL-10 and IL-10 Receptor Mutations in Very Early Onset Inflammatory Bowel Disease. Gastroenterology Res 10, 65–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena A et al. , Interleukin-10 paradox: A potent immunoregulatory cytokine that has been difficult to harness for immunotherapy. Cytokine 74, 27–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardoso A et al. , The Dynamics of Interleukin-10-Afforded Protection during Dextran Sulfate Sodium-Induced Colitis. Front Immunol 9, 400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colombel JF et al. , Interleukin 10 (Tenovil) in the prevention of postoperative recurrence of Crohn’s disease. Gut 49, 42–46 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buruiana FE, Sola I, Alonso-Coello P, Recombinant human interleukin 10 for induction of remission in Crohn’s disease. Cochrane Database Syst Rev, CD005109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saraiva M, Vieira P, O’Garra A, Biology and therapeutic potential of interleukin-10. J Exp Med 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steidler L et al. , Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Shigemori S, Shimosato T, Applications of Genetically Modified Immunobiotics with High Immunoregulatory Capacity for Treatment of Inflammatory Bowel Diseases. Front Immunol 8, 22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braat H et al. , A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol 4, 754–759 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Oft M, Immune regulation and cytotoxic T cell activation of IL-10 agonists - Preclinical and clinical experience. Semin Immunol 44, 101325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue FY et al. , Interleukin-10 is a growth factor for human melanoma cells and down-regulates HLA class-I, HLA class-II and ICAM-1 molecules. Int J Cancer 71, 630–637 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Mannino MH et al. , The paradoxical role of IL-10 in immunity and cancer. Cancer Lett 367, 103–107 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Emmerich J et al. , IL-10 Directly Activates and Expands Tumor-Resident CD8(+) T Cells without De Novo Infiltration from Secondary Lymphoid Organs. Cancer Res 72, 3570–3581 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Mumm JB et al. , IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell 20, 781–796 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Mumm JB, Oft M, Pegylated IL-10 induces cancer immunity: the surprising role of IL-10 as a potent inducer of IFN-γ-mediated CD8(+) T cell cytotoxicity. Bioessays 35, 623–631 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Naing A et al. , Pegilodecakin combined with pembrolizumab or nivolumab for patients with advanced solid tumours (IVY): a multicentre, multicohort, open-label, phase 1b trial. Lancet Oncol 20, 1544–1555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logsdon NJ, Jones BC, Josephson K, Cook J, Walter MR, Comparison of interleukin-22 and interleukin-10 soluble receptor complexes. J Interferon Cytokine Res 22, 1099–1112 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Wilmes S et al. , Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J Cell Biol 209, 579–593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter D et al. , Ligand-induced type II interleukin-4 receptor dimers are sustained by rapid re-association within plasma membrane microcompartments. Nat Commun 8, 15976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Josephson K et al. , Design and analysis of an engineered human interleukin-10 monomer. J Biol Chem 275, 13552–13557 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Walter MR, The molecular basis of IL-10 function: from receptor structure to the onset of signaling. Current topics in microbiology and immunology 380, 191–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao G et al. , Isolating and engineering human antibodies using yeast surface display. Nat Protoc 1, 755–768 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Mendoza JL et al. , The IFN-λ-IFN-λR1-IL-10Rβ Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity. Immunity 46, 379–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirchhofer A et al. , Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17, 133–U162 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Fabregas J et al. , Kinetics of cytokine receptor trafficking determine signaling and functional selectivity. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilmes S et al. , Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 367, 643–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moraga I et al. , Instructive roles for cytokine-receptor binding parameters in determining signaling and functional potency. Science Signaling 8, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moraga I et al. , Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands. Cell 160, 1196–1208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shouval DS et al. , Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40, 706–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frede N et al. , Evidence for non-neutralizing autoantibodies against IL-10 signalling components in patients with inflammatory bowel disease. BMC Immunol 15, 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wehinger J et al. , IL-10 induces DNA binding activity of three STAT proteins (Stat1, Stat3, and Stat5) and their distinct combinatorial assembly in the promoters of selected genes. FEBS Lett 394, 365–370 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Finbloom DS, Winestock KD, IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol 155, 1079–1090 (1995). [PubMed] [Google Scholar]
- 38.Mittal SK, Roche PA, Suppression of antigen presentation by IL-10. Curr Opin Immunol 34, 22–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito S et al. , Interleukin-10 inhibits expression of both interferon alpha- and interferon gamma- induced genes by suppressing tyrosine phosphorylation of STAT1. Blood 93, 1456–1463 (1999). [PubMed] [Google Scholar]
- 40.Dallagi A et al. , The activating effect of IFN-gamma on monocytes/macrophages is regulated by the LIF-trophoblast-IL-10 axis via Stat1 inhibition and Stat3 activation. Cell Mol Immunol 12, 326–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oft M, IL-10: master switch from tumor-promoting inflammation to antitumor immunity. Cancer Immunol Res 2, 194–199 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Naing A et al. , PEGylated IL-10 (Pegilodecakin) Induces Systemic Immune Activation, CD8. Cancer Cell 34, 775–791.e773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bengsch B et al. , Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity 48, 1029–1045 e1025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rollings CM, Sinclair LV, Brady HJM, Cantrell DA, Ross SH, Interleukin-2 shapes the cytotoxic T cell proteome and immune environment-sensing programs. Sci Signal 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warda W et al. , CML Hematopoietic Stem Cells Expressing IL1RAP Can Be Targeted by Chimeric Antigen Receptor-Engineered T Cells. Cancer Res 79, 663–675 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A, Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19, 683–765 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Ip WKE, Hoshi N, Shouval DS, Snapper S, Medzhitov R, Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang XP et al. , Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol 12, 247–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa-Pereira AP et al. , Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc Natl Acad Sci U S A 99, 8043–8047 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ray JP et al. , Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity 40, 367–377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrero C et al. , Reprogramming of IL-10 activity and signaling by IFN-gamma. J Immunol 171, 5034–5041 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Ding L, Linsley PS, Huang LY, Germain RN, Shevach EM, IL-10 inhibits macrophage costimulatory activity by selectively inhibiting the up-regulation of B7 expression. J Immunol 151, 1224–1234 (1993). [PubMed] [Google Scholar]
- 53.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE, Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med 174, 1209–1220 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao S, Wu D, Wu P, Wang Z, Huang J, Serum IL-10 Predicts Worse Outcome in Cancer Patients: A Meta-Analysis. PLoS One 10, e0139598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan JC, Indelicato SR, Narula SK, Zavodny PJ, Chou CC, Characterization of interleukin-10 receptors on human and mouse cells. J Biol Chem 268, 21053–21059 (1993). [PubMed] [Google Scholar]
- 56.Liu Y et al. , The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J Immunol 158, 604–613 (1997). [PubMed] [Google Scholar]
- 57.Yoon SI, Jones BC, Logsdon NJ, Walter MR, Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain. Structure 13, 551–564 (2005). [DOI] [PubMed] [Google Scholar]
- 58.LaPorte SL et al. , Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132, 259–272 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spangler JB, Moraga I, Jude KM, Savvides CS, Garcia KC, A strategy for the selection of monovalent antibodies that span protein dimer interfaces. J Biol Chem 294, 13876–13886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krutzik PO, Nolan GP, Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods 3, 361–368 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Boder ET, Wittrup KD, Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 15, 553–557 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Moraga I et al. , Instructive roles for cytokine-receptor binding parameters in determining signaling and functional potency. Sci Signal 8, ra114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morissette MC, Parent J, Milot J, Perforin, granzyme B, and FasL expression by peripheral blood T lymphocytes in emphysema. Respir Res 8, 62 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang da W, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Serge A, Bertaux N, Rigneault H, Marguet D, Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods 5, 687–694 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Yeast surface display selection for high affinity monomeric IL-10 variants and recombinant expression of WT and high affinity IL-10 monomeric and dimeric variants.
Supplementary Figure 2. Biophysical characterization of the high affinity IL-10 variants
Supplementary Figure 3. Single molecule imaging of IL-10 receptors by TIRF microscopy.
Supplementary Figure 4. Extended kinetics of IL-10 and variants in human monocytes.
Supplementary Figure 5. Analysis of gene expression profiles induced by WT IL-10 and high affinity variants in human monocytes.
Supplementary Figure 6. Characterization of the IL-10-treated CD8+ T cell phenotype.
Supplementary Figure 7. Analysis of gene expression profiles induced by WT IL-10 and high affinity variants in human CD8+ T cells.
Data file S1. RNA sequencing data set for monocytes.
Data file S2. RNA sequencing data set for CD8+ T cells.