Abstract
Objectives
To review the current knowledge of biomolecular factors surrounding otorhinolaryngeal illnesses and analyze their presence in COVID-19 virulence. Emphasis was placed on cytokines and vitamin D for determining susceptibility of illness.
Methods
A primary literature search of PubMed and Google Scholar for articles published between January 1, 2002 to May 31, 2020, was performed without language restrictions from May 8, 2020 to May 31, 2020. A focused second search was conducted from October 31, 2020 to November 2, 2020 for articles published between January 1, 2002 to October 31, 2020. Eligible articles were selected after evaluation of titles, abstracts, and references. A total of 45 were included in this review.
Results
Differing endotype classification schemes are used to determine cytokines present in chronic rhinosinusitis, asthma, and allergies. While immunologic responses and biomarkers are primary methods of differentiation, recent literature has also implicated geographic distribution of chronic rhinosinusitis patients in accounting for cytokine variations. The cytokines of interest (IL-4, IL-13, and INF-γ) present in the endotypes of these conditions may point towards protective mechanisms against COVID-19 through downregulation of the ACE2 receptor. These cytokines and Vitamin D highlight new areas of study for factors affecting SARS-CoV-2 virulence.
Conclusions
Further research is needed to understand the effects of Vitamin D and the various cytokines prevalent among endotypes of nasal/pharyngeal illnesses on COVID-19 pathogenesis. Findings may point towards epidemiologic trends of SARS-CoV-2 transmission and have future therapeutic indications.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Chronic rhinosinusitis endotypes, Asthma endotypes, Allergies ACE2
1. Introduction
The recent viral pandemic that started in Wuhan, China in December 2019 has been linked to a novel strain of the coronavirus family. The etiologic agent has been identified as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and causes COVID-19 [1,2]. Initial reports described the sale of live animals in Wuhan markets as the location where primary zoonotic infection in human hosts took place [2]. The mechanism of respiratory droplet transmission between individuals allowed widespread dissemination of the virus and caused a large spectrum of clinical symptoms ranging from mild to severe [1,2]. As further noted in the comprehensive literature review done by Krajewska et al. [2], fever, fatigue and dry cough are common manifestations of COVID-19. However, dyspnea, sore throat, rhinorrhea, nasal congestion, enlarged cervical lymph nodes, and sudden anosmia may be of interest in otolaryngologic practice as well [2].
Based on Center for Disease Control (CDC) advisories during the early onset of the pandemic, asthma patients remained classified as being at increased risk for adverse outcomes, similar to COPD [3]. Viral infections are a driver of flares in asthma patients under normal circumstances, however early data from China suggested that asthma patients accounted for fewer than 1% of COVID-19 patients compared to the 4.2% of asthmatic individuals in the population [4,5]. This data suggested that asthma patients may have lower rates of severe complications following SARS-CoV-2 transmission. Although this data did not confirm a protective mechanism among this population it raised intriguing questions about the underlying disease pathophysiology and if inflammatory pathways of other otorhinolaryngeal illnesses could play a role in decreased COVID-19 virulence.
The purpose of this study is to review current knowledge of the cytokines surrounding endotypes of various otorhinolaryngeal illnesses and discuss their potential effects on COVID-19 virulence.
2. Methods
The databases of PubMed and Google Scholar were searched without language restriction between May 8th, 2020 and May 31st, 2020 for terminology such as “COVID-19 pathogenesis review,” “ACE2 SARS coronavirus”, “Vitamin D in COVID-19,” “Age in coronavirus mortality,” “Immune system changes in aging review,” “Chronic rhinosinusitis endotype review,” “IL-5 CRS endotypes,” “Asia CRSsNP,” “United States CRSsNP,” “Asia CRSwNP,” “United States CRSwNP,” “Asthma endotypes review”, “Allergies and ACE2 expression,” “IL-4 SARS ACE2,” and “Coronavirus cytokines.” Articles published between January 1st, 2002 and May 31st, 2020 were initially screened. These filters yielded 2621 unique results. To provide new updates regarding ACE2 expression patterns in the head and neck, a second literature search was performed between October 31, 2020 and November 2, 2020 for terminology including “ACE2 head and neck expression.” Articles published between January 1st, 2002 and October 31st, 2020 were initially screened. These filters yielded 24 unique results.
After reviewing the article titles, abstracts, and references, further screening was completed independently. Inclusion criteria was determined based on discussion of endotypes, cytokines, vitamin D and COVID-19 pathogenesis. Reasons for exclusion included a lack of biomolecular profile and disease pathogenesis discussion, no endotype correlation to geographic populations, no relevance to the topic, and untranslatable source material to the English language. A total of 45 articles were included in this review. This study was completed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) reporting guideline and is outlined in the supplement.
To better assess the quality and risk of bias in each study, the level of evidence was scored based on the Oxford Centre for Evidence Based Medicine guide [6], listed below:
-
1.
Systematic review of inception cohort studies.
-
2.
Inception cohort studies.
-
3.
Cohort study or control arm of randomized trial.
-
4.
Case series or case-control studies or poor-quality prognostic cohort study.
A summary and scoring of select articles are included in Table 1 .
Table 1.
Evidence table summarizing results for selected studies.
| Source | Sample size | Age range (years) | Geographic distribution | Sample Localization | Pertinent results | OECBM score |
|---|---|---|---|---|---|---|
| Zhang et al. [4] | 140 | 25-87 | Wuhan, China | Oropharyngeal | No COVID-19 patients identified with asthma comorbidity | 4 |
| Dong et al. [5] | 11 | 2-69 | Wuhan, China | Oropharyngeal | Suggested that asthma and allergy do not appear as aggravating factors of COVID-19 | 4 |
| Zhou et al. [10] | 7 | 32-62 | Wuhan, China | Oropharyngeal, anal, blood, BALF | ACE2 is the target receptor for SARS-CoV-2 | 4 |
| Harmer et al. [11] | 3 | N/A | United Kingdom | All major organ systems | ACE2 expression found in renal, cardiovascular, and GI tissues | 4 |
| Hamming et al. [12] | 93 | N/A | Netherlands | Major Organ Systems previously affected by SARS-CoV | ACE2 expression found in lung alveolar cells and enterocytes of small intestine | 4 |
| Descamps et al. [15] | 70 | N/A | Belgium | Sinus, Tonsil, Salivary Gland, Glottic Larynx (Vocal Cord), Supraglottic Larynx, Oral Cavity, Pharynx | Ciliated respiratory epithelia of the sinuses, oral mucosa, vocal cords, and salivary glands display the highest expression patterns of ACE2 | 3 |
| Li et al. [16] | 15 | N/A | United Kingdom | Lung Tissue, BALF, Bronchial epithelial cells, SARS infected cells | ACE2 upregulated in chronic smokers; SARS-CoV infection leads to cytokine storm | 4 |
| Huang et al. [18] | 41 | 41-58 | Wuhan, China | Respiratory, Blood, Feces | SARS-CoV-2 patients demonstrate elevated IL1B, IFN-γ, IP10, and MCP1 | 3 |
| Zhou et al. [19] | 191 | >18 | Wuhan, China | Respiratory, Blood, Feces | Increased age associated with death in SARS-CoV-2 patients | 4 |
| Bunyavanich et al. [22] | 305 | 4-60 | United States | Nasal epithelium | ACE2 gene expression increases with age | 4 |
| Tomassen et al. [28] | 262 | 32.8-42.8* | Europe | Nasal polyps, sinus mucosa, inferior turbinates | The presence or absence of IL-5 is predictive of nasal polyposis | 4 |
| Van Zele et al. [29] | 48 | 3-70 | Belgium | Sinonasal mucosa | CRSsNP characterized by type 1 response in Belgian populations | 4 |
| Wang et al. [30] | 573 | 13-82 | Europe, Asia, Oceania | Sinonasal mucosa, ethmoidal mucosa, inferior turbinate mucosa | Geographic distribution of CRSsNP and CRSwNP patients affects type reactions | 3 |
| Tan et al. [31] | 376 | N/A | United States | Inferior turbinate, ethmoid mucosa, nasal polyps | United States CRSsNP patients displayed type 2 inflammatory patterns | 3 |
| Stevens et al. [32] | 255 | 19-76 | United States | Ethmoid mucosa, nasal polyps | United States CRSsNP patients displayed type 2 inflammatory patterns | 4 |
| Milonski et al. [33] | 104 | 42.8-48.78 | Poland | Paranasal sinus mucosa, nasal polyps | CRSwNP and CRSsNP patients displayed increased IL-4 and IL-13 | 3 |
| Shen and Hong. [34] | 4+ | N/A | China | Middle turbinate nasal mucosa | CRSwNP and CRSsNp patients displayed increased IL-4 and IL-12 | 3 |
| Nabavi et al. [35] | 80 | 18-70 | Iran | Nasal mucosa | CRSwNP patients displayed increased IL-4 but not IL-13 | 4 |
| Wenzel et al. [37] | 66 | 31-36 | United States | Endobronchial, BALF | Asthma can be defined into Type 2 high and Type 2 low endotypes | 4 |
| Stevens et al. [39] | 1059 | 52-54 | United States | Nasal polyps | AERD is common among CRSwNP patients | 4 |
| Raundhal et al. [41] | 66 | 32-47 | United States | BALF | Type 2 low patients with SA show elevated INF-γ 50% of the time | 4 |
| Peters et al. [44] | 729 | 35-47 | United States | Sputum | Systemic IL-6 inflammation and obesity are linked with more SA | 3 |
| de Lang et al. [45] | N/A | N/A | United States | Vero E6 cells | INF-γ and IL-4 decrease ACE2 cell surface expression and lower mRNA levels | 4 |
BALF = Bronchoalveolar lavage fluid; N/A = Not applicable; SA = Severe asthma; * = mean age.
3. Results
3.1. Coronavirus - SARS-CoV and SARS-CoV-2 nasal/oral infectivity
SARS-CoV-2 is an enveloped RNA virus belonging to the Coronaviridae family [7]. Structurally, coronaviruses are comprised of membrane (M), envelope (E), nucleocapsid (N) and spike (S) proteins [1,7]. Infection begins when the protruding surface S protein interacts with a cell specific host receptor and mediates the process of membrane fusion [1,7]. In the last two decades the prominent zoonotic coronaviruses SARS-CoV and MERS-CoV have been identified to interact with angiotensin converting enzyme 2 (ACE2) and dipeptidyl peptidase 4 (DPP4) receptors, respectively [1,8]. Interestingly, the aptly named SARS-CoV-2 uses an entry pathway involving ACE2 [1,9,10]. The cell surface receptor form has been found to be expressed in renal tissues, cardiovascular tissues, nasal goblet cells, ciliated airway cells, type II alveolar pneumocytes, enterocytes of the small intestine, and cardiovascular cells [11,12,13].
Moreover, recent literature has further supported expression of ACE2 in the head and neck as being highly relevant for SARS-CoV-2 pathogenesis. Examination of diverse single-cell RNA sequence datasets found that nasal epithelial, goblet, and ciliated cells show the highest expression patterns of ACE2 among different tissues of the respiratory tree. [14] Immunohistochemistry studies done by Descamps et al. [15] investigated expression patterns in seven different locations of the upper aerodigestive tract. Ciliated respiratory epithelia of the sinuses, oral mucosa, vocal cords, and salivary glands displayed the highest expression patterns of ACE2 [15]. Weaker distributions were found in the tonsils, pharyngeal, and supra-glottic laryngeal epithelia. [15]. Blood vessel endothelial cells of sinus and larynx samples showed moderate levels of expression [15].
Due to the similarity between SARS-CoV and SARS-CoV-2 in using ACE2, understanding the receptor's pathogenic pathway has been paramount. In Li et al. [16], the expression of ACE2 receptors was found to be upregulated under certain pre-existing conditions. The authors examined samples taken from healthy volunteers and patients with a history of smoking, COPD, and asthma [16]. While COPD and asthma cohorts had baseline ACE2 expression, levels of ACE2 were markedly upregulated in long term smokers [16]. Investigators also exposed airway epithelial cells to SARS-CoV in order to analyze possible post-exposure expression effects [16]. It was discovered that expression of ACE2 dramatically increased after 24 hours and indicated a possibility of post-infection regulation [16]. They also found that infection led to a significant activation of neutrophils, natural killer cells, Th17 cells, Th1 cells, dendritic cells and increased levels of IL-1, IL-6, and IL-10 [16]. These findings further led to the postulation that high levels of ACE2 expression after infection were involved with immune system dysfunction and followed the clinical course of a cytokine storm [16]. Supporting this idea in COVID-19 patients, high levels of IL-1B, IL-4, IL-10, INF- γ, TNF-α, interferon gamma-induced protein 10, and monocyte chemoattractant protein 1 have similarly been found [2,17,18].
In studying COVID-19 patients, recent efforts have uncovered that differences in the cytokine induced immune response can determine an individual's prognosis following exposure. A retrospective study done in Wuhan, China found that increased age was associated with a higher likelihood of severe complication and death in these patients [19]. Compared to younger individuals, the elderly express higher counts of ACE2 receptors and weakened innate and adaptive immune system responses [17,19,20,21,22]. Specifically, observations include decreased T cell memory, exhaustion of the naïve T cell population, and declines in B cell production [20]. Aging exerts significant negative effects on cells of the innate immune system including the function of neutrophils, macrophages, mast cells and eosinophils [20]. In Daneshkhah et al. [17], it is suggested that this weakening of the innate immune system leads to a higher SARS-CoV-2 viral count, overactivation of the adaptive immune system, and a consequent cytokine storm. In their study, the authors analyzed COVID-19 patients from around the globe and used the connection between C-reactive protein (CRP) levels and Vitamin D to determine a patient's Vitamin D status [17]. It was found that Vitamin D deficiency may exacerbate the reaction to the cytokine storm implicated in coronavirus patients [17,23]. Current literature supports this premise as vitamin D has been found to inhibit antigen presenting cells in cell-mediated (adaptive) immunity, inflammatory cytokine (IL-1α, IL-1β, TNF-α) expression, and pro-Th1 lymphocyte differentiation via IL-12 [24]. Thus, with a decreased inhibition of IL-12, a Vitamin D deficiency could potentially skew towards overexpression of Th1 cytokines involved in a cytokine storm.
3.2. CRS - The endotypes and cytokines involved in chronic rhinosinusitis
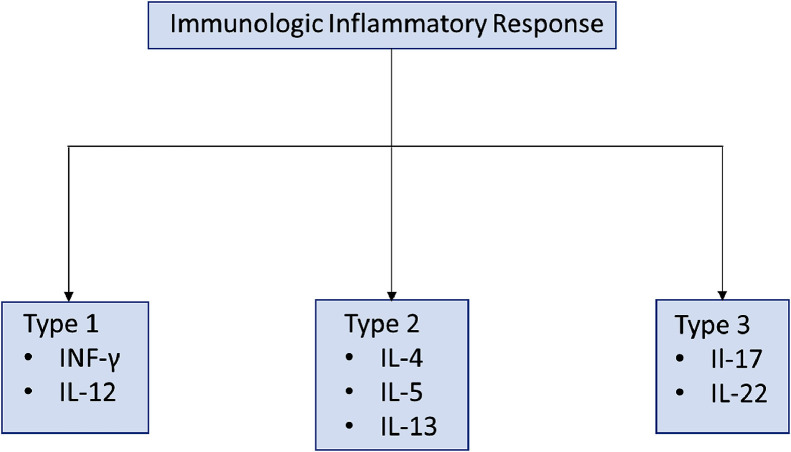
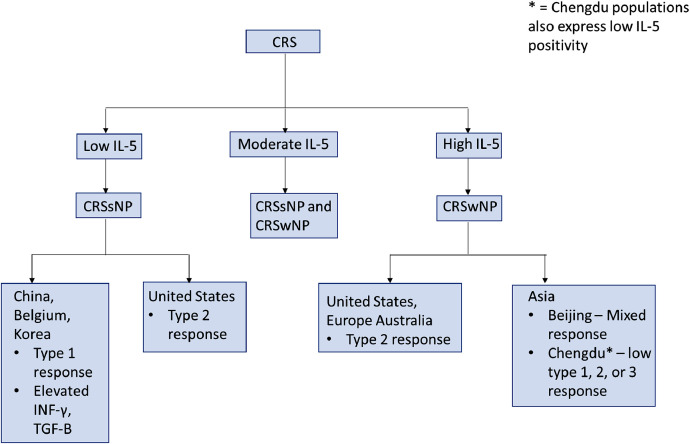
Chronic rhinosinusitis (CRS) is a chronic condition marked by sinonasal and mucosal inflammation affecting 3 to 6% of the population globally [25,26,27]. This condition is commonly further differentiated by the phenotypic presence of nasal polyps (CRSwNP) or their absence (CRSsNP) [25]. Recent efforts have led to the process of endotyping – a process focused on the use of immunohistologic biomarkers to create subtypes [25]. In their literature review, Dennis et al. [25] presented four distinct approaches to distinguish the development of nasal polyposis in CRS from its counterpart. Although they presented approaches using type 2 cytokines, eosinophils, immunoglobulins (IgE), and cysteinyl leukotrienes (CysLT), this study explored a cytokine-based methodology to provide an understanding of how this may be of value for SARS-CoV-2 [25] (Fig. 1, Fig. 2 ).
Fig. 1.
Immunologic endotypes and primary cytokines involved in inflammatory responses [26].
Fig. 2.
Differentiation and Endotypes of Chronic Rhinosinusitis [25,26,27,28,30,31,32]. Legend: * = Chengdu populations also express low IL-5 positivity.
In their hallmark paper, Tomassen et al. [28] performed biomarker analysis of 10 clusters using 14 different cytokine markers. The authors found that it was possible to match clusters to phenotypes using differentiation via the cytokine IL-5 [25,26,28]. Patients with low IL-5 were implicated in no development of nasal polyps (CRSsNP); high IL-5 was resembled in nasal polyposis (CRSwNP); and moderate IL-5 was indicative of both phenotypes (CRSsNP and CRSwNP) [25,26,28].
Current literature defines these endotypes further by using geography to account for cytokine differences. In their study, Van Zele et al. [29] sampled sinonasal mucosal tissue from cohort groups in Belgium and found that CRSsNP was characterized by a type 1 response based on elevated INF-γ and TGF-B. These findings have been supported by similar findings in Chinese and Korean CRSsNP populations [27,30]. In a study done by Tan et al. [31], CRSsNP specimens from patients in Chicago, Illinois were analyzed with type 2 (>40%) and type 3 (>15%) reactions being present, while some did not display any reaction (>40%). These findings in United States CRSsNP patients were replicated with patients from a separate study also displaying type 2 reaction dominance (55%) [32]. Therefore, the conclusion was made that CRSsNP typically displays a Type 2 inflammatory pattern in United States based populations [31]. These findings have gone against traditional conventions and have introduced the idea that CRSsNP can no longer be generalized as a Type 1 inflammatory condition due to geographical differences amongst populations [27,31].
CRSwNP characterization by high IL-5 levels can be further differentiated using similar geographic logic [25,27,28,32]. In studies conducted in the United States, results have shown a large type 2 dominant inflammatory response (>87%) with very little other type pathways present [26,27,32]. Wang et al. [26,27,30] looked at cytokine profiles of CRS patients across Europe, Asia, and Australia and were able to replicate these findings in Europe and Australia cohorts only. Surprisingly, Asian individuals displayed varying patterns and did not seem to follow the general principles outlined in literature [26,27,30]. Although patients from Beijing exhibited type 2 reactions (>61%), their inflammatory responses were usually mixed with Type 1 and 3 patterns present [26,30]. Findings in Chengdu populations stood out as an exception to the cited CRSwNP characteristics with low IL-5 positivity and low type 1, 2, or 3 responses [26].
Additional cytokines of interest in CRS patients display variances with multiple findings across the current literature. In patients from Poland, reverse transcription-polymerase chain assays found significantly increased IL-4 and IL-13 mRNA in comparison to controls in both CRSwNP and CRSsNP patients [33]. Enzyme linked immunosorbent assay (ELISA) results from China supported the findings of increased of IL-4 in both CRS types, but also indicated that IL-12 was elevated in their groups [34]. In a third study done in Tehran, ELISA serum studies implicated that IL-13, but not IL-4 or INF-γ were elevated in CRSwNP patients [35].
3.3. Asthma –the endotypes and cytokines involved
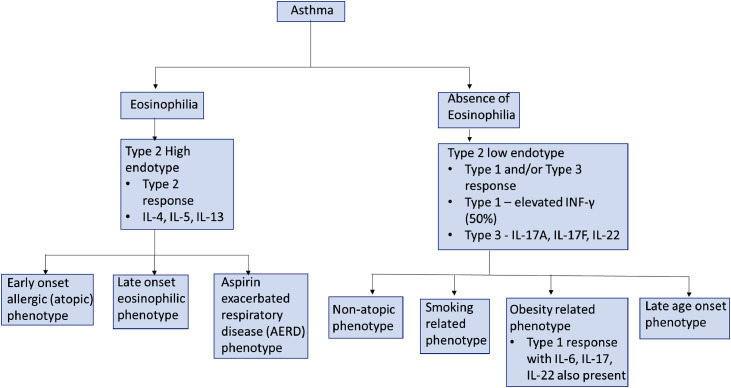
Asthma is a broad term used to describe inflammatory airway pathways with distinct endotypes and phenotypic presentations [36]. Asthma differentiation first appears on an endotype basis, with further characterization occurring on a phenotype basis [36,37]. Wenzel et al. [37], first described the endotype division by looking for pathologic differences in asthmatic patients. They proposed that the presence or absence of airway eosinophilia defines Type 2 high (eosinophilic) and Type 2 low (non-eosinophilic) endotypes [36,37]. The further cytokine involvement of asthma pathophysiology, rehashed below in this study, has previously been examined in greater detail in a literature review by Kuruvilla et al. [36] (Fig. 3 ).
Fig. 3.
Differentiation and Endotypes of Asthma [36].
In the Type 2 high endotype, eosinophilic innate immunity has an early and key role in the pathophysiology of the inflammatory response [36,37]. The primary immune response begins with airway epithelial cells (AEC) releasing cytokines or “alarmins” such as thymic stromal lymphopoietin (TSLP), IL-25, and IL-33 [34]. IL-25 and IL-33 primarily activate Group 2 innate lymphoid cells (ILC2) while TSLP activates antigen-presenting cells [36]. ILC2 cells are important inflammatory mediators that release IL-5 and IL-13 to begin an early Type 2 inflammatory response, while antigen presenting cells induce Th2 cells for additional production of IL-4, IL-5, and IL-13 [36]. Unique to this response, eosinophils are recruited to the site of inflammation via chemokines and they further exacerbate responses by releasing cytokines (IL-4, IL-5, IL-13) and cysteinyl leukotrienes (cysLT) [36]. CysLTs are bronchoconstrictors that work cooperatively with alarmin IL-33 to promote additional ILC2 release of type 2 cytokines [36]. IL-4 and IL-13 promote goblet cell overexpression of mucus and airway hyperreactivity, while IL-5 plays a role in the maturation/development of IL-5Rα + eosinophil progenitors or other type 2 cells [36]. Subtypes of this endotype can be defined phenotypically as early onset allergic (atopic) asthma, late onset eosinophilic asthma, and aspirin exacerbated respiratory disease (AERD). The European Academy of Allergy and Clinical Immunology has recently published a position paper in favor of utilizing NSAID-Exacerbated Respiratory Disease (N-ERD) as a more encompassing term to replace AERD, but this literature review will use the latter for consistency [38]. Although these clinical presentations are not explored in greater detail, it is worth noting that AERD can lead to the development of nasal polyps contributing to its complex relationship with CRSwNP patients [36,39,40]. In their study, Stevens et al. [39,40] summarized that all AERD patients have CRSwNP, but not all CRSwNP patients have AERD. The prevalence of AERD among CRSwNP is estimated in <10% of patients, however its true incidence remains unknown due to lack of epidemiological data [39].
The type 2 low endotype of asthma is marked by neutrophilic or paucigranulocytic inflammation and the absence of eosinophilia [36,37]. Inflammation typically follows type 1 and/or type 3 pathways in comparison to its counterpart [36]. Patients with severe asthma typically present with a skewed type 1 response with elevated IFN-γ reported in 50% of patients [36,41]. Individuals with skewed type 3 inflammation are characterized by production of IL-17A, IL-17F, IL-22 [36,42]. IL-17 produces neutrophilic inflammation while IL-8 attracts neutrophils to tissues [36,43]. Phenotypically this endotype can be classified into non-atopic, smoking related, obesity related, and very late age clinical presentations [3,36]. Of interest, obese populations demonstrate biases towards Th1 cell differentiation, but IL-6, IL-17, and IL-22 have been prevalent in these patients [36,44]. In their cross-sectional analysis of two cohorts, Peters et al. [44] linked systemic IL-6 inflammation and obesity with more severe asthma.
3.4. Factors affecting ACE2 expression and coronavirus transmission—cytokines and ACEI
During the early stages of the global pandemic, the CDC listed asthma and other respiratory viruses as significant risk factors for serious disease in COVID-19 patients [3]. Early data from China suggested that asthma and respiratory allergies were erroneously classified, and in a novel study done by Jackson et el. [3], these findings were examined in greater detail. The authors sampled three cohorts of children and adults to elucidate if asthma or allergies reduced ACE2 receptor expression, and likely SARS-CoV-2 transmission. In the first cohort, asthmatic children with moderate to high levels of allergic sensitization displayed a higher reduction in ACE2 expression than their nonallergic counterparts [3]. In the second cohort, adults with known cat allergies were exposed to stimuli and subsequently demonstrated reduction of ACE2 expression. In the third cohort, mildly asthmatic adults were exposed to allergic stimuli and demonstrated reduced ACE2 expression as a result of the allergen challenges [3]. The authors further differentiated that the non-atopic asthma phenotype was not associated with reduced receptor levels [3]. Interestingly, these findings led investigators to identify that IL-13, a cytokine strongly present in allergic asthma, was an important factor in reduced ACE2 expression in airway tissues [3].
The premise that cytokines can downregulate ACE2 expression has previously been cited in literature during studies done during the SARS-CoV outbreak. In a study done by de Lang et al. [45], INF-γ and IL-4 were found to decrease ACE2 cell surface expression and lower mRNA levels. To determine this, they exposed Vero E6 cells to various combinations of cytokines (IL-4, IL-10, INF-γ, and TNF-α) and tested for antiviral activity [45]. By treating cells with IL-4 medium prior to infection, it was determined that IL-4 is most likely only effective in early coronavirus transmission [45]. Further testing revealed that antiviral activity reaches a maximal at 48 hours, and these findings indicated that the gradual reduction of ACE2 most likely occurs via progressive downregulation of mRNA levels [45]. Similar downregulation of ACE2 receptors was also found with IL-13, INF-γ, and enhanced in an INF-γ/TNF-α medium [45].
The mechanism of action of ACE2 is known to oppose that of ACE. As ACE converts angiotensin I to angiotensin II, ACE2 converts angiotensin II to angiotensin-(1-7) affecting Mas receptors in vascular endothelial cells shifting to vasodilation from angiotensin II mediated vasoconstriction [46]. Early on there were concerns that hypertensive patients prescribed with angiotensin-converting enzyme inhibitors (ACEI) or angiotensin II receptor blockers (ARBs) could potentially demonstrate increased expression of ACE2 receptors or inhibit function of ACE2 itself (ACEI) – both of which could facilitate SARS-CoV entry into host cells [46]. Also, it has been noted that high levels of serum ACE2 with other cytokines is a poor prognostic sign in COVID-19 patients who require hospitalizations [46]. Since there has been no concrete clinical evidence to support or dispute the above concerns, the American Heart Association, the American Heart Failure Society and the American College of Cardiology have all advised the continuation of prescribed ACEI and ARB [46].
4. Discussion
In this literature review, we sought to examine the pathophysiology of COVID-19 virulence and the cytokines involved in classification schemes of otorhinolaryngeal illnesses. SARS-CoV and SARS-CoV-2 both use the ACE2 receptor for entry into cells so correlating the cytokines found in these illnesses may provide insight into susceptibility of illness. In CRS, it was demonstrated that geography plays a role in the prevalence of CRS endotypes and cytokine differences. Except for certain Chinese populations, the classification based on IL-5 levels appears to be a notable way to endotype CRSwNP and CRSsNP patients [28,30]. Typical CRSwNP and United States CRSsNP patients displaying type 2 responses correlate with increased IL-4, IL-5, and IL-13. As previous SARS-CoV studies have found that IL-4 and IL-13 decreased ACE2 expression, this raises the question whether enhancement of these cytokines can serve as protective mechanisms from SARS-CoV-2 in patient populations [45]. CRSsNP patients in Belgium, China, and Korea displaying type 1 reactions may similarly show protection due to their increased INF-γ levels [45].
Although the elderly or long-term smokers are high risk populations due to high expression of ACE2 receptors, atopic asthma and allergy data have suggested their exclusion as risk factors due to lowered ACE2 rates. In the type 2 high endotype of asthma, IL-4 and IL-13 were identified again as having a significant role in the cytokine responses of these illnesses [36]. In atopic allergies, IL-13 only was determined to be a key player in reducing ACE2 expression [3]. Interestingly, severe Type-2 low asthma patients which skew towards Type 1 reactions may express elevated INF-γ [36,37,41]. These findings again raise the question of whether a protective mechanism exists in these populations to reduce comorbidity with COVID-19. More studies are needed to investigate the implications of these cytokines of interest on COVID-19 transmission rates. Although it has been shown that asthma accounted for a low percentage of COVID-19 patients in China compared to controls, no study to our knowledge has looked at either asthma or allergy COVID-19 patient epidemiology globally [4,5]. Further consequences of these findings may correlate with SARS-CoV-2 hotspots throughout the world.
The cytokines of interest (IL-4, IL-13, INF-γ) and vitamin D create questions about possible treatment options for COVID-19. In SARS-CoV patients, only IL-4 was shown to demonstrate antiviral activity if administered prior to infection [45]. More work is needed to correlate this finding to possible prophylaxis for SARS-CoV-2 infection and its efficacy. The prominent cytokine storm that occurs after infection may point post-infection treatment towards examining vitamin D levels. Current literature states that vitamin D deficiency skews towards an overproduction of type 1 cytokines, however as observed in Li et al. [16], Type 2 cytokines may remain elevated in COVID-19 patients. Per Ebadi et al. [23], no study to date has performed high dose vitamin D treatment in COVID-19 patients. We support their suggestion that serum testing be done at various stages of the disease to further understand and classify risk of progression [23]. The implications of the above findings may point towards new therapeutic options against COVID-19.
4.1. Limitations
This literature review has important limitations that should be considered. The SARS-CoV-2 pandemic represents a recent ongoing event with most published literature taking place within the past 8 months. Although many studies were examined to gain a broad background for analysis, new findings describing COVID-19 transmission may affect this study. In addition, no language filters were applied when searching public databases. With a large amount of data reporting originating in China, literature that remained untranslated to the English language was excluded. This may have led to a screening bias and the exclusion of relevant data.
5. Conclusion
Continued research is needed to understand the effects of vitamin D and the various cytokines prevalent among endotypes of nasal/pharyngeal illnesses on COVID-19 pathogenesis. Findings may point towards epidemiologic trends of SARS-CoV-2 transmission and new treatment options.
Authors’ Contributions
All authors were involved with the design of the study, acquisition and interpretation of data, drafting the article, revising it, and giving approval of the version to be submitted.
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Funding/Support
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit-sectors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.anl.2020.11.006.
Appendix. Supplementary materials
Supplement: PRISMA Diagram showing the process of article selection for this review.
References
- 1.Shanmugaraj B, Siriwattananon K, Wangkanont K, Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac J Allergy Immunol. Mar 2020;38(1):10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 2.Krajewska J, Krajewski W, Zub K, Zatoński T. COVID-19 in otolaryngologist practice: a review of current knowledge. Eur Arch Otorhinolaryngol. Jul 2020;277(7):1885–1897. doi: 10.1007/s00405-020-05968-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson DJ, Busse WW, Bacharier LB, Kattan M, O'Connor GT, Wood RA. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J Allergy Clin Immunol. Jul 2020;146(1):203–206. doi: 10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. Jul. [DOI] [PubMed] [Google Scholar]
- 5.Dong X, Cao YY, Lu XX, Zhang JJ, Du H, Yan YQ. Eleven faces of coronavirus disease 2019. Allergy. 2020;75(7):1699–1709. doi: 10.1111/all.14289. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durieux N, Vandenput S, Pasleau F. Médecine factuelle: la hiérarchisation des preuves par le Centre for Evidence-Based Medicine d'oOford [OCEBM levels of evidence system] Rev Med Liege. Dec 2013;68(12):644–649. [PubMed] [Google Scholar]
- 7.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. Nov 27, 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. Mar 17, 2020;94(7):e00127. doi: 10.1128/JVI.00127-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. Mar, 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. Dec 4, 2002;532(1-2):107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 12.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. Jun 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sungnak W, Huang N, Bécavin C, Berg M. HCA lung biological network. SARS-CoV-2 entry genes are most highly expressed in nasal goblet and ciliated cells within human airways. Nat Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. May 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Descamps G, Verset L, Trelcat A, Hopkins C, Lechien JR, Journe F. ACE2 protein landscape in the head and neck region: the conundrum of SARS-CoV-2 infection. Biology (Basel) Aug 18, 2020;9(8):235. doi: 10.3390/biology9080235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, He X, Zhang L, Ran Q, Wang J, Xiong A. Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J Autoimmun. Aug 2020;112 doi: 10.1016/j.jaut.2020.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daneshkhan A., Agrawal V., Eshein A., Subramanian H., Roy H.K., Backman V. The possible role of Vitamin D in suppressing cytokine storm and associated mortality in COVID-19 patients. MedRxiv. 2020 [Google Scholar]
- 18.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. Feb 15, 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. Mar 28, 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez CR, Nomellini V, Faunce DE, Kovacs EJ. Innate immunity and aging. Exp Gerontol. Aug, 2008;43(8):718–728. doi: 10.1016/j.exger.2008.05.0168.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc Biol Sci. Dec 22, 2015;282(1821) doi: 10.1098/rspb.2014.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. Jun 16, 2020;323(23):2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebadi M, Montano-Loza AJ. Perspective: improving vitamin D status in the management of COVID-19. Eur J Clin Nutr. Jun, 2020;74(6):856–859. doi: 10.1038/s41430-020-0661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes DA, Norton R. Vitamin D and respiratory health. Clin Exp Immunol. Oct, 2009;158(1):20–25. doi: 10.1111/j.1365-2249.2009.04001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis SK, Lam K, Luong A. A review of classification schemes for chronic rhinosinusitis with nasal polyposis endotypes. Laryngosc Investig Otolaryngol. Oct, 2016;1(5):130–134. doi: 10.1002/lio2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staudacher AG, Peters AT, Kato A, Stevens WW. Use of endotypes, phenotypes, and inflammatory markers to guide treatment decisions in chronic rhinosinusitis. Ann Allergy Asthma Immunol. Apr 2020;124(4):318–325. doi: 10.1016/j.anai.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahern S, Cervin A. Inflammation and endotyping in chronic rhinosinusitis-a paradigm shift. Medicina (Kaunas) Apr 5, 2019;55(4):95. doi: 10.3390/medicina55040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. May 2016;137(5):1449–1456. doi: 10.1016/j.jaci.2015.12.1324. [DOI] [PubMed] [Google Scholar]
- 29.Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. Nov 2006;61(11):1280–1289. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Zhang N, Bo M, Holtappels G, Zheng M, Lou H. Diversity of TH cytokine profiles in patients with chronic rhinosinusitis: a multicenter study in Europe, Asia, and Oceania. J Allergy Clin Immunol. Nov 2016;138(5):1344–1353. doi: 10.1016/j.jaci.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 31.Tan BK, Klingler AI, Poposki JA, Stevens WW, Peters AT, Suh LA. Heterogeneous inflammatory patterns in chronic rhinosinusitis without nasal polyps in Chicago, Illinois. J Allergy Clin Immunol. Feb 2017;139(2):699–703. doi: 10.1016/j.jaci.2016.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens WW, Peters AT, Tan BK, Klingler AI, Poposki JA, Hulse KE. Associations between inflammatory endotypes and clinical presentations in chronic rhinosinusitis. J Allergy Clin Immunol Pract. Nov-Dec 2019;7(8):2812. doi: 10.1016/j.jaip.2019.05.009. -2820.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milonski J, Zielinska-Blizniewska H, Majsterek I, Przybyłowska-Sygut K, Sitarek P, Korzycka-Zaborowska B. Expression of POSTN, IL-4, and IL-13 in chronic rhinosinusitis with nasal polyps. DNA Cell Biol. May 2015;34(5):342–349. doi: 10.1089/dna.2014.2712. [DOI] [PubMed] [Google Scholar]
- 34.Shen J, Hong S. [Serum levels of IL-12, IL-4 and pathologic changes by scanning electron microscope of nasal mucous inflammation] Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. Oct 2010;24(20):913–917. [PubMed] [Google Scholar]
- 35.Nabavi M, Arshi S, Bahrami A, Aryan Z, Bemanian MH, Esmaeilzadeh H. Increased level of interleukin-13, but not interleukin-4 and interferon-γ in chronic rhinosinusitis with nasal polyps. Allergol Immunopathol (Madr) Sep-Oct 2014;42(5):465–471. doi: 10.1016/j.aller.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Kuruvilla ME, Lee FE, Lee GB. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol. Apr 2019;56(2):219–233. doi: 10.1007/s12016-018-8712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med. Sep 1999;160(3):1001–1008. doi: 10.1164/ajrccm.160.3.9812110. [DOI] [PubMed] [Google Scholar]
- 38.Kowalski ML, Agache I, Bavbek S, Bakirtas A, Blanca M, Bochenek G. Diagnosis and management of NSAID-exacerbated respiratory disease (N-ERD)-a EAACI position paper. Allergy. Jan 2019;74(1):28–39. doi: 10.1111/all.13599. [DOI] [PubMed] [Google Scholar]
- 39.Stevens WW, Peters AT, Hirsch AG, Nordberg CM, Schwartz BS, Mercer DG. Clinical characteristics of patients with chronic rhinosinusitis with nasal polyps, asthma, and aspirin-exacerbated respiratory disease. J Allergy Clin Immunol Pract. Jul-Aug 2017;5(4):1061. doi: 10.1016/j.jaip.2016.12.027. -1070.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens WW, Schleimer RP. Aspirin-exacerbated respiratory disease as an endotype of chronic rhinosinusitis. Immunol Allergy Clin North Am. Nov 2016;36(4):669–680. doi: 10.1016/j.iac.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raundhal M, Morse C, Khare A, Oriss TB, Milosevic J, Trudeau J. High IFN-γ and low SLPI mark severe asthma in mice and humans. J Clin Invest. Aug 3 2015;125(8):3037–3050. doi: 10.1172/JCI80911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bickel M. The role of interleukin-8 in inflammation and mechanisms of regulation. J Periodontol. May 1993;64(5 Suppl):456–460. [PubMed] [Google Scholar]
- 44.Peters MC, McGrath KW, Hawkins GA, Hastie AT, Levy BD, Israel E. Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: a cross-sectional analysis of two cohorts. Lancet Respir Med. Jul 2016;4(7):574–584. doi: 10.1016/S2213-2600(16)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Lang A, Osterhaus AD, Haagmans BL. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. Sep 30, 2006;353(2):474–481. doi: 10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brojakowska A, Narula J, Shimony R, Bander J. Clinical implications of SARS-CoV-2 interaction with renin angiotensin system: JACC review topic of the week. J Am Coll Cardiol. Jun 23, 2020;75(24):3085–3095. doi: 10.1016/j.jacc.2020.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement: PRISMA Diagram showing the process of article selection for this review.