Highlights
-
•
Replication and tropism of SARS-CoV-2 in cattle, sheep, and pigs using EVOCs, were investigated.
-
•
Respiratory tissues of cattle and sheep, but not those of pigs, are able to sustain viral replication.
-
•
A SARS-CoV-2 isolate harbouring mutation D614 G in the S protein has greater replication capabilities.
-
•
SARS-CoV-2 binds to ACE2-expressing cells of the respiratory tract of cattle and sheep.
Keywords: SARS-CoV-2, Respiratory ex vivo organ cultures, Cattle, Sheep, Pigs, ACE2, Tropism, Replication, D614G
Abstract
There is strong evidence that severe acute respiratory syndrome 2 virus (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic, originated from an animal reservoir. However, the exact mechanisms of emergence, the host species involved, and the risk to domestic and agricultural animals are largely unknown. Some domestic animal species, including cats, ferrets, and minks, have been demonstrated to be susceptible to SARS-CoV-2 infection, while others, such as pigs and chickens, are not. Importantly, the susceptibility of ruminants to SARS-CoV-2 is unknown, even though they often live in close proximity to humans. We investigated the replication and tissue tropism of two different SARS-CoV-2 isolates in the respiratory tract of three farm animal species - cattle, sheep, and pigs - using respiratory ex vivo organ cultures (EVOCs). We demonstrate that the respiratory tissues of cattle and sheep, but not of pigs, sustain viral replication in vitro of both isolates and that SARS-CoV-2 is associated to ACE2-expressing cells of the respiratory tract of both ruminant species. Intriguingly, a SARS-CoV-2 isolate containing an amino acid substitution at site 614 of the spike protein (mutation D614G) replicated at higher magnitude in ex vivo tissues of both ruminant species, supporting previous results obtained using human cells. These results suggest that additional in vivo experiments involving several ruminant species are warranted to determine their potential role in the epidemiology of this virus.
1. Introduction
Coronavirus disease 2019 (COVID-19) was recognized as a public health emergency of international concern on January 30, 2020 and acknowledged as a pandemic on March 11, 2020 (Zhou et al., 2020). COVID-19 is caused by a novel human coronavirus (CoV) now referred to as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, species SARS-related coronavirus, subgenus Sarbecovirus, genus Betacoronavirus, subfamily Orthocoronavirinae, family Coronaviridae) (Decaro and Lorusso, 2020; Gorbalenya et al., 2020). COVID-19 was first reported in December 2019 in the city of Wuhan, Hubei Province, China, in humans largely connected to the Huanan seafood wholesale market and where different species of farm and wild animals are commonly sold (Decaro and Lorusso, 2020; 2020a), although the role of the market in virus emergence is uncertain. COVID-19 is mainly a respiratory infection, with the most common symptoms comprising fever, dry cough, and shortness of breath. About 20 % of infected patients may develop severe disease, and a small percentage (5%) may become critically ill. Patients with severe COVID-19 disease usually develop pneumonia or acute respiratory distress syndrome (ARDS), a condition that may require mechanical ventilation and intensive care unit treatment (Yang et al., 2020). ARDS is often fatal (Matthay et al., 2019).
The recognition of and interaction with host cellular receptors are critical initial steps in the virus life cycle and play a key role in shaping host range, tissue tropism and viral pathogenesis. The numerous functions of viral receptors enable the virus to target the correct tissues for infection and breach cellular barriers. Interactions with viral receptors are usually mediated by specific viral attachment proteins expressed on the surface of the virion. SARS-CoV-2, like the earlier SARS-CoV that emerged in 2002/2003, uses the receptor binding domain (RBD) of the viral S1 portion of the spike (S) protein to bind to the angiotensin-converting enzyme 2 (ACE2) receptor that is widespread in several tissues of mammals (Zhou et al., 2020; Hoffmann et al., 2020; Wang et al., 2020; Damas et al., 2020). It is reasonable to think that mutations in this viral protein could potentially have dramatic effects on viral pathogenesis, replication, and tropism. Analyses of over 28,000 S gene sequences in May 2020 revealed a D614G (23403A > G) amino acid substitution that was rare before March but increased in frequency as the pandemic spread, reaching over 74 % of all published sequences by June 2020 (Yurkovetskiy et al., 2020). Notably, it has been proposed that D614G increases infectivity in cell assays in vitro and is also proposed to enhance viral transmissibility in nature (Plante et al., 2020; Korber et al., 2020). The D614G substitution is accompanied by a set of mutations including 241C > T in the leader sequence, 3037C > T, and 14408C > T. While 3037C > T causes a synonymous mutation in nsp3 (F105 F), 14408C > T is responsible for a substitution in the RNA primase (nsp 12, P323L) (Lorusso et al., 2020a).
Although it is clear that SARS-CoV-2 has an animal origin, the exact mechanisms of emergence and which animals were involved in its genesis are currently unknown. A human pandemic poses also risks for domestic animals, particularly given their close contacts with humans, and they potentially act as an additional spill-over source for human infection (Gollakner and Capua, 2020; Patterson et al., 2020). Previous studies have shown that SARS-CoV-2 replicates poorly in dogs, chickens and ducks, while cats, ferrets, and minks are highly permissive to infection (Enserink, 2020; Shi et al., 2020; Manes et al., 2020; Molenaar et al., 2020; Oreshkova et al., 2020; Schlottau et al., 2020; Gaudreault et al., 2020). Additionally, by previous analysis, it was suggested that SARS-CoV-2 may efficiently bind to the ACE2 moieties of different animal species (Damas et al., 2020, Zhai et al., 2020). In particular, it was proposed that the ACE2 of cattle and sheep can function as a SARS-CoV-2 receptor, while that of pigs, although structurally permissive to SARS-CoV-2 binding, is poorly expressed in the respiratory tract (Zhai et al., 2020). The latter finding may in part explain the absence of SARS-CoV-2 clinical signs following experimental infection in pigs (Meekins et al., 2020). Importantly, however, the susceptibility of farm animals to SARS-CoV-2 remains largely unknown as well as the effect of D614G in animals.
By utilizing SARS-CoV-2 infection of ex vivo organ cultures (EVOCs) we demonstrate here that the respiratory tissues of cattle and sheep, but not of pigs, sustain viral replication, and that SARS-CoV-2 was strictly related to ACE2-expressing cells of the respiratory tract of both ruminant species. Moreover, we show that a SARS-CoV-2 isolate which contains D614G in the S protein, mutation now widespread in the vast majority of globally SARS-CoV-2 currently circulating strains, replicates to higher titers than earlier SARS-CoV-2 variants that contain D614.
2. Materials and methods
2.1. Animals
Three 12-month old pigs, three 18-month old cattle and three 10-month-old sheep were used to obtain respiratory EVOCs. Sampling was performed in accordance with internal guidelines of the slaughterhouse (Centro Carni Val Tordino, Mosciano S. Angelo, Teramo-Italy) where the animals were officially slaughtered. All animals originated from herds located in the Abruzzi region of central Italy. All procedures, including handling and processing of tissues, were performed according to the internal guidelines of the Istituto Zooprofilattico Sperimentale dell’Abruzzo e Molise (IZSAM).
2.2. Viruses
Four virus isolates at low cell passage were used to infect the respiratory EVOCs: (i) SARS-CoV-2/INMI1-Isolate/2020/Italy (kindly donated by the Istituto Nazionale Malattie Infettive Lazzaro Spallanzani, Rome, Italy) and nucleotide sequence of which is available on the GISAID website (accession BetaCoV/Italy/INMI1-isl/2020: EPI_ISL_410545); (ii) SARS-CoV-2/IZSAM/46419 (GISAID, hCoV-19/Italy/ABR-IZSGC-TE46419/2020; (iii) bovine parainfluenza virus 3 (BPIV-3, species Bovine respirovirus 3, genus Respirovirus, family Paramyxoviridae, T1 strain, Weybridge, UK) and (iiii) suid herpes virus 1 (SuHV-1, species Suid alphaherpesvirus 1, genus Varicellovirus, subfamily Alphaherpesvirinae, family Herpesviridae, strain ADV32751/Italy2014 (Pizzurro et al., 2016)). Experiments with SARS-CoV-2 were performed in BSL-3 conditions as described previously (b). Both SARS-CoV-2 isolates have been isolated on VERO E6 cells (monkey kidney epithelial cells). SARS-CoV-2/IZSAM/46419 sequence differs from that of SARS-CoV-2/INMI1-Isolate/2020/Italy for 241C > T, 3037C > T, 14408C > T and 23403A > G. This latter leads to the non-synonymous mutation in the S protein encoding gene, D614G. Therefore, SARS-CoV-2/INMI1-Isolate/2020/Italy is indicated as D614 throughout the manuscript, while SARS-CoV-2/IZSAM/46419 as D614G.
2.3. Screening of serum and tissue samples
Serum samples were collected from animals before tissue collection and tested for the presence of neutralizing antibodies against SuHV-1, BPIV-3, bovine coronavirus (BCoV, species Betacoronavirus 1, subgenus Embecovirus, genus Betacoronavirus, subfamily Orthocoronavirinae, family Coronaviridae, strain 438/06, (Decaro et al., 2008), and SARS-CoV-2 by serum neutralization (IZSAM). Swab (respiratory and rectal) samples were also collected and purified nucleic acids extracted from them (using the MagMAX viral Pathogen II Nucleic Acid isolation kit, Thermofisher) and from a portion of collected tissues were tested for the presence of SARS-CoV-2 (TaqManTM 2019-nCoV Assay Kit v2, Thermofisher), SuHV-1 (Adiavet PRV Real Time, Adiagene), swine coronaviruses (VetMAX™ PEDV/TGEV/SDCoV Kit (Applied Biosystems), BCoV (LSI VetMAX™ Ruminant Rotavirus & Coronavirus Kit (Applied Biosystems)) and BPIV-3 virus (LSI VetMAX™ Triplex BRSV & PI3 Kit (Applied Biosystems)).
2.4. Culture and infection of respiratory EVOCs
Tracheal and lung EVOCs culture and viability were assessed as described previously (Di Teodoro et al., 2018, 2019). Briefly, at the slaughterhouse, the cranial portion of trachea and the right lung were immediately collected after the stunning and bleeding of the animals. Samples were stored at 4 °C and delivered to the laboratory.
To obtain tracheal EVOCs, the tracheal mucosa was stripped from the cartilage rings, cut in squares of about 25 mm2 and washed with warm phosphate buffered saline (pH 7.4, PBS). Tissue samples were then transferred into 24-well plates. To obtain lung EVOCs, the main bronchus was catheterized, and the lung parenchyma embedded with 1% low gelling temperature agarose (type VII-A agarose, Sigma-Aldrich) pre-warmed at 37 °C. The lung lobe was then cooled to 4 °C for 20 min and cut into 1 mm thick slices. Lung tissue samples of 25 mm2 were then transferred into 24-well plates. All EVOCs were cultured in an air–liquid interface system, slightly immersed in 1 ml of tissue culture medium (DMEM addicted with 1% of l-glutamine, 1% of Penicillin/Streptomycin/Amphotericin B Solution, Thermo-Fisher). EVOCs were incubated at 37 °C with 5% CO2 for up to 72 h.
Tracheal and lung EVOCs were infected with 103 TCID50/mL of both SARS-CoV-2 isolates, BPIV-3, and SuHV-1 virus by submerging the explanted tissues in 1 ml of each virus dilution. After 1 h at 37 °C in 5% CO2, EVOCs were washed three times with warm PBS to remove non-attached virus particles and cultured in 1 ml of fresh tissue culture media at 37 °C in 5% CO2 up to 72 h. Virus-free culture medium was used for mock-infected EVOCs. To study viral replication, 300 μl of supernatant was collected at 1, 24, 48 and 72 h post infection (hpi). The supernatants were titrated by TCID50 assay on VERO E6 cells. In addition, to determine the number of viral genome copies over time, viral RNA purified from EVOCs supernatants was tested by TaqMan™ 2019-nCoV Assay Kit v2. The absolute quantification of viral RNA was determined by a standard curve method using an RNA standard (Twist Synthetic SARS-CoV-2 RNA Controls, Twist Bioscence, San Francisco-CA, USA). As the adopted molecular assay detects three genes of SARS-CoV-2 genome namely ORF1ab, S and N protein encoding genes, for practical reasons, quantification has been performed only for the N protein encoding gene.
2.5. Immunohistochemistry for angiotensin converting enzyme 2 (ACE2)
To study the ACE2 immunohistochemical expression, trachea and lung specimens of the same animals included in the study were immediately fixed in 10 % neutral buffered formalin at the slaughterhouse and processed for histology. Three μm-thick sections were dewaxed, rehydrated, and incubated overnight at 4 °C with a rabbit anti-ACE2 polyclonal antibody (Abcam; ab15348). Immunoreactions were visualized using a biotin-streptavidin amplification method and 3−3′-diaminobenzidine as chromogen (Dako REAL™ detection system). According to the manufacturer, this polyclonal antibody binds to human ACE2, although it is also predicted to bind to the ACE2 of cat, dog, monkey, and orangutan.
2.6. Western blotting for detection of angiotensin converting enzyme 2 (ACE2)
The specificity of the anti-ACE2 antibody used in this study and its capability to detect ACE2 of different animal species was assessed by western blotting. For this purpose, bovine, ovine and swine kidney tissues (Hamming et al., 2004) from the same animals included in this study, were lysed with celLyticMT reagent (Sigma) with a 1:20 ratio. Caco-2 cell line was used as positive control as reported in the manufactured instructions of the adopted antibody. Lysates/proteins at 15 μg per lane were separated by NuPAGE 4–12 % Bis-Tris gel (Novex, Life Technologies) at 200 V and then transferred onto iBlot2 NC stacks nitrocellulose membranes (Life Technologies) by iBlot2® Dry Blotting System (Life Technologies). Membranes were blocked with PBS containing 0.05 % Tween 20 (PBST) and 5% skimmed milk for 2 h at room temperature. Membranes were incubated overnight at room temperature with rabbit anti ACE2 polyclonal antibody (Abcam; ab15348) at a concentration of 2 μg/mL diluted in PBST containing 2.5 % skimmed milk. After washing with PBST, membranes were incubated for 1 h at room temperature with goat anti-rabbit IgG-HRP (Bio-Rad) diluted 1:3000 in PBST containing 2.5 % skimmed milk. Antigen-antibody reactions were visualized by adding chemiluminescent substrates (GE Healthcare). Images were acquired using the ChemiDoc MP (Bio-Rad) and the Image Lab Software, version 4.0.1 (Bio-Rad).
2.7. Immunohistochemistry, double labelling indirect immunofluorescence and confocal microscopy
To study the cellular tropism of both SARS-CoV-2 isolates, three replicates of virus- and mock-infected respiratory EVOCs were fixed in 10 % neutral buffered formalin at 1, 24, 48 and 72 hpi and processed for immunohistochemistry (IHC). For IHC, three μm-thick sections of EVOCs were dried at 37 °C, dewaxed and rehydrated by standard procedures. Antigen retrieval was performed by autoclaving at 121 °C for 10 min in 0.01 M citrate buffer, pH 6. Sections were incubated overnight at 4 °C with a primary rabbit anti-SARS-nucleoprotein antibody (NovusBio; NB100-56576). Immunoreactions were visualized using a biotin-streptavidin amplification method and 3−3′-diaminobenzidine as chromogen (Dako REAL™ detection system).
For DLIIF and CM investigations three μm-thick tissue sections of SARS-CoV-2-infected bovine and ovine EVOCs were incubated overnight at 4 °C with a monoclonal anti-SARS-CoV-2 spike antibody (clone 1A9, GeneTex; GTX632604) and, simultaneously, with a rabbit anti-ACE2 polyclonal antibody (Abcam; ab15348) as primary antibodies. Immune reactions were visualized using anti-mouse and anti-rabbit IgG biotinylated as secondary antibodies (Vector Laboratories, Inc.), respectively, followed by incubation with streptavidin conjugate Alexa Fluor™ 546 (Invitrogen) and Fluorescein Avidin D FITC (Vector Laboratories, Inc.) as fluorochromes. Sections were mounted using an antifade medium with 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, Inc.), and imaged using a Leica TCS SP5 II confocal laser microscope. Suitable positive and negative controls were included in each IHC and DLIIF run.
2.8. Sequence analysis
RNAs purified from the supernatants collected at 72 hpi from EVOCs of both ruminant species infected with the two SARS-CoV-2 isolates were used for next generation sequencing (NGS). The two isolate stocks used for infection were also sequenced. RNA samples were amplified by ARTIC amplicon sequencing protocol (Itokawa et al., 2020) and then library preparation was carried out by using Illumina® DNA Prep, (M) Tagmentation (24 Samples) (Illumina Inc., San Diego, CA USA). Deep sequencing was performed on the MiniSeq (Illumina Inc., San Diego, CA) by the MiniSeq Mid Output Kit (300-cycles) and standard 150 bp paired-end reads. Quality control was performed using FASTQC quality control software v0.11.9. Analysis was performed by means of the SeqMan NGen of the Lasergene Genomics tool suite (DNASTAR, Inc., Madison-WI, USA).
2.9. Statistical analysis
Sample sizes were calculated using the G*Power software (Heinrich-Heine Universität Düsseldorf, Germany). All statistical analyses were performed using GraphPad Prism software, version 8.4.3 (GraphPad Software Inc., San Diego, USA). Results were expressed as means ± standard deviation (SD) derived from three replicates of three independent experiments (one animal for each experiment). Multiple Student t-test were performed to compare sets of data. Differences were considered statistically significant when p value was ≤ 0.05.
3. Results
3.1. Animals tested negative for the viral pathogens studied
All animals included in the study tested virologically negative for BPIV-3 virus, SuHV-1, BCoV, swine coronaviruses, and SARS-CoV-2 prior to the start of the EVOC experiments. In addition, serum samples tested negative for BPIV-3, SuHV-1, BCoV, and SARS-CoV-2 by serum neutralization.
3.2. BPIV-3 and SuHV-1 replication confirms the viability of respiratory EVOCs
The viability of explanted respiratory tissues was confirmed by the marked replication of common respiratory pathogens - BPIV-3 and SuHV-1 – that comprised positive control respiratory viruses. BPIV-3 replicated at the approximately the same magnitude and exhibited the same temporal trend in both ruminant respiratory EVOCs, with titers slightly higher in lung EVOCs compared to those from the trachea. Virus titers of BPIV-3 and SuHV-1 at 24-, 48- and 72- hpi were significantly higher (indicating viral growth and tissue viability) in all tissues with respect to those at 1hpi (p < 0.05, Supplementary Material).
3.3. SARS-CoV-2 does not replicate in swine EVOCs
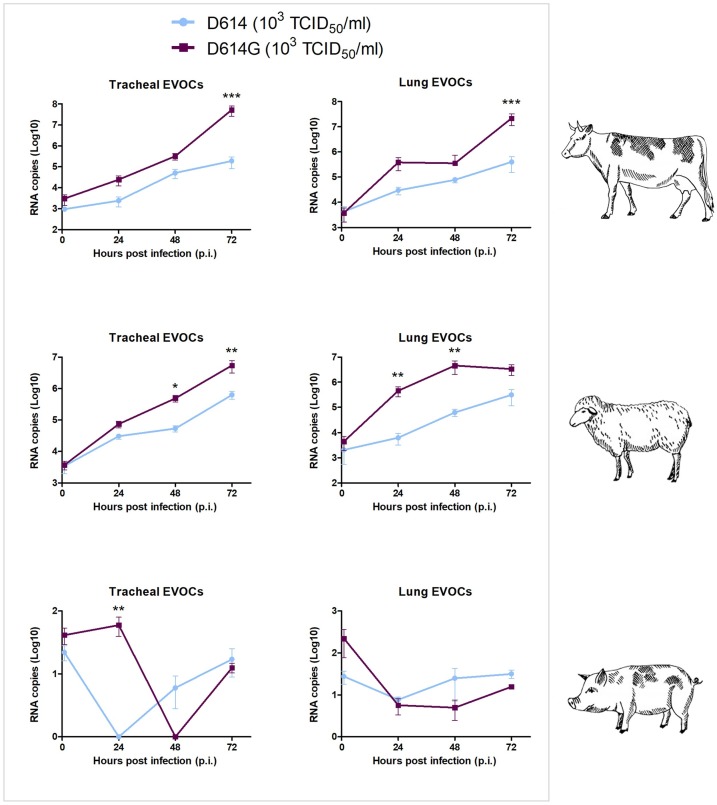
Although the viability of swine EVOCs was confirmed by the growth of SuHV-1, both SARS-CoV-2 isolates failed to replicate in lung and tracheal EVOCs from pigs as statistically significant differences were not observed between either infectious viral titers on cells or RNA copies at 1 hpi and 72 hpi (p > 0.05) (Figs. 1,2 ). Differences between the two SARS-CoV-2 isolates were observed only at 24 hpi in tracheal EVOCs as D614G virus showed a slight, but statistically significant, higher number of RNA copies.
Fig. 1.
SARS-CoV-2 kinetics in bovine, ovine, and swine respiratory EVOCs. Viral titers expressed as Log10 TCID50/mL of SARS-CoV-2 increased significantly in bovine and ovine EVOCs, suggesting viral growth, from 1 to 72 hpi (p < 0.05). In porcine EVOCs, SARS-CoV-2 titers did not change over time. In bovine tissues, D614G replicated significantly at higher magnitude compared to D614 at 48 hpi (p < 0.05) in tracheal EVOCs; 48 hpi (p <0.01) and 72 hpi (p < 0.05) in lung EVOCs. In ovine tissues, both viruses replicated at comparable levels. EVOCs were infected with a dose of 103 TCID50/mL of each SARS-CoV-2 isolate. Titer values represent the mean of three replicates of three independent experiments. Bars indicate standard deviation (SD). * (p < 0.05), ** (p < 0.01), *** (p < 0.001).
3.4. SARS-CoV-2 with D614G exhibits greater replication
SARS-CoV-2 isolates showed marked replication in bovine and ovine EVOCs as demonstrated by the increase of viral titers (Fig. 1) and RNA copies (Fig. 2) over time. More in details, in bovine and ovine respiratory tissues, titers of both viruses were significantly higher at 72 hpi when compared to the earliest time point (p < 0.05). As for bovine tissues, viral titers of both SARS-CoV-2 isolates were higher in lung than trachea (up to 2 Log10), whereas titers of both viruses were of comparable levels in ovine tissues. An analysis of viral titers revealed that D614G replicated in bovine tissues at higher magnitude than D614 at 48 hpi (p < 0.05) in tracheal EVOCs; at 48 hpi (p< 0.01) and 72 hpi (p < 0.05) in lung EVOCs. In ovine tissues, both viruses replicated at comparable levels. Quantification of RNA copies in EVOCs supernatants revealed more apparent differences between D614 and D614G. In particular, in bovine EVOCs, differences were significant at 72 hpi in both tissues (p <0.001), while in ovine EVOCs differences were significant at 48 (p < 0.05) and 72 (p < 0.01) hpi in trachea, 24 (p < 0.01) and 48 (p < 0.01) hpi in lung (Fig. 2).
Fig. 2.
Quantification of SARS-CoV-2 RNA in bovine, ovine and swine respiratory EVOCs. Number of RNA copies of SARS-CoV-2 in EVOC supernatants infected with a dose of 103 TCID50/mL of the D614 and D614G strains increased significantly in bovine and ovine EVOCs from 1 to 72 hpi (p < 0.05). Quantification of RNA copies in EVOC supernatants revealed more evident differences between D614 and D614G. Specifically, in bovine EVOCs, differences were significant at 72 hpi in both tissues (p < 0.001), while in ovine EVOCs, differences were significant at 48 (p < 0.05) and 72 hpi (p < 0.01) in trachea, 24 and 48 hpi in lung (p < 0.01). In swine EVOCs, SARS-CoV-2 genome copies did not change over time. Values represent the mean of three replicates of three independent experiments. Bars indicate standard deviation (SD). * (p < 0.05), ** (p < 0.01), *** (p < 0.001).
3.4.1. Immunoreactivity for ACE2 is mainly present in ovine and bovine respiratory epithelial cells
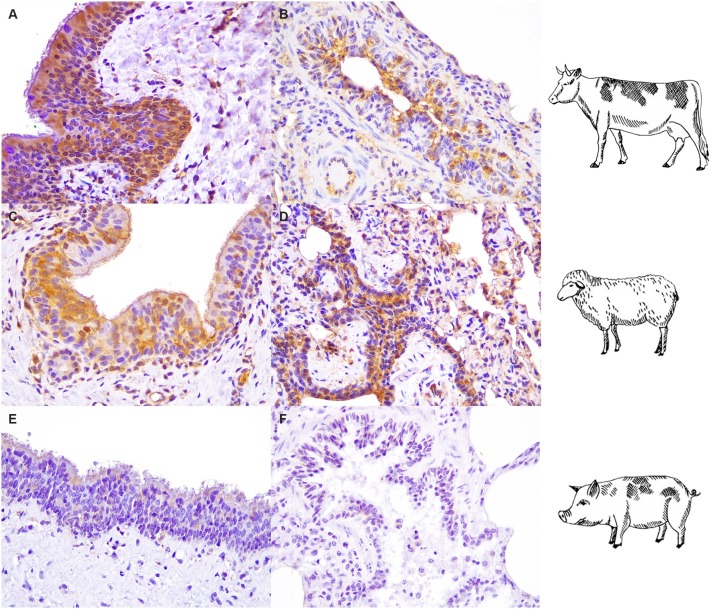
The polyclonal antibody analysis revealed the presence of ACE2 expressing cells in the tracheal (Fig. 3 A, C) and bronchiolar (Fig. 3B, D) epithelia of sheep and cattle. Notably, in our experimental settings, immunoreactivity was not observed in both porcine tissues (Fig. 4 E, F).
Fig. 3.
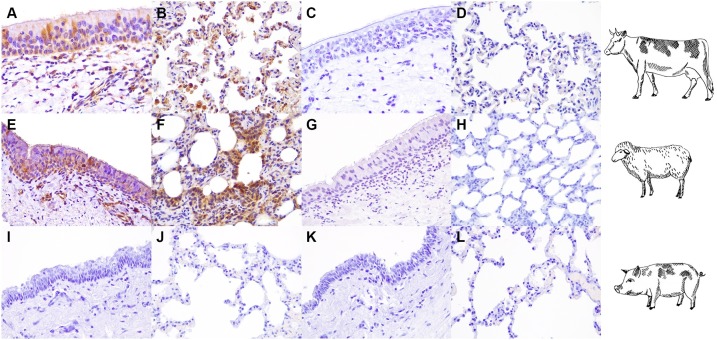
Angiotensin converting enzyme-2 (ACE2) immunohistochemical expression in the bovine, ovine and swine respiratory tracts. Strong immunoreactivity for ACE2 (brown) was visible in tracheal epithelial cells of cattle (A) and sheep (C). Bronchiolar epithelial cells were also positive stained with ACE2 antibody (brown) in bovine and ovine lung (B, D). No specific immunoreactions were observed in swine EVOCs (E, F). All tissues were counterstained with Mayer's Hematoxylin (blue). All pictures were kept at final magnification of X400 (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
Fig. 4.
Western blot analysis of ACE2 in kidney lysates. Western blotting showed a band of molecular weight of approximately 90-100 kDa (expected molecular weight of ACE2 is 97 kDa) in bovine kidney (lane 1), ovine kidney (lane 2) and swine kidney (lane 3). Caco2 cell lysate (lane 4) was used as positive control. Lysates/proteins at 15 μg per lane.
3.4.2. The anti-ACE2 polyclonal antibody recognizes bovine, ovine and swine ACE2
Western blotting analysis revealed high intensity and specific bands of approximately 90−100 kDa (predicted molecular weight of ACE2 is 97 kDa) in bovine, ovine, and porcine kidney lysates. The predicted band was also observed in the Caco-2 cell line lysate (Fig. 4). This indicates that the adopted antibody is able to bind to porcine ACE2.
3.4.3. SARS-CoV-2 has tropism for bovine and ovine respiratory ACE2-expressing cells
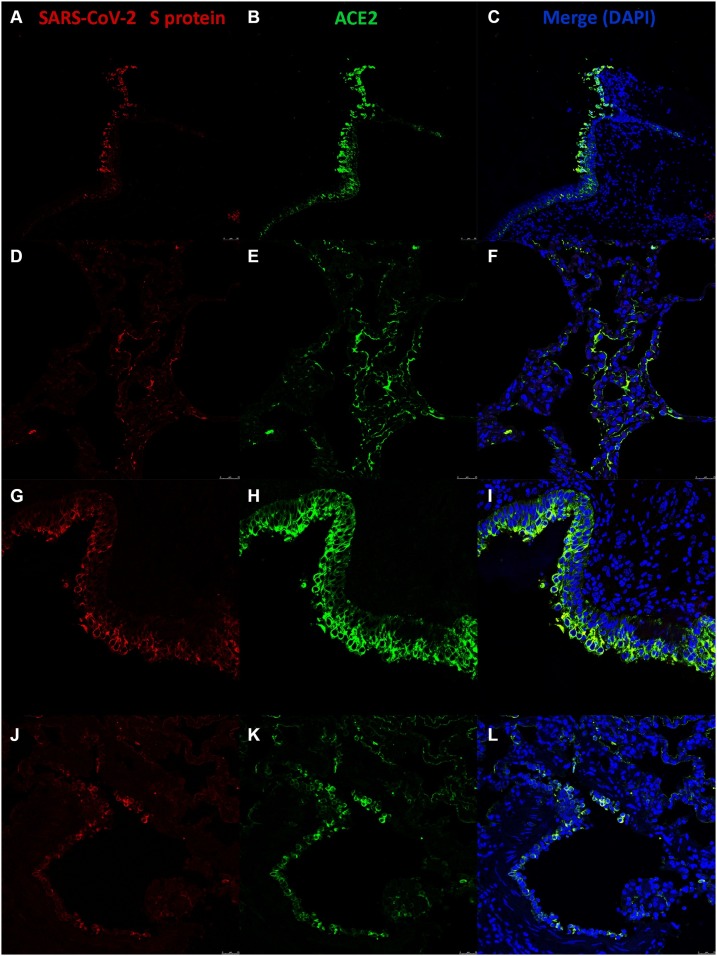
SARS-CoV-2 antigens were visualized by IHC in cells of the tracheal epithelia of both ruminant species (Fig. 5 A, E). In lung parenchyma, SARS-CoV-2 was detected in the morphologically spindled epithelial type I pneumocytes and in bronchiolar cells (Fig. 5B, F). In addition, immunoreactivity for SARS−COV-2 was observed in alveolar macrophages of bovine lung (Fig. 5B). Immunoreactivity was not detected in swine respiratory and in mock infected EVOCs (Fig. 5I, J, K, L, C, D, G, H). Double labelling indirect immunofluorescence (DLIIF) and confocal microscopy (CM) analyses performed in bovine and ovine SARS-CoV-2 infected EVOCs, revealed viral S protein in association with tracheal and pulmonary ACE2 positive cells (Fig. 6 ) of both species. CM and DLIIF analyses were not performed in porcine tissues because of the absence of viral replication and SARS-CoV-2/ACE2 immunoreaction in these EVOCs. No immunoreactivity for viral S protein was observed in mock infected EVOCs (data not shown). No differences were observed in terms of tropism between the two SARS-CoV-2 isolates.
Fig. 5.
Tissue tropism of SARS-CoV-2 in bovine, ovine and swine respiratory EVOCs. Immunoreactivity at 48 hpi for SARS-CoV-2 D614-nucleocapsid (N) protein (brown) was identified in the tracheal epithelium of infected bovine and ovine EVOCs (A, E). Viral antigens were also present in alveolar epithelial cells of both ruminant species (B, cattle and F, sheep) and in alveolar macrophages of bovine EVOCs (B). Mock-infected (C, D, G, H, K, L) and swine EVOCs (I-L) did not show specific immunolabeling for SARS-CoV-2 N protein. All tissues were counterstained with Mayer's Hematoxylin (blue). All pictures were kept at final magnification of X400 (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
Fig. 6.
Confocal microscopy investigation on SARS-CoV-2 infected bovine and ovine EVOCs. Immunofluorescence at 48 hpi for SARS-CoV-2 D614-spike (S) protein (red) were visualized in bovine (A, D) and ovine (G, J) tracheal and lung EVOCs, respectively, while ACE2 immunolabeling is visible in green in bovine (B, E) and ovine (H, K) tracheal and lung EVOCs, respectively. Merging of fluorochromes indicated the correlation of ACE2 positive cells and SARS-CoV-2 S protein in bovine and ovine tracheal (C, I) and lung (F, L) EVOCs. Nuclei were counterstained with DAPI in blue (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
3.5. Mutations in the consensus sequences were not observed
In this round of experiments, consensus sequences obtained from viruses collected from EVOCs were identical to those obtained from viral inocula.
4. Discussion
EVOCs of the respiratory tract from different animal species have been developed to study the cellular tropism and infectivity of respiratory viral pathogens (Pena et al., 2012; Di Teodoro et al., 2019). In this setting, the three-dimensional structure and cell diversity of the respiratory mucosa and lung parenchyma are preserved along with other important physiological features, including the normal expression levels of cell receptors, effective mucus production and ciliary activity.
EVOCs have been used also to characterize innate immune responses, host range, and tropism of different viruses, including SARS-CoV-2 (Nicholls et al., 2007; Niesalla et al., 2009; Chan et al., 2013; Gonzalez et al., 2014; Chan et al., 2016; Hui et al., 2020; Chu et al., 2020). Herein, we demonstrated that bovine and ovine respiratory tissues were permissive to SARS-CoV-2 infection as viral infectious titers and viral genome copies significantly increased over time. This is suggestive of viral replication, although clearly needs to be confirmed with additional studies. Importantly, we demonstrated that a SARS-CoV-2 isolate containing the D614G mutation in the S protein possessed enhanced replicative capabilities compared to earlier strains, represented here by a SARS-CoV-2 strain collected from a Chinese tourist who visited Rome, Italy, in January 2020. Because the SARS-CoV-2/IZSAM/46419 isolate with D614G is accompanied by three additional mutations in the viral genome, it can be reasonably argued that the enhanced replicative capabilities observed here are not necessarily associated with D614G. However, the effects of D614G on viral replication have recently been demonstrated in both human cells and hamsters (Plante et al., 2020). We therefore speculate that D614G played a role by enhancing the replicative capabilities of SARS-CoV-2/IZSAM/46419 in the experiments we performed. Nevertheless, additional studies are required to confirm the higher fitness and infectivity of D614G viruses in animal tissues.
Although pig tissues were viable as demonstrated by the viral burden of SuHV-1, both SARS-CoV-2 isolates did not show replication, confirming results previously obtained following in vivo experimental infection (Schlottau et al., 2020; Shi et al., 2020; Meekins et al., 2020). As demonstrated by western blotting analysis, the polyclonal antibody was able to recognize and bind to porcine ACE2 from kidney. Hence, the absence of viral replication in respiratory tissues is likely explained by the absence of ACE2 moieties in porcine respiratory tissues as suggested by the absence of immunohistochemical reactivity for this receptor. In turn, ACE2 immunoreactivity was present in respiratory tissues of cattle and sheep. Finally, SARS-CoV-2 was associated with ACE2-expressing cells of lung and trachea of both ruminant species, suggesting the affinity of this virus for the ACE2 of these animals as previously proposed (Zhai et al., 2020).
SARS-CoV-2 is the most recent example of an emerging zoonotic virus that has converted its ‘pandemic potential’ into reality (Gollakner and Capua, 2020), and that likely has an origin in another mammalian species (Andersen et al., 2020). Although SARS-CoV-2 is able to infect several animal species in close contact with humans, such as minks (Oreshkova et al., 2020), the potential of SARS-CoV-2 to become established in other animal species is unclear.
4.1. Conclusions
Our results strongly suggest that in vivo experiments involving ruminant species are warranted to help determine the clinical outcome of infection in these species and their potential role as host animals. Importantly, however, EVOCs experiments are only suggestive that these animal species may be susceptible to infection, and more confirmatory work is clearly warranted. More generally, our study highlights the potential for diverse animal species to play a role in the epidemiology of SARS-CoV-2 as well as the need for a One Health approach to tackle this major global problem (2020b).
5. Funding
IZSAM funding was provided by the Italian Ministry of Health (Ricerca Corrente 2015 “Strategie innovative per la riduzione della sperimentazione animale: colture di espianti di tessuti e virus-istochimica”, recipient Giovanni Savini and Ricerca Corrente 2020 “PanCO: epidemiologia e patogenesi dei coronavirus umani ed animali”, recipient Alessio Lorusso). ECH is supported by an Australian Research Council (ARC) Australian Laureate Fellowship (FL170100022). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the IZSAM.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgement
We gratefully thank Dr Paola Di Giuseppe (IZSAM) for support in formatting figures.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2020.108933.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M.C.W., Chan R.W.Y., Chan L.L.Y., Mok C.K.P., Hui K.P.Y., Fong J.H.M., Tao K.P., Poon L.L.M., Nicholls J.M., Guan Y., Peiris J.S.M. Tropism and innate host responses of a novel avian influenza A H7N9 virus: an analysis of ex-vivo and in-vitro cultures of the human respiratory tract. Lancet Respir. Med. 2013;1:534–542. doi: 10.1016/S2213-2600(13)70138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L.L.Y., Bui C.T.H., Mok C.K.P., Ng M.M.T., Nicholls J.M., Peiris J.S.M., Chan M.C.W., Chan R.W.Y. Evaluation of the human adaptation of influenza A/H7N9 virus in PB2 protein using human and swine respiratory tract explant cultures. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep35401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F.W., Wang Y., Yuen T.T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X., Zhang X., Cai J.P., Zhou J., Yuan S., Kok K.H., To K.K.W., Chan I.H.Y., Zhang A.J., Sit K.Y., Au W.K., Yuen K.Y. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.P., Pfenning A.R., Zhao H., Genereux D.P., Swoford R., Pollard K.S., Ryder O.A., Nweeia M.T., Linblad-Toh K., Teeling E.C., Karlsson E.K., Lewin H.A. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. PNAS. 2020;117(36):22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Campolo M., Desario C., Cirone F., D’Abramo M., Lorusso E., Greco G., Mari V., Colaianni M.L., Elia G., Martella V., Buonavoglia C. Respiratory disease associated with bovine coronavirus infection in cattle herds in Southern Italy. J Vet Diagn Invest. 2008:28–32. doi: 10.1177/104063870802000105. [DOI] [PubMed] [Google Scholar]
- Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet. Microbiol. 2020 doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Teodoro G., Marruchella G., Di Provvido A., Orsini G., Ronchi G.F., D’Angelo A.R., D’Alterio N., Sacchini F., Scacchia M. Respiratory explants as a model to investigate early events of contagious bovine pleuropneumonia infection. Vet. Res. 2018;49:5. doi: 10.1186/s13567-017-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Teodoro G., Bortolami A., Teodori L., Leone A., D’Alterio N., Malatesta D., Rosamilia A., Colaianni M.L., Petrini A., Terregino C., Savini G., Bonfante F., Lorusso A. Replication kinetics and cellular tropism of emerging reoviruses in sheep and swine respiratory ex vivo organ cultures. Vet. Microbiol. 2019;234:119–127. doi: 10.1016/j.vetmic.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Enserink M. Coronavirus rips through Dutch mink farms, triggering culls. Science. 2020;368:1169. doi: 10.1126/science.368.6496.1169. [DOI] [PubMed] [Google Scholar]
- Gaudreault N.N., Trujillo J.D., Carossino M., Meekins D.A., Morozov I., Madden D.W., Indran S.W., Bold D., Balaraman W., Kwon T., Artiaga B.L., Cool K., Garcia-Sastre A., Ma W., Wilson W.C., Henningson J., Balasuriya U.B.R., Richt J.A. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerging Microbes and Infections. 2020:2322–2332. doi: 10.1080/22221751.2020.1833687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollakner R., Capua I. Is COVID-19 the first pandemic that evolves into a panzootic? Vet. Ital. 2020;56:7–8. doi: 10.12834/VetIt.2246.12523.1. [DOI] [PubMed] [Google Scholar]
- Gonzalez G., Marshall J.F., Morrell J., Robb D., McCauley J.W., Perez D.R., Parrish C.R., Murcia P.R. Infection and pathogenesis of canine, equine, and human influenza viruses in canine tracheas. J. Virol. 2014;88:9208–9219. doi: 10.1128/JVI.00887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., Van Goor H. issue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004 doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 271-280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K.P.Y., Cheung M.C., Perera R.A.P.M., Ng K.C., Bui C.H.T., Ho J.C.W., Ng M.M.T., Kuok D.I.T., Shih K.C., Tsao S.W., Poon L.L.M., Peiris M., Nicholls J.M., Chan M.C.W. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itokawa K., Sekizuka T., Hashino M., Tanaka R., Kuroda M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplextiling PCR. PLOS ONE. 2020 doi: 10.1371/journal.pone.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Calistri P., Petrini A., Savini G., Decaro N. Novel coronavirus (SARS-CoV-2) epidemic: a veterinary perspective. Vet. Ital. 2020;56:5–10. doi: 10.12834/VetIt.2173.11599.1. [DOI] [PubMed] [Google Scholar]
- Lorusso A., Calistri P., Mercante M.T., Monaco F., Portanti O., Marcacci M., Cammà C., Rinaldi A., Mangone I., Di Pasquale A., Iommarini M., Mattucci M., Fazii P., Tarquini P., Mariani R., Grimaldi A., Morelli D., Migliorati G., Savini G., Borrello S., D’Alterio N. A “One-Health” approach for diagnosis and molecular characterization of SARS-CoV-2 in Italy. One Health. 2020 doi: 10.1016/j.onehlt.2020.100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes C., Gollakner R., Capua I. Could Mustelids spur COVID-19 into a panzootic? Vet. Ital. 2020 doi: 10.12834/VetIt.2375.13627.1. [DOI] [PubMed] [Google Scholar]
- Matthay M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., Herridge M., Randolph A.G., Calfee C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 2019 doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins D.A., Morozov I., Trujillo J.D., Gaudreault N.N., Bold D., Carossino M., Artiaga B.L., Indran S.V., Kwon T., Balaraman V., Madden D.W., Feldmann H., Henningson J., Ma W., Balasuriya U.B.R., Richt J.A. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerging Microbes and Infections. 2020:2278–2288. doi: 10.1080/22221751.2020.1831405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar R.J., Vreman S., Hakze-van der Honing R.W., Zwart R., de Rond J., Weesendorp E., Smit L.A.M., Koopmans M., Bouwstra R., Stegeman A., van der Poel W.H.M. Clinical and Pathological Findings in SARS-CoV-2 Disease Outbreaks in Farmed Mink (Neovison vison) Vet. Pathol. 2020;57:653–657. doi: 10.1177/0300985820943535. [DOI] [PubMed] [Google Scholar]
- Nicholls J.M., Chan M.C.W., Chan W.Y., Wong H.K., Cheung C.Y., Kwong D.L.W., Wongm M.P., Chui W.H., Poon L.L.M., Tsao S.W., Guan Y., Peiris J.S.M. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat. Med. 2007;13:147–149. doi: 10.1038/nm1529. [DOI] [PubMed] [Google Scholar]
- Niesalla H.S., Dale A., Slater J.D., Scholes S.F.E., Archer J., Maskell D.J., Tucker A.W. Critical assessment of an in vitro bovine respiratory organ culture system: a model of bovine herpesvirus-1 infection. J. Virol. Methods. 2009;158:123–129. doi: 10.1016/j.jviromet.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Oreshkova N., Molenaar R.J., Vreman S., Harders F., Oude Munnink B.B., Hakze-van der Honing R.W., Gerhards N., Tolsma P., Bouwstra R., Sikkema R.S., Tacken M.G., de Rooij M.M., Weesendorp E., Engelsma M.Y., Bruschke C.J., Smit L.A., Koopmans M., van der Poel W.H., Stegeman A. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson E.I., Elia G., Grassi A., Desario C., Medardo M., Smith S.L., Anderson E.R., Prince T., Patterson G.T., Lorusso E., Lucente M.S., Lanave G., Lauzi S., Bonfanti U., Stranieri A., Martella V., Solari Basano F., Barrs V.R., Radford A.D., Agrimi U., Hughes G.L., Paltrinieri S., Decaro N. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nature communication. 2020 doi: 10.1038/s41467-020-20097-0. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena L., Vincent A.L., Loving C.L., Henningson J.N., Lager K.M., Lorusso A., Perez D.R. Restored PB1-F2 in the 2009 pandemic H1N1 influenza virus has minimal effects in swine. J. Virol. 2012;86:5523–5532. doi: 10.1128/JVI.00134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzurro F., Mangone I., Zaccaria G., De Luca E., Malatesta D., Innocenti M., Carmine I., Cito F., Marcacci M., Di Sabatino D., Lorusso A. Whole-genome sequence of a suid Herpesvirus-1 strain isolated from the brain of a hunting dog in Italy. Genome Announc. 2016 doi: 10.1128/genomeA.01333-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K., Rissmann M., Graaf A., Schön J., Sehl J., Wylezich C., Höper D., Mettenleiter T.C., Balkema-Buschmann A., Harder T., Grund C., Hoffmann D., Breithaupt A., Beer M. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 894-904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:75–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B., Rafique A., Barve A., Sabeti P.C., Kyratsous C.A., Dudkina N.V., Shen K., Luban J. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X., Sun J., Yan Z., Zhang J., Zhao J., Zhao Z., Gao Q., He W.-T., Veit M., Su S. Comparison of SARS-CoV-2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020 doi: 10.1128/JVI.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X. Lou, Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.Di, Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.