Abstract
Genetic polymorphisms in the signalling pathway of estrogen receptor (ER) could modify the risk of breast cancer. A variable number of tandem repeats (VNTR) polymorphism in the promoter of PTTG1IP, pituitary tumor transforming gene binding factor targeted by estrogen receptor α (ERα) in endocrine neoplasia, has been shown to be functional, but its relevance to cancer etiology was unknown. We investigated its association with breast cancer risk by genotyping in 658 patients and 866 controls and further analysed its differential interaction with ERα. We found nine types of alleles ranging from 2 to 9 and 11 repeats that form 29 distinct genotypes and 11 different biallelic repeat numbers. Subjects who carry the six‐repeats allele (odds ratio [OR], 1.45; 95% confidence interval [CI], 1.17–1.79), long alleles (≥6 repeats) (OR, 1.55; 95% CI, 1.17–2.05) or a high dose of biallelic repeats (OR, 1.38; 95% CI, 1.07–1.77) were at significantly increased risk of cancer. In stratification analysis, these associations consistently manifested in ER‐positive breast cancer: in ER positive, PR‐positive subtype, genotypes with the six‐repeats allele (OR, 1.42; 95% CI, 1.06–1.90), long alleles (OR, 1.77; 95% CI, 1.17–2.67) or a high dose of biallelic repeats (OR, 1.67; 95% CI, 1.19–2.33) were associated with cancer risk; in ER positive, HER2‐negative subtype, they were susceptible factors with the ORs being 1.46 (95% CI, 1.06–2.02), 2.06 (95% CI, 1.28–3.32) and 1.85 (95% CI, 1.26–2.71), respectively. Furthermore, functional analysis revealed that an increase in the number of tandem repeats enhances the binding affinity of ERα. The present study provides the first epidemiological evidence that functional regulatory variants of PTTG1IP were associated with the risk of ER‐positive breast cancer, further supporting its relevance as one proto‐oncogene in breast cancer. (Cancer Sci 2012; 103: 1121–1128)
Breast cancer is the most prevalent cancer and the leading cause of cancer death among women worldwide.1 The incidence of breast cancer increased rapidly in China in recent years with a sharp rise of 38.5% from 2000 to 2005.2 Breast cancer is a multifactorial common disease caused by inherited and environmental factors including endocrinal status, especially endogenous estrogens. Estrogen exerts its roles in breast tumorigenesis through binding to estrogen receptor α (ERα) and triggering cell growth and transformation.3 The activated ERα binds to estrogen response elements (ERE) in the proximal or distal promoters of a variety of ER target genes, activating or repressing their transcription and consequently resulting in tumorigenic effects.3
Parallel to the well defined biological functions of estrogen and ERα in breast cancer, etiological implications of genetic polymorphisms in estrogen metabolism and ERα signalling have also been addressed. In Chinese women, genetic susceptibility to breast cancer is associated with variants of CYP19A1 4, 5 and CYP17A1,5 the coding genes for key enzymes in estrogen biosynthesis, and the ERα coding gene ESR1.5, 6 Similar associations have also been observed in other ethnic populations.7, 8 But there have been less reported efforts to define biologically functional and etiologically relevant genetic variants in the downstream of ERα signalling. ERα is among the few key transcription factors whose cistromes, the whole genome‐binding sites of a trans‐acting factor, have been intensively characterized with high throughput functional genomics strategies such as ChIP‐Chip and ChIP‐Seq.9 Yu et al.10 blasted the dbSNP database with ERα cistrome, screened out 21 regulatory single nucleotide polymorphisms (SNP) in ERα target genes and found that one common SNP in the ERE of NRCAM, which codes for a cell adhesion molecule, was associated with the risk of breast cancer. This pilot study suggested that novel ERα target genes and their allele‐differential interaction might be new modifiers of breast cancer susceptibility.
Recently, pituitary tumor transforming gene (PTTG) binding factor (PTTG1IP; PBF), a relatively uncharacterized proto‐oncogene, was reported to be one novel target of ERα in endocrine neoplasia.11 PTTG1IP expression is low or absent in normal breast tissue, but it is strongly expressed in epithelial cells of ERα‐positive breast tumor.11 Furthermore, PTTG1IP is a secreted protein and its secretion modulates induction of estrogen‐mediated invasion of breast cancer cells.11 Interestingly, the ERE in PTTG1IP promoter was found to be located in a polymorphic region (−399 to −291 relative to the translational start site) that contains a variable number of tandem repeats (VNTR) of an 18‐nucleotides sequence housing a putative ERE half‐site (gcccctcGGTCAcgcctc); greater numbers of repeats correlates with higher mRNA expression in normal breast tissue and tumor specimen.11 This report represented PTTG1IP as an entirely novel gene of direct relevance to breast cancer, but genetic epidemiology evidence for its implications in cancer development and progression was lacking.
In the present study, we reasoned that the VNTR variation in the PTTG1IP promoter might modify the risk of breast cancer development and metastasis. To test this hypothesis, we furthered functional characterization of the differential binding affinity of ERα to its response elements in the variable tandem repeats and investigated the association between the cis‐regulatory polymorphism and breast cancer risk in a case–control study.
Materials and Methods
Study subjects
The present study included a total of 658 breast cancer cases and 866 cancer‐free controls from two panels. We recruited 474 cases with a response rate of 92.1% between January 2005 and October 2006 from the Department of Breast Surgery at the Qilu Hospital of Shandong University in the city of Jinan in Shandong Province. We recruited 184 cases with a response rate of 89.5% between July 2006 and May 2007 from the Department of Surgical Oncology at the First Hospital of Xi'an Jiaotong University in the city of Xi'an in Shanxi Province. All cases were incident patients with histopathologically confirmed breast cancer. The patients who reportedly had previous cancer, other metastasized cancer and previous radiotherapy or chemotherapy were excluded. The status of estrogen receptor (ER), progesterone receptor (PR), epidermal growth factor receptor 2 (HER2) and axillary lymph node metastasis (ALNM) were obtained from the medical records of the hospitals. Six hundred and forty‐eight and 218 cancer‐free control women, frequency matched to the cases by age and residential area, were selected from the subjects who participated in a medical inspection conducted at the two hospitals, respectively, during the same time periods as the respective cases were recruited. All participants were genetically unrelated, ethnic Han Chinese women. After informed consent and demographic information (age and age at menarche) were obtained, a 5‐mL venous blood sample was collected from each subject. The study was approved by the Ethics Committees of Qilu Hospital of Shandong University, the First Hospital of Xi'an Jiaotong University and the School of Life Sciences of Fudan University.
Variable number of tandem repeats genotype analyse
Genomic DNA of controls and cases were extracted from peripheral blood leukocytes. Genotyping of the rs72416745 VNTR polymorphism in the promoter region of PTTG1IP was performed using direct sequencing of PCR products amplified with primers 5′‐CGCCTCGTCCAGGGCTCACTT‐3′ (forward) and 5′‐GCGTTACAACTCCGACTCCAGCA‐3′ (reverse) using the protocol previously described.12 The PCR products were sequenced using the forward primer with ABI PRISM Dye Terminator Sequencing kits (Applied Biosystems, Foster City, CA, USA) and loaded onto an ABI 3730XL sequencer. We identified the VNTR polymorphism genotypes with the Chromas 2.31 program (Technelysium Pty Ltd, Sydney, Australia). Allelic status for heterozygotes was carefully confirmed by manual evaluation of two reviewers (C.X. and H.W.).
Electrophoretic mobility shift assay (EMSA)
We used the EMSA assay to analyze allele‐differential binding of ERα to the PTTG1IP promoter. Six allelic double‐stranded oligonucleotides containing 3, 4, 5, 6, 7 and 8 copies of the 18‐bp repeat were prepared using the amplifying sequence from −420 to −277 relative to the translational start site with primers 5′‐CAGGCGCAGGAGCCGC‐3′ (forward) and 5′‐GCGGCGCGGATGGGC‐3′ (reverse) from the genomic DNA of subjects with homozygous genotypes for the respective repeat alleles. The oligonucleotide probes were then labelled with biotin. The EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific, Waltham, MA, USA). Briefly, a biotin‐labelled probe was combined with nuclear protein extract from MCF‐7 cells. Respective unlabelled probes for various tandem repeats and a consensus ERα binding site probe (5′‐CAAAGTCAGGTCACAGTGACCTGATCAAAG‐3′) were used for competition assays. After electrophoresis of a reaction mixture, the gel was transferred to a nylon membrane and cross‐link was performed with a UV cross‐linker (HL‐2000 Hybrolinker, UVP, Upland, CA, USA). Biotin‐labelled DNA on membrane was detected with stabilized Streptavidin–horseradish peroxidase conjugate.
Statistical analyses
We used the chi‐square test to examine differences in the demographic variables, alleles and genotypes between the cases and controls. We used GENEPOP 4.1 software13 to test deviations of the VNTR genotype frequencies in the controls and in cases from those expected under Hardy–Weinberg equilibrium (HWE). We estimated the cancer risk associated with genotypes by calculating the odds ratio (OR) and their 95% confidence interval (CI) adjusted for age and age at menarche with unconditional logistic regression models. The probability level of < 0.05 was used as the criterion of statistical significance, and all statistical tests were two sided. These statistical analyses were performed using Stata software (version 8.0, StataCorp LP, College Station, TX, USA).
Results
The characteristics of the cases and controls enrolled in the present study are shown in Table 1. There was no significant difference in the mean age between cases and controls (P = 0.568). Compared with the control subjects, the breast cancer cases had a significantly younger age at menarche (P < 0.05). Age and age at menarche were then taken as adjustment factors in the following association analysis. Among the 658 patients overall, ER status was available in 514 (78.1%) patients, with 316 (61.5%) positive and 198 (38.5%) negative; PR status was available in 514 (78.1%) patients, with 350 (68.1%) positive and 164 (31.9%) negative; HER2 status was available in 503 (76.4%) patients, with 198 (39.4%) positive and 305 (60.6%) negative; and ALNM status was available in 529 (80.4%) patients, with 284 (53.7%) positive and 245 (46.3%) negative.
Table 1.
Distributions of select characteristics by case–control status
| Variables | Cases (N = 658) | Controls (N = 866) | P valuea |
|---|---|---|---|
| Mean age (SD) (years) | 52.8 (11.1) | 52.5 (11.6) | 0.568 |
| Mean age at menarche (SD) (years)b | 15.1 (4.9) | 15.8 (4.6) | <0.05 |
| Estrogen receptor statusc | |||
| Positive | 316 (61.5) | ||
| Negative | 198 (38.5) | ||
| Progesterone receptor statusc | |||
| Positive | 350 (68.1) | ||
| Negative | 164 (31.9) | ||
| HER2 statusc | |||
| Positive | 198 (39.4) | ||
| Negative | 305 (60.6) | ||
| ALNM statusc | |||
| Positive | 284 (53.7) | ||
| Negative | 245 (46.3) | ||
Student's t‐test for the difference between cases and controls.
Age at menarche information was available in 632 breast cancer cases and 849 controls.
Information on estrogen receptor, progesterone receptor, epidermal growth factor receptor 2 (HER2) and axillary lymph node metastasis (ALNM) status was available in 514 (78.1%), 514 (78.1%), 503 (76.4%) and 529 (80.4%) patients, respectively.
We genotyped the PTTG1IP VNTR polymorphism for 1524 subjects in the case–control study. The genotypic and allelic frequencies in the cases and controls are shown in Table 2. We observed length variation ranging from 2 to 9 and 11 repeats; these nine different types of alleles form 29 distinct genotypes and 11 different biallelic repeat numbers. The genotypic frequencies in the controls and cases did not deviate from HWE. The most common genotype in this population is 5/6 (heterozygote with alleles of five and six repeats), followed by 6/6, 6/7 and 5/5. The frequencies of 5/5 (10.2% vs 4.7%, P < 0.001) and 6/7 (11.4% vs 14.9%, P = 0.046) significantly differed between the controls and cases, respectively. Meanwhile, among the four common alleles (4, 5, 6 or 7 repeats), the five‐repeats allele was significantly overrepresented in the controls than in the cases (29.9% vs 25.0%, P = 0.003). Allelic frequency of six repeats in the controls, on the contrary, was significantly lower than in the cases (35.2% vs 40.7%, P = 0.002). Furthermore, when alleles were accordingly combined into two groups, the short alleles (denoted as S) with ≤5 repeats and the long alleles (L) with ≥6 repeats, we found that the long allele was significantly underrepresented in the controls than in the cases (56.7% vs 61.8%, P = 0.005). In contrast, the number of biallelic repeats in the population ranges from 6 to 16 repeats with an overall significant difference in frequency between the cases and controls (P = 0.032), among which the biallelic 10 repeat was significantly overrepresented in the controls than in the cases (18.7% vs 13.2%, P = 0.004). When the 11 types of biallelic repeats were accordingly combined into low (≤10 repeats) and high (>10 repeats) repeat doses, consistent with the allele‐based observation, we found that the frequency of a high dose of biallelic repeats in the cases was significantly higher than that in the controls (75.7% vs 70.2%, P = 0.018). These results demonstrate that PTTG1IP VNTR variation is highly polymorphic with unbalanced genotype distribution between breast cancer patients and controls.
Table 2.
Genotype and allele distribution of PTTG1IP variable number of tandem repeats polymorphism between cases and controls
| Genotype or allele | Control (N = 866, n = 1732) | Case (N = 658, n = 1316) | P valuea | ||
|---|---|---|---|---|---|
| N or n | % | N or n | % | ||
| Genotypes | |||||
| 2/4 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| 2/5 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| 2/6 | 1 | 0.12 | 1 | 0.15 | 1.000 |
| 2/7 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| 3/3 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| 3/4 | 4 | 0.46 | 2 | 0.30 | 0.704 |
| 3/5 | 3 | 0.35 | 8 | 1.22 | 0.065 |
| 3/6 | 13 | 1.50 | 15 | 2.28 | 0.336 |
| 3/7 | 7 | 0.81 | 4 | 0.61 | 0.766 |
| 3/8 | 2 | 0.23 | 2 | 0.30 | 1.000 |
| 4/4 | 12 | 1.39 | 10 | 1.52 | 0.832 |
| 4/5 | 58 | 6.70 | 37 | 5.62 | 0.454 |
| 4/6 | 67 | 7.74 | 52 | 7.90 | 0.923 |
| 4/7 | 34 | 3.93 | 27 | 4.10 | 0.895 |
| 4/8 | 10 | 1.15 | 4 | 0.61 | 0.295 |
| 5/5 | 88 | 10.16 | 31 | 4.71 | <0.001 |
| 5/6 | 179 | 20.67 | 152 | 23.10 | 0.260 |
| 5/7 | 84 | 9.70 | 58 | 8.81 | 0.594 |
| 5/8 | 15 | 1.73 | 12 | 1.82 | 1.000 |
| 5/9 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| 6/6 | 107 | 12.36 | 92 | 13.98 | 0.358 |
| 6/7 | 99 | 11.43 | 98 | 14.89 | 0.046 |
| 6/8 | 34 | 3.93 | 32 | 4.86 | 0.377 |
| 6/9 | 2 | 0.23 | 1 | 0.15 | 1.000 |
| 6/11 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| 7/7 | 28 | 3.23 | 17 | 2.58 | 0.542 |
| 7/8 | 11 | 1.27 | 3 | 0.46 | 0.112 |
| 7/9 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| 8/8 | 1 | 0.12 | 0 | 0.00 | 1.000 |
| Allele status of tandem repeat number | 0.033 | ||||
| 2 | 4 | 0.23 | 1 | 0.08 | 0.398 |
| 3 | 31 | 1.79 | 31 | 2.36 | 0.301 |
| 4 | 198 | 11.43 | 142 | 10.79 | 0.601 |
| 5 | 517 | 29.85 | 329 | 25.00 | 0.003 |
| 6 | 610 | 35.22 | 535 | 40.65 | 0.002 |
| 7 | 293 | 16.92 | 224 | 17.02 | 0.961 |
| 8 | 74 | 4.27 | 53 | 4.03 | 0.784 |
| 9 | 4 | 0.23 | 1 | 0.08 | 0.398 |
| 11 | 1 | 0.06 | 0 | 0.00 | 1.000 |
| Allele length status | |||||
| Short allele (≤5) | 750 | 43.3 | 503 | 38.2 | 0.005 |
| Long allele (≥6) | 982 | 56.7 | 813 | 61.8 | |
| Number of biallelic repeats | 0.032 | ||||
| 6 | 2 | 0.23 | 0 | 0.00 | 0.509 |
| 7 | 6 | 0.69 | 2 | 0.30 | 0.478 |
| 8 | 16 | 1.85 | 19 | 2.89 | 0.227 |
| 9 | 72 | 8.31 | 52 | 7.90 | 0.850 |
| 10 | 162 | 18.71 | 87 | 13.22 | 0.004 |
| 11 | 215 | 24.83 | 181 | 27.51 | 0.239 |
| 12 | 201 | 23.21 | 154 | 23.40 | 0.951 |
| 13 | 114 | 13.16 | 110 | 16.72 | 0.058 |
| 14 | 63 | 7.27 | 49 | 7.45 | 0.921 |
| 15 | 13 | 1.50 | 4 | 0.61 | 0.139 |
| 16 | 2 | 0.23 | 0 | 0.00 | 0.509 |
| Biallelic‐repeats dose | |||||
| Low (≤10) | 258 | 29.79 | 160 | 24.32 | 0.018 |
| High (>10) | 608 | 70.21 | 498 | 75.68 | |
P for Fisher's exact test with other genotypes as the reference group.
By using multivariate logistic regression analysis, we further assessed the association between breast cancer risk and PTTG1IP VNTR genotypes (Table 3). We observed a significant reduction in cancer risk among subjects carrying the 5/5 genotype (OR, 0.42; 95% CI, 0.27–0.65) compared with those who do not carry the five‐repeats allele. On the contrary, we found that subjects who carry the six‐repeats alleles (OR, 1.45; 95% CI, 1.17–1.79) or ≥6 repeats alleles (OR, 1.55; 95% CI, 1.17–2.05) were, in the dominant genetic model, at significantly increased cancer risk compared with their respective reference groups. Likewise, when compared with homozygote of short alleles (≤5 repeats, S/S), genotypes with long alleles (≥6 repeats, L) were significantly associated with increased cancer risk, with the OR for heterozygous S/L and homozygous L/L genotypes being 1.51 (95% CI, 1.12–2.04) and 1.61 (95% CI, 1.18–2.20), respectively. Gene dose effects were apparent for these associations. Consistently, a high (>10 repeats) dose of biallelic repeats was significantly associated with cancer susceptibility compared with a low dose (≤10 repeats) (OR, 1.38; 95% CI, 1.07–1.77).
Table 3.
Association of genotype and allele of PTTG1IP variable number of tandem repeats with risk of breast cancer
| Genotype† | Control (N = 866) | Case (N = 658) | P value‡ | OR (95% CI)§ | P value§ | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| Status with 5 repeats | |||||||
| */* | 437 | 50.46 | 360 | 54.71 | <0.001 | 1.00 | |
| 5/* | 341 | 39.38 | 267 | 40.58 | 0.95 (0.77–1.18) | 0.662 | |
| 5/5 | 88 | 10.16 | 31 | 4.71 | 0.42 (0.27–0.65) | <0.001 | |
| 5/* + 5/5 | 429 | 49.54 | 298 | 45.29 | 0.100 | 0.84 (0.69–1.03) | 0.099 |
| Status with 6 repeats | |||||||
| #/# | 363 | 41.92 | 215 | 32.67 | 0.001 | 1.00 | |
| 6/# | 396 | 45.73 | 351 | 53.34 | 1.46 (1.17–1.82) | 0.001 | |
| 6/6 | 107 | 12.36 | 92 | 13.98 | 1.42 (1.02–1.97) | 0.035 | |
| 6/# + 6/6 | 303 | 58.08 | 443 | 67.33 | <0.001 | 1.45 (1.17–1.79) | 0.001 |
| Status with repeat length | |||||||
| Short (S; ≤5)/S | 168 | 19.40 | 88 | 13.37 | 0.006 | 1.00 | |
| Long (L; ≥6)/S | 414 | 47.81 | 327 | 49.70 | 1.51 (1.12–2.04) | 0.006 | |
| L/L | 284 | 32.79 | 243 | 36.93 | 1.61 (1.18–2.20) | 0.002 | |
| S/L + L/L | 698 | 80.60 | 570 | 86.63 | 0.002 | 1.55 (1.17–2.05) | 0.002 |
| Biallelic repeat dose | |||||||
| Low (≤10) | 258 | 29.79 | 160 | 24.32 | 0.018 | 1.00 | |
| High (>10) | 608 | 70.21 | 498 | 75.68 | 1.38 (1.07–1.77) | 0.013 | |
Bold values indicate statistical significance.†*, other repeat alleles rather than five repeats; #, other repeat alleles rather than six repeats; S, short (≤5); L, long (≥6). ‡Pearson's χ2 test for the difference between cases and controls.§Odds ratio (OR) and their 95% confidence interval (CI) and P values were calculated using unconditional logistic regression with the */*, #/#, S/S and low dose of biallelic‐repeats genotypes as reference groups, respectively, and adjusted for age and age at menarche.
Because PTTG1IP was reported to be one of the transcriptional targets of ER,11 we further conducted stratification analyses by ER status as a primary factor and PR and HER2 as two related immunochemical indexes. We found that the association between PTTG1IP VNTR genotypes and cancer risk significantly and consistently manifested in only ER‐positive breast cancer but not in ER‐negative breast cancer (Table 4). Genotypes with the six‐repeats allele were associated with increased risk of breast cancer in the primary strata of ER positive (OR, 1.45; 95% CI, 1.09–1.92), especially in the subgroups of PR positive, that is, ER(+)‐PR(+) (OR, 1.42; 95% CI, 1.06–1.90) and ER(+)‐HER2(−) (OR, 1.46; 95% CI, 1.06–2.02). Similarly, we observed a pronounced association between increased cancer risk and genotypes containing long alleles (OR, 1.98; 95% CI, 1.32–2.99) or a high dose of biallelic repeats (OR, 1.73; 95% CI, 1.25–2.39) only in the strata of ER‐positive patients. Consistently, genotypes containing long alleles were associated with increased cancer risk in the subgroups of ER(+)‐PR(+) (OR, 1.77; 95% CI, 1.17–2.67) and ER(+)‐HER2(−) (OR, 2.06; 95% CI, 1.28–3.32), and a high dose of biallelic repeats was associated with increased cancer risk in ER(+)‐PR(+) (OR, 1.67; 95% CI, 1.19–2.33) and ER(+)‐HER2(−) (OR, 1.85; 95% CI, 1.26–2.71) patients. These observations in the association study clearly demonstrate that an increase in the repeat number of PTTG1IP VNTR might be one susceptible factor of ER‐positive breast cancer predisposition.
Table 4.
Association of PTTG1IP variable number of tandem repeats (VNTR) with breast cancer risk stratified by immunochemical index (ER, PR and HER2)
| Characters | Genotype of PBF VNTR (cases/controls) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Status with 5 repeats | Status with 6 repeats | Status with repeat length | Biallelic‐repeats dose | |||||||||
| */* | 5/* + 5/5 | OR (95% CI) | #/# | 6/# + 6/6 | OR (95% CI) | S/S | S/L + L/L | OR (95% CI) | Low | High | OR (95% CI) | |
| ER (+) | 182/437 | 134/429 | 0.79 (0.61–1.04) | 103/363 | 213/503 | 1.45 (1.09–1.92) | 34/168 | 282/698 | 1.98 (1.32–2.99) | 66/258 | 250/608 | 1.73 (1.25–2.39) |
| PR (+) | 157/437 | 121/429 | 0.81 (0.61–1.07) | 92/363 | 186/503 | 1.42 (1.06–1.90) | 34/168 | 244/698 | 1.77 (1.17–2.67) | 60/258 | 218/608 | 1.67 (1.19–2.33) |
| PR (−) | 25/437 | 13/429 | 0.72 (0.34–1.54) | 11/363 | 27/503 | 1.76 (0.77–4.05) | 0/168 | 38/698 | 1.02 (1.00–1.05) | 6/258 | 32/608 | 2.59 (0.89–7.54) |
| HER2 (+) | 61/437 | 34/429 | 0.67 (0.42–1.07) | 29/363 | 66/503 | 1.61 (0.97–2.65) | 10/168 | 85/698 | 1.68 (0.85–3.34) | 24258 | 71/608 | 1.35 (0.79–2.32) |
| HER2 (−) | 118/437 | 96/429 | 0.84 (0.62–1.14) | 70/363 | 144/503 | 1.46 (1.06–2.02) | 24/168 | 190/698 | 2.06 (1.28–3.32) | 42/258 | 172/608 | 1.85 (1.26–2.71) |
| ER (−) | 103/437 | 95/429 | 0.89 (0.64–1.25) | 68/363 | 130/503 | 1.25 (0.88–1.78) | 32/168 | 166/698 | 1.18 (0.76–1.85) | 53/258 | 145/608 | 1.01 (0.70–1.46) |
| PR (+) | 38/437 | 34/429 | 0.92 (0.56–1.49) | 33/363 | 39/503 | 0.81 (0.49–1.32) | 15/168 | 57/698 | 0.88 (0.48–1.59) | 21/258 | 51/608 | 0.98 (0.57–1.67) |
| PR (−) | 65/437 | 61/429 | 0.87 (0.57–1.33) | 35/363 | 91/503 | 1.80 (1.12–2.89) | 17/168 | 109/698 | 1.53 (0.83–2.83) | 32/258 | 94/608 | 1.03 (0.64–1.64) |
| HER2 (+) | 52/437 | 51/429 | 0.98 (0.63–1.53) | 43/363 | 60/503 | 0.95 (0.60–1.50) | 20/168 | 83/698 | 1.00 (0.57–1.76) | 31/168 | 72/698 | 0.90 (0.56–1.46) |
| HER2 (−) | 47/437 | 44/429 | 0.85 (0.53–1.37) | 25/363 | 66/503 | 1.66 (0.99–2.79) | 12/168 | 79/698 | 1.39 (0.72–2.70) | 22/258 | 69/608 | 1.08 (0.64–1.83) |
Odds ratios (OR) and 95% confidence intervals (CI) were calculated using unconditional logistic regression with the */*, #/#, S/S and low dose of biallelic‐repeats genotypes as reference groups, respectively, and adjusted for age and age at menarche. Bold values indicate statistical significance. *, other repeat alleles rather than 5 repeats; #, other repeat alleles rather than 6 repeats; +, positive; −, negative; ER, estrogen receptor; HER2, epidermal growth factor receptor 2; L, long (≥6); PR, progesterone receptor; S, short (≤5).
Because overexpression and secretion of PTTG1IP modulates induction of breast cancer cell invasion11 and overexpression of its interacting protein PTTG1 promotes lymph node metastasis in esophageal squamous cell carcinoma,14 we evaluated a possible correlation between the PTTG1IP VNTR genotype and ALNM status in the case cohort (Table 5). In ER‐positive and HER2‐negative cases, we found that genotypes with the five‐repeats allele were associated with significantly reduced metastasis risk compared with those who do not carry the five‐repeats allele (OR, 0.53; 95% CI, 0.30–0.94). This preliminary observation suggests that PTTG1IP VNTR polymorphism might be a possible genetic modifier of ALNM metastasis risk in patients with ER‐positive and HER2‐negative breast cancer.
Table 5.
Association of PTTG1IP variable number of tandem repeats (VNTR) with metastasis risk stratified by immunochemical index
| Characters | Genotype of PBF VNTR (metastasis/no metastasis) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Status with 5 repeats | Status with 6 repeats | Status with repeat length | Biallelic‐repeats dose | |||||||||
| */* | 5/* + 5/5 | OR (95% CI) | #/# | 6/# + 6/6 | OR (95% CI) | S/S | S/L + L/L | OR (95% CI) | Low | High | OR (95% CI) | |
| ER (+) | 94/84 | 54/77 | 0.66 (0.41–1.06) | 44/55 | 104/106 | 1.19 (0.72–1.96) | 15/19 | 133/142 | 0.97 (0.46–2.04) | 33/32 | 115/129 | 0.81 (0.45–1.46) |
| PR (+) | 76/78 | 47/71 | 0.69 (0.42–1.14) | 36/52 | 87/97 | 1.30 (0.77–2.23) | 15/19 | 108/130 | 0.89 (0.42–1.90) | 29/30 | 94/119 | 0.73 (0.39–1.35) |
| PR (−) | 18/6 | 7/6 | 0.39 (0.08–2.00) | 8/3 | 17/9 | 0.35 (0.05–2.39) | 0/0 | 25/12 | n.d | 4/2 | 21/10 | 1.09 (0.11–10.58) |
| HER2 (+) | 33/27 | 18/16 | 0.95 (0.37–2.47) | 19/10 | 32/33 | 0.69 (0.24–1.96) | 6/4 | 45/39 | 0.82 (0.20–3.36) | 12/11 | 39/32 | 1.48 (0.48–4.57) |
| HER2 (−) | 59/56 | 33/60 | 0.53 (0.30–0.94) | 22/44 | 70/72 | 1.76 (0.95–3.26) | 9/15 | 83/101 | 1.15 (0.47–2.83) | 21/21 | 71/95 | 0.70 (0.35–1.42) |
| ER (−) | 58/36 | 61/31 | 1.23 (0.64–2.37) | 43/21 | 76/46 | 0.82 (0.41–1.65) | 18/12 | 101/55 | 1.29 (0.55–3.06) | 31/16 | 88/51 | 0.89 (0.42–1.87) |
| PR (+) | 19/18 | 18/15 | 1.14 (0.44–2.96) | 17/15 | 20/18 | 0.95 (0.36–2.50) | 8/7 | 29/26 | 0.93 (0.29–2.94) | 12/9 | 25/24 | 0.74 (0.26–2.10) |
| PR (−) | 39/18 | 43/16 | 1.32 (0.52–3.35) | 26/6 | 56/28 | 0.42 (0.13–1.39) | 10/5 | 72/29 | 1.48 (0.39–5.62) | 19/7 | 63/27 | 0.88 (0.29–2.70) |
| HER2 (+) | 29/20 | 37/12 | 1.86 (0.74–4.68) | 29/13 | 37/19 | 0.86 (0.34–2.17) | 14/5 | 52/27 | 0.86 (0.26–2.84) | 21/7 | 45/25 | 0.68 (0.24–1.96) |
| HER2 (−) | 27/15 | 24/19 | 0.76 (0.29–2.01) | 14/8 | 37/26 | 0.95 (0.30–3.02) | 4/7 | 47/27 | 2.93 (0.73–11.84) | 10/9 | 41/25 | 1.35 (0.45–4.02) |
Odds ratios (OR) and 95% confidence intervals (CI) were calculated using unconditional logistic regression with the */*, #/#, S/S and low dose of biallelic‐repeats genotypes as reference groups, respectively, and adjusted for age and age at menarche. Bold values indicate statistical significance. *, other repeat alleles rather than five repeats; #, other repeat alleles rather than six repeats; +, positive; −, negative; ER, estrogen receptor; HER2, epidermal growth factor receptor 2; L, long (≥6); n.d, no data; PR, progesterone receptor; S, short (≤5).
To clarify the functional significance that underlines the observed association between breast cancer risk and the VNTR polymorphism at the PTTG1IP promoter, we compared the binding affinity of ERα to the variable cis‐elements with EMSA in ERα‐positive MCF‐7 cells (Fig. 1). All of the six biotin‐labelled oligonucleotide probes, corresponding to cis‐regulatory alleles with 3, 4, 5, 6, 7 or 8 tandem repeats, generated a specific shift band of DNA/nuclear protein complex, which could be competed out by incubation with unlabelled competitor probes of PTTG1IP ERE oligonucleotide (Fig. 1A). Furthermore, we observed an obvious correlation between the thickness of bands for different probes and the number of repeats they contain (Fig. 1B). The binding specificity could be evidenced by the observation that the consensus ERα binding site competitor probe could abolish the binding shift band in a dose‐dependent manner: the higher the dose of ERE competitor probe, the larger the amount of free probes without binding and less pronounced shift band with bound probes (Fig. 1C, take the eight tandem repeats probe as an example). These results clearly reveal that an increase in the number of tandem repeats enhances the binding affinity of ERα to its response elements in the PTTG1IP promoter. When the EMSA results were superimposed with previously reported observations that the VNTR containing cis‐element confers oestrogen responsiveness, and the tumor specimen and normal breast tissue from subjects with greater numbers of ERE repeats showed higher mRNA expression,11 we can conclude that a lengthened element with more repeats can enhance transactivation of the transforming gene PTTG1IP and is thus associated with increased risk of ER‐positive breast cancer.
Figure 1.
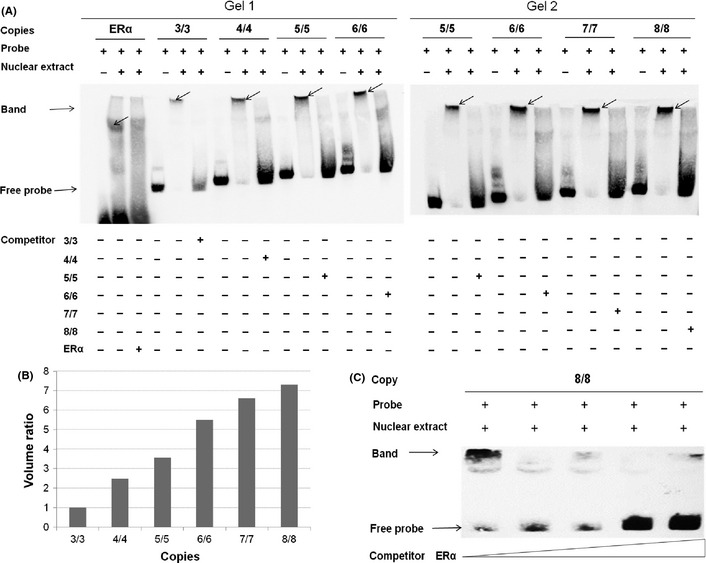
Electrophoretic mobility shift assays of variable number of tandem repeats (VNTR)‐containing region in the PTTG1IP promoter. (A) Electrophoretic mobility shift assay (EMSA) with biotin‐labelled oligonucleotides containing 3, 4, 5, 6, 7 and 8 repeats, respectively, at the rs72416745 site and nuclear extracts from the MCF‐7 cell line. Lanes 4, 7, 10 and 13 in gel 1 and lanes 1, 4, 7 and 10 in gel 2 show mobilities of labelled oligonucleotides without nuclear extracts; lanes 2, 5, 8, 11 and 14 in gel 1 and lanes 2, 5, 8 and 11 in gel 2 represent those (estrogen receptor α [ERα] consensus, 3, 4, 5 and 6 repeats in gel 1, and 5, 6, 7 and 8 repeats in gel 2, respectively) with nuclear extracts in the absence of competitor; specificity of nuclear protein binding (Band) was demonstrated by competition with unlabelled oligonucleotides (lanes 3, 6, 9, 12 and 15 in gel 1 and lanes 3, 6, 9 and 12 in gel 2). Due to a significant difference in the sequence length between the positive control oligonucleotides of ERα consensus (30 bp, lane 2 in gel 1) and cis‐oligonucleotides of 3/3 repeats (90 bp, lane 5 in gel 1), the band sizes of complexes respectively derived from ERα and the two cis‐elements vary in gel shift. (B) The thickness and intensity of DNA/nuclear protein complex bands for respective repeats were measured in BioRad Gel Doc EQ System using Quantity One software and compared with 3/3 repeats as a reference. (C) ERα‐specific binding was demonstrated by competition with ERα consensus unlabeled oligonucleotides in a dose‐dependent manner; eight repeats for example.
Discussion
Breast cancer is a common complex disease caused by genetic and environmental factors. So far, linkage analyses, candidate gene‐based and especially recent genome‐wide association studies (GWAS) have uncovered three types of susceptible genes according to the penetrance or the extent of effect on the phenotype of their alleles: rare but high‐penetrance susceptible genes such as BRCA1 and BRCA2; rare moderate‐penetrance genes functionally related to BRCA1 and/or BRCA2 such as CHEK2, ATM, BRIP1 and PALB2; and around 20 common but low‐penetrance variants in 19 genes or loci.15, 16 Although these genes and loci significantly broadened our understanding of the genetic architecture of breast cancer, they are estimated to account for only approximately one‐third of the inherited cause of the disease.16 The present study epidemiologically characterizes, for the first time to the best of our knowledge, the relevance of a functional regulatory VNTR variation in PTTG1IP to breast cancer etiology, adding into the disease allelic spectrum this polymorphic transforming gene as a new candidate susceptible factor.
PTTG1IP was first identified through its ability to interact with PTTG1, a multifunctional protein overexpressed in several endocrine and endocrine‐related tumors, including pituitary, thyroid, breast and ovarian carcinomas.17 Overexpression of PTTG1 induces aneuploidy, adversely affects DNA repair and genetic instability, and thus induces cellular transformation in vitro and tumorigenesis in vivo.18 PTTG1IP binds to PTTG1, facilitates its nuclear translocation and transcriptional activation function, which might augment the transforming ability of PTTG1.17 Meanwhile, PTTG1IP shows cellular transforming and tumorigenic ability in its own right and is proposed as a novel proto‐oncogene in thyroid and breast cancers.11, 19 Interestingly, both PTTG1 and PTTG1IP genes are transcriptional targets of ERα; parallel overexpression of their mRNA and protein has been observed in endocrine tumor compared with normal tissues.18 As functionally characterized in MCF‐7 cells by Watkins et al.11 and our group, PTTG1IP mRNA and protein expression are induced by estrogen, and ERα binds to PTTG1IP promoter that confers transactivational estrogens responsiveness. The naturally occurring VNTR polymorphism at the ERE‐containing promoter of PTTG1IP manifests differential transactivational ability, an increase in the number of repeats correlated with enhanced binding affinity of ERα and higher PTTG1IP mRNA expression. A biophysical mechanism could be invoked to explain the enhanced ERα binding associated with a greater number of ERE repeats. When a transcription factor is bound to one repeat unit of a VNTR, it will dynamically stay in the same region of DNA even after initial disassociation due to sliding over and attaching to another copy of the binding site, resulting in enhanced interaction with its response element.20 In keeping with these findings, the present case–control study revealed that individuals carrying long alleles with more repeats and of higher transactivational capability were at increased risk of developing breast cancer. Our results are therefore biologically plausible and provide epidemiological evidence to further support the hypothesis proclaiming PTTG1IP as one novel proto‐oncogene in breast cancer.
Breast cancer is an etiologically heterogeneous disorder with specific histopathological subtypes defined by several biomarkers such as ER, PR and HER2 used in routine clinical management. In primary stratification analyses by ER status (Table 4), we found that the modification of cancer risk consistently conferred by genotypes of PTTG1IP VNTR regarding six repeats, long length of more repeats, or a high dose of biallelic repeats, were all confined to ER‐positive but not ER‐negative status. Furthermore, when the status of PR and HER2 were taken into account, these pronounced associations were observed only in the ER(+)‐PR(+) and ER(+)‐HER2(−) cases. Some explanations could partly account for the heterogeneity in the strength of association with respect to the risk of tumor subtypes. Approximately 65% of ER‐positive breast cancers are also PR positive, and there is a high correlation between ER and PR expression.21 PTTG1IP is a proto‐oncogene in breast cancer directly targeted by ERα and the VNTR hypervariable sequence contains cis‐element that confers responsiveness to estrogen.11 In the meantime, the estrogen–ER complex can also directly bind to a cis‐element within ERBB2, the coding gene of HER2, but repress its transcription,22 resulting in a strong inverse correlation between HER2 and ERα expression.23 It is thus conceivable that the association of functional PTTG1IP VNTR polymorphism was consistently more pronounced in the ER(+)‐PR(+) and ER(+)‐HER2(−) subtypes. However, we also observed that genotypes with the six‐repeats allele were associated with cancer risk in the ER(−)‐PR(−) subtype. It is possible that PR even have a part in tumor etiology beyond its role as a co‐expressor with ER;24 we do not know the reason for this discrepancy. Therefore, further studies with a larger sample size and combined markers classification are needed to confirm the heterogeneous associations of PTTG1IP VNTR with breast cancer risk in relation to specific tumor subtypes.
The current GWAS of complex diseases are almost solely focused on two forms of sequence variations, SNP and copy number variations, but are of relative paucity of research interest in VNTR, the third form of sequence variation. In fact, there are more than one million VNTR dispersed in the human genome, some of which occur in obviously functional positions, such as gene promoters or enhancers targeted by trans‐acting factors, and possibly affect gene regulation and thereby potentially contribute to disease etiology,20 as exemplified in our previous studies25, 26 and the present investigation. It is notable that the functional VNTR variation at the PTTG1IP promoter is highly polymorphic in humans and differences in allelic types and frequencies among populations are apparent. In the previous report by Watkins et al.11, the PTTG1IP promoter contains between 1 and 6 repeats, as found in 122 British subjects, and the allele of three repeats is the only one prevalent with a high frequency (approximately 80%). However, in our Chinese population, alleles of 6 (35%) and 5 (30%) repeats were most common, alleles of 7 (17%) and 4 (11%) repeats were relatively common, but alleles with smaller or a greater number of repeats were significantly underrepresented, especially the allele of three repeats, which was remarkably rare (approximately 2%). The pronounced difference in allelic frequencies between populations indicated that the extent of functionality of PTTP1IP in ERα signalling and its etiological role in breast cancer might vary in different ethnic groups, suggesting that replication of association studies in other populations are highly warranted.
We should acknowledge that this study is the first genetic association report on PTTG1IP and cancer with some limitations. First, we did not genotype other neutral VNTR polymorphisms as genomic control to address potential population stratification between the two panels and between the respective cases and controls. Whereas in our previous genomic dissection of population substructure of Han Chinese,27 subjects from the two cities in the north of China were clearly grouped into one cluster of northern Han. The fact that allelic frequencies in the two panels are comparable and genotypic frequencies among the controls and cases fit the HWE further support the randomness of subject selection. Therefore, it is at least possible that confounding factors such as population stratification and ascertainment bias could account for the observed association. Second, as an exploratory effort, we also genotyped two common linked SNP (rs1862391 and rs1862392) at the PTTG1 promoter, but did not observe a significant association with cancer risk (data not shown), which is in line with a previous report in a Swedish prospective study.28 Even though cis‐variants of PTTG1IP and PTTG1 were the focus in the present study due to their lack of apparently functional common non‐synonymous SNP, multiple tagSNP and haplotype‐based association studies are needed to clarify possible gene–gene interaction in the etiology of breast cancer.18 Third, we revealed significant and consistent associations between breast cancer risk and genotypes of long alleles with more repeats as well as a high dose of biallelic repeats; however, in the present study we did not observe a significant trend of association for the number of repeats and cancer risk due to the relatively small sample size of subgroups of this highly variable PTTG1TIP VNTR polymorphism (data not shown), which is to be tested further with statistically well‐powered association study with larger sample size.
In conclusion, the present work confirmed differential functionality of PTTG1IP promoter VNTR polymorphism, and provided epidemiological evidence that its regulatory variants were associated with the risk of ER‐positive breast cancer, further supporting the hypothesis proclaiming PTTG1IP as one novel proto‐oncogene in breast cancer. Given that PTTG1IP is a secreted protein implicated in the development and metastasis of breast cancer, genotyping the VNTR polymorphism at the promoter of its coding gene and measuring its serum concentration particularly for patients with ER‐positive cancer (using ELISA, for example) may provide a potential informative biomarker for the prevention and prognosis of breast cancer. Because this is the first genetic association study on PTTG1IP polymorphism, replication investigations and large population‐based prospective studies in ethnically diverse groups are needed to verify these findings.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
The authors thank Jianshi Liu and Xiaochen Kou for their experimental assistance with this work. This study was supported by grants from the National Natural Science Foundation of China (30971593, 81172093 and 30890034), grants from the National Science and Technology Major Project (2008ZX10002‐019 and 2012ZX10002‐011), Program for New Century Excellent Talents in University (NCET‐07‐0204), Shanghai Rising‐Star Program (07QA14006), Shanghai Pujiang Program (11PJD005) and Scientific Research Program of Shanghai Municipal Health Bureau (2007Y53).
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Yang L, Parkin DM, Ferlay J, Li L, Chen Y. Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer Epidemiol Biomarkers Prev 2005; 14: 243–50. [PubMed] [Google Scholar]
- 3. Cheskis BJ, Greger JG, Nagpal S, Freedman LP. Signaling by estrogens. J Cell Physiol 2007; 213: 610–7. [DOI] [PubMed] [Google Scholar]
- 4. Chen C, Sakoda LC, Doherty JA et al Genetic variation in CYP19A1 and risk of breast cancer and fibrocystic breast conditions among women in Shanghai, China. Cancer Epidemiol Biomarkers Prev 2008; 17: 3457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang L, Gu L, Qian B et al Association of genetic polymorphisms of ER‐alpha and the estradiol‐synthesizing enzyme genes CYP17 and CYP19 with breast cancer risk in Chinese women. Breast Cancer Res Treat 2009; 114: 327–38. [DOI] [PubMed] [Google Scholar]
- 6. Ding SL, Yu JC, Chen ST et al Diverse associations between ESR1 polymorphism and breast cancer development and progression. Clin Cancer Res 2010; 16: 3473–84. [DOI] [PubMed] [Google Scholar]
- 7. Dunning AM, Healey CS, Baynes C et al Association of ESR1 gene tagging SNPs with breast cancer risk. Hum Mol Genet 2009; 18: 1131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kristensen VN, Harada N, Yoshimura N et al Genetic variants of CYP19 (aromatase) and breast cancer risk. Oncogene 2000; 19: 1329–33. [DOI] [PubMed] [Google Scholar]
- 9. Welboren WJ, Sweep FC, Span PN, Stunnenberg HG. Genomic actions of estrogen receptor alpha: what are the targets and how are they regulated? Endocr Relat Cancer 2009; 16: 1073–89. [DOI] [PubMed] [Google Scholar]
- 10. Yu JC, Hsiung CN, Hsu HM et al Genetic variation in the genome‐wide predicted estrogen response element‐related sequences is associated with breast cancer development. Breast Cancer Res 2011; 13: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watkins RJ, Read ML, Smith VE et al Pituitary tumor transforming gene binding factor: a new gene in breast cancer. Cancer Res 2010; 70: 3739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H, Hao B, Zhou K et al Linkage disequilibrium and haplotype architecture for two ABC transporter genes (ABCC1 and ABCG2) in Chinese population: implications for pharmacogenomic association studies. Ann Hum Genet 2004; 68: 563–73. [DOI] [PubMed] [Google Scholar]
- 13. Rousset F. genepop'007: a complete re‐implementation of the genepop software for Windows and Linux. Mol Ecol Resour 2008; 8: 103–6. [DOI] [PubMed] [Google Scholar]
- 14. Yan S, Zhou C, Lou X et al PTTG overexpression promotes lymph node metastasis in human esophageal squamous cell carcinoma. Cancer Res 2009; 69: 3283–90. [DOI] [PubMed] [Google Scholar]
- 15. Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet 2008; 9: 321–45. [DOI] [PubMed] [Google Scholar]
- 16. Zhang B, Beeghly‐Fadiel A, Long J, Zheng W. Genetic variants associated with breast‐cancer risk: comprehensive research synopsis, meta‐analysis, and epidemiological evidence. Lancet Oncol 2011; 12: 477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chien W, Pei L. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor‐transforming gene product. J Biol Chem 2000; 275: 19422–7. [DOI] [PubMed] [Google Scholar]
- 18. Smith VE, Franklyn JA, McCabe CJ. Pituitary tumor‐transforming gene and its binding factor in endocrine cancer. Expert Rev Mol Med 2010; 12: e38. [DOI] [PubMed] [Google Scholar]
- 19. Stratford AL, Boelaert K, Tannahill LA et al Pituitary tumor transforming gene binding factor: a novel transforming gene in thyroid tumorigenesis. J Clin Endocrinol Metab 2005; 90: 4341–9. [DOI] [PubMed] [Google Scholar]
- 20. Breen G, Collier D, Craig I, Quinn J. Variable number tandem repeats as agents of functional regulation in the genome. IEEE Eng Med Biol Mag 2008; 27: 103–4. [DOI] [PubMed] [Google Scholar]
- 21. Yang XR, Pfeiffer RM, Garcia‐Closas M et al Hormonal markers in breast cancer: coexpression, relationship with pathologic characteristics, and risk factor associations in a population‐based study. Cancer Res 2007; 67: 10608–17. [DOI] [PubMed] [Google Scholar]
- 22. Hurtado A, Holmes KA, Geistlinger TR et al Regulation of ERBB2 by oestrogen receptor‐PAX2 determines response to tamoxifen. Nature 2008; 456: 663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ali S, Coombes RC. Endocrine‐responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2002; 2: 101–12. [DOI] [PubMed] [Google Scholar]
- 24. Broeks A, Schmidt MK, Sherman ME et al Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet 2011; 20: 3289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Liu Y, Tan W et al Association of the variable number of tandem repeats polymorphism in the promoter region of the SMYD3 gene with risk of esophageal squamous cell carcinoma in relation to tobacco smoking. Cancer Sci 2008; 99: 787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang Y, Wang W, Wang J et al Functional regulatory variants of MCL1 contribute to enhanced promoter activity and reduced risk of lung cancer in nonsmokers: implications for context‐dependent phenotype of an antiapoptotic and antiproliferative gene in solid tumor. Cancer 2011; doi: 10.1002/cncr.26502. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 27. Xu S, Yin X, Li S et al Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet 2009; 85: 762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brendle A, Brandt A, Johansson R et al Single nucleotide polymorphisms in chromosomal instability genes and risk and clinical outcome of breast cancer: a Swedish prospective case–control study. Eur J Cancer 2009; 45: 435–42. [DOI] [PubMed] [Google Scholar]


