Abstract
Cancer stem cells (CSCs) are predicted to be critical drivers of tumor progression due to their “stemness”, but the molecular mechanism of CSCs in regulating metastasis remains to be elucidated. Epithelial‐mesenchymal transition (EMT), hypoxia‐inducible factor (HIF)‐1α, and miR‐21, all of which contribute to cell migration for metastasis, are interrelated with CSCs. In the present study, third‐sphere forming (3‐S) CSC‐like cells, which showed elevated CSC surface markers (ALDH1+ and CD44+/CD24−/low) and sphereforming capacity as well as migration and invasion capacities, were cultured and isolated from breast cancer MCF‐7 parental cells, to evaluate the role of miR‐21 in regulating the CSC‐like cell biological features, especially EMT. EMT, which was assessed by overexpression of mesenchymal cell markers (N‐cadherin, Vimentin, alpha‐smooth muscle actin [α‐SMA]) and suppression of epithelial cell marker (E‐cadherin), was induced in 3‐S CSC‐like cells. Moreover, both of HIF‐1α and miR‐21 were upregulated in the CSC‐like cells. Interestingly, antagonism of miR‐21 by antagomir led to reversal of EMT, downexpression of HIF‐1α, as well as suppression of invasion and migration, which indicates a key role of miR‐21 involved in regulate CSC‐associated features. In conclusion, we demonstrated that the formation of CSC‐like cells undergoing process of EMT‐like associated with overexpression of HIF‐1α, both of which are regulated by miR‐21. (Cancer Sci 2012; 103: 1058–1064)
Cancer stem cells (CSCs) are predicted to be the cell origin of the tumor and responsible for tumor progression, relapse and metastasis due to their self‐renewal capacity and limitless proliferative potential, as well as invasion and migration capacity.1, 2, 3, 4, 5 Therefore, the development of successful cancer therapeutic regimen requires targeting the CSCs, such as the elucidation of molecular pathways, which regulate CSC features.
Recently, breast cancer cells forming mammospheres in suspension cultures were generally acknowledged as breast cancer CSCs (bCSCs) due to their self‐renewal capacity,6, 7 while mammospheres were also accepted as bCSC‐like cell models, enriching bCSCs. Besides, bCSCs also could be identified and isolated according to cell surface markers such as aldehyde dehydrogenase 1+ (ALDH1+; ALDHbright)2 and the phenotype of CD44+/CD24−/low.1 But now, the isolation and culture of high‐purity bCSC model is still one of the “choke points” in bCSC research.
For most epithelial tumors, including breast cancer, progression toward malignancy is accompanied by a process of epithelial‐mesenchymal transition (EMT), which is characterized by a loss of epithelial differentiation and a shift towards mesenchymal phenotype.8 The EMT towards a more mesenchymal phenotype involves downexpression of epithelial markers (e.g. E‐cadherin and Keratins) and upexpression of mesenchymal markers (e.g. N‐cadherin, Vimentin, alpha‐smooth muscle actin [α‐SMA]), as well as increased cell mobility and invasive phenotype.9, 10, 11 Accumulating evidence demonstrated that the induction of EMT in vitro in transformed mammary epithelial cells12, 13 or in vivo in epithelial breast cancer cells in mouse models14 generates cells with bCSC characteristics. This suggests that through the EMT program breast epithelial cells can gain bCSC characteristics. Moreover, bCSCs enriched from breast tumors and metastatic breast pleural effusions express markers similar to cells that have undergone a process of EMT.12, 15, 16 Similarly, EMT and bCSC markers are frequently associated with breast cancers that have a propensity to metastasis, such as basal‐like17 and metaplastic18 breast cancers.
Increasingly evidence suggests that a pivotal role of hypoxic niche in CSCs development in a variety of human cancers,19, 20 and the role of hypoxia‐inducible factor (HIF) in these cells has become an important focus in understanding tumor malignant behavior.21 Hypoxia‐inducible factor is a heterodimeric transcription factor, composed of HIF‐1α and HIF‐1β. Hypoxia‐inducible factor‐1α is induced by hypoxia, growth factors and oncogenes; whereas HIF‐1β protein is constitutively expressed in cells.22 A previous study demonstrated that culture in hypoxia promotes expansion of the sub‐population of cells positive for CSC marker CD133 through activation of HIF‐1α in glioma.23 Another study identified a direct molecular link between HIF‐1α and Notch signal pathway,24 and the latter is one of the CSC‐associated signal pathways.25 These results indicate that HIF‐1α may be an important factor associated with bCSCs.
MicroRNA‐21 (miR‐21), serve as an oncomiR, has been found to be frequently overexpressed in different solid tumors, including breast cancer.26, 27 Previous studies have demonstrated that miR‐21 played a key role in enhancing tumor malignant behavior including invasion and migration,28, 29, 30 as well as the potential importance of miR‐21 expression as a marker of poor prognosis in breast cancer.31 Corresponding to the fact that miR‐21 plays a critical role in EMT32, 33 associated with CSC signatures,32 it is suggested that miR‐21 plays a key role in the process of EMT and the formation of CSC‐like cells.
On the basis of these findings, we believe that all or partly of miR‐21, EMT and HIF‐1α are involved in the formation process of CSC‐like cells. So we undertook to associate miR‐21, EMT markers and HIF‐1α with bCSCs, with the hope that such associations might provide insights into the causal mechanisms of bCSC‐like cell invasion and metastasis.
Materials and Methods
Cell culture
Human breast cancer MCF‐7 parental (P) cells were grown with high‐glucose Dulbecco's modified eagle medium (DMEM) (Gibco, Grand Island, NY, USA), supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin and 10 μg/mL insulin, and incubated in a humidified atmosphere with 5% CO2 at 37°C.
Mammosphere culture
Mammosphere culture was performed as previously described.6, 7 Single‐cell suspensions were suspended in a serum‐free DMEM/F12 (Gibco) supplemented with 2% B27 (Invitrogen, Carlsbad, CA, USA), 20 ng/mL EGF (PeproTech, London, UK), 20 ng/mL bFGF (PeproTech), 0.4% bovine serum albumin (BSA) (Sigma, St. Louis, MO, USA), and 5 μg/mL insulin, and seeded into six‐well non‐adherent plates at a density of 5 × 103–1 × 104 cells/mL in primary culture and 1 × 103–5 × 103 cells/mL in the following passages. Cultures were fed weekly and passaged every 2 weeks. We named the first generation mammosphere, second generation mammosphere, third generation mammosphere, fourth generation mammosphere as “first‐sphere (1‐S),” “second‐sphere (2‐S),” “third‐sphere (3‐S),” “fourth‐sphere (4‐S),” respectively.
ALDEFLUOR assay
The ALDEFLUOR kit (Aldagen, Durham, NC, USA) was used to isolate the cell population with high ALDH enzyme activity (ALDHbright), enriching bCSCs, as previously described.2 Briefly, cells were incubated in ALDEFLUOR assay buffer containing ALDH substrate (BAAA, 1 μmol/L) for 30 min at 37°C. In each experiment a sample of cells was stained under identical conditions with 50 mmol/L of diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor, as negative control. Flow cytometry (FACS) analysis was used to measure ALDHbright cell subpopulation.
FACS of CD44+/CD24−/low cell subpopulation
Flow cytometry of CD44+/CD24−/low cell subpopulation was performed as previously described.34 Briefly, unfixed cells were incubated with fluorescein isothiocyanate (FITC)‐labeled anti‐CD44 antibody and phycoerthrin (PE)‐labeled anti‐CD24 antibody (BioLegend, San Diego, CA, USA) at 4°C in darkness for 1 h. Cells were analyzed by FACS using CellQuest software (BD Biosciences, Franklin Lakes, NJ, USA).
Western blot analysis and antibodies
Western blot analysis was conducted as per our previous study.35 Briefly, the protein was separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) (8%, 10% or 12%) and transferred to polyvinylidene fluoride (PVDF) membranes. Non‐specific binding sites were blocked by incubating with tris buffered saline with tween 20 (TBST) containing 5% (w/v) non‐fat dried milk. Then incubated with primary antibodies and horse radish peroxidase (HRP)‐conjugated anti‐rabbit immunoglobulin (IgG) secondary antibody in order, and visualized by enhanced chemiluminescence (ECL). β‐actin or glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as loading control.
Primary antibodies including rabbit antihuman ALDH1 antibody (No: ab51028; 1:1000; Abcam, Cambridge, UK), CD44 antibody (No: BA0321; 1:500; Boster, Wuhan, China), N‐cadherin antibody (No: BS2224; 1:750; Bioworlde, St. Louis, MO, USA), E‐cadherin antibody (No: BS1098; 1:1000; Bioworlde), Vimentin antibody (No: BS1776; 1:750; Bioworlde), α‐SMA antibody (No: ab5694; 1:700; Abcam), HIF‐1α antibody (No: sc‐10790; 1:600; Santa Cruz, CA, USA), HIF‐1β antibody (No: sc‐5580; 1:600; Santa Cruz), β‐actin antibody (No: BA2305; 1:1000; Boster) and GAPDH antibody (No: AP0063; 1:1000; Bioworlde), and HRP‐conjugated anti‐rabbit IgG secondary antibody (No: BA1055; 1:2500; Boster) were used for western blot analysis.
Immunofluorescence
P cells were grown on glass chamber slides while 3‐S cells were suspended in eppendorf (EP) tubes. Both cells were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X‐100 and blocked with 0.5% BSA. Then the cells were incubated with rabbit antihuman ALDH1 antibody (No: ab51028; 1:100; Abcam) or CD44 antibody (No: BA0321; 1:50; Boster), and FITC‐conjugated AffiniPure Goat Anti‐Rabbit IgG (H+L) secondary antibody (No: BS10950; 1:200; Bioworlde), and then stained nuclei with 4′, 6‐diamidino‐2‐phenylindole (DAPI).
Real time RT‐PCR analysis
All primers (Data S1) involve in real time RT‐PCR analysis were synthesized by Sangon Biotech (Shanghai, China). For miR‐21, miRNAs were extracted from cells using a mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). SYBR Green‐based real time RT‐PCR was performed using SYBR PrimeScript miRNA RT‐PCR Kit (Takara, Dalian, China) to measure the expression of mature miR‐21 in cells by a MiniOpticon Two‐Color Real‐Time PCR Detection System (Bio‐Rad, Hercules, CA, USA). U6 was used as endogenous control.
For mRNAs, total RNA from cells was isolated using TRIzol reagent (Invitrogen). Real time RT‐PCR reactions were carried out using SYBR Premix Ex Taq II (Takara). β‐actin was used as endogenous control.
Migration and invasion assay
Cell migration and invasion assays were performed as we described previously.35 For invasion assay, 2.5 × 104 cells were seeded on an 8‐μm‐pore size Transwell filter insert (Corning Inc, Corning, NY, USA) coated with extracellular matrix (ECM) (1:7.5) (Sigma), while cell migration assay did not coat with ECM. After 48 h of incubation at 37°C and 5% CO2, cells adherent to the upper surface of the filter were removed. Cells were stained with 0.4% crystal violet dissolved in methanol, and the numbers of cells on the bottom were counted.
Hsa‐miR‐21 antagomir transfection
Cells (5 × 105) were seeded in six‐well plates and grown to 60% confluence. Human has‐miR‐21 (MIMAT0000076) antagomir or its negative control (Ribobio, Guangzhou, China) was directly transfected into 3‐S cells in free of serum Opti‐MEM I (Invitrogen) at a final concentration of 50 nmol, according to the manufacturer's protocol.
Statistical analysis
Statistical analysis was performed using the Student's t‐test.
Results
3‐S cells serve as high‐purity bCSC‐like cell models with superior mammosphere forming efficiency, ALDH bright activity and CD44+/CD24−/low phenotype
To establish a high‐purity bCSC‐like cell model, all of 1‐S, 2‐S, 3‐S, and 4‐S cells were prolonged cultured from breast cancer MCF‐7 P cells, then the mammosphere forming efficiency (MSFE), ALDHbright activity and CD44+/CD24−/low were measured in the above cells. We found that the MSFE gradually increased in 1‐S, 2‐S, and 3‐S cells in turn (2‐S vs 1‐S, P < 0.0001; 3‐S vs 2‐S, P < 0.0001;), except for 4‐S cells (vs 3‐S cells; P = 0.3664), by suspended culture (Fig. 1); accompanied with that ALDHbright activity gradually increased in P, 1‐S, 2‐S and 3‐S cells in turn (1‐S vs P, P = 0.0004; 2‐S vs 1‐S, P < 0.0001; 3‐S vs 2‐S, P < 0.0001), but not for 4‐S cells (vs 3‐S cells, P = 0.2151), by ALDEFLUOR assay (Fig. 1); as well as CD44+/CD24−/low phenotype gradually increased in P, 1‐S, 2‐S, 3‐S and 4‐S cells in turn (1‐S vs P, P = 0.0018; 2‐S vs 1‐S, P = 0.0008; 3‐S vs 2‐S, P < 0.0001; 4‐S vs 3‐S, P = 0.0497), by FACS (Fig. 1). These data demonstrated that the CSC‐like characteristics of 3‐S cells were the highest among these in all of the cell groups based on an overall consideration of various factors, which was once again proved by western blot analysis (Fig. 1) and immunofluorescence assay (Fig. 1), indicating that 3‐S could serve as a high‐purity bCSC‐like cell model is selected for further study.
Figure 1.
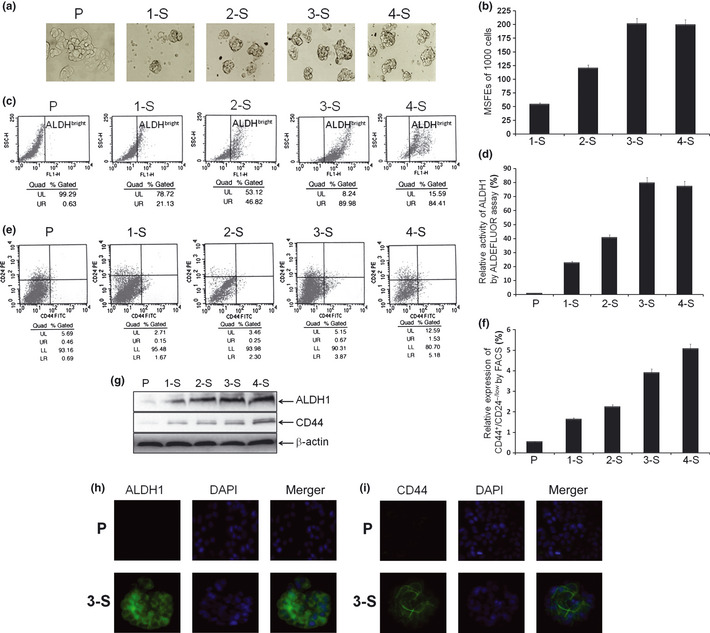
Culture and identification of mammospheres. (a) Typical images of 1‐S, 2‐S, 3‐S, 4‐S. (b) The mammosphere forming efficiencies (MSFEs) were calculated and analysis based on 1000 single cells that were initially seeded. (c,d) Examples and analysis of ALDH bright cell subpopulation in indicated cells, by ALDEFLUOR assay. (e,f) Examples and analysis of CD44+/CD24−/low phenotype cell subpopulation in indicated cells, by fluorescence‐activated cell sorting (FACS). (g) The protein levels of ALDH1 and CD44 in indicated cells by western blot analysis. (h) Immunofluorescence images of the P cells and 3‐S cells stained with antibody against ALDH1 (green), and cell nuclei were stained with 4′6′‐diamidino‐2‐phenylindole dihydrochloride (DAPI) (blue). (i) Immunofluorescence images stained with antibody against CD44 (green) and DAPI (blue).
The bCSC‐like cells underwent EMT‐like process
To examine the differences on EMT phenotype between bCSC‐like cells and P cells, the mRNA and protein levels of EMT markers were measured. The relative mRNA levels of N‐cadherin, Vimentin and α‐SMA were significantly elevated in 3‐S cells compared with P cells (P = 0.0007, P = 0.0016, and P = 0.0459, respectively; Fig. 2), whereas the relative mRNA level of E‐cadherin was significantly decreased (P = 0.0055; Fig. 2), as assessed by real time RT‐PCR assay. The results from western blot analysis demonstrated that the relative protein levels of N‐cadherin, Vimentin and α‐SMA were also increased (Fig. 2), whereas that of E‐cadherin was diminished in the bCSC‐like cells, as compared with P cells (Fig. 2). These results indicate that the expressions of mesenchymal phenotype cell biomarkers were elevated, while expression of epithelial phenotype cell biomarker was decreased, further suggesting that EMT phenotype was acquired in the bCSC‐like cells as compared to P cells.
Figure 2.
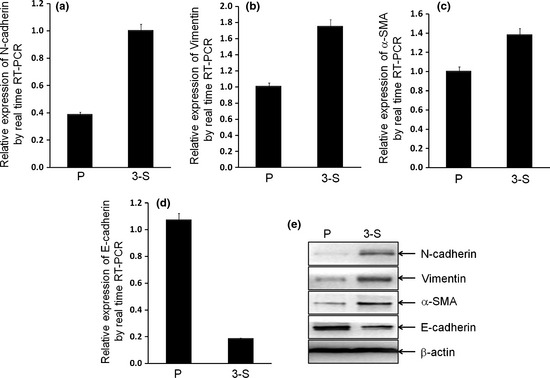
Breast cancer stem cell (bCSC)‐like cells induced epithelial‐mesenchymal transition (EMT) phenotype. (a–c) The mRNA levels of mesenchymal markers (N‐cadherin, Vimentin and alpha‐smooth muscle actin [α‐SMA]) in 3‐S bCSC‐like cells were significantly overexpressed (P = 0.0007, P = 0.0016, and P = 0.0459, respectively), while (d) the mRNA level of epithelial marker (E‐cadherin) was significantly decreased (P = 0.0055), by real time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis. (e) The relative protein levels of EMT markers were shown, by western blotting analysis.
Overexpression of HIF‐1α in the bCSC‐like cells
To investigate the role of HIF‐1 in the process of P cells to the bCSC‐like cells, the relative mRNA and protein levels of HIF‐1α and HIF‐1β were measured. The relative mRNA level of HIF‐1α was significantly increased in bCSC‐like cells as compared with P cells (P = 0.0079; Fig. 3), but not for HIF‐1β (P = 0.187; Fig. 3), by real time RT‐PCR assay. The results from western blot analysis demonstrated that the relative protein levels of HIF‐1α and HIF‐1β accorded with the corresponding mRNA level above (Fig. 3). The results suggested that HIF‐1α, but not HIF‐1β, is involved in the formation of the bCSC‐like cells.
Figure 3.
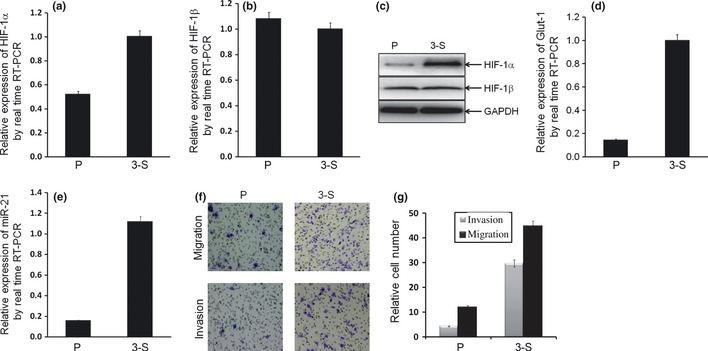
Breast cancer stem cell (bCSC)‐like cells showed hypoxia‐inducible factor (HIF)‐1α and miR‐21 overexpression, as well as more invasion and migration properties. (a,b) The relative mRNA expression of HIF‐1α in 3‐S bCSC‐like cells were significantly upregulated (a; P = 0.0079), but not for HIF‐1β (b; P = 0.187). (c) The relative protein levels of HIF‐1α and HIF‐1β were shown, by western blotting analysis. (d) The relative mRNA expression of Glut‐1 in 3‐S bCSC‐like cells were significantly upregulated (P = 0.0017). (e) The relative expression of miR‐21 in 3‐S bCSC‐like cells were significantly upregulated (P = 0.0018). (f,g) Cell invasion and migration properties in bCSC‐like cells significantly increased (P < 0.0001 and P < 0.0001, respectively), by Transwell assay.
To show if Hif‐1α is actually working and active in the bCSC‐like cells, the relative mRNA level of Glut‐1, a direct downstream gene of HIF‐1α,36 was measured. The relative level of Glut‐1 mRNA was significantly increased in the bCSC‐like cells as compared with P cells (P = 0.0017; Fig. 3), by real time RT‐PCR assay. The result indicated that HIF‐1α is actually working and active in the bCSC‐like cells.
Overexpression of miR‐21 in the bCSC‐like cells
To investigate the role of miR‐21, an EMT‐associated miRNA, in the process of MCF‐7 P cells to the bCSC‐like cells, the relative expression of miR‐21 was examined by real time RT‐PCR assay. As expected, the relative expression of miR‐21 was significantly increased in the bCSC‐like cells as compared to P cells (>sevenfold; P = 0.0018; Fig. 3), which suggested that miR‐21 is involved in the formation of the bCSC‐like cells, even as it is involved in the regulation of EMT‐like process in the bCSC‐like cells.
The bCSC‐like cells promoted invasion and migration property
To confirm the cell invasion and migration property in the bCSC‐like cells, Transwell assay was performed. The relative cell numbers of invasion and migration were significantly increased in the bCSC‐like cells as compared with P cells (P < 0.0001, P < 0.0001, respectively; Fig. 3), which indicated a positive role of bCSC‐like cells in breast cancer invasion and metastasis.
Antagonism of miR‐21 reversed EMT phenotype, HIF‐1α expression consistent with invasion and migration in the bCSC‐like cells
To determine whether miR‐21 regulates the bCSC‐like cells associated EMT phenotype, 3‐S bCSC‐like cells with mesenchymal phenotype were selected for further study. Has‐miR‐21 antagomir was transfected into 3‐S cells that led to a 92% reduction of mature miR‐21 expression (Fig. 4; P = 0.0027). The antagomir treatment decreased protein levels of N‐cadherin, α‐SMA and Vimentin, whereas the increased level of E‐cadherin protein in the bCSC‐like cells (Fig. 4), which suggested that antagonism of miR‐21 led a reversal of EMT phenotype in the bCSC‐like cells. Moreover, antagomir treatment also decreased protein expression of HIF‐1α in the bCSC‐like cells (Fig. 4), which suggested that HIF‐1α is one of the downstream molecular of miR‐21. More interestingly, antagomir treatment significantly decreased the relative cell numbers of invasion and migration (P < 0.0001, P < 0.0001, respectively; Fig. 4), as well as the MSFE (P < 0.0001, Fig. 4), which indicated a positive role for miR‐21 in invasion and migration, and even “stemness” in the bCSC‐like cells.
Figure 4.
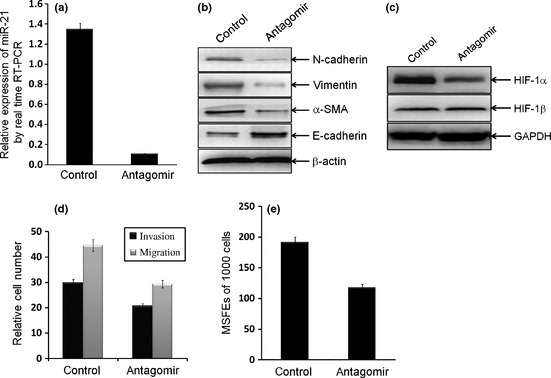
Antagonism of miR‐21 reversed epithelial‐mesenchymal transition (EMT) phenotype and hypoxia‐inducible factor (HIF)‐1α expression, as well as invasion and migration in breast cancer stem cell (bCSC)‐like cells. (a) Antagonism of miR‐21 was established in 3‐S bCSC‐like cells by transfected with hsa‐miR‐21 antagomir. (b,c) Antagomir reversed EMT phenotype (b) and HIF‐1α expression (c), but not for HIF‐1β (c), in bCSC‐like cells. The relative protein levels of EMT markers, HIF‐1α and HIF‐1β were shown. (d) Antagonism of miR‐21 significantly decreased cell invasion and migration properties in bCSC‐like cells (P < 0.0001 and P < 0.0001, respectively), by Transwell assays. (e) Antagonism of miR‐21 significantly decreased the mammosphere forming efficiency (MSFE) of bCSC‐like cells (P < 0.0001).
Discussion
In this study, we prolonged cultured a 3D bCSC‐like cell model – 3‐S cells arise from MCF‐7 P cells. The bCSC‐like cells showed an EMT phenotype and overexpression of HIF‐1α and miR‐21. Functionally, antagonism of miR‐21 led to reversal of EMT‐like process and HIF‐1α expression, even with the migratory and self‐renewal abilities in the bCSC‐like cells.
We found that, for the first time, both of MSFE and ALDHbright activity gradually increased with a high rate in 1‐S, 2‐S and 3‐S cells in turn, but not for 4‐S cells (vs 3‐S cells) (Fig. 1); while the other bCSC phenotype biomarker CD44+/CD24−/low gradually increased with a low rate in 1‐S, 2‐S, 3‐S and 4‐S cells (Fig. 1). Moreover, the bCSC‐like cells also showed overexpression of ALDH1 and CD44, by western blot analysis (Fig. 1) and immunofluorescence assay (Fig. 1), which could be associated with cancer progression because overexpression of ALDH1 and CD44 have been found in poorly differentiated tumors.1, 2, 37, 38, 39 On the basis of these results, we believed that the bCSC‐like characteristics of 3‐S cells is more significant than the other cell groups in this study, and selected 3‐S to serve as a high‐purity bCSC model to further study. More interestingly, the difference of expression between ALDHbright and CD44+/CD24−/low indicates that the bCSC‐like cells with bearing ALDHbright and CD44+/CD24−/low have different molecular mechanisms, even as cell origin, suggesting that both ALDHbright and CD44+/CD24−/low may not serve as the best universal markers for bCSC identification and isolation.
Next, a characteristic EMT phenotype was evidenced, by decreased epithelial specific markers and increased mesenchymal markers, in the bCSC‐like cells. This process involves a disassembly of cell–cell junctions, including downregulation of E‐cadherin and upregulation of mesenchymal molecular markers such as N‐cadherin, Vimentin and α‐SMA (Fig. 2), in accordance with EMT‐related theory.9, 10, 11 The results suggested that MCF‐7 P cells with epithelial cell phenotype acquired mesenchymal cell phenotype with 3‐D mammosphere morphology in the bCSC‐like cells, indicating EMT is a process that is reminiscent of bCSC‐like cell characteristics, which was consistent with bCSCs being similar to cells that have undergone a process of EMT.12, 15, 16 In addition, mRNA and protein expression of HIF‐1α, which is known to contribute to hypoxia “niche” as well as cell invasion and migration, was also elevated in the bCSC‐like cells (Fig. 3). These findings indicated that HIF‐1α plays some role in bCSC‐like cells, which was similar to that activation of HIF‐1α promoted expansion of the CSC‐like cells in glioma.23
In accordance with the expression of EMT markers, miR‐21 was markedly increased in the bCSC‐like cells with EMT phenotype (Fig. 3), which was similar to miR‐21 links to CSC characteristics as well as EMT phenotype in pancreatic cancer cells.32 These findings indicated that miR‐21 plays an important role in the bCSC‐like cells. Functionally, cell invasion and migration abilities were increased in the bCSC‐like cells (Fig. 3), which was consistent in that the mesenchymal cells have less adhesion between cells than their epithelial counterparts, allowing for more motile and invasive characteristics, which contribute to cancer cell invasion and metastasis,9, 10, 11 the results are also explained by using ALDH1 overexpression in the bCSC‐like cells, because ALDH1+ bCSCs promote breast cancer invasion and metastasis.5
More interestingly, antagonism of miR‐21 in the bCSC‐like cells showed partial reversal of EMT phenotype as documented by increased expression of E‐cadherin and decreased expression of N‐cadherin, Vimentin and α‐SMA (Fig. 4). These results suggest that miR‐21 regulates the expression of E‐cadherin, N‐cadherin, Vimentin and α‐SMA, and that antagonism of miR‐21 could be useful for the reversal of EMT phenotype to mesenchymal‐epithelial transition (MET) in the bCSC‐like cells, further indicating that antagonism of miR‐21 might partly eliminate bCSC‐like cells by reversing EMT phenotypic cells, but this needs further study. Moreover, antagonism of miR‐21 also decreased the expression of HIF‐1α as well as cell invasion, migration, and self‐renewal abilities in the bCSC‐like cells (Fig. 4). These findings provided a new insight into the regulation of EMT and HIF‐1α, and demonstrated that miR‐21 plays a key role in invasion and migration, even as “stemness” of the bCSC‐like cells. We also found that antagonism of miR‐21 could suppress the expression of endogenous miR‐21 almost completely (~92% reduction; Fig. 4), but the reduction efficiencies of cell migration and invasion properties are not correspondingly high (both ~30% reduction; Fig. 4). These findings raise the possibility that there are additional candidate genes that are not regulated by miR‐21 (e.g. Twist or Snail) to promote cell migration and invasion, and the correlated mechanism needs further investigation.
In summary, we cultured MCF‐7 3‐S cells and identified them as having cellular and molecular characteristics of bCSC‐like cells shared EMT phenotype as well as overexpression of HIF‐1α, while demonstrating that miR‐21 plays a critical role in linking bCSC signatures with EMT phenotype and HIF‐1α. We firmly believe that strategies of miR‐21 could either reverse the EMT to MET phenotype in bCSC‐like cells, and could even selectively kill bCSC‐like cells, which would become a novel approach for the prevention of tumor progression and metastasis.
Disclosure statement
The authors have no conflict of interest.
Supporting information
Data S1. Primers representations in Doc S1–Doc S10.
Acknowledgments
This work was supported in part by National Natural Science Foundation of China (NSFC 81072147, NSFC 31171336), the Natural Science Foundation of Chongqing Science & Technology Commission, China. We also thank the members of the Department of Endocrine Surgery, the First Affiliated Hospital of Chongqing Medical University, and the Institute of Pediatrics, Children's Hospital of Chongqing Medical University, for technical support.
References
- 1. Al‐Hajj M, Wicha MS, Benito‐Hernandez A et al Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 2003; 100: 3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ginestier C, Hur MH, Charafe‐Jauffret E et al ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007; 1: 555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell 2007; 1: 241–2. [DOI] [PubMed] [Google Scholar]
- 4. Sottoriva A, Verhoeff JJ, Borovski T et al Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res 2010; 70: 46–56. [DOI] [PubMed] [Google Scholar]
- 5. Charafe‐Jauffret E, Ginestier C, Iovino F et al Aldehyde dehydrogenase 1‐positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res 2010; 16: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dontu G, Abdallah WM, Foley JM et al In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 2003; 17: 1253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ponti D, Costa A, Zaffaroni N et al Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell progerties. Cancer Res 2005; 65: 5506–11. [DOI] [PubMed] [Google Scholar]
- 8. Thiery JP. Epithelial‐mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2: 442–54. [DOI] [PubMed] [Google Scholar]
- 9. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial‐mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7: 131–42. [DOI] [PubMed] [Google Scholar]
- 10. Lee JM, Dedhar S, Kalluri R et al The epithelial‐mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 2006; 172: 973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeisberg M, Neilson EG. Biomarkers for epithelial‐mesenchymal transitions. J Clin Invest 2009; 119: 1429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mani SA, Guo W, Liao MJ et al The epithelial‐mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morel AP, Lievre M, Thomas C et al Generation of breast cancer stem cells through epithelial‐mesenchymal transition. PLoS ONE 2008; 3: e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santisteban M, Reiman JM, Asiedu MK et al Immune‐induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Cancer Res 2009; 69: 2887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aktas B, Tewes M, Fehm T et al Stem cell and epithelial‐mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 2009; 11: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shipitsin M, Campbell LL, Argani P et al Molecular definition of breast tumor heterogeneity. Cancer Cell 2007; 11: 259–73. [DOI] [PubMed] [Google Scholar]
- 17. Sarrio D, Rodriguez‐Pinilla SM, Hardisson D et al Epithelial‐mesenchymal transition in breast cancer relates to the basal‐like phenotype. Cancer Res 2008; 68: 989–97. [DOI] [PubMed] [Google Scholar]
- 18. Hennessy BT, Gonzalez‐Angulo AM, Stemke‐Hale K et al Characterization of a naturally occurring breast cancer subset enriched in epithelial‐to‐mesenchymal transition and stem cell characteristics. Cancer Res 2009; 69: 4116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heddleston JM, Li Z, McLendon RE et al The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009; 8: 3274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bar EE, Lin A, Mahairaki V et al Hypoxia increases the expression of stem‐cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol 2010; 177: 1491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hill RP, Marie‐Egyptienne DT, Hedley DW. Cancer stem cells, hypoxia and metastasis. Semin Radiat Oncol 2009; 19: 106–11. [DOI] [PubMed] [Google Scholar]
- 22. Semenza GL. Targeting HIF‐1 for cancer therapy. Nat Rev Cancer 2003; 3: 721–32. [DOI] [PubMed] [Google Scholar]
- 23. Soeda A, Park M, Lee D et al Hypoxia promotes expansion of the CD133‐positive glioma stem cells through activation of HIF‐1alpha. Oncogene 2009; 28: 3949–59. [DOI] [PubMed] [Google Scholar]
- 24. Gustafsson MV, Zheng X, Pereira T et al Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell 2005; 9: 617–28. [DOI] [PubMed] [Google Scholar]
- 25. Jögi A, Øra I, Nilsson H et al Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest‐like phenotype. Proc Natl Acad Sci USA 2002; 99: 7021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iorio MV, Ferracin M, Liu CG et al MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65: 7065–70. [DOI] [PubMed] [Google Scholar]
- 27. Volinia S, Calin GA, Liu CG et al A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Si ML, Zhu S, Wu H et al miR‐21‐mediated tumor growth. Oncogene 2007; 26: 2799–803. [DOI] [PubMed] [Google Scholar]
- 29. Yan LX, Wu QN, Zhang Y et al Knockdown of miR‐21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res 2011; 13: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang TH, Wu F, Loeb GB et al Upregulation of miR‐21 by HER2/neu signaling promotes cell invasion. J Biol Chem 2009; 284: 18515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan LX, Huang XF, Shao Q et al MicroRNA miR‐21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008; 14: 2348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bao B, Wang Z, Ali S et al Notch‐1 induces epithelial‐mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett 2011; 307: 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Braun J, Hoang‐Vu C, Dralle H et al Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 2010; 29: 4237–44. [DOI] [PubMed] [Google Scholar]
- 34. Liu M, Casimiro MC, Wang C et al p21CIP1 attenuates Ras‐ and c‐Myc‐dependent breast tumor epithelial mesenchymal transition and cancer stem cell‐like gene expression in vivo . Proc Natl Acad Sci USA 2009; 106: 19035–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu M, Ju X, Willmarth NE et al Nuclear factor‐kappaB enhances ErbB2‐induced mammary tumorigenesis and neoangiogenesis in vivo . Am J Pathol 2009; 174: 1910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sapra P, Kraft P, Pastorino F et al Potent and sustained inhibition of HIF‐1α and downstream genes by a polyethyleneglycol‐SN38 conjugate, EZN‐2208, results in anti‐angiogenic effects. Angiogenesis 2011; 14: 245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morimoto K, Kim SJ, Tanei T et al Stem cell marker aldehyde dehydrogenase 1‐positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci 2009; 100: 1062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neumeister V, Agarwal S, Bordeaux J et al In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am J Pathol 2010; 176: 2131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kai M, Onishi H, Souzaki M et al Semi‐quantitative evaluation of CD44(+)/CD24(‐) tumor cell distribution in breast cancer tissue using a newly developed fluorescence immunohistochemical staining method. Cancer Sci 2011; 102: 2132–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Primers representations in Doc S1–Doc S10.


