SUMMARY
Protein isoforms generated by alternative splicing contribute to proteome diversity. Because of the lack of effective techniques, the isoform-specific function, expression, localization, and signaling of endogenous proteins are unknown for most genes. Here, we report a genetic method, isoTarget, for multi-purpose studies of targeted isoforms in select cells. Applying isoTarget to two isoforms of Drosophila Dscam, Dscam[TM1] and [TM2], we found that, in neurons, endogenous Dscam[TM1] is in dendrites, whereas Dscam[TM2] is in both dendrites and axons. We demonstrate that the difference in subcellular localization, rather than biochemical properties, leads to the two isoforms’ functional differences. Moreover, we show that the subcellular enrichment of functional partners results in a DLK/Wallenda-Dscam[TM2]-Dock signaling cascade in axons. We further apply isoTarget to study two isoforms of a GABA receptor to demonstrate its general applicability. isoTarget is an effective technique for studying how alternative splicing enhances proteome complexity.
Graphical Abstract
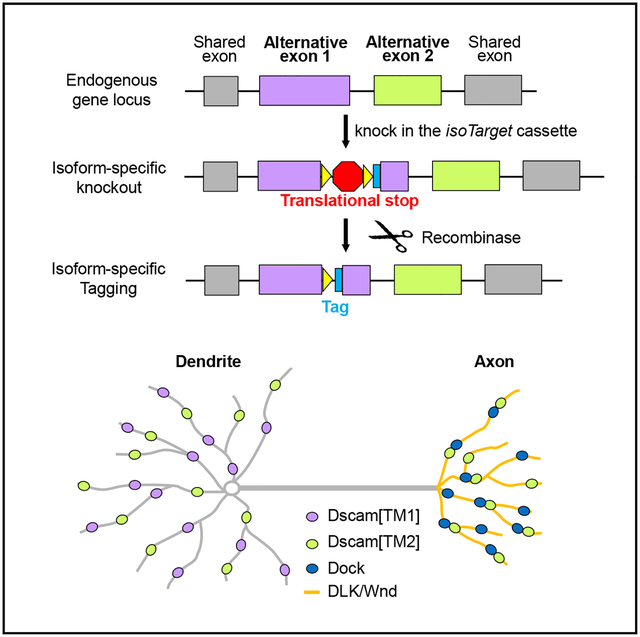
In Brief
Liu et al. develop a genetic method that enables the investigation of isoform-specific function, expression, localization, and signaling of endogenous proteins in select cells. Using this method, they demonstrate that the difference in subcellular localization of two isoforms of Down syndrome cell adhesion molecule leads to functional differences between them.
INTRODUCTION
Alternative splicing is a fundamental biological process that expands proteome diversity in eukaryotes. Genome-wide transcriptome analyses have shown that 90%–95% of human genes encode two or more isoforms (Baralle and Giudice, 2017). The percentage of multi-exonic genes that undergo alternative splicing is estimated to be 63% in mice, 45% in Drosophila, and 25% in Caenorhabditis elegans (Lee and Rio, 2015). Alternative splicing is regulated by the coordination of RNA-binding proteins, RNA polymerase II, and epigenetic modifications of DNA (Baralle and Giudice, 2017). Perturbations of RNA splicing cause neurodevelopmental, cardiovascular, and other diseases (Baralle and Giudice, 2017; Scotti and Swanson, 2016). Despite the importance of alternative splicing, how distinct protein isoforms resulting from alternative splicing differ in their functions and regulations is poorly understood. In fact, the cellular functions, endogenous expression and localization, and signaling cascades of individual splicing isoforms are only known for a very few genes (Baralle and Giudice, 2017).
Protein isoforms encoded by alternative exons often differ in their structures and biochemical properties, which lead to the distinct functions of the isoforms (Kelemen et al., 2013). In addition, different types of cells may express different splicing variants (Baralle and Giudice, 2017), which further diversifies the biological functions of different isoforms. Intriguingly, different protein isoforms of some genes are localized to distinct subcellular compartments within a cell (Baralle and Giudice, 2017; Kelemen et al., 2013; Lee et al., 2016; Lerch et al., 2012; Yap and Makeyev, 2016). Compared with our understanding of how biochemical and expressional differences contribute to distinct functions of splicing isoforms, much less is known about whether and how isoform-specific subcellular localization contributes to distinct cellular functions. This is a challenging problem because solving it requires manipulating a specific isoform at its endogenous locus—without affecting other isoforms—in a cell-specific fashion.
Transgene-mediated overexpression of splicing variants of interest is widely used for studying isoform-specific functions and subcellular localization in specific cells. However, it is well documented that overexpressed proteins often do not mimic the endogenous proteins in their spatiotemporal expression, localization, and functions (Baralle and Giudice, 2017; Kelemen et al., 2013; Moriya, 2015; Prelich, 2012).
Here, we report a genetic method, termed isoTarget, for studying isoform-specific functions, localizations, and signaling of endogenous splicing isoforms of interest. This method allows us to knock out select isoforms for functional studies and to tag the endogenous proteins conditionally for multi-disciplinary analyses in specific cells without affecting other isoforms. To achieve that, we created a translational stop sequence, used it to generate a cleavable cassette that contained an epitope tag for conditional tagging, and inserted the cassette into the exon encoding the isoform of interest. As a proof-of-concept, we applied isoTarget to study two mutually exclusive isoforms of Drosophila Down syndrome cell-adhesion molecule (Dscam), Dscam[TM1] and [TM2] (Schmucker et al., 2000; Wang et al., 2004; Zhan et al., 2004). We demonstrate that isoform-specific functions for Dscam[TM1] and [TM2] result from the distinct endogenous subcellular localization patterns of these two isoforms. We further describe a compartment-specific signaling pathway in the axon terminals, which involves Dscam[TM2], but not [TM1], because of the differential localization of the two isoforms and their functional partners. Furthermore, we applied isoTarget to study two isoforms of the GABA receptor resistance to dieldrin (Rdl), illustrating the general applicability of the method. These findings illustrate the versatility of isoTarget in isoform studies and its effectiveness in uncovering mechanisms governing the expansion of proteome diversity by alternative splicing in vivo. In addition, they establish the causality between the subcellular localization and cellular function of Dscam splicing isoforms, demonstrating the critical role of subcellular localization in expanding the functional diversity of splicing isoforms.
RESULTS
The Design of isoTarget
The transcriptional stop cassettes commonly used for conditional knockouts or knockins (Lakso et al., 1992) do not specifically disrupt the expression of a select splicing isoform and are, hence, not applicable for isoform-specific studies. This is because RNA splicing occurs after transcription and, consequently, transcriptional stop cassettes disrupt the expression of all isoforms downstream of the targeted isoform (Figure 1A). We created a translational stop (tlstop) sequence by introducing multiple stop codons (TAA, TAG, or TGA) into the DNA sequence encoding a non-catalytic region of β-galactosidase (β-Gal) (Figures 1A and S1A). By design, the presence of a tlstop in an alternatively spliced exon would lead to isoform-specific protein truncation or nonsense-mediated mRNA decay (NMD) (Brogna and Wen, 2009; Smith and Baker, 2015), causing the targeted isoform to lose its function (Figure 1A). By contrast, other alternatively spliced exons would not be affected.
Figure 1. Design of isoTarget and Its Application to Studying the Functional Diversity of Splicing Isoforms.
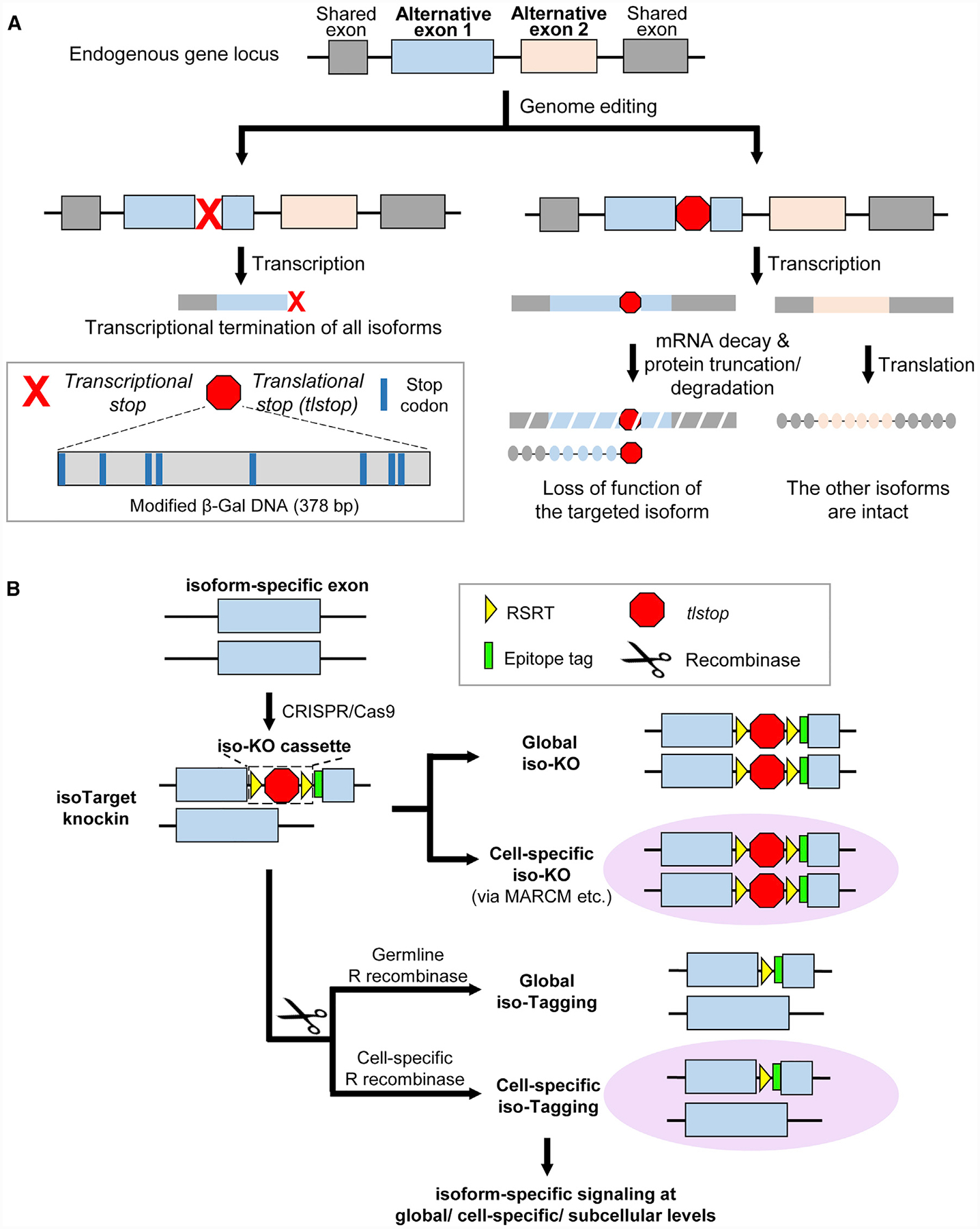
(A) Introducing translational stops into alternative exons allows isoform-specific manipulations of the gene. Inserting the commonly used transcriptional stop cassette into an alternative exon leads to transcriptional termination of all isoforms downstream of the targeted exon (left branch). To specifically manipulate one isoform (right branch), we engineered a translational stop (tlstop) cassette by introducing multiple stop codons into the DNA sequence encoding a non-catalytic region of β-Gal (bottom left). This tlstop cassette causes loss of function in targeted isoform by translational truncation or, possibly, mRNA decay.
(B) Combining tlstop with other genetic methods for multi-purpose studies of the targeted splicing isoform. The isoTarget cassette, which consists of an RSRT site, a translational stop (tlstop), a second RSRT site, and an epitope tag, is inserted into an isoform-specific exon by CRISPR/Cas9-mediated genome editing. The insertion is expected to generate a loss-of-function allele of the targeted isoform. Single cells that are homozygous for the targeted allele can be produced by genetic mosaic techniques, such as MARCM. The endogenous isoforms can be visualized in specific cells by selective expression of R recombinase to remove the RSRT-tlstop-RSRT cassette. Expression of R recombinase in female germline cells leads to tagging of the isoform in all cells that express that isoform in the progeny (“global iso-tagging”). Expression of R recombinase in specific cell types or single cells leads to tagging of the isoform in those cells. In the iso-tagged flies, upstream regulators and downstream effectors of specific isoforms can be identified through genetic, cell biological, and biochemical analyses.
See also Figures S1–S3 and S7.
To achieve cell-type-specific labeling of targeted isoforms, the tlstop sequence is flanked by two R recombinase recognition sites (RSRTs) and then followed by an epitope tag (Chen et al., 2014; Nern et al., 2011). When R recombinase is expressed to remove tlstop, the epitope tag is inserted in-frame into the targeted isoform, allowing the detection and biochemical analyses of endogenous proteins in cells of interest (Figure 1B). We refer to the RSRT-tlstop-RSRT as “iso-KO cassette” (for “isoTarget knockout cassette”), because its insertion into an isoform-specific exon is expected to create a loss-of-function mutant of that particular isoform. In Drosophila, the iso-KO alleles can be used in combination with mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999) to study the isoform-specific function in targeted single neurons (Figure 1B). We refer to the epitope tagging resulting from the excision of the tlstop sequence as “iso-tagging”. As shown below, the iso-tagging approach allows the investigation of upstream and downstream signaling mechanisms that involve targeted isoforms at the organismal, cell-type-specific, or subcellular levels (Figure 1B).
The Validation of isoTarget and Mitigation of Off-Target Effects of Translational-Stop Cassettes
For the designed applications, the isoTarget technique should meet the following requirements. First, the iso-KO cassette must abolish the function of the targeted isoform. Second, iso-KO or iso-tagging of one isoform should not impair the expression of other isoforms. Third, iso-tagging, which results from the excision of the iso-KO cassette, should restore the isoform functions disrupted by the iso-KO cassette. We determined whether isoTarget met these requirements by testing it on the Drosophila Dscam gene.
In Drosophila, exons 17.1 and 17.2 of the Dscam gene encode two different transmembrane and juxtamembrane domains (TM) (Schmucker et al., 2000). Alternative splicing of these two mutually exclusive exons produces two isoforms called Dscam[TM1] and Dscam[TM2] (Figure S1B). We inserted the iso-KO cassette into the juxtamembrane domain in exon 17.1 (TM1) and 17.2 (TM2) (Figures S1B and S1C). In homozygous Dscam[TM2] iso-KO (Dscam[TM2]iso-KO) larvae, the axon terminal growth was dramatically impaired in the classIV dendritic arborization (C4 da) neurons (Figures 2A, 2B, and 2H), a widely used model for studying dendrite and axon development (Grueber and Jan, 2004; Grueber et al., 2007; Jan and Jan, 2010; Ye et al., 2007). This is consistent with previous reports that Dscam is required for axon terminal growth in C4 da neurons (Kim et al., 2013) and suggests that the iso-KO cassette abolishes Dscam[TM2] functions. Excision of the iso-KO cassette allowed us to generate homozygous global Dscam[TM2] iso-tagging larvae (Dscam[TM2]iso-tagging) (Figures S1C). C4 da axon terminals of Dscam[TM2]iso-tagging larvae were indistinguishable from those of the wild type (Figures S2A and S2B), suggesting that tagging endogenous Dscam[TM2] with the V5-tag does not disrupt the function of the isoform.
Figure 2. IsoTarget Uncovers a Specific Role for the Dscam[TM2] Isoform in Axon Terminal Growth.
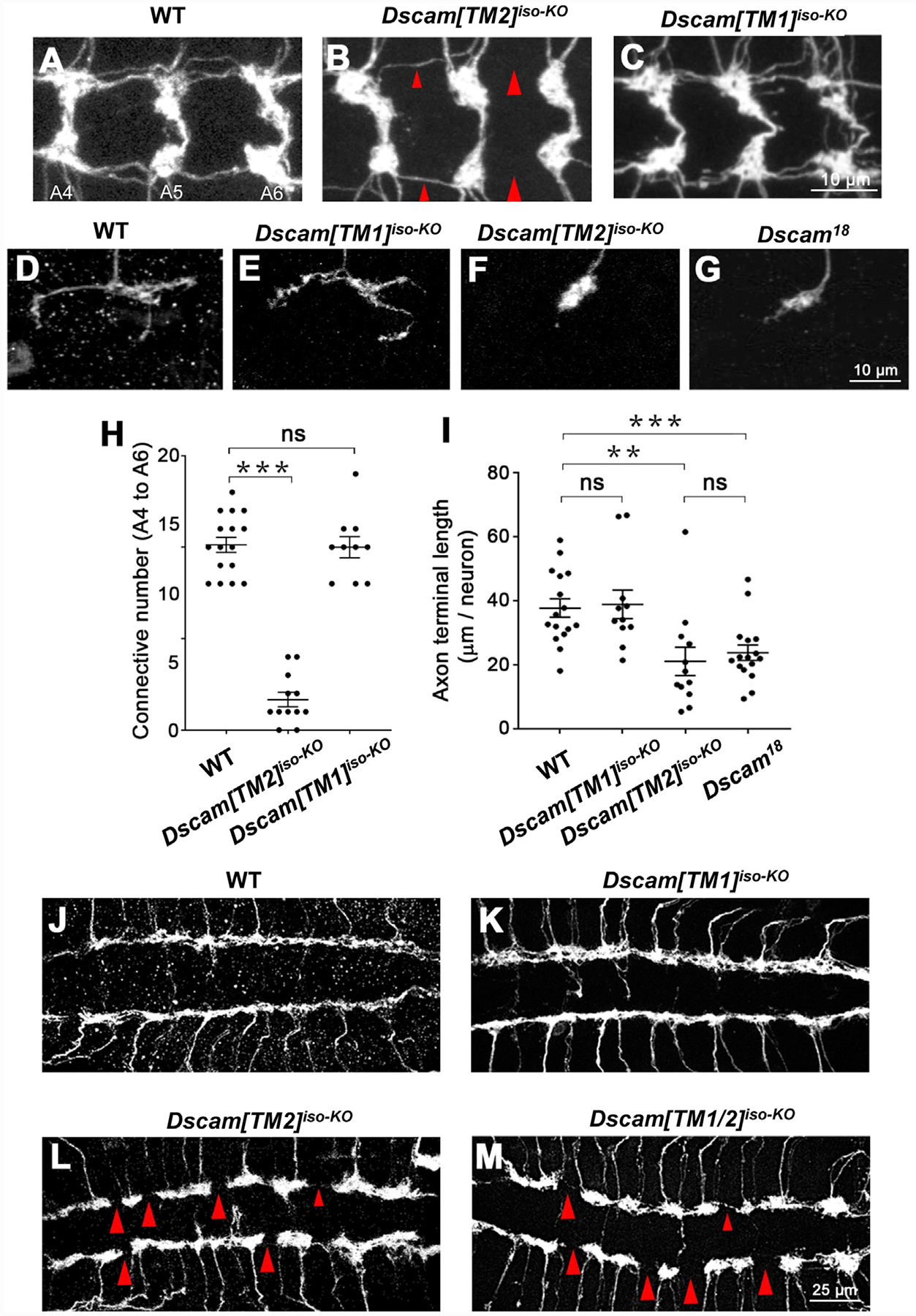
(A–C) Compared with wild-type (WT) (A), global Dscam[TM2]iso-KO (B), but not Dscam[TM1]iso-KO (C), impairs axon terminal growth in C4 da neurons. The C4 da-specific driver ppk-Gal4 was used to label all C4 da axon terminals in the CNS. Shown are representative images of abdominal segments 4 to 6 (A4–A6). The large red arrowheads point to the sites at which longitudinal axon tracts are broken, and the small arrowheads point to where the tracts are thinned. See also Figures S2 and S3.
(D–G) Dscam[TM2], but not [TM1], is required for the growth of axon terminals in single C4 da neurons. The MARCM technique was used to generate single GFP-labeled C4 da neurons that were homozygous of the indicated alleles. Dscam [TM1]iso-KO has no effect on axon terminal growth (D) and (E), whereas Dscam[TM2]iso-KO reduced the length of axon terminals to the same level as the loss of both isoforms in Dscam18 (F) and (G).
(H) Quantification of the number of axon connectives (i.e., the longitudinal branches) from A4 to A6. Unless specified otherwise, means ± SEM are shown in all figures. Comparisons among multiple groups were performed by one-way ANOVA, followed by Student’s t test, and those between two groups were performed with a Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001; ns: not significant (p > 0.05).
(I) Quantification of presynaptic terminal length in the C4 da neuron ddaC.
(J–M) Dscam[TM2], but not [TM1], is required for the growth of axon terminals in C3 da neurons. The longitudinal axon tracts of C3 da neurons in the CNS remain intact in Dscam[TM1]iso-KO (J) and (K), but are disrupted in Dscam[TM2]iso-KO (L) or in mutants lacking both isoforms (Dscam[TM1/2]iso-KO) (M). The large red arrowheads point to the sites at which longitudinal axon tracts are broken; the small red arrowheads point to the sites where longitudinal axon tracts are thinned. Sample numbers (VNCs): WT, 10; Dscam[TM1]iso-KO, 11; Dscam[TM2]iso-KO, 14; Dscam[TM1/2]iso-KO, 12.
Next, we determined whether the isoTarget of one isoform affected the expression of another isoform by using reverse-transcription real-time PCR and immunohistochemistry on isoTarget samples. Dscam[TM1] mRNA levels were not affected in the central nervous system (CNS) of homozygous Dscam [TM2]iso-KO or Dscam[TM2]iso-tagging third-instar larvae (Figure S2C). Unexpectedly, inserting the iso-KO cassette into Dscam[TM1] abolished Dscam[TM2] mRNA and protein expression, creating a loss-of-function mutant for both Dscam[TM1] and [TM2] (Dscam[TM1/2]iso-KO) (Figures S2D, S2E, and S2F). By contrast, the expression of Dscam[TM2] mRNA was not affected in Dscam[TM1]iso-tagging (Figure S2D). These results suggest that inserting a long piece of DNA in the TM1-encoding exon disrupts the splicing of the TM2-encoding exon of Dscam premRNA (Figure S2I), possibly by overly extending the distance between exon 16 and the TM2-encoding exon (Anastassiou et al., 2006; Yue et al., 2013). To test that possibility, we reduced the size of the iso-KO cassette from 561 bp to 285 bp by cutting a 276-bp fragment from the tlstop sequence. Strikingly, insertion of the short iso-KO cassette in [TM1] did not impair Dscam [TM2] expression, including both mRNA and protein expression (Figures S2D, S2G, and S2H). These studies demonstrate that the effect of an isoTarget cassette on off-target isoforms could be mitigated by reducing the cassette length and found a new translational stop sequence for achieving that.
We further assessed the possibility that iso-KO cassettes introduce aberrant exons in the mRNAs. Nested PCRs were applied to enhance the detection sensitivity of possible aberrant exons between Dscam exon 16 and 18 in cDNA samples from larval CNS (Figure S3). No aberrant band was observed in either Dscam[TM1]iso-KO or Dscam[TM2]iso-KO (Figures S3B), suggesting that the iso-KO cassette is unlikely to introduce aberrant exons during alternative splicing.
Using reverse-transcription real-time PCR, we observed that, although iso-KO cassettes did not affect the mRNAs of the isoform that lacks the cassette, they reduced the mRNA levels of the targeted isoform. Both the regular and short iso-KO cassettes reduced the mRNA levels of targeted Dscam isoforms to around 50% (Figure S3C). This is likely caused by NMD, which degrades the transcripts harboring premature termination codons after alternative splicing (Brogna and Wen, 2009; Smith and Baker, 2015). It is noteworthy that the reduction of mRNAs of the targeted isoforms does not affect the utility of iso-KO cassettes for loss-of-function studies because it also reduces the expression of the targeted isoform.
To assess the stability of the potentially truncated Dscam isoforms caused by the insertion of iso-KO cassettes, we performed western blotting with an antibody against an epitope in the common ectodomain of Dscam (Hattori et al., 2009). We did not observe any extra band around the predicated size of truncated polypeptides (180 kDa) or at other molecular weights in either iso-KO mutant (Figure S3D). These results suggest that the truncated Dscam proteins caused by iso-KO cassettes are unstable. Alternatively, it is possible that the translation of the targeted isoform is suppressed because of selective suppression of the translation of mRNAs containing premature termination codons (You et al., 2007).
Taken together, these results validate the use of isoTarget for studying endogenous Dscam[TM1] and [TM2] isoforms.
Studying the Functions of Dscam Isoforms in Axon Terminals with isoTarget
We previously demonstrated that, in larval peripheral nervous system (PNS) neurons, Dscam instructs the presynaptic terminal growth, which is a function that is distinct from Dscam’s role in neurite self-avoidance (Kim et al., 2013). However, whether both Dscam[TM1] and [TM2] contribute to that process is unknown. With Dscamiso-KO larvae, we found that knocking out the [TM2] isoform impaired axon terminal growth in larval C4 da neurons (Figures 2A, 2B, and 2H). By contrast, the axonal development remained intact in Dscam[TM1]iso-KO larvae (Figures 2C and 2H). Next, we combined isoTarget with the mosaic analysis with a repressible cell marker (MARCM) technique to determine whether the Dscam isoforms functioned cell-autonomously to regulate C4 da axon terminal growth. Single C4 da neurons that were homozygous for Dscam[TM1]iso-KO had normal axon terminal growth (Figures 2D, 2E, and 2I). By contrast, loss of Dscam[TM2] in C4 da neurons significantly impaired axon terminal growth to the same extent as loss of Dscam (Figures 2F, 2G, and 2I).
Similar to C4 da neurons, whereas C3 da neurons in homozygous Dscam[TM1]iso-KO larvae showed normal growth of axon terminals, those in homozygous Dscam[TM2]iso-KO displayed dramatically reduced axon terminal growth, as evident in the gaps in longitudinal axon tracts (Figures 2J–2L). The disruption in longitudinal axon tracts was also observed in Dscam[TM1/2]KO larvae (Figure 2M).
These results suggest that Dscam[TM2], but not Dscam[TM1], regulates the growth of axon terminals.
Studying the Functions of Dscam Isoforms in Dendrites with isoTarget
Dscam has been shown to mediate dendritic self-avoidance without affecting dendritic growth in the PNS neurons of Drosophila larvae (Hughes et al., 2007; Matthews et al., 2007; Soba et al., 2007). Again, it is unknown whether both Dscam [TM1] and [TM2] isoforms are responsible for this process. We applied isoTarget to answer this question. As expected from previous studies, homozygous Dscam[TM1/2]iso-KO mutant larvae, which lack both [TM1] and [TM2] functions, exhibited increased dendritic crossing in both C3 da mechanosensors (Figures 3A, 3B, and 3E) and C4 da nociceptors (Figures S4A, 4B, and 4E), indicating defective avoidance among dendrites of the same neuron. Different from what we observed in axons, neither Dscam[TM1]iso-KO nor Dscam[TM2]iso-KO caused any defect in dendritic self-avoidance in these neurons (Figuress 3C–3E and S4C–S4E).
Figure 3. IsoTarget Uncovers Redundant Functions for Dscam[TM1] and [TM2] in Dendrite Self-Avoidance.
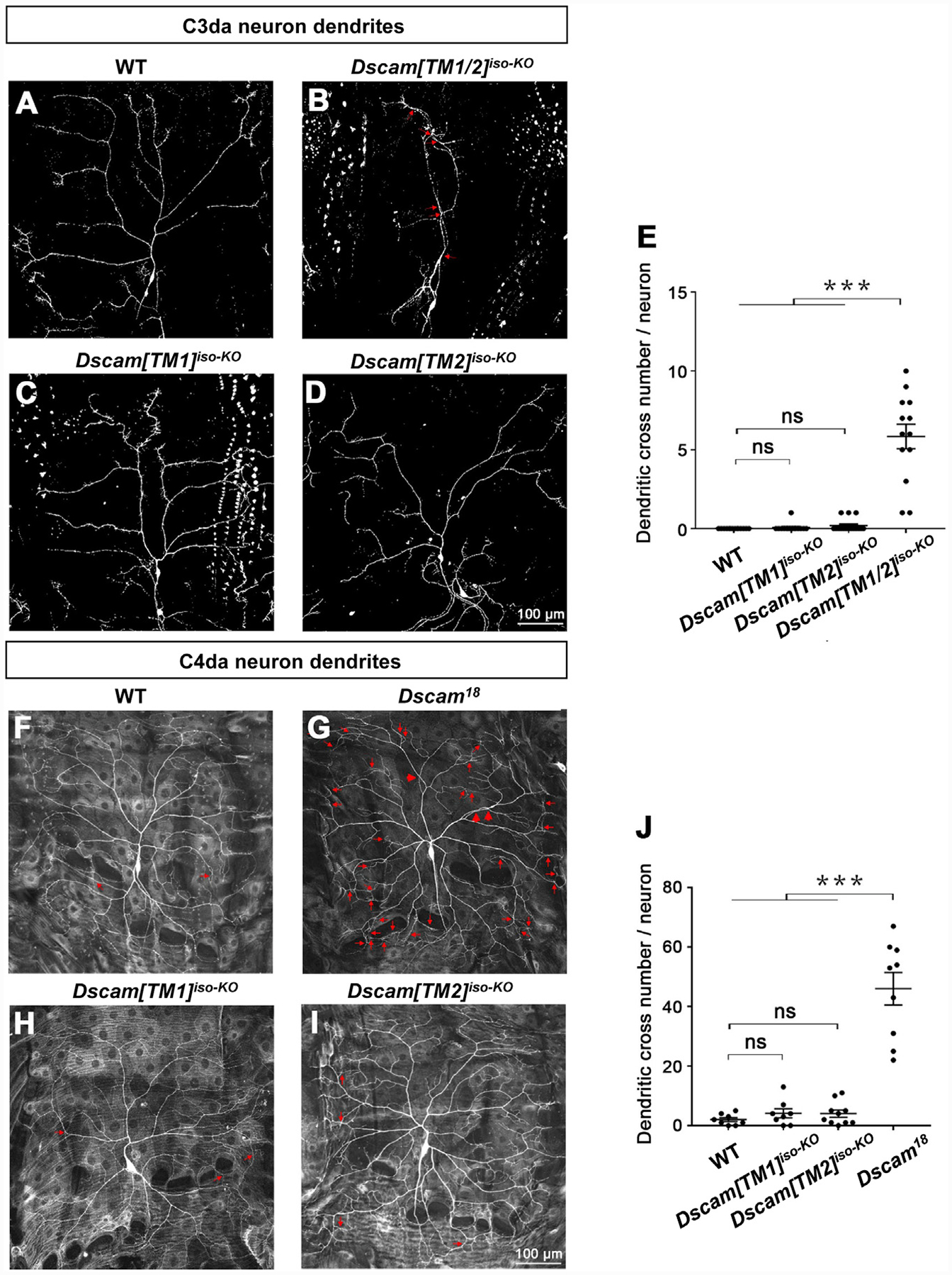
(A–D) Dscam[TM1] and [TM2] function redundantly in mediating dendritic self-avoidance in C3 da neuron. In third-instar larvae, C3 da dendrites rarely fasciculate or intersect with each other in WT (A), whereas the loss of both Dscam[TM1] and [TM2] isoforms (Dscam[TM1/2]iso-KO) significantly impairs self-avoidance (B). Loss of either Dscam [TM1] (C) or [TM2] (D) does not affect dendritic self-avoidance. The red arrows point to dendritic crossing sites.
(E) Quantification of dendritic branch crossings in the C3 da neuron ddaF.
(F–I) Dscam[TM1] and [TM2] function redundantly in mediating dendritic self-avoidance in C4 da neurons. The MARCM technique was used. Small red arrows point to crossings of fine dendritic branches, and large red arrows point to crossings of major dendritic branches, which are only observed when both isoforms are lost.
(J) Quantification of dendritic branch crosses in the C4 da neuron ddaC.
See also Figure S4.
Figure 4. Using isoTarget to Identify the Subcellular Localizations of Endogenous Dscam Isoforms.
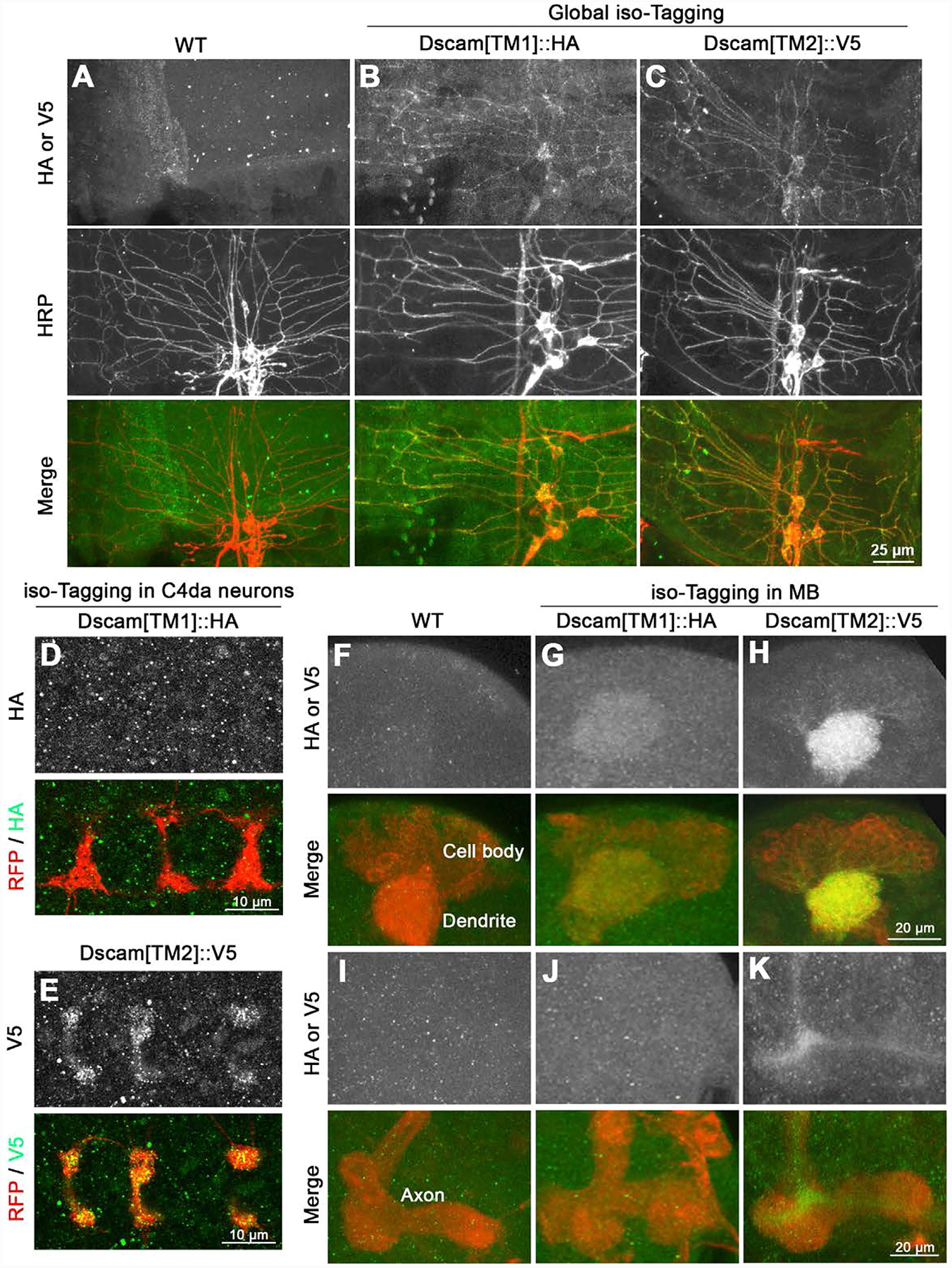
(A–C) Both Dscam[TM1] and [TM2] are localized in the dendrites of larval PNS neurons. At early second-instar stage, compared with control (A), global iso-tagging reveals the localization of endogenous [TM1] and [TM2] in the dendrites of PNS da neurons, which is double-labeled by the PNS neuron marker anti-HRP.
(D and E) Iso-tagging shows that endogenous Dscam[TM2], but not [TM1], is in the axon terminals of C4 da neurons in early second-instar larvae. The C4 da-specific driver ppk-Gal4 was used to tag endogenous Dscam[TM1] or [TM2] by driving the expression of UAS-R-recombinase and to label C4 da axon terminals by driving the expression of UAS-mCD8::RFP.
(F–H) Both Dscam[TM1] and [TM2] are localized in the dendrites of CNS neurons. Iso-tagging in mushroom body (MB) neurons shows that both endogenous [TM1] and [TM2] are present in the MB calyx, which is a cluster of dendrites in third-instar larvae. The MB driver OK107-Gal4 was used to drive the expression of R recombinase for tagging endogenous Dscam[TM1] or [TM2] and the expression of mCD8::RFP for labeling MB morphology.
(I–K) Dscam[TM2], but not [TM1], is localized in MB axons. Compared with the WT control, endogenous Dscam[TM2], but not [TM1], is detected in the axon peduncles of MB in third-instar larvae.
See also Figure S5.
We further combined MARCM with isoTarget to study cell-autonomous functions of targeted isoforms in single cells. Single C4 da neurons that were homozygous of Dscam18, which abolishes both [TM1] and [TM2] isoforms, showed a significant self-avoidance defect (Figures 3G and 3J). By contrast, loss of either Dscam[TM1] or [TM2] in these neurons did not cause any defect in dendritic self-avoidance (Figures 3H–3J).
The heterozygous mutants of Dscamiso-KO, Dscam[TM1]iso-KO/+, and Dscam[TM2]iso-KO/+ did not exhibit any defect in dendritic self-avoidance or axonal growth (Figures S4F–S4L), demonstrating that these mutants are not dominant negative.
These results suggest that Dscam[TM1] and [TM2] function redundantly in dendritic self-avoidance. Thus, these two isoforms function differently in dendrite and axon development.
Using isoTarget to Identify the Subcellular Localizations of Endogenous Protein Isoforms
Previous studies have shown that, in CNS neurons, transgenic Dscam[TM1] is restrained in somatodendritic compartments, whereas [TM2] is in both dendrites and axons (Wang et al., 2004; Yang et al., 2008; Zhan et al., 2004). However, transgenic Dscam[TM1] and [TM2] were both found to be ubiquitously present in PNS neurons (Soba et al., 2007) (Figures 6B and 6D and data not shown). Does this discrepancy indicate that the subcellular localizations of Dscam isoforms vary in different neuronal types? To answer that question, we applied iso-tagging to examine the subcellular localizations of endogenous Dscam isoforms in PNS and CNS neurons. We found that, in PNS neurons, both endogenous Dscam[TM1] and [TM2] were localized in the dendrites (Figures 4A–4C). Notably, different from Dscam transgenes, only endogenous Dscam[TM2] was localized in the presynaptic terminals of PNS neurons (Figures 4D and 4E). Similar isoform-specific localizations were observed in CNS neurons. By iso-tagging in larval mushroom body (MB) neurons, we observed endogenous Dscam[TM1] and [TM2] in the MB calyx (Figures 4F–4H), which is a cluster of dendritic branches, but only Dscam[TM2] in the core fibers of axonal peduncles (Figures 4I–4K). The finding that only Dscam[TM2] is localized in axons was further supported by the observation of Dscam proteins in axonal shafts that connect the PNS and CNS. In iso-tagging larvae, despite a substantial amount of Dscam[TM1] signals in the neuropil region of the ventral nerve cord (VNC), no [TM1] signal was observed in axonal shafts (Figures S5A and S5B). By contrast, Dscam[TM2] puncta were abundant in axonal shafts in global Dscam[TM2]iso-tagging larvae (Figure S5C). Similar results were observed in transgenic flies that expressed Dscam [TM1]::GFP or [TM2]::GFP by the endogenous Dscam promoter (Wang et al., 2004) (Figures S5D–S5F). Taken together, in the PNS and CNS, both endogenous Dscam[TM1] and [TM2] isoforms are present in dendrites, whereas only [TM2] is in axons.
Figure 6. Dscam[TM1] and [TM2] Exhibit Similar Biochemical Properties and Cellular Functions When They Are Localized in the Same Subcellular Compartment.
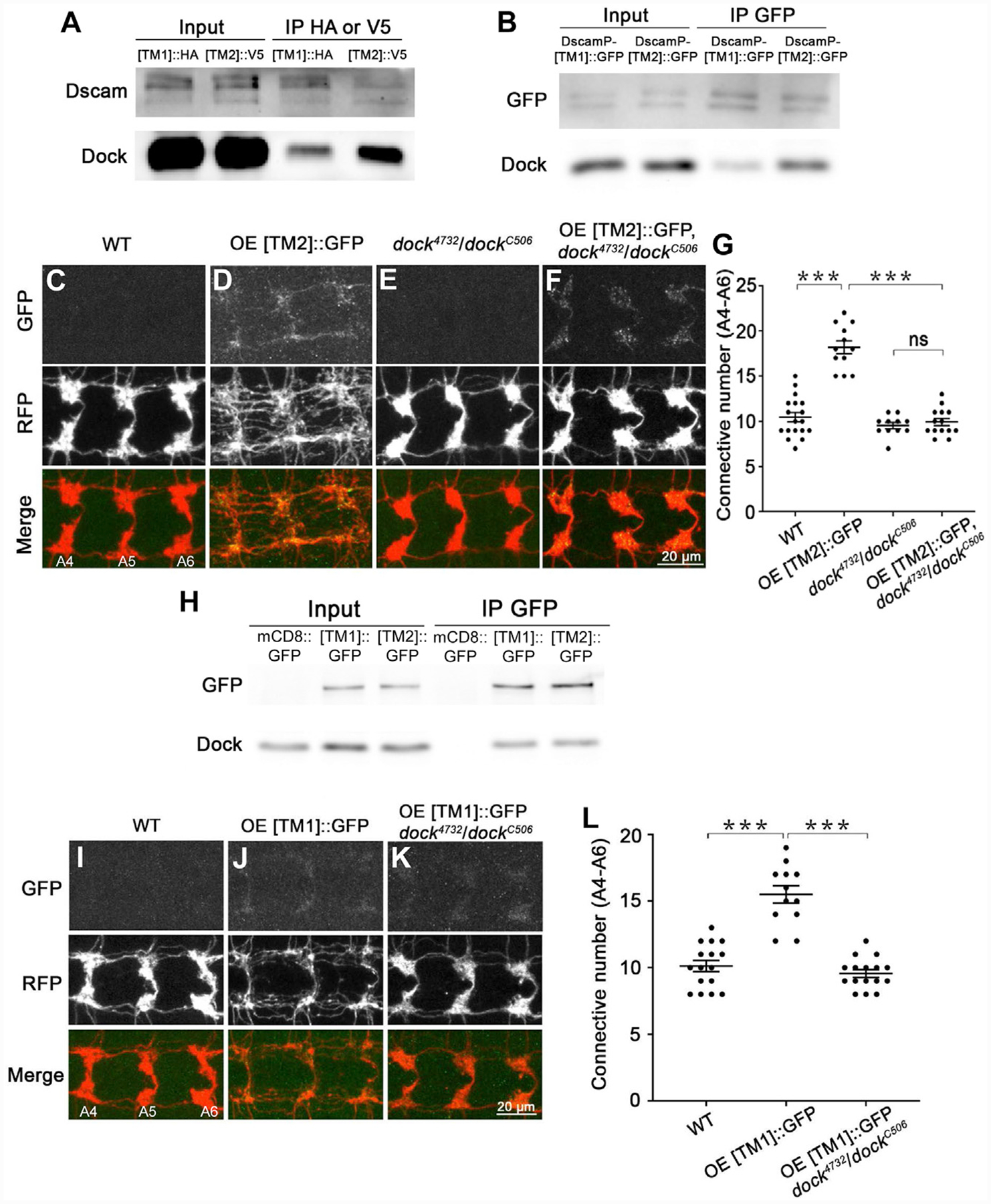
(A and B) Dock preferentially interacts with endogenous Dscam[TM2] in vivo. (A) Brain lysates of larvae with global iso-tagging of [TM1]::HA or [TM2]::V5 were immunoprecipitated by anti-HA or V5 beads, respectively, and immunoblotted with an anti-Dscam antibody that recognizes both [TM1] and [TM2] and an anti-Dock antibody. This experiment was repeated twice. (B) Brain lysates of larvae that expressed [TM1]::GFP or [TM2]::GFP through the endogenous Dscam promoter were immunoprecipitated by an anti-GFP antibody. The immunoprecipitates were immunoblotted with anti-GFP and anti-Dock antibodies. This experiment was repeated three times.
(C–F) Dscam[TM2] requires Dock to promote presynaptic terminal growth. Shown are representative images of A4–A6. Overexpressing a Dscam[TM2]::GFP transgene significantly promotes axonal growth in C4 da neurons (C) & (D). Although the loss of dock does not affect C4 da axon terminals (E), it completely abolishes the overgrowth caused by Dscam[TM2]::GFP overexpression (F).
(G) Quantification of the number of C4 da axon connectives.
(H) Dock binds to [TM1] and [TM2] in similar affinity in cultured S2 cells. Lysates of S2 cells expressing mCD8::GFP, Dscam[TM1]::GFP or [TM2]::GFP were immunoprecipitated with an anti-GFP antibody. Inputs and immunoprecipitates were blotted by anti-GFP and anti-Dock antibodies. This experiment was repeated three times.
(I–K) Transgenic Dscam[TM1] requires Dock to promote presynaptic terminal growth. Compared with WT (I), overexpressing [TM1]::GFP transgene significantly promotes axonal growth in C4 da neurons (J), which is completely abolished by loss of dock (K).
(L) Quantification of the number of C4 da axon connectives from A4–A6.
See also Figure S6.
Endogenous Dscam expression seemed to be enriched in developing, but not mature, neurons. Although both endogenous Dscam[TM1] and [TM2] were observed in PNS neurons at the early second-instar stage (Figures 4A–4C), no signal was detectable in third-instar larvae (data not shown). In larval MB, endogenous Dscam[TM2] was detected only in the core fiber, which is the axonal projections of nascent developing neurons. These observations are consistent with previous immunostaining results with an anti-Dscam antibody (Zhan et al., 2004). The discrepancies of spatiotemporal expression pattern between transgenes and endogenous Dscam isoforms underscore the importance of studying protein isoforms at the native level.
Dendrite-Specific Localization Restrains Endogenous Dscam [TM1] from Functioning in Axons
The studies above show that endogenous Dscam[TM1] is required for dendritic, but not axonal, development. Although this could be a result of the compartmentalized localization of Dscam[TM1] in dendrites, trans-compartmental communication has been shown for membrane proteins (Terenzio et al., 2017). It is thus essential to determine whether the dendrite-specific localization of Dscam [TM1], instead of its biochemical properties, restrains it from functioning in axons.
We determined whether forced localization of Dscam[TM1] in axon terminals was sufficient to rescue the axon phenotype caused by Dscam[TM2]iso-KO. Overexpression of a Dscam [TM1] transgene in C4 da neurons led to a modest level of Dscam[TM1] in axon terminals and significantly mitigated axonal growth defects caused by Dscam[TM2]iso-KO (Figures 5A, 5B, and 5F). As expected, two different Dscam[TM2] transgenes (#1 and #2, with #2 being expressed at a higher levels than #1) both caused higher levels of [TM2] in axon terminals and stronger rescue than the Dscam[TM1] transgene (Figures 5C and 5E–5G). Strikingly, when we used two copies of the C4-da-specific driver ppk-Gal4 to increase the axonal level of Dscam[TM1] to that expressed by Dscam[TM2] transgenes, the rescue effects were comparable (Figures 5C, 5D, 5F, and 5G). By quantitatively assessing transgenic Dscam::GFP levels in C4 da axon terminals (Figure 5G), we found a strong correlation between presynaptic Dscam isoform levels and their rescue effects (Figure 5H). These results suggest that, when localized in the same subcellular compartment, Dscam[TM1] functions equally as Dscam[TM2] in promoting axonal growth. Thus, the dendrite-specific function of endogenous Dscam[TM1] is a result of its compartmentalized localization.
Figure 5. Dendrite-Specific Localization Restrains Endogenous Dscam[TM1] from Functioning in Axons.
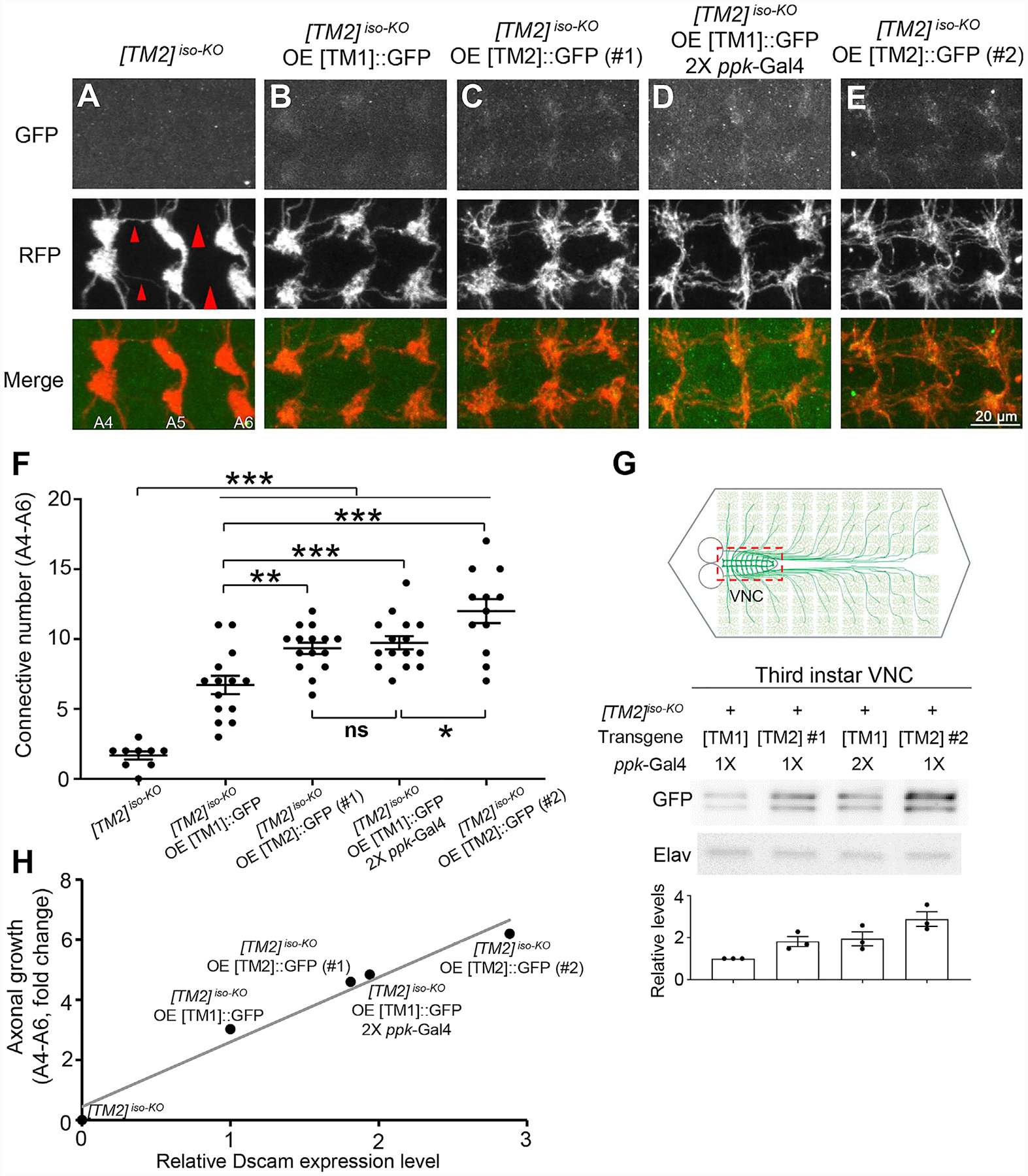
(A) Dscam[TM2]iso-KO dramatically impairs the axon terminal growth in C4 da neurons. The large red arrowheads point to the sites where longitudinal axon tracts are broken, and the small arrowheads point to where the tracts are thinned.
(B–E) Overexpression of a Dscam[TM1], Dscam[TM2]#1, or Dscam[TM2]#2 transgenes with the ppk-Gal4 driver leads to axon localization of Dscam isoforms at different levels and mitigates the axonal defect in Dscam[TM2]iso-KO to different extents.
(F) Quantification of the number of C4 da axon connectives in segments A4-A6.
(G) Quantification of transgenic Dscam::GFP levels in C4 da axon terminals. The experiments were done in Dscam[TM2]iso-KO larvae. As shown in the schematic, UAS-Dscam::GFP transgenes were expressed in C4 da neurons with the ppk-Gal4 driver. VNCs (indicated by the dashed red box in the top panel), which contained transgenic Dscam::GFP in C4 da axon terminals, were dissected out from third-instar larvae for western blotting.
(H) The rescue of the axonal defect caused by the loss of Dscam is proportional to the level of transgenic Dscam in C4 da axon terminals, regardless of the isoform. The relative Dscam expression levels were determined by western blotting of CNS lysates from larvae overexpressing [TM1]::GFP or [TM2]::GFP by ppk-Gal4 and plotted against the rescue effect of the transgene. Linear regression, R2 = 0.969.
Axonally Enriched Dscam[TM2]-Dock Signaling Is Essential for the Axonal Function of Dscam[TM2]
Although the study above demonstrates that the dendrite-specific localization of Dscam[TM1] prevents it from functioning in axon development, it remains unknown whether Dscam[TM1] and [TM2] exhibit any biochemical difference that might also explain the difference in their cellular functions. To test that possibility, we took advantage of isoTarget to compare Dscam[TM1] and [TM2] for their interactions with functional partners. Among the molecules known to bind to the intracellular domain of Dscam and mediate its signaling (Kamiyama et al., 2015; Liu et al., 2009; Purohit et al., 2012; Schmucker et al., 2000; Sterne et al., 2015), we found that Dock, an SH2/SH3 adaptor protein that is preferentially localized in axons (Figure S6) (Desai et al., 1999; Fan et al., 2003; Hing et al., 1999), primarily associated with Dscam[TM2] in Drosophila larval CNS (Figure 6A). That finding was confirmed with transgenic flies that overexpress Dscam[TM1]::GFP or [TM2]::GFP through their endogenous promoters (Wang et al., 2004) (Figure 6B). Consistent with that notion, we found that Dock is required for Dscam[TM2] to promote axon terminal growth. Although overexpressing Dscam [TM2] promoted axon terminal growth in C4 da neurons, its effect was completely eliminated by the loss of dock (Figures 6C–6G). Interestingly, Dock bound to Dscam[TM1] and Dscam[TM2] isoforms with equal affinity in cultured Schneider 2 (S2) cells, which do not have dendrite-axon compartmentalization (Figure 6H). This result is consistent with the previous finding that the Dock-binding sites, including PxxP sites and polyproline motifs, are located in the cytoplasmic region shared by the two isoforms (Schmucker et al., 2000). In addition, the axon terminal overgrowth resulting from Dscam[TM1] overexpression was also eliminated by loss of dock (Figures 6I–6L). Thus, when they are in the same subcellular compartment, Dscam[TM1] and [TM2] are biochemically comparable and share the same signaling mechanism. Moreover, the ectopic localization, function, and signaling of transgenic Dscam[TM1] in axons underscore the importance of studying splicing isoforms at physiological levels of expression.
Axonal Enrichment of Wallenda (Wnd) Compartmentalizes the Wnd-Dscam[TM2] Signaling
We showed previously that the signaling pathway involving the E3 ubiquitin ligase highwire (Hiw) and the dual leucine zipper kinase Wnd promotes Dscam expression (Kim et al., 2013). How ever, whether the Hiw-Wnd pathway regulates the expression of both Dscam[TM1] and [TM2] isoforms is unknown. To address that, we determined whether loss of hiw or overexpression of Wnd changed the levels of endogenous Dscam[TM1] and [TM2]. To test the effects of loss of hiw, western blotting was performed on CNS lysates from trans-heterozygous Dscam [TM1]::HA / [TM2]::V5 larvae that were generated through global iso-tagging. Loss of hiw led to increased expression of both Dscam[TM1] and [TM2] in the CNS (Figure 7A). As global overexpression of Wnd caused larval lethality, we co-expressed Wnd and R recombinase (for iso-tagging of endogenous Dscam [TM1] and [TM2]) in a subset of CNS neurons with GAL44−77 (Kim et al., 2013). Interestingly, Wnd overexpression only increased the level of Dscam[TM2] (Figure 7B), but not that of [TM1], suggesting that, unlike Hiw, Wnd preferentially regulates the expression of Dscam[TM2]. That finding from biochemical studies was confirmed in C4 da neurons by immunostaining of the endogenous Dscam[TM2]::V5. Immunostaining of Dscam [TM2]::V5iso-tagging showed that endogenous Dscam[TM2] was undetectable in C4 da axon terminals in third-instar larvae, but detectable at this developmental stage when Wnd was overexpressed in these neurons (Figure 7C).
Figure 7. Axonal Enrichment of Wnd Compartmentalizes Wnd-Dscam[TM2] Signaling.
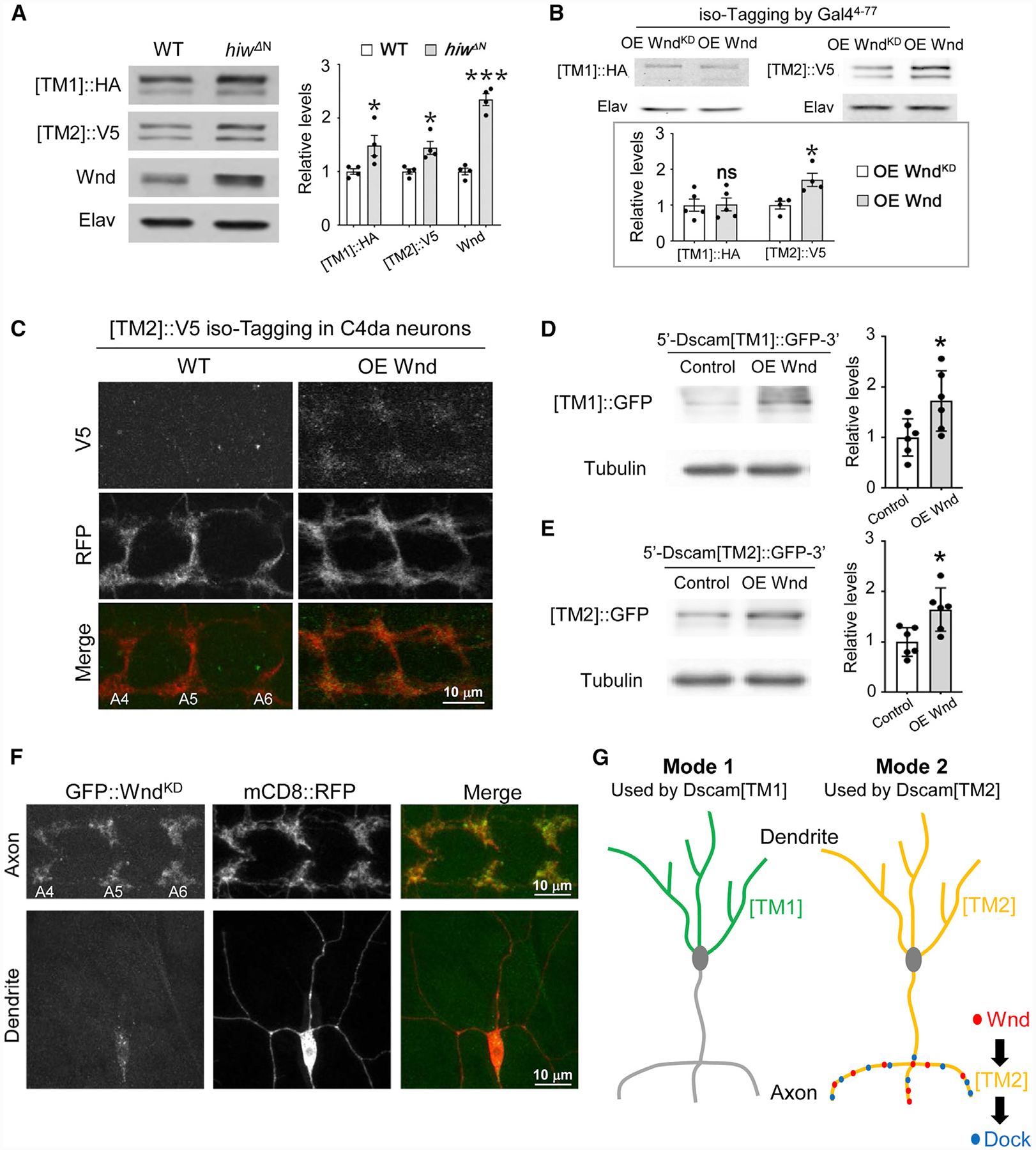
(A) Hiw suppresses the expression of both Dscam[TM1] and [TM2]. In the brains of third-instar larvae that were trans-heterozygotes of global iso-tagging of Dscam[TM1]::HA and [TM2]::V5, loss of hiw (hiwΔN) elevated the levels of both endogenous Dscam[TM1] and [TM2]. Left: western blots; right: quantification of western blots. Each dot represents the result from one independent experiment.
(B) Overexpression of Wnd increases the levels of Dscam[TM2], but not that of [TM1]. Top: the Gal44−77, which is expressed in a small set of PNS neurons and a number of CNS neurons, was used to drive the expression of R recombinase for iso-tagging and the overexpression of Wnd. Each dot represents the result of one independent experiment.
(C) Overexpression of Wnd increases endogenous Dscam[TM2] levels in the presynaptic terminals of C4 da neurons. At the third-instar larval stage, Dscam [TM2]::V5iso-tagging is no longer detectable in C4 da axon terminals. Overexpression of Wnd elevates the level of Dscam[TM2]::V5iso-tagging in these terminals.
(D and E) Wnd promotes Dscam[TM1] and [TM2] expression to a similar extent in cultured S2 cells. S2 cells were transfected with plasmids expressing Wnd and Dscam[TM1]::GFP or [TM2]::GFP with endogenous Dscam 5′ and 3′ UTR. Lysates of S2 cells were blotted by anti-GFP and anti-tubulin antibodies. Each dot represents the result of one independent experiment.
(F) Wnd is enriched in axons. GFP-tagged kinase-dead Wnd (GFP::WndKD) was expressed in C4 da neurons by ppk-Gal4. mCD8::RFP was used to label the neurons. Although the GFP::WndKD signal is enriched in axon terminals, little is observed in dendrites. Yellow arrows point to major dendritic branches.
(G) Subcellular localization expands the functional diversity of splicing isoforms via two different modes. In mode 1, the localization of an isoform in a particular subcellular location (e.g., Dscam[TM1] in dendrites) restrains this isoform from functioning in other compartments. In mode 2, the enrichment of the functional partners (e.g., Wnd and Dock) for a ubiquitously localized isoform (e.g., Dscam[TM2]) leads to isoform-specific subcellular signaling and functions.
Next, we investigated the mechanism underlying the differential effect of Wnd on Dscam[TM1] and [TM2]. First, we determined whether Wnd was capable of increasing Dscam[TM1] expression in a cell type without the dendrite-axon compartmentalization. We overexpressed Dscam isoforms with endogenous 5′ and 3′ untranslated regions (UTRs) in S2 cells and found that Wnd similarly promoted Dscam[TM1] and [TM2] expression (Figures 7D and 7E). This result suggests that Dscam[TM1] and [TM2] are molecularly indistinguishable for the Wnd regulation. Second, we identified the distribution of Wnd in neurons and found that Wnd was enriched in axons but not dendrites. A GFP-tagged kinase-dead version of Wnd (GFP-WndKD) (Xiong et al., 2010) was expressed in C4 da neurons; substantial signals were detected in axon terminals but not in dendrites (Figure 7F). These data suggest that axonal localization of Wnd compartmentalizes the Wnd-Dscam[TM2] signaling.
Applying isoTarget to Study Rdl Isoforms
We tested the generality of isoTarget by applying it to study another gene with an exon-intron arrangement that is different from Dscam. The 17.1 and 17.2 exons of Dscam are separated by an intron of 252 bp, which is substantially shorter than that upstream of 17.1 (2,504 bp). The big size difference between the two introns flanking exon 17.1 exon might be the reason why the full-length tlstop cassette inserted in 17.1 affected the splicing of 17.2 (Figures S2D–S2F). To test that possibility, we applied isoTarget to the Rdl gene, which encodes a Drosophila GABAA receptor. The length of the intron between the mutually exclusive exons, RG and RF, is 1,531 bp (Figure S7A), which is similar to that upstream of the leading exon RG (1,516 bp).
Computational simulation of Drosophila Rdl with the structure of a human GABAA receptor (Miller and Aricescu, 2014) was used to select a putative surface-exposed loop region for inserting the full-length iso-KO cassette (see STAR Method) (Figure S7B), because insertion into this region is less likely to affect Rdl isoform function. Unlike its insertion into the 17.1 exon of Dscam (Figure S2D), insertion of the full-length iso-KO cassette into the upstream exon RG did not affect the mRNA levels of the downstream RF isoform (Figure S7C). Moreover, as expected from what we observed of Dscam (Figure S2C), insertion of the full-length iso-KO cassette into the downstream RF exon did not affect the mRNA levels of the upstream RG isoform. These results suggest that whether the full-length iso-KO cassette inserted in an alternative exon affects the splicing of the downstream alternative exons depends on the intron sizes around the targeted exon. Iso-tagging of Rdl[RG] and [RF] isoforms showed that these two isoforms exhibited similar expression patterns that were comparable with that of total Rdl proteins tagged at the C terminus with the V5-tag (Figure S7D), suggesting that iso-tagging was successful.
Consistent with what we observed in Dscam isoforms (Figure S3C), the iso-KO cassette reduced the mRNA levels of targeted isoforms in Rdl (Figure S7C). The reductions of Rdl isoforms (~75%) were greater than those of Dscam isoforms (~50%). Because the positions of premature stop codons affect the efficiency of NMD (Smith and Baker, 2015), the different extents of mRNA decays between Rdl and Dscam are possibly because the insertion sites of the iso-KO cassette is proximal to the 5′ end in Rdl while those in Dscam is closer to the 3′ end.
DISCUSSION
In this study, we developed isoTarget to generate splicing isoform-specific loss-of-function mutants and conditional tagging in specific neurons. As proofs of concept, we applied isoTarget to investigate isoforms of Dscam and Rdl. These findings highlight the versatility of isoTarget and the importance of studying splicing isoform at endogenous levels in vivo. In addition, they establish the causality between the subcellular localization and cellular function of two Dscam isoforms, demonstrating the critical role of subcellular localization in expanding the functional diversity of splicing isoforms. We anticipate this methodology to be useful for isoform studies in various cell types and organisms.
Subcellular Localization Defines the Cellular Functions of Alternative Splicing Isoforms That Have the Same Biochemical Functions
Alternative isoforms often differ in their protein structures and biochemical properties (Kelemen et al., 2013). Recent studies suggest the importance of subcellular localization in defining the cellular functions of alternative splicing isoforms. For example, in cultured neurons, the RBFOX1 gene generates a nuclear variant (by excluding exon 19) that regulates RNA splicing and a cytoplasmic variant (by including exon 19) that stabilizes mRNAs (Lee et al., 2016). However, whether subcellular localization is the only reason why the nuclear and cytoplasmic variants differ in their cellular functions remains to be determined because it is possible that the two isoforms have distinct biochemical functions. More broadly, a challenging question in the field is whether the difference in subcellular localization determines the compartmentalized functions for the isoform (i.e., the issue of causality), especially at the endogenous levels in vivo. By applying isoTarget in Drosophila sensory neurons, we established the causality between the subcellular localization and cellular functions of two Dscam isoforms.
Even if they are localized in different subcellular compartments, splicing isoforms with the same biochemical function might not exhibit distinct cellular functions because of mechanisms such as trans-compartmental communication (Terenzio et al., 2017). To achieve cellular functions specific to a compartment, one solution is to localize the functional partners shared by the isoforms to specific subcellular compartments. Indeed, we found that the compartmental enrichment of interactors leads to isoform-specific signaling. The Dscam[TM1] and [TM2] do not differ in their biochemical interactions with Wnd and Dock (Figures 6H, 7C, and 7D). However, the axonal enrichment of Wnd and Dock forms a compartmentalized Wnd-Dscam[TM2]-Dock signaling cascade in vivo (Figures 7C and S6), even though Dscam[TM2] is present in both dendrites and axons (Figures 4C, 4E, 4H, and 4K). These findings suggest that, for splicing isoforms with the same biochemical functions, specific cellular functions can be achieved by the compartment-specific colocalization of the isoforms and their functional partners (Figure 7G). Taken together, our study demonstrates that Dscam isoforms use two different modes to achieve distinct compartment-specific functions in vivo (Figure 7G). The dendrite-specific localization restrains Dscam[TM1] from functioning in axons; and axonal enrichment of functional partners forms a subcellular signaling pathway involving the ubiquitously distributed Dscam[TM2].
Sphingolipids are essential for the trafficking of dendritic and axonal cell adhesion molecules, including Dscam (Goyal et al., 2019). Loss of serine palmitoyltransferase (SPT), a key enzyme in sphingolipid biosynthesis, reduces the membrane localization of Dscam[TM1] and [TM2] in subsets of MB neurons, resulting in [TM1] aggregates in the soma and [TM2] aggregates in the axons. isoTarget would be instrumental for testing the function of sphingolipids in the trafficking of endogenous Dscam isoforms in select neuron populations. Moreover, the isoTarget-based approaches described in our article allowed us to address whether subcellular localization is the cause of the isoform-specific function. These approaches can also be used to determine the causality between alternations in subcellular localization (e.g., those caused by trafficking defects) and changes in cellular functions.
Advantages of isoTarget
The studies reported in this article demonstrate several advantages of isoTarget. First, it can be used to generate classic genetic mutants for analyzing the functions of specific splicing isoforms. Second, isoTaget enables the identification of isoform-specific localization at the subcellular level in cells of interest in vivo, which is otherwise challenging because of the difficulty in discerning immunofluorescent signals in subcellular compartments among neurons expressing the same protein in the vicinity. In addition, this method expresses tagged proteins at more physiological levels than transgenes do. Third, isoTarget allows us to study isoform-specific compartmentalized signaling. Using isoTarget, we identified a Wnd-Dscam[TM2]-Dock signaling pathway enriched at the presynaptic terminals of C4 da neurons. Complementing the in vivo studies with isoTarget, we performed biochemical studies in S2 cells and found that Dscam[TM1] and [TM2] did not differ in their biochemical interactions with Wnd and Dock (Figures 6H, 7C, and 7D). Consistent with that, forced localization of ectopic Dscam[TM1] in axons also increased axonal growth in vivo (Figures 5A–5G) through the same downstream effector Dock used by Dscam[TM2] (Figures 6I–6L). In this series of studies, isoTarget was essential for establishing the cellular functions and biochemical interactions in vivo.
Limitations of isoTarget
We see several limitations of the isoTarget technique. First, for the successful application of isoTarget, the isoform inserted with iso-KO cassette is expected to lose its function. This is not necessarily always the case, especially when the targeted exon encodes a fragment located near the C terminus of the protein. This problem is common in isoform studies by genetic modifications, including Cre-LoxP and isoEXPRESS (Gu et al., 2019). Developing an isoform-specific nanobody might be a way to solve that problem (Röth et al., 2019). Second, we discovered that the full-length iso-KO cassette may affect the splicing of downstream alternative exons and that such effects depend on the length of the cassette (Figures S2D–S2F and S2I). Thus, the expression of isoforms other than the targeted one should always be examined by techniques such as reverse transcription real-time PCR. Moreover, whether the full-length iso-KO cassette inserted in an exon affects the splicing of downstream alternative exons depends on the sizes of the introns flanking the targeted exon (Figure S7). To minimize the chance of affecting downstream exons, we recommend using the short iso-KO cassette for isoTarget applications. Third, the iso-KO cassettes may lead to truncated polypeptides of the targeted isoform. This could be tested by western blotting with an antibody against the N-terminal part of the targeted protein. The concern that the truncated polypeptides may cause dominant-negative effects could be addressed by examining the phenotypes in heterozygotes (Figures S4F–S4L). Fourth, successful uses of isoTarget require that epitope tagging preserves the function of the splicing isoform. Structural information would help choose the proper inserting site. For example, inserting the isoTarget cassette into the loop region of a polypeptide is likely to increase the chance of success.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Bing Ye (bingye@umich.edu).
Materials Availability
The Drosophila lines and DNA constructs generated in this study are available from the Lead Contact upon request.
Data and Code Availability
The datasets generated in the study are available from the corresponding authors on reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Male and female 2nd and 3rd-instar Drosophila larvae were used. Experiments were done on age-matched larvae.
Cell lines
S2 cells were maintained in Drosophila Schneider’s medium supplemented with 10% heat-inactivated fetal bovine serum at 25°C in a humidified chamber.
METHOD DETAILS
Generation of DNA constructs
The design of isoTarget is described in Figure S1A. The translational stop cassette was amplified by PCR from non-catalytic region of lacZ with frameshift to introduce multiple premature termination codons. The loxP-dsRed-loxP sequence is described previously (Gratz et al., 2014). The pBluescript-based donor plasmid, which contains the isoTarget cassette (GS linker-RSRT-tlstop-loxP-dsRed-loxP-RSRT-epitope-G3 linker) and Dscam or Rdl isoform homologous sequences with mutated PAM, was generated with the In-Fusion HD Cloning Kit (Clontech Laboratories, Inc.). The pCFD3-dU63 gRNA plasmid was used to produce gRNA in fly embryos (Ran et al., 2013). Iso-tagging of total Rdl proteins tagged at the C terminus with the V5-tag is reported in a separate paper (Hu et al., 2020). Briefly, the isoTarget cassette followed by the V5-epitope tag was inserted right before the Rdl stop codon.
The pUAST-Dscam5′UTR-Dscam[TM1]::GFP-Dscam3′UTR (4.3–6.36–9.25) plasmid was made by modifying pUAST-Dscam5′UTR- Dscam[TM2]::GFP-Dscam3′UTR (4.3–6.36–9.25) (Kim et al., 2013). Specifically, the fragment containing Dscam5′UTR and exons 1–16 and the fragment containing exons 18–24 plus Dscam3′UTR were amplified from pUAST-Dscam5′UTR- Dscam [TM2]::GFP-Dscam3′UTR by PCR. The fragments, together with PCR products of Dscam exon 17.1, were fused with the pUASTattB vector (linearized by EcoRV) by In-Fusion HD Cloning (Clontech Laboratories, Inc.).
To make the pUAST-Dock-OLLAS construct, the Dock-OLLAS fragment was amplified by PCR from pBluescript-Dock (Ang et al., 2003) with the OLLAS tag (Park et al., 2008) added to C terminus through a PCR primer. This fragment was then inserted into the pUASTattB vector by In-Fusion HD Cloning (Clontech Laboratories, Inc.).
Sequences of isoTarget cassettes
tlstop (in-frame stop codons that we introduced are underlined):
TAACGTAAGCTAGCTAGACCGGTCCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGGCCCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGGCGCTTTGCCTGGTTTCCGGCACCAGAAGCGGTGCCGGAAAGCTGGCTGGAGTGCGATCTTCCTGAGGCCGATACTGTCGTCGTCCCCTCAAACTGGCAGATGCACGGTTACGATGCGCCCATCTACACCAACGTAACCTATCCCATTACGGTCAATCCGCCGTTTGTTCCCACGGAGAATCCGACGGGTTGTTACTCGCTCACATTTAATGTTGATGAAAGCTGGCTACAGGAAGGCCACGCGTA
Short tlstop (in-frame stop codons that we introduced are underlined):
TAACGTAAGCTAGCTAGACCGGTTTCCCACGGAGAATCCGACGGGTTGTTACTCGCTCACATTTAATGTTGATGAAAGCTGGCTACAGGAAGGCCACGCGTA
GS-linker: GGTGGCGGCGGAAGCGGAGGTGGAGGCTCC
RSRT: CTTGATGAAAGAATAACGTATTCTTTCATCAAG
loxP: ATAACTTCGTATAATGTATGCTATACGAAGTTAT
dsRed box (consisting of the 3xP3 enhancer/HSP70 promoter (Gratz et al., 2014), the cDNA encoding the dsRed fluorescent protein [underlined], and the SV40 3′ UTR [italicized])
CGTACGGGATCTAATTCAATTAGAGACTAATTCAATTAGAGCTAATTCAATTAGGATCCAAGCTTATCGATTTCGAACCCTCGACCGCCGGAGTATAAATAGAGGCGCTTCGTCTACGGAGCGACAATTCAATTCAAACAAGCAAAGTGAACACGTCGCTAAGCGAAAGCTAAGCAAATAAACAAGCGCAGCTGAACAAGCTAAACAATCGGCTCGAAGCCGGTCGCCACCATGGCCTCCTCCGAGGACGTCATCAAGGAGTTCATGCGCTTCAAGGTGCGCATGGAGGGCTCCGTGAACGGCCACGAGTTCGAGATCGAGGGCGAGGGCGAGGGCCGCCCCTACGAGGGCACCCAGACCGCCAAGCTGAAGGTGACCAAGGGCGGCCCCCTGCCCTTCGCCTGGGACATCCTGTCCCCCCAGTTCCAGTACGGCTCCAAGGTGTACGTGAAGCACCCCGCCGACATCCCCGACTACAAGAAGCTGTCCTTCCCCGAGGGCTTCAAGTGGGAGCGCGTGATGAACTTCGAGGACGGCGGCGTGGTGACCGTGACCCAGGACTCCTCCCTcCAGGACGGCTCCTTCATCTACAAGGTGAAGTTCATCGGCGTGAACTTCCCCTCCGACGGCCCCGTAATGCAGAAGAAGACTATGGGCTGGGAGGCgTCCACCGAGCGCCTGTACCCCCGCGACGGCGTGCTGAAGGGCGAGATCCACAAGGCCCTGAAGCTGAAGGACGGCGGCCACTACCTGGTGGAGTTCAAGTCCATCTACATGGCCAAGAAGCCCGTGCAGCTGCCCGGCTACTACTACGTGGACTCCAAGCTGGACATCACCTCCCACAACGAGGACTACACCATCGTGGAGCAGTACGAGCGCGCCGAGGGCCGCCACCACCTGTTCCTGTAGGGGCCGCGACTCTAGATCATAATCAGCCATACCACATTTGTAGAGGTTTTACTTGCTTTAAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCAATTGTTGTTGTTAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTAACCGGT
V5: GGCAAGCCCATCCCAAACCCACTGCTCGGCCTGGATAGCACC
HA: TACCCATACGATGTTCCAGATTACGCT
OLLAS: AGCGGCTTCGCCAACGAGCTGGGCCCCCGCCTGATGGGCAAG
G3 linker: GGTGGCGGC
Generation of isoTarget flies
The donor plasmid (750 ng/μl) and gRNA plasmid (250 ng/μl) were co-injected into fly embryos to generate mosaic G0 flies, which were crossed to white1118 flies to get G1 heterozygous knock-in flies that express the selection marker dsRed in their eyes. G1 flies with red fluorescence in their eyes were crossed to Cre-expressing flies (BL1092) to generate iso-KO flies. To generate global iso-Tagging flies, iso-KO flies were mated with the germline driver nos-Gal4 (BL4442) and 20x UAS-R recombinase (BL55795). In our experience, around 20% of fertile G0 flies produced G1 flies with fluorescent red eyes. In the progeny of each G0 fly, 1%–30% of G1 were knock-in flies. To generate cell-type-specific iso-Tagging, iso-KO flies were mated with cell-type-specific Gal4 drivers, such as ppk-Gal4, and 20x UAS-R recombinase.
S2 cell transfection
Plasmids were transfected into cultured S2 cells with polyethylenimine (PEI) (Ehrhardt et al., 2006). Cells were harvested for western blotting two days after transfection.
Co-immunoprecipitation and western blotting
To perform co-immunoprecipitation with neural tissues, the CNS of 3rd-instar larvae (~120 per experimental condition) were dissected out and placed in ice-cold 1x PBS containing 2 mM sodium vanadate. After a pulse of centrifugation, larval CNS were isolated and then lysed on ice with the lysis buffer (50 mM Tris-HCl/pH 7.4, 150 mM NaCl, 2 mM sodium vanadate, 10 mM sodium fluo-ride, 1% Triton X-100, 10% glycerol, 10 mM imidazole and 0.5 mM phenylmethylsulfonyl fluoride). Lysates were centrifuged at 15,000 g for 20 min at 4°C. The resulting supernatants were either saved as inputs or incubated with magnetic beads conjugated with appropriate antibodies for 4 hours at 4°C. After washing once with the lysis buffer, twice with lysis buffer containing 0.1% deoxycholate, and 3 times with lysis buffer lacking Triton X-100, the immunoprecipitates and total lysates were resolved on 7.5% SDS-PAGE gels followed by western blotting as previously described (Kim et al., 2013). Protein samples were transferred to nitrocellulose membranes and detected by chemiluminescence (Catalog# 32106, Pierce ECL Western Blotting Substrate) with either a BIO-RAD ChemiDoc Touch Imaging system or a Kodak X-OMAT 2000 film processor.
The procedure of co-immunoprecipitation with lysates from S2 cells was similar except that cultured S2 cells were re-suspended in ice-cold 1x PBS before lysis.
Imaging and image analysis
Larvae immunostaining was described previously (Ye et al., 2011). Confocal imaging was done with a Leica SP5 confocal system with 20x or 63x glycerol immersion lenses. To minimize the variation, we only imaged neuronal dendrites and presynaptic terminals of C3 da or C4 da neurons in abdominal segments 4, 5, and 6. Images were collected with z stacks of 1-μm-step size for dendrites and 0.3-μm-step size for axons. The resulting three-dimensional images were projected into two-dimensional images by maximum projection. The same imaging settings were applied throughout the imaging process.
The Neurolucida software was used to quantify axon terminals of single C4 da neurons. Branches shorter than 5 μm were excluded from the analysis. For quantifying the number of longitudinal axonal branches visible between abdominal segment 4–6 (i.e., the connectives) of C4 da neurons, only complete connectives spanning neighboring segments were quantified. Fasciculated connectives were counted as two.
To eliminate experimenter’s bias, these experiments were carried out in double-blind fashion. The images acquired by the primary experimenter were coded and randomized by another lab member. After the primary experimenter quantified the data, the data were decoded for statistical analysis.
Reverse-transcription real-Time PCR
The procedure was as described before (Kim et al., 2013). Briefly, mRNA was extracted from around 20 3rd-instar larval CNS with a TRIzol method by following manufacturer’s guide (Invitrogen). cDNA was synthesized with Invitrogen SuperScript III First-Strand Synthesis SuperMix (Invitrogen). 10 ng cDNA was used as the template for each real-time PCR reaction by SYBR Green mix (Thermo Scientific) with the Applied Biosystems QuantStudio 6 Flex. To normalize Dscam transcripts to those of reference genes, we calculated ΔCt (Dscam) = Ct (Dscam) – Ct (reference gene). Statistical analysis was performed to compare ΔCt (Dscam)(wild-type) and ΔCt (Dscam)(mutant) to determine whether there is the expression of test gene is different between wild-type and mutants (Yuan et al., 2006). Relative mRNA levels are calculated as: 2−ΔΔCt, where ΔΔCt (Dscam) = ΔCt (Dscam)(mutant) – ΔCt (Dscam)(wild-type). We used elav as the reference gene and Chmp1 as the internal control. Following primers were used: Chmp1: 5′-AAAGGCCAAGAAGGCGATTC-3′ and 5′-GGGCACTCATCCTGAGGTAGTT-3′; elav: 5′-CTGCCAAAGACGATGACC-3′ and 5′-TAAAGCC TACTCCTTTCGTC-3′; Dscam[TM1]: 5′- CGTTACCGGAGGCACTATCG-3′ and 5′- ATCGTCTTTGTGGTGA TTGCC-3′; Dscam [TM2]: 5′- CGTTACCGGAGGCACCATT-3′ and 5′- ACTACATCG TAGTACACATCCTTT-3′.
Nested PCR for detecting aberrant exons in Dscam mRNAs
To detect potential aberrant exons between exon 16 and 18 in Dscam mRNAs, 10 ng of cDNA from 3rd instar larval CNS of desired genotypes was used as the templates for nested PCR. Primers F1 and R1 were used for the first round of amplification of the cDNA. Subsequently, 1 μL of the PCR products from the first round were used as the templates for the second round of PCR with primers F2 and R2. The primer sequences are: F1: 5′-TCGAACAACG TGAAGCCCGATAACA-3′; R1: 5′-CCTCGCTTAATCCGGTCACAGGTA-3′; F2: 5′-TGGTACAACCTCCGCATCAC-3′; R2: 5′-TATAGTTGGATCCGGGAACGG-3′.
Computational simulation of Rdl structure
PyMOL was used to simulate protein structures (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger LLC). To predict the structures of Rdl isoforms, the amino acid sequences of RG and RF were aligned with that of the human β3 GABAA receptor, whose crystal structure has been resolved (Miller and Aricescu, 2014). We then selected the insertion sites of the epitope-tags within a predicted surface-exposed loop.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± SEM. Comparisons among multiple groups were performed by One-way ANOVA followed by Student’s t test. Comparisons between two groups were done by Student’s t test. Linear regression was used in Figure 5H. p < 0.05 was considered to be statistically significant. For all quantification, *p < 0.05; **p < 0.01; ***p < 0.001; ns: not significant (p > 0.05).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-RFP | Rockland | RRID: AB_2209751 |
| mouse anti-V5 | Invitrogen | RRID: AB_2556564 |
| chicken anti-GFP | Aves Laboratories | RRID: AB_2307313 |
| mouse anti-GFP | Invitrogen | RRID: AB_221568 |
| rat anti-HA | Roche | RRID: AB_390918 |
| rat anti-Elav | DSHB | RRID: AB_528218 |
| mouse anti-Dscam (cytoplasmic domain) | Shi et al., 2007 | N/A |
| mouse anti-Dscam (ectodomain) | Hattori et al., 2009 | N/A |
| rabbit anti-Dock | Gift from Jack Dixon; Schmucker et al., 2000 | N/A |
| rabbit anti-Wnd | Collins et al., 2006 | N/A |
| anti-rabbit Rhodamine RX | Jackson ImmunoResearch | RRID: AB_2340613 |
| anti-mouse Alexa Fluor 488 | Jackson ImmunoResearch | RRID: AB_2340846 |
| anti-chicken Alexa Fluor 488 | Jackson ImmunoResearch | RRID: AB_2340375 |
| anti-rat Alexa Fluor 488 | Jackson ImmunoResearch | RRID: AB_2749829 |
| anti-mouse HRP | Jackson ImmunoResearch | RRID: AB_10015289 |
| anti-rat HRP | Jackson ImmunoResearch | RRID: AB_2338128 |
| anti-rabbit HRP | Jackson ImmunoResearch | RRID: AB_10015282 |
| Experimental Models: Drosophila Strains | ||
| Dscam[TM1]-RSRT-tlstop-RSRT-HA | This study | N/A |
| Dscam[TM1]-RSRT-short tlstop-RSRT-HA | This study | N/A |
| Dscam[TM2]-RSRT-tlstop-RSRT-V5 | This study | N/A |
| RDL[RG]-RSRT-tlstop-RSRT-V5 | This study | N/A |
| RDL[RF]-RSRT-tlstop-RSRT-V5 | This study | N/A |
| RDL-RSRT-tlstop-RSRT-V5 | Hu et al., 2020 | N/A |
| UAS-Dock-OLLAS | This study | N/A |
| ppk-GAL4 | Grueber et al., 2007 | N/A |
| GMR83B04-GAL4 | Pfeiffer et al., 2008 | N/A |
| UAS-Dscam[TM2]::GFP (#1, 3.36.25, chromosome X) | Wang et al., 2004 | N/A |
| UAS-Dscam[TM2]::GFP (#2, 3.36.25, chromosome III) | Wang et al., 2004 | N/A |
| UAS-Dscam[TM1]::GFP (3.36.25, chromosome X) | Wang et al., 2004 | N/A |
| DscamP-Dscam[TM1]::GFP (3.36.25) | Wang et al., 2004 | N/A |
| DscamP-Dscam[TM2]::GFP (3.36.25) | Wang et al., 2004 | N/A |
| Dscam18 | Wang et al., 2002 | N/A |
| HiwΔN | Wu et al., 2005 | N/A |
| UAS-Wnd | Collins et al., 2006 | N/A |
| UAS- GFP-WndKD | Xiong et al., 2010 | N/A |
| GAL4-OK107 | Bloomington | BL854 |
| dockC506 | Bloomington | BL43376 |
| dock4732 | Bloomington | BL10444 |
| Oligonucleotides | ||
| Dscam[TM1] forward primer: 5’-CGTTACCGGAGGCACTATCG-3’ | This study | N/A |
| Dscam[TM1] reverse primer: 5’- ATCGTCTTTGTGGTGATTGCC-3’ | This study | N/A |
| Dscam[TM2] forward primer: 5’- CGTTACCGGAGGCACCATT-3’ | This study | N/A |
| Dscam[TM2] reverse primer: 5’- ACTACATCGTAGTACACATCCTTT-3’. | This study | N/A |
| Chmp1 forward primer: 5’-AAAGGCCAAGAAGGCGATTC-3’ | Kim et al., 2013 | N/A |
| Chmp1 reverse primer: 5’-GGGCACTCATCCTGAGGTAGTT-3’ | Kim et al., 2013 | N/A |
| Elav forward primer: 5’-CTGCCAAAGACGATGACC-3’ | Kim et al., 2013 | N/A |
| Elav reverse primer: 5’-TAAAGCCTACTCCTTTCGTC-3’ | Kim et al., 2013 | N/A |
| Dscam exon 16–18 forward primer 1: 5’-TCGAACAACG TGAAGCCCGATAACA-3’ | This study | N/A |
| Dscam exon 16–18 Reverse primer 1: 5’-CCTCGCTTAATCCGGTCACAGGTA −3’ | This study | N/A |
| Dscam exon 16–18 forward primer 2: 5’-TGGTACAACCTCCGCATCAC-3’ | This study | N/A |
| Dscam exon 16–18 Reverse primer 2: 5’-TATAGTTGGATCCGGGAACGG-3’ | This study | N/A |
| Software and Algorithms | ||
| Prism 8 | GraphPad | RRID:SCR_002798 |
| Fiji | NIH | RRID:SCR_002285 |
| Leica Microsystems LAS AF (version 2.6.3) | Leica Microscopy | RRID:SCR_008960 |
| Neurolucida | MBF Bioscience | RRID:SCR_001775 |
| Neurolucida Explorer | MBF Bioscience | RRID:SCR_017348 |
Highlights.
Translational stop cassettes allow isoform-specific manipulations of genes
isoTarget enables multi-purpose studies of a targeted isoform in select cells
The subcellular localizations of Dscam isoforms determine their cellular functions
isoTarget is generalizable for studying alternative splicing isoforms
ACKNOWLEDGMENTS
We thank Drs. Kenneth Kwan, Ken Inoki, Yukiko Yamashita, and Dawen Cai for helpful discussions and Dr. Jung Hwan Kim for teaching H.L. and for critical suggestions on isoTarget validation. We also thank Drs. Tzumin Lee, Larry Zipursky, Yi Chen, Jack Dixon, Catherine Collins, Ryan Insolera, Huey Hing, Scott Barolo, and David Lorberbaum for sharing reagents. We thank Drs. Catherine Collins, Laura Smithson, and Elizabeth Cebul for their critiques on an earlier version of the manuscript. This work was supported by grants from NIH (R01 MH112669 and R01 NS104299 to B.Y.) and the Protein Folding Disease Initiative of the University of Michigan to B.Y. and F31NS100391 and T32GM007315 to M.W.V. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108361.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anastassiou D, Liu H, and Varadan V (2006). Variable window binding for mutually exclusive alternative splicing. Genome Biol. 7, R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang LH, Kim J, Stepensky V, and Hing H (2003). Dock and Pak regulate olfactory axon pathfinding in Drosophila. Development 130, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Baralle FE, and Giudice J (2017). Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol 18, 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S, and Wen J (2009). Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol 16, 107–113. [DOI] [PubMed] [Google Scholar]
- Chen Y, Akin O, Nern A, Tsui CY, Pecot MY, and Zipursky SL (2014). Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, and DiAntonio A (2006). Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron 51, 51–69. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Garrity PA, Keshishian H, Zipursky SL, and Zinn K (1999). The Drosophila SH2-SH3 adapter protein Dock is expressed in embryonic axons and facilitates synapse formation by the RP3 motoneuron. Development 126, 1527–1535. [DOI] [PubMed] [Google Scholar]
- Ehrhardt C, Schmolke M, Matzke A, Knoblauch A, Will C, Wixler V, and Ludwig S (2006). Polyethylenimine, a cost-effective transfection reagent. Signal Transduct. 6, 179–184. [Google Scholar]
- Fan X, Labrador JP, Hing H, and Bashaw GJ (2003). Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron 40, 113–127. [DOI] [PubMed] [Google Scholar]
- Goyal G, Zheng J, Adam E, Steffes G, Jain M, Klavins K, and Hummel T (2019). Sphingolipid-dependent Dscam sorting regulates axon segregation. Nat. Commun 10, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and O’Connor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, and Jan YN (2004). Dendritic development: lessons from Drosophila and related branches. Curr. Opin. Neurobiol 14, 74–82. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, and Jan YN (2007). Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development 134, 55–64. [DOI] [PubMed] [Google Scholar]
- Gu P, Gong J, Shang Y, Wang F, Ruppell KT, Ma Z, Sheehan AE, Freeman MR, and Xiang Y (2019). Polymodal nociception in Drosophila requires alternative splicing of TrpA1. Curr. Biol 29, 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori D, Chen Y, Matthews BJ, Salwinski L, Sabatti C, Grueber WB, and Zipursky SL (2009). Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature 461, 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hing H, Xiao J, Harden N, Lim L, and Zipursky SL (1999). Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell 97, 853–863. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang C, Pan G, Liu H, Yu G, and Ye B (2020). A Neural Basis for Categorizing Sensory Stimuli to Enhance Decision Accuracy. Curr Biol 30 10.1016/j.cub.2020.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Bäumer P, Kondo M, Uemura T, and Schmucker D (2007). Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron 54, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, and Jan LY (2010). Branching out: mechanisms of dendritic arborization. Nat. Rev. Neurosci 11, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama D, McGorty R, Kamiyama R, Kim MD, Chiba A, and Huang B (2015). Specification of dendritogenesis site in Drosophila aCC motoneuron by membrane enrichment of Pak1 through Dscam1. Dev. Cell 35, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, and Stamm S (2013). Function of alternative splicing. Gene 514, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Wang X, Coolon R, and Ye B (2013). Dscam expression levels determine presynaptic arbor sizes in Drosophila sensory neurons. Neuron 78, 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B Jr., Lee EJ, Manning RW, Yu SH, Mulder KL, and Westphal H (1992). Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl. Acad. Sci. USA 89, 6232–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, and Luo L (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- Lee Y, and Rio DC (2015). Mechanisms and regulation of alternative premrna splicing. Annu. Rev. Biochem 84, 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, Geschwind DH, Black DL, and Martin KC (2016). Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron 89, 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JK, Kuo F, Motti D, Morris R, Bixby JL, and Lemmon VP (2012). Isoform diversity and regulation in peripheral and central neurons revealed through RNA-Seq. PLoS ONE 7, e30417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li W, Wang L, Kar A, Guan KL, Rao Y, and Wu JY (2009). DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc. Natl. Acad. Sci. USA 106, 2951–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, and Grueber WB (2007). Dendrite self-avoidance is controlled by Dscam. Cell 129, 593–604. [DOI] [PubMed] [Google Scholar]
- Miller PS, and Aricescu AR (2014). Crystal structure of a human GABAA receptor. Nature 512, 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H (2015). Quantitative nature of overexpression experiments. Mol. Biol. Cell 26, 3932–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Svoboda K, and Rubin GM (2011). Multiple new site-specific recombinases for use in manipulating animal genomes. Proc. Natl. Acad. Sci. USA 108, 14198–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Cheong C, Idoyaga J, Kim JY, Choi JH, Do Y, Lee H, Jo JH, Oh YS, Im W, et al. (2008). Generation and application of new rat monoclonal antibodies against synthetic FLAG and OLLAS tags for improved immunodetection. J. Immunol. Methods 331, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TTB, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. (2008). Tools for neuro-anatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G (2012). Gene overexpression: uses, mechanisms, and interpretation. Genetics 190, 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit AA, Li W, Qu C, Dwyer T, Shao Q, Guan KL, and Liu G (2012). Down syndrome cell adhesion molecule (DSCAM) associates with uncoordinated-5C (UNC5C) in netrin-1-mediated growth cone collapse. J. Biol. Chem 287, 27126–27138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röth S, Fulcher LJ, and Sapkota GP (2019). Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci 76, 2761–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, and Zipursky SL (2000). Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684. [DOI] [PubMed] [Google Scholar]
- Scotti MM, and Swanson MS (2016). RNA mis-splicing in disease. Nat. Rev. Genet 17, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Yu H-H, Yang JS, and Lee T (2007). Specific Drosophila Dscam Juxtamembrane Variants Control Dendritic Elaboration and Axonal Arborization. J Neurosci 27, 6723–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, and Baker KE (2015). Nonsense-mediated RNA decay—a switch and dial for regulating gene expression. BioEssays 37, 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, and Jan YN (2007). Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 54, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne GR, Kim JH, and Ye B (2015). Dysregulated Dscam levels act through Abelson tyrosine kinase to enlarge presynaptic arbors. eLife 4, e05196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzio M, Schiavo G, and Fainzilber M (2017). Compartmentalized signaling in neurons: from cell biology to neuroscience. Neuron 96, 667–679. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CHJ, and Lee T (2004). Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron 43, 663–672. [DOI] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CHJ, and Lee T (2002). Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron 33, 559–571. [DOI] [PubMed] [Google Scholar]
- Wu CL, Wairkar YP, Collins CA, and DiAntonio A (2005). Highwire function at the Drosophila neuromuscular junction: Spatial, structural, and temporal requirements. J Neurosci 25, 9557–9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, Diantonio A, and Collins CA (2010). Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J. Cell Biol 191, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JSJ, Bai JM, and Lee T (2008). Dynein-dynactin complex is essential for dendritic restriction of TM1-containing Drosophila Dscam. PLoS ONE 3, e3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, and Makeyev EV (2016). Functional impact of splice isoform diversity in individual cells. Biochem. Soc. Trans 44, 1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, and Jan YN (2007). Growing dendrites and axons differ in their reliance on the secretory pathway. Cell 130, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Kim JH, Yang L, McLachlan I, Younger S, Jan LY, and Jan YN (2011). Differential regulation of dendritic and axonal development by the novel Krüppel-like factor Dar1. J. Neurosci 31, 3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KT, Li LS, Kim NG, Kang HJ, Koh KH, Chwae YJ, Kim KM, Kim YK, Park SM, Jang SK, and Kim H (2007). Selective translational repression of truncated proteins from frameshift mutation-derived mRNAs in tumors. PLoS Biol. 5, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, and Stewart CN Jr. (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Yang Y, Zhang W, Pan H, Chen R, Shi F, and Jin Y (2013). Regulation of Dscam exon 17 alternative splicing by steric hindrance in combination with RNA secondary structures. RNA Biol 10, 1822–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, and Zipursky SL (2004). Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron 43, 673–686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in the study are available from the corresponding authors on reasonable request.


