Abstract
Background
Long noncoding RNAs play influential roles in the progression of many types of human malignancies. The present study aimed to explore the prognostic value of long noncoding RNA FTX (FTX) on patients with glioma.
Methods
FTX expression in glioma specimens and matched adjacent non‐neoplasm specimens was examined by a quantitative real‐time polymerase chain reaction assay. Furthermore, assays of the relationships between FTX expression and clinicopathologic characteristics of patients with glioma were also performed. Kaplan–Meier methods were applied for the assays of the overall survival (OS) and progression‐free survival (PFS) of patients and Cox regression assays were used to analyze the clinical value of FTX used as a possible biomarker.
Results
FTX levels were significantly up‐regulated in glioma specimens compared to the paired non‐neoplasm specimens (p < 0.01). Furthermore, high FTX expression in neoplasm tissues was dramatically associated with World Health Organization grade (p = 0.001) and Karnofsky Performance Score (p = 0.009). Kaplan–Meier assays with 187 patients revealed that patients with high level of FTX expression displayed poorer OS (p = 0.002) and PFS (p = 0.000). Subsequently, multivariable Cox regression analysis identified FTX expression as an independent prognostic factor of unfavorable survivals in glioma (OS: p = 0.001; PFS: p = 0.002).
Conclusions
These findings indicated that FTX may be a novel predictor for prognostic assessment of glioma patients. However, studies conducted with larger numbers of patients are essential to confirm our findings.
Keywords: glioma tumor, molecular genetics, neuroscience, oncology
1. INTRODUCTION
Gliomas are the most common and deadly brain neoplasms of the central nervous system, accounting for 2.3% of all original tumor cases and 2.8% of cancer‐associated deaths worldwide. 1 , 2 According to the World Health Organization (WHO) criteria, glioma can be classified into four grades: diffuse astrocytoma, pilocytic astrocytoma, anaplastic astrocytoma and glioblastoma multiform (GBM). 3 Despite new improvement to the standard of care therapy for glioma, the 5‐year survival rate of GBM patients is less than 3%. 4 The invasiveness and the chemoresistance of GBM are the main barriers to an effect therapy. Therefore, it is imperative to exploit novel prognostic biomarkers, which can guide new treatment modalities.
Long non‐coding RNAs (lncRNAs) are a large class of non‐coding transcripts that are >200 bases in length. 5 It has been confirmed that lncRNAs displayed critical roles in cellular processes, such as cellular growth, survival, migration, invasion and differentiation. 6 , 7 , 8 Recently, more and more evidence has indicated that lncRNAs are capable of performing as oncogenes or tumor suppressors, or as a predictor of prognosis in various tumors, including glioma. 9 , 10 For example, Liu et al. 11 reported that overexpression of LOC730101 promoted the proliferation of lung cancer cell by regulation of Wnt/β‐catenin signaling. Wang et al. 12 demonstrated that the deregulated expression of lncRNA HULC may influence the clinical outcome of cervical cancer. A recent study by Chao et al. 13 reported that low lncRNA TUSC7 expression predicted a significantly shorter survival time in glioma patients, and its overexpression suppressed glioma migration and invasion by sponging miR‐23b. However, to date, only a few lncRNAs have been functionally annotated in glioma.
lncRNA FTX (FTX), located within the X‐inactivation center, is a newly identified lncRNA. 14 Previous studies reported that FTX was evidently highly expressed in glioma 15 and renal cell carcinoma, 16 whereas down‐regulation of FTX was observed in hepatocellular carcinoma. 17 These results suggest FTX may exerted different function in different types of tumors. To the best of our knowledge, whether FTX had a clinical influence on the long‐term survivals of glioma patients had not been reported. The present study aimed to explore the expression pattern and clinical involvement of FTX in glioma patients.
2. MATERIALS AND METHODS
2.1. Patients and specimens
In tota, 187 paired primary glioma specimens and matched non‐neoplasm tissues were acquired from patients with glioma undergoing surgery in the First Hospital of Shandong First Medical University. Informed consent was obtained from all patients. The diagnosis of all specimens was based on pathological evidence. All samples were immediately frozen and stored at −80°C for further experiments. Based on the standard of the WHO classification and grading system, staging and grading was pathologically determined. Before surgery, all of the patients had not received chemotherapy, radiotherapy or immunotherapy. The study was approved by the Research Ethics Committee of the first affiliated hospital of Shandong First Medical University.
2.2. Quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNA was extracted from glioma samples and matched adjacent normal samples using TRIzol1 reagent (Invitrogen, Jiangning, Nanjing, China). Total RNA was quantified using a Nanodrop ND‐1110 spectrophotometer (NanoDrop, Hangzhou, Zhejiang, China). We purchased a Reverse Transcription Kit (Takara, Dalian, Niaoling, China), which was used to reverse transcribe the isolated RNA into cDNA. A qRT‐PCR was conducted with SYBR Premixs Ex# Taq (TaKaRa, Otsu, Shiga, Japan) on a CFX96 Real‐Time PCR Examination System. GAPDH was used as an endogenous control. Table 1 shows the primers designed by Gema Technology (Pudong, Shanghai, China). The relative expression of FTX was calculated by the use of the 2−ΔΔCt methods. The assays were performed in triplicate.
TABLE 1.
Primers used in the present study
| Gene | Primer sequences |
|---|---|
| FTX | Forward primer: 5'‐TATGCCACCTAGCCTTTCTACA‐3' |
| Reverse primer: 5'‐ATCTCTTCA AAAGCGGCATAAT‐3' | |
| GAPDH | Forward primer: 5'‐GCACCGTCAAGGCTGAGAAC‐3' |
| Reverse primer: 5'‐TGGTGAAGACGCCAGTG GA‐3' |
2.3. Statistical analysis
All data quantification and statistical analysis were performed using SPSS, version 19.0 software (IBM Corp., Armonk, NY, USA). The differences between groups were analyzed using Student's t‐test. The correlations of FTX with the clinical factors of patients was determined by a chi‐squared test. Overall survival (OS) and progression‐free survival (PFS) were evaluated by Kaplan–Meier analyses with a log‐rank test. The prognostic value of FTX was examined by a Cox regression assay. p < 0.05 was considered a statistically significant.
3. RESULTS
3.1. FTX up‐regulation in glioma specimens
To explore the possible role of FTX in glioma development, the expression of FTX was determined in 187 pairs of glioma tissues and adjacent non‐neoplasm tissues. This revealed that FTX expression was increased in specimen tissues compared to matched adjacent non‐neoplasm tissues (p < 0.01) (Figure 1). Our results indicated that FTX up‐regulation may contribute to glioma development.
FIGURE 1.
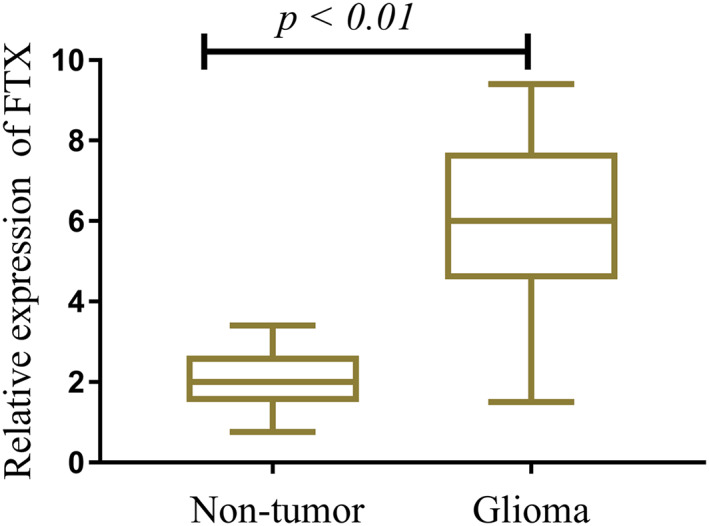
FTX expression in 187 glioma tissues samples and corresponding normal tissues
3.2. Relationship between FTX and clinicopathologic features of glioma patients
Given the distinct overexpression of FTX in glioma, we further considered whether FTX had possible clinical relevance in glioma patients. We categorized the patients into low and high expression groups according to the mean value of FTX expression. Subsequently, data assays with a chi‐squared test revealed that the expression of FTX was inversely correlated with progressive clinicopathologic features, including WHO grade (p = 0.001) and Karnofsky Performance Score (KPS) (p = 0.009) (Table 2). However, correlations of FTX expression with other parameters, including age, gender and extent of resection, were not observed (Table 2).
TABLE 2.
Correlations between FTX expression and clinicopathologic features in glioma patients
| Variable | n | FTX expression | p value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.533 | |||
| ≥ 45 | 116 | 55 | 61 | |
| < 45 | 71 | 37 | 34 | |
| Gender | 0.127 | |||
| Male | 89 | 49 | 40 | |
| Female | 98 | 43 | 55 | |
| Extent of resection | 0.534 | |||
| Partial | 57 | 30 | 27 | |
| Total | 130 | 62 | 68 | |
| WHO grade | 0.001 | |||
| I/II | 111 | 66 | 45 | |
| III/IV | 76 | 26 | 50 | |
| KPS score | 0.009 | |||
| ≥ 80 | 123 | 69 | 54 | |
| < 80 | 64 | 23 | 41 | |
3.3. FTX overexpression was a clinical regulatory factor in glioma patients
The prognostic value of FTX expression with respect to glioma patient survival was evaluated via Kaplan–Meier survival assays. Our results suggested that patients with high FTX expression showed significantly reduced 5‐year OS and PFS (p = 0.002 and p = 0.000, respectively) (Figures 2 and 3). Then, Cox regression assays were applied to evaluate the prognostic value of FTX expression in glioma. As shown in Table 3, FTX was identified as an independent prognostic factors for both OS (p = 0.001) and PFS (p = 0.001).
FIGURE 2.
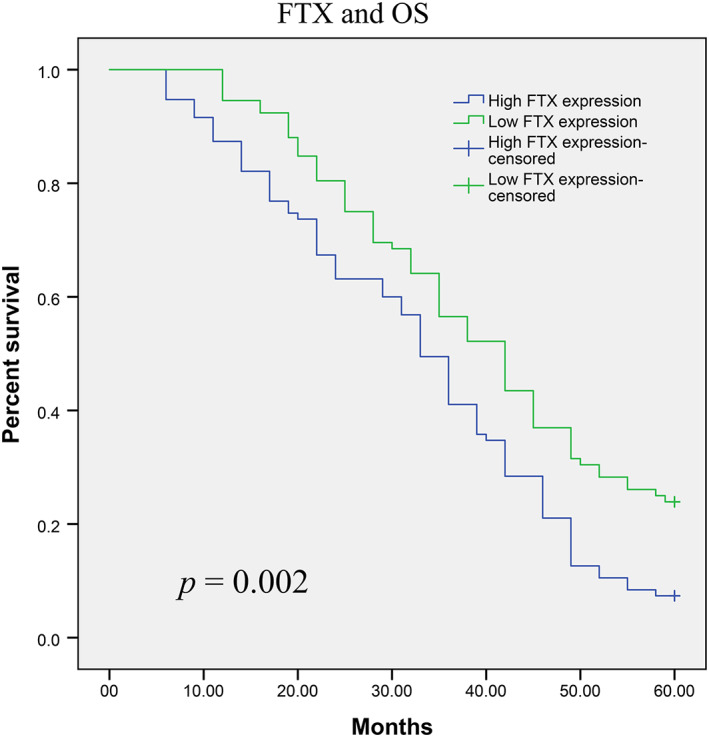
Glioma patients with high FTX expression had a significantly shorter OS than those with low FTX expression (p = 0.002, log‐rank test)
FIGURE 3.
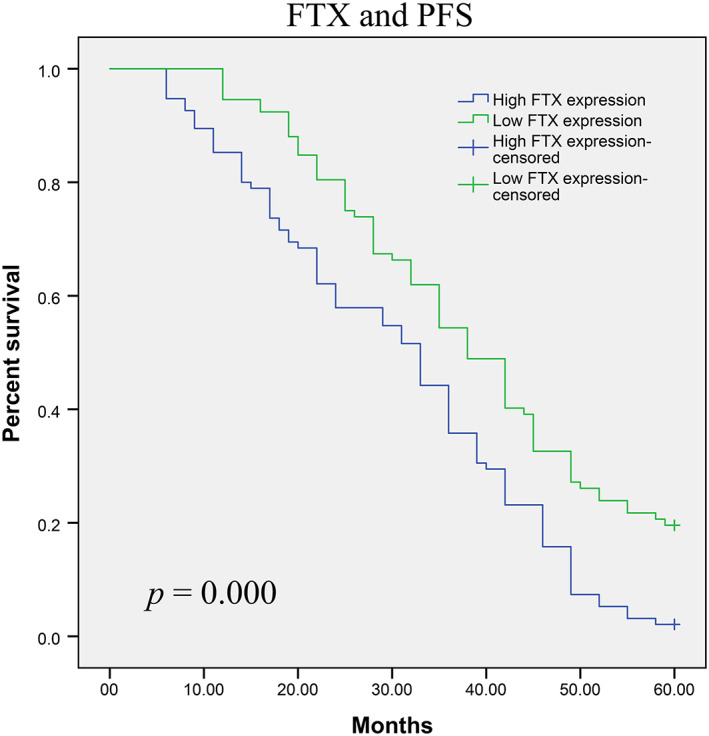
Glioma patients with high FTX expression had a significantly shorter PFS than those with low FTX expression (p = 0.000, log‐rank test)
TABLE 3.
Cox regression model for multivariate analysis of glioma prognostic factors
| Variable | Overall survival | Progression‐free survival | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |
| Age | 1.423 | 0.832–1.849 | 0.317 | 1.631 | 0.944–2.213 | 0.186 |
| Gender | 1.679 | 0.927–2.432 | 0.144 | 1.832 | 0.774–2.632 | 0.084 |
| Extent of resection | 1.321 | 0.755–1.934 | 0.138 | 1.583 | 0.893–1.737 | 0.155 |
| WHO grade | 3.674 | 1.663–6.343 | 0.001 | 4.362 | 1.993–7.822 | 0.001 |
| KPS score | 3.228 | 1.342–5.489 | 0.004 | 3.893 | 1.542–6.138 | 0.007 |
| FTX expression | 5.358 | 1.775–8.569 | 0.001 | 6.323 | 2.213–9.934 | 0.001 |
CI, confidence interval; HR, hazard ratio.
4. DISCUSSION
Considering that the survival rate of glioma is still low, novel and more effective prognostic biomarkers are immediately essential for guiding the individual treatment of glioma patients. 18 To date, traditional staging systems may not be effective for predicting outcome in individual patients. 19 , 20 Recently, because of advances in the investigation of glioma carcinogenesis, more and more key molecular events are being identified. 21 Many scientists try to find effective biomarkers such as tumor‐related genes, microRNAs and long noncoding RNAs. 22 , 23 , 24 Compared with other biomarkers, 25 lncRNAs are considered to be ideal biomarkers because of their important regulation effect in glioma progression.
Metastasis and recurrence are the main cause of mortality in tumor patients. Because tumor progression is very complex, the underlying mechanism for tumor metastasis and recurrence is also very unclear. Previous studies have reported that FTX was involved in the regulation of metastasis and clinical prognosis in various types of human malignancies. For example, Liu et al. 17 found that FTX was decreased in liver cancer and indicated a poor prognosis. Furthermore, forced FTX expression inhibited the proliferation and metastasis of tumor cells by modulating miR‐374a/MCM2. He et al. 16 reported that FTX may function as a neoplasm promoter in renal cell carcinoma and its knockdown could suppress proliferation, migration and invasion. More importantly, recent findings by Zhang et al. 15 highlighted the promoting role of FTX in glioma through negatively regulating miR‐342‐3p. Although the tumor‐promotive role of FTX in glioma was confirmed by an in vitro assay, its prognostic significance in glioma remains unknown.
In the present study, to explore the biological implication of FTX in glioma patients, our group examined the levels of FTX in glioma tissues and matched normal tissues. Unsurprisingly, in accordance with previous reports, the results of the present study showed that significant up‐regulation of FTX was observed in glioma tissues. In addition, we found a similar phenomenon in which high expression of FTX in glioma was significantly associated with various clinicopathologic characteristics, such as WHO grade and KPS score. These results indicated that FTX is a biomarker for a poor prognosis in glioma patients. The subsequent survival assay confirmed our hypothesis. Finally, multivariate assays confirmed that FTX expression was an independent prognostic marker for PFS and OS in glioma patients.
5. CONCLUSIONS
In conclusion, for the first time, the current data offer convincing evidence for FTX being frequently up‐regulated in glioma and it comprise a useful poor prognostic biomarker for glioma patients. In the future, the molecular mechanisms of FTX that are involved in glioma must be investigated further.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
Liang Y, Lu H. Long noncoding RNA FTX is associated with prognosis of glioma patients. J Gene Med. 2020;22:e3237 10.1002/jgm.3237
Yongjuan Liang and Hongzhen Lu contributed equally to this work.
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323‐342. [DOI] [PubMed] [Google Scholar]
- 2. Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;100:2235‐2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thurnher MM. 2007 World Health Organization classification of tumours of the central nervous system. Cancer Imaging. 2009;9(Special issue A):S1‐S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lv D, Lu L, Hu Z, et al. Nestin expression is associated with poor Clinicopathological features and prognosis in glioma patients: an association study and meta‐analysis. Mol Neurobiol. 2017;54:727‐735. [DOI] [PubMed] [Google Scholar]
- 5. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629‐641. [DOI] [PubMed] [Google Scholar]
- 6. Gibb EA, Brown CJ, Lam WL. The functional role of long non‐coding RNA in human carcinomas. Mol Cancer. 2011;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non‐coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063‐1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z, Zhang LW. Identification of key long non‐coding RNAs as competing endogenous RNAs for miRNA‐mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20:2285‐2295. [PubMed] [Google Scholar]
- 9. Zheng J, Li XD, Wang P, et al. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR‐186. Oncotarget. 2015;6:25339‐25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non‐coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu L, Zhang Y, Cao W. Highly expressed lncRNA LOC730101 promotes lung cancer cell growth through Wnt canonical pathway. Biochem Biophys Res Commun. 2017;493:992‐997. [DOI] [PubMed] [Google Scholar]
- 12. Wang YF, Zhang S, Li XQ, Wang Y. Expression of lncRNA HULC in cervical cancer and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20:3987‐3991. [PubMed] [Google Scholar]
- 13. Shang C, Guo Y, Hong Y, Xue YX. Long non‐coding RNA TUSC7, a target of miR‐23b, plays tumor‐suppressing roles in human gliomas. Front Cell Neurosci. 2016;10:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chureau C, Chantalat S, Romito A, et al. Ftx is a non‐coding RNA which affects Xist expression and chromatin structure within the X‐inactivation center region. Hum Mol Genet. 2011;20:705‐718. [DOI] [PubMed] [Google Scholar]
- 15. Zhang W, Bi Y, Li J, et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR‐342‐3p. Lab Invest. 2017;97:447‐457. [DOI] [PubMed] [Google Scholar]
- 16. He X, Sun F, Guo F, et al. Knockdown of long noncoding RNA FTX inhibits proliferation, migration, and invasion in renal cell carcinoma cells. Oncol Res. 2017;25:157‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu F, Yuan JH, Huang JF, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR‐374a. Oncogene. 2016;35:5422‐5434. [DOI] [PubMed] [Google Scholar]
- 18. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842‐1850. [DOI] [PubMed] [Google Scholar]
- 19. Grant R, Kolb L, Moliterno J. Molecular and genetic pathways in gliomas: the future of personalized therapeutics. CNS Oncol. 2014;3:123‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kang JH, Adamson C. Novel chemotherapeutics and other therapies for treating high‐grade glioma. Expert Opin Investig Drugs. 2015;24:1361‐1379. [DOI] [PubMed] [Google Scholar]
- 21. Kondo Y, Katsushima K, Ohka F, Natsume A, Shinjo K. Epigenetic dysregulation in glioma. Cancer Sci. 2014;105:363‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang YA, Zhou Y, Luo X, et al. SHOX2 is a potent independent biomarker to predict survival of WHO grade II‐III diffuse gliomas. EBioMedicine. 2016;13:80‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jing SY, Jing SQ, Liu LL, Xu LF, Zhang F, Gao JL. Down‐expression of miR‐373 predicts poor prognosis of glioma and could be a potential therapeutic target. Eur Rev Med Pharmacol Sci. 2017;21:2421‐2425. [PubMed] [Google Scholar]
- 24. Li Y, Wang X. Role of long noncoding RNAs in malignant disease (review). Mol Med Rep. 2016;13:1463‐1469. [DOI] [PubMed] [Google Scholar]
- 25. Lan F, Yue X, Xia T. Exosomal microRNA‐210 is a potentially non‐invasive biomarker for the diagnosis and prognosis of glioma. Oncol Lett. 2020;19:1967‐1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.


