Graphical abstract
Keywords: SARS-CoV-2, COVID, PLpro, Mpro, RdRp
Abstract
The unprecedented scale of the ongoing COVID-19 pandemic has catalyzed an intense effort of the global scientific community to unravel different aspects of the disease in a short time. One of the crucial aspects of these developments is the determination of more than three hundred experimental structures of SARS-CoV-2 proteins in the last few months. These include structures of viral non-structural, structural, and accessory proteins and their complexes determined by either X-ray diffraction or cryo-electron microscopy. These structures elucidate the intricate working of different components of the viral machinery at the atomic level during different steps of the viral life cycle, including attachment to the host cell, viral genome replication and transcription, and genome packaging and assembly of the virion. Some of these proteins are also potential targets for drug development against the disease. In this review, we discuss important structural features of different SARS-CoV-2 proteins with their function, and their potential as a target for therapeutic interventions.
Introduction
The coronavirus disease 2019 (COVID-19), caused by the virus SARS-CoV-2, has emerged as one of the most widespread and devastating pandemics in the recorded history of mankind. To mitigate the impact of the pandemic, intense efforts are being put in by researchers from academia and industry to develop diagnostic, therapeutic, and vaccine candidates. Also, remarkable are the efforts of the scientific community to visualize and understand the complex biology driving the pandemic through structure–function studies of different SARS-CoV-2 proteins. The efforts began soon after the publication of the viral genome sequence in January 2020. The crystal structure of SARS-CoV-2 main protease (PDB ID: 6LU7) was determined and released in the first week of February 2020.1 Thereafter, the cryo-electron microscopy (cryo-EM) structure of the virus’s spike protein was released. This was followed by several other structures of viral non-structural, structural and accessory proteins. As on September 3, 2020, there are 369 structures of various SARS-CoV-2 proteins in the apo form or in complex with ligands and other proteins in the PDB (www.rcsb.org). Visualizing these structures to atomic details provided important insights into the molecular basis of different steps in the viral life cycle. Many of these structures are also useful as a target for the structure-based drug design against COVID-19. This review attempts to put together all the available structural information on SARS-CoV-2 proteins.
Genome organization and lifecycle of SARS-CoV-2
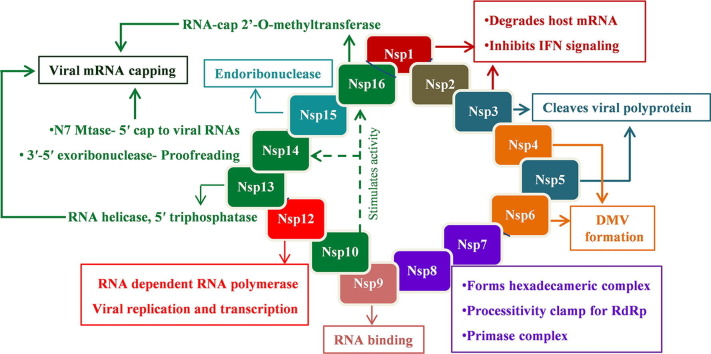
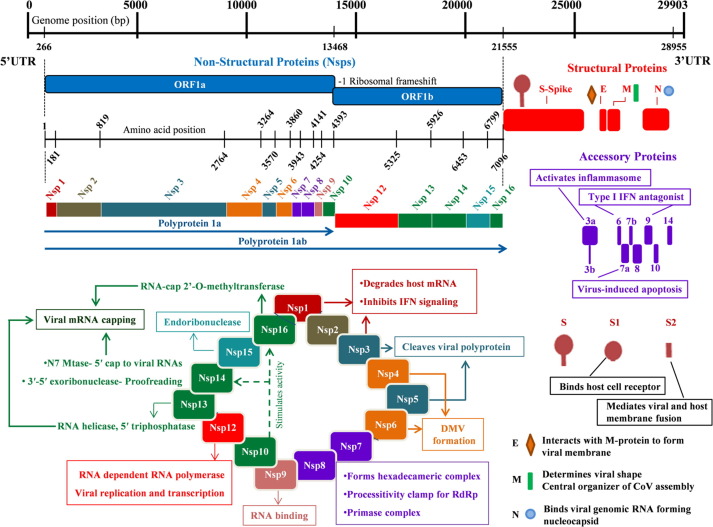
Coronaviruses are the enveloped, single-stranded RNA viruses. In recent years, some of the coronaviruses belonging to the genus betacoronavirus, namely the Severe acute respiratory syndrome coronavirus (SARS-CoV), the Middle East respiratory syndrome coronavirus (MERS-CoV), and the Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have caused serious illnesses in humans. SARS-CoV-2 has ~30 kb positive-sense single-stranded RNA genome which shares ~80% sequence identity with that of SARS-CoV. The genomic RNA (gRNA) of SARS-CoV-2 comprises 14 open reading frames (ORFs) (Fig. 1 ). Two main ORFs, ORF1a and ORF1b, overlapping with a (-1) ribosomal frame-shift, encompasses two-thirds of the genome and are translated to polyproteins pp1a and pp1ab, respectively.[2], [3] These polyproteins are processed by viral proteases (papain-like protease and main-protease) to produce nonstructural proteins (Nsps), Nsp1 to Nsp16.4 Some of the Nsps along with a few host factors form a replication-transcription complex (RTC) inside a double-membrane vesicles (DMV). The RTCs are the central hub for viral genome replication and transcription.[3], [5] The remaining one-third of the genome has overlapping ORFs, encoding four major structural proteins: spike (S), Membrane (M), Envelope (E) and Nucleocapsid (N), and some accessory proteins including ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF9 and ORF10 (Fig. 1).
Fig. 1.
Genome organization of the SARS-CoV-2. The viral genome encodes 16 Non-structural proteins (Nsps) required for replication/transcription along with the structural proteins required for the assembly of new virions. The proteins are marked below the genome with their respective coding regions. A short description of the functions of different proteins is also shown.
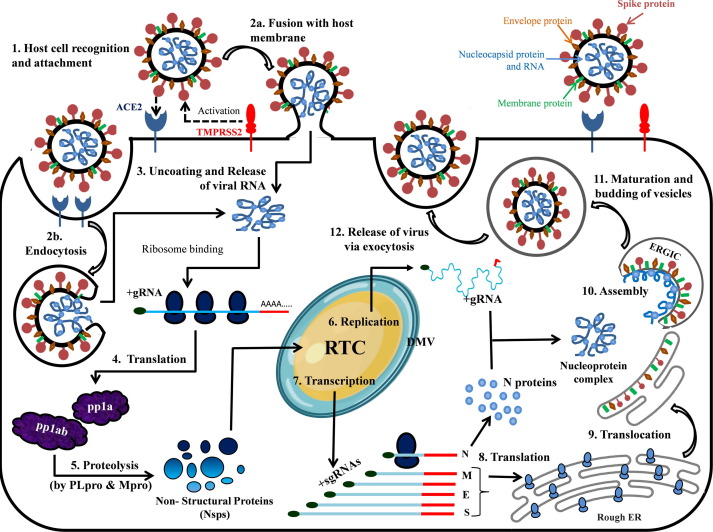
During the viral infection, SARS-CoV-2 injects its genome into the host cell via endosomes or direct fusion of the viral envelope with the host cell membrane, mediated by binding of the spike (S) protein to the human angiotensin-converting enzyme 2 (ACE2) at the cell surface (steps 1–2, Fig. 2 ).[6], [7] Following the entry into the host cell, the viral gRNA is uncoated and released into the host cell cytoplasm and translated by the host ribosomes (steps 3–4). The translation products, polyproteins pp1a and pp1ab, are proteolytically cleaved into nonstructural proteins Nsp1 − 16 by the viral proteases, PLpro and Mpro (step 5). Several Nsps (Nsp2-16) along with other factors assemble together to form RTC complex inside the infected host cell. While Nsp2-11 are supposed to play a supporting role, Nsp12-16 provide the required enzymatic function for viral genome replication/transcription inside the RTC. The RNA (+) strand first gets replicated to the RNA (-) strand and then the negative-strand is used either for replication to the RNA (+) strand for new virion assembly (step 6) or transcription of sub-genomic mRNAs (steps 7). These sub-genomic mRNAs are translated to the structural proteins – S, M, E, N, and the accessory proteins (step 8). The S, M, and E proteins enter the endoplasmic reticulum (ER), and the N protein attaches to the genomic RNA (+) strand to produce nucleoprotein complex.[8], [9] The nucleoprotein complex and the structural proteins move to the ER-Golgi intermediate compartment (ERGIC) where the virions assemble, mature, and bud off from the Golgi in the form of small vesicles (steps 9–11). These vesicles travel to the host cell membrane where they are released into the extracellular region through exocytosis (step 12) (Fig. 2). The released virions infect a new set of cells leading to disease progression.[4], [5]
Fig. 2.
The infection cycle of SARS-CoV-2 inside the host cell. The sequence of events, from host cell recognition through the release of new virion, is represented graphically as steps 1 to 12.
Nonstructural proteins (NSPs)
Nsp1
Nsp1 is the N-terminal cleavage product of polyprotein precursors, pp1a and pp1ab, produced through proteolysis by viral papain-like protease (PLpro). Nsp1 interferes with the host cell protein synthesis by binding to the 40S ribosomal subunit and endonucleolytic cleavage of host mRNA.10 The translational slowdown prevents adequate expression of several host factors in response to the viral infection and subsequent clearance by the innate immune system. Although Nsp1 hinders the host protein expression, it doesn’t prevent the expression of viral proteins.[11], [12] The conserved stem-loop region in 5′-UTR of viral mRNAs seems to interact with Nsp1 in such a way that it allows their translation through unknown mechanisms.13
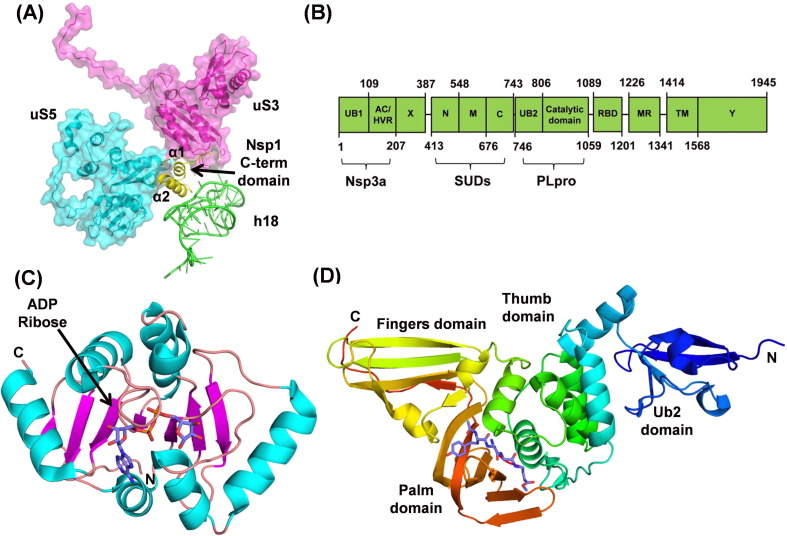
Recent cryo-EM structures of Nsp1 in complex with the 40S ribosomal subunit provided the structural basis of translation inhibition.[11], [12] It was shown that the C-terminal domain of Nsp1 blocks the mRNA entry channel. The C-term has a short α-helix joined to a larger α-helix through a short loop. The shorter α-helix interacts with the ribosomal proteins uS3 and uS5 while the loop interacts with 18s rRNA helix h18 (Fig. 3 (A)). The loop contains the conserved residues, K164 and H165, which make key interactions with h18. Mutations K164A and H165A lead to the loss of 40S ribosomal subunit binding and thus translational inhibition.[10], [11], [12] The larger α-helix also interacts with rRNA h18 and uS5. Nsp1 of SARS-CoV-2 shows 84% amino acid sequence identity with SARS-CoV Nsp1, suggesting a similar structure. The structure of SARS-CoV Nsp1 has been determined earlier by NMR.14 In their structure, C-term is flexibly disordered. Residues 13–128 at the N-term of Nsp1 form a novel ɑ/β fold consisting of a mixed six-stranded β-barrel. The opening on one side of the barrel is covered by an α-helix, and a 310-helix is present alongside the barrel. However, in the cryo-EM structures of SARS-CoV-2 Nsp1-ribosome complexes, the N-terminal region is not unambiguously determined. Though the virus encodes several other factors to evade immune responses, the therapeutic agents targeting the interaction between Nsp1 and the 40S ribosomal subunit would support the innate immune system to destroy the virus.[11], [12]
Fig. 3.
(A) Nsp1 C-terminal domain interacting with the ribosomal proteins (uS3 and uS5) and 18s rRNA helix h18. (B) Domain organization of the SARS-CoV-2 Nsp3 (C) Cartoon representation of the SARS-CoV-2 Macro-X domain with bound ADP-ribose (PDB ID: 6W02). (D) Cartoon representation of PLpro (PDB ID: 6WX4) showing different subdomains and an inhibitor bound in the catalytic site.
Nsp3
Nsp3, a multi-domain membrane-bound protein (Fig. 3(B)), is the largest protein (1945 amino acid residues) encoded by the coronavirus genome.15 It acts as a membrane-anchored scaffold that interacts with the other Nsps and the host proteins to form the viral replication-transcription complex.[16], [17] Nsp3 consists of the N-terminal Nsp3a domain (includes ubiquitin-like domain 1 (UB1) and acidic domain (Ac) or hypervariable region (HVR)), Macrodomain-X, SARS unique domains (SUDs), papain-like protease domain (includes ubiquitin-like domain 2 (UB2) and catalytic core domain), RNA binding domain (RBD), marker domain (MR), transmembrane domains (TM), and Y-domain (Fig. 3(b)). The papain-like protease (PLpro) domain cleaves Nsp3 from polyproteins pp1a and pp1ab.18 Currently, crystal structures of the Macrodomain-X and the PLpro domain of Nsp3 from SARS-CoV-2 are available in the PDB.
Macrodomain-X
Earlier named as the “X” domain, it is also known as Macrodomain-X because of its similarity with the macrodomain part of the human histone 2A variant, macroH2A.[19], [20] Although the precise role of the Macrodomain-X in viral replication and pathogenesis remains elusive, several pieces of evidence show that it interferes with the host innate immune response.[21], [22], [23], [24], [25], [26] Similar to the Macrodomain from other coronaviruses, structural and biophysical data have shown that SARS-CoV-2 Macrodomain-X binds to ADP-ribose[27], [28] and removes ADP-ribose from the proteins through its hydrolase activity.[27], [28], [29] ADP-ribosylation is a post-translational regulatory modification involved in signal transduction, immune response, DNA damage repair, and cellular stresses.[30], [31] However, specific molecular targets for de-ADP-ribosylation by the viral macrodomains are not known yet.
Recently, several apo and liganded crystal structures of Macrodomain-X of SARS-CoV-2 have been deposited in the PDB. Macrodomain-X adopts a three-layered ɑ/β/ɑ fold.[20], [27], [28] The structure consists of a seven-stranded mixed β-sheet flanked on either side by three helices each (Fig. 3(C)). The ADP-ribose binding pocket lies between the helices and the C-terminal edge of the central four parallel strands of the β-sheet. Surface exposed loops, β3-α2 and β6-α5 take part in the ligand binding (Fig. 3(c)). Since Macrodomain-X has a high affinity to bind to ADP-ribose, the binding site could be targeted to develop therapeutics against SARS-CoV-2.
Papain-like protease (PLpro)
The PLpro, proteolytically cleaves the viral polyprotein precursors, pp1a and pp1ab, at three sites to produce non-structural proteins Nsp1, Nsp2, and Nsp3.18 The consensus sequence of recognition for proteolysis by PLpro is LXGG↓X (P4P3P2P1↓P1′). Interestingly, both ubiquitin (Ub) and ISG15 (interferon-stimulated gene product 15) carry the sequence LXGG at their C-terminus. Consequently, PLpro has both deubiquitinating and deISG15ylating activities.[32], [33], [34], [35] The post-translational modifications of signaling molecules, in the form of ubiquitination and ISGylation, are known to activate the innate immune responses[36], [37], [38] and the enzymatic activity of PLpro may significantly hinder such responses.
There is a high overall sequence similarity between SARS-CoV and SARS-CoV-2 Plpros (83% identity). Similar to SARS-CoV PLpro, the enzyme consists of N-terminus ubiquitin-like domain which is well separated from the catalytic core domain.[39], [40] The catalytic core domain adopts an open right-hand architecture with the thumb, the palm, and the fingers subdomains (Fig. 3(D)). The thumb subdomain is formed by four ɑ-helices, while a six-stranded β-sheet makes the palm. The finger subdomain is formed by a four-stranded, twisted, anti-parallel β-sheet. A zinc ion is coordinated by four cysteine residues in the fingertips region with tetrahedral geometry which is essential for structural integrity and activity of the enzyme.39
PLpro is a cysteine protease with its active site located in the cleft between thumb and palm subdomains. The active site contains a catalytic triad of cysteine, histidine, and aspartic acid residues. A flexible β-loop (BL2 loop) of the palm domain, present near the entrance of the active site, acts as a flap/gate. The flap is observed in an open conformation in the unliganded PLpro structures. It closes over the catalytic cleft entrance upon inhibitor binding, via an induced-fit mechanism, forming intermolecular interactions with the ligand. The loop can assume multiple conformations depending upon the size and type of the inhibitor molecule.[40], [41], [42] Consideration of the conformation of the flap will be very crucial for future structure-based drug design efforts against SARS-CoV-2 PLpro.
Amino acids forming S4-S1 subsites are identical in SARS-CoV and SARS-CoV-2 PLpros. The S1 and the S2 subsites are rather restrictive and can accommodate only glycine residues. The S3 pocket has a preference for positive and hydrophobic residues. The S4 pocket can accommodate hydrophobic residues only. Using a hybrid combinatorial substrate library approach, it was shown that both SARS-CoV and SARS-CoV-2 PLpro recognize natural and unnatural amino acids in a very similar fashion.[40], [43] The enzyme kinetics studies with the tetrapeptide substrates for SARS-CoV and SARS-CoV-2 PLpros show that the catalytic efficiencies of both the enzymes are similar. The same S4-S1 subsites architecture, substrate preferences, and catalytic efficiencies of PLpros from both these viruses indicate that previously gained knowledge about the inhibitors of SARS-CoV PLpro can be a good start point for drug discovery against SARS-CoV-2 PLpro. Recently it has been shown that naphthalene based inhibitors developed for SARS-CoV PLpro are also effective against SARS-CoV-2 PLpro.[44], [45], [46]
PLpro possesses two distinct binding subsites (SUb1 and SUb2) for Ub and ISG15, distant from the catalytic site.[46], [47] The SUb1 subsite, located at the boundaries of palm and fingers subdomains, is the primary binding site for Ub and the C-terminal ubiquitin-like domain of ISG15. The SUb2 subsite is the binding site for the second Ub molecule of the K48-linked di-Ub chain, and the N-terminal ubiquitin-like domain of ISG15. It is located in a ridge region of the thumb subdomain. SARS-CoV-2 PLpro preferentially cleaves ISG15 and its ability to hydrolyze K48-linked Ub chains is significantly reduced when compared to SARS-CoV PLpro.[40], [44], [45], [46] In contrast to the S4-S1 and SUb1 subsites, the SUb2 subsite of SARS-CoV-2 PLpro is much less conserved (67% identity) when compared with SARS-CoV PLpro. The subtle structural variations in the SUb2 subsite of SARS-CoV-2 PLpro affect the ability of the enzyme to bind and process the K48-linked polyUb and preference for ISG15.[40], [46] How these differences in the biochemical activities of SARS-CoV and SARS-CoV-2 PLpro influence the virus biology is not yet certain.
Nsp5/Mpro/3CLpro
Nsp5 is a 33 kDa cysteine protease, also known as the main protease (Mpro), or 3C-like protease (3CLpro, named after the 3C proteases of the Picornaviridae). Mpro from SARS-CoV-2 shares 96% sequence similarity with SARS-CoV. It cleaves viral polyproteins, pp1a and pp1ab, at 11 distinct sites with cleavage sequences LQ↓(S/A/G),48 generating 12 functional proteins.[48], [49] Inhibitors targeting this enzyme would block the viral replication making MPro an attractive target for anti-CoV drug design.
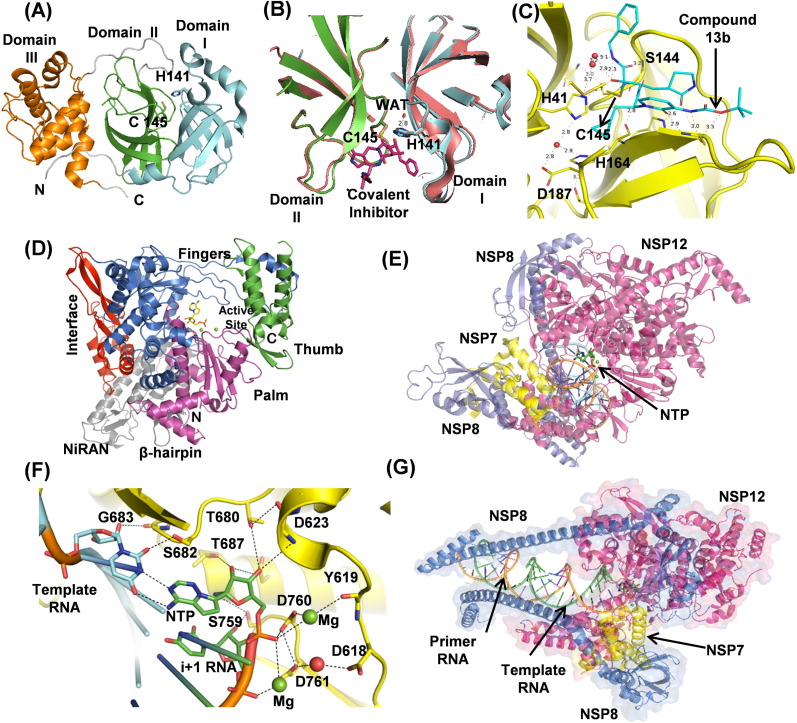
Crystal structures of SARS-CoV-2 Mpro show that it functions as an active homodimer with an approximate C2 symmetry, similar to other CoV Mpros, and has an estimated dissociation constant of dimerization ~2.5 μM.[50], [51] Each protomer has three domains- domains I, II, and III (Fig. 4 (A)). Domains I (residues 10–99) and II (residues 100–182) adopt a chymotrypsin-like (and picornavirus 3C protease–like) six-stranded antiparallel β-barrel fold, which is common to the structures of transmissible gastroenteritis CoV (TGEV), HCoV-229E, and SARS-CoV Mpros.52 A deep cleft between domains I and II forms the substrate-binding site, where the N-terminal finger (domain I, residues 1–7) plays an important role in catalysis.[52], [53] The C-terminal globular domain III (residues 198–303), comprising five antiparallel α-helices, is involved in dimerization through an intermolecular salt-bridge interaction between E290 and R4.[54], [55] Domains II and III are connected by a loop (residues 183–198) which exhibits two distinct conformations, in some, it assumes a fairly extended conformation while in others, residues 186–190 form a short helix. Dimerization of the enzyme is necessary for higher catalytic activity because the N-finger of each protomer interacts with E166 of the other protomer shaping the S1 pocket of the substrate-binding site.[51], [52] The dimer of SARS-CoV-2 Mpro is tighter and closer than that of SARS-CoV Mpro due to a substitution (T285A) in domain III, resulting in increased catalytic efficiency.51
Fig. 4.
(A) The cartoon representation of Mpro (PDB ID: 6YB7). The domain I (cyan), domain II (green), and domain III (orange) are shown with the catalytic dyad C15 and H41 in sticks. The N-terminal loop of domain III is shown as a grey loop. (B) Cartoon representation of unliganded Mpro (PDB ID: 6YB7) shows the active site cleft formed between domains I (cyan) and II (green). Unliganded Mpro is superposed with Mpro (beige) complexed to covalent N3 inhibitor (purple sticks) (PDB ID: 7BQY). The conserved water molecule, shown as a red sphere, is strongly hydrogen-bonded to H41 at the catalytic site. (C) Compound 13b (cyan), a structure-based inhibitor bound in the Mpro active site (PDB ID: 6Y2F). (D) Nsp12 cartoon representation (PDB ID: 7BV2) with the domains shown in different colors. (E) Nsp12 (red) cartoon representation showing bound Nsp7 (yellow), two Nsp8s (blue), and RNA. (F) Nsp12 active site showing the polymerase residues involved in NTP (as Remdesivir), template RNA (green), and primer RNA (orange) recognition (G) Nsp7 interacts with primer RNA (orange).
Each protomer contains one substrate binding site contributed mainly by itself, but the two protomers swap their N-termini to stabilize their S1 pockets. The substrate-binding site has a catalytic dyad formed by H41 and C145 lying at the center of this cleft (Fig. 4(B)), and a conserved water molecule making a hydrogen bond with H41 supports catalysis.56 The superposition of the crystal structures of Mpro with and without ligands has rmsds ranging from 0.26 to 0.38 Å, indicating the binding pocket is pre-shaped. The substrate, from P6 to P1 positions, adopts an extended conformation with its backbone forming an antiparallel β-sheet with the Mpro residues. The P1 position of the substrate invariably has a conserved glutamine residue making two strong hydrogen bonds with side chain atom of H163 and the main-chain carbonyl oxygen of F140. The P2 to P4 residues bind to the enzyme in extended beta-sheet conformation with specific side chains occupying the S2-S4 MPro pockets.57 The substrate residues at P5 and P6 positions interact with the enzyme through van der Waals interactions. On the C-terminal side of the substrate, a shallow S1′ subsite accommodates only small residues, namely Ser, Gly, or Ala, at P1′ position. The S2′ subsite is narrow but deep and can accommodate a long side chain, like Lys, at the P2′ position. The side chain at P3′ position is solvent-exposed.
Several attempts have been made to design and develop inhibitors against SARS-CoV-2 Mpro. More than 170 crystal structures of the enzyme in complex with the inhibitors and the PanDDA fragments have been deposited in the PDB. The structures under the PanDDA project provide crucial input to fragment-based drug discovery research. A library of ~10,000 compounds consisting of approved drugs, preclinical drug candidates, and natural products was recently screened using FRET assay and a few approved drugs (disulfiram and carmofur) and preclinical candidates (ebselen, shikonin, tideglusib, and PX-12) showed good IC50 values.1 Tandem MS/MS data showed that carmofur, PX-12, and ebselen bind covalently to catalytic cysteine, C145, of Mpro. The FDA approved HIV-1 protease inhibitors (Lopinavir, Ritonavir, and Darunavir), and the antibiotic Azithromycin also target Mpro, but the results from clinical trials are not encouraging. Structure-guided ab initio inhibitors, called mechanism-based inhibitors, have also been designed against Mpro, where a vinyl or α-keto-amide moieties make a covalent bond with the S-atom of catalytic cysteine, C145, forming an irreversible and a semi-reversible covalent complexes with the enzyme.[1], [51] The other structure-based drug (13b) designed using pyridone-moiety (Fig. 4(C)) blocked viral RNA replication in human lung cells and exhibited an extended half-life in blood plasma.51 Further modifications are being made in these inhibitors to enhance their efficacies. FDA-approved Hepatitis C medication, an HCV protease inhibitor, boceprevir was shown to be a potent Mpro inhibitor and also inhibits replication of SARS-CoV-2 in cell culture with EC50 values in the μM range.58 Another HCV protease inhibitor, telaprevir, has also been shown to inhibit Mpro.59 Crystal structures of Mpro in complex with boceprevir and telaprevir have been determined (PDB IDs 6ZRU, 6WNP, 6ZRT, 7C7P). These structures of Mpro in complex with various inhibitors will allow understanding the molecular basis of their interaction with Mpro and will help in designing more potent inhibitors.
Nsp12, Nsp7, and Nsp8
Nsp12 is a 103 kDa multi-subunit RNA-dependent RNA polymerase (RdRp). Along with Nsp7 and Nsp8, it forms a replicase complex for replication and transcription of the viral RNA genome. SARS-CoV-2 Nsp12 is very similar to SARS-CoV having 96% sequences identity. Several single-particle cryo-EM structures of SARS-CoV and SARS-CoV-2 Nsp12 have been determined,[60], [61], [62], [63], [64] including 160 kDa complex of SARS-CoV-2 Nsp12 bound to Nsp7 and Nsp8 (PDB IDs: 7BTF, 6M71) and to primer-template RNAs along with Remdesivir (GS-5734) (PDB IDs: 7BV2, 6YYT). SARS-CoV-2 Nsp12 contains an N-terminal extension nidovirus-unique RdRp-associated nucleotidyltransferase (NiRAN) domain (residues 60–249) and an RdRp domain (residues 366–920), interconnected by an interface domain. The exact role of the NiRAN domain in the nidoviral lifecycle is unknown, however, it exhibits nucleotidylation activity in equine arteritis virus (EAV).65 NiRAN domain possesses a kinase-like fold, and an intermediate N-terminal β-hairpin region (residues 31–50) positioned in a groove between NiRAN and RdRp palm domains.60 There are two Zn2+ ions located distally from RdRp catalytic site in the NiRAN domain and fingers domain co-ordinated with highly conserved residues possibly required for structural stability.
The RdRp domain adopts a typical right-hand fold with fingers (residues 366–581; 621–679), palm (residues 582–620; 680–815) and thumb (residues 816–920) subdomains (Fig. 4(D)).[60], [61] The palm subdomain forms the polymerase catalytic core accommodating two Mg2+ ions coordinated to D618, D760, D761, and NTP (Fig. 4(E), (F)).64
At the RdRp catalytic site, the ribose moiety of NTP to be incorporated is first positioned properly by a conserved S682 located in motif F. S682 performs the fidelity checks of the incoming NTPs through hydrogen bonds with the 2′-OH group of the ribose ring. The incoming NTPs form a base pair with the template RNA strand while the ribose 2′- and 3′-OHs form hydrogen bonds with the polymerase residues (Fig. 4(F)). The 2′- OH of the incoming NTP forms hydrogen bonds with T680 and N691 in motif B. D623 in motif A also interacts with the incoming NTP, and their proper positioning is facilitated by stacking of the i + 1 template RNA base with V557 hydrophobic side chain in the motif (Fig. 4(F)). The NTP is incorporated at the 3′-end of primer RNA where 3′-OH acts as a nucleophile to the NTP α–phosphate. The two Mg2+ ions act as an oxyanion hole to stabilize the NTP phosphates and pyrophosphate leaving group, reducing the activation energy barrier (Fig. 4(F)).66
Nsp12 is known to function alongside Nsp7 and Nsp8. The Nsp7-Nsp8 complex performs the RNA primase activity during the viral RNA synthesis.[67], [68] The crystal structure of the Nsp7-Nsp8 complex was first determined to 2.4Ả resolution (PDB ID: 2AHM) showing Nsp8 adopts a unique golf-club-like fold (Fig. 4(E), (G)).69 The complex forms a cylindrical hexadecameric architecture, composed of eight copies of Nsp7 and eight copies of Nsp8. The hexadecamer central channel has optimal dimensions with a positively charged surface to encircle the negatively charged double-stranded RNA and help RdRp in processing. The structure of Nsp7 has also been determined in the free unbound form by NMR.70 It has been shown that Nsp12 also interacts with Nsp5, Nsp9, and Nsp13,71 while Nsp8 can interact with Nsp7, Nsp9, Nsp10, Nsp13, and Nsp14 forming a multimeric replication complex.72
In the recent cryo-EM structures (PDB ID: 7BV2, 6YYT), it is observed that Nsp7-Nsp8 heterodimer binds to the polymerase thumb subdomain of Nsp12 facing the NTP entry channel (Fig. 4(E)). The Nsp12 polymerase finger loop is sandwiched between Nsp7-Nsp8 and the polymerase thumb subdomain. The second subunit of Nsp8 interacts with the Nsp12 interface domain close to the fingers subdomain (Fig. 4(E)) and the RNA template-binding channel (Fig. 4(G)). The binding of the Nsp7-Nsp8 heterodimer to the finger loop stabilizes the polymerase domain, enabling higher affinity with template RNA. The second subunit of Nsp8 is expected to play an important role in polymerase activity, possibly through binding to the template RNA to provide an extended interaction surface, thereby holding the RNA strand in position (Fig. 4(E), (G)). In fact, in the recent pre- and post-translocated complexes, the second Nsp8 N-terminal extension swings ~45° toward duplex RNA forming a more rigid complex.73 The binding affinity of Nsp12 to template-primer RNA increased significantly in the presence of both Nsp7 and Nsp8 and the polymerase activity is enhanced.[60], [64] Recent cryo-EM structure (PDB ID: 6XEZ) reveals RdRp-Nsp8-Nsp7 can form complex with two helicases (Nsp13) with double-stranded RNA helix.74 The two helicases interact with two Nsp8 molecules and one helicase is closer to the RNA strand. The proposed function of helicase is to unwind stalled+-strand primer RNA from 3′ to 5′ during template-switching while transcribing sub-genomic RNA. As a result, the RdRp complex backtracks, freeing the primer RNA at 3′-end paving way for another RdRp complex continuing transcription. But further experimental evidence is required to prove the hypothesis.
Many RdRp inhibitors are nucleotide/nucleoside analogs (NAs), similar to RT-RH inhibitors used against HIV. The antiviral effects of NAs are caused by either chain termination, or through increased/decreased rate of their incorporation in nascent RNA, leading to lethal mutations and non-viable genomes.[75], [76] Developing NAs as drugs is challenging mainly due to the appearance of NA-resistant mutations impairing NAs efficacy, and the possibility of removal of the incorporated NAs from RNA at 3′-end by the exonuclease (ExoN) activity.[63], [77] The NA, Remdesivir (GS-5734), which is an investigational broad-spectrum antiviral, received the “emergency use authorization” recently by FDA to treat SARS-CoV-2 infections. Remdesivir has been shown to get incorporated by SARS-CoV RdRp at a higher rate than the excision rate of SARS-CoV ExoN and /or is removed less efficiently by the SARS-CoV ExoN.78 In murine hepatitis virus, related to SARS-CoV, drug resistance mutations, F480L and V557L appeared against Remdesivir.78 The active site residue, V557, modulates polymerase fidelity, and the bulkier V557L mutant rejects Remdesivir.79 F480L mutation alters active site hydrophobic pocket reducing Remdesivir incorporation dynamics.80 Several other inhibitors currently in clinical trials, targeting the nucleotide site of RdRp, are Favipravir, Ribavirin, Galedesvir, etc.
Nsp9
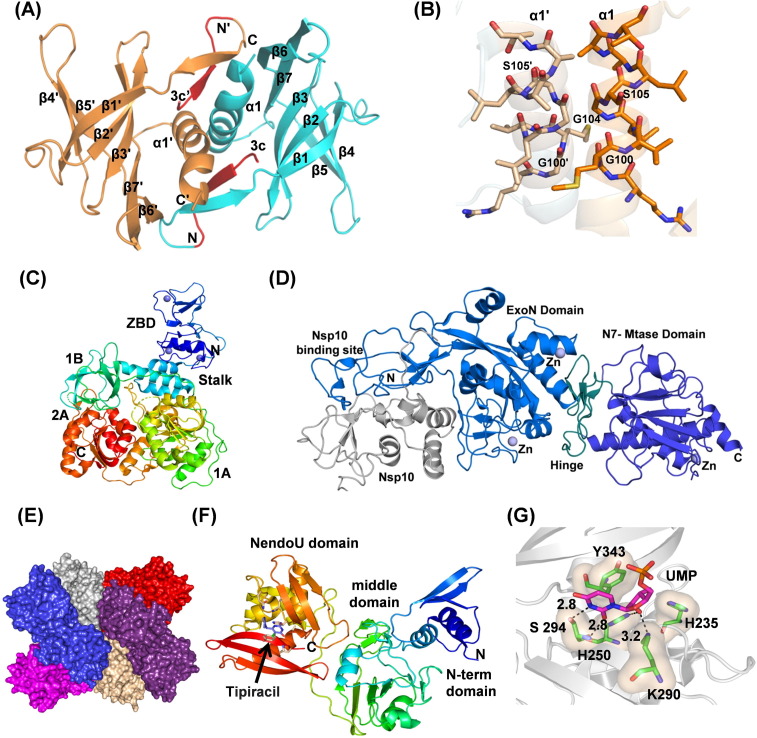
Nsp9 is a single-stranded RNA-binding protein implicated in the virulence of the virus.81 Nsp9 of SARS-CoV-2 shares 97% sequence identity with SARS-CoV ortholog. Nsp9 structure consists of a series of extended loops projecting outward from a 6-stranded enclosed β-barrel core (Fig. 5 (A)).82 Two positively charged, glycine-rich loops β2-β3 and β3-β4 are supposed to participate in RNA-binding (Fig. 5(A)). Nsp9 forms a dimer with the N-terminal β-strand and C-terminal α1-helix contributing to the dimer interface (Fig. 5(A)). The GxxxG motif formation makes the 14-residue helix of the respective monomer to cross each other (Fig. 5(A), (B)). The C-terminal portion of the helix, with hydrophobic residues, forms funnel-like hydrophobic cavities on either side of the interfacing helices (Fig. 5(A)). In the structure of Nsp9 containing an N-terminal tag with rhinovirus 3C protease sequence (LEVL), these cavities are filled by these 3C sequences (Fig. 5(A)). The 3C sequence formed new β-sheet interactions with the N-terminal strand from the neighboring monomer (Fig. 5(A)). The structural information suggests the compounds disrupting the dimer interface of Nsp9 may be developed as drugs against coronavirus diseases.82
Fig. 5.
(A) Cartoon representation of Nsp9 (PDB ID: 6W9Q) showing dimer interface and peptide (shown in red) bound in the hydrophobic cavity. (B) Nsp9 showing protein–protein interacting helices with the GxxxG motif in stick representation. (C) The structure of Nsp13 (PDB ID: 6ZSL) showing zinc-binding domain (blue), stalk (blue-green), 1B (green), 1A (green-yellow) and 2A (red) domains. Three zinc atoms are shown as dark blue spheres. (D) Cartoon representation of SARS-CoV Nsp14-Nsp10 complex structure (PDB Id 5C8U) showing Nsp10 (gray), Nsp14 ExoN domain with nsp10-binding site (marine blue), hinge region (deep petal cyan), and the C-terminal N7-MTase domain (tv-blue). Zinc atoms are shown as spheres (slate). (E) Hexameric assembly of SARS-CoV-2 Nsp15 in surface representation. Each monomer is shown in different colors. (F) The structure of Nsp15 showing different domains (PDB ID: 6WXC). The active site residues are shown with the drug Tipiracil bound in the active site. (G) Binding of uridine monophosphate to the Nsp15 active site (PDB ID: 6WLC). Key conserved residues, H235, H250, K290, S294, and W343 are shown in a stick representation. S294 provides uridylate specificity. The hydroxyl group of S294 binds to the nitrogen atom of 5′ uracil while the main chain nitrogen atom interacts with the carbonyl oxygen.
Nsp10
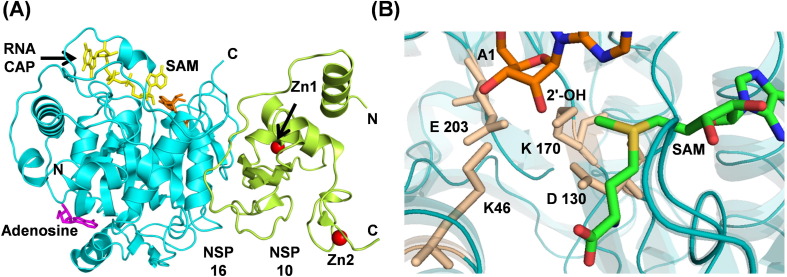
Nsp10 is a small, 139 amino acids long, single-domain protein having 99% sequence identity with SARS-CoV Nsp10. Nsp10 acts as a scaffold protein to form the mRNA cap methylation complex with Nsp14 (exonuclease and N7-methyltransferase) and Nsp16 (2′-O-methyltransferase).5 The crystal structure of Nsp10 (PDB ID: 6ZCT) shows that it consists of an anti-parallel β-sheet, a helical domain, and two binding sites for Zn2+ ions (Fig. 6 (A)). Nsp10 physically interacts with Nsp14 and Nsp16 to enhance their activities. For SARS-CoV-2, the crystal structures of Nsp10 in complex with Nsp16 have been determined.[83], [84] Since the interaction of Nsp10 with Nsp14 and Nsp16 is essential for their optimal activity,85 disruption of Nsp10/Nsp14 or Nsp10/Nsp16 interface could be a useful drug target strategy. Interestingly, it has been reported previously that an Nsp10 derived peptide target Nsp16/Nsp10 methyltransferase activity of coronaviruses in vivo and in vitro.86
Fig. 6.
(A) Heterodimer (PDB ID: 6WKS) of Nsp16 (cyan) and Nsp10 (lemon). Proteins and nucleotides are shown in cartoon and stick modes, respectively. Zn2+ are shown in red spheres and adenosine is in color magenta. (B) The SAM MTase motif is formed by residues K46, D130, K170, and E203 (catalytic tetrad).
Nsp13
Nsp13 is a superfamily 1B helicase with NTPase, duplex RNA/DNA unwinding, and 5′-RNA capping activities.[74], [87], [88] The structure of SARS-CoV-2 helicase (PDB ID: 6ZSL) is very similar to SARS-CoV Nsp13 helicase (PDB ID: 6JYT). The overall structure consists of five different domains arranged in a triangular pyramidal shape. Three domains, 1A, 2A, and 1B form the base of this pyramid which is connected through a stalk domain to the N-terminal zinc-binding domain (ZBD) forming the apex of the pyramid (Fig. 5(C)). The structural assembly is very well arranged to allow all the five domains of Nsp13 to participate in helicase activity in a coordinated manner.89
Nsp13 has distinct NTP and DNA binding sites. NTPase active site is present in the cleft between the 1A and 2A domains. The mutagenesis experiment in SARS-CoV helicase has shown that six residues, K288, S289, D374, E375, Q404, and R567 are crucial for NTPase activity.89 Based on the docking, H/D exchange, and mutagenesis experiments, it has been found that the residues involved in dsDNA binding are:176–186 (1B domain), 209–214 (1B domain), 330–350 (1A domain), and 516–541 (2A domain).89 SARS-CoV-2 Nsp13 has identical residues at these positions. Recently, the cryo-EM structure of Nsp13 in complex with SARS-CoV-2 replication-transcription complex consisting of Nsp7, Nsp8, and Nsp12 (PDB ID: 6XEZ) has been determined to 3.5 Å resolution. The structure has shown that Nsp13 interacts with Nsp8 and Nsp12 with potential implications for helicase activity and regulation.74
Due to its essential role in viral replication and conservation across all CoV species, Nsp13 is an important target for antiviral drug development. The NTPase, DNA binding, and/or helicase translocation activities of Nsp13 can be targeted by small-molecule inhibitors.90 Among several inhibitors reported in the literature, SSYA10-001 is the most promising candidate which prevents the helicase activity of Nsp13.[88], [91], [92] However, no inhibitor bound structures of Nsp13 is available till date.
Nsp14
Nsp14 from coronavirus is a bifunctional protein having an N-terminal exonuclease (ExoN) domain implicated in proofreading function and a C-terminal guanine-N7 methyl transferase (N7-MTase) domain involved in the methylation of viral RNA cap.93 The proofreading activity of Nsp14 has been suggested to be responsible for the evolution and maintenance of large genomes of coronavirus.94 The ExoN knockout mutants of SARS-CoV and MHV show accumulation of a large number of mutations.[95], [96] The mRNA capping activity of Nsp14 is important for the stability of viral mRNA by evading its degradation from the host immune response.97
Though the structure of SARS-CoV-2 Nsp14 has not yet been determined, it shares 99.1% sequence similarity with SARS-CoV Nsp14. Therefore, the structural inferences drawn here are based on the structure of SARS-CoV Nsp14 (PDB Ids 5NFY, 5C8T, 5C8S, 5C8U). Nsp14 has four structural regions: the Nsp10 binding site, an ExoN domain, a flexible hinge region and a C-terminal N7-MTase domain (Fig. 5(D)).98 The N-terminal residues 1–76 and 119–145 participate in the interaction with Nsp10 (Fig. 5(D)).[93], [98] Though Nsp14 alone has ExoN activity, its binding to Nsp10 enhances the activity by more than 35-fold. It has been suggested that the binding of cofactor, Nsp10, stabilizes the ExoN active site in the correct conformation for the catalysis.[85], [93], [99] The ExoN domain of Nsp14 has a usual α/β fold, typical of DEDD superfamily of exonucleases. The active site residues (D90, E92, E191 and D273) form a “DEED outlier” in the DEDD superfamily.93 Moreover, the recent identification of a conserved histidine residue (H268) in the active site places it in DEDDh subfamily.100 The catalytic residues in ExoN domain coordinate to two Mg2+ ions, which are involved in the activation of a water molecule for nucleophilic attack and the subsequent product release.100 The ExoN domain is linked to the C-terminal N7-MTase domain by a highly conserved flexible hinge region101 as shown in the Fig. 5D. The N7-MTase domain functions as an S-adenosyl-L-methionine (SAM) dependent methyl transferase to methylate the N7-position of the guanylate cap of the mRNA. It contains a canonical SAM-binding motif (DxG) and a second pocket for holding GTP of the mRNA cap in close proximity to SAM.102. Additionally, Nsp14 also has three zinc-finger motifs shown to be crucial for its ExoN activity and the interaction with Nsp10.[93], [98]
The crucial role of Nsp14 in proofreading function during genome replication and viral mRNA-capping activity makes it an important target for antiviral drug developments. It has been suggested that the proofreading function of Nsp14 acts as a barrier for the development of NAs as antivirals against SARS-CoV-2.100 NAs that avoid detection by the exonuclease activity of Nsp14 or outcompete the exonuclease activity are more likely to be successful.103 Moreover, the simultaneous inhibition of RdRp and ExoN activities by small-molecule inhibitors could provide a synergistic effect.
Nsp15
Nsp15 is an endoribonuclease which cleaves RNA specifically at the 3′-end of uridylates. Its endoribonuclease domain is unique to vertebrate-infecting nidovirales, including coronaviruses.104 The endonuclease activity of Nsp15 helps the virus evade immune system by preventing the detection of viral dsRNA by the host. It cleaves the 5′-polyuridines from the negative strand of viral RNAs, which are pathogen-associated molecular patterns (PAMPs) for MDA-5,105 major sensors of RNA viral infection. Nsp15 activity limits the accumulation of these PAMP and therefore dampens the host immune response. Crystal structures of SARS-CoV-2 Nsp15 are similar to the structure of SARS-CoV as they share 88.7% identity. The structures show that Nsp15 assembles as a compact hexamer and such an assembly is essential for its activity (Fig. 5(E)).[106], [107], [108] The Nsp15 protomer consists of three domains: a small N-terminal domain, a middle domain, and a large C-terminal catalytic NendoU domain (Fig. 5(F)). The N-terminal domain and the middle domain provide most of the interface area for oligomeric assembly while the C-terminal domain faces outwards providing six active sites on the surface. The middle domain also creates concave surfaces for potential interaction with other proteins and RNA.109 The endonuclease active site is structurally similar to RNase A and small molecule inhibitors of RNase A have been shown to inhibit Nsp15.106 Based on the similarity with the active site of RNase A, H235, H250, and K290 have been proposed to act as a catalytic triad with H235 playing the role of a general acid, H250 acting as a base, and S294 together with Y343 have been suggested to provide uridine specificity (Fig. 5(G)). The role of S294 in determining uridine specificity has been shown in the structures of Nsp15 in complex with nucleotides 5′ UMP, 3′UMP, and 5′ GpU (Fig. 5(G)).109
Mutations in Nsp15 have been shown to impact viral replication leading to greatly attenuated disease in mice. Hence modulating Nsp15 activity and/or stability may be a promising strategy for developing therapeutics.[110], [111] The extensive interactions between the subunits make the hexameric assembly sensitive to mutations and small molecule inhibitors capable of disrupting oligomeric assembly could be developed as therapeutics. Recently, it has been shown that Tipracil, an FDA approved anti-cancer drug, inhibits Nsp15 endoribonuclease activity. The structures of Nsp15 in complex with tipiracil have been determined109 (Fig. 5(F)) and it shows the molecular interactions of the drug in the Nsp15 binding site. In another study, it has also been shown that ciclesonide, an inhaled corticosteroid, suppressed human coronavirus replication in cultured cells by targeting Nsp15.112 However, the exact mechanism of ciclesonide mediated inhibition of Nsp15 remains unknown. Combining currently available broad-spectrum antivirals with Nsp15 inhibitors has been recently suggested as a potential approach against Covid-19.113
Nsp16
The Nsp16 is an m7GpppA (Cap-1)-specific, S-adenosyl methionine (SAM) dependent, nucleoside 2′-O-methyltransferase and catalyzes the 5′-methyl capping of viral mRNA.[114], [115] The 5′-methyl capping protects viral RNA from degradation by host 5′-exoribonucleases and serves to evade the induction of innate immune response. Genetic disruption of SARS-CoV Nsp16 leads to a ten-fold reduction in the synthesis of viral RNA.116 Therefore, Nsp16 is another attractive drug target for Covid-19. SARS-CoV-2 Nsp16 shares 93.3% identity with SARS-CoV Nsp16. It adopts a canonical SAM-MTase fold with eight stranded central twisted β-sheet flanked by two ɑ-helices on one side and three helices on the other side (Fig. 6(A)). For mRNA capping, Nsp16 transfers a methyl group from the donor SAM to the ribose 2′-OH of the first nucleotide, i.e., adenosine in SARS-CoV-2 RNA. The acceptor RNA cap and the donor SAM reside in separate cavities formed by the loops originating from different strands (Fig. 6(A), (B)). The purine ring of SAM and the target adenine (A1) are separated by a four amino acid long stretch (M131 to P134), helping in the proper orientation of ribose sugar of adenine and the methyl group of SAM for methylation.84 The highly conserved catalytic tetrad for methyl transfer is formed by K46, D130, K170, and E203, where K170 serves as a general base (Fig. 6(D)).117 The catalytic pocket makes a cone-like formation with the catalytic tetrad residues at the bottom and the target atom (2′-OH) of the A1 adenine at the tip.84 Nsp16 alone is inactive and requires Nsp10 for the activity.[114], [118] Nsp10 interacts with Nsp16 through hydrophobic and charge interactions to stabilize the SAM binding site.119 Due to the conserved nature of the SAM binding site, inhibitors targeting this pocket may be developed as pan-antiviral inhibitors. Structural studies in complex with sinefungin, a pan-MTase inhibitor binding in the SAM binding pocket will help in this direction.83 Structural studies have also revealed the presence of an allosteric ligand-binding site at the back of catalytic pocket occupied by either adenosine (PDB ID: 6WKS) or β-D-fructopyranose (PDB ID: 6W4H). This allosteric site has also been proposed as a target for drug development.84
Structural Proteins
Spike protein (S)
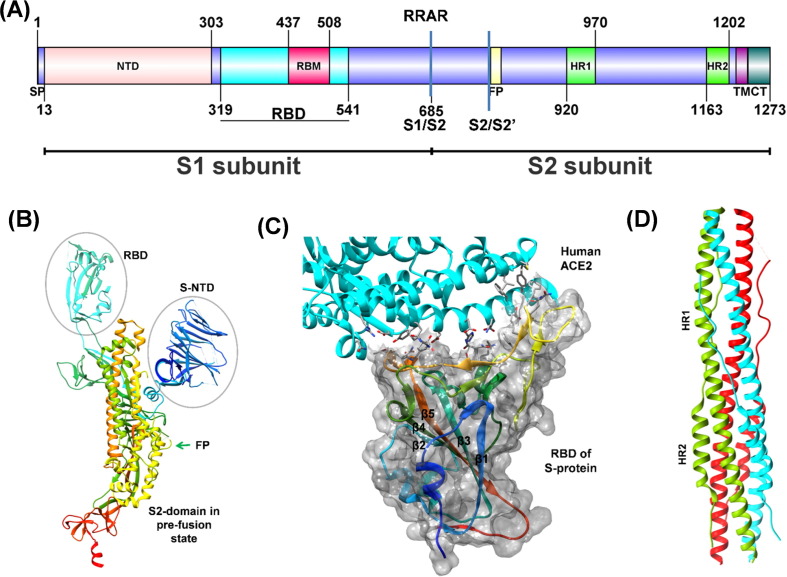
The homo-trimeric spike glycoprotein (S protein), protruding from the viral surface, is the first anchoring point of the virus to the host cell. SARS-CoV-2 S protein shares ~77% sequence identity with SARS-CoV S protein, and binds to the human ACE2 receptor for virion’s entry into the host cells.[120], [121] The 1273 residues long S protein has two major subunits, S1 and S2 (Fig. 7 (A)). The distal S1 subunit plays a role in receptor recognition and binding while the membrane-anchored S2 subunit mediates fusion of the viral and the host cell membranes. The S2 subunit exists in two structurally distinct conformations, prefusion, and postfusion.[122], [123] The S1 subunit has two well defined structural domains, the receptor binding domain (RBD, residues 319–541) and the N-terminal galectin like domain (S-NTD). The cryo-EM structure of full length S protein (PDB ID: 6VYB) and crystal structure of RBD bound with human ACE2 receptor (PDB ID: 6M0J) are available (Fig. 7(B), (C)).[122], [124] The RBD structure has a five-stranded antiparallel β-sheet (β1-β3-β5-β4-β2) core, flanked on either side by a short helix. A loop (the receptor-binding motif, RBM, residues 440–506) extends out of the main core (joining β4 and β5) forming a cradle-like structure for receptor binding.[124], [125] The RBD undergoes hinge motions that transiently hide (down) or expose (up) the determinants of receptor binding. The down confirmation refers to receptor inaccessible while the up corresponds to the receptor accessible, which are likely to be intrinsically unstable.[122], [123] This conformational variability is thought to be conserved among the Coronaviridae family.126 The RBM, stabilized by a disulfide bond, does not have a regular secondary structure except for two small β-sheets, each having two small β-strands (Fig. 7(C)). The interaction of RBM with the N-terminal helix of the ACE2 is stabilized by 14 hydrogen bonds and one salt bridge.124 The S-NTD has a galectin-like domain with a sugar-binding pocket and a ceiling-like structure over it, very similar to S-NTD of SARS-CoV. The role of SARS-CoV-2 S-NTD has been proposed to stabilize the S2 subunit in a constrained prefusion state.127 However, in many beta coronaviruses, like the mouse hepatitis virus (MHV), the S-NTD binds to its host receptor directly.[126], [128]
Fig. 7.
(A) Schematic representation of the domain structure of spike monomer: N-terminal domain (NTD), receptor-binding domain (RBD), receptor-binding motif (RBM), fusion peptide (FP), heptad repeat 1/2 (HR1/HR2), transmembrane region (TM), and cytoplasmic tail (CT). The furin protease site (RRAR) and two distinct protease sites (S1/S2 and S2/S2′) are indicated. (B) The S protein monomer in the open conformation (PDB ID: 6VYB) with labeled domains. The peptide chain is in rainbow color from N- to C-terminus (blue to red color). (C) Heteromeric complex (PDB ID: 6M0J) of RBD of Sprotein (rainbow colors from N- to C-terminus; blue to red) with its receptor human ACE2 (Cyan). The interface residues are displayed as sticks. The molecular surface of RBD (gray) forms a cradle-like structure for receptor binding. (D) Three pairs of HR1/HR2 of trimeric S protein (colored green, cyan, and red) form a six-helix bundled coiled-coil structure in the post-fusion state after cleavage at S1/S2 boundary (PDB ID: 6LXT).
The S2 subunit has four conserved structural regions; a fusion peptide (FP), two heptad repeats (HR1, HR2), and a transmembrane region. In the prefusion state, the HR1 region forms four helices as a part of the main helical stalk of S2, the HR2 region is disordered and FP forms a short helix and a loop with hydrophobic residues buried inside.[123], [129] Once the S1 subunit binds to the receptor, the S2 subunit undergoes molecular maturation leading to postfusion conformation. First, S1 and S2 subunits dissociate by the proteolytic action of host proteases (serine protease-TMPRSS2 and cysteine proteases-cathepsin B/L), and then three pairs of HR1/HR2 of trimeric S2 form coiled-coil structure of six-helix bundle (PDB ID: 6LXT, Fig. 7(D)).[6], [130], [131] The transition brings the viral membrane closer to the host membrane and the hydrophobic FP gets exposed on either side of the HR helical bundle making the region accessible for another proteolytic cleavage upstream of FP. After cleavage, the fusion peptides from each protomer of the trimeric assembly associate, and fuse into the host membrane leading to the insertion of viral RNA into the host cell.[126], [130], [132].
The sequence of SARS-CoV-2 S protein has an insertion of four residues (RRAR) at the S1/S2 boundary, creating an additional furin protease cleavage site. The recent articles have demonstrated that S1/S2 subunits are cleaved during virion assembly in the host cell but remain associated with each other through non-covalent interactions.122 Though the deletion of the furin protease site does not inhibit fusion with the host cell, an additional cleavage site might have a significant role in the rapid spread and higher infection rate of SARS-CoV-2.122 Notably, the reproduction number (Ro) for COVID-19 is ~3, much higher than other coronavirus diseases.133 Receptor recognition by coronaviruses serves as a critical determinant of cross-species infectivity, pathogenesis, and host tropism.126 Studies have demonstrated that antibodies raised against spike glycoprotein can inhibit the binding of RBD to the host cell receptors.[134], [135] In fact, the SARS-CoV antibodies, CR3022 and 47D11, are able to cross-react with SARS-CoV-2 RBDs.[134], [136] Thus, the RBD of S-protein represents an important target for antibody-mediated neutralization, vaccine development, and other antiviral strategies.137
Nucleocapsid Protein (N)
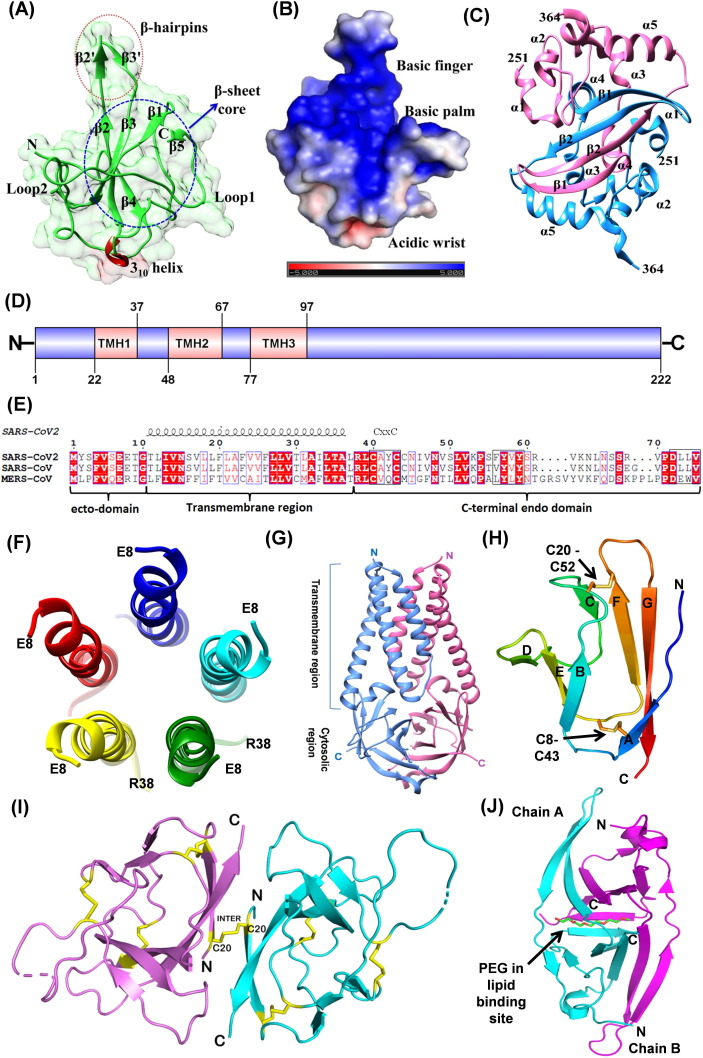
Nucleocapsid (N) protein is an important structural protein that packages the viral RNA into helical ribonucleocapsid (RNP) and interacts with the other structural proteins during virions’ assembly leading to genome encapsidation.[138], [139], [140]. The N protein is produced in high abundance during infection and is highly immunogenic, thus a potent target for vaccine development.[141], [142], [143]. The SARS-CoV-2 N protein consists of two highly conserved domains: the N-terminal RNA binding domain (N-NTD; 46–174) and the C-terminal dimerization domain (N-CTD; 247–364)[144], [145] separated by an intrinsically disordered and highly phosphorylated linker region rich in serine/arginine (184–196, SR motif). The N- and C- terminal ends of the protein (residues 1–42 and 365–419) are disordered.144
Recently reported crystal structures of SARS-CoV-2 N-NTD and N-CTD resemble with corresponding SARS-CoV structures with 0.8 Å and 0.44 Å RMSD, respectively. SARS-CoV-2 N-NTD comprises five antiparallel β-sheets (β4-β2-β3-β1-β5) sandwiched between two loops and a protruding β-hairpin (β2 and β3) between β2 and β5 (Fig. 8 (A), (B)).[144], [146] Overall N-NTD has a right-hand fold, where β-hairpin and β-sheet core are reminiscent of the finger and the palm, respectively, and are enriched in basic and aromatic amino acid residues. The positively charged residues of the palm and finger regions grasp the negatively charged backbone of ssRNA, whereas aromatic residues from the palm region provide additional base stacking interaction.[147], [148], [149] In IBV (Infectious bronchitis virus), it has been shown that the N- and the C-terminal regions, and not the central region, interact with the specific 3′-UTR (excluding the region lying between 215 and 78 nt from the 3′ end) of the viral gRNA.150 The N-terminal residues (1–91) are essential but not sufficient for binding to RNA.150 Fan et al. have suggested that the conserved residues R76 and K78 are involved in electrostatic interactions, and residues Y92 and Y94 stack against RNA bases (PDB ID: 2BXX and 2BTL).151 Additional interactions might occur between N-NTD and RNA upon closure of the β2-β3 hairpin.148 The residues in this region can be targeted for drug design strategy.
Fig. 8.
(A) N-terminal RNA binding domain of N protein (NTD; PDB ID: 6M3M) showing a right-hand fold structure. (B) The electrostatic surface of N NTD (surface colored based on charge distribution) highlighting the accessible region for RNA binding (C) C-terminal Dimerization domain of N protein (CTD; PDB ID: 6WZO). (D) Domain analysis of Membrane protein of SARS-CoV-2. TMH indicates the transmembrane helices. (E) Sequence alignment of Envelope proteins of coronaviruses, and their sequence features. (F) Pentameric oligomerization of transmembrane region (E8-R38) of SARS-CoV-2 E protein, resembling to viroporins (PDBID 7K3G). Only one of the 10 NMR models has been represented here. (G) The dimeric ORF3a protein (PDB ID: 6XDC). The six transmembrane helices form ion channel in the host membrane. (H) Compact seven-stranded β sandwich fold of ORF7a with disulfide bonds shown in stick representation. (I) Cartoon representation of the SARS-CoV-2 ORF8 covalent dimer (PDB ID: 7JTL). Disulfide bonds are shown as yellow sticks representing both inter- and intramolecular S-S bonds. (J) Cartoon representation of ORF9b illustrating the interlocked dimer with PEG (shown as sticks) bound at the interface (PDB ID: 6Z4U). Two chains are colored cyan and magenta.
The oligomerization of N protein is mediated by N-CTD that is essential for packaging of the gRNA into the capsid. The N-CTD monomer is composed of two β-strand, five α-helices, and three 310 helices along with β-bridges (PDB ID: 6WZO, Fig. 8(C)).145 Two such monomers are intertwined to form functional dimer such that the dimeric interface contains four-stranded intermolecular antiparallel β-sheets and two α-helices (two β-strands and one α-helix contributed by each monomer) (Fig. 8(C)).[147], [152] The disordered C-terminal region (365–419) and the linker SR-motif are required for higher-order assembly of the N-protein that may lead to helical filament formation with bound RNA during virions assembly.145 The SR-motif, in SARS-CoV N-protein, also interacts with the endodomain of Membrane (M) protein, for anchoring and condensing of RNP.138 The N-protein has also been implicated in RNA replication via its interaction with Nsp3, a component of viral replication.153
The oligomerization of N-NTD and N-CTD, and its implication on nucleocapsid formation has been studied extensively for IBV N protein.[151], [154] In one of the high resolution crystal structures, N-NTD of IBV forms a very stable dimer with interlocking monomeric subunits arranged in head-to-tail fashion extending into a fibril-like structure with the palm region, rich in basic residues, exposed to the surface.154 Similarly, N-CTD were found to self-associate in multiple ways forming either rod-like or bended fibril structures. It has been proposed that such multiple self-association might be required to induce curvature in nucleocapsid for tight packaging inside the virion. Moreover, the flexible linker region might bring NTD and CTD fibrils facing each other such that the viral gRNA is engulfed in these subunits.154.
Membrane (M) and Envelope (E) proteins
The coronaviruses assemble/bud at the lumens of the ERGIC and are released by exocytosis (steps 9–12, Fig. 2).155 The structural proteins, M, E and S, possess the trafficking signal sequences and accumulate in the ER. The efficient incorporation of these proteins and the ribonucleoprotein complex is essential for the maturation and budding of new virion particles.156 The M and E proteins play major role in regulation of the virion’s assembly. Both proteins are conserved in β-coronaviruses and share more than 90% sequence identity with their SARS-CoV homologs.
The SARS-CoV-2 M protein is a 222 amino acid long transmembrane glycoprotein of type III and is the most abundant structural protein. The M protein has three major domains: N-terminal ecto-domain followed by three transmembrane helices (TMH1-TMH3) and the C-terminal endo-domain (Fig. 8(D)). The M protein interacts with itself (homotypic) as well as other structural proteins, S, E and N (hetrotype interaction). These interactions are essential for inducing the membrane bending (budding) and serve as a checkpoint for the assembly of new virions.155 The residues required for homotypic interactions are present throughout the sequence of M-protein including the TM regions.157 Its interaction with E and N proteins are through C-terminal endo-domain. The residues L218 and L219 of SARS-CoV M protein are required for the packaging of nucleocapsid.157 The M protein induces a strong humoral response and the antigenic epitopes have been identified in the TM1 and TM2 regions of SARS-CoV M protein.158 Thus, the M protein can be a potential immunogen in therapeutic applications.
The E protein of SARS-CoV-2 is the smallest transmembrane structural protein of 75 amino acids comprising three domains: an N-terminal hydrophilic ecto-domain, a hydrophobic transmembrane domain (TMD) followed by a long hydrophilic C-terminal endo-domain (Fig. 8(E)). Recent NMR structure of TMD of SARS-CoV-2 E protein displayed a pentameric helix bundle surrounding a narrow cationic hydrophilic central pore, similar to viroporins (Fig. 8(F)).[159], [160] These ion channels cause the loss of membrane potential and activate host inflammasome.161 Three cysteines in the C-terminal domain undergo palmitoylation that aid in subcellular trafficking and membrane binding.[162], [163] The conserved residue, P54, is essential for its localization to the ERGIC.156 The last four amino acids (DLLV) of E protein are implicated in its interaction with host junction associated proteins (PALS1 and syntenin) that may facilitate the viral dissemination.[156], [164] This event together with the viroporins activity of the E protein are proposed to induce the cytokine storm. Recombinant CoVs lacking E protein display reduced viral titres, impaired viral maturation and incompetent viral propagation and thus has been suggested as a good vaccine candidates.163
Accessory Proteins
ORF3a
The ORF3a is a transmembrane protein of the viroporin family that forms ion channels in the host membrane. It shares ~72% sequence identity with SARS-CoV orthologue. The protein has been implicated in inducing apoptosis, pathogenicity, and virus release.[165], [166] The induction of cytokine storms in COVID-19 patients might be linked to ORF3a mediated activation of inflammasome.
The recent cryo-EM structure of SARS-CoV-2 ORF3a is the first structure of the viroporin family of coronavirus proteins (PDB ID: 6XDC, Fig. 8(G)).167 The structure has three transmembrane helices, (TM1-TM3, residues 41–132) followed by a cytosolic domain of two anti-parallel β-sheets forming a β-sandwich. ORF3a forms a dimer and the six transmembrane helices of the dimer form an ion channel with polar/charged residues in the interior of the channel capable of conducting cations. The pore size at its narrower side is ~1 Å in the reported structure, suggesting that the structure represents the closed conformation of the ion channel.167 The ion channel has been reported to have a higher preference for Ca2+/K+ cations compared to Na+ ion. The cytosolic domains of two protomers interact with a large continuous buried hydrophobic core creating a narrow-bifurcated pore. The ORF3a protein also contains a TRAF-binding domain at the N-terminus that activates NF-κB and the NLRP3 inflammasome. However, this domain is absent in the reported structure.[167], [168].
ORF3a induces an extrinsic apoptotic pathway initiated by cleavage of caspase-8, which in turn cleaves Bid, leading to the release of cytochrome c from mitochondria, formation of the apoptosome, and activation of caspase-9.166 The strength of apoptotic activity is, however, weaker in SARS-CoV-2 compared to SARS-CoV that might be linked with the difference in their pathogenicity. Mutations in C130/C133/Y160 result in loss of pro-apoptotic activity and membrane localization which contribute to the pathogenicity of the virus.166 The suppression of ORF3a expression in SARS-CoV resulted in decreased virus release and less morbidity.[165], [169] Moreover, ion channels are important therapeutic targets and many ion-channel drugs have already been developed. ORF3a, therefore, can be another potent drug target for disease management owing to its role in pathogenicity.
ORF7a
The ORF7a of the SARS-CoV-2 encodes a type I transmembrane protein, known as accessory protein 7a, protein U122, or protein X4 and shows 85.2% sequence identity to SARS-CoV ORF7a. It was shown that the ORF7a protein of SARS-CoV is expressed and retained intracellularly in the infected cells.170 Though it is involved in virus-host interaction, the exact function is not yet known. It has been suggested that ORF7a plays a role in protein trafficking within the endoplasmic reticulum and the Golgi complex. Sequence analysis shows that ORF7a encodes a protein of 121 residues with an N-terminal signal peptide (15 residues), an ectodomain, a transmembrane region, and a five-residues cytoplasmic di-lysine motif (KRKTE) for ER localization. The structure (PDB ID: 6 W37) shows a compact seven-stranded β-sandwich fold similar to the immunoglobulin superfamily (Fig. 8(H)). The ORF7a structure also shows the presence of two disulfide bonds connecting the BED and AGFC sheets (Fig. 8(H)). Due to these disulfide bonds, a deep hydrophobic pocket is created near the middle of the A strand.170 In this arrangement, the peptide backbone of the B strand remains free and along with the hydrophobic pocket on A strand, it may be involved in the interaction with other proteins. A deep groove between the C-D and the E-F loops may also be an adaptation for interactions with different ligands.170
ORF8
SARS-CoV-2 ORF8 is an accessory protein of 121 residues having an N-terminal signal sequence (residues 1–17) for transport to endoplasmic reticulum, and a β-strand core comprising residues 18–121, resembling immunoglobulin (Ig)-like fold. It lacks the C-terminal transmembrane domain present in other Ig superfamily proteins. SARS-CoV-2 ORF8 has less than 20% sequence similarity with SARS-CoV ORF8a/b. Multiple functions of ORF8 have been proposed. When ORF8 is exogenously overexpressed in cells, it disrupts IFN-I signaling.171 Unlike ORF8a/b of SARS-CoV, the SARS-CoV-2 ORF8 downregulates MHC-I in cells.172
Crystal structures of SARS-CoV-2 ORF8 show the asymmetric unit contains a dimer with its 60 residues monomeric core. Though the core structure has an Ig-like fold, similar to SARS-CoV ORF7a, the sequence similarity between them is only ~14%. Each monomer has eight antiparallel β-sheets held by three intramolecular disulphide bonds. The Pro85 residue, near the disulfide-forming Cys83, adopts cis-conformation. The reported crystal structures of SARS-CoV-2 ORF8 (PDB ID: 7JTL and 7JX6) show that it has two distinct protein–protein interfaces with significant buried surface area corresponding to two potential dimers/oligomers.172 The covalent-dimer has ~1320 Å2 buried surface area and is linked by intermolecular disulfide bond between two conserved Cys20 residues from each monomer located near their N-terminals (Fig. 8(I)). The covalent-dimer is also stabilized by complimentary surface charges with several hydrophobic interactions and multiple hydrogen bonds. The second potential non-covalent oligomeric interface has a buried surface area of ~1700 Å2. It is formed by SARS-CoV-2 specific 73YIDI motif that makes non-covalent interaction with Leu95, Ile58, Val49 and Pro56. These SARS-CoV-2 specific interfaces in ORF8 indicate it is capable of forming large-scale assemblies. However, the biological significance of these oligomeric interfaces and the exact biological function of ORF8 remains unclear, and requires further investigations.
ORF9b
ORF9b, a 97 amino acids long accessory protein, is encoded by an alternative open reading frame within the nucleocapsid gene. ORF9b suppresses IFN-I response through association with an adapter protein, TOM70.173 IFN-I has a central role in the immune response against viral infections. Due to this critical role of ORF9b, targeting the interaction between ORF9b-TOM70 has been suggested as an effective therapeutic option against COVID-19. ORF9b of SARS-CoV-2 shows ~70.45% identity with ORF9b of SARS-CoV. The structure of SARS-CoV-2 ORF9b (PDB ID: 6Z4U) superposes very well with the structure of SARS-CoV ORF9b (PDB ID: 2CME) (rmsd 1.14 Å). These structures show that ORF9b is dimeric and is primarily a β-sheet protein (Fig. 8(J)). β-strands from both the monomers interact in an interlocked architecture to form a twisted antiparallel β- sheet at the interface. The dimer has a central hydrophobic cavity that can accommodate lipid molecules and is suggested to participate in the membrane attachment.174 Understanding the molecular details of ORF9b’s interaction with the host proteins, like TOM70, will help in designing inhibitors against it.
Conclusion
Remarkable progress has been made by the structural biology community in the last few months to elucidate the structures of different SARS-CoV-2 proteins providing an atomic view of different molecular processes during the viral life cycle. These structures are important not only to understand the virus biology but also to design and develop molecules that would act as potential therapeutic agents. Most of the therapeutics currently under clinical trials are the result of repurposing the existing drugs. Though these drugs show efficacy in vitro, their clinical usefulness has not yet been established unequivocally. The lack of safe and effective therapeutics against COVID-19 will continue to fuel the structural studies of viral targets. The new structures of SARS-CoV-2 proteins and their complexes with inhibitors, RNA, and other proteins will continue to enrich our understanding of the working of different components of the virus during different steps of its life cycle.
CRediT authorship contribution statement
Rimanshee Arya: Writing - review & editing. Shweta Kumari: Writing - review & editing. Bharati Pandey: Writing - review & editing. Hiral Mistry: Writing - review & editing. Subhash C. Bihani: Conceptualization, Writing - review & editing. Amit Das: Writing - review & editing. Vishal Prashar: Writing - review & editing. Gagan D. Gupta: Writing - review & editing. Lata Panicker: Writing - review & editing. Mukesh Kumar: Conceptualization, Writing - review & editing.
Acknowledgements
We thank Dr. VP Venugopalan, former director, BSG, BARC, for critically reading the manuscript and providing useful comments, and the Department of Atomic Energy, Government of India for funding our research.
Edited by M.F. Summers
References
- 1.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020:1–5. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 2.Alanagreh L., Alzoughool F., Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9:331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata A.A. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9:1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:1–5. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou G., Chen S., Chen Z. Advances in COVID-19: the virus, the pathogenesis, and evidence-based control and therapeutic strategies. Front. Med. 2020;14:117–125. doi: 10.1007/s11684-020-0773-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nature Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., Leibundgut M., Thiel V. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nature Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012;86:11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wüthrich K. Novel β-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4:e00524–13. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imbert I., Snijder E.J., Dimitrova M., Guillemot J.-C., Lécine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 20.Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eriksson K.K., Cervantes-Barragán L., Ludewig B., Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1’’-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2014;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7:e01721–16. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuri T., Eriksson K.K., Putics A., Züst R., Snijder E.J., Davidson A.D., Siddell S.G., Thiel V. The ADP-ribose-1’’-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- 26.Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R. A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication–transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frick D.N., Virdi R.S., Vuksanovic N., Dahal N., Silvaggi N.R. Molecular basis for ADP-ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry. 2020;59:2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalska K., Kim Y., Jedrzejczak R., Maltseva N.I., Stols L., Endres M., Joachimiak A. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ. 2020;7:814–824. doi: 10.1107/S2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alhammad Y.M.O., Kashipathy M.M., Roy A., Gagné J.-P., Nonfoux L., McDonald P., Gao P., Battaile K.P. The SARS-CoV-2 conserved macrodomain is a highly efficient ADP-ribosylhydrolase. BioRxiv. 2020 doi: 10.1101/2020.05.11.089375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alhammad Y.M.O., Fehr A.R. The viral macrodomain counters host antiviral ADP-ribosylation. Viruses. 2020;12:384. doi: 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford K., Bonfiglio J.J., Mikoč A., Matic I., Ahel I. Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit. Rev. Biochem. Mol. Biol. 2018;53:64–82. doi: 10.1080/10409238.2017.1394265. [DOI] [PubMed] [Google Scholar]
- 32.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. Deubiquitinating activity of the SARS-CoV papain-like protease. Adv. Exp. Med. Biol. 2006;581:37–41. doi: 10.1007/978-0-387-33012-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Ménard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas A.L., Ahrens P., Bright P.M., Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 37.Jiang X., Chen Z.J. The role of ubiquitylation in immune defence and pathogen evasion. Nature Rev. Immunol. 2011;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 39.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. PNAS. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., Oualid F.E., Huang T.T. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci. Adv. 2020;6:eabd4596. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Báez-Santos Y.M., Barraza S.J., Wilson M.W., Agius M.P., Mielech A.M., Davis N.M., Baker S.C., Larsen S.D. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. PNAS. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poreba M., Salvesen G.S., Drag M. Synthesis of a HyCoSuL peptide substrate library to dissect protease substrate specificity. Nature Protoc. 2017;12:2189–2214. doi: 10.1038/nprot.2017.091. [DOI] [PubMed] [Google Scholar]
- 44.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6(8):2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 45.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39 doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 49.Lee H.J., Shieh C.K., Gorbalenya A.E., Koonin E.V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M.M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. PNAS. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J., Wei Z., Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004;279:24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kneller D.W., Phillips G., O’Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L. Structural plasticity of SARS-CoV-2 3CL M pro active site cavity revealed by room temperature X-ray crystallography. Nature Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.B.J. Anson, M.E. Chapman, E.K. Lendy, S. Pshenychny, R.T. D’Aquila, Broad-spectrum inhibition of coronavirus main and papain-like proteases by HCV drugs, (2020). doi: 10.21203/rs.3.rs-26344/v1.
- 60.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nature Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDonald S.M. RNA synthetic mechanisms employed by diverse families of RNA viruses. Wiley Interdiscip. Rev. RNA. 2013;4:351–367. doi: 10.1002/wrna.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. PNAS. 2014;111:E3900–3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das A., Gerlits O., Parks J.M., Langan P., Kovalevsky A., Heller W.T. Protein kinase A catalytic subunit primed for action: time-lapse crystallography of michaelis complex formation. Structure. 2015;23:2331–2340. doi: 10.1016/j.str.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 67.Imbert I., Guillemot J., Bourhis J., Bussetta C., Coutard B., Egloff M., Ferron F., Gorbalenya A.E. A second, non-canonical RNA-dependent RNA polymerase in SARS Coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.te Velthuis A.J.W., Arnold J.J., Cameron C.E., van den Worm S.H.E., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nature Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peti W., Johnson M.A., Herrmann T., Neuman B.W., Buchmeier M.J., Nelson M., Joseph J., Page R. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 2005;79:12905–12913. doi: 10.1128/JVI.79.20.12905-12913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77:10515–10527. doi: 10.1128/jvi.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Andres E.L., Bluemling G.R., Lockwood M.A. Small-molecule antiviral β-d-N4-hydroxycytidine Inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93 doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z., Andino R., Cameron C.E. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nature Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 77.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. PNAS. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sexton N.R., Smith E.C., Blanc H., Vignuzzi M., Peersen O.B., Denison M.R. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J. Virol. 2016;90:7415–7428. doi: 10.1128/JVI.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Campagnola G., McDonald S., Beaucourt S., Vignuzzi M., Peersen O.B. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 2015;89:275–286. doi: 10.1128/JVI.01574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miknis Z.J., Donaldson E.F., Umland T.C., Rimmer R.A., Baric R.S., Schultz L.W. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 2009;83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Littler D.R., Gully B.S., Colson R.N., Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp 9. IScience. 2020;23 doi: 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nature Commun. 2020;11:1–7. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., Park J.-G., Oladunni F. Structural basis of RNA cap modification by SARS-CoV-2. Nature Commun. 2020;11:3718. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ke M., Chen Y., Wu A., Sun Y., Su C., Wu H., Jin X., Tao J. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012;167:322–328. doi: 10.1016/j.virusres.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neuman B.W., Chamberlain P., Bowden F., Joseph J. Atlas of coronavirus replicase structure. Virus Res. 2014;194:49–66. doi: 10.1016/j.virusres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shum K.T., Tanner J.A. Differential inhibitory activities and stabilisation of DNA Aptamers against the SARS Coronavirus Helicase. ChemBioChem. 2008;9:3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47:6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Habtemariam S., Nabavi S.F., Banach M., Berindan-Neagoe I., Sarkar K., Sil P.C., Nabavi S.M. Should we try SARS-CoV-2 helicase inhibitors for COVID-19 therapy? Arch. Med. Res. 2020;51:733–735. doi: 10.1016/j.arcmed.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adedeji A.O., Singh K., Kassim A., Coleman C.M., Elliott R., Weiss S.R., Frieman M.B., Sarafianos S.G. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome mouse hepatitis, and middle east respiratory syndrome coronaviruses. Antimicrobial Agents Chemother. 2014;58:4894–4898. doi: 10.1128/AAC.02994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.Á., Urquiza J., Ramírez D. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 93.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. PNAS. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koonin E.V., Moss B. Viruses know more than one way to don a cap. Proc. Natl. Acad. Sci. USA. 2010;107:3283–3284. doi: 10.1073/pnas.0915061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ferron F., Subissi L., Morais A.T.S.D., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. PNAS. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.-L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ogando N.S., Ferron F., Decroly E., Canard B., Posthuma C.C., Snijder E.J. The curious case of the nidovirus exoribonuclease: its role in RNA synthesis and replication fidelity. Front. Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. PNAS. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deng X., Baker S.C. An “Old” protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–163. doi: 10.1016/j.virol.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ricagno S., Egloff M.-P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. PNAS. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim Y., Wower J., Maltseva N., Chang C., Jedrzejczak R., Wilamowski M., Kang S., Nicolaescu V. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.06.26.173872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Züst R., Hwang M. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Matsuyama S., Kawase M., Nao N., Shirato K., Ujike M., Kamitani W., Shimojima M., Fukushi S. The inhaled corticosteroid ciclesonide blocks coronavirus RNA replication by targeting viral NSP15. BioRxiv. 2020 doi: 10.1101/2020.03.11.987016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Senanayake S.L. Overcoming nonstructural protein 15-nidoviral uridylate-specific endoribonuclease (nsp15/NendoU) activity of SARS-CoV-2. Future Drug Discov. 2020;2(3) doi: 10.4155/fdd-2020-0012. [DOI] [Google Scholar]
- 114.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In Vitro Reconstitution of SARS-Coronavirus mRNA Cap Methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N. Crystal structure and functional analysis of the SARS-coronavirus RNA Cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Almazán F., DeDiego M.L., Galán C., Escors D., Álvarez E., Ortego J., Sola I., Zuñiga S. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bollati M., Milani M., Mastrangelo E., Ricagno S., Tedeschi G., Nonnis S., Decroly E., Selisko B. Recognition of RNA cap in the wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in flavivirus. J. Mol. Biol. 2009;385:140–152. doi: 10.1016/j.jmb.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 118.von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA Cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. BioRxiv. 2020 doi: 10.1101/2020.04.17.047498. [DOI] [Google Scholar]
- 120.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C. Structural and functional basis of SARS-CoV-2 Entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 125.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.-J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. PNAS. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bartlam M., Yang H., Rao Z. Structural insights into SARS coronavirus proteins. Curr. Opin. Struct. Biol. 2005;15:664–672. doi: 10.1016/j.sbi.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Y.-T., Landeras-Bueno S., Hsieh L.-E., Terada Y., Kim K., Ley K., Shresta S., Saphire E.O. Spiking pandemic potential: structural and immunological aspects of SARS-CoV-2. Trends Microbiol. 2020;28:605–618. doi: 10.1016/j.tim.2020.05.012. S0966842X20301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L. A human monoclonal antibody blocking SARS-CoV-2 infection. Nature Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhu Z., Chakraborti S., He Y., Roberts A., Sheahan T., Xiao X., Hensley L.E., Prabakaran P. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV – a target for vaccine and therapeutic development. Nature Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang C., Chen C.-M.M., Chiang M., Hsu Y., Huang T. Transient oligomerization of the SARS-CoV N protein–implication for virus ribonucleoprotein packaging. PLoS ONE. 2013;8:e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., Li Y., Bastien N. Characterization of protein–protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 2004;105:121–125. doi: 10.1016/j.virusres.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao X., Nicholls J.M., Chen Y.-G. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with smad3 and modulates transforming growth factor-β signaling. J. Biol. Chem. 2008;283:3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Padron-Regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infectious Dis. Therapy. 2020:1–20. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shang B., Wang X.-Y., Yuan J.-W., Vabret A., Wu X.-D., Yang R.-F., Tian L., Ji Y.-Y., Deubel V., Sun B. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem. Biophys. Res. Commun. 2005;336:110–117. doi: 10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527:618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29:1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dinesh D.C., Chalupska D., Silhan J., Veverka V., Boura E. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. BioRxiv. 2020 doi: 10.1101/2020.04.02.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen C.-Y., Chang C., Chang Y.-W., Sue S.-C., Bai H.-I., Riang L., Hsiao C.-D., Huang T. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 149.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhou M., Collisson E.W. The amino and carboxyl domains of the infectious bronchitis virus nucleocapsid protein interact with 3′ genomic RNA. Virus Res. 2000;67:31–39. doi: 10.1016/S0168-1702(00)00126-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yu I.-M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona-and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V.V. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51:482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tseng Y.-T., Chang C.-H., Wang S.-M., Huang K.-J., Wang C.-T. Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F., Li T., Yan J. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202:1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.M. Hong, V. Mandala, M. McKay, A. Shcherbakov, A. Dregni, A. Kolocouris, Structure and Drug Binding of the SARS-CoV-2 Envelope Protein in Phospholipid Bilayers, In Review, 2020. doi: 10.21203/rs.3.rs-77124/v1. [DOI] [PMC free article] [PubMed]
- 160.Sarkar M., Saha S. Structural insight into the role of novel SARS-CoV-2 E protein: A potential target for vaccine development and other therapeutic strategies. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0237300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mukherjee S., Bhattacharyya D., Bhunia A. Host-membrane interacting interface of the SARS coronavirus envelope protein: Immense functional potential of C-terminal domain. Biophys. Chem. 2020;266 doi: 10.1016/j.bpc.2020.106452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schoeman D., Fielding B.C. Is there a link between the pathogenic human coronavirus envelope protein and immunopathology? A review of the literature. Front. Microbiol. 2020;11:2086. doi: 10.3389/fmicb.2020.02086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Toto A., Ma S., Malagrinò F., Visconti L., Pagano L., Stromgaard K., Gianni S. Comparing the binding properties of peptides mimicking the Envelope protein of SARS-CoV and SARS-CoV -2 to the PDZ domain of the tight junction-associated PALS1 protein. Protein Sci. 2020;29:2038–2042. doi: 10.1002/pro.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9(3) doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.-Y. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kern D.M., Sorum B., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Brohawn S.G. Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs. BioRxiv. 2020 doi: 10.1101/2020.06.17.156554. [DOI] [Google Scholar]
- 168.Siu K., Yuen K., Castano-Rodriguez C., Ye Z., Yeung M., Fung S., Yuan S., Chan C. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lu W., Zheng B.-J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., Ge X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., Yang T., Yu F. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. BioRxiv. 2020 doi: 10.1101/2020.05.24.111823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jiang H., Zhang H., Meng Q., Xie J., Li Y., Chen H., Zheng Y., Wang X. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J.C., Grimes J.M., Stuart D.I. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]