Abstract
Theileria annulata is a tick‐transmitted apicomplexan parasite that infects and transforms bovine leukocytes into disseminating tumours that cause a disease called tropical theileriosis. Using comparative transcriptomics we identified genes transcriptionally perturbed during Theileria‐induced leukocyte transformation. Dataset comparisons highlighted a small set of genes associated with Theileria‐transformed leukocyte dissemination. The roles of Granzyme A (GZMA) and RAS guanyl‐releasing protein 1 (RASGRP1) were verified by CRISPR/Cas9‐mediated knockdown. Knocking down expression of GZMA and RASGRP1 in attenuated macrophages led to a regain in their dissemination in Rag2/γC mice confirming their role as dissemination suppressors in vivo. We further evaluated the roles of GZMA and RASGRP1 in human B lymphomas by comparing the transcriptome of 934 human cancer cell lines to that of Theileria‐transformed bovine host cells. We confirmed dampened dissemination potential of human B lymphomas that overexpress GZMA and RASGRP1. Our results provide evidence that GZMA and RASGRP1 have a novel tumour suppressor function in both T. annulata‐infected bovine host leukocytes and in human B lymphomas.
Keywords: GZMA, human cancer cell atlas, RASGRP1, Theileria annulata, transcriptome, tumour suppressor
Summarising the main findings of this study that identified new players in dissemination and oxidative stress regulation of Theileria‐transformed leukocytes and provided evidence for similar roles for GZMA and RASGRP1 in transcriptionally matched human B lymphoma cell lines.
1. INTRODUCTION
Theileria annulata is a tick‐transmitted apicomplexan parasite that infects and transforms bovine leukocytes into disseminating tumours that cause a widespread disease called tropical theileriosis. In countries endemic for tropical theileriosis live attenuated vaccines are produced by multiple in vitro passages of virulent, transformed macrophages and vaccination with attenuated macrophages protects animals from severe disease (Nene & Morrison, 2016). The transformation is reversed and leukocytes revert to a quiescence state and die upon drug‐induced parasite death making Theileria‐infected leukocytes a powerful cellular model to identify genes regulating cellular transformation and dissemination (Tretina, Gotia, Mann, & Silva, 2015). This parasite‐based reversible model of leukocyte transformation has allowed the identification of several cell signalling pathways associated with the virulence of Theileria‐transformed leukocytes such as c‐Jun NH2‐terminal kinase/c‐Jun/PI3 kinase signalling (Lizundia et al., 2006), protein kinase A (PKA) (Haidar, Echebli, Ding, Kamau, & Langsley, 2015), transforming growth factor beta 2 (TGF‐β2) (Chaussepied et al., 2010) (Haidar, Whitworth, et al., 2015) and SMYD3/MMP9 (Cock‐Rada et al., 2012). MMP‐9 and c‐Jun are associated with proliferation, angiogenesis and dissemination/metastasis in Theileria‐induced host cell transformation and human cancer (Adamson, Logan, Kinnaird, Langsley, & Hall, 2000; Hofmann, Westphal, Van Muijen, & Ruiter, 2000; Lizundia et al., 2006; Vleugel, Greijer, Bos, van der Wall, & van Diest, 2006). Epigenetic changes also contribute to Theileria‐induced leukocyte transformation (Robert McMaster, Morrison, & Kobor, 2016). OncomiR addiction has been described as being generated by a miR‐155 feedback loop in T. annulata‐transformed B cells (Marsolier et al., 2013). Similarly, miR‐126‐5p contributes to infected macrophage dissemination through JNK‐Interacting Protein‐2 (JIP2)/JNK1/AP1‐mediated MMP9 transcription (Haidar et al., 2018).
It is well established that T. annulata infection hijacks key leukocyte signalling cascades to modulate host cell gene expression. For example, RNA extracted from T. annulata‐transformed B cells was used to screen bovine microarrays demonstrating that infection had reconfigured host cell gene expression (Kinnaird et al., 2013). Nonetheless, a systematic and genome scale transcriptional comparison of B cells and macrophages transformed by T. annulata has been lacking. In this study, we used RNA‐seq to define the transcriptional landscapes of two T. annulata‐transformed B cell lines and a virulent T. annulata‐transformed macrophage line (Ode) and the attenuated live vaccine directly derived from it. High stringency bioinformatic comparisons of the transcriptional landscapes identified four candidate genes (MMP9, GZMA, RASGRP1 and SEPP1), as potential players in the dissemination of virulent T. annulata‐transformed macrophages.
Theileria annulata infection confers on its host leukocyte properties largely similar to human cancer, most notably immortalization, independence of exogenous growth factors, uncontrolled proliferation and dissemination. The similarity between Theileria‐transformed leukocytes and human leukaemia suggests that Theileria‐induced transformation could be a powerful model to elucidate common mechanisms underpinning tumour virulence. In order to generalise our Theileria‐based observations we compared the transcriptome maps of 934 human cancer cell lines to the transcriptomes of T. annulata‐transformed B lymphocytes and provide functional evidence for shared roles for GZMA and RASGRP1 in controlling dissemination of both human B lymphomas and Theileria‐transformed leukocytes.
2. RESULTS
2.1. Differentially expressed bovine genes in T. annulata‐transformed leukocytes
The infection and full transformation of the BL20 cell line with T. annulata causes profound transcriptional changes (Kinnaird et al., 2013). We have compared the transcriptome of the T. annulata‐transformed TBL3 and TBL20 B lymphocytes to their non‐infected counterparts, BL3 and BL20, respectively. The quality of the sequencing results is shown in Figure S1. Infection of BL3 and BL20 lymphocytes with T. annulata provoked significant changes in host cell gene expression (1,179 and 1,517 differentially expressed genes respectively, with fold change >2 and padj < .05). Transcriptional changes between virulent compared to attenuated Ode macrophages are less profound (76 genes, with fold change >2 and padj < .05), likely because the infected and transformed macrophages only appear to differ in dissemination potential (Figure S2A).
To identify bovine genes whose transcription is perturbed by transformation and attenuation of dissemination of T. annulata‐transformed leukocytes we concentrated on the most differentially expressed genes (DEGs) (Figure 1 and Table S1). Many of these genes are annotated as being implicated in cell proliferation and metastasis. Among the top five up‐regulated transcripts in TBL20 is MMP9 (matrix metallopeptidase 9), a gene highly expressed in different cancer types and linked to metastasis and angiogenesis (Yu & Stamenkovic, 2000). WC1‐8 is the third most up‐regulated gene in TBL20 lymphocytes and has been described as being also up‐regulated in ovarian carcinoma cells (Mangala et al., 2009). The most down‐regulated transcripts in TBL20 cells include LAIR1 (leukocyte‐associated immunoglobulin‐like receptor 1) and VPREB (pre‐B lymphocyte 1). LAIR1 is a strong inhibitor of natural killer cell‐mediated cytotoxicity and an inhibitory receptor, which down‐regulates B lymphocyte immunoglobulin and cytokine production (Merlo et al., 2005). Relative down‐regulation of LAIR‐1 was not unexpected, as its loss of expression is observed during B cell proliferation (van der Vuurst de Vries et al., 1999). This is because TBL20 lymphocytes possess a higher proliferative capacity compared to their uninfected BL20 counterpart. ZBTB32 (zinc finger and BTB domain containing 32), IL21R (interleukin 21 receptor), and MMP9 are among the top five up‐regulated transcripts in infected TBL3 B lymphocytes. MMP9 and ANXA5 are among the top 10 up‐regulated genes in both TBL3 and TBL20, reflecting a common role of these genes in the promotion of the Theileria‐induced transformation phenotype. ANXA5 was previously reported to promote proliferation, migration and invasion in renal cell cancer. The five most down‐regulated transcripts in TBL3 are KRT6C (keratin 6C), MATK (megakaryocyte‐associated tyrosine kinase), IGSF9B (immunoglobulin superfamily member 9B), A2M (alpha‐2‐macroglobulin) and H2AFY2 (H2A histone family, member Y2). The biological functions of these genes include inhibition of cell growth and proliferation (Kim et al., 2004), repression of DNA transcription (Perche et al., 2000) and inhibition of cell adhesion and migration (Kurz et al., 2017), functions that are often dampened to allow continuous proliferation and survival of transformed cells. Out of the top 10 up‐ and down‐regulated genes in TBL3, 8 are up and 3 are down‐regulated in TBL20. Similarly, out of the top 10 up‐ and down‐regulated genes in TBL20 cells, 5 are up‐ and 4 are down‐regulated in TBL3. This shows that the transcriptional changes induced by T. annulata are not identical between the two lymphocyte cell lines. We confirmed by qRT‐PCR biologically reproducible differential expression patterns of 21 randomly selected genes from the BL3/TBL3 and BL20/TBL20 RNA‐seq datasets (Figure S2B).
FIGURE 1.
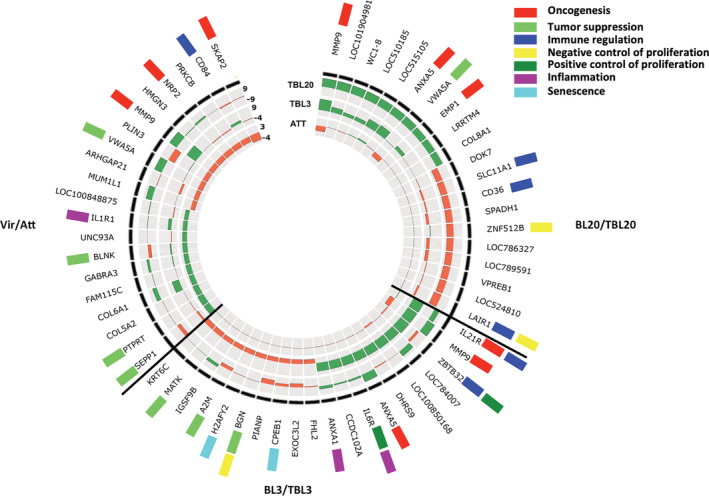
Top 20 DEGs in Theileria‐transformed bovine host cells. Circos plot showing the top 10 up‐ and down‐regulated DEGs in BL3/TBL3, BL20/TBL20 and attenuated versus virulent (Att/Vir) Ode macrophages. The circular heatmap represents the FC of the top DE genes in BL20/TBL20, BL3/TBL3 and Att/Vir Ode in the outer, middle and inner rings, respectively, where green reflects the level of up‐regulation and orange down‐regulation. The genes with biological functions related to tumorigenesis and immune regulation are tagged with coloured rectangles. Genes with no tag are hypothetical genes or have no known function in tumorigenesis and immune regulation. DE, differentially expressed; DEG, differentially expressed gene; FC, fold change
2.2. Identification of key genes potentially involved in Theileria‐mediated macrophage dissemination
The most down‐regulated transcripts in attenuated Ode macrophages are SKAP2 (src kinase associated phosphoprotein 2), a gene known to promote tumour dissemination through the regulation of podosome formation in macrophages (Tanaka et al., 2016) and NRP2 that regulates tumour progression by promoting TGF‐β‐signalling (Grandclement et al., 2011). Down‐regulation of these genes correlates with decreased dissemination of attenuated macrophages, as previously we have described loss of TGF‐β‐signalling as being associated with decreased dissemination (Chaussepied et al., 2010). By contrast, the most highly up‐regulated transcripts include SEPP1 (selenoprotein P) and PTPRT (protein tyrosine phosphatase, receptor type T). Both SEPP1 and PTPRT have been previously described as tumour suppressor genes (Scott & Wang, 2011; Short et al., 2016). Taken together, the identity of the most strongly up‐ and down‐regulated genes argues that our differential transcription screen could identify novel genes regulating Theileria‐transformed macrophage dissemination.
To define the genes likely playing important roles in transformation and dissemination, we compared genes differentially expressed (DE) in TBL3, TBL20 and attenuated Ode macrophages. We assumed that the genes most likely to play a key role are up‐regulated after infection and down‐regulated upon attenuation, and vice versa, that is, the genes down‐regulated after infection and up‐regulated after attenuation. To identify these genes we have intersected the obtained DEG lists of BL3/TBL3, BL20/TBL20 and Vir/Att Ode. This approach identified four genes with potential to play key roles in the dissemination of Theileria‐transformed leukocytes (Figure 2a).
FIGURE 2.
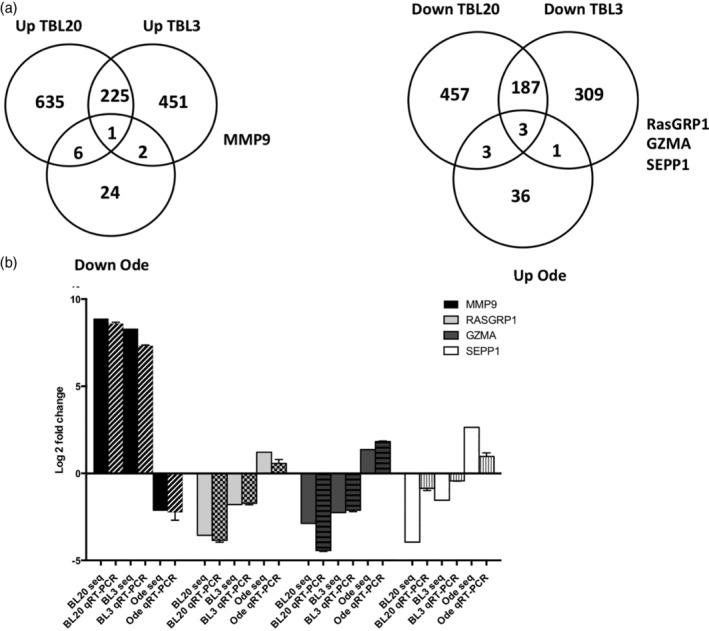
Inversely DEGs in TBL20, TBL3 and Att Ode leukocytes. (a) Venn diagrams illustrating the genes inversely DE in TBL3, TBL20 and attenuated Ode macrophages. (b) qRT‐PCR confirmation of DEGs potentially playing key roles in leukocyte transformation and dissemination. Seq and qRT‐PCT refer to sequencing and real‐time quantitative reverse transcription PCR, respectively. The reactions were set in three biological replicates and the fold change calculated with the 2ΔΔct method. The error bars represent SEM. DE, differentially expressed; DEG, differentially expressed gene
The genes are MMP9, SEPP1, GZMA and RASGRP1. MMP9 is the only gene up‐regulated after T. annulata‐mediated transformation and down‐regulated after attenuation. Inversely, SEPP1, GZMA and RASGRP1 are down‐regulated after transformation and up‐regulated after attenuation. The biological functions of these genes have been implicated in metastasis and cell invasion (Yu & Stamenkovic, 2000), selenium transport (Burk et al., 1995), peptide cleavage by immune cells (Chowdhury & Lieberman, 2008) and regulation of B cell‐development and homeostasis and differentiation (Priatel et al., 2007), respectively (Table 1). Differential expression of these genes was confirmed by qRT‐PCR (Figure 2b). We focused on GZMA, RASGRP1 and SEPP1, as the role of MMP9 is associated with the virulence of a number of T. annulata‐infected cell lines (Somerville, Adamson, Brown, & Hall, 1998). CRISPR/Cas9‐mediated loss of SEPP1 in attenuated macrophages resulted in a lethal phenotype. It is possible that SEPP1 plays an essential role in transformed macrophage survival, and so the role of SEPP1 in Theileria‐induced transformation was not further evaluated (Figure S3).
TABLE 1.
Biological functions of DEGs potentially playing key roles in T. annulata‐mediated leukocyte transformation and dissemination
| Gene symbol | Log2 FC (TBL20) | Adj. p‐value | Log2 FC (TBL3) | Adj. p‐value | Log2 FC (ATT) | Adj. p‐value | Biological functions | References |
|---|---|---|---|---|---|---|---|---|
| MMP9 | 8.87 | 0 | 8.29 | 0 | −2.13 | 5.14 E‐94 | Metastasis formation, cancer cells invasion | PMID: 10652271 |
| SEPP1 | −3.95 | 8.74 E‐174 | −1.54 | 3.75 E‐248 | 2.63 | 1.01 E‐140 | Transports selenoprotein, tumour suppressor | PMID: 26053663 PMID: 27314080 PMID: 8884283 |
| GZMA | −2.87 | 1.45 E‐05 | −2.25 | 1.97 E‐41 | 1.02 | 9.14 E‐21 | Plays a role in killing pathogen‐infected cells and cancer cells | PMID: 18304003 PMID: 15780992 |
| RASGRP1 | −3.55 | 0 | −1.78 | 1.97 E‐41 | 1.2 | 2.15 E‐15 | Required for correct functioning of lymphocytes in chronic infections | PMID: 17675473 |
Abbreviations: DEG, differentially expressed gene; FC, fold change.
2.3. Ablation of GZMA and RASGRP1 by CRISPR/Cas9 knockdown
To investigate the biological functions and molecular mechanisms of GZMA and RASGRP1, we knocked down their expression by CRISPR/Cas9. Although the macrophage transfection efficiency is generally not higher than 30%, analysis of the mixed population by qRT‐PCR revealed a 3.4‐ and 1.4‐fold reduction in gene expression for RASGRP1 and GZMA, respectively (Figure 3a,b). All subsequent phenotypical analyses were performed on this mixed population. Also, as controls we transfected a CRISPR/Cas9 plasmid encoding an irrelevant guide RNA and demonstrated specificity of knockdown by showing that CRISPR/Cas9 targeting of RASGRP1 does not knockdown GZMA (Figure S4A).
FIGURE 3.
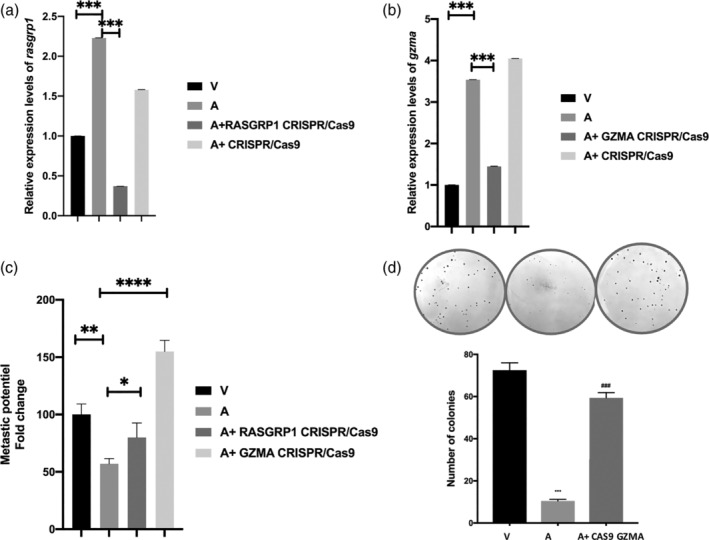
Colony formation and invasiveness of Ode macrophages after RASGRP1 and GZMA knockdown. (a,b) qRT‐PCR confirmation of GZMA and RASGRP1 knockdown, respectively. (c) Matrigel chamber assay showing a regain in matrigel traversal after RASGRP1 and GZMA knockdown. (d) Increased colony formation in soft agar following RASGRP1 knockdown. Non‐transfected virulent disseminating Ode macrophages are indicated by V, and non‐transfected poorly disseminating attenuated Ode macrophages by A. Error bars represent SEM of three biological replicates. ****p < .0005, ***p < .001, **p < .005 and *p < .05
Next, we tested whether GZMA and RASGRP1 can regulate the capacity of attenuated macrophages to traverse Matrigel. The inhibition of their expression led to a regain in dissemination potential, as estimated in matrigel traversal assays (Figure 3c). No effect was observed in traversal capacity when infected macrophages were transfected with the control CRISPR/Cas9 Plasmid (Figure S4B). Although only a 1.4‐fold decrease in GZMA expression was observed following CRISPR/Cas9, this lead to a 3.2‐fold increase in matrigel traversal. A reduction of RASGRP1 expression by 3.4‐fold lead to a 1.9‐fold increase in matrigel traversal. Both GZMA and RASGRP1 have therefore, the potential to function as suppressors of tumour dissemination and consistently, knockdown of GZMA also led to a regain in the ability of attenuated macrophages to form colonies in soft agar (Figure 3d). Taken together it strongly suggests that GZMA and RASGRP1 function as tumour suppressors.
2.4. GZMA and RASGRP1 dampen in vivo dissemination of Ode macrophages
Similar to metastatic tumour cells T. annulata‐transformed leukocytes also disseminate in immunodeficient mice to distant organs and form proliferative foci (Fell, Preston, & Ansell, 1990). Dissemination of Theileria‐transformed leukocytes has been previously attributed to increased production of matrix metaloproteinases (MMPs) (Somerville et al., 1998). As GZMA and RASGRP1 knockdown led to a regain in matrigel traversal we used Rag2γC immunodeficient mice to test for a regain in dissemination in vivo. The CRISPR/Cas9‐induced ablation of expression of GZMA gave rise to an increase in the number of Theileria‐containing tumours in kidney, while knockdown of RASGRP1 increased the number of tumours in the lung and mesentary (Figure 4). Thus, loss of RASGRP1 and GZMA expression led to a regain in the organ/tissue invasive capacity of T. annulata‐transformed macrophages into these organs.
FIGURE 4.
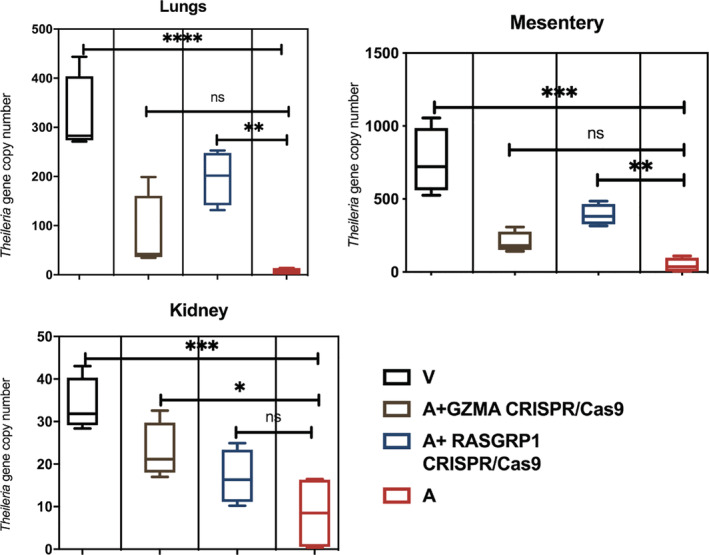
Effect of GZMA and RASGRP1 knockdown on transformed macrophage dissemination in vivo. Panels represent the copy number of the single copy T. annulata gene (ama‐1, TA02980) in the lung, mesentery and left kidney. Transformed macrophages were injected into five Rag2γC immunodeficient mice and plotted values represent the mean of obtained T. annulata‐specific ama1 gene copy number. There was no obvious difference in proliferation rate between transfected cells and control in vitro, and no obvious difference in number of nuclei per schizont between knockdown and control cells. Error bars represent SD of five biological replicates. ***p < .001, **p < .005 and *p < .05 compared to virulent and attenuated Ode macrophages
2.5. RASGRP1 knockdown reduces GZMA expression
RASGRP1‐activated Ras family proteins possess both pro‐ and anti‐oncogenic properties, depending on the downstream effector pathway and cellular context; reviewed in (Cox & Der, 2003). Our transcription profiling showed that most members of the RASGRP gene family (RASGRP1, 2 and 4) are significantly down‐regulated in TBL3 and TBL20 (Figure 5a). In order to explain the higher dissemination of RASGRP1‐knocked down macrophages in vivo, we investigated whether loss of RASGRP1 could perhaps also provoke a drop in GZMA expression rendering attenuated macrophages more deficient in dissemination. As hypothesised, we found that expression of GZMA decreased after RASGRP1 knockdown (Figure 5b). Moreover, GZMA and RASGRP1 expression is repressed by TGF‐β (Takami, Cunha, Motohashi, Nakayama, & Iwashima, 2018; Thomas & Massague, 2005) and the role of TGF‐β in regulating dissemination of Theileria‐transformed macrophages is well established (Chaussepied et al., 2010; Haidar, Echebli, et al., 2015; Haidar, Whitworth, et al., 2015). Indeed, expression of GZMA and RASGRP1 was significantly decreased in attenuated Ode macrophages treated with TGF‐β (Table S2). Taken together, it suggests that one way TGF‐β promotes dissemination of Theileria‐transformed leukocytes could be via repression of both GZMA and RASGRP1 transcription and their impact on dissemination that we confirmed in vivo in mice.
FIGURE 5.
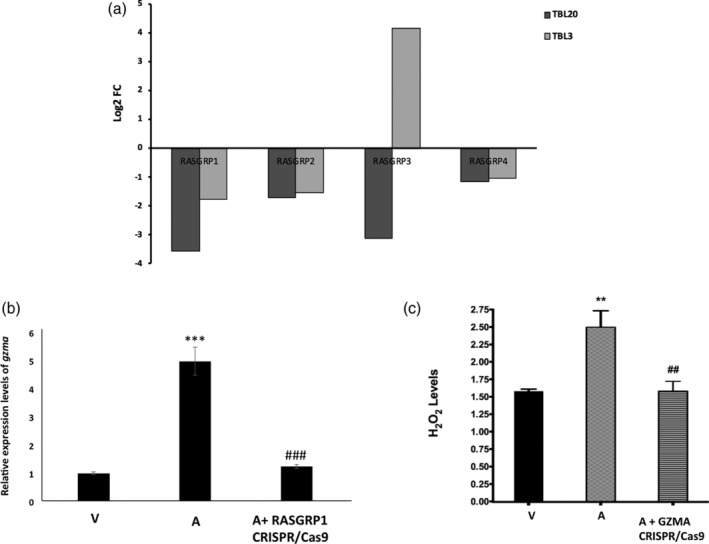
RASGRP1 and GZMA knockdown reduces GZMA expression and dampening of H2O2 levels, respectively. (a) Log2FC values of RASGRP1‐4 in TBL3 and TBL20 RNAseq Log2FC values from DESeq2 of TBL3 and TBL20 compared to BL3 and BL20, respectively. (b) Effect of RASGRP1 knockdown on GZMA expression. qRT‐PCR of GZMA in virulent (V), attenuated (A) and attenuated Ode macrophages after CRISPR/Cas9‐mediated RASGRP1 knockdown. Error bars represent SD of three biological replicates *** and ### represent p < .001 compared to virulent and attenuated Ode macrophages, respectively. (c) H2O2 output by virulent (V), attenuated (A), and attenuated Ode macrophages after CRISPR/Cas9‐mediated GZMA knockdown. Error bars represent SD of three biological replicates. ** and ## represent p < .01 compared to virulent and attenuated Ode macrophages, respectively. FC, fold change
2.6. GZMA loss of expression dampens H2O2 levels
GZMA is a serine protease that contributes to killing of both tumours and pathogen‐infected cells via a caspase‐independent pathway (Chowdhury & Lieberman, 2008). GZMA expression induces reactive oxygen species (Martinvalet, Zhu, & Lieberman, 2005) and attenuated macrophages are known to be more oxidatively stressed than virulent macrophages (Metheni et al., 2014). Indeed, H2O2 output was reduced in attenuated macrophages following CRISPR/Cas9‐mediated GZMA knockdown (Figure 5c).
2.7. Induced expression of GZMA and RASGRP1 reduces human B lymphoma cell dissemination
In order to extend the roles of GZMA and RASGRP1 to human cancer we sought human tumour cells displaying transcriptional signatures similar to T. annulata‐transformed leukocytes. To this end, the transcriptional profiles of 934 human cancer cells were obtained from the EBI cancer cell line expression atlas (Papatheodorou et al., 2018) and their profiles compared to those of Theileria‐transformed TBL3 and TBL20 B lymphocytes (Figure 6a). A total of 46 out of 53 and 76 out of 79 of the lymphoma and leukaemia cell lines respectively present in the dataset, clustered in the subset of T. annulata‐infected B cell lines and their uninfected counterparts, representing 86 and 96% of the total number of lymphoma/ leukaemia cell lines in the entire dataset, respectively. The closely clustered cell line OCI‐LY19, along with the B cell non‐Hodgkin lymphoma cell line RI‐1, were used to test if GZMA and RASGRP1 can act as suppressors of dissemination in certain types of human cancer. CRISPR‐mediated transcriptional activation of GZMA and RASGRP1 resulted in decreased matrigel traversal of the OCI‐LY19 B lymphoma. By contrast, only up‐regulation of GZMA showed a statistically significant decrease in traversal of the RI‐1 B lymphoma (Figure 6c). Moreover, elevated expression of GZMA and RASGRP1 decreased the ability of both RI‐1 and OCI‐LY19 B lymphoma cells to form colonies on soft agar (Figure 6d), providing further functional evidence that GZMA and RASGRP1 have the potential to act as suppressors in two independent human B lymphomas.
FIGURE 6.
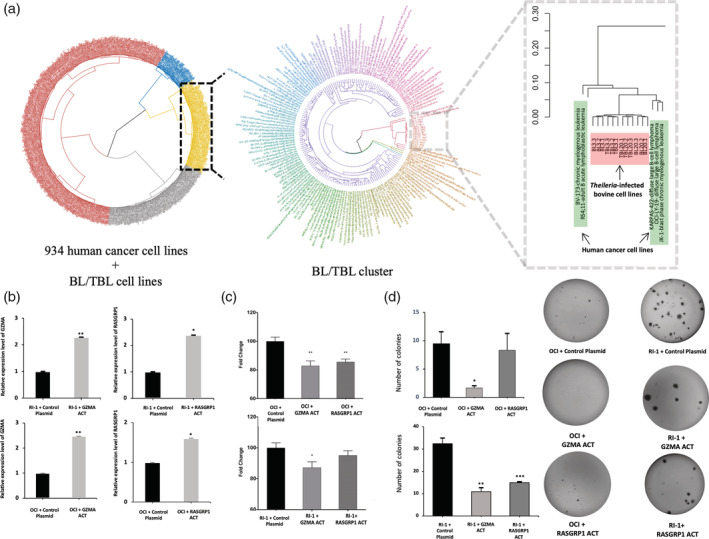
Effect of GZMA and RASGRP1 activation on human B lymphomas. (a) (Left panel) PCA based hierarchical clustering of 934 human cancer cell lines and T. annulata‐transformed bovine B cells and their non‐infected counterparts. The cluster containing bovine cells mainly contains leukaemic human cancer cell lines. The samples are coloured by cluster ID. (Middle cluster) The samples from the BL3/TBL3 and BL20/TBL20 sub‐cluster were reclustered by comparing the similarity of their gene expression profiles. The sample labels are coloured by their similarity to each other. (Right panel) The sub‐cluster containing bovine cell lines and human cancer cell lines with similar gene expression profile. (b) qRT‐PCR determination of GZMA and RASGRP1 expression after CRISPR‐mediated gene activation in RI‐1 (top panel) and OCI‐LY19 (bottom panel). The error bars represent SEM of three biological replicates. (c) Matrigel chamber assay illustrating the dissemination potential of RI‐1 and OCI‐LY19 following activation of RASGRP1 and GZMA transcription. (d) Decrease in colony formation in soft agar of OCI‐LY19 and RI‐1 following GZMA and RASGRP1 activation. Errors bars represent SEM values of three biological replicates *, ** and *** represent student t test p < .05, p < .005 and p < .0005, respectively
3. DISCUSSION
In this study, we provide a holistic view of the transcriptional landscape of two T. annulata‐transformed B cell lines, TBL3 and TBL20, and in addition the landscape of virulent versus attenuated Ode macrophages. In order to find genes with commonly perturbed transcription the different datasets were compared using three independent pipelines to identify just four genes, as potential regulators of tumour dissemination. In addition to MMP9 three other genes (SEPP1, GZMA and RASGRP1) were identified as potentially having a role in dissemination. SEPP1 is a major selenoprotein involved in selenium transport and cellular defence against oxidative stress (Hill, Dasouki, Phillips 3rd, & Burk, 1996). Attenuated macrophages did not survive CRISPR/Cas9‐knockdown of SEPP1 implying death might be due to a failure to control excessive oxidative stress, since attenuated macrophages display high levels of H2O2 (Metheni et al., 2014).
GZMA is known to cleave APEX1 (apurinic/apyrimidinic endodeoxyribonuclease 1) after Lys31 and cleavage destroys its oxidative repair functions. APEX1 is involved in NK‐cell‐mediated killing via GZMA (Fan et al., 2003; Martinvalet et al., 2005) and can suppress the activation of PARP1 during repair of oxidative DNA damage (Peddi, Chattopadhyay, Naidu, & Izumi, 2006), and prevent oxidative stress by negatively regulating Rac1/GTPase activity (Ozaki, Suzuki, & Irani, 2002). Additionally, APEX1 directly reduces the redox‐sensitive cysteine residues of target transcription factors, enhancing their DNA binding and transcriptional activity. Analysis of our deep RNA‐seq data revealed that APEX1 is down‐regulated in attenuated macrophages and its expression increases after TGF‐β treatment, along with an important down‐regulation of GZMA and RASGRP1 (Table S2). APEX1 is over‐expressed in many cancers (Kakolyris et al., 1998; Moore, Michael, Tritt, Parsons, & Kelley, 2000; Yang, Irani, Heffron, Jurnak, & Meyskens, 2005) and has been implicated in growth, migration and invasion of colon cancer both in vitro and in vivo (Kim, Kim, et al., 2013 ). Interestingly, APEX‐1 protects melanoma cells from H2O2 induced apoptosis (Yang et al., 2005). The established function of APEX1 in human cancer sustains our novel observations on the tumour suppressor roles of GZMA and RASGRP1. Published data and our results suggest that reduced TGF‐β2 signalling leads to an increase in the transcription of GZMA which likely leads to APEX1 cleavage and, hence, an increase in oxidative stress because of H2O2 accumulation. This is consistent with our data shown in Figure 5c, where CRISPR‐Cas9‐mediated knockdown of GZMA caused a decrease in H2O2 levels.
In order to investigate the expression levels of MMP9, GZMA, RASGRP1 and SEPP1 in other human cancer types, we interrogated the meta‐analysis of lung cancer across 19 independent studies (Cai et al., 2019). Interestingly, the findings suggest a striking similarity in the differential expression profiles of three out of the four key genes. MMP9 shows an up‐regulation in lung cancer tissue [Effect size (ES): 15.22; p = 2.7E−052], while GZMA (ES: −4.99; p = 5.9E−07) and SEPP1 (ES: −11.4; p = 4.40E−30) show a marked down‐regulation in expression profiles in comparison to healthy tissues (Cai et al., 2019). The strong correlation in the differential expression profiles of these genes reflects their relevance in at least two unrelated cancer types.
This study has revealed new players in dissemination and oxidative stress regulation of Theileria‐transformed leukocytes and has provided evidence for similar roles for GZMA and RASGRP1 in transcriptionally matched human B lymphoma cell lines. The similarity between Theileria‐induced B cell transformation and human B lymphomas opens novel targets for intervention against tumour dissemination. This is a potent illustration of the benefits of trans‐disciplinary research in general, and of the use of infectious agents as providers of untapped perspectives on fundamental cellular processes ‐ in this case, processes associated with cancer progression.
4. EXPERIMENTAL PROCEDURES
4.1. Cell lines
The BL3 (Theilen et al., 1968), TBL3, BL20 (Morzaria, Roeder, Roberts, Chasey, & Drew, 1984), TBL20 B lymphocytes, and Ode macrophages (Singh et al., 2001) were cultured in RPMI 1640 medium supplemented with 2 mM of l‐glutamine (Lonza, catalogue number 12‐702F) and 10 mM Hepes (Lonza, catalogue number 17‐737E), 10% heat‐inactivated FBS (Gibco, catalogue number 10082147), 100 units/mL of Penicillin and 100 μg/mL of streptomycin (Lonza, catalogue number 17‐602E) and 10 mM b‐mercaptoethanol (Sigma‐Aldrich, catalogue number M6250) for BL3/TBL3 and BL20/TBL20. The virulent (Vir) hyper‐disseminating Ode cell line was used at low passage (53–71), while its attenuated (Att) poorly disseminating vaccine counterpart corresponded to passages 309–317. The OCI‐LY19 cell line (DSMZ, ACC 528) was cultured in Minimum Essential Medium Eagle ‐ Alpha Modification (Gibco, catalogue number 12000063) supplemented with 2.2 g/L of sodium bicarbonate (Thermofisher Scientific, catalogue number 25080094), 20% FBS, 10 mM Hepes and 100 units/mL of Penicillin and 100 ug/ml of streptomycin. The RI‐1 cell line (DSMZ, ACC 585) was cultured in RPMI 1640 and supplemented with 10% FBS, 100 units/mL of Penicillin and 100 ug/ml of streptomycin and 10 mM Hepes. All cell lines were incubated at 37°C with 5% CO2. All cell lines were regularly tested for mycoplasma contamination.
4.2. RNA extraction
Cells were seeded in three biological replicates at a density of 2.5 × 105 cell/ml. RNA extraction was performed using the PureLink RNA Mini Kit (Life technologies, catalogue number 12183018A) following the manufacturer's instructions. Briefly, cells were pelleted, lysed and homogenised using a 21‐gauge needle, then 70% ethanol was added to the cell lysates and the samples were loaded on spin cartridges to bind RNA. After three washes, RNA was eluted in RNase‐free water. The quality of extracted RNA was verified using a Bioanalyzer 2100 and quantification carried using Qubit (Invitrogen, catalogue number Q10210).
4.3. Illumina library preparation and sequencing
Strand‐specific RNA‐sequencing (ssRNA‐seq) libraries were prepared using the illumina Truseq Stranded mRNA Sample Preparation Kit (Illumina, catalogue number RS‐122‐2101) following the manufacturer's instructions. Briefly, 1ug of total RNA was used to purify mRNA using poly‐T oligo‐attached magnetic beads. mRNA was then fragmented and cDNA was synthesised using SuperScript III reverse transcriptase (Thermofisher, catalogue number 18080044), followed by adenylation on the 3′ end, barcoding and adapter ligation. The adapter ligated cDNA fragments were then enriched and cleaned with Agencourt Ampure XP beads (Agencourt, catalogue number A63880). Libraries validation was conducted using the 1000 DNA kit on 2100 Bioanalyzer (Agilent Technologies, catalogue number 5067‐1504) and quantified using qubit (Thermofisher, catalogue number Q32850). ssRNA libraries were sequenced on Illumina Hiseq2000 and Hiseq4000. The sequenced reads were mapped to the Bos taurus genome Btau 4.6.1. The quality of the sequenced libraries is shown in Figure S1.
4.4. Sequencing data analysis
The quality of sequence reads and other parameters were checked using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The raw RNA‐seq reads were processed for adaptor trimming by Trimmomatic (Bolger, Lohse, & Usadel, 2014). The strand‐specific reads were mapped on to Bovine genome (bosTau7; Btau_4.6.1; GCF_000003205.5) using Tophat2 (‐g 1 ‐‐library‐type fr‐firststrand). The samples with respective replicates were analysed further for differential gene expression by three different tools, baySeq (Hardcastle & Kelly, 2010), DESeq2 (Kim, Pertea, et al., 2013) (fitType = ‘local’) and CuffDiff2 (Trapnell et al., 2013) with default parameters unless mentioned specifically. The count values for DESeq2 and baySeq were calculated from BAM files using HTSeq‐count tool (Anders, Pyl, & Huber, 2015). The transcriptome quality plots were generated by cummeRbund package (v2.14.0) in R (http://bioconductor.org/packages/release/bioc/html/cummeRbund.html). The sequencing data is available in the NCBI Gene Expression Omnibus, GEO ID: GSE135377.
4.5. Identification of differentially expressed genes after infection and attenuation by comparative transcriptome analysis
The transcriptome data was analysed with baySeq, DESeq2 and CuffDiff2. A gene was considered as a DEG if it has a padj < .05 and a fold change (FC) > 2. The final list of DEGs contained genes commonly differentially expressed in CuffDiff2, DESeq2 and baySeq. This approach minimalizes the total number of DEGs for further analysis and allows stringent selection of the most significant and reproducible DEGs.
4.6. qRT‐PCR
Total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, catalogue number 4368814) as follows: 2 μg of total RNA, 2 μL of RT buffer, 0.8 μL of 100 mM dNTP mix, 2.0 μL of 10× random primers, 1 μL of MultiScribe reverse transcriptase and Nuclease‐free water to a final volume of 20 μL. The reaction was incubated 10 min at 25°C, 2 hr at 37°C then the enzyme inactivated at 85°C for 5 min. Real‐time PCR was performed in a 10 μL reaction containing 20–30 ng cDNA template, 5 μL 2× Fast SYBR Green Master Mix and 500 nM of forward and reverse primers. The reaction was run on the 7500 HT Fast Real‐Time PCR System (Applied Biosystems). GAPDH was used as a housekeeping gene and the results were analysed by the 2−∆∆CT method. The error bars represent the SEM of three biological replicates. Primers were designed and assessed for secondary structures using the Primer Express Software v3.0. The primers of all genes are listed in Table S3.
4.7. Transfection
Macrophages were transfected by electroporation using the Nucleofector system (Amaxa Biosystems). A total of 5 × 105 cells were suspended in 100 μL of Nucleofector V solution mix (Lonza, VCA‐1003) with 2 μg of GZMA (sc‐437323), RASGRP1 (sc‐437322) CRISPR/Cas9 KO plasmids, or a negative control plasmid harbouring non‐specific guide RNA (SCBT, sc‐418922) was used in nucleofection using the cell line‐specific program T‐O17. CRISPR/Cas9 KO plasmids consist of a pool of three plasmids, each encoding the Cas9 nuclease and a different target‐specific 20 nt guide RNA (gRNA) designed for maximum efficiency. The human B lymphoma cells were transfected with 2 μg of GZMA (SCBT, sc‐403958‐ACT) or RASGRP1 (SCBT, sc‐402120‐ACT) synergistic activation mediator (SAM) transcription activation system consisting of three plasmids encoding the deactivated Cas9 (dCas9) nuclease, the MS2‐p65‐HSF1 fusion protein and a target‐specific 20 nt guide RNA. A non‐specific control plasmid with a non‐specific guide RNA was used as a control (SCBT, sc‐437275). The transfection was done using the T‐O17 program. After transfection, cells were suspended in fresh complete medium and incubated at 37°C with 5% CO2.
4.8. Matrigel chamber assay
The invasive capacity of Ode macrophages was assessed in vitro using matrigel migration chambers, as described in (Lizundia et al., 2006). The CultureCoat Medium basement membrane extract (BME) 96‐wells cell invasion assay was performed according to Culturex instructions (Trevigen, catalogue number 3482‐096‐K). After 24 hr of incubation at 37°C, each well of the top chamber was washed once in buffer. The top chamber was placed back onto the receiver plate. One hundred microliters of cell dissociation solution‐Calcein AM was added to the bottom chamber of each well, and the mixtures were incubated at 37°C for 1 hr with fluorescently labelled cells to dissociate the cells from the membrane before reading at 485‐nm excitation and 520‐nm emission wavelengths.
4.9. Soft agar colony forming assay
A two‐layer soft agar culture system was used. Cell counts were performed by ImageJ software. A total of 2,500 cells were plated in a volume of 1.5 mL (0.7% bacto Agar + 2× RPMI 20% Foetal bovine Serum) over 1.5 mL base layer (1% bacto agar + 2× RPMI 20% Foetal bovine Serum) in six‐well plates. Cultures were incubated in humidified 37°C incubators with an atmosphere of 5% CO2 in air, and control plates were monitored for growth using a microscope. At the time of maximum colony formation final colony numbers were counted in image J after fixation with 0.005% Cristal Violet.
4.10. Intracellular levels of hydrogen peroxide (H2O2)
Cells were seeded at 1 × 105 cell/well in a 96 well plate and incubated in complete medium for 18 hr prior to the assay. Cells were then washed with PBS and incubated with 100 μL of 5 M H2‐DCFDA in PBS (Molecular Probes, catalogue number D399). H2O2 levels were assayed on a fusion spectrofluorimeter (PackardBell) at 485 and 530 nm excitation and emission wavelengths respectively.
4.11. In vivo mouse studies and quantification of T. annulata‐transformed macrophages load in mouse tissues
Theileria annulata‐infected macrophage cell lines (Virulent Ode passage 53, attenuated Ode passage 309, attenuated Ode transfected with RASGRP1 CRISPR/Cas9 knockout plasmid and attenuated Ode transfected with GZMA CRISPR/Cas9 knockout plasmid) were injected into four groups of five Rag2γC immunodeficient mice that were equally distributed on the basis of age and sex in each group. The injection site was disinfected with ethanol and one million cells (in 200 μL PBS) were injected under the skin after gentle shaking of the insulin syringe. The mice were kept for 3 weeks and then they were humanely sacrificed and dissected. Six internal organs including heart, lung, spleen, mesentery, left kidney and liver were taken and stored in 500 μL PBS in Eppendorf tubes at −20°C. The tissues were subjected to genomic DNA extraction using the QIAmp DNA mini kit (Qiagen, catalogue number 51304). DNA concentrations were measured by Nanodrop 1000 spectrophotometer (Thermo Fischer scientific) and before each quantitative PCR reaction samples were diluted to give a DNA concentration of 0.5 ng/μL. Absolute copy numbers of a single copy T. annulata gene (ama‐1, TA02980) that is representative of T. annulata‐infected macrophage load in each tissue were estimated by the method described in Gotia et al. (2016), with some modifications. Ama‐1 was cloned into pJET 1.2/blunt cloning vector using CloneJET PCR Cloning Kit (Thermo scientific, catalogue number K1232). The cloned plasmid was amplified in DH5‐Alpha cells and purified with QIAfilter Plasmid Maxi Kit (Qiagen, catalogue number 12243). Plasmid concentration was measured using Qubit (Thermofisher, catalogue number Q32850). The primers for cloning were: forward 5′‐GGAGCTAACTCTGACCCTTCG‐3′ and reverse 5′‐CCAAAGTAGGCCAATACGGC‐3′. Quantitative PCR primers were: forward 5′‐GACCGATTTCATGGCAAAGT‐3′ and reverse 5′‐TTGGGGTCATGATGGGTTAT‐3′.
4.12. Transcriptome‐based clustering of Theileria‐transformed bovine host cells and human cancer cell lines
The processed and quality trimmed reads from TBL20/BL20 and BL3/TBL3 samples were mapped to Bos taurus UMD3.1 genome using HISAT2 software (Kim, Langmead, & Salzberg, 2015) with default settings. The mapped reads were used for gene‐level TPM quantification using StringTie (Version 1.3.3b) (Pertea et al., 2015; Pertea, Kim, Pertea, Leek, & Salzberg, 2016). The quantified genes were converted into their human ortholog Ensemble gene ID by finding one‐to‐one orthologs between human and B. taurus genomes using OMA browser (Altenhoff et al., 2018). Subsequently, TPM values of transcripts expressed across 934 human cancer cell lines were obtained from EBI cancer cell line Expression Atlas (Papatheodorou et al., 2018). The redundant transcripts in the cancer cell line expression set were collapsed using collapseRow function from the WGCNA R package (Langfelder & Horvath, 2008). Using the common human ensemble gene ID, gene expression matrices of B. taurus and human cell lines were merged together, which was then subjected to hierarchical clustering of samples using HCPC (Lê, Josse, & Husson, 2008) with three Principal Components (nPCs), which resulted in four broad clusters. The sub‐cluster containing the human cancer cell lines along with TBL20/BL20 and BL3/TBL3 were shortlisted for further analysis. Whereby, samples were scanned for similar gene expression profiles by computing the adjacency of each shortlisted sample with the rest of the samples, using adjacency function (method = ‘Distance’) from WGCNA R package. The resultant adjacency matrix was then subjected to flashClust (Lê et al., 2008) program for computing the dendrogram for manual inspection of TBL20/BL20 and BL3/TBL3 containing sub‐cluster. A schematic of the used pipeline is presented in Figure S5. The complete dendrogram of the 934 human cancer cell lines and Theileria‐transformed lymphocytes can be viewed in Figure S6.
4.13. Ethics statement
The protocol (Burk et al., 1995; Grandclement et al., 2011; Haidar et al., 2018; Kim et al., 2004; Kinnaird et al., 2013; Kurz et al., 2017; Mangala et al., 2009; Marsolier et al., 2013; Merlo et al., 2005; Perche et al., 2000; Scott & Wang, 2011; Short, Whitten‐Barrett, & Williams, 2016; Tanaka et al., 2016; van der Vuurst de Vries, Clevers, Logtenberg, & Meyaard, 1999; Yu & Stamenkovic, 2000) was approved by the ethics committee for animal experimentation of the University of Paris‐Descartes (CEEA34.GL.03312). The university ethics committee is registered with the French National Ethics Committee for Animal Experimentation that itself is registered with the European Ethics Committee for Animal Experimentation. The right to perform the mice experiments was obtained from the French National Service for the Protection of Animal Health and satisfied the animal welfare conditions defined by laws (R214‐87 to R214‐122 and R215‐10) and GL was responsible for all animal experimentation, as he holds the French National Animal Experimentation permit with the authorisation number (B‐75‐1249). This project is also covered by the KAUST IBEC number 19IBEC12.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Arnab Pain and Gordon Langsley conceived and designed the study. Zineb Rchiad prepared the ssRNAseq libraries. Sara Mfarrej, Malak Haidar and Zineb Rchiad ran qRT‐PCR reactions. Malak Haidar performed the soft agar colony formation assay and Matrigel chamber for Theileria‐infected macrophages and Zineb Rchiad and Sara Mfarrej for human cancer cell lines. Malak Haidar performed the intracellular H2O2 levels and Matrigel chamber assays. Shahin Tajeri performed the mouse dissemination assays. Hifzur Rahman Ansari, Abhinav Kaushik and Zineb Rchiad performed data analysis. Zineb Rchiad and Malak Haidar prepared the figures. Zineb Rchiad prepared the first draft of the manuscript with input from Fathia Ben Rached that was then edited by Malak Haidar, Gordon Langsley and Arnab Pain.
Supporting information
Appendix S1: Supporting Information
ACKNOWLEDGEMENTS
This study was supported by a Competitive Research Grant from the Office for Sponsored Research (OSR‐2015‐CRG4‐2610) at King Abdullah University of Science and Technology (KAUST) awarded to A. P. and G. L. Z. R. acknowledges KAUST for awarding her PhD studentship. S. T. was supported by a post‐doctoral fellowship from ParaFrap (ANR‐11‐LABX‐0024) and in addition to ANR‐11‐LABX‐0024, G. L. also acknowledges core support from INSERM and the CNRS. We thank members of the Bioscience Core Laboratory (BCL) at KAUST for producing the raw sequencing datasets. We also thank Franck Letourneur of the genomics platform at the Cochin Institute (GENOM'IC) for quantifying the pJET‐ama‐1 plasmid.
Rchiad Z, Haidar M, Ansari HR, et al. Novel tumour suppressor roles for GZMA and RASGRP1 in Theileria annulata‐transformed macrophages and human B lymphoma cells. Cellular Microbiology. 2020;22:e13255 10.1111/cmi.13255
Zineb Rchiad and Malak Haidar are co‐first authors.
Hifzur Rahman Ansari and Shahin Tajeri are co‐second authors.
Funding information Competitive Research Grant, King Abdullah University of Science and Technology, Grant/Award Number: OSR‐2015‐CRG4‐2610; ParaFrap, Grant/Award Number: ANR‐11‐LABX‐0024
Contributor Information
Gordon Langsley, Email: gordon.langsley@inserm.fr.
Arnab Pain, Email: arnab.pain@kaust.edu.sa.
REFERENCES
- Adamson, R. , Logan, M. , Kinnaird, J. , Langsley, G. , & Hall, R. (2000). Loss of matrix metalloproteinase 9 activity in Theileria annulata‐attenuated cells is at the transcriptional level and is associated with differentially expressed AP‐1 species. Molecular and Biochemical Parasitology, 106(1), 51–61. [DOI] [PubMed] [Google Scholar]
- Altenhoff, A. M. , Glover, N. M. , Train, C. M. , Kaleb, K. , Warwick Vesztrocy, A. , Dylus, D. , … Dessimoz, C. (2018). The OMA orthology database in 2018: Retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Research, 46(D1), D477–D485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P. T. , & Huber, W. (2015). HTSeq – A python framework to work with high‐throughput sequencing data. Bioinformatics, 31(2), 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, R. F. , Hill, K. E. , Awad, J. A. , Morrow, J. D. , Kato, T. , Cockell, K. A. , & Lyons, P. R. (1995). Pathogenesis of diquat‐induced liver necrosis in selenium‐deficient rats: Assessment of the roles of lipid peroxidation and selenoprotein P. Hepatology, 21(2), 561–569. [PubMed] [Google Scholar]
- Cai, L. , Lin, S. , Girard, L. , Zhou, Y. , Yang, L. , Ci, B. , … Xie, Y. (2019). Correction: LCE: An open web portal to explore gene expression and clinical associations in lung cancer. Oncogene, 38, 2551–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussepied, M. , Janski, N. , Baumgartner, M. , Lizundia, R. , Jensen, K. , Weir, W. , … Langsley, G. (2010). TGF‐b2 induction regulates invasiveness of Theileria‐transformed leukocytes and disease susceptibility. PLoS Pathogens, 6(11), e1001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, D. , & Lieberman, J. (2008). Death by a thousand cuts: Granzyme pathways of programmed cell death. Annual Review of Immunology, 26, 389–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock‐Rada, A. M. , Medjkane, S. , Janski, N. , Yousfi, N. , Perichon, M. , Chaussepied, M. , … Weitzman, J. B. (2012). SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP‐9. Cancer Research, 72(3), 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, A. D. , & Der, C. J. (2003). The dark side of Ras: Regulation of apoptosis. Oncogene, 22(56), 8999–9006. [DOI] [PubMed] [Google Scholar]
- Fan, Z. , Beresford, P. J. , Zhang, D. , Xu, Z. , Novina, C. D. , Yoshida, A. , … Lieberman, J. (2003). Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nature Immunology, 4(2), 145–153. [DOI] [PubMed] [Google Scholar]
- Fell, A. H. , Preston, P. M. , & Ansell, J. D. (1990). Establishment of Theileria‐infected bovine cell lines in scid mice. Parasite Immunology, 12(3), 335–339. [DOI] [PubMed] [Google Scholar]
- Gotia, H. T. , Munro, J. B. , Knowles, D. P. , Daubenberger, C. A. , Bishop, R. P. , & Silva, J. C. (2016). Absolute quantification of the host‐to‐parasite DNA ratio in Theileria parva‐infected lymphocyte cell lines. PLoS One, 11(3), e0150401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandclement, C. , Pallandre, J. R. , Valmary Degano, S. , Viel, E. , Bouard, A. , Balland, J. , … Borg, C. (2011). Neuropilin‐2 expression promotes TGF‐beta1‐mediated epithelial to mesenchymal transition in colorectal cancer cells. PLoS One, 6(7), e20444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar, M. , Echebli, N. , Ding, Y. , Kamau, E. , & Langsley, G. (2015). Transforming growth factor beta2 promotes transcription of COX2 and EP4, leading to a prostaglandin E2‐driven autostimulatory loop that enhances virulence of Theileria annulata‐transformed macrophages. Infection and Immunity, 83(5), 1869–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar, M. , Rchiad, Z , Ansari, H. R. , Ben‐Rached, F. , Tajeri, S. , Latre De Late, P. , … Pain, A. (2018). miR‐126‐5p by direct targeting of JNK‐interacting protein‐2 (JIP‐2) plays a key role in Theileria‐infected macrophage virulence. PLOS Pathogens, 14(3). 10.1371/journal.ppat.1006942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar, M. , Whitworth, J. , Noe, G. , Liu, W. Q. , Vidal, M. , & Langsley, G. (2015). TGF‐beta2 induces Grb2 to recruit PI3‐K to TGF‐RII that activates JNK/AP‐1‐signaling and augments invasiveness of Theileria‐transformed macrophages. Scientific Reports, 5, 15688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle, T. J. , & Kelly, K. A. (2010). baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics, 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K. E. , Dasouki, M. , Phillips, J. A., 3rd , & Burk, R. F. (1996). Human selenoprotein P gene maps to 5q31. Genomics, 36(3), 550–551. [DOI] [PubMed] [Google Scholar]
- Hofmann, U. B. , Westphal, J. R. , Van Muijen, G. N. , & Ruiter, D. J. (2000). Matrix metalloproteinases in human melanoma. The Journal of Investigative Dermatology, 115(3), 337–344. [DOI] [PubMed] [Google Scholar]
- Kakolyris, S. , Kaklamanis, L. , Engels, K. , Fox, S. B. , Taylor, M. , Hickson, I. D. , … Harris, A. L. (1998). Human AP endonuclease 1 (HAP1) protein expression in breast cancer correlates with lymph node status and angiogenesis. British Journal of Cancer, 77(7), 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. , & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12(4), 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. , & Salzberg, S. L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14(4), R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. H. , Kim, H. B. , Yoon, S. P. , Lim, S. C. , Cha, M. J. , Jeon, Y. J. , … You, H. J. (2013). Colon cancer progression is driven by APEX1‐mediated upregulation of jagged. The Journal of Clinical Investigation, 123, 3211–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. O. , Avraham, S. , Jiang, S. , Zagozdzon, R. , Fu, Y. , & Avraham, H. K. (2004). Differential expression of Csk homologous kinase (CHK) in normal brain and brain tumors. Cancer, 101(5), 1018–1027. [DOI] [PubMed] [Google Scholar]
- Kinnaird, J. H. , Weir, W. , Durrani, Z. , Pillai, S. S. , Baird, M. , & Shiels, B. R. (2013). A bovine lymphosarcoma cell line infected with Theileria annulata exhibits an irreversible reconfiguration of host cell gene expression. PLoS One, 8(6), e66833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, S. , Thieme, R. , Amberg, R. , Groth, M. , Jahnke, H. G. , Pieroh, P. , … Birkenmeier, G. (2017). The anti‐tumorigenic activity of A2M‐A lesson from the naked mole‐rat. PLoS One, 12(12), e0189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê, S. , Josse, J. , & Husson, F. (2008). FactoMineR: An R package for multivariate analysis. Journal of Statistical Software, 25(1), 18. [Google Scholar]
- Lizundia, R. , Chaussepied, M. , Huerre, M. , Werling, D. , Di Santo, J. P. , & Langsley, G. (2006). c‐Jun NH2‐terminal kinase/c‐Jun signaling promotes survival and metastasis of B lymphocytes transformed by Theileria. Cancer Research, 66(12), 6105–6110. [DOI] [PubMed] [Google Scholar]
- Mangala, L. S. , Zuzel, V. , Schmandt, R. , Leshane, E. S. , Halder, J. B. , Armaiz‐Pena, G. N. , … Sood, A. K. (2009). Therapeutic targeting of ATP7B in ovarian carcinoma. Clinical Cancer Research, 15(11), 3770–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier, J. , Pineau, S. , Medjkane, S. , Perichon, M. , Yin, Q. , Flemington, E. , … Weitzman, J. B. (2013). OncomiR addiction is generated by a miR‐155 feedback loop in Theileria‐transformed leukocytes. PLoS Pathogens, 9(4), e1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinvalet, D. , Zhu, P. , & Lieberman, J. (2005). Granzyme A induces caspase‐independent mitochondrial damage, a required first step for apoptosis. Immunity, 22(3), 355–370. [DOI] [PubMed] [Google Scholar]
- Merlo, A. , Tenca, C. , Fais, F. , Battini, L. , Ciccone, E. , Grossi, C. E. , & Saverino, D. (2005). Inhibitory receptors CD85j, LAIR‐1, and CD152 down‐regulate immunoglobulin and cytokine production by human B lymphocytes. Clinical and Diagnostic Laboratory Immunology, 12(6), 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metheni, M. , Echebli, N. , Chaussepied, M. , Ransy, C. , Chereau, C. , Jensen, K. , … Langsley, G . (2014). The level of H(2)O(2) type oxidative stress regulates virulence of Theileria‐transformed leukocytes. Cellular Microbiology, 16(2), 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, D. H. , Michael, H. , Tritt, R. , Parsons, S. H. , & Kelley, M. R. (2000). Alterations in the expression of the DNA repair/redox enzyme APE/ref‐1 in epithelial ovarian cancers. Clinical Cancer Research, 6(2), 602–609. [PubMed] [Google Scholar]
- Morzaria, S. P. , Roeder, P. L. , Roberts, D. H. , Chasey, D. , & Drew, T. W. (1984). Characteristics of a continuous suspension cell line (BL20) derived from a calf with sporadic bovine leukosis.
- Nene, V. , & Morrison, W. I. (2016). Approaches to vaccination against Theileria parva and Theileria annulata . Parasite Immunology, 38(12), 724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, M. , Suzuki, S. , & Irani, K. (2002). Redox factor‐1/APE suppresses oxidative stress by inhibiting the rac1 GTPase. The FASEB Journal, 16(8), 889–890. [DOI] [PubMed] [Google Scholar]
- Papatheodorou, I. , Fonseca, N. A. , Keays, M. , Tang, Y. A. , Barrera, E. , Bazant, W. , … Petryszak, R. (2018). Expression atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Research, 46(D1), D246–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddi, S. R. , Chattopadhyay, R. , Naidu, C. V. , & Izumi, T. (2006). The human apurinic/apyrimidinic endonuclease‐1 suppresses activation of poly(adp‐ribose) polymerase‐1 induced by DNA single strand breaks. Toxicology, 224(1–2), 44–55. [DOI] [PubMed] [Google Scholar]
- Perche, P. Y. , Vourc'h, C. , Konecny, L. , Souchier, C. , Robert‐Nicoud, M. , Dimitrov, S. , & Khochbin, S . (2000). Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Current Biology, 10(23), 1531–1534. [DOI] [PubMed] [Google Scholar]
- Pertea, M. , Kim, D. , Pertea, G. M. , Leek, J. T. , & Salzberg, S. L. (2016). Transcript‐level expression analysis of RNA‐seq experiments with HISAT, StringTie and Ballgown. Nature Protocols, 11(9), 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M. , Pertea, G. M. , Antonescu, C. M. , Chang, T. C. , Mendell, J. T. , & Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nature Biotechnology, 33(3), 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priatel, J. J. , Chen, X. , Zenewicz, L. A. , Shen, H. , Harder, K. W. , Horwitz, M. S. , & Teh, H. S. (2007). Chronic immunodeficiency in mice lacking RasGRP1 results in CD4 T cell immune activation and exhaustion. Journal of Immunology, 179(4), 2143–2152. [DOI] [PubMed] [Google Scholar]
- Robert McMaster, W. , Morrison, C. J. , & Kobor, M. S. (2016). Epigenetics: A new model for intracellular parasite‐host cell regulation. Trends in Parasitology, 32(7), 515–521. [DOI] [PubMed] [Google Scholar]
- Scott, A. , & Wang, Z. (2011). Tumour suppressor function of protein tyrosine phosphatase receptor‐T. Bioscience Reports, 31(5), 303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short, S. P. , Whitten‐Barrett, C. , & Williams, C. S. (2016). Selenoprotein P in colitis‐associated carcinoma. Molecular & Cellular Oncology, 3(3), e1075094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Khatri, N. , Manuja, A. , Sharma, R. D. , Malhotra, D. V. , & Nichani, A. K. (2001). Impact of field vaccination with a Theileria annulata schizont cell culture vaccine on the epidemiology of tropical theileriosis. Veterinary Parasitology, 101(2), 91–100. [DOI] [PubMed] [Google Scholar]
- Somerville, R. P. , Adamson, R. E. , Brown, C. G. , & Hall, F. R. (1998). Metastasis of Theileria annulata macroschizont‐infected cells in scid mice is mediated by matrix metalloproteinases. Parasitology, 116 (Pt 3), 223–228. [DOI] [PubMed] [Google Scholar]
- Takami, M. , Cunha, C. , Motohashi, S. , Nakayama, T. , & Iwashima, M. (2018). TGF‐beta suppresses RasGRP1 expression and supports regulatory T cell resistance against p53‐induced CD28‐dependent T‐cell apoptosis. European Journal of Immunology, 48(12), 1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M. , Shimamura, S. , Kuriyama, S. , Maeda, D. , Goto, A. , & Aiba, N. (2016). SKAP2 promotes podosome formation to facilitate tumor‐associated macrophage infiltration and metastatic progression. Cancer Research, 76(2), 358–369. [DOI] [PubMed] [Google Scholar]
- Theilen, G. H. , Rush, J. D. , Nelson‐Rees, W. A. , Dungworth, D. L. , Munn, R. J. , & Switzer, J. W. (1968). Bovine leukemia: Establishment and morphologic characterization of continuous cell suspension culture, BL‐1. Journal of the National Cancer Institute, 40(4), 737–749. [PubMed] [Google Scholar]
- Thomas, D. A. , & Massague, J. (2005). TGF‐beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell, 8(5), 369–380. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Hendrickson, D. G. , Sauvageau, M. , Goff, L. , Rinn, J. L. , & Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA‐seq. Nature Biotechnology, 31(1), 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretina, K. , Gotia, H. T. , Mann, D. J. , & Silva, J. C. (2015). Theileria‐transformed bovine leukocytes have cancer hallmarks. Trends in Parasitology, 31(7), 306–314. [DOI] [PubMed] [Google Scholar]
- van der Vuurst de Vries, A. R. , Clevers, H. , Logtenberg, T. , & Meyaard, L. (1999). Leukocyte‐associated immunoglobulin‐like receptor‐1 (LAIR‐1) is differentially expressed during human B cell differentiation and inhibits B cell receptor‐mediated signaling. European Journal of Immunology, 29(10), 3160–3167. [DOI] [PubMed] [Google Scholar]
- Vleugel, M. M. , Greijer, A. E. , Bos, R. , van der Wall, E. , & van Diest, P. J. (2006). c‐Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Human Pathology, 37(6), 668–674. [DOI] [PubMed] [Google Scholar]
- Yang, S. , Irani, K. , Heffron, S. E. , Jurnak, F. , & Meyskens, F. L., Jr. (2005). Alterations in the expression of the apurinic/apyrimidinic endonuclease‐1/redox factor‐1 (APE/Ref‐1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref‐1 inhibitor. Molecular Cancer Therapeutics, 4(12), 1923–1935. [DOI] [PubMed] [Google Scholar]
- Yu, Q. , & Stamenkovic, I. (2000). Cell surface‐localized matrix metalloproteinase‐9 proteolytically activates TGF‐beta and promotes tumor invasion and angiogenesis. Genes & Development, 14(2), 163–176. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information


