SUMMARY
Tissues undergoing morphogenesis impose mechanical effects on one another. How developmental programs adapt to or take advantage of these effects remains poorly explored. Here, using a combination of live imaging, modeling, and microsurgical perturbations, we show that the axial and paraxial tissues in the forming avian embryonic body coordinate their rates of elongation through mechanical interactions. First, a cell motility gradient drives paraxial presomitic mesoderm (PSM) expansion, resulting in compression of the axial neural tube and notochord; second, elongation of axial tissues driven by PSM compression and polarized cell intercalation pushes the caudal progenitor domain posteriorly; finally, the axial push drives the lateral movement of midline PSM cells to maintain PSM growth and cell motility. These interactions form an engine-like positive feedback loop, which sustains a shared elongation rate for coupled tissues. Our results demonstrate a key role of inter-tissue forces in coordinating distinct body axis tissues during their co-elongation.
eTOC Blurb
Multiple tissues undergo distinct morphological changes during body axis formation yet their elongation rate is similar. Xiong et al. reveal that this co-elongation is coordinated through a mechanical feedback loop between paraxial and axial tissues in the chicken embryo. This type of inter-tissue mechanical coupling provides robustness for complex morphogenesis.
Graphical Abstract
INTRODUCTION
During development, the morphogenesis of one tissue can produce forces that directly impact the dynamics of its neighboring tissues (Savin et al., 2011; Lye et al., 2015; Morita et al., 2017; Bailles et al., 2019). These interactions may play a role not only in controlling the correct development of each tissue and organ but also in coordinating them into an integrated system. Such role of inter-tissue forces is poorly explored due to the challenge of experimentally detecting and applying forces and the lack of a framework that bridges developmental signals, cell dynamics and tissue mechanics.
Avian embryos are an ideal vertebrate system to address these challenges because of their large size and accessibility. The forming body axis provides a striking example of coordinated morphogenesis of multiple distinct tissues (Bénazéraf and Pourquié, 2013). The axis forms in a head to tail (anterior to posterior, AP) sequence where tissues derived from the three germ layers are progressively added from a posterior growth zone (e.g. primitive streak, tailbud). The posterior movement of the growth zone lays down in its trail a conserved pattern of tissues which include the axial organs: neural tube (NT) and notochord (NC), and the flanking paraxial tissues: presomitic mesoderm (PSM), intermediate mesoderm and lateral plates (Figures 1A–B). While these tissues achieve similar lengths at the same pace, they exhibit distinct cellular organization and elongation dynamics (Keller and Danilchik, 1988; Shih and Keller, 1992; Steventon et al., 2016; Colas and Schoenwolf, 2001; Glickman et al., 2003; Sausedo and Schoenwolf, 1993; Yang, et al. 2002; Bénazéraf et al., 2010; Dray et al., 2013; Bénazéraf et al., 2017). First, the elongation of the NC and NT largely relies on intrinsic cell polarity-driven medio-lateral intercalation promoting convergence and extension along the AP axis. In contrast, the adjacent PSM shows limited convergence in the posterior body axis region (Shih and Keller, 1992; Bénazéraf and Pourquié, 2013). Second, in NC and NT, cells exhibit an epithelial organization, move coherently and only limited cell rearrangements are seen, while in the PSM cells are mesenchymal, show random cell movements and extensive cell mixing (Colas and Schoenwolf, 2001; Bénazéraf et al., 2010). Third, in tailbud stages, both NT and PSM rely on new cell addition from a specific subdomain of the growth zone called the chordo-neural hinge (or PD for progenitor domain) at the posterior end of the axis (Figure 1A). To enter the PSM, newly specified PSM cells on the ventral side of the PD move laterally away from the midline (Denans et al., 2015). In contrast, NC lengthens mainly through cell rearrangement and differentiation with little addition of new cells (Sausedo and Schoenwolf, 1993; Bénazéraf et al., 2017). These differences in cell behaviors lead to counterintuitive cellular dynamics such as sliding between PSM and NC cells where NC cells appear to move faster posteriorly than PSM cells relative to the somites, despite that the two tissues elongate hand-in-hand (Figure 1B, Yang et al., 2002; Glickman et al., 2003; Bénazéraf et al., 2017). These observations raise the question of how do the PSM, NC and NT, which employ such distinct cellular processes to lengthen, coordinate their elongation during body axis formation?
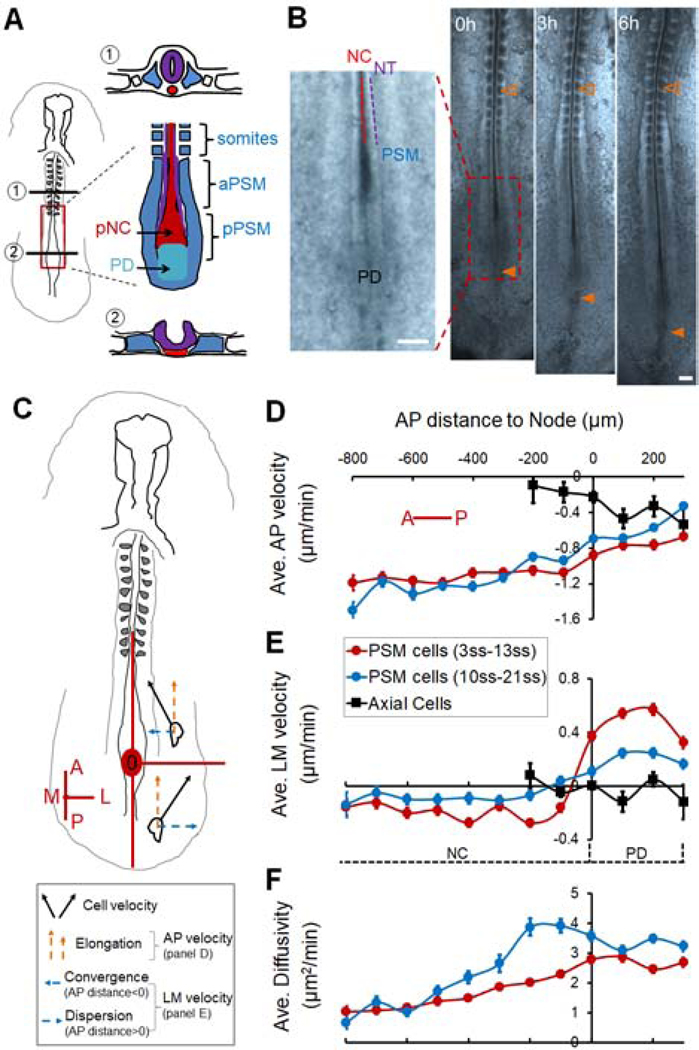
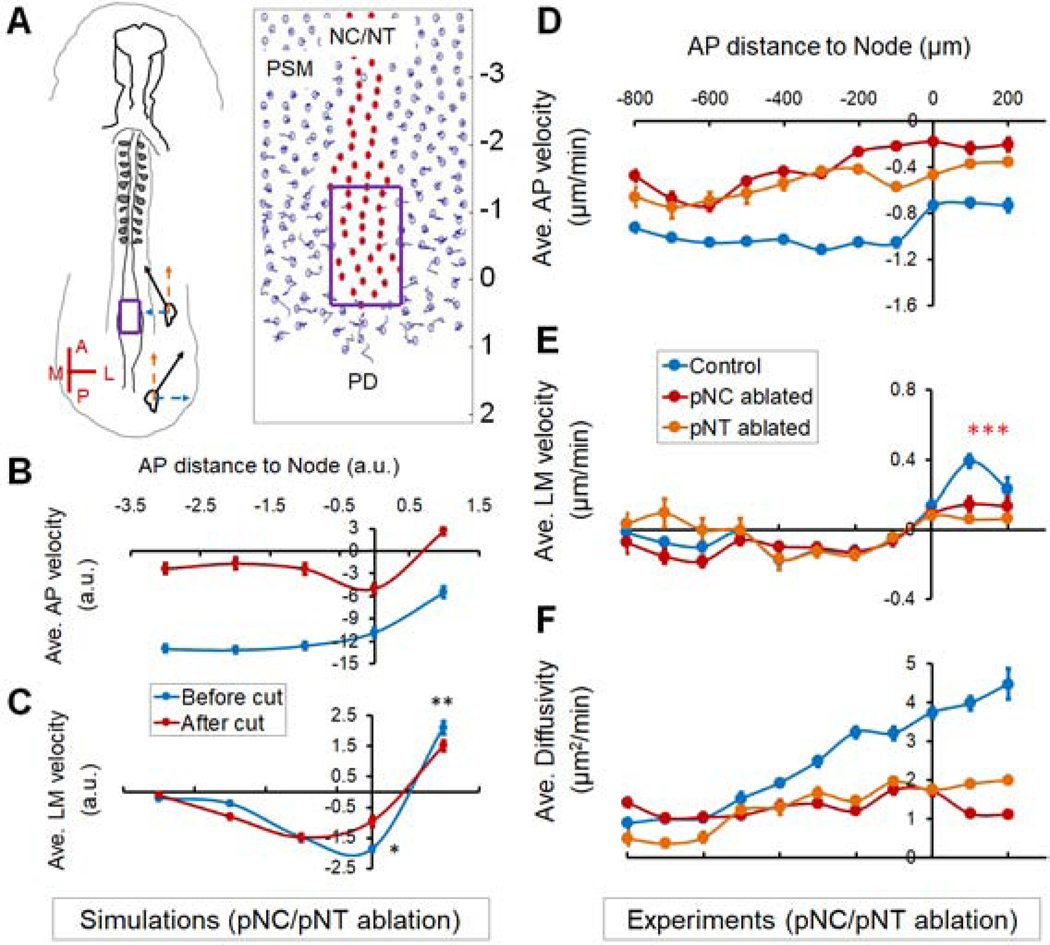
Figure 1.
Avian posterior body axis tissues and their cellular dynamics during elongation
(A) Diagram of the avian body axis (Left) and its composing tissues (Right). Cross-section (①) shows the formed section of the body axis including a closed neural tube (NT, purple), rod-like notochord (NC, red) and somites (blue), with significant interstitial space between them. Cross-section (②) shows the newly formed section in the posterior near the progenitor domain (PD) including a folding neural plate (purple), a wider NC (red), and presomitic mesoderm (PSM, blue), which are in close contact. In ①, the uncolored small squares by the somites represent the intermediate mesoderm, the further lateral uncolored blocks represent the lateral plate mesoderm. The red box marks the posterior body axis shown in a detailed ventral view of the tissue arrangement. The NT and NC are in the middle anterior to the PD, flanked by two stripes of PSM and columns of somites. A lowercase letter “p” (or “a”) in front of NC/NT/PSM indicates “posterior” (or “anterior”), same in other figures.
(B) The elongating posterior body axis of a chicken embryo. On the left the magnified area shows the spatial arrangement of the PD and newly formed NC, NT and PSM, as illustrated in (A). On the right axis elongation is observed over time. Empty arrowheads mark the same somite as a reference; filled arrowheads marks the PD. Scale bars: 200μm.
(C) Schematic view of the reference axes used for plotting cell dynamics data. The Node area (red circle) is set as the origin (0,0). A, anterior; P, posterior; M, medial; L, lateral. In this reference frame body axis elongation is represented by posterior to anterior movement of PSM cells as they are left behind by the node (orange arrows). The posterior area to the Node (PD) has a positive AP position while the anterior area to the Node (NC) has a negative AP position.
(D-E) Average cell speed along the AP (D) and LM (E) axes. Along AP, negative indicates P to A movement relative to the Node (0,0). Along LM, negative and positive indicate convergent (L to M) and dispersive (M to L) movements, respectively. Each bin integrates 100μm along the AP axis averaging 100–200 speed measurements from all time points of the movie. Error bars: ±SEM. Each movie is repeated at least 3 times. In each embryo, around 50 cells were tracked through ~200 time points.
(F) Cell “Diffusivity” obtained by fitting the mean square displacement (MSD) of single cell tracks with a drifting speed (directional, v) and a diffusion component (random, D) where MSD=4Dt+v2t2. t is time.
Mechanical interactions between tissues might offer an answer to this question. The close proximity and lack of interstitial space between the PSM, NC, NT and the PD at the posterior end of the body axis (Schoenwolf 1979) means that the deformation of one would likely have a direct mechanical effect on the others. Indeed, it was found that ablating the posterior PSM (pPSM) greatly reduces the elongation rate of all tissues (Bénazéraf et al., 2010). This finding suggests that the pPSM may mechanically contribute to axial elongation. It was further hypothesized that cell motility in the PSM produces tissue expansion forces (Bénazéraf et al., 2010; Regev et al., 2017). To test these possibilities, tissue and cell behaviors need to be observed in vivo with readout of tissue forces.
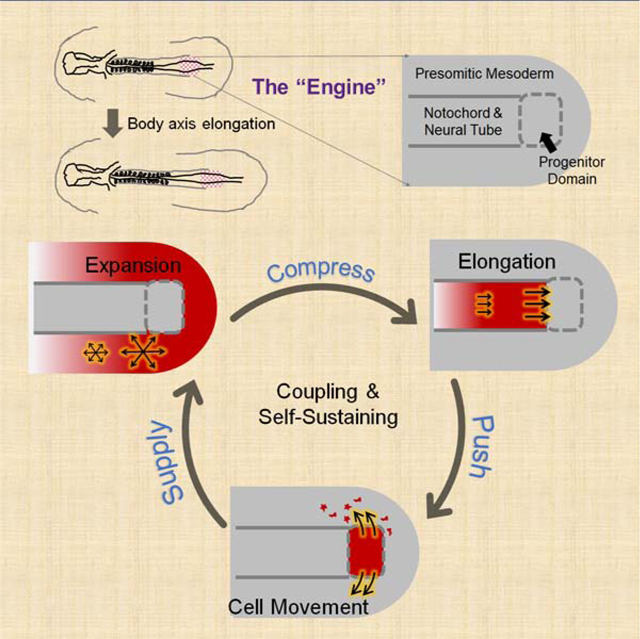
Here we used soft gels to replace different tissues in the elongating posterior body axis of chicken embryos to track the forces applied by the neighboring tissues. We found that the pPSM compresses the axial tissues. This extrinsic force adds to the intrinsic process of cell intercalation to promote convergence and elongation of the NT and NC. The compression requires FGF dependent cell motility in the pPSM while the NT cell intercalation requires Wnt/PCP (planar cell polarity) mediated cell polarity. Using an agent(cell)-based model that incorporates cell motility and polarity, we faithfully recapitulate the dynamics of cells and tissues under different experimental conditions. The model predicts that the advancing axial tissues exert a pushing force on the PD, which we validated by gel deformation. Surprisingly, the model also predicts that the lateral movement of the PSM cells from the ventral midline requires this axial pushing force. To test this hypothesis, we inserted a magnet-controlled pin in place of the posterior NC/NT (pNC/pNT) region to apply a posteriorly oriented force to the PD. This mechanical signal partially restores PSM cell movements and posterior movement of the PD (i.e. axial elongation). Together our findings reveal mechanical coupling between the PSM and axial tissues: the left and right pPSMs generate a bilateral compression of the axial tissues promoting their elongation. The axial tissues in turn push on the PD promoting PSM growth by driving cell flow. These interactions form a self-sustaining positive feedback loop that prevents elongation offset from occurring between different tissues. Just like an internal combustion engine that self-refills fuel for the next cycle, expanding pPSM (combustion) forces axial tissues (piston) to drive PSM progenitors (fuel) out of the PD (fuel reserve). Our model thus provides a simple physical mechanism and an intuitive explanation of the striking robustness and coordination between tissues observed in chicken embryonic body axis elongation.
RESULTS
Cellular dynamics of axial and paraxial elongation
To set up a quantitative baseline of tissue and cell behaviors during body axis elongation, we performed live imaging of Tg(CAG:GFP) chicken (McGrew et al., 2008) embryos after labeling the new PSM cells and progenitors in the primitive streak and tailbud with DiI (Figures S1A–B, Movie S1) between Hamburger Hamilton (HH) stages 8 and 14. During these stages, body elongation first follows the regression of the primitive streak and then the posterior movement of the tailbud. These movements result in the progressive formation of the NT, NC and PSM (among other tissues) at the posterior end of the embryo (Hamburger 1992).
Using cell tracking (Figure S1C), we analyzed the movement of cells relative to the Node (Figure 1C). Both axially-localized cells (including NC cells and new PSM cells from the PD) and paraxially-localized PSM cells are left behind by the Node but at increasingly different speeds (Figure 1D, note that while some axial cells move posteriorly with the Node, others are left behind, resulting in a net negative average speed at the Node [position 0]). This speed offset reflects the rate of the posterior movement of the PD (i.e. the elongation rate). In early embryos, at the PD level, cells move away from the midline while in the PSM flanking the NC, cells converge toward the midline (Figure 1E, positive indicates medial to lateral movement). Similar dynamics occur in later stage embryos but at markedly reduced average speeds (Figure 1E). The medial to lateral dispersion of new PSM cells at the PD level results in their joining the PSM on both sides of the axis (Figures S1D). This dispersion speed is an indirect measurement of the PSM cell addition rate as the PD and PSM form a continuum of cell flow. To measure the motility of cells (Figures S1E), we fit the tracks with a diffusion model to separate the directional and non-directional components (Regev et al., 2017). This allows us to assess the local motility or “cell diffusivity” in the PSM. A posterior to anterior motility gradient is observed in both early and later stages of elongation (Figure 1F). These cellular dynamics are consistent with previous studies (Kulesa and Fraser, 2002; Yang et al., 2002; Manning and Kimelman, 2015; Lawton et al., 2013; Bénazéraf et al., 2010, 2017).
Posterior PSM exerts a lateral to medial compression on the axial tissues
To investigate how the pPSM affects the axial tissues, we performed bilateral ablations of the pPSM (Figure 2A). Interestingly, both pNC and pNT remained wider even after PSM cuts closed, suggesting impaired tissue convergence (Figures 2B–C, S2L). Correspondingly, the elongation rate of the operated embryos reduces drastically (Figures S2A–C). These data show that the pPSM is required for the convergence and elongation of the axial tissues. To test if pPSM supplies a biochemical cue to promote axial elongation, we inserted tailored aluminum foil pieces between pPSM and pNC/NT to cut off signal exchange (Figure 2D). Unlike pPSM ablation, foil insertion does not slow down elongation (Figure 2E), suggesting the requirement of pPSM is not through diffusible signals. To test if the pPSM passively confines pNC/pNT to prevent their lateral expansion, we replaced the pPSM with a stiff alginate gel (1% alginate at 100mM Ca2+) as a barrier. The gel-flanked portions of pNC/pNT remained wider (Figure S2D, n=4/4), suggesting that the pPSM actively compresses the axial tissues. To detect this compression, we replaced the pNC with a soft alginate gel containing fluorescent beads (Figure S2F, 1% alginate at 10mM Ca2+, order of 100 Pa [Banerjee et al., 2009], comparable to embryonic tissues [Zhou et al., 2009]), which showed ~5% narrowing indicating that pPSM produces a compressive stress in the order of 10s of Pa. We further lowered the gel stiffness by injecting 1% alginate solution into ablation sites allowing gelation in vivo. Using live confocal imaging, we found that these implants are more deformable as a paste and round up edges quickly under surface tension (Figure 2F, Movie S2). The pNC/pNT implant narrows significantly over time along the LM axis (Figures 2F–H). Similar deformation dynamics were observed in replacement experiments of the pNC (Figures S2G–I) or the pNT (Figure S2J–K) alone. These results together show that pPSM contributes to axial convergence and elongation through lateral tissue compression.
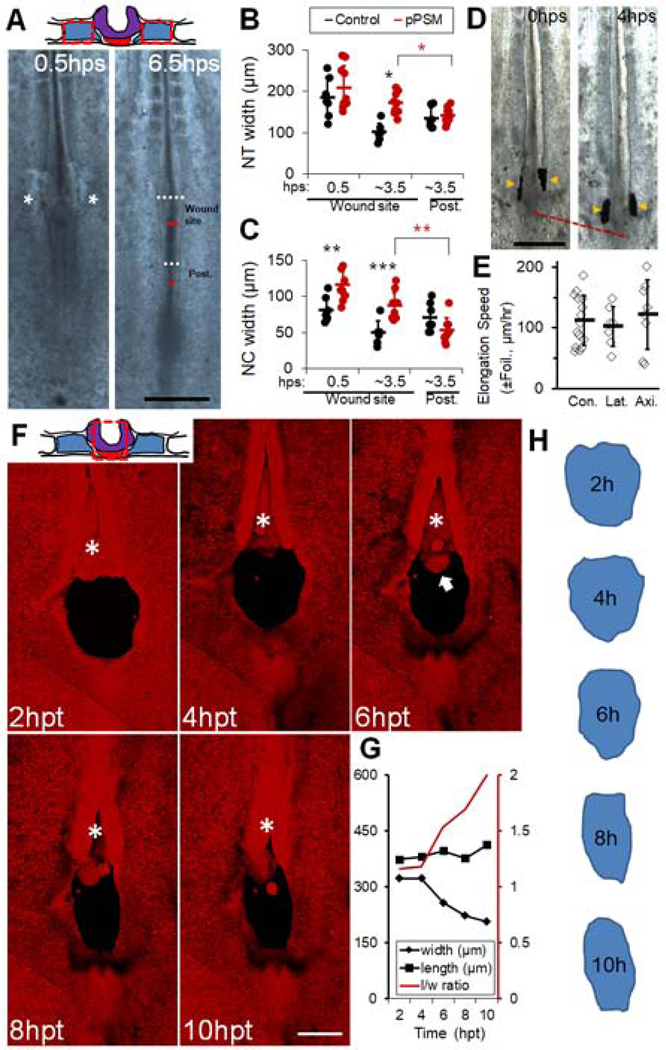
Figure 2.
Posterior PSM exerts a lateral compression on axial tissues
(A) Ablation of the pPSM. The top schematic shows cross-section of the operated area. Red dashed boxes mark ablated pPSM (blue). Embryo images show axial tissue dynamics after pPSM ablation. Asterisks indicate the closing PSM ablation sites shortly after ablation (hps, hours-post-surgery). The dark line in the midline of the embryo delineates NC (the short red lines measure NC’s width). Lighter regions besides the NC show the neural folds on the further dorsal side (the white dashed lines measure NT’s width). The widths were measured for both the wound sites and a control area posterior to the wound sites (Post.). Scale bar: 500μm.
(B-C) NT and NC width after pPSM ablation (Control n=7, pPSM n=10). Asterisks indicate significant difference (p<0.01, t-tests, unpaired (black), paired (red)). “Post.” is a control area posterior to the wound site as annotated in panel (A).
(D) Foil insertion between pPSM and pNC/pNT. Foils (arrowheads) are vertically inserted between the tissues and are observed to move along during elongation. Scale bar: 500μm.
(E) Elongation speeds in Foil insertion experiments. Con., Control (no insertion); Lat., Lateral insertion between pPSM and pNC/NT; Axi., axial cross-sectional insertion between pNC and PD (See Figure S2E). The average speeds of these groups are not significantly different (p>0.05, t-tests).
(F) Gel implant followed by confocal timelapse. The top schematic shows cross-section of the operated area. Red dashed boxes mark ablated pNC (red) and pNT (purple). Images are maximum projections of GFP (shown in red) z-stacks (dorsal view). Asterisk shows the convergence and closure of NT anterior to the implant site. Arrow marks protruding NC underneath the NT. Scale bar: 200μm. See also Movie S2.
(G) Quantified shape changes of the gel implant in (F). Length (l) is measured along the midline, width (w) is measured following the widest line perpendicular to the midline. Consistent narrowing was observed in n=3 movies.
(H) Contours of the gel implant in (G).
Cellular basis of PSM compression, axial convergence, and elongation
To investigate how pPSM creates compression on axial tissues, we constructed a 2D agent (i.e. cell)-based computational model (Figure 3A, Movie S3). The model contains a field of PSM cells subdivided by the axial tissue in the midline, and the posterior PD that supplies new PSM cells (for the feasibility of simulation the number of cells is small compared to the real tissues but has a similar paraxial-axial proportion). All cells occupy a volume in the tissue and thus push against each other upon close contact. Cell movement is countered by a resistance mimicking the effect of the viscous extracellular environment (Figures S3A–B, methods). When a motility gradient is added across the field of PSM cells, the high motility region (corresponding to the pPSM) shows a diffusion-like expansion (Figure S3C) on the tissue level that reproduces observations in isolated PSM explants (Palmeirim et al., 1998). When this expansion encounters a lateral boundary, such as the axial tissues, a pressure gradient parallel to the motility gradient is recorded (Figure S3D). To test this pressure gradient in vivo, we replaced the aNC with gels (Figures S3E–F). In contrast to the pNC control, gels replacing aNC show minimal deformations (Figures S3G–H). These data are consistent with the pPSM (but not the aPSM) generating compression in the region of maximal cell motility (Figure 1F). Our model also generates the cell density gradient observed in the PSM in vivo (Figures S3I–J), which is another consequence of the motility gradient and tissue expansion. To model axial cell behaviors, we implemented a polarity parameter that controls the shape of the axial cells. A polarized axial cell is elongated along the medio-lateral axis and intercalates with other axial cells more easily in the model, mimicking the cell shape and behavior observed in vivo such as neural progenitors in the NT (Colas and Schoenwolf, 2001). To model the PD production of new PSM cells, we periodically added cells posterior to the axial end in the simulation iterations. This mimics the in vivo configuration where the anterior primitive streak or tailbud adds PSM cells ventrally at the axis end by ingression and proliferation (for simplicity, we only consider the PD placement at the posterior end of the midline, which is similar to the actual geometrical arrangement between different stages and species. In addition, the newly added cells behave identically to PSM cells as their in vivo counterparts have undergone PSM specification and epithelial to mesenchymal transition [EMT, Denans et al., 2015] before moving to the ventral midline). A combination of the PSM motility gradient, the polarization of axial cells and the cell addition from PD produces sustained axial convergence and elongation in silico (Figure 3B, Movie S3).
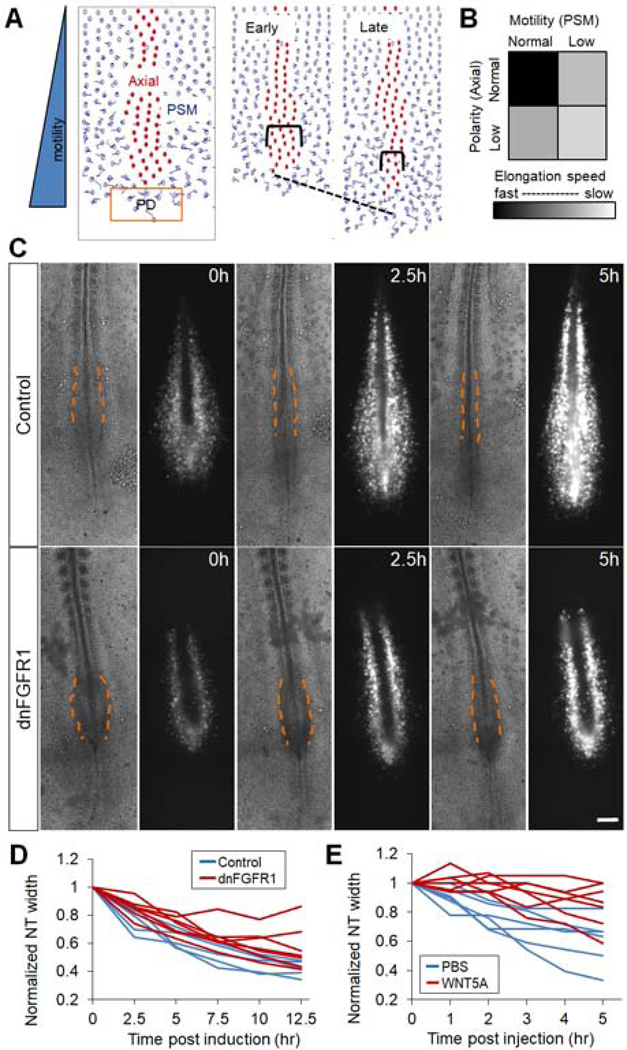
Figure 3.
PSM cell motility and axial cell polarity contribute to axial elongation
(A) Agent-based 2D model for posterior body axis elongation. Cell centroids but not cell shapes are shown for the ease of visualization. NC/NT cells (red) are flanked by PSM cells (blue). New PSM cells are added from the PD domain (orange box) periodically on the midline. The simulation recapitulates axis elongation (dashed line) and axial convergence (brackets). See also Movie S3.
(B) Elongation speed as a function of PSM cell motility and axial cell polarity in the model. Results were averaged from 10 simulation runs (4000 time-steps each) for each parameter pair.
(C) Inhibition of FGF signaling by electroporating a dominant negative FGFR1 and a GFP construct into the PSM. Bright field and GFP fluorescence images of representative embryos (ventral view) at three different time points are shown. Dashed lines contour the narrowing pNT behind the pPSM, dark strips in the fluorescent images are the NC tissue flanked by electroporated PSM cells. Scale bars: 200μm.
(D-E) Convergence of pNT after dnFGFR1 induction and after WNT5A injection, respectively. Each line represents one embryo. The width is normalized to the width at the first time point. The perturbed convergence rate was significantly lower compared to control in both cases (p<0.05, t-tests).
To test the key cell behavior assumptions (motility and polarity) of the model, we considered the molecular signals known to control these cell properties. FGF signaling is known to control the cell motility gradient in the PSM (Bénazéraf et al., 2010). We reduced cell motility by electroporating an inducible dominant-negative FGF receptor (dnFGFR1) construct and a fluorescent reporter in the PD (the anterior primitive streak, Iimura and Pourquie, 2008). The electroporation allows PSM-specific expression of the transgenic construct (Figure S3K). Embryos in which FGF signaling is inhibited in the PSM exhibit a wider pNT as seen after pPSM ablation, together with slower pNT convergence and axis elongation (Figures 3C–D, Bénazéraf et al., 2010). Similar results on pNT convergence and elongation were obtained by electroporation of a dominant negative MAP kinase (dnMKK1, Figures S3L–M, Delfini et al., 2005) and by embryo treatment with the FGF inhibitor PD173074 (data not shown). Furthermore, embryos treated with Blebbistatin to directly inhibit cell movement show no medial-lateral compression on implanted gels and delayed axial convergence (Figures S3O,4G). These data support the hypothesis that pPSM cell movement downstream of FGF signaling collectively generates compression on the axial tissues.
pNT and pNC cells exhibit cell polarity which promotes active medio-lateral intercalation driving intrinsic tissue elongation (Keller et al., 2008; Williams et al., 2014). In amphibian embryos, interfering with cell polarity by disrupting the posterior WNT5 gradient by either up- or down-regulating the pathway reduces axial convergence and elongation (Moon et al., 1993; Wallingford and Harland, 2001). We tested whether disrupting medio-lateral cell intercalation by WNT5A also interferes with axis elongation in the chicken embryo. We injected WNT5A protein into the anterior portion of the pPSM or the pNT, where endogenous expression is low (Chapman et al., 2004). This efficiently slowed down elongation while control injections of buffer, WNT10 at the same location, or WNT5A at the Node (where endogenous expression is high) do not have significant impacts (Figures S4A–C). Consistently, the pNT in WNT5A injected embryos maintains a larger width compared to controls (Figures 3E, S4A‘-B‘) indicating impaired convergence. To analyze cell polarity changes following WNT5A perturbation, we electroporated a membrane tagged GFP in the NT which allows mosaic labeling of pNT cells that highlight cell shapes (Figure S5A). We also compared NT cell nuclei shapes following DAPI staining (Figure S5B). This analysis reveals that WNT5A causes a reduction of the lateral-medial polarization of NT cells in terms of cell shape (Figure S5D), while PSM cells do not exhibit changes in cell shape or density in the WNT5A condition (Figures S5C,E). Furthermore, the PSM cell motility gradient is unaffected (Figures S5F). Using live imaging and cell tracking, we found that the medial-lateral intercalation of pNT cells is greatly reduced following WNT5A injection (Figures S5G–H). These results suggest that WNT5A perturbation delays convergence and elongation by disrupting NT cell polarity, consistent with previous studies. Taken together, our data suggest that FGF signaling in the PSM and WNT/PCP signaling in the NT provide extrinsic and intrinsic forces of axial elongation through motility and polarity, respectively. These results add confidence to our model assumptions, allowing us to utilize the model to further dissect the role of these mechanical interactions between tissues.
Elongating axial tissues produce a pushing force on the progenitor domain
Our perturbations show that posterior movement of the PD (which marks elongation) greatly slows down whenever axial tissues are perturbed (Figures S2B–C, S4), suggesting that the movement of the PD is not autonomous but rather requires pNT/pNC. Inserting a piece of aluminum foil to separate pNT/pNC with the PD does not affect elongation (Figures 2D, S2E), suggesting that the pNT/pNC requirement is not due to a diffusible molecular signal. To understand how axial elongation promotes PD movement, we resorted to our simulation. Counter-intuitively, ablating PD is predicted to slow down elongation more mildly compared to pPSM or axial ablations, which agrees with experiments (Figures 4A–B, Movie S4, compare to Figures S2A–C). We followed the average AP forces that two representative cells experience in the simulations (arrows in Figure 4A): one axial cell in the midline bordering the PD thus detecting the net force from the axial tissue and the PD, the other a paraxial PSM cell that starts off at the same AP level for comparison. The PSM cell is initially pushed posteriorly (by other pPSM cells) but then anteriorly as it transitions to become an aPSM cell, consistent with the model of pPSM expansion (Figure 4C). In contrast, the axial cell (and the PD) is continuously pushed towards the posterior. To test whether this predicted pushing force exists in vivo, we replaced the PD with a soft alginate gel. Strikingly, the gel thins along AP especially in the middle where it is in direct contact with the axial structures (Figure 4D–F, Movie S5). This is in sharp contrast to the lateral to medial compressive deformations seen in gels that replace the pNT or pNC (Figures 2F–H, S2F–K, Movie S2), showing that elongating axial tissues indeed push on the PD. This pushing force is also dependent on cell movement and the pPSM (Figures 4G). Notably, as the PD is the source that supplies new cells to the pPSM, the axial push might impact the dynamics of new PSM cells on the midline. In some PD gel replacement experiments, as the axial tissues advance the gel is seen to move aside from the midline and into the paraxial pPSM (data not shown), raising the possibility that new PSM cells might respond to the axial push in a similar fashion. To test this, we replaced the PD and midline PSM domain of a transgenic host embryo by a different colored donor (Figure 4H; Huss et al., 2015). The transplant appears flattened on the anterior similar to gel deformations, suggesting tissues respond to the high stress at the axial-PD interface. Cells at the anterior lateral sides of the transplant start to move medial-laterally into the pPSM. These observations suggest that the axial push may be required for driving the medial-lateral PSM cell flow.
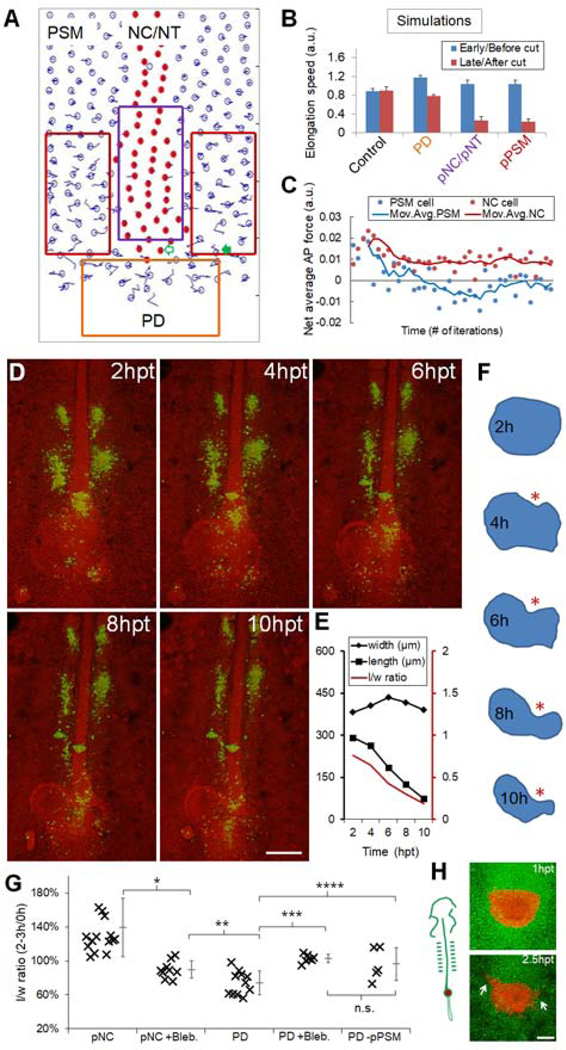
Figure 4.
Axial tissues exert a pushing force on the PD
(A) Layout of the model showing simulated ablations (boxes) corresponding to results in (B). Empty arrow, an axial cell; filled arrow, a PSM cell. See also Movie S4.
(B) Axial elongation speeds (±SEM) for both before and after tissue ablation. For PD, cell addition was disabled in the beginning of simulation. Speeds were averaged over 2000 time points in each simulation. Each experiment includes n=40 simulations. p=0.93, control; p<0.05, other groups (paired t-tests).
(C) Net forces along AP experienced by an axial cell and a PSM cell. The cells start at the same AP level as marked in panel (A). Positive indicates A to P. Data points are binned by 200 iteration windows. Curves: 4 times moving average.
(D) Gel replacement of PD followed by confocal timelapse. Green indicates spots of DiI injection to follow cell movements. Spots in the pPSM can be seen expanding while the axial tissues narrow. Scale bar: 200μm.
(E) Quantified shape changes of the gel implant in (D). Consistent observations in n=3 embryos.
(F) Contours of the gel implant in (D). Asterisk highlights the indentation where the gel is in contact with pNC.
(G) Summary of gel replacement experiments. The l/w ratios after 2–3 hours are plotted as a percentage of the initial aspect ratio. Tissue names indicate implant locations (e.g. pNC, PD), “+/−“ indicates further perturbations (e.g. Blebbistatin treatment [+Bleb.], pPSM ablation [-pPSM]). Asterisks: p<0.05, t-tests. n.s.: p=0.4. See also Figures S3O–P.
(H) Shape change and cell dispersion of PD transplant. A group of GFP+ progenitors (shown in red) is transplanted posterior to the pNC of cherry+ host (shown in green). Arrows: cells in the transplant disperse into host pPSM. Anterior flattening of the transplant and cell dispersion were observed in n=4/6 similar experiments. Scale bar: 100μm.
Axial push is required for new PSM cell addition to the posterior PSM
To test the effect of the axial push on the midline PSM cells, we analyzed the average PSM cell movement in the simulation. The model recapitulates the long-term trajectories of PSM cells including their medial to lateral movement from the midline (Figures 5A–C, see also Figure 1E). In the simulation, there are no additional assumptions beyond cell motility and mechanical pushes between cells. Therefore, the medial to lateral movement is promoted simply as the elongating axial tissue pushes the PD posteriorly and occupies the midline space (Movie S4). Indeed, our cell tracking (both in simulations and imaging experiments) shows that, following axial ablation, the medio-lateral (ML) speed of PSM cells exiting the midline is reduced along with the elongation rate (Figures 5B–E, S5I, Movie S6), indicating that the rate of PSM growth is slowed down. Furthermore, the motility gradient is not sustained (Figure 5F) due to reduced addition of motile cells. These results suggest that the axial tissues not only push the PD posteriorly, but in doing so also promote cell addition into the pPSM thus extending PSM length and sustaining the motility gradient. However, a more definitive test of this hypothesis would require direct application of pushing forces on the PD in place of the axial tissues.
Figure 5.
Axial push is required for cell addition from the PD to the PSM
(A) Axial ablation in the embryo and the model. Purple boxes label the surgically and digitally ablated areas, respectively. The AP axis labels of the simulation field (−3 to 2) correspond to the AP distances to Node in (B) and (C). See also Movies S4.
(B) Modeled AP velocity (compare to experimental measurements in [D]), recapitulating elongation (indicated by a negative AP speed) and slow-down of elongation after axial ablation.
(C) Modeled PSM cell LM velocity (compare to experimental measurements in [E]), recapitulating convergence, midline-exit and reduction of midline-exit after axial ablation (n=40 simulations. Asterisks, p<0.05, paired t-tests).
(D-F) Average PSM cell AP (D) and LM (E) velocity and diffusivity (F) under different surgical ablations (n=3 each). Measured and plotted similarly as in Figures 1D–F. See also Movie S6.
Externally applied axial push rescues PD posterior movement and cell addition to the PSM
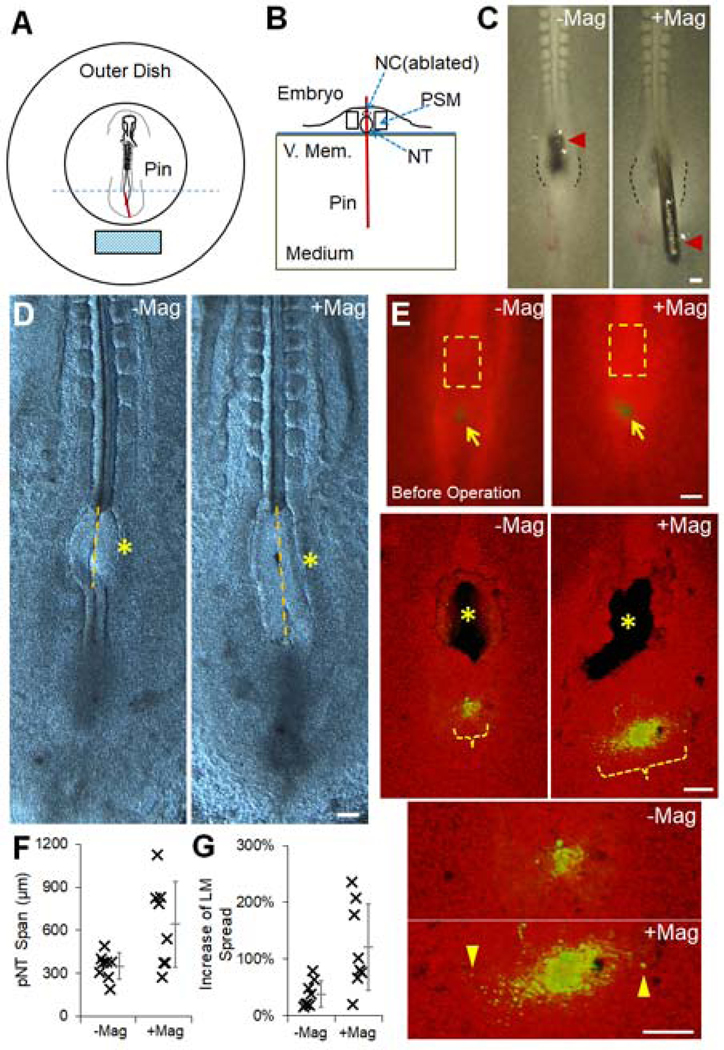
To apply an ectopic pushing force on the PD, we first performed an ablation of the pNC and then inserted a steel pin through the NT at the ablation site immediately anterior to the PD (Figure 6A). The main portion of the pin resides in the semi-liquid culture medium while the remaining tip portion stays at the embryonic tissue level. This tip can rotate in the opposite direction of the main portion at the vitelline membrane entry point (Figure 6B). In this way, the pin acts as a lever which can be controlled remotely using magnets, allowing a pushing force to be applied on the tissue towards a specified direction (Figure 6C). Using alginate gels (1–10kPa) in place of embryos under the same experimental set-up (Figure S6A), we found that the pin creates strains comparable to those induced by pNC in softer gels (Figure S6B), suggesting the ectopic pin stress is higher than the endogenous forces. Nonetheless, the NT and PD stayed largely intact despite some tear near the pin insertion site (Figure 6C). Embryos with clean and minimally invasively inserted pins were incubated for 4 hours with or without the magnets and then imaged without the pin (live imaging was not feasible with the pin and magnet setup). In the absence of the magnet, the pin stands straight and axis elongation and PSM cell addition are slowed down as expected in pNC ablation experiments, indicated by a reduction of both PD posterior movement and medial to lateral spread of midline PSM cells (Figures 6D–G). Strikingly, when the magnetic field was present to let the pin push along the posterior direction onto the PD (Figure 6C), both the posterior movement of the PD and the medial lateral spread of PSM cells were partially restored (Figures 6D–G). This rescue experiment shows that an external mechanical push can mimic the axial pushing force to drive the PD to move posteriorly and to increase lateral addition of new PSM cells.
Figure 6.
Axial push can drive PD movement and cell addition
(A-B) Schematics of the magnetic pin experiment set up. Top (A) and cross-sectional (B, corresponding to the section marked by the blue dashed line in [A]) views, respectively. Red line indicates the inserted pin and the cyan block shows the magnet (movable). The pin is perpendicularly inserted in the initial position. It rotates around the entry point of the vitelline membrane (V.Mem.). The medium is the semi-liquid albumin agar mix of the embryonic culture allowing pin rotation. See also Figure S6A.
(C) Example of pNC replacement with pin (left) and the push along the posterior direction after magnet is moved in range (right). With magnet, the embryo can be seen stretched and NT walls (marked by dashed lines) at the insertion site show expansion under stress. Scale bars: 100μm.
(D) Elongation after pNC ablation by the pin. Examples of phenotype after 4 hours with or without magnet. Pin was removed and the embryo was cleaned before imaging. Asterisks mark the pNC ablation and pin insertion site. Yellow lines mark the pNT span over the ablation site to measure PD posterior displacement.
(E) Cell dispersion followed by pNC ablation and pin insertion. Top images show the labeled spot (DiI, green) of ventral midline PSM cells (arrows). Before the microsurgery and incubation, they occupy a narrow LM range compared to the pNC (dashed boxes). Middle images are 3D projected confocal images showing the ablation/insertion sites (asterisks) and cell dispersion span (Orange bracket) after 4 hours. Note that the cells have dispersed to wider LM ranges than pre-operation in both experiments. Bottom images are enlarged views showing cells that have migrated significantly laterally (arrowheads).
(F-G) Quantification of elongation (F, p=0.002) and cell dispersion (G, p=0.011) corresponding to (D) and (E), respectively. t-tests.
Coupling cell dynamics and tissue forces results in a self-sustaining elongation engine
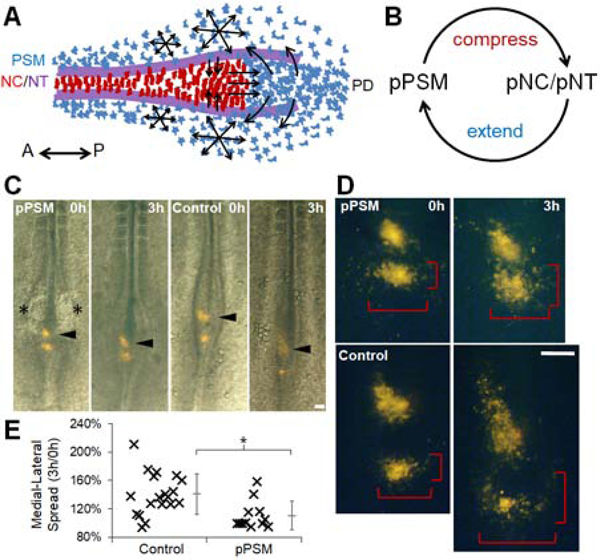
Our findings show two mechanical interactions between tissues in the elongating body axis: the medio-lateral compression of axial tissues by the pPSM and the antero-posterior push on the PD by axial tissues (Figure 7A). In addition, the axial push drives new PSM cell addition to sustain pPSM growth and cell motility, thus forming a positive feedback loop (Figure 7B). While these forces do not represent all processes that drive elongation in these tissues, their simple cross-talk uniquely ensures a tight coordination between axial and paraxial tissues, which would be difficult to achieve with separate tissue-intrinsic elongation programs. A closed loop implies that pPSM will feedback to itself. To test this, we looked at PSM cell addition in pPSM ablated embryos. Indeed, lack of pPSM delays the lateral movement of PSM cells even with intact axial tissues (Figures 7C–E, Movie S7), and the axial pushing force is reduced when pPSM is ablated or loses cell motility (Figure 4G). These results show a tight linkage between inter-tissue mechanics and cell dynamics in this system. Our model also predicts that co-elongation will be disrupted (i.e. length offsets between tissues will emerge) as a result of breaking the feedback loop (Figure S6C, Movie S4). Indeed, ablating the PD, while itself not causing any immediate tissue length offset and allowing continued elongation (as pPSM compression remains intact in short-term), results in NC and NT that elongate further than the PSM (Figures 4D, S6B). Conversely, ablating axial tissues causes the pPSM from both sides to merge on the midline and continue to grow from the PD. This results in an axial PSM that elongates further than the NC/NT (Figures S6D–F). Coupled together, these mechanical interactions produce local mechanical heterogeneity causing distinct spatial-temporal dynamics of different tissue ablation sites, which we reproduced both in silico and in vivo (Figure S7).
Figure 7.
A mechanical feedback loop couples elongating tissues
(A) Schematic ventral view of observed tissue geometry and cell behavior in PSM, NC, NT and PD. Red cells: NC cells (polarized shape). Blue cells: PSM cells (mesenchymal). Purple lines: NT folds on the dorsal side. Arrows show cell movement.
(B) A conceptual mechanical positive feedback loop that couples co-elongation of tissues.
(C) Movement of labeled midline PSM cells. Two DiI-labeled clusters immediately posterior to the pNC are followed in control and pPSM ablation experiments (asterisks indicate the ablation sites). After pPSM ablation, the labeled clusters move less posteriorly (arrowheads), and remain concentrated (cells did not disperse as much) as compared to the control. Scale bar: 100μm.
(D) A close-up view of labeled clusters showing dispersion of single cells. The brackets measure the AP and LM spread of the cells over time. Scale bar: 100μm. See also Movie S7.
(E) The LM spreads measured as in (D) are compared between control and pPSM ablation groups as a ratio between 3h and 0h. Asterisk: p=0.004. t-test.
DISCUSSION
Cellular responses to extrinsic morphogenetic forces
Our study identifies inter-tissue forces as a key element in coordinated morphogenesis. This opens new avenues of investigation of specific cellular mechanisms that respond to these forces, and potential cross-talks with other cellular changes taking place at the same time. For example, for new PSM cells, the exit from the PD and migration into the pPSM are not only cell movement but also accompanied by drastic changes of the cell state. During primitive streak regression, PSM progenitors located in the anterior primitive streak region upregulate Wnt and FGF signaling, undergo an epithelial to mesenchymal transition (EMT) and gain enhanced motility. After tail bud formation, the PD territory becomes located at the tip of the NC and NT (the posterior wall of the chordo-neural hinge) which expresses both Sox2 and Brachyury (a neural and a mesodermal marker, respectively) and continues to produce motile mesodermal precursors at a smaller spatial scale (Sun et al., 1999; Tzouanacou et al., 2009; Olivera-Martinez et al., 2012; Denans et al., 2015; Oginuma et al., 2017). In fish, the equivalent of the PD (called Dorso-Medial Zone) exhibits coherent cell movements (Lawton et al., 2013) suggesting an epithelial-like organization similar to that of the epiblast. More posteriorly in the tailbud, cells move incoherently, become specified to PSM and migrate laterally, causing increased tissue fluidity (Mongera et al., 2018), which may propagate mechanical information to anterior tissues (Das et al., 2019). We showed that the axial pushing force on the PD regulates this lateral migration of new PSM cells in chick. However, whether a mechanistic link exists between the axial force and the EMT of progenitors remains to be investigated as the movement does not directly measure PSM production. On one hand, we found that in the absence of axial tissues, the PSMs from both sides merge and continue to grow from the PD and eventually form somites along the midline (which are larger and mis-located) indicating normal PSM differentiation, suggesting that the axial force controls morphogenesis but not cell fate specification. On the other hand, we found a decrease of PSM cell motility in axial ablation experiments suggesting that the axial push may contribute to the upregulation FGF signaling in the PD, which is important for both EMT and PSM cell motility (Sun et al., 1999; Bénazéraf et al., 2010). Interestingly, mechanically driven FGF upregulation was recently reported in the context of branching morphogenesis in the embryonic lung explants (Nelson et al., 2017). Our simulations also predict the force experienced by single cells through their movement in the tissue which may influence their differentiation, in particular we predicted PSM cells to sense a reversing net force over time. Future studies involving imaging of force or signaling reporters and in vivo mechanical perturbations will be important to test these possibilities.
Force generation by cell dynamics in the posterior PSM
The physical mechanism linking the movements of PSM cells and the tissue-level expansion remains to be clarified. A jamming-transition model relates PSM cell movement to tissue mechanical properties (Mongera et al., 2018) but does not account for the source of internal stress that drives expansion. In our computational model, the force arises as highly motile cells collectively increase the space they occupy through agent repulsion. While this simple assumption proved effective for us to connect the observed compression and its dependency on cell motility, and to subsequently recapitulate tissue shape changes and cell trajectories, the process in vivo is likely different and involves the remodeling of the extracellular matrix (ECM) driven by the net sum of forces exerted by single cells. Consistent with this hypothesis, fibronectin fibers in the posterior PSM follow a movement pattern that is identical to the average movement of the cells (Bénazéraf et al., 2010). Unlike the cells, the fibers do not show local random movement. Our 2D model also does not include the surface and interface ECM between tissues, whose mechanical role is supported by the observations that knocking-down fibronectins leads to decoupling of PSM and axial tissues, causing defects in NC and NT morphogenesis in zebrafish (Dray et al., 2013; Guillon et al., 2020). These elements may be added in a future agent-mesh hybrid model and extended to 3D.
In addition to producing compression on the lateral sides, the pPSM expansion also creates an anterior to posterior pushing force alongside the axial tissues from the underlying random cell motility (Regev et al., 2017). The presence of this force is supported by the observation that PSM tissue can elongate on its own posteriorly without NC (Charrier et al., 1999; this study). The importance of this force in elongation is further evidenced by the ability of PSM to elongate to some extent even when PD is ablated (i.e. no new PSM cells, Bénazéraf et al., 2010; this study). While this pushing force appears weaker compared to the axial force as gel implants deform most in the midline, it is sufficient to continue elongation by pushing the PD posteriorly as a joint midline PSM (this study). To further dissect the contribution of axial and paraxial forces in elongation, it is important to design and adapt quantitative tools of mechanical measurement for very small (orders of 10–100μm) live embryonic tissues (Serwane et al., 2017; Mongera et al., 2018).
Robustness and self-maintenance of axis elongation in a coupled system
Our findings reveal a mechanical positive feedback loop coordinating elongation across germ layers. This situation is reminiscent of that observed during elongation of the drosophila embryo where extrinsic forces generated by the mesoderm and the invaginating gut, coupled with intrinsic forces from cell reorganization, lead to AP extension of the epithelial germ band (Butler et al., 2009; Lye et al., 2015; Collinet et al., 2015). Whether similar mechanical coupling occurs in the body axis of other vertebrates remains to be investigated, as there are many tissue and cellular differences between species and stages of elongation (reviewed in Mongera et al., 2019). It is tempting to speculate that our model may also play a role in other contexts because it provides some advantages in addition to coordinating elongation rates. For example, the FGF-dependent PSM cell motility sets a relationship between pPSM expansion and cell density, such that the compression on axial tissues increases when motile cells become more packed. This means that temporarily slower axial convergence will elicit stronger compression leading to correction. Such a mechanism could also play a role in ensuring the robustness of straight elongation (bilateral symmetry), as bending to one side will encounter increasing resistant pressure that increases with curvature, making it difficult to form large curvatures. This might provide a mechanical explanation to the recent observation that disordered cell motion in the posterior PSM is required for bilateral symmetry of paraxial mesoderm formation (Das et al., 2017).
Our model also explains the self-maintenance of axis elongation. Transplantation of the Node to an ectopic location such as the area opaca can induce a new embryonic axis which undergoes posterior elongation similar to that of the endogenous AP axis (Waddington 1932). Our simulation shows that the generation of elongation behavior does not necessarily require either the formed portion of the axis or any global cues that direct cell movement (while they could still contribute to elongation, Yang et al., 2002), but only a minimal set of tissues (namely pPSM, pNC/pNT, and PD) arranged around and inducible by the Node (the initial symmetry-breaking). As long as a PD (the fuel tank) that produces high motility PSM cells (combusting fuel) continues to function under the corresponding inductive signals, the positive feedback loop (the engine) can kick start locally and drive posterior Node movement, leaving a patterned body axis behind. Consistent with this analogy, elongation can terminate naturally when progenitors run out due to suppression of signals supporting PD proliferation (no more fuel, Denas et al., 2015), or in situations when signals (e.g., FGF) supporting the “combustion” are depleted (Bénazéraf et al., 2010).
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Olivier Pourquié (pourquie@hms.harvard.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes most datasets generated during this study. Additional replicas and source datasets are available upon request. The computational model is publicly accessible via the link provided in the Key Resource Table and methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| BactoAgar | BD Bioscience | Cat# 214050 |
| D-glucose | Sigma | Cat# G7021 |
| PD173074 | Sigma | Cat# P2499 |
| Blebbistatin | Sigma | Cat# B0560 |
| DiI | Sigma | Cat# 42364 |
| Recombinant Human/Mouse Wnt-5a Protein | R&D systems | Cat# 645-WN-010 |
| Recombinant Human Wnt-10b Protein | R&D systems | Cat# 7196-WN-010 |
| Alginic acid sodium salt | Sigma | Cat# 180947 |
| Experimental Models: Organisms/Strains | ||
| Wild Type Chicken Eggs | Charles River | Cat#10100331 |
| Tg(CAG-GFP) Chicken Eggs | Clemson university (originally by University of Edinburgh; McGrew et al., 2008) | n/a |
| Tg(PGK1:H2 B-chFP) Quail Eggs | Ozark egg company (Huss et al., 2015) | n/a |
| Recombinant DNA | ||
| pBI-EGFP | Clontech | Cat# 6154-1 |
| pBI-FGFRdn | Bénazéraf et al., 2010 | n/a |
| pCIRX-rtTA-Advanced | Bénazéraf et al., 2010; T. Iimura | n/a |
| pCIG-dnMKK1 | Delfini et al., 2005 | n/a |
| Software and Algorithms | ||
| Fiji | NIH | https://fiji.sc/ |
| Matlab (R2017a) | Mathworks | https://www.mathworks.com/products/matlab.html |
| FluoRender | SCI, Univ. of Utah | http://www.sci.utah.edu/software/fluorender.html |
| Agent-based mutli-tissue elongation model (in Matlab) | this paper | https://scholar.harvard.edu/files/fengzhu_xiong/files/fx_elongation20_modelcode.zip |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Chicken eggs
Wild type chicken eggs were supplied by Charles River Laboratories. Tg(CAG-GFP) (McGrew et al., 2008) chicken eggs were provided by Clemson university (originally by University of Edinburgh). Tg(PGK1:H2B-chFP) (Huss et al., 2015) quail eggs were provided by Ozark egg company. Eggs were kept in monitored 15°C fridge for storage and 37.5°C ~60% humidity egg incubators for incubation. HH stage 4–12 embryos were used. Sex as a variable is not relevant at the stages studied. All procedures have been approved by Harvard Medical Area (HMA) Standing Committee on Animals.
METHOD DETAILS
Embryo preparation
Whole embryo explant cultures were prepared using filter paper extraction (Chapman et al., 2001). Two adjacent 0.5cm holes are punched on a 2cm × 2cm piece of filter paper (Whatman). Eggs were opened into a petri dish where the embryo will be found near the top of the yolk. The thick albumin is swept aside gently with small filter paper pieces with a tweezer. The holed filter paper is then lowered to attach to the vitelline membrane with the holes allowing embryos to be viewed. The vitelline membrane is then cut around the filter paper to release the embryo. The filter culture embryo is then rinsed in PBS to remove excess yolk. Cleaned embryo is placed on a 3.5cm petri dish containing 2ml of semi-liquid culture media made with the following formula (per 100ml of culture media): Part A: 50ml Albumin (beaten for 15min) then supplement with 0.8ml 20% D-Glucose (Sigma); Part B: 0.3g BactoAgar (Sigma) solved in 50ml water in a microwave then supplement with 1.23ml 5M NaCl. Warm part A and cool part B to 55°C in a water bath. Mix thoroughly and add to petri dishes before gelation. The embryo cultures are maintained in a slide box with wet paper towels in the 37.5°C ~60% humidity egg incubators except when surgeries or snapshot imaging are performed.
Dye labeling, injections, and electroporation
3–14ss embryos were used for cell labeling. 2.5mg/ml DiI in ethanol was diluted in PBS to 0.5mg/ml before injection. The injection solution was loaded into a sharp-tipped glass needle and injected by mouth pipetting from the ventral side of the embryo into spots in different tissues (e.g., PSM, NC, the Node, and primitive streak). For over-expression of transgenes, DNA solutions (pBI-EGFP, pBI-FGFRdn and pCIRX-rtTA-Advanced, Bénazéraf et al., 2010) were injected from the ventral side into the anterior primitive streak of HH stage 4–5 embryos, followed by electroporation using a NEPA21 type II electroporator (NEPA GENE) in PBS. The pulse setting is at 6V, 150ms interval, 25ms duration per pulse, and 3 pulses per embryo. For PSM expression, the electrode targets the anterior primitive streak (the injection site). For NT expression, the electrode targets the area slightly anterior and flanking the Node. Embryos were checked for fluorescence and development 12hr later. Poorly developed and low/mis-expressed embryos were excluded from imaging and analysis. For the induction of dnFGFR1 expression in the PSM, embryos were incubated on plates with 2μg/ml of doxycyclin (Sigma). Imaging was started 2–3hrs after the incubation started to allow transgene induction. FGF signaling inhibitor PD173074 (Sigma) was solved in DMSO and diluted to 2μM in PBS before use. ~20μL solution or DMSO control was dropped on each cultured embryo on the ventral side. Blebbistatin (Sigma) was diluted to 70μM in PBS from DMSO stock (25μM) before use. ~50μL solution or control was dropped on the ventral side. For injections of PBS, WNT5A and WNT10 (R&D systems, recombinant human/mouse), 10μg/mL protein solution in PBS was loaded in a glass pipette and injected into the anterior portion of the pPSM close to both sides of the NT/NC, or into the NT apical side directly through the NC and floorplate.
Surgeries, gel implants, and pin experiments
8–12ss embryos were used for surgery experiments. Surgeries were performed under a Leica M90 dissecting scope. Cutting was performed with an electrolysis sharpened steel or tungsten needle from either the ventral or dorsal side, for PSM/NC or NT, respectively. In the ventral surgeries, endoderm was cut open first and gently peeled aside, followed by shallow cutting into the mesoderm while avoiding damaging the neural ectoderm and epiblast. In the dorsal surgeries, the vitelline membrane over the surgery site was first slit in the middle and gently peeled on either side to make a triangular shape opening, followed by cutting around the neural plate/NT area, care was taken to minimize damage to NC and endoderm. The wound areas were gently brushed by the needle to removal cells. For transplant, the comparable region of the donor was cut intact and moved out of wound with a tungsten rod. The tissue was then transferred with a micropipette loaded with PBS to the host. A smaller opening was cut open at the desired location and the transplant was gently pushed in. For gel implant, the cleared cut opening either received an injection of 1% (w/v) alginic acid sodium (Sigma Aldrich) solution, or a small piece of pre-formed soft alginate gel (40:1 mix with 5% (w/v) calcium chloride solution). Injected alginic solution formed gel with calcium ion in the embryo culture and integrated much better than transplant of pre-formed gel. The deformation results of both methods were qualitatively similar (e.g., transplanted: Figure S2D; injected: Figure S2E) even though the transplanted gel (higher stiffness, ~100 Pa, Banerjee et al., 2009) shows less deformation. Gels prepared with higher concentration of calcium ions (20:1 or 10:1 mix with 5% (w/v) calcium chloride solution) or agarose gels do not show significant deformation as tracked by the movement of encapsulated beads (For example, 0.5% agarose with a modulus in the order of 1k Pa [Markert et al., 2013], data not shown). Between 20 to 45min of healing and integration was allowed before mounting the host for imaging. For steel pin (MINUCIE, stainless, No.15, ~40μm) insertion, the tip of the pin is cut to around 1cm long. The pin is held with a tweezer and inserted between the neural folds into the culture medium from the ventral side after pNC ablation until only a small head remains level with the embryo. The embryo culture dish is placed in a bigger dish where the magnet is placed. Both distance and angle from the embryo to the magnet are adjusted by hand under a dissecting microscope to get optimal pin movement. The magnet and angle are fixed once proper pin rotation and tissue deformation under the pushing force can be seen and the configuration stays the same through a 4hr incubation.
Imaging
Bright field and fluorescent images of the embryos were taken with a Leica M205 fluorescent microscope. Embryo culture dishes were kept in a slide box with wet paper towel. The box was kept in the egg incubator at all times except image taking (~5 minutes for 10 embryos). For confocal timelapse imaging, the embryo (cultured as explant on filter paper) was mounted ventral or dorsal side down on a 50mm glass bottom dish (MatTek) pre-warmed to 37.5°C. A thin layer of albumin-agarose medium (200μl) was plated on the cover glass before mounting. The sample was then imaged on an inverted Zeiss confocal 780 with heating stage and environmental chamber to maintain temperature and humidity. Typical imaging set up for movies such as Movie S1: Imaging space: 1.4×1.4×0.15mm; voxel size: 1.4×1.4×2.5μm; time coverage: 15–20h; temporal resolution: 2–3min; lasers: 561nm 0.4%, 488nm 3%.
Image Analysis
Dissecting scope images were analyzed by drawing lines over features of interest (e.g., length, wound width, notochord width, etc.) in PowerPoint (Microsoft) or Fiji (ImageJ). For elongation, the AP level of somite 2–3 were chosen as the reference point (except where it is noted differently in Figure legends). The distance increase between the reference point and the Node was used to calculate elongation speed. For confocal timelapse movies, the original .lsm (Zeiss) files were rendered into 3D maximum projection movies in FluoRender and compiled to movies in Fiji. Cells were tracked on this movie with the Manual Tracking plugin in Fiji. For cell density measurements, region of interest was selected from PSM z-stack images for fluorescent intensity or nuclei count in Fiji. Tracking results and measurements were processed in Matlab (Mathworks) with custom scripts. A track of the moving Node and a track of the anterior midline were used to linear transform the coordinates of cells to anterior-posterior / lateral-medial axes before all position and speed calculation.
Modeling
All models were coded in Matlab. Detailed documentation is provided below. The model can be downloaded following the link in the Key Resource Table.
1. Model design and protocol
The goal of the model is to test what types of tissue behavior emerge from cells behaving under simple rules. Tissue geometry and cell activities break the homogeneity and symmetry and underlie these behaviors, although they act with a number of other interactions that should be accounted for in the model. For simplicity and feasibility, the tissue is modeled as a field of cells on a 2D plane with boundaries. The field is seeded with a finite number of cells (much fewer than in actual tissues for feasibility but geometry and proportions are kept) with user defined properties including position, velocity and cell type. Time is introduced as number of iterations for the field to evolve. At each iteration, a cell receives a net “force” defined by its position, previous velocity, type, and motility (eq.1, implementation of each term is explained in section 2). This force causes it to change position and velocity. Collectively the field evolves to a new pattern. Additionally, functionalities are build-in to allow new cells to be added or specific cells to be removed during simulation, to provide tools for assessing progenitor addition and tissue surgery experiments. A chemotaxis gradient generator is also included but was kept off for the present study.
| (1) |
To use the model, copy all provided scripts to the working folder of Matlab. Run simulation.m. In this script, a user can define the geometrical and interaction parameters (to be discussed in detail in section 2), as well as number of iterations in one simulation, rounds of independent simulations, and whether a perturbation cut on the tissue is performed (to be discussed in detail in the section after the next). To create a movie, specify a “moviename” string variable and use command:
“>>makemovie2(Data,N+NewN,chemoN,tipN,NCN,boundary,moviename)”
2. Model assumptions and implementation details
The model includes both symmetry breaking and non-symmetry breaking assumptions. The former dictates the emergent patterns of the field. The assumptions and their rationale, impact and implementation are described below:
Non-symmetry breaking:
1. Repulsion between cells:
when cells move close to each other, a repulsion force is generated as a function of distance. This assumption shows the basic volume constrain where no two cells could occupy the same position. This assumption provides the basis for force transduction in the system and is key for implementation of several other assumptions. It alone however does not cause any symmetry break or pattern evolution. The strength of repulsion is controlled by parameter alpha. The interaction term for a given cell (Ii) is calculated as eq.2:
| (2) |
In which rij = |xi – xj|. R is the cell diameter.
2. Viscosity/friction:
a resistance against the cell’s velocity is added at each simulation, to mimic the fact that cells move in a viscous tissue environment where inertia is not important. This force ensures that simulated cells do not project out of the tissues. The strength of viscosity is controlled by parameter beta. In eq.1, the calculation gives a force in the opposite direction proportional to the step of movement (v) from the previous time point.
3. Repulsion between cells and boundary:
when cells run into a boundary it is bounced back similarly as it runs into a cell (eq.3). This repulsion is not particularly important for the simulation. The parameter controlling which is set to equal alpha. In addition, the size of tissue field and cell field can be set by parameters boundary and boundaryc.
| (3) |
In which rib = |xi – xb|. xb is the location of the boundaries.
Symmetry breaking:
1. Geometrical layout:
in the standard configuration, the model places the NT/NC cells (analogously referred to as NC cells in subsequent descriptions and the model codes) in between two flanking PSM cell groups (controlled by parameter NCN assigning NC cell type to the designated cell ID numbers). The progenitor domain that adds new PSM cells is placed posterior to the end of NC cells (controlled by parameter tipN). In the standard configuration, the parameter tipN restricts a particular NC cell (the tipN cell) at the posterior (open) end of the tissue to only move anterior-posteriorly (no free medial lateral movement that could arise due to small number of cells). A medial-lateral line posterior to the tipN cell at a specified distance defines the entry point of the new cells, thereby serving as the PD domain in the simulation. This layout is analogous to the actual embryo.
2. New cell addition:
in the standard configuration, the model adds 80 new PSM cells to the existing field of 200 cells (controlled by parameters N, NewN). The cells are added periodically with parameters available to control the frequency, number and pattern of addition in gastrulation.m. At each time when new cells are to be added, their AP locations are determined by the location of the tipN cell, thereby following the movement of the PD. The tipN cell and the new cells otherwise follow the same mechanical interactions as any other cells in the field. This reflects the actual constant PSM cell production in the progenitor zone and the new PSM cells emerge from the dorsal side to the ventral midline. In the current study the cell addition is either stable throughout simulation or shut down completely following removal of the PD.
3. Random motility gradient across the field in the PSM cells:
in the standard configuration, the model implements a simple linear motility gradient as a function of the PSM cell’s distance to the PD/pNC boundary (as marked by the location of the tipN cell). The motility is produced with a random number to control direction and magnitude, with parameter gamma controlling overall magnitude (eq.4).
| (4) |
NC/NT cells are assumed to not have this motility. This assumption is justified by the observation of the exact type of motility gradient in the PSM in previous studies and the current study (Figure 1), and its known regulation by FGF signaling (also validated in this study, Figures 3,S3). This assumption includes cell property and a geometry asymmetry and is the key assumption of the model.
4. Polarity of NC cells:
in the standard configuration, the model takes into account NC cell’smedial-laterally polarized shape by using a parameter ncp to bias the intercalation preference, implemented as differential repulsion strengths along different axes. When the value of ncp is bigger than 1 it means a medial-laterally elongated cell that would intercalate more easily medial-laterally than anterior-posteriorly (eq.5). This assumption is justified by the polarized NT cell shapes in previous studies and its known regulation by WNT/PCP signaling (also validated in this study, Figures S4,S5).
| (5) |
5. Adhesion in the anterior:
an adhesion gradient from anterior to posterior in the PSM (implemented as reduced repulsion in the anterior) is used to mimic the same observed properties in vivo. The simulations however show that its impact on elongation is minimal. This is consistent with the observation that the main driver of elongation is the posterior PSM where adhesion is low. For this reason, adhesion is not further investigated in the model or tested in wet lab experiments in this instance.
3. Simulations
This section describes a more detailed account for the simulations. After specifying parameters described in the previous section. The user goes on to determine number of iterations (time). Sufficient time is often needed for the field to evolve into a stable state. The sum of parameters Tcut and Tcut2 is the total number of iterations. This is split into two parts as the algorithm provides an opportunity of tissue cutting perturbation in between, so that the model takes the specified removed cells out of the interactions during Tcut2. To specify a region to cut, the user can either select one that is pre-specified in the script (cutmatrix) or write a new one. Note that this region should be a non-zero-area box (or boxes) that are within the boundary parameter otherwise it will not have an effect. By controlling parameter sc, the user can also specify how many independent runs the simulation runs.
4. Result analysis
The output of simulaiton.m includes a cell array named “Data”, in which the cut perturbation info and the whole simulation are recorded. A user can obtain any cell’s position and transient velocity as a function of time (iteration number) and use logic index to obtain information of interest. The elongation speeds, time average cell velocities and regional density changes are computed from “Data” using simple custom scripts.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis summary
Quantitative results generated from the image analysis were sorted into experimental groups and Student’s t-test is performed with Excel (Microsoft) and Matlab (Mathworks) to compare the results with p<0.05 considered significant difference. The type of statistical tests, exact value of n, definition of center, and dispersion and precision measures are all precisely defined in each figure legend in the manuscript.
Statistical considerations in study design
Sample size: For qualitative yes/no tests where the distinction is clear (e.g., direction of gel deformation) and no exceptions were observed, sample size is at least 3. For quantitative comparisons (e.g., elongation speed), at least 3 samples per group are used per experiment. >=5 samples are used to in key groups. This sample size is in line with literature and is preferred for consistency and minimizes system errors that might be introduced between groups when total experiment time becomes too long (>2 hrs), taking into consideration the time consuming procedure of embryo preparation and individual surgical operations. Replications were performed to confirm the results. For cell behavior analysis, >30 individual cells were tracked through the duration of the movies (hundreds of time points). Before applying t-tests, the measurements were normalized to the mean and pooled for a chi square goodness of fit test. The tests suggest that the variabilities observed in cell speed, elongation speed, convergence speed, tissue width, and cell spread do not come from a distribution significantly different (p>0.05) from a normal distribution. Data exclusions: NO data point exclusions in any analysis. In sample preparation and before data collection, embryos that show unspecific developmental abnormalities or lack of labels (indicating failed electroporation or injection) were discarded. This health and technical screen is applied in an unbiased manner to all experimental and control groups. Replication: All wet experiments reported have been replicated at least once (key experiments 3 or more times) with a different batch of eggs (shipment in a different week) and new preparation of reagents (culture and imaging plates, injection needles and labeling reagents (different aliquots of the same source)). The same dissecting tools, imaging protocols and microscopes were used during replications. Modeling experiments were replicated at least 10 times for each type of simulation run. Randomization and Blinding: In each embryonic experimental test involving groups of embryos, all embryos were incubated, prepared and handled together except for the particular experimental condition(s) being tested. After an initial health screen of all embryos, the investigators do not use any criteria or variables when allocating embryos into control and experimental groups. Blinding is not possible for surgical perturbations on the embryos as data collection and analysis require the investigator to look at the images where the type of surgical perturbation is clearly visible. For chemical treatment and injections on the embryo, a look-up table (relating operation to sample number) is generated at the operation time and is not referred to during data collection and analysis. Only when analysis is complete is the look-up table linked to sort the results into different experimental groups. Blinding is not applicable in confocal movie experiment or modeling as the test conditions are clearly affiliated with the samples throughout data acquisition and analysis.
Supplementary Material
The movie on the left is a 3D maximum projection (ventral view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated duration (starting ~3 somite stage). Green (DiI labeled) cells start as clusters at the injection site. This movie is oriented to have the anterior on the left. NC in the center can be seen moving posterior over time, followed by somite formation on both sides from the PSM. DiI labeled PSM cells disperse from the midline progenitor domain (PD) and join pPSM on both sides. On the right side the movie shows the trajectories of tracked cells of the same dataset. The solid red sphere marks the end of NC. The tracks over time are shown in Figure S1C and two time points of the movie are used for the fate map in Figure S1D. Time stamp: hh:mm. Scale bar: 100 μm.
The movie is maximum projection (dorsal view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated duration. pNT and pNC were ablated and the opening was filled with alginate gel. Imaging started 20 minutes after the operation. Time stamp: hh:mm. Scale bar: 100 μm.
Movies show the 2D field (Anterior to the top) of cells (small spheres). Red cells are axial cells and Blue cells are PSM cells. In the standard simulation, new PSM cells are added at the posterior end of the axial tissues (PD). The cell addition grid is determined by one center advancing NC cell, which is allowed to move along the AP axis only to prevent bending from simulation fluctuations due to small cell number. From left to right, the simulations are: Control, Reduced axial cell polarity, Reduced motility gradient, Reduced both motility and polarity. All perturbations last the duration of the simulation (4000 iterations).
Similar to Movie S3. The cells matching different tissue areas were identified by their coordinates at the time of surgery and removed from the simulation. From left to right, the simulations are: Control, pNC/pNT ablation, pPSM ablation, No Axial PD. The surgeries take place at iteration 1001 instantly. The No Axial PD condition lasts the duration of the simulation (i.e., no new PSM cells were added over the 4000 iterations).
The movie is maximum projection (ventral view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated duration. Green (DiI labeled) cells start as clusters at the injection sites in the PSM and can be observed to spread in the pPSM over time. PD was ablated and the opening was filled with alginate gel. Imaging started 30 minutes after the operation. Time stamp: hh:mm. Scale bar: 100 μm.
Maximum projections (ventral view) of Tg(CAG:GFP) (shown in red) chicken embryos imaged for the indicated durations. Green (DiI labeled) cells start as clusters in the PD and can be seen to move laterally and show impaired movement in the axial tissue ablated conditions. Imaging started 25 minutes after the operation for the two ablated conditions (pNC and pNT as labeled). Time stamp: hh:mm. Scale bar: 100 μm.
The movie is a maximum projection (ventral view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated durations. Similar to Movie S6, green (DiI labeled) cells start as clusters in the PD show impaired movement after pPSM ablations. Imaging started 30 minutes after the operation. Time stamp: hh:mm. Scale bar: 100 μm.
Highlights.
Posterior presomitic mesoderm (PSM) compresses elongating body axis
Axial neural tube and notochord push the caudal progenitor domain
Axial push promotes new PSM cell supply
Tissues elongate hand-in-hand through mechanical coupling in an engine-like loop
ACKNOWLEDGEMENTS
We thank J Chung, C. Chan, C. Guillot, M. Oginuma, A. Hubaud, T. Hiscock, J. Young, N. Nerurkar, S. Megason, C. Tabin, C. Cepko, M. Kirschner, P. Lenne, Pourquie and Mahadevan lab members for equipment, reagents and comments. This work is supported by a Helen Hay Whitney Foundation (HHWF) fellowship funded by Howard Hughes Medical Institute (HHMI) and a NIH pathway to independence award (K99-HD092582) to F.X., and a NIH grant (R01-HD097068) to O.P. and L.M., F.X. also acknowledges Wellcome Trust and Royal Society (215439/Z/19/Z). L.M. also acknowledges NIH (R01-HD087234) and NSF (DMS-1764269).
Footnotes
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alvarez IS and Schoenwolf GC (1992). Expansion of surface epithelium provides the major extrinsic force for bending of the neural plate. Journal of Experimental Zoology Part A: Ecological Genetics and Physiology 261(3), 340–348. [DOI] [PubMed] [Google Scholar]
- Bailles A, Collinet C, Philippe JM, Lenne PF, Munro E and Lecuit T (2019). Genetic induction and mechanochemical propagation of a morphogenetic wave. Nature 572, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Arha M, Choudhary S, Ashton RS, Bhatia SR, Schaffer DV and Kane RS (2009). The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells. Biomaterials 30(27), 4695–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénazéraf B, Francois P, Baker RE, Denans N, Little CD and Pourquié O (2010). A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature 466, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benazeraf B, Beaupeux M, Tchernookov M, Wallingford A, Salisbury T, Shirtz A, Shirtz A, Huss D, Pourquie O, Francois P and Lansford R (2017). Multi-scale quantification of tissue behavior during amniote embryo axis elongation. Development 144(23), 4462–4472. [DOI] [PubMed] [Google Scholar]
- Bénazéraf B and Pourquié O (2013). Formation and segmentation of the vertebrate body axis. Annual Review of Cell and Developmental Biology 29, 1–26. [DOI] [PubMed] [Google Scholar]
- Butler LC, Blanchard GB, Kabla AJ, Lawrence NJ, Welchman DP, Mahadevan L, Adams RJ and Sanson B (2009). Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Nature Cell Biology 11(7), 859. [DOI] [PubMed] [Google Scholar]
- Charrier JB, Teillet MA, Lapointe F and Le Douarin NM (1999). Defining subregions of Hensen’s node essential for caudalward movement, midline development and cell survival. Development 126(21), 4771–4783. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Brown R, Lees L, Schoenwolf GC and Lumsden A (2004). Expression analysis of chick Wnt and frizzled genes and selected inhibitors in early chick patterning. Developmental Dynamics 229(3), 668–676. [DOI] [PubMed] [Google Scholar]
- Chapman SC, Collignon J, Schoenwolf GC and Lumsden A (2001). Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 220, 284–289. [DOI] [PubMed] [Google Scholar]
- Collinet C, Rauzi M, Lenne PF and Lecuit T (2015). Local and tissue-scale forces drive oriented junction growth during tissue extension. Nature Cell Biology 17(10), 1247. [DOI] [PubMed] [Google Scholar]
- Colas JF and Schoenwolf GC (2001). Towards a cellular and molecular understanding of neurulation. Developmental Dynamics 221(2), 117–145. [DOI] [PubMed] [Google Scholar]
- Das D, Chatti V, Emonet T and Holley SA (2017). Patterned disordered cell motion ensures vertebral column symmetry. Developmental Cell 42(2), 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D, Jülich D, Schwendinger-Schreck J, Guillon E, Lawton AK, Dray N, Emonet T, O’Hern CS, Shattuck MD and Holley SA (2019). Organization of embryonic morphogenesis via mechanical information. Developmental Cell 49(6), 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfini MC, Dubrulle J, Malapert P, Chal J and Pourquié O (2005). Control of the segmentation process by graded MAPK/ERK activation in the chick embryo. Proceedings of the National Academy of Sciences of the United States of America 102(32), 11343–11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denans N, Iimura T & Pourquié O (2015). Hox genes control vertebrate body elongation by collinear Wnt repression. Elife 4, e04379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray N, Lawton A, Nandi A, Jülich D, Emonet T and Holley SA (2013). Cell-fibronectin interactions propel vertebrate trunk elongation via tissue mechanics. Current Biology 23(14), 1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman NS, Kimmel CB, Jones MA and Adams RJ (2003). Shaping the zebrafish notochord. Development 130(5), 873–887. [DOI] [PubMed] [Google Scholar]
- Guillon E, Das D, Jülich D, Hassan AR, Geller H and Holley S (2020). Fibronectin is a smart adhesive that both influences and responds to the mechanics of early spinal column development. Elife 9, e48964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V (1992). The stage series of the chick embryo. Developmental Dynamics 195, 273–275. [DOI] [PubMed] [Google Scholar]
- Huss D, Benazeraf B, Wallingford A, Filla M, Yang J, Fraser SE and Lansford R (2015). A transgenic quail model that enables dynamic imaging of amniote embryogenesis. Development 142(16), 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iimura T and Pourquie O (2008). Manipulation and electroporation of the avian segmental plate and somites in vitro. Methods in Cell Biology 87, 257–270. [DOI] [PubMed] [Google Scholar]
- Keller R and Danilchik MIKE (1988). Regional expression, pattern and timing of convergence and extension during gastrulation of Xenopus laevis. Development 103(1), 193–209. [DOI] [PubMed] [Google Scholar]
- Keller R, Shook D and Skoglund P (2008). The forces that shape embryos: physical aspects of convergent extension by cell intercalation. Phys. Biol. 5, 015007. [DOI] [PubMed] [Google Scholar]
- Kulesa PM and Fraser SE (2002). Cell dynamics during somite boundary formation revealed by time-lapse analysis. Science 298, 991–995. [DOI] [PubMed] [Google Scholar]
- Lawton AK, Nandi A, Stulberg MJ, Dray N, Sneddon MW, Pontius W, Emonet T and Holley SA (2013). Regulated tissue fluidity steers zebrafish body elongation. Development 140(3), 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye CM, Blanchard GB, Naylor HW, Muresan L, Huisken J, Adams RJ and Sanson B (2015). Mechanical coupling between endoderm invagination and axis extension in Drosophila. PLoS Biology 13, e1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert CD, Guo X, Skardal A, Wang Z, Bharadwaj S, Zhang Y, Bonin K and Guthold M (2013). Characterizing the micro-scale elastic modulus of hydrogels for use in regenerative medicine. Journal of the Mechanical Behavior of Biomedical Materials 27, 115–127. [DOI] [PubMed] [Google Scholar]
- Manning AJ and Kimelman D (2015). Tbx16 and Msgn1 are required to establish directional cell migration of zebrafish mesodermal progenitors. Developmental Biology 406(2), 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew MJ, Sherman A, Lillico SG, Ellard FM, Radcliffe PA, Gilhooley HJ, Mitrophanous KA, Cambray N, Wilson V and Sang H (2008). Localised axial progenitor cell populations in the avian tail bud are not committed to a posterior Hox identity. Development 135, 2289–2299. [DOI] [PubMed] [Google Scholar]
- Mongera A, Michaut A, Guillot C, Xiong F and Pourquié O (2019). Mechanics of anteroposterior Axis formation in vertebrates. Annual Review of Cell and Developmental Biology 35, 259–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera A, Rowghanian P, Gustafson HJ, Shelton E, Kealhofer DA, Carn EK, Serwane F, Lucio AA, Giammona J and Campàs O (2018). A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, Campbell RM, Christian JL, McGrew LL, Shih J and Fraser S (1993). Xwnt-5A: a maternal Wnt that affects morphogenetic movements after overexpression in embryos of Xenopus laevis. Development 119(1), 97–111. [DOI] [PubMed] [Google Scholar]
- Morita H, Grigolon S, Bock M, Krens SG, Salbreux G and Heisenberg CP (2017). The physical basis of coordinated tissue spreading in zebrafish gastrulation. Developmental Cell 40(4), 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Gleghorn JP, Pang MF, Jaslove JM, Goodwin K, Varner VD, Miller E, Radisky DC and Stone HA (2017). Microfluidic chest cavities reveal that transmural pressure controls the rate of lung development. Development 144(23), 4328–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Honda H and Takeichi M (2012). Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149(5), 1084–1097. [DOI] [PubMed] [Google Scholar]
- Oginuma M, Moncuquet P, Xiong F, Karoly E, Chal J, Guevorkian K and Pourquié O (2017). A gradient of glycolytic activity coordinates FGF and Wnt signaling during elongation of the body axis in amniote embryos. Developmental Cell 40(4), 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera-Martinez I, Harada H, Halley PA & Storey KG (2012). Loss of FGF-dependent mesoderm identity and rise of endogenous retinoid signalling determine cessation of body axis elongation. PLoS Biology 10, e1001415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeirim I, Dubrulle J, Henrique D, Ish-Horowicz D and Pourquie O (1998). Uncoupling segmentation and somitogenesis in the chick presomitic mesoderm. Developmental Genetics 23(1), 77–85. [DOI] [PubMed] [Google Scholar]
- Regev I, Guevorkian K, Pourquie O and Mahadevan L (2017). Motility-gradient induced elongation of the vertebrate embryo. BioRxiv. 10.1101/187443 [DOI] [Google Scholar]
- Sausedo RA and Schoenwolf GC (1993). Cell behaviors underlying notochord formation and extension in avian embryos: quantitative and immunocytochemical studies. The Anatomical Record 237(1), 58–70. [DOI] [PubMed] [Google Scholar]
- Savin T, Kurpios NA, Shyer AE, Florescu P, Liang H, Mahadevan L and Tabin CJ (2011). On the growth and form of the gut. Nature 476, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenwolf GC (1979). Histological and ultrastructural observations of tail bud formation in the chick embryo. The Anatomical Record 193(1), 131–147. [DOI] [PubMed] [Google Scholar]
- Serwane F, Mongera A, Rowghanian P, Kealhofer DA, Lucio AA, Hockenbery ZM and Campàs O (2017). In vivo quantification of spatially varying mechanical properties in developing tissues. Nature Methods 14(2), 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J and Keller R (1992). Cell motility driving mediolateral intercalation in explants of Xenopus laevis. Development 116(4), 901–914. [DOI] [PubMed] [Google Scholar]
- Smith JL and Schoenwolf GC (1991). Further evidence of extrinsic forces in bending of the neural plate. Journal of Comparative Neurology 307(2), 225–236. [DOI] [PubMed] [Google Scholar]
- Steventon B, Duarte F, Lagadec R, Mazan S, Nicolas JF and Hirsinger E (2016). Species-specific contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 143(10), 1732–1741. [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M & Martin GR (1999). Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 13,1834–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell-Badge R, Papaioannou VE and Kondoh H (2011). Tbx6-dependent Sox2 regulation determines neural vs mesodermal fate in axial stem cells. Nature 470, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V and Nicolas JF (2009). Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Developmental Cell 17(3), 365–376. [DOI] [PubMed] [Google Scholar]
- Waddington CH (1932). Experiments on the development of the chick and the duck embryo cultivated in vitro. Proc. Trans. R. Soc. Lond. B 211, 179–230. [Google Scholar]
- Wallingford JB and Harland RM (2001). Xenopus Dishevelled signaling regulates both neural and mesodermal convergent extension: parallel forces elongating the body axis. Development 128(13), 2581–2592. [DOI] [PubMed] [Google Scholar]
- Williams M, Yen W, Lu X and Sutherland A (2014). Distinct apical and basolateral mechanisms drive planar cell polarity-dependent convergent extension of the mouse neural plate. Developmental Cell 29(1), 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Dormann D, Münsterberg AE and Weijer CJ (2002). Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Developmental Cell 3(3), 425–437. [DOI] [PubMed] [Google Scholar]
- Zhou J, Kim HY and Davidson LA (2009). Actomyosin stiffens the vertebrate embryo during crucial stages of elongation and neural tube closure. Development 136(4), 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie on the left is a 3D maximum projection (ventral view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated duration (starting ~3 somite stage). Green (DiI labeled) cells start as clusters at the injection site. This movie is oriented to have the anterior on the left. NC in the center can be seen moving posterior over time, followed by somite formation on both sides from the PSM. DiI labeled PSM cells disperse from the midline progenitor domain (PD) and join pPSM on both sides. On the right side the movie shows the trajectories of tracked cells of the same dataset. The solid red sphere marks the end of NC. The tracks over time are shown in Figure S1C and two time points of the movie are used for the fate map in Figure S1D. Time stamp: hh:mm. Scale bar: 100 μm.
The movie is maximum projection (dorsal view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated duration. pNT and pNC were ablated and the opening was filled with alginate gel. Imaging started 20 minutes after the operation. Time stamp: hh:mm. Scale bar: 100 μm.
Movies show the 2D field (Anterior to the top) of cells (small spheres). Red cells are axial cells and Blue cells are PSM cells. In the standard simulation, new PSM cells are added at the posterior end of the axial tissues (PD). The cell addition grid is determined by one center advancing NC cell, which is allowed to move along the AP axis only to prevent bending from simulation fluctuations due to small cell number. From left to right, the simulations are: Control, Reduced axial cell polarity, Reduced motility gradient, Reduced both motility and polarity. All perturbations last the duration of the simulation (4000 iterations).
Similar to Movie S3. The cells matching different tissue areas were identified by their coordinates at the time of surgery and removed from the simulation. From left to right, the simulations are: Control, pNC/pNT ablation, pPSM ablation, No Axial PD. The surgeries take place at iteration 1001 instantly. The No Axial PD condition lasts the duration of the simulation (i.e., no new PSM cells were added over the 4000 iterations).
The movie is maximum projection (ventral view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated duration. Green (DiI labeled) cells start as clusters at the injection sites in the PSM and can be observed to spread in the pPSM over time. PD was ablated and the opening was filled with alginate gel. Imaging started 30 minutes after the operation. Time stamp: hh:mm. Scale bar: 100 μm.
Maximum projections (ventral view) of Tg(CAG:GFP) (shown in red) chicken embryos imaged for the indicated durations. Green (DiI labeled) cells start as clusters in the PD and can be seen to move laterally and show impaired movement in the axial tissue ablated conditions. Imaging started 25 minutes after the operation for the two ablated conditions (pNC and pNT as labeled). Time stamp: hh:mm. Scale bar: 100 μm.
The movie is a maximum projection (ventral view) of a Tg(CAG:GFP) (shown in red) chicken embryo imaged for the indicated durations. Similar to Movie S6, green (DiI labeled) cells start as clusters in the PD show impaired movement after pPSM ablations. Imaging started 30 minutes after the operation. Time stamp: hh:mm. Scale bar: 100 μm.
Data Availability Statement
The published article includes most datasets generated during this study. Additional replicas and source datasets are available upon request. The computational model is publicly accessible via the link provided in the Key Resource Table and methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| BactoAgar | BD Bioscience | Cat# 214050 |
| D-glucose | Sigma | Cat# G7021 |
| PD173074 | Sigma | Cat# P2499 |
| Blebbistatin | Sigma | Cat# B0560 |
| DiI | Sigma | Cat# 42364 |
| Recombinant Human/Mouse Wnt-5a Protein | R&D systems | Cat# 645-WN-010 |
| Recombinant Human Wnt-10b Protein | R&D systems | Cat# 7196-WN-010 |
| Alginic acid sodium salt | Sigma | Cat# 180947 |
| Experimental Models: Organisms/Strains | ||
| Wild Type Chicken Eggs | Charles River | Cat#10100331 |
| Tg(CAG-GFP) Chicken Eggs | Clemson university (originally by University of Edinburgh; McGrew et al., 2008) | n/a |
| Tg(PGK1:H2 B-chFP) Quail Eggs | Ozark egg company (Huss et al., 2015) | n/a |
| Recombinant DNA | ||
| pBI-EGFP | Clontech | Cat# 6154-1 |
| pBI-FGFRdn | Bénazéraf et al., 2010 | n/a |
| pCIRX-rtTA-Advanced | Bénazéraf et al., 2010; T. Iimura | n/a |
| pCIG-dnMKK1 | Delfini et al., 2005 | n/a |
| Software and Algorithms | ||
| Fiji | NIH | https://fiji.sc/ |
| Matlab (R2017a) | Mathworks | https://www.mathworks.com/products/matlab.html |
| FluoRender | SCI, Univ. of Utah | http://www.sci.utah.edu/software/fluorender.html |
| Agent-based mutli-tissue elongation model (in Matlab) | this paper | https://scholar.harvard.edu/files/fengzhu_xiong/files/fx_elongation20_modelcode.zip |