Abstract
Asthma puts a tremendous overhead on healthcare. To enable effective preventive care to improve outcomes in managing asthma, we recently created two machine learning models, one using University of Washington Medicine data and the other using Intermountain Healthcare data, to predict asthma hospital visits in the next 12 months in asthma patients. As is common in machine learning, neither model supplies explanations for its predictions. To tackle this interpretability issue of black-box models, we developed an automated method to produce rule-style explanations for any machine learning model’s predictions made on imbalanced tabular data and to recommend customized interventions without lowering the prediction accuracy. Our method exhibited good performance in explaining our Intermountain Healthcare model’s predictions. Yet, it stays unknown how well our method generalizes to academic healthcare systems, whose patient composition differs from that of Intermountain Healthcare. This study evaluates our automated explaining method’s generalizability to the academic healthcare system University of Washington Medicine on predicting asthma hospital visits. We did a secondary analysis on 82,888 University of Washington Medicine data instances of asthmatic adults between 2011 and 2018, using our method to explain our University of Washington Medicine model’s predictions and to recommend customized interventions. Our results showed that for predicting asthma hospital visits, our automated explaining method had satisfactory generalizability to University of Washington Medicine. In particular, our method explained the predictions for 87.6% of the asthma patients whom our University of Washington Medicine model accurately predicted to experience asthma hospital visits in the next 12 months.
Index Terms—: Asthma, automated explanation, extreme gradient boosting, machine learning, patient care management, predictive model
I. INTRODUCTION
Background
Asthma affects 7.7% of American people, resulting in 3,441 deaths, 188,968 hospitalizations, and 1,776,851 emergency department (ED) visits annually [1]. Currently, the leading method for cutting down the number of asthma hospital visits involving hospitalizations and ED visits is to adopt a model to predict which asthma patients are most likely to incur poor outcomes at a later date. We then place these patients into a care management program, letting care managers follow up with them regularly and help them book services on healthcare and other related aspects. This method is used by numerous healthcare systems including University of Washington Medicine, Kaiser Permanente Northern California [2], and Intermountain Healthcare, as well as many health plans encompassing those in nine of 12 metropolitan communities [3]. With proper implementation, we can use this method to avoid as many as 40% of future hospital visits of the patients [4–7].
A care management program has the capacity to accommodate only a small fraction of the patients [8]. Hence, the predictive model’s accuracy puts an upper limit on the program’s efficacy. Yet, due to not including some important features in the model, none of the prior published models for predicting asthma hospital visits in asthma patients [2], [9–21] is accurate enough. Every prior published model misses over 50% of the patients who will experience future asthma hospital visits (i.e., sensitivity < 50%) and misclassifies many other patients to incur such visits. This limited model accuracy leads to suboptimal patient outcomes and unneeded healthcare costs. To tackle this prediction accuracy issue, we recently used extreme gradient boosting (XGBoost) [22] and extensive candidate features to create two machine learning models to predict asthma hospital visits in the next 12 months in asthma patients. Both models were more accurate than the prior published models. One model was built on Intermountain Healthcare data [23]. The other was built on University of Washington Medicine (UWM) data [24]. As is common in machine learning, neither model supplies explanations for its predictions, despite explanations are essential for care managers to understand the predictions, decide whether the patient should be enrolled in the care management program, and find appropriate tailored interventions for the patient. To tackle this interpretability issue of black-box models, we developed an automated method to produce rule-style explanations for any machine learning model’s predictions made on imbalanced tabular data and to recommend customized interventions without degrading the prediction accuracy [25]. Our method exhibited good performance in explaining the predictions made by our Intermountain Healthcare model [25].
Problem statement and objectives
Yet, it stays unknown how well our automated explanation method generalizes to academic healthcare systems, whose business practices and patient composition differ from those of Intermountain Healthcare. Usually, academic healthcare systems handle sicker and more complex patients than non-academic healthcare systems [26]. This study assesses how well our automated explanation method generalizes to UWM with regard to predicting asthma hospital visits.
II. METHODS
A. ETHICS APPROVAL AND STUDY DESIGN
This secondary analysis retrospective cohort study using administrative and clinical data was approved by UWM’s institutional review board.
B. PATIENT COHORT
UWM is the largest academic healthcare system in Washington State. Starting from 2011, its enterprise data warehouse began to collect complete clinical and administrative data from 12 clinics and 3 hospitals that mainly take care of adults. We employed the same patient cohort used in our prior UWM predictive model paper [24]: asthmatic adults (age≥18) who received care at any of these UWM facilities between 2011 and 2018. We regarded a patient to have asthma in a given year if in that year, the patient had at a minimum one asthma diagnosis code (International Classification of Diseases, Tenth Revision [ICD-10]: J45.x; International Classification of Diseases, Ninth Revision [ICD-9]: 493.0x, 493.1x, 493.8x, 493.9x) record in the encounter billing database [10], [27], [28]. The exclusion criterion was to drop the patients who died in that year.
C. PREDICTION TARGET (ALSO CALLED THE DEPENDENT VARIABLE)
We employed the same prediction target used in our prior UWM predictive model paper [24]. There, an asthma hospital visit was defined as an ED visit or hospitalization whose principal diagnosis is asthma (ICD-10: J45.x; ICD-9: 493.0x, 493.1x, 493.8x, 493.9x). For every patient whom we regarded to have asthma in a given index year, the outcome of interest was whether the patient experienced any asthma hospital visit in the next 12 months. In training and testing our UWM model and our automated explanation method, we took the patient’s data through the end of the year to predict the patient’s outcome in the next 12 months.
D. DATA SET
We employed the same structured, administrative and clinical data set from UWM’s enterprise data warehouse used in our prior UWM predictive model paper [24]. This data set covered the patient cohort’s visits at the 12 UWM clinics and 3 UWM hospitals between 2011 and 2019. Because the outcome of interest occurred in the next 12 months, our data set contained eight years of effective data (2011–2018) over the nine-year span of 2011–2019.
E. DATA PRE-PROCESSING, FEATURES (ALSO CALLED INDEPENDENT VARIABLES), AND PREDICTIVE MODEL
We employed the same data pre-processing method used in our prior UWM predictive model paper [24] to prepare, normalize, and clean the data. Our UWM model [24] uses 71 features and the XGBoost classification algorithm [22] to predict asthma hospital visits in the next 12 months in asthma patients. As detailed in Table 2 in our prior UWM predictive model paper’s [24] Appendix, these 71 features were calculated using the structured attributes in our data set and involve various aspects like medications, patient demographics, diagnoses, vital signs, visits, laboratory tests, and procedures. An example feature is the number of ED visits related to asthma that the patient had in the previous 12 months. Each input data instance entered into our UWM model contains these 71 features, pinpoints to an (index year, patient) pair, and is used to predict the patient’s outcome in the next 12 months. As in our prior UWM predictive model paper [24], we placed the cutoff point for binary classification at the top 10% of asthma patients who had the highest predicted risk.
F. REVIEW OF OUR AUTOMATED EXPLANATION METHOD
In 2016, we published an automated method to produce rule-style explanations for any machine learning model’s predictions made on tabular data and to recommend customized interventions without lowering the prediction accuracy. Our original method [29] was created for reasonably balanced data. We subsequently extended the method to cope with imbalanced data [25], in which one outcome value is much less prevalent than another. Imbalanced data appear in predicting asthma hospital visits in asthma patients because the two possible values of the outcome of interest have a skewed distribution. At UWM, ~2% of asthma patients would experience asthma hospital visits in the next 12 months. The following sections focus on the extended automated explanation method.
1). BASIC IDEA:
As Fig. 1 shows, the fundamental idea of our automated explanation method is to adopt two models at the same time to set producing and explaining predictions apart. Each model plays a distinct role in this system. We employ the first model to produce predictions. It can be any model taking categorical and continuous features and is usually set to the most accurate model. The second model is composed of class-based association rules [30], [31] mined from prior data. It is employed to explain the first model’s predictions instead of producing predictions. To create the second model, we first use an automated discretizing method [30], [32] to turn continuous features into categorical features. Then we use a standard method such as Apriori to mine the association rules [31]. Every generated rule expresses a feature pattern connecting to an outcome value c in the form of
Fig. 1.
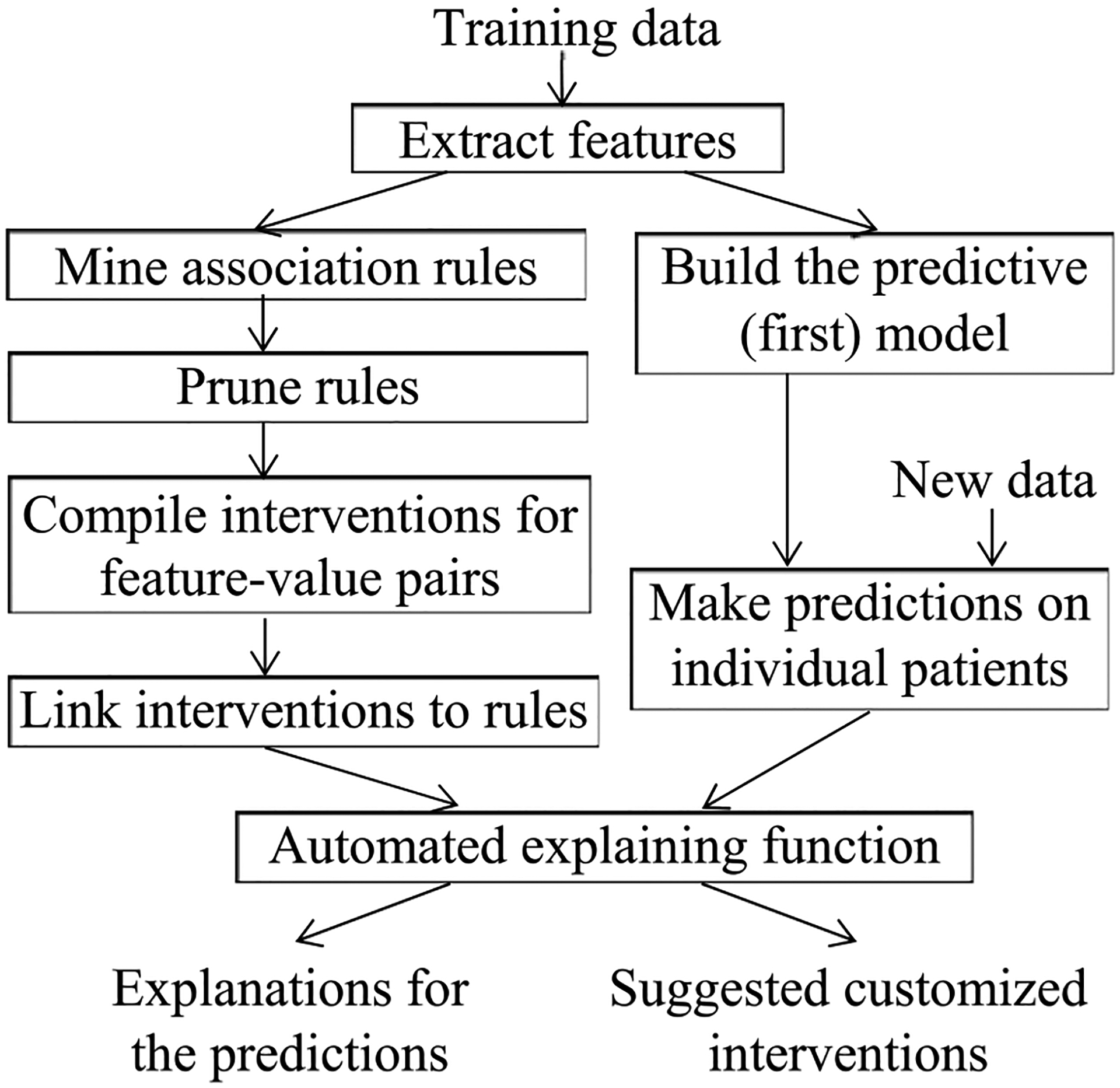
The flow chart of our automated explanation method.
For binary classification of good vs. poor outcomes, c is normally the poor outcome value. r’s and c’s values can vary across rules. Each item bi (1 ≤ i ≤ r) is a feature-value pair (h, w). When w is a value, bi expresses that feature h has value w. When w is a range, bi expresses that h’s value is in w. The rule denotes that when a patient satisfies all of b1, b2, …, and br, the patient’s outcome tends to be c. Here is an example rule to illustrate these relationships:
The patient received ≥20 diagnoses of asthma with (acute) exacerbation in the previous 12 months
AND cumulatively ≥4 systemic corticosteroid medications were ordered for the patient in the previous 12 months
→ the patient will experience asthma hospital visit in the next 12 months.
2). MINING AND PRUNING ASSOCIATION RULES:
Our automated explanation method has five parameters: the lower limit of commonality, the lower limit of confidence, the upper limit number of items allowed on the left hand side of a rule, the confidence difference bound, and the number of top features adopted to form rules. For a specific rule
its commonality gives its coverage in c’s context: among all of the data instances linked to c, the percentage of data instances fulfilling b1, b2, …, and br. Its confidence gives its precision: among all of the data instances fulfilling b1, b2, …, and br, the percentage of data instances linked to c. Our method employs the rules whose commonality is no less than commonality’s lower limit, whose confidence is no less than confidence’s lower limit, and each with at most the allowed upper limit number of items on the rule’s left hand side.
To avoid running into an exceedingly large number of association rules, we employ three techniques to drop rules. First, we drop every more specific rule when there is a more general rule whose confidence is lower by at most the confidence difference bound. Second, if the first model uses too many features, we form rules using just the top few features with the biggest importance values computed by, e.g., XGBoost [22]. Third, a clinician in the automated explaining function’s design team inspects all allowed values and value ranges of these features, and tags those that could be positively correlated with the poor outcome value. Rules are permitted to contain only the tagged feature values and value ranges.
For every feature-value pair item taken to form association rules, a clinician in the automated explaining function’s design team assembles zero or more interventions. We call an item actionable if it connects to one or more interventions. Every rule coming through the rule pruning process is auto-connected to the interventions linking to the actionable items on the rule’s left hand side. We call a rule actionable if it connects to one or more interventions.
3). THE EXPLANATION METHOD:
For every patient the first model predicts to incur a poor outcome value, we explain the prediction by displaying the association rules in the second model with that value on their right hand sides. Each rule presents a reason why the patient is predicted to incur that value. For every actionable rule displayed, we present next to it its linked interventions. From them, the automated explaining function’s user can pinpoint customized interventions appropriate for the patient. Normally, the rules in the second model give common reasons for incurring poor outcomes. Some patients will incur poor outcomes for other reasons. Hence, the second model can explain most, but not all, of the poor outcomes the first model accurately predicts.
G. PARAMETER SETTING
In our experiments, we set parameters in the same way as in our prior automated explanation paper [25]. Each association rule had at most five items on its left hand side. Our UWM model [24] adopted 71 features, all of which were used to form rules. We set confidence’s lower limit to 50%, the same as what we used on Intermountain Healthcare data [25].
For predicting asthma hospital visits, our UWM model [24] reached a higher area under the receiver operating characteristic curve on UWM data than our Intermountain Healthcare model on Intermountain Healthcare data [23]. Our prior automated explanation paper [25] points out that the harder the outcome is to predict, the smaller the lower limit of commonality needs to be. This ensures our automated explanation method can provide explanations for a large fraction of the patients the first model accurately predicts to incur a poor outcome value. Applying this strategy to UWM data, we set commonality’s lower limit to 1%. This lower limit is larger than the value 0.2% we used on Intermountain Healthcare data [25].
To set the confidence difference bound t, we plotted the number of association rules coming through the rule pruning process vs. t. Our prior paper [25] showed that this number first declines quickly as t elevates, and then declines slowly when t grows large enough. We set t’s value at the shift point.
H. DATA ANALYSIS
1). SPLIT OF THE TEST AND TRAINING SETS
We employed the same method used in our prior UWM predictive model paper [24] to split the full data set into the test and training sets. Because the outcome of interest occurred in the next 12 months, our data set contained eight years of effective data (2011–2018) over the nine-year span of 2011–2019. To match future use of our UWM model and our automated explanation method in clinical practice, we used the 2011–2017 data as the training set to mine the association rules employed by our automated explanation method and train our UWM model. We then used the 2018 data as the test set to measure our automated explanation method’s performance.
2). PERFORMANCE METRICS
To measure our automated explanation method’s performance, we employed the same performance metrics in our prior automated explanation paper [25]. One performance metric about our method’s explanation power is: among the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months, the fraction our method could produce explanations for. We calculated the average number of (actionable) rules applying to such a patient. If a patient satisfies all of the items on a rule’s left hand side, we say that the rule applies to the patient.
Our prior automated explanation paper [25] demonstrated that in many cases, several rules applying to a patient differ by only one item on their left hand sides. When a lot of rules apply to a patient, the amount of essential information they contain is normally much less than the number of rules in them. To fully quantify and present the amount of information embedded in the automated explanations offered on the patients, we graphed the following distributions of asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months: the distribution by the number of (actionable) rules applying to a patient, and the distribution by the number of unique actionable items pinpointed on a patient. The latter number refers to the number of unique actionable items contained in all of the rules applying to the patient.
III. RESULTS
A. OUR PATIENT COHORT’S DEMOGRAPHIC AND CLINICAL CHARACTERISTICS
Each data instance pinpoints to an (index year, patient) pair. Tables 2 and 3 in our prior UWM predictive model paper [24] display the demographic and clinical characteristics of our UWM patient cohort during 2011–2017 and 2018 separately. The two sets of characteristics are similar to each other. In 2011–2017, we had 68,244 data instances, 1.74% (1,184) of which linked to asthma hospital visits in the next 12 months. In 2018, we had 14,644 data instances, 1.49% (218) of which linked to asthma hospital visits in the next 12 months. Our prior UWM model paper [24] compared these two sets of characteristics in detail.
B. THE NUMBER OF LEFTOVER ASSOCIATION RULES
We mined 2,400,948 association rules from the training set. Fig. 2 presents the number of leftover rules vs. the confidence difference bound t. This number first declines quickly as t elevates, and then declines slowly when t grows ≥0.15. Correspondingly, we set t’s value to 0.15 and obtained 257,898 leftover rules.
Fig. 2.
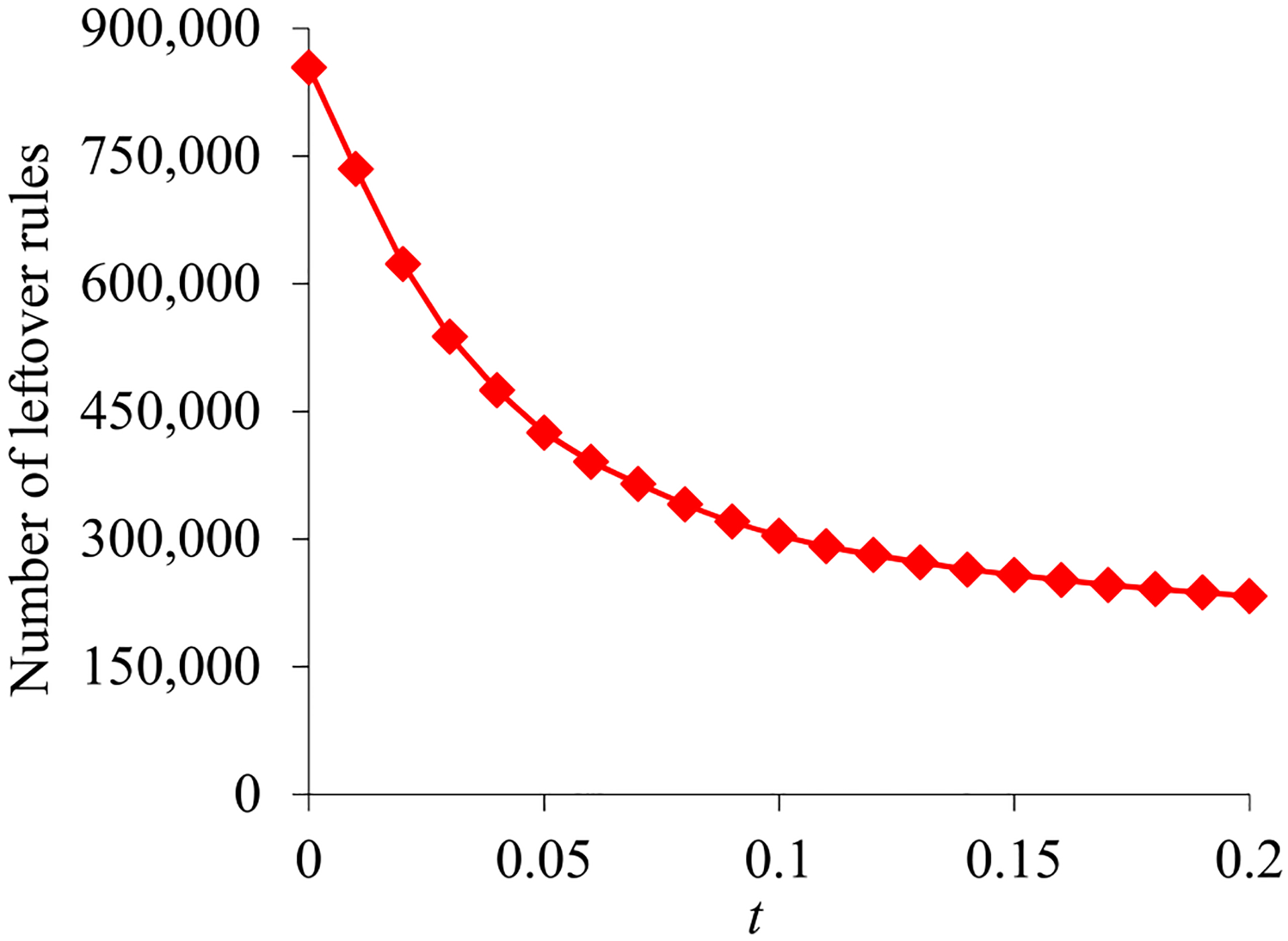
The number of leftover rules vs. the confidence difference bound t.
Our team’s asthma clinical expert (AIM) manually tagged the values and value ranges of the features that could be positively correlated with asthma hospital visits in the next 12 months. After removing the association rules involving other values or value ranges, we obtained 227,621 rules. Each of them was actionable and presented a reason why a patient was predicted to experience asthma hospital visits in the next 12 months.
C. EXEMPLAR ASSOCIATION RULES EMPLOYED BY THE SECOND MODEL
The following five exemplar rules are provided to help concretely explain the association rule concept that is central to the second model:
-
Rule 1: The patient encountered ≥7 ED visits related to asthma in the previous 12 months
→ the patient will experience asthma hospital visit in the next 12 months.
Encountering many ED visits related to asthma signifies bad asthma control. An intervention linked to the item “the patient encountered ≥7 ED visits related to asthma in the previous 12 months” is to employ control strategies to help the patient avoid requiring emergency care.
-
Rule 2: The patient received ≥20 diagnoses of asthma with (acute) exacerbation in the previous 12 months AND cumulatively ≥4 systemic corticosteroid medications were ordered for the patient in the previous 12 months
→ the patient will experience asthma hospital visit in the next 12 months.
Receiving many diagnoses of asthma with (acute) exacerbation signifies bad asthma control. An intervention linked to the item “the patient received ≥20 diagnoses of asthma with (acute) exacerbation in the previous 12 months” is to give the patient suggestions on ways to gain better asthma control.
Consuming lots of systemic corticosteroids signifies bad asthma control. An intervention linked to the item “cumulatively ≥4 systemic corticosteroid medications were ordered for the patient in the previous 12 months” is to advise the patient to better adhere to daily asthma control medications or to better avoid asthma triggers.
-
Rule 3: The patient received ≥28 asthma diagnoses in the previous 12 months
AND the patient had zero outpatient visit in the previous 12 months
→ the patient will experience asthma hospital visit in the next 12 months.
Receiving many asthma diagnoses signifies bad asthma control. An intervention linked to the item “the patient received ≥28 asthma diagnoses in the previous 12 months” is to give the patient suggestions on ways to gain better asthma control.
In many cases, having zero outpatient visit indicates that the patient has no primary care provider. Yet, as part of the asthma management process, an asthma patient is expected to visit the primary care provider from time to time. An intervention linked to the item “the patient had zero outpatient visit in the previous 12 months” is to ensure that the patient has a primary care provider and to encourage the patient to see that provider on a regular basis.
-
Rule 4: The patient received ≥18 primary or principal asthma diagnoses in the previous 12 months
AND the patient incurred ≥5 no shows in the previous 12 months
AND cumulatively ≥10 short-acting beta-2 agonist medications were ordered for the patient in the previous 12 months
→ the patient will experience asthma hospital visit in the next 12 months.
Receiving many primary or principal asthma diagnoses signifies bad asthma control. An intervention linked to the item “the patient received ≥18 primary or principal asthma diagnoses in the previous 12 months” is to give the patient suggestions on ways to gain better asthma control.
Incurring a large number of no shows is correlated with having bad outcomes. An intervention linked to the item “the patient incurred ≥5 no shows in the previous 12 months” is to provide social resources to address social chaos contributing to missed appointments.
Consuming lots of short-acting beta-2 agonists signifies bad asthma control. An intervention linked to the item “cumulatively ≥10 short-acting beta-2 agonist medications were ordered for the patient in the previous 12 months” is to advise the patient to better adhere to daily asthma control medications or to better avoid asthma triggers.
-
Rule 5: The patient is black
AND the patient received ≥28 asthma diagnoses in the previous 12 months
AND the patient’s mean respiratory rate in the previous 12 months is >16.89
AND the patient was a smoker according to the most recent record
→ the patient will experience asthma hospital visit in the next 12 months.
Black people are inclined to have worse asthma outcomes than other people in the U.S. Having a high respiratory rate signifies bad asthma control. An intervention linked to the item “the patient’s mean respiratory rate in the previous 12 months is > 16.89” is to improve asthma control to decrease respiratory rate.
Smoking is a strong trigger of asthma symptoms. An intervention linked to the item “the patient was a smoker according to the most recent record” is to suggest the patient to stop smoking.
D. OUR AUTOMATED EXPLANATION METHOD’S PERFORMANCE
As shown in our paper [24], our UWM model reached on the test set an area under the receiver operating characteristic curve of 0.902, an accuracy of 90.6%, a sensitivity of 70.2%, a specificity of 90.9%, a positive predictive value of 10.5%, and a negative predictive value of 99.5%.
Our automated explanation method was evaluated on the test set. Our method explained the predictions for 134 (87.6%) of the 153 asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months. For an average such patient, our method gave 5296.58 explanations, each from one rule, and pinpointed 26.62 unique actionable items.
For the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months, Fig. 3 and 4 plot their distribution by the number of actionable rules applying to a patient. This distribution is markedly skewed towards the left and has a long tail. As the number of rules applying to a patient grows, the number of patients, to each of whom so many rules apply, tends to fall non-monotonically. The maximum number of rules applying to a patient is quite large: 35,484. Nevertheless, there is just one patient to whom so many rules apply.
Fig. 3.
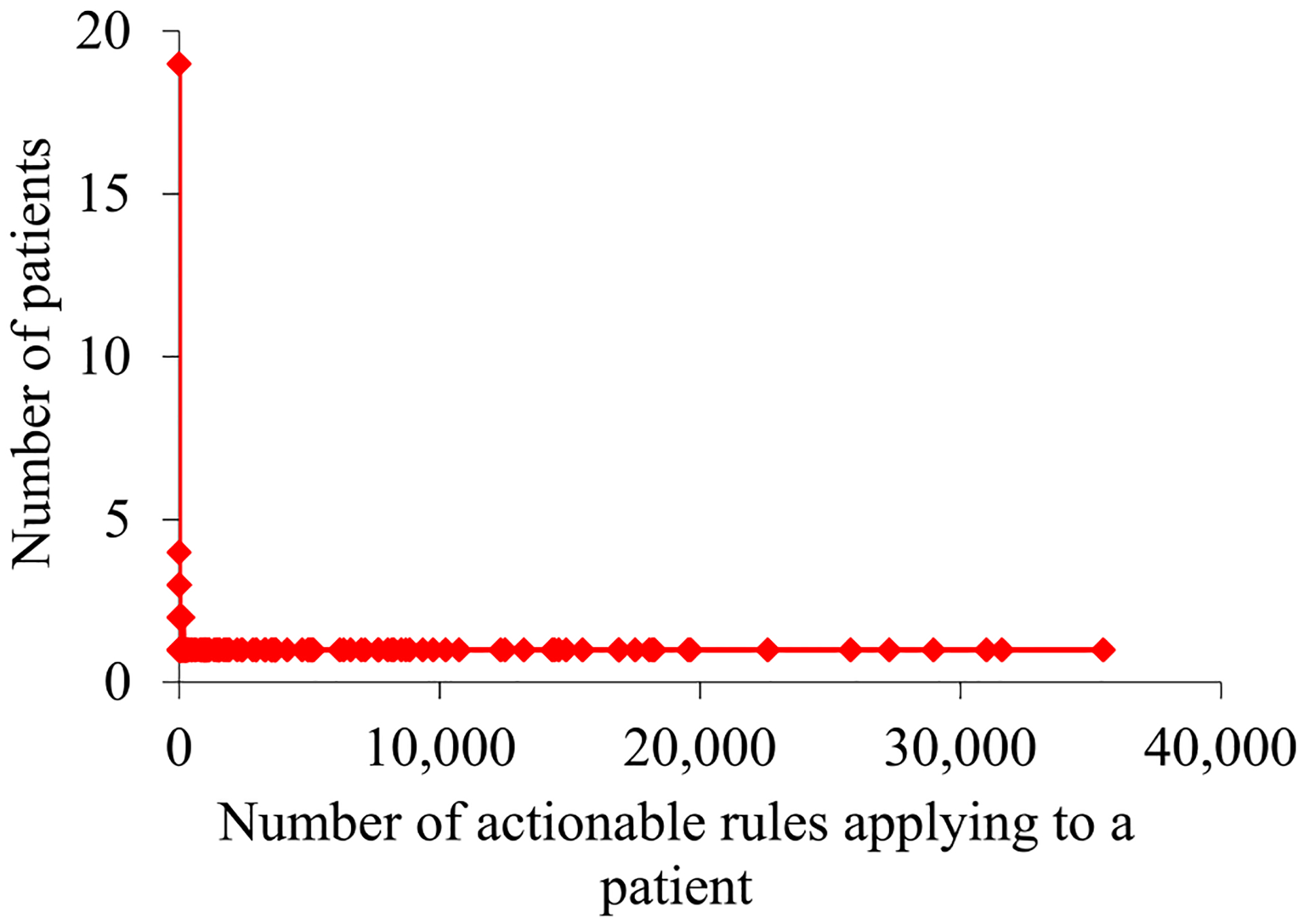
For the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months, their distribution by the number of actionable rules applying to a patient.
Fig. 4.
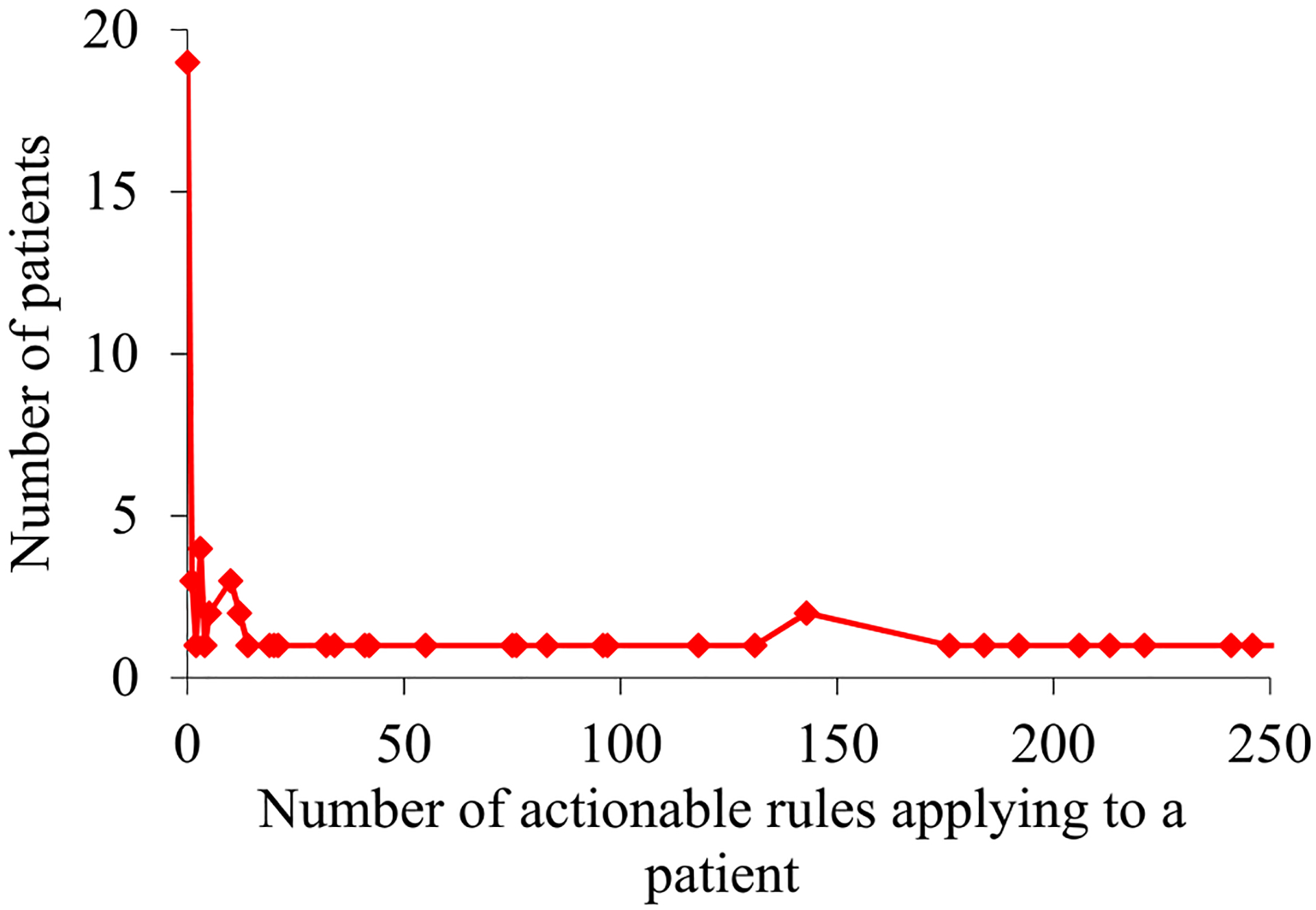
For the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months, their distribution by the number of actionable rules applying to a patient when this number is ≤250.
For the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months, Fig. 5 plots their distribution by the number of unique actionable items pinpointed on a patient. The maximum number of unique actionable items pinpointed on a single patient is 47, much smaller than the maximum number of actionable rules applying to a patient. As described in our prior automated explanation paper [25], several actionable items pinpointed on a patient often relate to the same intervention.
Fig. 5.
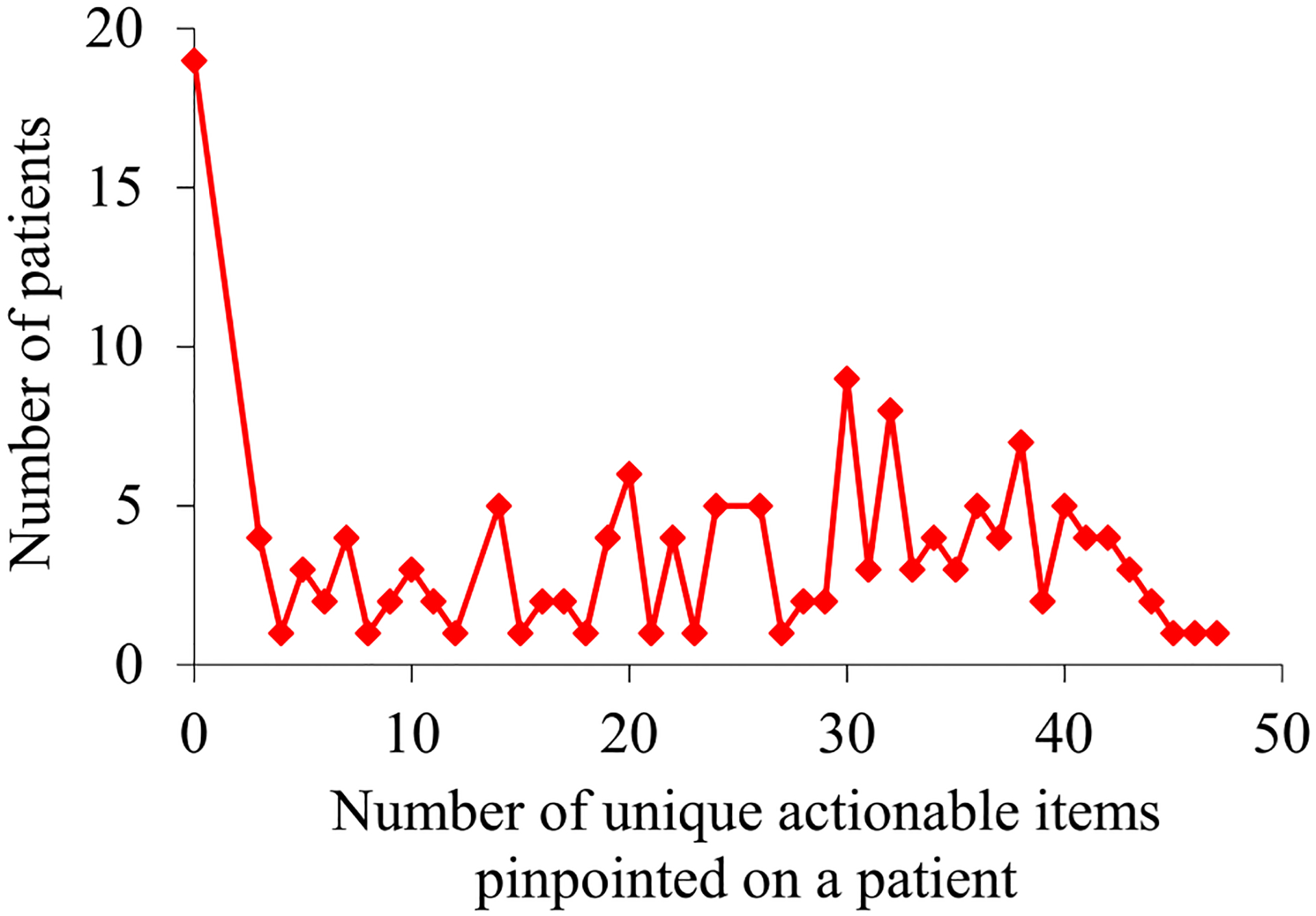
For the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months, their distribution by the number of unique actionable items pinpointed on a patient.
Our automated explanation method produced explanations for 147 (67.4%) of the 218 asthma patients who would experience asthma hospital visits in the next 12 months.
IV. DISCUSSION
A. MAIN RESULTS
The results in this paper are similar to those published in our prior automated explanation paper [25]. For predicting asthma hospital visits, our automated explanation method had satisfactory generalizability to UWM. Notably, our method explained the predictions for 87.6% (134/153) of the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months. This percentage is sufficiently high to allow deployment of our automated explanation method for routine clinical use, and is similar to the corresponding percentage (89.68%) our prior automated explanation paper [25] reported on Intermountain Healthcare data. After further optimization to raise its accuracy, our UWM model integrated with our automated explanation method could be deployed to help direct use of asthma care management resources to improve patient outcomes and cut healthcare costs.
Our automated explanation method produced explanations for 67.4% (147/218) of the asthma patients who would experience asthma hospital visits in the next 12 months. This percentage is less than our method’s 87.6% (134/153) success rate in explaining the predictions for the asthma patients our UWM model accurately predicted to experience asthma hospital visits in the next 12 months. As explained in our prior automated explanation paper [25], this pattern is likely due to correlation among distinct models’ computational results. If a patient’s future outcome is easier to predict, it is likely to be also easier to explain.
Overall, this work provides a crucial, rational, clinical explanation structure for our UWM model’s predictions on asthma outcomes, which also suggests customized interventions. This is vital for our UWM model’s acceptability by clinical users and future clinical adoption.
B. LIMITATIONS
Below are two limitations of this study that can direct future work:
This study assessed our automated explanation method’s generalizability to a single healthcare system for one outcome of a single chronic disease. It would be useful to assess our automated explanation method’s generalizability to other healthcare systems, diseases, and outcomes [33].
Our current automated explanation method is developed for non-deep-learning machine learning algorithms and structured data. It would be useful to extend our method to apply to deep learning models created on longitudinal data [33], [34].
V. RELATED WORK
Many other research groups have developed a range of methods to automatically produce explanations for machine learning models’ predictions, as recently surveyed by Guidotti et al. [35]. Such explanations are typically not presented as clear decision rules. None of these published methods could automatically suggest customized interventions. Also, many of these published methods lower the prediction accuracy, and/or are developed for a fixed machine learning algorithm. In comparison, our automated explanation method produces rule-style explanations for any machine learning model’s predictions made on tabular data, and simultaneously suggests customized interventions without lowering the prediction accuracy. Rule-style explanations are easier to understand and can be used to suggest customized interventions more directly than other types of explanations. This will facilitate adoption and clinical implementation of the predictive model by providing some logic and rationale that clinicians and other users can understand.
VI. CONCLUSIONS
In the first assessment of its generalizability to an academic healthcare system, our automated explanation method had satisfactory generalizability to UWM for predicting asthma hospital visits. After further optimization to raise its accuracy, our UWM model integrated with our automated explanation method could be deployed to help direct use of asthma care management resources to improve patient outcomes and cut healthcare costs.
ACKNOWLEDGMENT
We thank Katy Atwood for helping retrieve the UWM data set.
Gang Luo was partially supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number R01HL142503. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Yao Tong did the work at the University of Washington when she was a visiting PhD student there.
LIST OF ABBREVIATIONS
- ED
Emergency department
- ICD-9
International Classification of Diseases, Ninth Revision
- ICD-10
International Classification of Diseases, Tenth Revision
- UWM
University of Washington Medicine
- XGBoost
Extreme gradient boosting
Biographies

YAO TONG received the B.S. degree in electronic information engineering from Hohai University, Nanjing, Jiangsu Province, P.R. China, in 2015 and the M.S degree in software engineering from Hohai University, Nanjing, Jiangsu Province, P.R. China, in 2017. She is currently pursuing the Ph.D. degree in software engineering at Zhengzhou University, Zhengzhou, Henan Province, P.R. China.
Since 2019, she has been a Visiting Student in the Department of Biomedical Informatics and Medical Education at the University of Washington, Seattle, WA, USA. Her research interests include machine learning, computer vision, database systems, and clinical informatics.

AMANDA MESSINGER received the B.A. degree in medical anthropology from Cornell University, Ithaca, NY, USA, in 2007, and the M.D. degree from the University of Washington, Seattle, WA, USA, in 2012. She completed Pediatrics Residency at Northwestern University, Chicago, IL, USA, in 2015, and Pediatric Pulmonology Fellowship in 2017. She received the M.S. degree in clinical informatics from the University of Washington, Seattle, WA, USA, in 2020, and is currently board certified in Pediatrics and Pediatric Pulmonology.
She is an Assistant Professor in the School of Medicine at the University of Colorado, Denver and the Breathing Institute at Children’s Hospital Colorado in Aurora, CO, USA. Her research interests include pediatric asthma, applied clinical informatics, machine learning, clinical prediction, clinical decision support, and database systems.

GANG LUO received the B.S. degree in computer science from Shanghai Jiaotong University, Shanghai, P.R. China, in 1998, and the PhD degree in computer science from the University of Wisconsin-Madison, Madison, WI, USA, in 2004.
From 2004 to 2012, he was a Research Staff Member at IBM T.J. Watson Research Center, Hawthorne, NY, USA. From 2012 to 2016, he was an Assistant Professor in the Department of Biomedical Informatics at the University of Utah, Salt Lake City, UT, USA. He is currently an Associate Professor in the Department of Biomedical Informatics and Medical Education at the University of Washington, Seattle, WA, USA. He is the author of over 70 papers. His research interests include machine learning, information retrieval, database systems, and health informatics.
Footnotes
CONFLICTS OF INTEREST
None declared.
REFERENCES
- [1].Centers for Disease Control and Prevention. Most recent national asthma data. Accessed: Aug. 20, 2020 [Online]. Available: https://www.cdc.gov/asthma/most_recent_national_asthma_data.htm
- [2].Lieu TA, Quesenberry CP, Sorel ME, Mendoza GR, and Leong AB, “Computer-based models to identify high-risk children with asthma,” Am. J. Respir. Crit. Care Med, vol. 157, no. 4 Pt 1, pp. 1173–1180, 1998. [DOI] [PubMed] [Google Scholar]
- [3].Mays GP, Claxton G, and White J, “Managed care rebound? Recent changes in health plans’ cost containment strategies,” Health Aff, vol. 23, Suppl. Web Exclusives, pp. W4–427–W4–436, 2004. [DOI] [PubMed] [Google Scholar]
- [4].Caloyeras JP, Liu H, Exum E, Broderick M, and Mattke S, “Managing manifest diseases, but not health risks, saved PepsiCo money over seven years,” Health Aff, vol. 33, no. 1, pp. 124–131, 2014. [DOI] [PubMed] [Google Scholar]
- [5].Greineder DK, Loane KC, and Parks P, “A randomized controlled trial of a pediatric asthma outreach program,” J. Allergy Clin. Immunol, vol. 103, no. 3 Pt 1, pp. 436–440, 1999. [DOI] [PubMed] [Google Scholar]
- [6].Kelly CS, Morrow AL, Shults J, Nakas N, Strope GL, and Adelman RD, “Outcomes evaluation of a comprehensive intervention program for asthmatic children enrolled in Medicaid,” Pediatrics, vol. 105, no. 5, pp. 1029–1035, 2000. [DOI] [PubMed] [Google Scholar]
- [7].Axelrod RC, Zimbro KS, Chetney RR, Sabol J, and Ainsworth VJ, “A disease management program utilizing life coaches for children with asthma”. J. Clin. Outcomes Manag, vol. 8, no. 6, pp. 38–42, 2001. [Google Scholar]
- [8].Axelrod RC and Vogel D, “Predictive modeling in health plans,” Disease Management & Health Outcomes, vol. 11, no. 12, pp. 779–787, 2003. [Google Scholar]
- [9].Loymans RJ, Honkoop PJ, Termeer EH, Snoeck-Stroband JB, Assendelft WJ, Schermer TR, Chung KF, Sousa AR, Sterk PJ, Reddel HK, Sont JK, Ter Riet G, “Identifying patients at risk for severe exacerbations of asthma: development and external validation of a multivariable prediction model,” Thorax, vol. 71, no. 9, pp. 838–846, 2016. [DOI] [PubMed] [Google Scholar]
- [10].Schatz M, Cook EF, Joshua A, and Petitti D, “Risk factors for asthma hospitalizations in a managed care organization: development of a clinical prediction rule,” Am. J. Manag. Care, vol. 9, no. 8, pp. 538–547, 2003. [PubMed] [Google Scholar]
- [11].Eisner MD, Yegin A, and Trzaskoma B, “Severity of asthma score predicts clinical outcomes in patients with moderate to severe persistent asthma,” Chest, vol. 141, no. 1, pp. 58–65, 2012. [DOI] [PubMed] [Google Scholar]
- [12].Sato R, Tomita K, Sano H, Ichihashi H, Yamagata S, Sano A, Yamagata T, Miyara T, Iwanaga T, Muraki M, and Tohda Y, “The strategy for predicting future exacerbation of asthma using a combination of the Asthma Control Test and lung function test,” J. Asthma, vol. 46, no. 7, pp. 677–682, 2009. [DOI] [PubMed] [Google Scholar]
- [13].Osborne ML, Pedula KL, O’Hollaren M, Ettinger KM, Stibolt T, Buist AS, and Vollmer WM, “Assessing future need for acute care in adult asthmatics: the Profile of Asthma Risk Study: a prospective health maintenance organization-based study,” Chest, vol. 132, no. 4, pp. 1151–1161, 2007. [DOI] [PubMed] [Google Scholar]
- [14].Miller MK, Lee JH, Blanc PD, Pasta DJ, Gujrathi S, Barron H, Wenzel SE, Weiss ST; TENOR Study Group, “TENOR risk score predicts healthcare in adults with severe or difficult-to-treat asthma,” Eur. Respir. J, vol. 28, no. 6, pp. 1145–1155, 2006. [DOI] [PubMed] [Google Scholar]
- [15].Peters D, Chen C, Markson LE, Allen-Ramey FC, and Vollmer WM, “Using an asthma control questionnaire and administrative data to predict health-care utilization,” Chest, vol. 129, no. 4, pp. 918–924, 2006. [DOI] [PubMed] [Google Scholar]
- [16].Yurk RA, Diette GB, Skinner EA, Dominici F, Clark RD, Steinwachs DM, and Wu AW, “Predicting patient-reported asthma outcomes for adults in managed care,” Am. J. Manag. Care, vol. 10, no. 5, pp. 321–328, 2004. [PubMed] [Google Scholar]
- [17].Loymans RJB, Debray TPA, Honkoop PJ, Termeer EH, Snoeck-Stroband JB, Schermer TRJ, Assendelft WJJ, Timp M, Chung KF, Sousa AR, Sont JK, Sterk PJ, Reddel HK, and Ter Riet G, “Exacerbations in adults with asthma: a systematic review and external validation of prediction models,” J. Allergy Clin. Immunol. Pract, vol. 6, no. 6, pp. 1942–1952.e15, 2018. [DOI] [PubMed] [Google Scholar]
- [18].Lieu TA, Capra AM, Quesenberry CP, Mendoza GR, and Mazar M, “Computer-based models to identify high-risk adults with asthma: is the glass half empty or half full?,” J. Asthma, vol. 36, no. 4, pp. 359–370, 1999. [DOI] [PubMed] [Google Scholar]
- [19].Schatz M, Nakahiro R, Jones CH, Roth RM, Joshua A, and Petitti D, “Asthma population management: development and validation of a practical 3-level risk stratification scheme,” Am. J. Manag. Care, vol. 10, no. 1, pp. 25–32, 2004. [PubMed] [Google Scholar]
- [20].Grana J, Preston S, McDermott PD, and Hanchak NA, “The use of administrative data to risk-stratify asthmatic patients,” Am. J. Med. Qual, vol. 12, no. 2, pp. 113–119, 1997. [DOI] [PubMed] [Google Scholar]
- [21].Forno E, Fuhlbrigge A, Soto-Quirós ME, Avila L, Raby BA, Brehm J, Sylvia JM, Weiss ST, and Celedón JC, “Risk factors and predictive clinical scores for asthma exacerbations in childhood,” Chest, vol. 138, no. 5, pp. 1156–1165, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen T and Guestrin C, “XGBoost: A scalable tree boosting system,” in Proc. ACM SIGKDD Int. Conf. Knowl. Discovery Data Mining, 2016, pp. 785–794. [Google Scholar]
- [23].Luo G, He S, Stone BL, Nkoy FL, and Johnson MD, “Developing a model to predict hospital encounters for asthma in asthmatic patients: secondary analysis,” JMIR Med. Inform, vol. 8, no. 1, Art. no. e16080, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tong Y, Messinger AI, Wilcox AB, Mooney SD, Davidson GH, Suri P, and Luo G, “Building a model to forecast asthmatic patients’ future asthma hospital encounters in an academic healthcare system: secondary analysis.” http://pages.cs.wisc.edu/~gangluo/predict_hospital_use_for_asthma_UW.pdf. [DOI] [PMC free article] [PubMed]
- [25].Luo G, Johnson MD, Nkoy FL, He S, and Stone BL, “Automatically explaining machine learning prediction results on asthma hospital visits in asthmatic patients: secondary analysis.” http://pages.cs.wisc.edu/~gangluo/explain_predict_hospital_use_for_asthma.pdf. [DOI] [PMC free article] [PubMed]
- [26].Liu LL, Forgione DA, and Younis MZ, “A comparative analysis of the CVP structure of nonprofit teaching and for-profit non-teaching hospitals,” J. Health Care Finance, vol. 39, no. 1, pp. 12–38, 2012. [PubMed] [Google Scholar]
- [27].Desai JR, Wu P, Nichols GA, Lieu TA, and O’Connor PJ, “Diabetes and asthma case identification, validation, and representativeness when using electronic health data to construct registries for comparative effectiveness and epidemiologic research,” Med. Care, vol. 50, Suppl., pp. S30–S35, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wakefield DB and Cloutier MM, “Modifications to HEDIS and CSTE algorithms improve case recognition of pediatric asthma,” Pediatr. Pulmonol, vol. 41, no. 10, pp. 962–971, 2006. [DOI] [PubMed] [Google Scholar]
- [29].Luo G, “Automatically explaining machine learning prediction results: a demonstration on type 2 diabetes risk prediction,” Health Inf. Sci. Syst, vol. 4, no. 2, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu B, Hsu W, and Ma Y, “Integrating classification and association rule mining,” in Proc. 4th Int. Conf. Knowl. Discovery Data Mining, 1998, pp. 80–86. [Google Scholar]
- [31].Thabtah FA, “A review of associative classification mining,” Knowledge Eng. Rev, vol. 22, no. 1, pp. 37–65, 2007. [Google Scholar]
- [32].Fayyad UM and Irani KB, “Multi-interval discretization of continuous-valued attributes for classification learning,” in Proc. IJCAI, 1993, pp. 1022–1029. [Google Scholar]
- [33].Luo G, Stone BL, Koebnick C, He S, Au DH, Sheng X, Murtaugh MA, Sward KA, Schatz M, Zeiger RS, Davidson GH, and Nkoy FL, “Using temporal features to provide data-driven clinical early warnings for chronic obstructive pulmonary disease and asthma care management: protocol for a secondary analysis,” JMIR Res. Protoc, vol. 8, no. 6, Art. no. e13783, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Luo G, “A roadmap for semi-automatically extracting predictive and clinically meaningful temporal features from medical data for predictive modeling,” Glob. Transit, vol. 1, pp. 61–82, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guidotti R, Monreale A, Ruggieri S, Turini F, Giannotti F, and Pedreschi D, “A survey of methods for explaining black box models,” ACM Comput. Surv, vol. 51, no. 5, Art. no. 93, 2019. [Google Scholar]


