ABSTRACT
Aims:
Complete denture patients have a plethora of microorganisms inhabiting their complete dentures. Some bacteria are capable of causing systemic illness such as aspiration pneumonia and infective endocarditis. Hence, detection as well as the categorization of biofilms, which form on the denture surface is vital in the study of denture biofilm-associated local and systemic diseases. This study aimed at the detection and categorization of biofilm-forming Staphylococcus aureus, Viridans streptococcus, Klebsiella pneumoniae, and Escherichia coli isolated from complete dentures and visualization of biofilms using scanning electron microscopy (SEM).
Materials and Methods:
Thirty complete denture patients were selected for the study and swabs were collected from their complete denture surfaces. Isolation of the bacteria was done using selective media and confirmed using biochemical tests and 16SrRNA sequencing. The bacteria were subjected to biofilm assays via Microtiter plate assay. The biofilm-forming bacteria were categorized as weak, moderate, and strong biofilm formers based on optical density (OD) values. As a visual confirmation of the biofilms, scanning electron microscopic (SEM) images were taken for each of the strong biofilm-forming bacteria. Descriptive statistical analysis was carried out with the help of Statistical Package for the Social Sciences (SPSS) statistical package version 20.0.
Results:
The average OD of S. aureus was 1. 333±0. 015 and the average OD of V. streptococcus species was 1. 304 ± 0.023. The average OD value of K. pneumoniae was 0.8 ± 0.012 and the average value of E. coli was 1.014 ± 0.01.
Conclusions:
The study of biofilms especially the strongly biofilm formers is very useful to understand the potential pathogenic effect of biofilms. These biofilms cause the systemic spread of the planktonic bacteria which could lead to systemic diseases that are resistant to conventional treatment. This could be due to the inherent nature of the biofilm to offer drug resistance to existing antibacterial agents.
KEYWORDS: Biofilms, dental deposits, gram-negative bacteria, gram-positive bacteria, prosthesis-related infections
INTRODUCTION
The concept of biofilm structure is based on the pioneering studies done by the Bozeman Montana Center for Biofilm Engineering. They showed the biofilm as a thin basal layer on the substratum which was in contact with the acquired pellicle. Biofilms have columnar, mushroom-shaped multifactorial extensions into the lumen of the solution, separated by regions (“channels”) seemingly empty or filled with extracellular polysaccharide (EPS).[1] As the biofilm matures, lack of nutrients and space leads to detachment of bacteria from the biofilm colony.[2] The released bacteria may be transported to newer locations to restart the biofilm formation. This could lead to the initiation of metastatic infections like aspiration pneumonia and endocarditis.[1,3] With the advancement of microscopy, this surface-associated well-organized, cooperating communities of microorganisms came to be referred to as a “Biofilm,” coined by Bill Costerton in 1978.[1] Extracellular polymeric substance can also be called as glycocalyx, slime, or capsule.[1] Potential mechanisms by which biofilms could prove to be pathogenic have been proposed. These include (1) attachment to a solid surface, (2) “Division of labor” thereby increasing metabolic efficiency of the community, (3) evading host defenses such as phagocytosis, (4) repository of high density of microorganisms; exchange of genes that can result in emergence of more virulent strains of microorganisms, (5) production of large concentration of toxins, (6) protection from antimicrobial agents, and (7) detachment of microbial aggregates thereby transmitting microorganisms to other sites.[1,2,4]
Dentures create an appropriate habitat for a variety of microorganisms, especially bacteria.[5] These bacteria form biofilms on the denture surface, which mature and dissipate free planktonic bacteria into the body system. Hence, these biofilms have to be detected, studied, and categorized to know their pathogenic potential as well as their potential to provide drug resistance to the bacteria via biofilm formation. This study aimed to detect and categorize biofilms of selective bacteria isolated from complete denture patients and visualization of the biofilms by scanning electron microscopy (SEM) which would be helpful in adopting oral and denture disinfection measures for better oral and systemic health.
MATERIALS AND METHODS
The study was conducted at the Department of Prosthodontics, Annamalai University between June 2016 to December 2019 after obtaining the approval of the institutional ethics committee (Approval no.: IHEC/0142/2016). The subjects for the study were explained about the complete details of the study. The details are as follows: (1) purpose of the study, (2) sample collection method, (3) tests to be conducted with the sample, and (4) maintenance of patient privacy. This was followed by the procurement of informed consent from the patient.
SELECTION OF SUBJECTS
Inclusion criteria
Healthy completely edentulous patients with no history of previous systemic disease and wearing dentures for a minimum of 6 months were selected for the study.
Exclusion criteria
Patients with chronic systemic diseases with a history of medications.
Patients who have used local or systemic antibiotics within 30 days prior to sample collection.
Patients who have a smoking habit.
Hence, complete edentulous patients (n = 30 patients) who are wearing complete dentures at least for the past 6 months were selected for the study.
SAMPLE COLLECTION
A sterile swab (Hiculture Transport Swab, Himedia, Mumbai, India) was used to collect samples from the patient’s oral cavity as well as from all over the complete denture’s surface. Before the sample collection, the patient was instructed to wear the denture as well as not to rinse the mouth for at least 3 h. The patient’s denture surfaces, namely occlusal surfaces, tissue surfaces as well as polished surfaces were swabbed. After swabbing the denture surfaces, the palatal, buccal, and tonsillar surfaces of the intraoral cavity as well as the dorsal/lingual surfaces of the tongue were swabbed. The entire swabbing procedure was done by means of a single swab to avoid dilution of the sample. Then the swab was subjected to microbiological investigation in the laboratory.
ISOLATION OF BACTERIA
Selective media [Table 1] were prepared and sterilized using autoclave and poured in petri plates before isolation of the bacteria.
Table 1.
Details of selective media
| Bacteria | Selective media used | Lot number |
|---|---|---|
| Viridians streptococcus species | Mutans-Sanguis Agar | 0000237969 |
| Staphylococcus aureus | Mannitol salt agar (HiChrome Staph Agar) | 0000112805 |
| Klebsiella pneumoniae | HiChrome Klebsiella Selective Agar | 0000219388 |
| Escherichia coli | Eosin Methylene Blue HiChrome E. coli Agar) | 00001123036 |
PROCEDURE
About 10 µL of each sample was inoculated in sterile selective media plates by spread plate using Autoclavable L-shaped spreader (Himedia) and incubated at 37°C for 24 h initially. The plates were observed for changes until 48 h.[6] Based on colony morphology, the colonies were selected and inoculated on a sterile nutrient agar plate by quadrant streaking. The colony morphology for the selection of bacterial colonies were as follows: V. streptococcus species (white colony, mucoid with effervescence),Staphylococcus aureus (yellow colony: dotted, convex or spherical), K. pneumoniae (Purple colony), Escherichia coli (colonies with pink/violet with mucoid texture and/or Green metallic sheen). Around 10 colonies were detected and pure isolates were obtained and further Gram stained.[6]
GRAM STAINING
The pure isolate was smeared in a slide and stained using Gram stain Kit (Himedia Mumbai, India, Lot number: 0000187211). The stained slide was observed under 100x magnification oil immersion objective using Bright Field Microscope to study the morphology.[6] Based on similar characteristics (Gram-positive, Gram-negative, cocci, bacilli, single, cluster, chain) 10 colonies each of the bacteria were selected and further biochemical tests were done to ascertain the microorganism.
BIOCHEMICAL TEST
Biochemical tests were selected based on the characters identified from selective media and Gram staining. Tests namely Indole, Methyl Red, Voges Proskauer, Citrate utilization, Triple sugar iron agar (for K. pneumoniae and E. coli) and Carbohydrate fermentation tests, blood agar, Catalase, Coagluase test (for V. streptococcal species and S. aureus) were used.[6]
CONFIRMATION OF IDENTIFICATION OF MICROBIAL CULTURE USING 16s rRNA-BASED MOLECULAR TECHNIQUE
Steps involved
-
a.
Genomic deoxyribonucleic acid (DNA) was isolated and quantity was measured using Nano Drop Spectrophotometer 2000/2000c (Thermo-Fisher Scientific, USA) and the quality was determined using 2% agarose gel. A single band of high-molecular-weight DNA was observed.[7]
-
b.
16S rRNA (ribosomal ribonucleic acid) gene was amplified by forward and reverse 16sRNA primers. A single polymerase chain reaction (PCR) amplicon was observed using Agarose gel.
-
c.
The purified PCR amplicon was subjected to forward and reverse DNA sequencing reaction of PCR amplicon was carried by Bigdye Terminator (BDT) v3. 1 Cycle sequencing kit on Applied Biosystems (ABI) 3730xl Genetic Analyzer and the Consensus sequence of 16S rRNA gene was generated.[8,9]
-
d.
16S rRNA gene sequence was used to carry out basic local alignment search tool (BLAST) with the database of the National Centre for Biotechnogy Information (NCBI) GenBank database. Based on maximum identity score sequences were selected and alignment done by using software Clustal W. By using molecular evolutionary genetic analysis (MEGA 7)distance matrix and phylogenetic tree were constructed.[10]
Hence, after the isolation of the respective bacteria, they were subjected to biochemical tests and 16SrRNA sequencing [Flowchart 1] and 10 isolates each of S. aureus, V. streptococcus, K. pneumoniae, and E. coli were isolated. They were subjected to microtiter plate assay for biofilm quantification.
Flowchart 1.
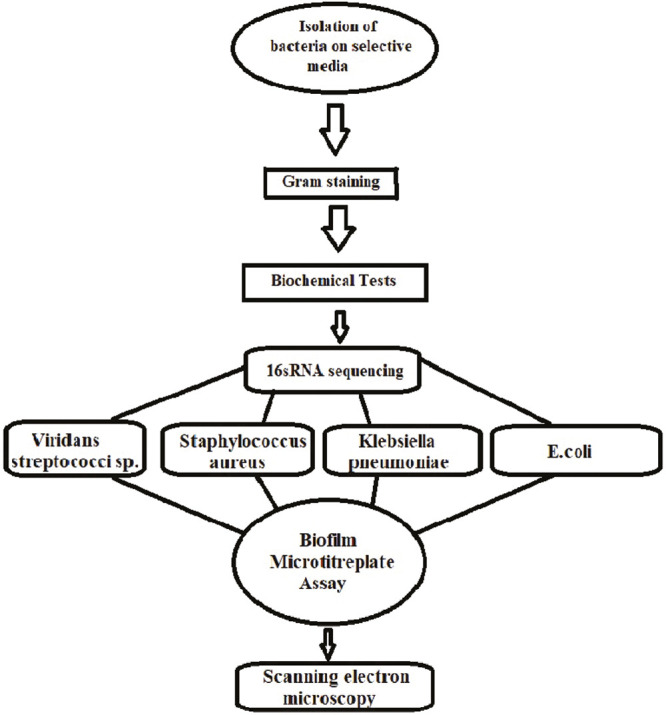
Sequence of procedures done
MICROTITER PLATE ASSAY FOR BIOFILM QUANTIFICATION
Biofilms were formed in pre-sterilized 24 well flat bottom polystyrene microtiter plates. Briefly, a 10 µL (microliter) of cell suspension having 0.5 optical density (OD) 600 was inoculated in 190 µL TSB (Trypticase soy broth) medium in each well and 200 µL of autoclaved distilled water was added in the peripheral wells as controls [Figure 1A]. Then microtiter plates were incubated for 16 h at 37oC. After aspiration of planktonic cells, biofilms were fixed with 99% methanol (used as fixer). Plates were washed twice with phosphate buffer saline and air-dried.[11] The phosphate buffer saline was used to maintain pH, ion concentration, and osmolarity.
Figure 1.
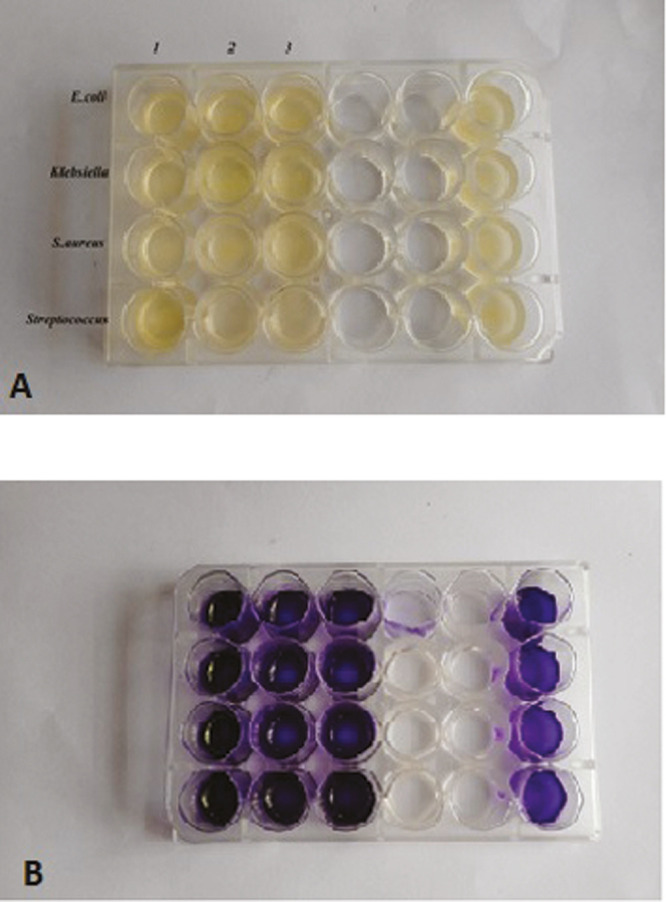
(A) Microtiter plate with trypticase soy broth. (B) Microtiter plate with crystal violet staining
Then, 200 μL of crystal violet solution (0.2%) was added to all the wells as a dye for the quantification of biofilm mass [Figure 1B]. After 5min, the excess crystal violet was removed and plates were washed twice and air dried. Finally, the cell-bound crystal violet was dissolved by using a dissolving agent which is 33% acetic acid. Biofilm growth was monitored in terms of OD 570nm (nanometer) using microprocessor photocolorimetry (Deep Vision, Model 1312,ESICO).[12,13]
The optical densities of specimens (OD) and negative controls (ODavg) were obtained. From these values the optical density cut-off (ODcut) was calculated using the following formula:[11]

Hence, 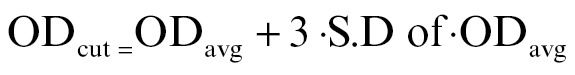 .
.
The criteria for the classification of the biofilms as nonbiofilm former, weak-biofilm former, moderate biofilm former, and strong biofilm former is illustrated in Table 2.
Table 2.
Classification criteria for biofilms along with optical densities obtained in this study
| Criteria for classification | Optical density values |
|---|---|
| OD ≤ ODcut = Nonbiofilm former (NBF) | OD is < 0.37 |
| ODcut < OD ≤ 2 × ODcut = Weak biofilm-former (WBF) | 0.38< OD ≤ 0.74 |
| 2 ODcut < OD ≤ 4 × adequate = Moderate biofilm-former (MBF) | 0.74 < OD ≤ 1.48 |
| OD >4 × ODcut = Strong biofilm-former. | OD >1.48 |
SCANNING ELECTRON MICROSCOPE STUDY
The biofilms were grown overnight on a sterilized acrylic strip of length and breadth of 8 mm x 8 mm and thickness of 2 mm. The samples were then rinsed in 0.1 M buffered sodium cacodylate and dehydrated by serial transfers of ethyl alcohol.[14] After 24 h at room temperature, the specimens were mounted on metal stubs, coated with a layer of gold under vacuum by sputter coating machine (JEOL, JPC 1600, JEOL companies, Japan) [Figure 2]. After the gold coating the specimens are visualized by scanning electron microscope (JEOL, JEOL Ltd, Tokyo, Japan) operated at 15Kilovolts.
Figure 2.

Gold sputter coating of the specimens.
RESULTS
In this study, ODavg of negative control was 0.32 ±0.02, Hence, ODcut was found to be 0.37. After the determination ODavg and ODcut, the ODs of the specimens were subjected to the classification according to the criteria listed in Table 2.
It was found that among 10 isolates of S. aureus 10% were weak biofilm formers, 30% were moderate biofilm formers, whereas 60% were strong biofilm formers having OD of more than 1.48. None of the S. aureus isolates were nonbiofilm formers [Table 3].
Table 3.
Biofilm optical density values for Staphylococcus aureus, Viridans streptococcus species, Klebsiella pneumoniae, and Escherichia coli
| Bacteria | Optical density (mean ± standard deviation) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Staphylococcus aureus | 0.57 (±0.31) | 0.77 (±0.25) | 1.11 (±0.04) | 1.48 (±0.01) | 1.50 (±0.27) | 1.52 (±0.09) | 1.56 (±0.09) | 1.57 (±0.10) | 1.59 (±0.39) | 1.66 (±0.19) |
| Viridans streptococcus species | 0.85 (±0.20) | 1.51 (±0.23) | 1.52 (±0.26) | .95 (±0.28) | 1.54 (± 0.38) | 1.50 (±0.52) | 1.50 (±0.35) | 1.53 (±0.22) | 1.16 (±0.09) | 0.98 (±0.37) |
| Klebsiella pneumoniae | 0.45 (±0.05) | 1.51 (±0.02) | .56 (±0.01) | .40 (±0.05) | 1.49 (± 0.01) | .76 (±0.04) | .80 (±0.02) | .47 (±0.01) | 0.91 (±0.02) | 0.65 (±0.07) |
| Escherichia coli | 1.50 (±0.02) | 0.49 (±0.03) | 0.40 (±0.05) | 1.58 (±0.01) | 1.62 (± 0.06) | 0.80 (±0.1) | 0.56 (±0.05) | 1.47 (±0.03) | 0.62 (±0.06) | 1.1 (±0.02) |
Among the V. streptococcus species, 40% were moderate biofilm formers with OD values greater than 0.74 but not more than 1.48, whereas 60% isolates were strong biofilm formers [Table 3].
With regard to K. pneumoniae isolates, 50% isolates were weak biofilm formers with OD values more than 0.38 but less than 0.74, whereas 30% isolates were moderate biofilm formers and only 20% isolates were strong biofilm formers [Table 3].
When it comes to E. coli, 40%isolates were weak biofilm formers, 30% isolates were moderate biofilm formers and 30% isolates were strong biofilm formers [Table 3].
It has been found that among all the selected bacteria, none of the isolates were nonbiofilm formers with OD values of less than 0.37 [Graph 1].
Graph 1.
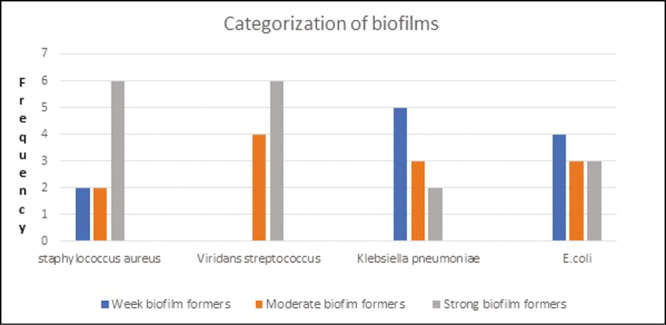
Categorization of biofilms of various bacteria
From the strong biofilm formers, specimens were selected to visualize under scanning electron microscope (SEM). The representative images of the bacteria are illustrated in [Figures 3–6].
Figure 3.

Biofilm of Staphylococcus aureus
Figure 6.

Biofilm of Escherichia coli
DISCUSSION
This study was undertaken to detect as well as categorize selective biofilm-forming bacteria from complete denture patients. The core purpose for carrying out the study was that these selective bacteria are common causative agents of life-threatening nosocomial infections among critically ill and immunocompromised individuals and are part of cocktail of bacteria called ESKAPE bacteria.[15] In this study, the average OD of S. aureus was 1.333±0.015 and average OD of V. streptococcus species was 1.304±0.023. The average OD value of K. pneumoniae was 0.8±0.012 and average value of E. coli was 1.014±0.01[Graph 1]. It was found that, 6(60%) isolates of S. aureus, 6(60%) isolates of V. streptococcus species, 2 (20%) isolates of K. pneumoniae and 3(30%) isolates of E. coli showed strong biofilm-forming abilities.
Figure 5.

Biofilm of Klebsiella pneumoniae
Manandharet al.[12] have evaluated the biofilm-forming bacteria by using Microtiter plate biofilm assay method and classified strains as high 22 (14.47%), moderate 60 (39.4%), and weak biofilm producers 70 (46.0%) in S. aureus. This finding is in sharp contrast to our study wherein 60% S. aureus isolates were strong biofilm formers. This could be due to nonoral isolates in contrast to oral-denture isolate in this study.
This study is much similar to the test settings of the microtiter plate biofilms assays by Coenye and Nelis[13] wherein the wells of the tissue culture plates are inoculated with a bacterial suspension along with positive and negative controls and these are incubated for 24–48 h. Coenye and Nelis[13] have used TSB similar to this study, but also have classified biofilms are nonadherent (score 0), weakly adherent (score 1), moderately adherent (score 3) and strongly adherent (score 4). They have also indicated that OD of strong biofilm formers was 1.5 very much similar to 4x OD cut which is 1.48 in this study.
Altieri et al.[16] have done a study involving the Methicillin-resistant S. aureus, which had OD value of 1.58 at 492nm whereas in our study the strong biofilm formers had OD values of above 1.48 at absorbance at 570nm. This is could be due to difference in the measurement of absorbance (OD) at 492nm.
Singh et al.[11] have conducted in Streptococcus epidermidis and noted that the average OD value was 0.986±0.019 when the media used was trypticase soy broth. But have observed the average OD value of 1.452 ±0.019 when BHI broth was used, but had average OD value of 1.961±0.017 when BHI agar was used. They have also noted that the OD values increased drastically when agar was used instead of broth as well as with increased time duration. This implies that for various constituents, consistencies, and ingredients of the chosen media, different OD values would be obtained. However, the researcher has to judiciously employ and compare similar test settings and material characteristics to interpret the result of the Microtiter plate biofilm assay for a particular bacterium of study.
SEM has emerged as a visualization aid for viewing and detection of the biofilm morphology. In this study, SEM was used for visualization of Biofilms in this study and the images [Figures 3–6] obtained are comparable to results obtained by Brentel et al.[14]
Figure 4.

Biofilm of Viridans streptococcus species
CONCLUSION
The study of biofilms especially the strongly biofilm forming is very useful to understand the potential pathogenic effect of these biofilms in causing the systemic spread of the planktonic bacteria which could cause systemic diseases which are resistant to conventional treatment due to the inherent nature of the biofilm to confer drug resistance to existing antibacterial agents. The importance in classifying biofilms as nonformers, week formers, moderate formers, and strong formers will be a valuable tool to execute adequate therapeutic measures in dealing with the biofilm-forming bacteria. The advancement in SEM helps us to visualize the strong biofilm former and concur with the microtiter plate method of quantification of biofilms.
ACKNOWLEDGEMENT
We wish to thank Mr. Srinivasan, microbiologist, Refsyn Biosciences, Pondicherry and Mrs. Saritha, Royal Bioresearch Institute, Chennai for all the laboratory support. We are also thankful to Dr. Rajasigamani, former Dean, and Dr. S. Senthikumar, Dean, Rajah Muthiah Dental College and Hospital for all the support and encouragement during the course of the research.
FINANCIAL SUPPORT AND SPONSORSHIP
Self-supported.
CONFLICTS OF INTEREST
There are no conflicts of interest.
AUTHORS CONTRIBUTIONS
(LA) Research work, data collection,manuscript preparation and revisions. (SK) Research guidance, manuscript revisions. (SAA) Research guidance and manuscript revisions. (FAJW) Data analysis and statistics.
ETHICAL POLICY AND INSTITUTIONAL REVIEW BOARD STATEMENT
The research was conducted after obtaining the approval of the institutional ethics committee, Rajah Muthiah Medical college (approval no.: IHEC/0142/2016) dated 15/03/2016.
PATIENT DECLARATION OF CONSENT
Informed consent was obtained from all individual participants included in the study.
DATA AVAILABILITY STATEMENT
The data that support the study results are available from the author (Dr. Leoney, A., e-mail: antony.leoney@gmail.com) on reasonable request.
REFERENCES
- 1.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–32. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan JB. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–18. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Donnell LE, Robertson D, Nile CJ, Cross LJ, Riggio M, Sherriff A, et al. The oral microbiome of denture wearers is influenced by levels of natural dentition. PLoS One. 2015;10:e0137717. doi: 10.1371/journal.pone.0137717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS. 2013;121:1–54. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 5.Garcia O, Gabriela K, Julio BR. Oral microbiota in edentulous patients. Int J Odontostomatol. 2015;9:79–84. [Google Scholar]
- 6.Jorgensen JH, Pfaller MA, Carroll KC. Manual of Clinical Microbiology. 11th ed. Washington, DC: ASM Press; 2015. [Google Scholar]
- 7.Price MN, Dehal PS, Arkin AP. Fasttree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformat. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol. 2018;35:1550–52. doi: 10.1093/molbev/msy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh AK, Prakash P, Achra A, Singh GP, Das A, Singh RK. Standardization and classification of In vitro biofilm formation by clinical isolates of Staphylococcus aureus. J Glob Infect Dis. 2017;9:93–101. doi: 10.4103/jgid.jgid_91_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manandhar S, Singh A, Varma A, Pandey S, Shrivastava N. Evaluation of methods to detect in vitro biofilm formation by staphylococcal clinical isolates. BMC Res Notes. 2018;11:714. doi: 10.1186/s13104-018-3820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coenye T, Nelis HJ. In vitro and in vivo model systems to study microbial biofilm formation. J Microbiol Methods. 2010;83:89–105. doi: 10.1016/j.mimet.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Brentel AS, Kantorski KZ, Valandro LF, Fúcio SB, Puppin-Rontani RM, Bottino MA. Confocal laser microscopic analysis of biofilm on newer feldspar ceramic. Oper Dent. 2011;36:43–51. doi: 10.2341/10-093-LR. [DOI] [PubMed] [Google Scholar]
- 15.Rice LB. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol. 2010;31:S7–10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 16.Altieri KT, Sanitá PV, Machado AL, Giampaolo ET, Pavarina AC, Jorge JH, et al. Eradication of a mature methicillin-resistant Staphylococcus aureus (MRSA) biofilm from acrylic surfaces. Braz Dent J. 2013;24:487–91. doi: 10.1590/0103-6440201302289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the study results are available from the author (Dr. Leoney, A., e-mail: antony.leoney@gmail.com) on reasonable request.


