Abstract
Purpose
Hepatocellular carcinoma (HCC) is a common malignant tumor with limited treatment. Our previous studies demonstrated that Huaier enhanced chemotherapy sensitivity and restrained HCC proliferation. This study aimed to identify differentially expressed proteins with Huaier treatment in HCC cells, providing molecular targets for future targeted therapy of HCC.
Materials and Methods
The effects of Huaier on the cell cycle were determined by flow cytometry and Western blot (WB). Xenograft models were used to verify the effects of Huaier on tumor growth. Then, proteomics was performed to identify the potential proteins regulated by Huaier. The enrichment analysis of GO and KEGG was performed for the differentially expressed proteins. Western blot (WB) and immunohistochemistry (IHC) were used to detect the levels of proteins after Huaier treatment. After that the correlation of differentially expressed proteins with pathological stages was analyzed via the GEPIA database. We also analyzed candidate expression after Huaier treatment in HCC cells by WB and qRT-PCR. Furthermore, siRNA was performed to verify the targeted regulation of Huaier on candidate proteins.
Results
First, the proteomics data showed that a total of 160 proteins were identified as differentially expressed proteins, among which six minichromosome maintenance (MCM) family members were enriched in the tumor-associated pathways after Huaier treatment. Moreover, MCM proteins were highly expressed in HCC and closely correlated with the survival of HCC patients. Finally, we confirmed that MCM proteins were targets of Huaier treatment in HCC cells.
Conclusion
Huaier treatment was closely associated with the activation and inhibition of cancer-related pathways, and the MCM family was identified as a potential target in the antitumor process of Huaier. This study is helpful in understanding the molecular alterations and clinical relevance of HCC after Huaier treatment, which is beneficial for finding new targets and designing effective chemotherapy regimens for the future treatment of HCC.
Keywords: MCM proteins, proteomics, cell cycle, Huaier, HCC
Introduction
Hepatocellular carcinoma (HCC) is one of the most common types of malignant tumors, and it is the third leading cause of cancer death.1 The therapeutic options for HCC patients are extremely limited, and one of the important reasons is that effective chemotherapy drugs are still lacking. Chemotherapeutic drugs commonly used in HCC clinically include fluorouracil, doxorubicin, cisplatin, and mitomycin.2 Recently, some new drugs have also achieved effective programs in HCC, including gemcitabine, capecitabine, and irinotecan. The combined chemotherapy regimens for HCC are oxaliplatin plus 5-fluorouracil (5Fu), gemcitabine plus oxaliplatin, capecitabine plus oxaliplatin, capecitabine plus cisplatin, and adriamycin plus oxaliplatin.3,4 However, the efficacy of chemotherapy regimens is not ideal, and toxic effects are large. Sorafenib, as a new targeted therapy, has a better survival rate for HCC patients, but the overall efficacy is still not satisfactory.5 In addition, regorafenib optimizes the efficacy of sorafenib by adding a fluorine atom to the central benzene ring of sorafenib. At present, research on the combination treatment protocol based on regorafenib is still in the in vitro experimental stage, and further basic and clinical studies are needed for the clinical efficacy of regorafenib.5
In China, adjuvant chemotherapy based on traditional Chinese medicine (TCM) has emerged as an increasing method in the systemic treatment of HCC patients.6 Trametes robiniophila murr (Huaier), a traditional Chinese herbal medicine, has been confirmed to have successful antitumor effects in clinical trials and is widely used in adjuvant therapies of HCC.7 Accumulating evidence has reported that Huaier is able to induce HCC cell apoptosis, inhibit angiogenesis, and enhance immunity.8 In addition, our previous study also found that Huaier effectively inhibited the biological functions and enhanced the chemotherapeutic sensitivity of HCC cells by targeting the Yes-associated protein (YAP)-Cyr61 axis.9,10 However, Huaier has multiple targets in vivo, and its antitumor mechanisms are not fully understood, which greatly limits the clinical application and promotion of drugs. Thus, there is an urgent need to explain the mechanisms by which Huaier enhances chemosensitivity and to predict more molecular targets in this antitumor process.
Bioinformatics analysis based on proteomics has been widely used to identify differential proteins after chemotherapy in cancers.11,12 The present study aimed to further screen differential proteins before and after Huaier treatment and to predict their underlying functions by bioinformatics analysis. We then explored the clinicopathologic correlation of the differential proteins. Moreover, our data indicated that Huaier could effectively inhibit the expression of these differentially expressed proteins, thus laying a foundation for the development of targeted therapy strategies based on Huaier.
Materials and Methods
HCC Tissue Specimens
Ten pairs of HCC tissues and adjacent normal tissues were collected from patients with HCC in our previous study.13,14 Patients with other comorbidities, such as liver cirrhosis or hepatitis, were excluded from that study; only those with primary liver cancer alone were included. The specimens were frozen immediately after surgery in liquid nitrogen. None of the patients received radiotherapy, chemotherapy, or other treatments before surgery. All patients provided written informed consent for use of the samples during their hospitalization. This study was approved by the Ethics Committee of Shanghai Municipal Hospital of Traditional Chinese Medicine and conducted in accordance with the Declaration of Helsinki.
Cell Culture and Transfection
HL-02 cell lines were obtained from the Chinese Academy of Sciences (Shanghai, China), Bel-7404, SK-Hep1, Bel-7402, SMMC-7721, and HepG2 were obtained from American Type Culture Collection (ATCC, USA), which also used in our previous studies.15,16 Cells were cultured in DMEM (HyClone, USA) with 10% fetal bovine serum and 1% penicillin-streptomycin in a 37°C cell culture incubator with 5% CO2. Short interfering RNAs (siRNAs) were synthesized by GenePharma (Shanghai, China). Lipofectamine 2000 (Invitrogen, USA) was used to transfect the MCM and negative control siRNAs into the cells according to the manufacturer’s suggestions. The specific siRNA primers are listed in Table S1.
Huaier
The granules of Huaier were obtained from Qidong Gaitianli Medicine Co., Ltd. The drug was dissolved in enzyme-free water to a final concentration of 300 mg/mL. Then, the supernatant was collected after centrifugation at 1000 rpm for 1 min and stored at 4°C for short-term use.
Flow Cytometry and qRT-PCR
For flow cytometry, the cell cycle was detected by a detection kit (KeyGen, KGA512) and was analyzed by ModFit LT 5.0 software.
For qRT-PCR, total RNA was extracted using TRIzol Reagent, and cDNA was synthetized with All-in-One cDNA Synthesis SuperMix (Biotool, B24403). Then, qPCR was performed using 2×SYBR Green qPCR Master Mix (Biotool, B21202). The specific primers are listed in Table S2.
Western Blot (WB) and Immunohistochemistry (IHC)
For WB, the detailed procedures were performed as described previously [8] using the following primary antibodies: anti-MCM2 (Abcam, ab4461), anti-MCM3 (Abcam, ab128923), anti-MCM4 (Abcam, ab4459), anti-MCM5 (Abcam, ab75975), anti-MCM6 (Abcam, ab201683), anti-MCM7 (Abcam, ab2360), anti-CDK9 (Abcam, ab76320), anti-CyclinD1 (Abcam, ab16663), anti-CyclinE1 (Abcam, ab33911), anti-p53 (Abcam, ab241566), anti-p21 (Abcam, ab109502), anti-p27 (Abcam, ab32034), and anti-GAPDH (Abcam, ab181602).
For IHC, the standard method was described previously.14 Slides were incubated with primary antibodies (Abs) against MCM2 (Immuway, YM6642), MCM3 (Abcam, ab128923), MCM4 (Immuway, YT2681), MCM5 (Abcam, ab75975), MCM6 (Abcam, ab190948), and MCM7 (Abcam, ab2360).
Xenograft Mouse Model
A total of 5×106 Bel-7404 cells were subcutaneously injected into the left armpit of athymic nude mice (SLAC, Shanghai, China). One week after injection, Huaier (100 μL of solution containing 50 mg) was administered via gavage daily for 35 days. The control group received normal saline in the same volume by the same route. Inhaled carbon dioxide was used to kill the animals while under deep anesthesia with sodium pentobarbital (250 mg/kg, intraperitoneally) at 35 days. All animal experiments were conducted in the Experimental Center of Shanghai Municipal Hospital of Traditional Chinese Medicine and approved by the Ethics Committee of Shanghai Municipal Hospital of Traditional Chinese Medicine and performed according to the NIH Guide for the Care and Use of Laboratory Animals.
Bioinformatics Analysis
The expression levels of MCM2, MCM3, MCM4, MCM5, MCM6 and MCM7 in normal and liver cancer tissues, overall survival curves with high or low MCM2, MCM3, MCM4, MCM5, MCM6 and MCM7 expression, and correlations with pathological stages were analyzed from the GEPIA database.
Proteomics, GO and KEGG Enrichment Analysis
The differentially expressed proteins were analyzed using iTRAQ proteomics. The expression profiles are provided for HCC Bel-7404 cells, which were divided into the control group and Huaier group (15 mg/mL). For the iTRAQ proteomic analysis, lysis buffer (2% SDS, 7 M urea, 1x Protease Inhibitor Cocktail (Roche Ltd. Basel, Switzerland) was added to the samples. The lysis was performed by sonication on ice for 3 min (5 min for bacteria) and kept on ice for 30 min. After centrifugation at 15,000 rpm for 15 min at 4°C, the supernatant was collected and transferred to a new Eppendorf tube. Proteins were redissolved in 500 mM TEAB (triethylammonium bicarbonate). The protein concentration of the supernatant was determined using the BCA protein assay, and then100 µg proteins per condition were transferred into a new tube and adjusted to a final volume of 100 µL with 8 M urea. Then, 11 µL of 1 M DTT was added, the sample was incubated at 37°C for 1 hour, and then it was transferred into a 10K ultrafiltration tube (Millipore). To remove urea, samples were centrifuged by adding 100 mM TEAB three times. Then, 120 µL of 55 mM iodoacetamide was added to the sample and incubated for 20 min protected from light at room temperature. Then, proteins were tryptically digested with sequence-grade modified trypsin (Promega, Madison, WI). Each sample was labeled with the corresponding iTRAQ reagents from the 8plex kit according to the manufacturer’s protocol. Then, the labeled samples of each iTRAQ analysis were pooled and mixed in equal proportions, dried by vacuum centrifugation, and prepared for fractionation by IEF separation. Then, liquid chromatography and tandem mass spectrometry (LC-MS/MS) analysis was performed according to the manufacturer’s protocol. Different expressed proteins were filtered if their fold change was over 1.5-fold and contained 2 unique peptides with −10 lgP.
For GO, we performed enrichment analysis on biological processes (BP), molecular functions (MF), and cell components (CC). Significance level was p < 0.05.
Pathway enrichment analysis used the KEGG pathway as a unit and applied a hypergeometric test to find pathways that compared to the entire proteomic background, were significantly enriched in candidate target proteins. Significance level was considered p < 0.05.
Statistical Analysis
All data and error bars are presented as the mean ± SD from at least three independent experiments. All differences between two groups were analyzed with Student’s t-test using IBM SPSS Statistics 21. Significance level was considered p < 0.05.
Results
Identify Differentially Expressed Proteins After Huaier Treatment in HCC Cells
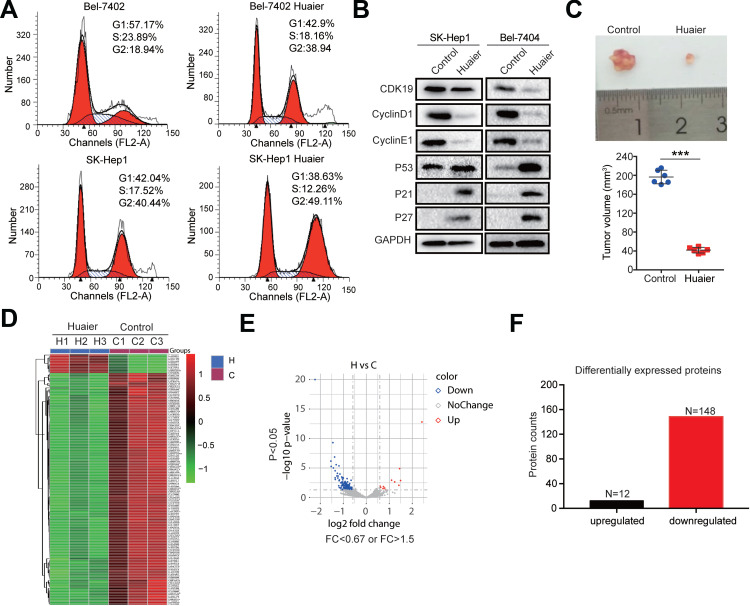
Huaier has significant inhibitory effects on tumors. The results from flow cytometry indicated that Huaier inhibited the G1 to S phase transition in Bel-7402 and SK-Hep1 cells, suggesting that Huaier could inhibit the cell cycle of HCC cells (Figure 1A). WB analysis was also consistent with the results of flow cytometry. As shown in Figure 1B, the cell cycle-associated proteins, CDK9, CyclinD1, and CyclinE1, were significantly decreased, but the CDK inhibitors, p53, p21, and p27, were increased after Huaier treatment in both SK-Hep1 and Bel-7404 cells (Figure 1B). Then, in vivo experiments showed that Huaier was able to inhibit the growth of xenografts effectively (Figure 1C). To explore the targets of Huaier, we then performed proteomics analysis with or without Huaier treatment. According to the proteomics results, we performed a hierarchical cluster analysis on the protein expression profile. A large number of differentially expressed proteins were observed between the control group and Huaier group in Bel-7404 cells (Figure 1D). As shown in Figure 1E, the volcano map indicated that differentially expressed proteins had a significant effect between the control group and Huaier group. According to the statistical analysis, there were 160 differentially expressed proteins before and after Huaier treatment, among which 12 were upregulated and 148 were downregulated (Figure 1F). Therefore, these results demonstrated that Huaier can inhibit the HCC cell cycle.
Figure 1.
Identification of proteins regulated by Huaier in HCC cells. (A) Cell cycle of Bel-7404 and SK-Hep1 cells was determined by flow cytometry in the control group and Huaier group (15 mg/mL). (B) The expression levels of CDK9, CyclinD1, CyclinE1, p53, p21, and p27 in SK-Hep1 and Bel-7404 cells treated with or without Huaier (15 mg/mL) for 24 h measured by WB assay. (C) Representative tumor pictures and tumor volume with or without Huaier (100μL of solution containing 50 mg) treatment. Pictures were captured 35 days after subcutaneous injection of Bel-7404 cells. N=6/group. (D) Heat map showing the differentially expressed proteins between the control group (n = 3) and Huaier group (n = 3) detected using proteomics. (E) Volcano map showing the down-regulated, up-regulated and unchanged proteins after Huaier treatment. (F) Histograms showing the number of differentially expressed proteins after Huaier treatment.
Bioinformatics Analysis After Huaier Treatment in HCC Cells
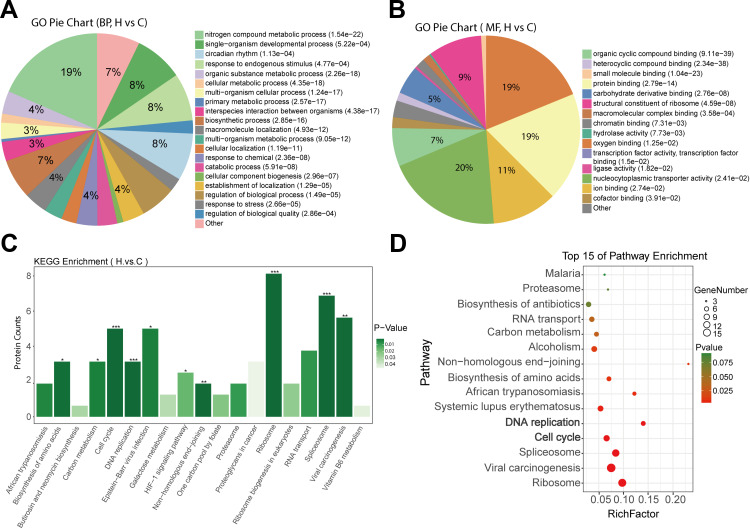
First, we performed GO enrichment analysis on BP, MF, and CC for the 160 differentially expressed proteins. The results showed that the BP of differentially expressed proteins was mainly enriched in nitrogen compound metabolic process (19%), single-organism developmental process (8%), circadian rhythm (8%), and response to an endogenous stimulus (8%) (Figure 2A). In MF analysis, differentially expressed proteins were mainly enriched in organic cyclic compound binding (20%), oxygen binding (19%), protein binding (19%), and ion binding (11%) (Figure 2B). In addition, in CC analysis, differentially expressed proteins were mainly enriched in virion part (19%), ribonucleoprotein complex (18%), catalytic complex (16%), and anchoring junction (10%) (Supplementary Fig. S1A).
Figure 2.
Bioinformatics analysis of differentially expressed proteins after Huaier treatment. (A and B) Pie diagram of GO enrichment of differential proteins. The three basic classifications of GO term are biological process (BP) (A), molecular function (MF) (B) and cellular component (CC) (Data shown in Figure S1). (C) Histograms showing KEGG enrichment of differentially expressed proteins. *p < 0.05, **p < 0.01, ***p < 0.001. (D) Top 15 of pathway enrichment.
KEGG enrichment revealed that these differentially expressed proteins were significantly enriched in ribosome, spliceosome, viral carcinogenesis, vitamin B6 metabolism, cell cycle, and DNA replication pathways (Figure 2C). Figure 2D shows the bubble maps of the top 15 of the pathway enrichment analysis of differentially expressed proteins (standard cutoff p < 0.05). Significant changes were observed in the cell cycle and DNA replication pathways, which were closely related to tumorigenesis, after Huaier treatment (Figure 2D). Notably, differential expression levels of MCM family members, such as MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7, were also enriched in the cell cycle and DNA replication pathways, suggesting the importance of the MCM family in the cell cycle and DNA replication processing with Huaier treatment (Table 1).
Table 1.
KEGG Enrichment Top 11 Signaling Pathway
| Pathway Description | NO. | P value | GENE ID |
|---|---|---|---|
| Ribosome | 13 | 1.54787E-10 | RPLP2/RPL17/RPL3/RPL27/RPS6/RPL19/RPL21/RPL13/RPS8/RPL18/RPL35/RPS2/RPL8 |
| Spliceosome | 11 | 1.88672E-08 | HNRNPK/HNRNPU/HNRNPA3/DDX46/PCBP1/HNRNPA1/SRSF3/HNRNPC/DDX5/HNRNPM/SRSF2 |
| Cell cycle | 8 | 1.99324E-05 | MCM5/MCM7/MCM4/CDK1/MCM6/YWHAG/MCM2/PRKDCa |
| DNA replication | 5 | 2.23215E-05 | MCM5/MCM7/MCM4/MCM6/MCM2b |
| Viral carcinogenesis | 9 | 0.000326665 | CDK1/PSMC1/HNRNPK/YWHAG/DDX3X/SND1/CHD4/RANBP1/HIST1H4A;HIST1H4B;HIST1H4C; |
| HISTIH4D/HISTIH4E/HISTIH4F/HIST1H4H/HISTIH4J;HISTIH4K;HIST1H4L;HIST2HA;HIST2H4B;HIST4H4 | |||
| Biosynthesis of amino acids | 5 | 0.000848813 | PYCR1/IDH3A/CPS1/PFKP/PHGDH |
| Epstein-Barr virus infection | 8 | 0.003970254 | CDK1/PSMD12/PSMC1/PSMC3/YWHAG/SND1/XPO1/HSPB1 |
| Carbon metabolism | 5 | 0.004100065 | CPS1/IDH3A/PHGDH/PFKP/HK2 |
| RNA transport | 6 | 0.007360002 | EIF4G1/EEF1A1/EIF4G2/XPO1/RANGAP1/EIF4A1 |
| HIF-1 signaling pathway | 4 | 0.016269678 | HMOX1/ELOC/RPS6/HK2 |
| Proteoglycans in cancer | 5 | 0.044286847 | CTTN/DDX5/PDCD4/FLNA/RPS6 |
Notes: aDifferential expression levels of MCM family members were enriched in the cell cycle. bDifferential expression levels of MCM family members were enriched in DNA replication pathways.
MCM Family Members are Highly Expressed in HCC Cells and Tissues
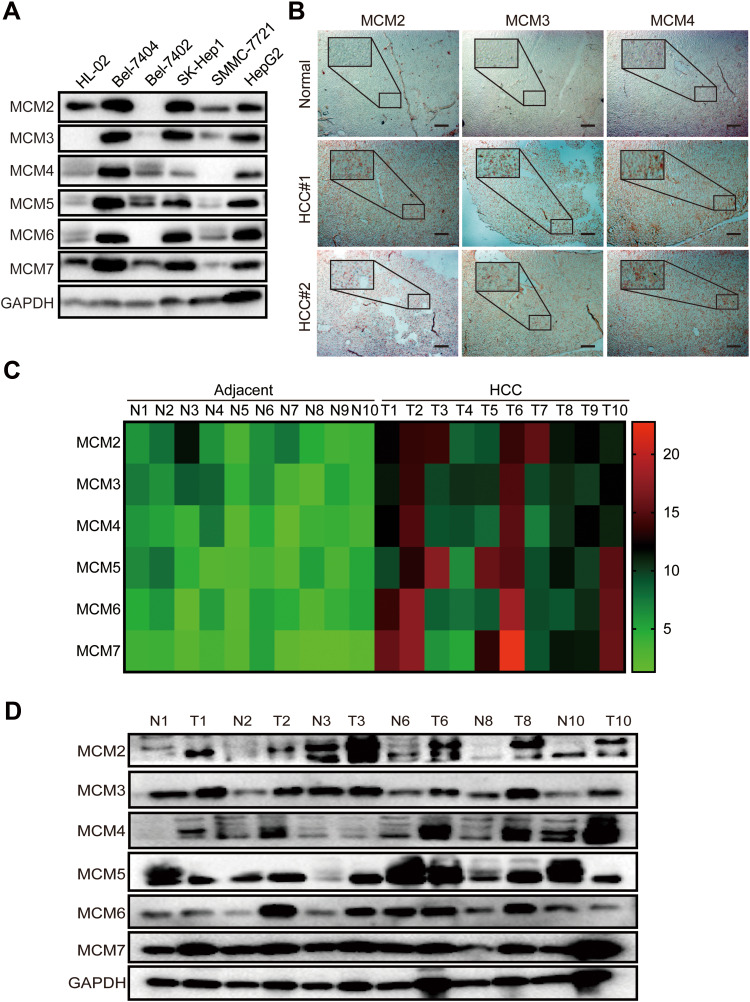
The above results suggested that the MCM family might be involved in the process of the cell cycle and DNA replication in HCC cell lines. Unexpectedly, the results from WB revealed that all MCM family members were obviously upregulated in HCC cell lines, especially in BeL-7404, SK-Hep1, and HepG2 cells, whereas their expression levels were low in normal liver epithelial HL-02 cell lines (Figure 3A). IHC results also revealed that compared with normal tissues, MCM family members were highly expressed in HCC tissues (Figure 3B, Supplementary Fig. S1B). Next, we tested HCC samples from 10 patients and matched adjacent tissues and found that the expression of MCM family members was aberrantly elevated at the mRNA level (Figure 3C). In addition, compared with paired adjacent tissues, the protein expression levels of MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7 were also enhanced in HCC tissues (Figure 3D). Therefore, these findings indicated that the expression of the MCM family was increased in HCC tissues and cell lines.
Figure 3.
MCM proteins were up-regulated in HCC cell lines and tissues. (A) Western blots assay showed the expression levels of MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7 in HL-02, Bel-7404, SK-Hep1, Bel-7402, SMMC-7721, and HepG2 cell lines. (B) Representative IHC images of MCM2, MCM3, and MCM4 staining from normal liver and HCC tissues, scar bar 100μm. (C) The heat-map exhibits the differentially expressed MCM between 10 pairs of HCC tissues and adjacent normal tissues measured by RT-qPCR assay. (D) Western blots of the expression levels of MCM proteins between 6 pairs of HCC tissues and adjacent normal tissues.
The MCM Family is Crucial for HCC Patient Survival and is Inhibited by Huaier
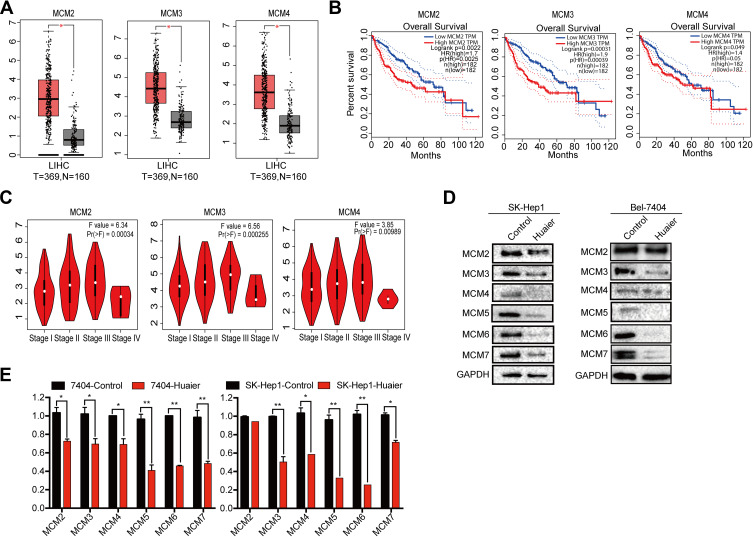
To further investigate the correlation of MCM levels with the survival of HCC patients, we searched the GEPIA database that contained 160 normal liver tissue samples and 369 HCC tissue samples. First, the results indicated that MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7 were expressed at higher levels in HCC tissues than in normal liver tissues (Figure 4A, Supplementary Fig. S2A). Additionally, we found that high expression levels of these MCM family members were associated with shorter overall survival of HCC patients (Figure 4B, Supplementary Fig. S2B), and their expression levels were significantly associated with higher tumor stage (Figure 4C, Supplementary Fig. S2C).
Figure 4.
Up-regulated of MCM proteins were closely correlated with shorter survival time and inhibited by Huaier. (A) Relative expression levels of MCM2, MCM3, and MCM4 in 369 HCC tissues and 160 normal tissues got form GEPIA database. (B) Kaplan-Meier survival curves for HCC patients with low (n = 182) or high (n = 182) MCM expressions (MCM2, MCM3, and MCM4) which extracted from GEPIA database. (C) The correlations of MCM2, MCM3 and MCM4 proteins with clinicopathologic stages got from GEPIA database. (D) The expression levels of MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7 in SK-Hep1 and Bel-7404 cell lines treated with Huaier (15 mg/mL) for 24 h measured by WB assay. (E) Relative mRNA levels of MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7 in SK-Hep1 and Bel-7404 cell lines treated with Huaier (15 mg/mL) for 24h (Student’s t-test, n = 3).
Furthermore, we also found that MCM protein (Figure 4D) and mRNA expression (Figure 4E) were inhibited by Huaier in Bel-7404 and SK-Hep1 cells. These results strongly suggested that MCM family members were upregulated in HCC cells, and that they might be potential targets of Huaier.
Huaier Suppressed the HCC Cell Cycle by Regulating MCM Proteins
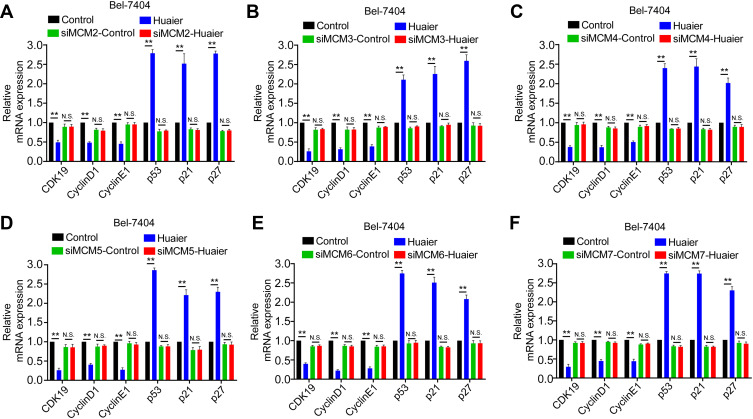
To further explore whether Huaier targeted MCM to inhibit the cell cycle and exerted its antitumor effects, specific siRNAs were used to mediate the downregulation of MCM proteins in Bel-7404 and SK-Hep1 cells (Supplementary Fig. S3). As shown in Figure 5 and Supplementary Fig. S4, Huaier effectively inhibited the cycle-related proteins, CDK19, CyclinD1, and CyclinE1, and enhanced p53, p21, and p27 levels. However, after knocking down MCM proteins, the effects of Huaier on these proteins were significantly reversed, indicating that Huaier suppressed the HCC cell cycle in an MCM-dependent manner.
Figure 5.
Huaier suppressed Bel-7404 cell cycle via regulating MCM. (A–F) The mRNA levels of cell cycle related genes in SK-Hep1 cell lines treated with Huaier (0 and 15 mg/mL) for 24 h with or without a knockdown of indicate.
Discussion
According to global cancer statistics from Globocan, there were 841,080 cases of liver cancer worldwide (HCC accounted for 85%) in 2018, accounting for 4.7% of the total cancer incidence in the same period. At the same time, the number of deaths attributable to liver cancer worldwide was 781,631, thus accounting for 8.2% of the total cancer deaths in the same period.17 In China, adjuvant chemotherapy based on TCM is a new direction for HCC therapy. A large number of randomized trials have continuously confirmed the clinical efficiency of traditional herbal medicine.18 Of these, Huaier has been widely used for HCC treatment. Recent studies recommended that Huaier was able to prevent intrahepatic metastasis (IM) or multicentric occurrence (MO) to improve survival in patients with multifocal HCC.19 Additionally, a nationwide multicenter study involving 1044 patients proved that Huaier could improve recurrence-free survival (RFS) and reduce extrahepatic recurrence after curative liver resection.7 Our study also reported that the combination treatment of Huaier and oxaliplatin could improve the intrinsic or acquired resistance of oxaliplatin in HCC.10 Although related clinical research has made promising progress, the molecular mechanisms of Huaier treatment for HCC remain unclear. One of the pivotal reasons is that the effective ingredients of herbal medicine are relatively complex and have multiple mechanisms of action in vivo.20 The active ingredient of Huaier is a polysaccharide composed of 6 kinds of monosaccharides, namely, L-fucose, L-arabinose, D-xylose, D-mannose, D-galactose, and D-glucose combined with 18 amino acids.21 Related studies have revealed that Huaier could inhibit the initiation and progression of HCC by regulating the PI3K/AKT,22 YAP/CREB,8 c-Myc/Bmi1,23 JAK/STAT3,24 and tumor-associated miRNA signaling pathways.22 Hence, Huaier also has multiple targets in the antitumor process.
Our previous studies and other more recent studies have shown that the Bel-7404 cell line is highly malignant and has multidrug resistance.9,10 Here, this study identified 160 differentially expressed proteins before and after Huaier treatment in Bel-7404 cells. Notably, all the MCM family members (MCM2/3/4/5/6/7) were differentially expressed after Huaier treatment via bioinformatics analysis. KEGG pathway enrichment analysis showed that these differentially expressed MCM proteins are mainly enriched in tumor-associated pathways, suggesting that the MCM family might be important targets in the treatment of HCC by Huaier. The six subunits of the MCM family are essential for DNA replication and extension.23 In eukaryotic cells and mammals, the MCM family belongs to a class of replication licensing factors (RLFs) that can bind to the origin of replication. In the early G1 phase, the six proteins (MCM2/3/4/5/6/7) are involved in the formation of the preinitiation complex (PIC) of replication. In this process, MCM proteins can interact with each other to form different complexes. MCM4/6/7 has helicase activity and DNA-dependent ATPase activity, which is the catalytic core of MCM complexes. MCM3/5 can inhibit the activity of MCM4/6/7 helicase, while MCM2 stabilizes the interaction between MCM3/5 and MCM4/6/7.23,24 In addition to participating in DNA replication, MCM proteins also play an important role in RNA transcription, chromatin reconstruction, and maintaining gene homeostasis.25,26 In the clinic, MCM proteins have been identified as diagnostic markers of cancers. Liu et al,27 found that MCM2/3/4/5/6/7 mRNAs were significantly upregulated in HCC patients. Overexpression of MCM2, MCM6, and MCM7 was closely associated with adverse tumor features and poorer outcomes. Additionally, the results of other similar studies were basically consistent with ours. This study also found that MCM proteins were upregulated in HCC cells and tissues and that overexpression of MCM proteins was crucial for the overall survival of HCC patients. At the cellular level, Huaier decreased the protein and mRNA levels of all MCM family members. Hence, given the important role of the MCM family in tumorigenesis, we suggest that MCM proteins are the key targets of Huaier treatment for HCC. A related study reported that knockdown of MCM proteins could induce a delay in S/G2-phase progression by restraining the expression levels of CDK4, CyclinD1, and CyclinE in HCC cells.28,29 Additionally, MCMs are regulated in the cell cycle through phosphorylation by ATM (ataxia telangiectasia mutated), and phosphorylation of MCM proteins is closely associated with oncogenesis and cancer progression.30 Importantly, recent studies also demonstrated that both CDK and ATM were regulated by Huaier and finally inhibited cancer cell proliferation.31 Furthermore, Huaier plus 5-Fu had significant inhibitory effects on the cell cycle by targeting CDK9, CyclinD1, and CyclinE1.10 This study also demonstrated that Huaier suppressed the MCM-dependent HCC cell cycle. Therefore, we suggest that there may be a molecular network between Huaier and cell cycle-related proteins, which is mediated by MCM proteins.
In conclusion, this study demonstrated alterations in expression after Huaier treatment in HCC cells. Huaier treatment was closely correlated with the activation and inhibition of cancer-related molecular pathways, and MCM family members were identified as potential targets in the antitumor process of Huaier. These findings will also be beneficial for the clinical application of Huaier and related therapeutic targets.
Acknowledgments
This study was sponsored by Shanghai Sailing Program (Grant: 18YF1421800), funded by a Municipal Human Resources Development Program for Outstanding Young Talents in Medical and Health Sciences in Shanghai (Grant: 2018YQ15), “Chen Guang” project supported by the Shanghai Municipal Education Commission and Shanghai Education Development Foundation (Grant: 17CG43), supported by the Natural Science Foundation of China (Grant: 81774291 and 81902315).
Highlights
1. Identifying differentially expressed proteins before and after Huaier treatment in HCC cells via proteomics data.
2. Minichromosome maintenance (MCM) family members are highly expressed in HCC and closely correlated with the survival of HCC patients.
3. MCM proteins are targets of Huaier treatment in HCC cells.
Author Contributions
Yongjie Niu and Liang Shan researched, analysed data, and wrote manuscript. Han Gao, Congcong Zhang, and Zijun Qian collected and analysed clinicopathological information. Zhixian Wang and Xin Xu researched and analysed data. Xiao Zhang and Jiayi Wang contributed to discussion. Lifang Ma wrote the manuscript and designed the study. Liyun Chen and Yongchun Yu designed the study. All authors read and approved the final manuscript. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors declare they have no conflicts of interest.
References
- 1.Ozakyol A. Global epidemiology of hepatocellular carcinoma (HCC Epidemiology). J Gastrointest Cancer. 2017;48:238–240. doi: 10.1007/s12029-017-9959-0 [DOI] [PubMed] [Google Scholar]
- 2.Boucher E, Corbinais S, Brissot P, et al. Treatment of hepatocellular carcinoma (HCC) with systemic chemotherapy combining epirubicin, cisplatinum and infusional 5-fluorouracil (ECF regimen). Cancer Chemother Pharmacol. 2002;50(4):305–308. doi: 10.1007/s00280-002-0503-x [DOI] [PubMed] [Google Scholar]
- 3.Mohamed B, Neil M, Aiwu R, et al. Systemic treatment for advanced hepatocellular carcinoma. Liver Cancer. 2019;8(10):341–358. doi: 10.1159/000496439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cinnamon E, Pikarsky E. Are we ready for targeted therapy combinations in HCC? Gut. 2020;69(4):613–614. doi: 10.1136/gutjnl-2019-319780 [DOI] [PubMed] [Google Scholar]
- 5.da Fonseca LG, Reig M, Bruix J. Tyrosine kinase inhibitors and hepatocellular carcinoma. Clin Liver Dis. 2020;244(4):719–737. doi: 10.1016/j.cld.2020.07.012 [DOI] [PubMed] [Google Scholar]
- 6.Ting CT, Li WC, Chen CY, et al. Preventive and therapeutic role of traditional Chinese herbal medicine in hepatocellular carcinoma. J Chin Med Assoc. 2015;78(3):139–144. doi: 10.1016/j.jcma.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 7.Chen Q, Shu C, Laurence AD, et al. Effect of Huaier granule on recurrence after curative resection of HCC: a multicentre, randomised clinical trial. Gut. 2018;67(11):2006–2016. doi: 10.1136/gutjnl-2018-315983 [DOI] [PubMed] [Google Scholar]
- 8.Ren J, Zheng C, Feng G, et al. Inhibitory effect of extract of fungi of Huaier on hepatocellular carcinoma cells. J Huazhong Univ Sci Technolog Med Sci. 2009;29(2):198–201. doi: 10.1007/s11596-009-0212-3 [DOI] [PubMed] [Google Scholar]
- 9.Liang S, Yan L, Jiang H, et al. Huaier restrains proliferative and migratory potential of hepatocellular carcinoma cells partially through decreased yes-associated protein 1. J Cancer. 2017;8(19):4087–4097. doi: 10.7150/jca.21018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao Y, Shan L, Xu X, et al. Huaier augmented the chemotherapeutic sensitivity of oxaliplatin via downregulation of yap in hepatocellular carcinoma. J Cancer. 2018;9(21):3962–3970. doi: 10.7150/jca.25909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang XB, Li J, Zheng L, et al. Bioinformatics analysis reveals potential candidate drugs for HCC. Pathology Oncology Research Por. 2013;19(2):251–258. doi: 10.1007/s12253-012-9576-y [DOI] [PubMed] [Google Scholar]
- 12.Min N, Cao J, Zhang J, et al. Identification of hub genes involved in the occurrence and development of hepatocellular carcinoma via bioinformatics analysis. Oncol Lett. 2020;20(2):1695–1708. doi: 10.3892/ol.2020.11752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 14.Xi SY, Minuk GY. Role of traditional Chinese medicine in the management of patients with hepatocellular carcinoma. World J Hepatol. 2018;10(11):799–806. doi: 10.4254/wjh.v10.i11.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JY, Ma LF, Weng WH, et al. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 2013;58(3):1011–1020. doi: 10.1002/hep.26420 [DOI] [PubMed] [Google Scholar]
- 16.Yang ZG, Tao YQ, Xu X, et al. Bufalin inhibits cell proliferation and migration of hepatocellular carcinoma cells via APOBEC3F induced intestinal immune network for IgA production signaling pathway. Biochem Biophys Res Commun. 2018;503(3):2124–2131. doi: 10.1016/j.bbrc.2018.07.169 [DOI] [PubMed] [Google Scholar]
- 17.Xie DY, Fan HK, Ren ZG, et al. Identifying clonal origin of multifocal hepatocellular carcinoma and its clinical implications. Clin Transl Gastroenterol. 2019;10(2):e00006. doi: 10.14309/ctg.0000000000000006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Martin RC. Herbal medicine and hepatocellular carcinoma: applications and challenges. Evid Based Complement Alternat Med. 2011;541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song X, Li Y, Zhang H, et al. The anticancer effect of Huaier (Review). Oncol Rep. 2015;34(1):12–21. doi: 10.3892/or.2015.3950 [DOI] [PubMed] [Google Scholar]
- 20.Zheng J, Li C, Wu X, et al. Astrocyte elevated gene-1 (AEG-1) shRNA sensitizes Huaier polysaccharide (HP)-induced anti-metastatic potency via inactivating downstream P13K/Akt pathway as well as augmenting cell-mediated immune response. Tumor Biology. 2004;35(5):4219–4224. doi: 10.1007/s13277-013-1552-y [DOI] [PubMed] [Google Scholar]
- 21.Li C, Wu X, Zhang H, et al. A Huaier polysaccharide restrains hepatocellular carcinoma growth and metastasis by suppression angiogenesis. Int J Biol Macromol. 2015;75:115–120. doi: 10.1016/j.ijbiomac.2015.01.016 [DOI] [PubMed] [Google Scholar]
- 22.Zou Y, Xiong H, Xiong H, et al. A polysaccharide from mushroom Huaier retards human hepatocellular carcinoma growth, angiogenesis, and metastasis in nude mice. Tumor Biology. 2014;36(4):2929–2936. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Wang X, Chen T, et al. Huaier extract inhibits breast cancer progression through a LncRNA-H19/MiR-675-5p Pathway. Cell Physiol Biochem. 2017;44(2):581–593. doi: 10.1159/000485093 [DOI] [PubMed] [Google Scholar]
- 24.Tye, Bik K. MCM PROTEINS IN DNA REPLICATION. Annu Rev Biochem. 1999;68(6):649–686. doi: 10.1146/annurev.biochem.68.1.649 [DOI] [PubMed] [Google Scholar]
- 25.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91(1):59–69. doi: 10.1016/S0092-8674(01)80009-X [DOI] [PubMed] [Google Scholar]
- 26.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol mol biol rev. 2004;68(1):109–131. doi: 10.1128/MMBR.68.1.109-131.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Li J, Chen J, et al. MCM family in HCC: MCM6 indicates adverse tumor features and poor outcomes and promotes S/G2 cell cycle progression. Bmc Cancer. 2018;18(1):200. doi: 10.1186/s12885-018-4056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pozo PN, Matson JP, Cole Y, et al. Cdt1 variants reveal unanticipated aspects of interactions with cyclin/CDK and MCM important for normal genome replication. Mol Biol Cell. 2018;29(25):2989–3002. doi: 10.1091/mbc.E18-04-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H, Xiong Y, Zhang G, et al. Elevated expression of minichromosome maintenance 3 indicates poor outcomes and promotes G1/S cell cycle progression, proliferation, migration and invasion in colorectal cancer. Biosci Rep. 2020;40(7):BSR20201503. doi: 10.1042/BSR20201503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fei L, Xu H. Role of MCM2-7 protein phosphorylation in human cancer cells. Cell Biosci. 2018;8:43. doi: 10.1186/s13578-018-0242-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao S, Li X, Ding X, et al. Huaier extract restrains the proliferative potential of endocrine-resistant breast cancer cells through increased ATM by suppressing miR-203. Sci Rep. 2017;7(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]