Abstract
Background
The viral load and tissue distribution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remain important questions. The current study investigated SARS-CoV-2 viral load, biodistribution and anti-SARS-CoV-2 antibody formation in patients suffering from severe corona virus disease 2019 (COVID-19) induced acute respiratory distress syndrome (ARDS).
Methods
This is a retrospective single-center study in 23 patients with COVID-19-induced ARDS. Data were collected within routine intensive care. SARS-CoV-2 viral load was assessed via reverse transcription quantitative polymerase chain reaction (RT-qPCR). Overall, 478 virology samples were taken. Anti-SARS-CoV-2-Spike-receptor binding domain (RBD) antibody detection of blood samples was performed with an enzyme-linked immunosorbent assay.
Results
Most patients (91%) suffered from severe ARDS during ICU treatment with a 30-day mortality of 30%. None of the patients received antiviral treatment. Tracheal aspirates tested positive for SARS-CoV-2 in 100% of the cases, oropharyngeal swabs only in 77%. Blood samples were positive in 26% of the patients. No difference of viral load was found in tracheal or blood samples with regard to 30-day survival or disease severity. SARS-CoV-2 was never found in dialysate. Serologic testing revealed significantly lower concentrations of SARS-CoV-2 neutralizing IgM and IgA antibodies in survivors compared to non-survivors (p = 0.009).
Conclusions
COVID-19 induced ARDS is accompanied by a high viral load of SARS-CoV-2 in tracheal aspirates, which remained detectable in the majority throughout intensive care treatment. Remarkably, SARS-CoV-2 RNA was never detected in dialysate even in patients with RNAemia. Viral load or the buildup of neutralizing antibodies was not associated with 30-day survival or disease severity.
Introduction
The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was declared a pandemic by the WHO on March 11, 2020 [1]. As of September 01, 2020 more than 25 million people had been tested positive and more than 850000 people had died [2]. The proportion of patients requiring hospital treatment significantly increases with age [3]. Approximately 14–17% of hospitalized patients require intensive care due to an acute respiratory distress syndrome (ARDS) [4, 5].
In the wake of this pandemic, hospitals and intensive care units (ICU) are overwhelmed by the number of patients and health-care workers bear a high risk of SARS-CoV-2 infection themselves [6]. Containment strategies have focused on preventing airborne and droplet transmission, as viral loads are highest in respiratory secretions and seem to correlate with Corona virus disease 2019 (COVID-19) severity [7]. However, there is growing evidence for viral shedding within non-respiratory tissues. Recent studies reported SARS-CoV-2 RNA in blood (RNAemia) [7, 8], feces [7, 9, 10], urine [11], and peritoneal fluid [12, 13]. Some authors suggested a correlation between RNAemia and critical illness [14] whereas others found RNAemia in mild cases as well [11, 14]. Moreover, the serologic response to COVID-19 has recently received increasing attention. SARS-CoV-2 antibody levels increase between days eight to 14 which correlates with a slow but steady decline in viral load [15]. Prior studies further suggested that antibody titers are higher in severe clinical cases [16]. However, in many COVID-19 studies disease severity was largely undefined and the term severe disease included a wide range of clinical presentations.
The current retrospective study investigated SARS-CoV-2 viral load, biodistribution and anti-SARS-CoV-2 neutralizing antibody formation in critical ill patients suffering from moderate to severe COVID-19 induced ARDS during ICU treatment. We not only provide novel insights into COVID-19 in a high-risk population with over 50% extracorporeal membrane oxygenation (ECMO), but also important implications regarding the protection of ICU personnel from SARS-CoV-2.
Methods
Study design
This is a retrospective single-center cohort study adhering to the STROBE-Guidelines [17]. Approval by the Institutional Review Board of the University Hospital Wuerzburg, Germany and informed consent were waived due to sole retrospective chart review (63/20-kr; 25 March 2020).
Patient selection
All patients (≥ 18 years) treated in the Intensive Care Unit of the Department of Anesthesiology and Critical Care at the University Hospital Wuerzburg (local ICU) between March 19, 2020 and May 06, 2020 with a confirmed diagnosis of COVID-19 were consecutively included in the study. Severity of ARDS was defined according to the Berlin definition [18].
All data were retrospectively extracted from the patient data management system between June 1th - 7th, 2020 (COPRA6 RM1.0, COPRA System GmbH, Berlin, Germany), written records or communication with close relatives and general practitioners. Survival was defined as the survival 30 days after admission to the ICU. For patients discharged to general wards digital records were observed in order to determine the 30-day mortality.
Sample collection
During ICU treatment oropharyngeal swabs, tracheobronchial aspirates, blood (serum or EDTA), urine, dialysate and anal swabs were longitudinally obtained to assess the viral load of COVID-19 patients. Sample collection was summarized for days 1 (ICU admission), 4, 7, 10 and 14. Samples that had been taken ± 1 day were assigned to the defined time points, respectively. Samples taken between days 20–28 were summarized as day 21. All specimens were analyzed during routine clinical care, specific treatment protocols were not defined. However, local standard protocols during the pandemic included a close monitoring of all potentially contaminated specimens. Overall, 478 virology samples were taken. Among those were 86 oropharyngeal swabs, 94 tracheobronchial aspirates, 96 blood samples, 85 urine samples, 33 dialysate samples, 83 anal swabs and 1 cerebrospinal fluid sample. Respiratory samples were obtained from all patients. Blood and urine samples were only available from 22, anal swabs from 21 and dialysate samples from 13 patients on continuous renal replacement therapy. In addition, one sample was obtained from cerebrospinal fluid at a single time point (day 3). Routine laboratory parameters included complete blood counts, markers of inflammation, immunoglobulin M, G and A levels as well as liver and renal function. Arterial blood gas samples were recorded multiple times per day.
Reverse transcription quantitative polymerase chain reaction for SARS-CoV-2
Detection of SARS-CoV-2-RNA by Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was carried out according to the recommendations of the World Health Organization [19] and the manufacturer’s instructions. Primers located in the SARS-CoV-2 E-gene as described by Corman et al. [20] and the test kit FTD SARS-CoV-2 (Siemens Healthineers, Erlangen, Germany) were used. Cycle thresholds (Ct) ≥40 were considered negative.
Anti-SARS-CoV2-Spike-receptor binding domain (RBD) antibody detection
Serologic testing of blood samples was performed with an Enzyme-linked immunosorbent assay (ELISA) using the receptor-binding domain (RBD) of the SARS-CoV-2 Spike protein [21]. RBD was recombinantly expressed in Expi293F HEK cells (Expi293F HEKTM cells, ThermoFisher Scientific#A14527, Lot# 1900411, authenticated by the provider) using the RBD_6His expression plasmid [22, 23]. The ELISA protocol was carried out as previously published [22, 23] with the following reagents: RBD was immobilized on NUNC (Nunc, Roskilde, Denmark) Maxisorp plates, blocking was done using 1x Roti-Block (Carl Roth; A151.1, Karlsruhe, Germany. Detection antibodies goat anti-human IgG [Thermo Fischer; 31410] and goat anti-human IgM [YO-Proteins; Ronninge, Sweden; ABIN301951] were diluted 1:10000, goat anti-human IgA [KPL International Limited, New Delhi, India, 5220–0360] was diluted 1:5000. For detection, ready-to-use s(hs)TMB (SDT-Reagents; sTMB, Baesweiler, Germany) was incubated for 10 minutes. Readout was performed on a Tecan sunrise (Tecan, Maennedorf, Switzerland) at 450 nm, corrected at 620 nm. An internal negative control pool was used to define the background signal. Since a reference antibody against RBD was not available at the time of the analyses, quantification was based on serum derived IgG (Sigma-Aldrich GmbH Munich, Germany; I2511), IgM (Sigma-Aldrich GmbH Munich, Germany; I8260) and IgA (Sigma-Aldrich; I4036) antibodies immobilized on NUNC Maxisorp plates. Standard curves were fitted using Origin (OriginLab Corporation, Northhampton, USA). R2 values for IgM: 0.99992, for IgG: 0.99964, and for IgA: 0.99965, respectively.
Statistics
Data are reported as medians and interquartile range (IQR) (continuous data) or absolute numbers and percentages (categorical data). Normality of the data could not be assumed. We performed Fisher’s exact test for categorical variables and a Mann-Whitney-rank sum test for metric data. Two-sided statistical significance was set at p<0.05. Data were collected in Microsoft Excel. Statistical analyses were performed with SPSS 26 (IBM, Armonk, NY, USA) and Prism 8 (GraphPad Software, San Diego, CA, USA).
Results
Baseline characteristics and clinical course
A total of 23 patients met the inclusion criteria. Median age was 58 (49–69) years and the majority was male (65%). Baseline characteristics and co-morbidities are shown in Table 1. Two patients had a history of rheumatologic diseases and current medication with immunosuppressant (Prednisolone or Leflunomide), respectively.
Table 1. Demographic and baseline characteristics.
| Overall | Survivors | Non-Survivors | p-value | |
|---|---|---|---|---|
| (N = 23) | (n = 16) | (n = 7) | ||
| Median age (IQR)–years | 58 (49–69) | 54 (48–69) | 66 (56–69) | 0.198 |
| Sex–no. (%) | ||||
| Male | 15 (65) | 12 (75) | 3 (43) | 0.182 |
| Female | 8 (35) | 4 (25) | 4 (57) | |
| Median BMI (IQR)–kg/m2 | 29 (25–31) | 28 (24–31) | 31 (26–35) | 0.193 |
| Smoker–no. (%) | ||||
| Never | 13 (57) | 10 (62.5) | 3 (43) | 0.650 |
| Former | 10 (43) | 6 (37.5) | 4 (57) | |
| Active | 0 (0) | 0 (0) | 0 (0) | |
| Co-morbidities–no. (%) | ||||
| Chronic obstructive pulmonary disease | 9 (39) | 7 (44) | 2 (29) | 0.657 |
| Arterial hypertension | 15 (65) | 10 (63) | 5 (71) | 1.0 |
| Coronary artery disease | 3 (13) | 1 (6) | 2 (29) | 0.209 |
| Diabetes mellitus | 5 (22) | 3 (19) | 2 (29) | 0.621 |
| Chronic renal insufficiency | 2 (9) | 1 (14) | 1 (6) | 0.526 |
| Symptoms–no. (%) | ||||
| Dyspnea | 12 (52) | 9 (56) | 3 (43) | 0.667 |
| Fever | 22 (96) | 16 (100) | 6 (86) | 0.304 |
| Cough | 18 (78) | 12 (75) | 6 (86) | 1.0 |
| Nausea | 7 (30) | 3 (19) | 4 (57) | 0.137 |
| Diarrhea | 7 (30) | 3 (19) | 4 (57) | 0.137 |
| Median duration (IQR)–days | ||||
| Symptom onset—hospital admission | 6 (3–7) | 6 (4–13) | 5 (2–7) | 0.281 |
| Symptom onset–ICU admission | 8 (5–10) | 9 (5–14) | 5 (5–9) | 0.138 |
| ARDS–no. (%) | ||||
| moderate | 2 (9) | 2 (12.5) | 0 (0) | 1.0 |
| severe | 21 (91) | 14 (87.5) | 7 (100) | |
| Median oxygenation index on admission (IQR)—PaO2/FiO2 | 140 (88–190) | 134 (88–181) | 164 (64–246) | 0.769 |
| Median worst oxygenation index during ICU treatment (IQR)—PaO2/FiO2 | 69 (54–97) | 73 (59–97) | 59 (49–82) | 0.149 |
| Duration of ICU treatment (local ICU)–days (IQR) | 23 (13–41) | 38 (17–43) | 15 (8–20) | 0.021 |
| Duration of Mechanical ventilation–days (IQR) | 20 (17–41) | 39 (18–44) | 18 (9–20) | 0.028 |
| Extracorporeal membrane oxygenation (ECMO)–no. (%) | 15 (65) | 10 (63) | 5 (71) | 1.0 |
| Renal replacement therapy–no. (%) | 17 (74) | 10 (63) | 7 (100) | 0.124 |
Chronic obstructive pulmonary disease = chronic pulmonary obstructive disease or bronchial asthma; IQR = interquartile range; ICU = intensive care unit; BMI = body mass index; PaO2/FiO2 = arterial pressure of oxygen/fraction of inspired oxygen; Duration of Mechanical ventilation = time from intubation to extubation/decannulation or pressure support of or below 5 cm H2O for at least 24h; ECMO = extracorporeal membrane oxygenation; CVVHD = continuous venous-venous hemodialysis
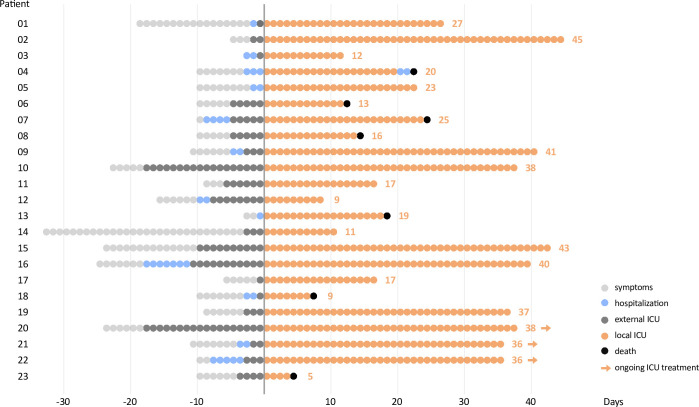
All Patients had been transferred either from external ICUs (78%) or general wards (22%) due to worsening of pulmonary symptoms or increasing COVID-19-induced ARDS severity. Median duration (IQR) from symptom onset to hospital admission and ICU were 6 (3–7) and 8 (5–10) days, respectively (Fig 1).
Fig 1. Clinical course of each COVID-19 ARDS patient within the study cohort.
The figure starts with symptom onset and also displays hospital and ICU treatment prior transfer to the University Hospital of Wuerzburg (local ICU). Treatment at the ICU of the University Hospital Wuerzburg was defined as day 1. Patient 04 was transferred from the ICU to a general ward and died 3 days later.
Median PaO2/FiO2 oxygenation index (IQR) on admission was 140 mmHg (88–190). All 23 patients suffered from moderate (9%) or severe (91%) ARDS during ICU treatment with a median worst PaO2/FiO2 (IQR) of 69 (54–97). All patients were mechanically ventilated, 15 patients (65%) received extracorporeal membrane oxygenation (ECMO) and 17 patients (74%) were treated with continuous veno-venous hemodialysis (CVVHD). None of the patients received antiviral treatment. Mortality within 30 days after admission to the local ICU was 30%.
SARS-CoV-2 viral load
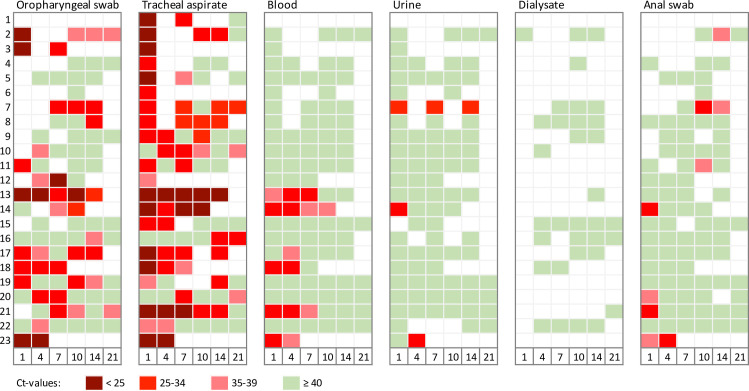
SARS-CoV-2 viral loads upon admission and during the course of ICU treatment are depicted in Fig 2. Tracheal aspirates tested positive for SARS-CoV-2 in all 23 cases (100%). Pharyngeal swabs tested positive for SARS-CoV-2 in 17 of 22 patients (77%). Simultaneous tracheal and oropharyngeal testing resulted in a negative oropharyngeal swab and a positive tracheal aspirate 15 times. The proportion of positive SARS-CoV-2 tests was much lower in non-respiratory samples: blood samples were positive in 26% of the patients. Urine tested positive in three (13%) and anal swabs in seven patients (30%). However, nearly half of the anal swabs provided positive test results for the first time after 10 days of ICU treatment. All samples of dialysate were negative. Cerebrospinal fluid was negative in one patient with concomitantly positive respiratory samples.
Fig 2. Viral load during treatment in local ICU (days 1, 4, 7, 10, 14, and 21 after ICU admission).
Values of Cycle thresholds (Ct) are graduated. Lower values reflect higher viral loads. Values ≥ 40 were considered negative.
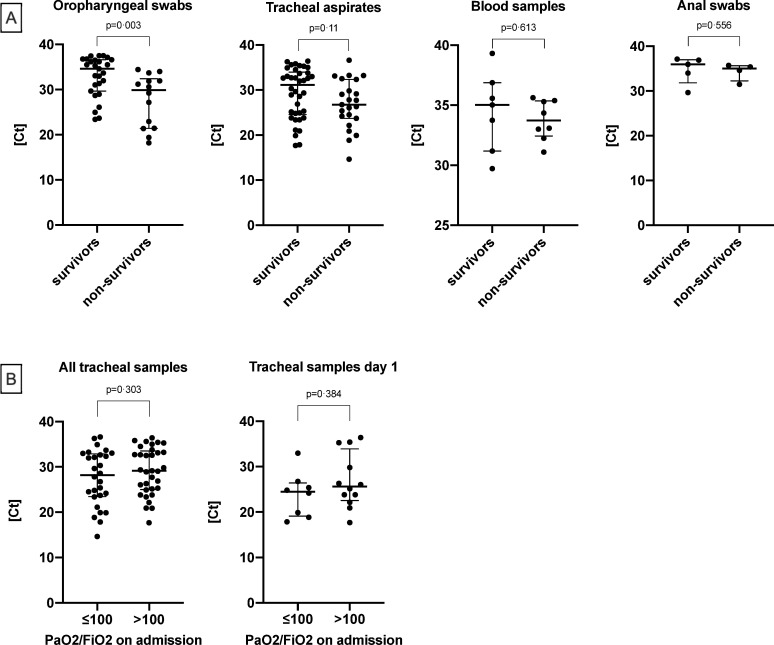
Cycle thresholds of positive samples differed between survivors and non-survivors: Median cycles of oropharyngeal swabs were 35 (30–37) in survivors and 30 (21–32) in non-survivors (p = 0.003). However, no significant difference was observed in tracheal swabs with 31 (25–34) in survivors and 27 (24–32) in non-survivors (p = 0.11). Median cycles (IQR) in blood samples were equal in survivors and non-survivors (p = 0.613). The same applied to median cycles of anal swabs (p = 0.556) (Fig 3A). Cycle thresholds of urine and dialysate samples are only graphically depicted (Fig 2) due to a low number or absence of positive results. Moreover, cycle thresholds in tracheal aspirates did not differ according to disease severity. No significant difference was found in samples from patients with moderate or severe ARDS (Fig 3B).
Fig 3.
Median RT-qPCR cycles for SARS-CoV-2 RNA in all positive oropharyngeal swabs, tracheal aspirates, blood samples and anal swabs (A). Values were compared between survivors and non-survivors. Ct-values of tracheal aspirates of all time-points as well as day 1 in the local ICU based on the oxygenation index on admission (B). Ct = cycle thresholds.
Anti-SARS-CoV-2 neutralizing antibody formation
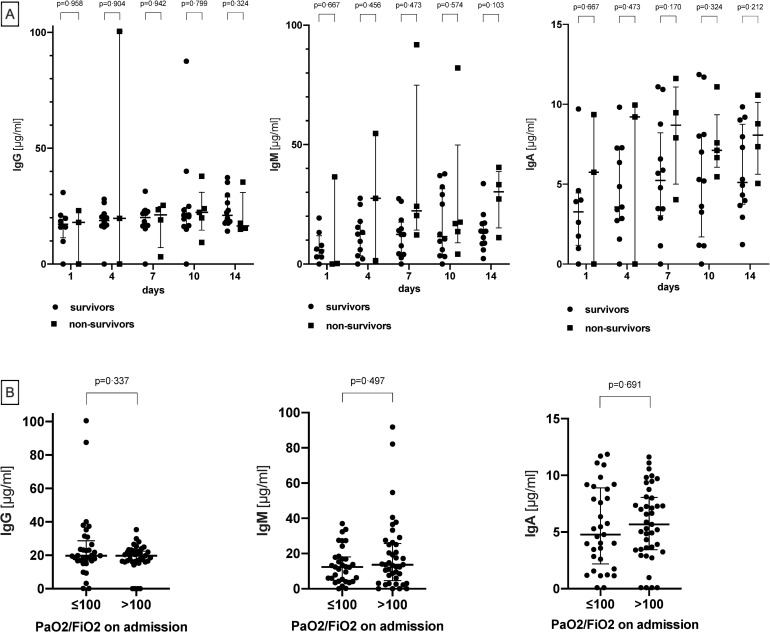
Serologic tests were based on the detection of antibodies which bind to the receptor-binding domain of the SARS-CoV-2 Spike protein. In line with the biological function of this domain, an internal validation confirmed that such antibodies have neutralizing potential (data not shown). Serologic results of 21 patients revealed the presence of IgG, IgM and IgA neutralizing antibodies against SARS-CoV-2. All three immunoglobin classes were present in the same ten patients (43%) on ICU admission. In all but two patients (in which samples were not available) anti-SARS-CoV-2 IgG, IgM and IgA antibodies were detectable during the course of therapy (Fig 4). Median overall-concentrations of IgG were similar in survivors and non-survivors (20 [17–23] vs. 20 [15–24]; p = 0.99), whereas overall-concentrations of IgM and IgA were lower in survivors than in non-survivors (12 [4–18] vs. 20 [11–37]; p = 0.026 and 5 [3–7] vs. 8 [5–9]; p = 0.009). No difference in antibody concentrations was found when breaking down disease severity into moderate vs. severe ARDS on ICU admission (Fig 5).
Fig 4. Levels and distribution of immunoglobulins (IgG, IgM, IgA) detected in blood samples (ELISA) on days 1–14.
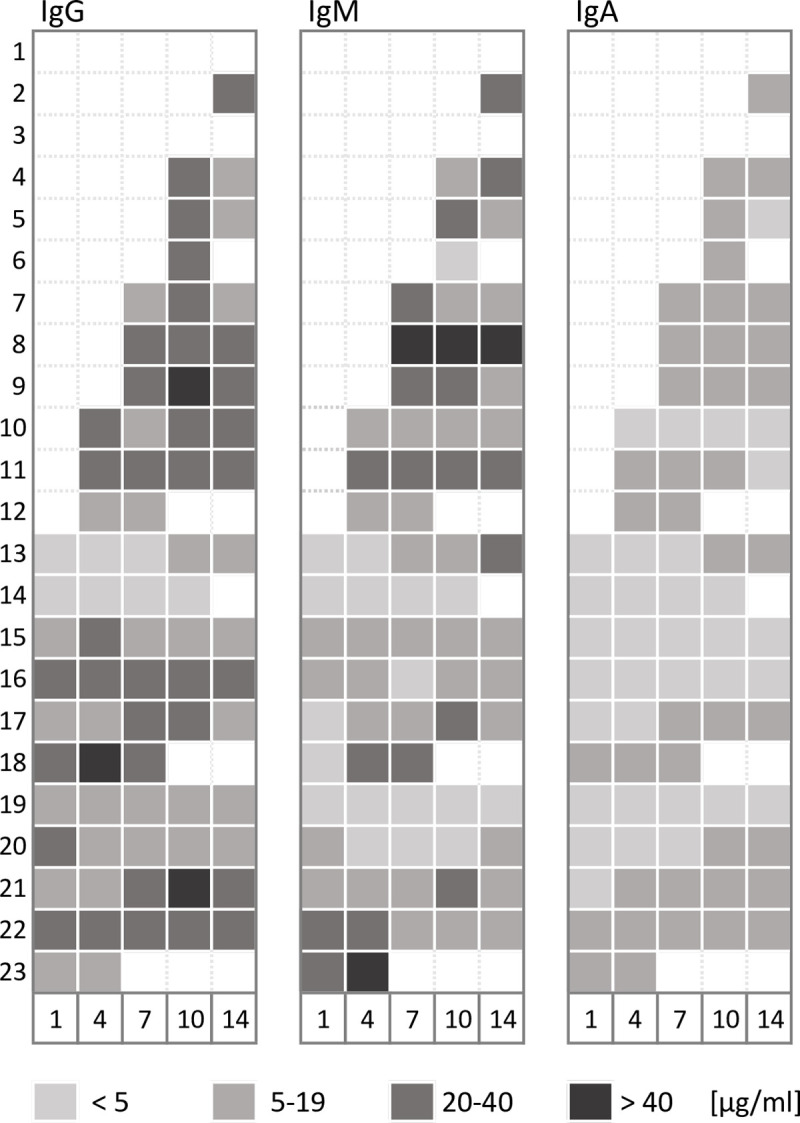
Serologic testing started later than virologic testing of blood samples, which explains discrepancies in the availability of results. White fields indicate unavailable results.
Fig 5.
Comparisons of immunoglobulin serum concentrations (IgG, IgM and IgA) of survivors and non-survivors (A). Comparisons of immunoglobulin serum concentrations (IgG, IgM and IgA) according to ARDS severity on ICU admission (B).
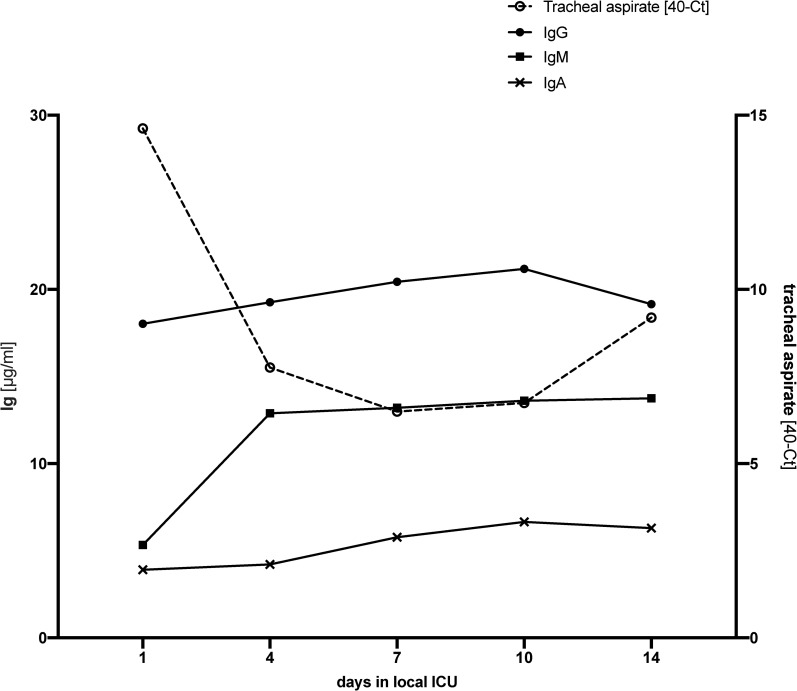
The associations of tracheal viral load and neutralizing serum antibody concentrations are depicted in Fig 6. Tracheal viral load rapidly decreased within the first seven days of ICU treatment, whereas immunoglobin concentrations increased. As expected, IgM peaked earlier than IgG or IgA, nevertheless, on ICU admission IgG already was the most abundant immunoglobulin.
Fig 6. Immunoglobulin (Ig) G, M and A concentrations (medians) in relation to tracheal SARS-CoV-2 viral load over the course of the first 14 days of ICU therapy.
SARS-CoV-2 loads in tracheal aspirate are displayed as medians of 40-Ct resulting from RT-qPCR.
Discussion
The infectiousness of SARS-CoV-2, viral load, clearance and severity of COVID-19 remain important and puzzling questions since the emergence of the pandemic. Our data indicate that severe COVID-19-induced ARDS is accompanied by a high viral load of SARS-CoV-2 in tracheal aspirates, which decreased over the course of ICU treatment. In the majority of patients (57%) tracheal virus RNA remained detectable throughout the course of ICU treatment. Neither viral load nor neutralizing antibody titers distinguished between moderate or severe ARDS. Moreover, both parameters were not different when comparing survivors and non-survivors. As such, our data do not allow a clear-cut distinction between these parameters and treatment or prognosis of COVID-19. We believe that this is an important finding itself and pinpoints towards pathophysiological mechanisms other than viral elimination contributing to disease severity and outcome. These mechanisms e.g. may include an impeded cytotoxic immune response [24] or microthrombosis due to platelet hyperactivation [25]. Oropharyngeal swabs expressed a similar pattern, however, results were less reliable and indicate that oropharyngeal swabs are prone to false negative results. Our data certainly emphasize that in patients receiving invasive mechanical ventilation a lower tracheal aspirate or bronchoalveolar lavage should be collected. In these patients, oropharyngeal swabs are likely not the most suitable specimens. Overall, detection of the highest viral load in respiratory samples confirms previous studies [7, 26].
Importantly, SARS-CoV-2 RNAemia was present in 26% of the patients, a finding contradictory to studies suggesting a direct correlation of RNAemia and severity of COVID-19. SARS-CoV2 RNAemia has been associated with more severe COVID-19, including a higher probability of ICU admission and mechanical ventilation. However, in these studies disease severity was either heterogenous [27] and ARDS severity remained undefined, respectively [28]. Only a minority of patients enrolled in these studies actually required intensive care, whereas our study only included critically ill ICU patients. Hence, although we did not find associations between viral load, RNAemia and survival it might be possible that RNAemia increases in-hospital mortality when considering all severities of COVID-19 [29]. Anal swabs and urine infrequently tested positive for SARS-CoV-2 RNA. Nevertheless, urine samples were positive in more cases than previously reported [9, 26, 30]. This could be an indication of a postulated renal tropism of SARS-CoV-2 [31]. The proportions of positive anal samples were in line with previous findings [9, 10], whereas a study from China reported no differences in blood and fecal viral load between mild and severe cases [7]. Positive anal swabs had previously been suggested as a potential indicator of critical disease, whereas the duration of virus RNA detection of the virus outlasted respiratory samples [32]. This potentially allows SARS-CoV2 transmission in other ways than respiratory droplets over an extended time-period. However, virus RNA detection within the digestive tract was not correlated with gastrointestinal symptoms [33]. It remains unclear why SARS-CoV2 RNA detection was delayed by up to ten days after ICU admission in some of our patients. However, impaired gastrointestinal mobility and delayed defecation is common in critically ill patients and may explain these findings.
Remarkably, SARS-CoV-2 RNA was never detected in dialysate from renal replacement therapy even in patients with RNAemia, indicating that the virus does not cross the dialysis membrane. Hence, dialysis fluid can probably be safely handed and disposed by the ICU personnel. This is an important finding, as a high proportion of severely ill ICU patients require renal replacement therapy, which is in turn associated with handling a high volume of potentially infectious fluid.
Antibodies against SARS-CoV-2 can be detected approximately 7–14 days after symptom onset [34]. The SARS-CoV2 spike protein recognizes and binds to the host receptor angiotensin-converting enzyme (ACE) 2, subsequently mediating viral cell membrane fusion. It is pivotal for SARS-CoV2 infection and a potential therapeutic target including vaccine development. Antibodies against the spike protein are immunodominant and can neutralize SARS-CoV-2 [35]. However, antibody kinetics, acquired immunity, duration of protection, as well as the potential occurrence of mutations disrupting neutralizability remain unknown. This also applies to its relation to disease severity. Patients may be protected from future SARS-CoV2 infection by their acquired immunity, suffer from moderate COVID-19 or even experience increased severity of infection due to immunopathogenesis. The latter may be relevant to both first and second exposure, particularly in an ICU study population [36].
Considering a median of 8 days between symptom onset and ICU admission in our patients, the presence of these antibodies in the majority of patients is expected. In two patients, however, antibody formation was not observed. Both of these patients had been on long-term immunosuppressive medication prior to COVID-19. One of those patients survived ICU treatment. Three classes of antibodies, i.e. IgM, IgA and IgG, could be found at the time of ICU admission and during the course of ICU therapy. IgG antibody titers did not differ between survivors and non-survivors, while IgM and IgA were significantly higher in non-survivors. Studies have found high virus specific antibody titers correlating with enhanced virus elimination [16, 37]. The observed difference might reflect different stages of disease or differences in the interplay between the humoral and cytotoxic immune defense. The observed lower anti-SARS-CoV-2-Spike-RBD IgM levels in survivors may indicate clinical recovery as IgM is the antibody class which predominates in the early stages of an immune response and is the first to be produced. However, other data imply that higher titers might be associated with more severe disease [16, 37]. This finding was also present in patients dying from SARS-Co-V1 [38]. Of course, all patients were critically ill in our cohort, and there was no general association between survival and antibody titers. Interestingly, antibody titers plateaued between days 7 to 14, which likely indicates that antibody formation was fully developed at that time point. In some cases, viral RNA remained detectable even after seroconversion. A lack of complete viral clearance was previously reported and is confirmed by the present results [39]. However, the detection of SARS-CoV-2 RNA alone is not sufficient to diagnose active disease and always need to be interpreted within the clinical context.
In this cohort of ICU patients, most patients (91%) suffered from severe ARDS during the course of therapy with a survival of 70%. Moreover, 65% were supported with ECMO and 74% with renal replacement therapy. None of the patients received antiviral treatment. Median age, prevalence of co-morbidities and duration between symptom onset and ICU admission were in line with previous studies [7, 40]. Hospitalization and ICU treatment were significantly longer [41] and compared to a previous investigation our cohort exhibited higher rates of invasive ventilation, need for ECMO therapy, as well as a higher mortality [30].
When interpreting the results of the current study it is important to emphasize the severity of disease in our patient population. Our data represent ICU patients surviving the worst clinical course of COVID-19 while receiving maximum ICU care. The majority of studies which have assessed SARS-CoV-2 viral loads to date, mainly reported mild cases [7, 9, 42, 43]. Moreover, viral load, virus elimination and neutralizing antibody formation are only some components of the complex COVID-19 pathophysiology. Further limitations include incomplete data in a few patients due to the retrospective design and collection of data within standard care. Generally, all findings regarding virus detection must be interpreted with caution since viral load as assessed via RT-qPCR may also reflect differences in sampling. Furthermore, RT-qPCR can only detect virus RNA but not infective particles. The latter would require cell culture, which is time-consuming and not sensitive enough [44] to be of use in acute clinical care. Being aware that the detection of virus RNA is not proof of intact virus or infectivity, RT-qPCR still remains the standard of care in SARS-CoV-2 diagnostics and the best surrogate for viral load [5]. Moreover, based on an early decision to closely monitor biodistribution and the serologic response, we are confident that the reported results in general represent the SARS-CoV-2 biodistribution and viral loads in the described cohort of severely ill COVID-19 patients.
Supporting information
(DOCX)
Acknowledgments
All investigators would very much like to thank all nurses, physicians and supporting staff for their dedication and relentless work in order to achieve the best possible outcome for their patients.
Florian Krammer (Icahn School of Medicine at Mount Sinai, New York, NY, USA) kindly provided the RBD_6His expression plasmid.
Viktoria Ruecker (Institute of Clinical Epidemiology and Biometry, University of Wuerzburg, Wuerzburg, Germany) kindly provided statistical advice.
Abbreviations
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- Covid-19
corona virus disease 2019
- ICU
intensive care unit
- ARDS
acute respiratory distress syndrome
- RBD
receptor binding domain
- ECMO
extracorporeal membrane oxygenation
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- ELISA
enzyme linked immunosorbent assay
- Ct
cycle thresholds
- RNAemia
ribonucleid acid detected in blood
Data Availability
Data contain potentially identifying and sensitive patient information. According to local requirements, anonymized individual patient level data can only be shared upon request and prior written permission from the Ethics Committee of the University of Wuerzburg (Versbacher Str. 9, 97078 Wuerzburg, Germany, ethikkommission@uni-wuerzburg.de) and the data protection officer of the University hospital of Wuerzburg (Versbacher Str. 2, 97080 Wuerzburg, Germany, datenschutz@ukw.de).
Funding Statement
None. Commercial affiliations did not play a role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript and did not provide any financial support of the current work.
References
- 1.World Health Organization (WHO): WHO Director-General's opening remarks at the media briefing on COVID-19. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020. Published March 11, 2020. Accessed May 11, 2020.
- 2.Center for Systems Science and Engineering (CSSE)—Johns Hopkins University Medicine. Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html. Accessed July 01, 2020.
- 3.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. The Lancet Infectious Diseases. 2020;20(6):669–77. 10.1016/S1473-3099(20)30243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Docherty AB, Harrison EM, Green CA, Hardwick HE, Pius R, Norman L, et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020;369:m1985 10.1136/bmj.m1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO): Clinical management of COVID-19—interim guidance. https://www.who.int/publications/i/item/clinical-management-of-covid-19. Published May 27, 2020. Accessed June 11, 2020.
- 6.Fagiuoli S, Lorini FL, Remuzzi G. Adaptations and Lessons in the Province of Bergamo. New England Journal of Medicine. 2020;382(21):e71 10.1056/NEJMc2011599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. Bmj. 2020;369:m1443 10.1136/bmj.m1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323(18):1843–4. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. The Lancet Infectious Diseases. 2020;20(4):411–2. 10.1016/S1473-3099(20)30113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng L, Liu J, Xu W, Luo Q, Chen D, Lei Z, et al. SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gisella V, Silvia D, Giuseppe G, Maria DF. SARS-CoV2 in the peritoneal waste in a patient treated with peritoneal dialysis. Kidney Int. 2020. [Google Scholar]
- 13.Coccolini F; Dario T; Puglisi Adolfo; Giordano Cesira; Pistello Mauro; Lodato Marianna; et al. SARS-CoV-2 is present in peritoneal fluid in COVID-19 patients. Annals of Surgery. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–74. 10.1016/S1473-3099(20)30196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–9. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases. 2020. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenbroucke JP, von Elm E, Altman DG, Gøtzsche PC, Mulrow CD, Pocock SJ, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. Ann Intern Med. 2007;147(8):W163–94. 10.7326/0003-4819-147-8-200710160-00010-w1 [DOI] [PubMed] [Google Scholar]
- 18.Force ADT, Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307(23):2526–33. 10.1001/jama.2012.5669 [DOI] [PubMed] [Google Scholar]
- 19.World Health Organzation (WHO): Laboratory testing for coronavirus disease (COVID-19) in suspected human cases. Interim guidance 19 March 2020. https://apps.who.int/iris/bitstream/handle/10665/331501/WHO-COVID-19-laboratory-2020.5-eng.pdf?sequence = 1&isAllowed = y. Accessed June 30, 2020.
- 20.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A, et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 2020;5(48). 10.1126/sciimmunol.abc8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr Protoc Microbiol. 2020;57(1):e100 10.1002/cpmc.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–6. 10.1038/s41591-020-0913-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notz Q, Schmalzing M, Wedekink F, Schlesinger T, Gernert M, Herrmann J, et al. Pro- and Anti-Inflammatory Responses in Severe COVID-19-Induced Acute Respiratory Distress Syndrome—An Observational Pilot Study. Frontiers in Immunology. 2020;11(2631). 10.3389/fimmu.2020.581338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hottz ED, Azevedo-Quintanilha IG, Palhinha L, Teixeira L, Barreto EA, Pão CRR, et al. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136(11):1330–41. 10.1182/blood.2020007252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, et al. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin Infect Dis. 2020. 10.1093/cid/ciaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberhardt KA, Meyer-Schwickerath C, Heger E, Knops E, Lehmann C, Rybniker J, et al. RNAemia Corresponds to Disease Severity and Antibody Response in Hospitalized COVID-19 Patients. Viruses. 2020;12(9). 10.3390/v12091045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogan CA, Stevens BA, Sahoo MK, Huang C, Garamani N, Gombar S, et al. High Frequency of SARS-CoV-2 RNAemia and Association With Severe Disease. Clin Infect Dis. 2020. 10.1093/cid/ciaa1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu D, Zhou F, Sun W, Chen L, Lan L, Li H, et al. Relationship Between serum SARS-CoV-2 nucleic acid(RNAemia) and Organ Damage in COVID-19 Patients: A Cohort Study. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z, et al. SARS-CoV-2 Viral Load in Clinical Samples from Critically Ill Patients. Am J Respir Crit Care Med. 2020;201(11):1435–8. 10.1164/rccm.202003-0572LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, et al. Multiorgan and Renal Tropism of SARS-CoV-2. New England Journal of Medicine. 2020. 10.1056/NEJMc2011400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin W, Xie Z, Li Y, Li L, Wen C, Cao Y, et al. Association between detectable SARS-COV-2 RNA in anal swabs and disease severity in patients with coronavirus disease 2019. J Med Virol. 2020. 10.1002/jmv.26307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parasa S, Desai M, Thoguluva Chandrasekar V, Patel HK, Kennedy KF, Roesch T, et al. Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. JAMA Netw Open. 2020;3(6):e2011335 10.1001/jamanetworkopen.2020.11335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52(6):910–41. 10.1016/j.immuni.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei C, Qian K, Li T, Zhang S, Fu W, Ding M, et al. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nature Communications. 2020;11(1):2070 10.1038/s41467-020-16048-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick LC, Rattigan SM, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11(1):4704 10.1038/s41467-020-18450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis. 2020;26(7):1478–88. 10.3201/eid2607.200841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Zhang F, Yu W, He T, Yu J, Yi CE, et al. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78(1):1–8. 10.1002/jmv.20499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yongchen Z, Shen H, Wang X, Shi X, Li Y, Yan J, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microbes Infect. 2020;9(1):833–6. 10.1080/22221751.2020.1756699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–20. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dreher M, Kersten A, Bickenbach J, Balfanz P, Hartmann B, Cornelissen C, et al. The Characteristics of 50 Hospitalized COVID-19 Patients With and Without ARDS. Dtsch Arztebl International. 2020;117(16):271–8. 10.3238/arztebl.2020.0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clinical Infectious Diseases. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Yan L-M, Wan L, Xiang T-X, Le A, Liu J-M, et al. Viral dynamics in mild and severe cases of COVID-19. The Lancet Infectious Diseases. 2020;20(6):656–7. 10.1016/S1473-3099(20)30232-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joynt GM, Wu WKK. Understanding COVID-19: what does viral RNA load really mean? The Lancet Infectious Diseases. 2020;20(6):635–6. 10.1016/S1473-3099(20)30237-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Data contain potentially identifying and sensitive patient information. According to local requirements, anonymized individual patient level data can only be shared upon request and prior written permission from the Ethics Committee of the University of Wuerzburg (Versbacher Str. 9, 97078 Wuerzburg, Germany, ethikkommission@uni-wuerzburg.de) and the data protection officer of the University hospital of Wuerzburg (Versbacher Str. 2, 97080 Wuerzburg, Germany, datenschutz@ukw.de).