c-di-GMP has been widely recognized for its essential role in the production of exopolysaccharides in bacteria, such as alginate produced by Pseudomonas and Azotobacter spp. This study reveals that the levels of c-di-GMP also affect the physical properties of alginate, favoring the production of high-molecular-mass alginates in response to lower OTRs. This finding opens up new alternatives for the design of tailor-made alginates for biotechnological applications.
KEYWORDS: Azotobacter, c-di-GMP, alginate, MucG, molecular mass, oxygen transfer rate
ABSTRACT
Azotobacter vinelandii produces the linear exopolysaccharide alginate, a compound of significant biotechnological importance. The biosynthesis of alginate in A. vinelandii and Pseudomonas aeruginosa has several similarities but is regulated somewhat differently in the two microbes. Here, we show that the second messenger cyclic dimeric GMP (c-di-GMP) regulates the production and the molecular mass of alginate in A. vinelandii. The hybrid protein MucG, containing conserved GGDEF and EAL domains and N-terminal HAMP and PAS domains, behaved as a c-di-GMP phosphodiesterase (PDE). This activity was found to negatively affect the amount and molecular mass of the polysaccharide formed. On the other hand, among the diguanylate cyclases (DGCs) present in A. vinelandii, AvGReg, a globin-coupled sensor (GCS) DGC that directly binds to oxygen, was identified as the main c-di-GMP-synthesizing contributor to alginate production. Overproduction of AvGReg in the parental strain phenocopied a ΔmucG strain with regard to alginate production and the molecular mass of the polymer. MucG was previously shown to prevent the synthesis of high-molecular-mass alginates in response to reduced oxygen transfer rates (OTRs). In this work, we show that cultures exposed to reduced OTRs accumulated higher levels of c-di-GMP; this finding strongly suggests that at least one of the molecular mechanisms involved in modulation of alginate production and molecular mass by oxygen depends on a c-di-GMP signaling module that includes the PAS domain-containing PDE MucG and the GCS DGC AvGReg.
IMPORTANCE c-di-GMP has been widely recognized for its essential role in the production of exopolysaccharides in bacteria, such as alginate produced by Pseudomonas and Azotobacter spp. This study reveals that the levels of c-di-GMP also affect the physical properties of alginate, favoring the production of high-molecular-mass alginates in response to lower OTRs. This finding opens up new alternatives for the design of tailor-made alginates for biotechnological applications.
INTRODUCTION
Alginates are linear polysaccharides composed of variable proportions of (1–4)-linked β-d-mannuronic acid (M) and its C-5 epimer α-l-guluronic acid (G). This polymer is produced by brown algae and by bacteria of the genera Azotobacter and Pseudomonas (1). Bacterial alginates may be O-acetylated at some of the C-2 or C-3 carbons of the mannuronic acid residues (1). Alginates are important biopolymers with broad applications in the medical and industrial fields, where they are used as stabilizing, thickening, and gelling agents. Alginate microspheres have also been used in therapeutic delivery, releasing drugs, proteins, vaccines, and cells (2). It is well known that the properties of the polymer are determined by the ratio and distribution of M and G residues, by the acetylation degree, and by the overall molecular mass of the polymer (1, 3). Currently, this polymer is manufactured from brown algae but, given the intrinsic variation in their composition, Azotobacter vinelandii has been proposed as a source for the production of tailor-made alginates (1).
In Pseudomonas aeruginosa, alginate acts as a virulence factor mediating the formation of biofilms, which provide a thick protective layer against host immune attacks and antimicrobials (4, 5); in Azotobacter species, which are free-living bacteria, alginate is produced in copious amounts during vegetative growth, where it may serve as a diffusion barrier against heavy metals and oxygen (6). This polymer is also produced during the developmental process of A. vinelandii, leading to the formation of dormant cells called cysts. These differentiated cells are surrounded by an alginate coat that is essential to resist harsh environmental conditions (6).
The alginate biosynthetic pathway is well conserved between the Azotobacter and Pseudomonas genera (1, 7). The activated cytosolic precursor GDP-mannose is polymerized to alginate in a synthase-dependent pathway by the action of the membrane-anchored glycosyltransferase Alg8 and the copolymerase Alg44. A multiprotein scaffold complex that is composed of 10 subunits and includes Alg8-Alg44 spans the cytoplasmic membrane, the periplasm, and the outer membrane and guides the nascent polymannuronate chain for secretion (1, 8, 9). Translocation of the nascent polymannuronate chain across the periplasm is coupled with modifications, such as O-acetylation (conducted by AlgX) and epimerization of β-d-mannuronic acid moieties (conducted by AlgG). A periplasmic alginate lyase (AlgL) forms part of the multiprotein scaffold complex and is necessary to degrade alginate chains that have been misguided into the periplasm or to degrade the nascent alginate chain (10, 11). In A. vinelandii, the secreted alginate is further modified by a family of extracellular mannuronan C-5 epimerases (AlgE1 to AlgE7), which introduce various amounts of α-l-guluronic acid residues in different patterns and distributions (12).
The molecular basis of polymer chain length determination in alginate is not totally understood. In P. aeruginosa, however, the process of alginate polymerization and the modification events (i.e., acetylation and epimerization) show a clear relationship. While the polymer acetylation rate directly correlates with the molecular mass, the rate of alginate epimerization by AlgG inversely correlates with the molecular mass of the alginate produced, presumably as a result of AlgG-mediated polymer degradation (9). Furthermore, it has been suggested that AlgG, as a scaffold subunit, might be critical for processivity of alginate polymerization. In A. vinelandii, it has been proposed that both the activity of the alginate polymerase complex Alg8-Alg44 and that of alginate lyases contribute to the polymer chain length of alginate (13). AlgL in A. vinelandii was shown to reduce the molecular mass of the alginate during vegetative growth at late stationary phase (14). In addition to AlgL, A. vinelandii contains five other alginate lyases (AlyA1, AlyA2, AlyA3, AlgE7, and AlyB), but their role in determining the alginate molecular mass has not been investigated (15). AlyA1 to AlyA3 belong to the PL7 family of alginate lyases. AlyA2 is necessary for normal growth in the laboratory, while AlyA1 is dispensable for growth. AlyA3 and AlgE7 are extracellular Ca2+-dependent alginate lyases that are necessary for the rupture of the A. vinelandii cyst during germination and the release of the nascent alginate chain from the cell surface, respectively. AlyB is an exolyase whose function is still unknown (11, 15).
The biosynthesis of a great variety of exopolysaccharides is regulated at multiple levels by the ubiquitous second messenger bis(3′,5′)-cyclic dimeric GMP (c-di-GMP) (16, 17). c-di-GMP is produced from two GTP molecules by the action of diguanylate cyclases (DGCs) with a GGDEF domain and is degraded by the action of specialized c-di-GMP phosphodiesterases (PDEs) with either EAL or HD-GYP domains (18). Genomic analyses have shown that GGDEF and EAL domains can coexist in the same polypeptide chain and also may be fused to sensory domains such as PAS, GAF, and REC domains, among others. Therefore, the enzymatic activities of a particular DGC or PDE could be regulated by environmental signals (18, 19). To exert its function, c-di-GMP must bind to an effector/receptor component that is responsible for mounting a cellular response. Several classes of c-di-GMP receptors have been reported and include transcription factors, riboswitches, degenerate GGDEF and EAL domains, and proteins containing a c-di-GMP-binding PilZ domain (18, 20).
Alginate polymerization in P. aeruginosa is regulated by c-di-GMP at the posttranslational level by allosteric binding to the PilZ domain of the copolymerase Alg44 (21, 22). The DGC MucR has been found to be the source for c-di-GMP-dependent regulation of Alg44 in P. aeruginosa (23). The molecular basis of the Alg8 activation mechanism upon binding of c-di-GMP to the PilZ domain of Alg44 was investigated (24). This mechanism does not involve the stabilization of the polymerase complex or its interaction with other scaffold subunits (i.e., AlgX, AlgG, or AlgK). Analysis of critical residues in Alg8 for the c-di-GMP-mediated activation of alginate polymerization suggests that such a mechanism is dissimilar from the autoinhibitory mechanism proposed for cellulose polymerization (24). To date, the effect of different levels of c-di-GMP on the molecular mass of the alginate has not been investigated.
A. vinelandii has orthologues of Alg44, which has a conserved PilZ domain (22), and of MucR, raising the question of whether c-di-GMP is involved in the regulation of alginate polymerization in this organism. Furthermore, we showed previously that a hybrid GGDEF-EAL protein named MucG, which is not present in P. aeruginosa, negatively controls A. vinelandii alginate production and molecular mass in response to the oxygen transfer rate (OTR) (25). All of these observations strongly suggest that c-di-GMP plays an important role in alginate biosynthesis in A. vinelandii, and the goal of this work was to investigate the molecular basis of this regulation.
Our results indicate that the mechanism by which MucG and the OTR modulate alginate biosynthesis and molecular mass in A. vinelandii involves c-di-GMP signaling. Artificially induced accumulation of c-di-GMP had effects on alginate biosynthesis similar to those of the absence of mucG. In contrast to what happens in P. aeruginosa, the absence of the MucR orthologue did not impair alginate production. Our results revealed that AvGReg, a GCS DGC, is the main contributor to c-di-GMP-dependent activation of alginate production in A. vinelandii.
RESULTS
MucG is an active c-di-GMP PDE.
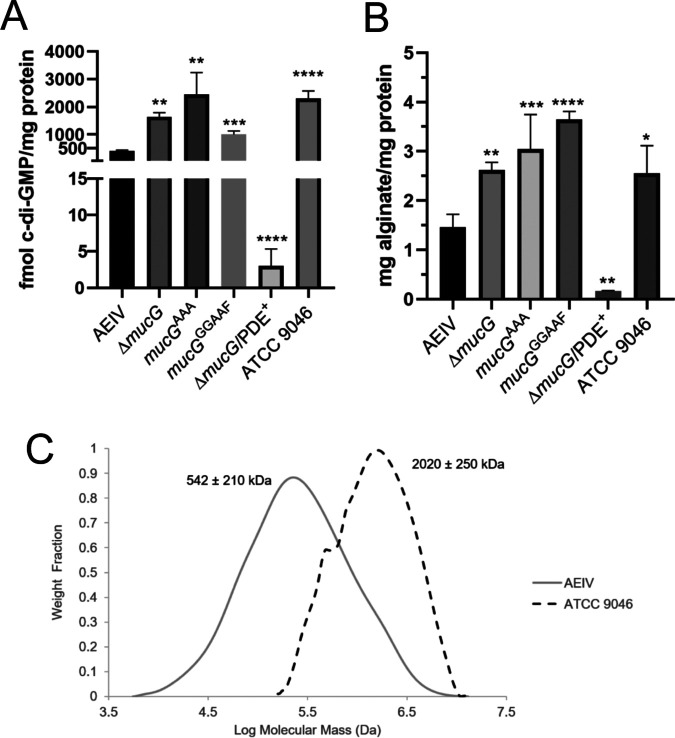
The GGDEF-EAL MucG protein Avin_07910 (GenBank accession number ACO77036; also referred to as AVIN_RS03680) is implicated in determining the total amount and mean molecular mass of alginate, and these effects depend on the presence of an intact EAL domain (25). These observations strongly suggest that MucG has c-di-GMP PDE activity; however, this activity has not been demonstrated experimentally. To begin to address this point, we quantified c-di-GMP in the wild-type (WT) strain AEIV, a ΔmucG strain (CLAM04), and a strain with a mutated allele of mucG (mucGAAA) in the chromosome that produces a MucG protein lacking conserved amino acids of the EAL catalytic site. The absence of MucG or the production of a putatively inactive version of MucG resulted in increased c-di-GMP accumulation, compared to the WT strain (Fig. 1A). Together, these results strongly suggest that MucG is an active PDE capable of modulating the abundance of c-di-GMP in A. vinelandii.
FIG 1.
MucG negatively controls c-di-GMP accumulation and alginate production. (A) c-di-GMP quantification in the WT strain AEIV, in its derivative mutants CLAM04 (ΔmucG), CLAM01 (mucGAAA), CLAM05 (ΔmucG/PDE+), and CLAM06 (mucGGGAAF), and in the alginate-overproducing strain ATCC 9046. (B) Alginate quantification in the same A. vinelandii strains used in panel A. Means and standard deviations from three independent experiments are shown. Significant differences were analyzed by t test. Statistical significance is indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Graphic showing the molecular mass distribution of the alginates produced by strains AEIV and ATCC 9046. A representative curve for each strain is shown. Cells were grown for 48 h in liquid Burk’s-sucrose medium.
MucG is a hybrid protein containing both GGDEF and EAL conserved domains. To begin to address the contribution of the GGDEF domain of MucG to the intracellular c-di-GMP pool, we quantified c-di-GMP in a strain with a mutant mucG allele that produces a MucG variant with a GGAAF motif. These amino acid changes typically result in loss of DGC activity in GGDEF-containing enzymes. c-di-GMP accumulation in the mucGGGAAF strain was greater than that in the WT strain, clearly suggesting that the GGDEF domain is required for the PDE activity of MucG. This result is not unprecedented, as in other GGDEF-EAL proteins the GGDEF domain modulates the PDE activity in response to the availability of GTP (26–28).
c-di-GMP levels are linked to the regulation of alginate abundance by MucG.
Although c-di-GMP has been shown to control alginate production in P. aeruginosa (21, 23), its role in A. vinelandii has not been addressed previously. To determine the importance of the cellular concentration of c-di-GMP for alginate production in the ΔmucG strain, we analyzed the effect of the overproduction of a putative A. vinelandii PDE (Avin_50640 [GenBank accession number ACO81152], also referred to as AVIN_RS23150). Avin_50640 has a HD-GYP domain with conserved catalytic residues with respect to that of the active PDE PmGH from Persephonella marina (29). Overproduction of Avin_50640 in the ΔmucG strain (ΔmucG/PDE⁺) resulted in severely reduced c-di-GMP levels, compared to both the WT strain and the ΔmucG strain (Fig. 1A). While alginate levels were higher in the ΔmucG, mucGAAA, and mucGGGAAF strains, compared to the WT strain, the production of this polymer was negligible in the ΔmucG strain overproducing Avin_50640 (Fig. 1B). These results indicate that increased alginate accumulation in the ΔmucG strain can be suppressed by ectopically expressing an active PDE.
The A. vinelandii strain ATCC 9046 has been extensively studied due to its natural high-molecular-mass alginate-overproducing phenotype (13, 30). We hypothesized that this strain would accumulate higher levels of c-di-GMP, compared to our WT strain AEIV. The quantification of c-di-GMP in the ATCC 9046 strains revealed that the c-di-GMP concentration was approximately 5-fold higher in the ATCC 9046 strain than in the AEIV strain (Fig. 1A). Under our tested growth conditions, this strain synthesized about 2-fold more alginate than the AEIV strain, a level comparable to that of the ΔmucG and mucGAAA mutants (Fig. 1B). The mean molecular mass of the synthesized alginate was 2,020 ± 250 kDa, approximately 4 times greater than that of AEIV (Fig. 1C). The nucleotide sequence of the mucG gene in these two strains is conserved. Furthermore, strain ATCC 9046 showed comparable mucG expression levels, relative to strain AEIV, as deduced by quantitative reverse transcription-PCR (qRT-PCR) analysis conducted at both 24 and 48 h of cultivation (0.98 ± 0.06 and 1.01 ± 0.10 relative units, respectively). The gyrA mRNA levels were used as an internal control, allowing normalization of the qRT-PCR data. This result implies that other factors are responsible for the observed differences in c-di-GMP concentrations between these strains.
MucG is the only PDE implicated in the control of alginate production under our test conditions.
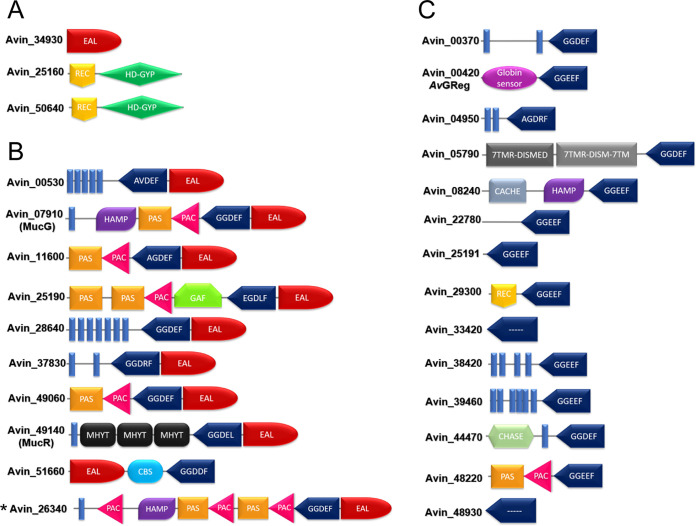
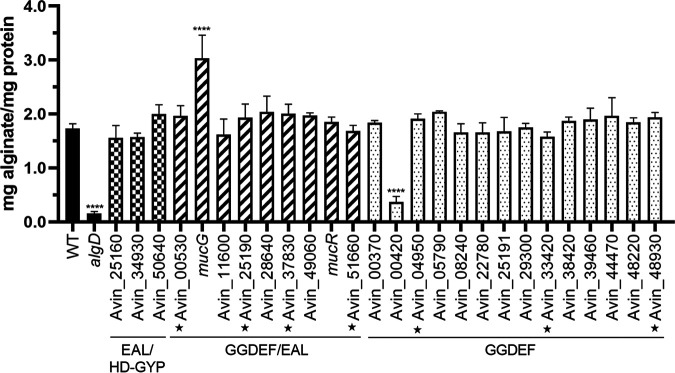
The genome of A. vinelandii DJ strain is predicted to encode 27 proteins with putative domains for the synthesis or degradation of c-di-GMP (Fig. 2), including 3 PDEs (1 EAL type and 2 with HD-GYP domains), 10 tandem GGDEF/EAL domain proteins, and 14 GGDEF domain proteins. To investigate whether additional PDEs (besides MucG) control alginate production, we constructed insertional mutants in genes encoding proteins with predicted EAL or HD-GYP domains. The mutants were evaluated for their capacity to stimulate the production of alginate in liquid Burk’s-sucrose medium. As can be seen in Fig. 3, except for the MucG-deficient strain, the mutants with genes encoding EAL/HD-GYP or GGDEF-EAL proteins showed alginate levels similar to those of the parental strain AEIV. This result confirmed the central role of the MucG PDE activity in the control of alginate production in A. vinelandii under our test conditions.
FIG 2.
Depiction of the A. vinelandii proteins containing GGDEF, EAL, or HD-GYP domains in the genome of strain DJ. Prediction of the domains was according to Pfam and SMART. Models are not drawn to scale. Large amino acid regions without a predicted domain are also depicted. (A) EAL and HD-GYP domain-containing proteins. (B) Dual GGDEF-EAL domain-containing proteins. (C) GGDEF domain-containing proteins. The star denotes a protein encoded by the A. vinelandii DJ genome but not the AEIV genome. The GGDEF-EAL protein Avin_25190 was originally annotated as having an additional GGDEF domain at the C terminus. However, analysis of the DNA sequence of this open reading frame in both strain DJ and strain AEIV indicated to us that this GGDEF domain constitutes a different open reading frame, which we have named Avin_25191 (for details, see Fig. S1 in the supplemental material).
FIG 3.
Effects on alginate production of mutations in genes involved in synthesizing or degrading c-di-GMP. Alginate quantification in the parental AEIV strain (WT) and in strains with deletion-insertion mutations in 3 putative PDEs (checkered), 9 hybrid DGC-PDEs (striped), and 14 DGCs (dotted). The stars denote proteins with degenerate GGDEF domains. A mutant unable to produce alginate was included as a negative control (algD); means and standard deviations from three independent experiments are shown. Significant differences were analyzed by one-way analysis of variance with Dunnett's post hoc test. Statistical significance is indicated. ****, P < 0.0001. Cells were grown for 48 h in liquid Burk’s-sucrose medium.
The globin-coupled sensor DGC AvGReg is the main c-di-GMP contributor for alginate production in A. vinelandii.
The ATCC 9046 strain and the ΔmucG mutant accumulate higher levels of c-di-GMP and produce alginates of higher molecular mass, compared to the alginate produced by strain AEIV (25) (Fig. 1). These data suggest that c-di-GMP levels may control alginate polymerization.
A. vinelandii contains an orthologue of the DGC MucR from P. aeruginosa (31) that in the latter bacterium provides the c-di-GMP pool for activation of the Alg8-Alg44 polymerase complex. We evaluated the effect of MucR (Avin_49140 [GenBank accession number ACO81013], also referred to as AVIN_RS22475) on alginate biosynthesis in A. vinelandii. A strain carrying a deletion of mucR (ICM01) produced alginate levels similar to those of the parental strain AEIV, indicating that under the conditions tested MucR is not involved in alginate production (Fig. 3).
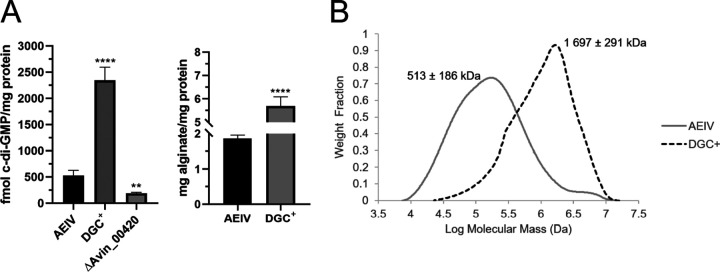
To identify potential DGCs implicated in alginate production, we constructed strains carrying individual deletions in genes coding for proteins with GGDEF domains (Fig. 2). A total of 14 mutants were generated and tested for the production of alginate in liquid Burk’s-sucrose medium. As can be seen in Fig. 3, Avin_00420 was the only DGC affecting alginate production; its absence reduced by 70% the accumulation of this polysaccharide and by 60% the amount of c-di-GMP detected, relative to the parental AEIV strain (Fig. 4A). The role of Avin_00420 in alginate production was also tested in the ATCC 9046 genetic background. An ATCC 9046 derivative carrying a deletion of Avin_00420 showed a reduction of about 40% in the production of alginate, compared to the WT strain (see Fig. S2 in the supplemental material).
FIG 4.
Effect of AvGReg (Avin_00420) overproduction on the alginate produced by A. vinelandii. (A) Quantification of the amount of c-di-GMP (left) and alginate (right) production in strain AEIV and in its derivative overexpressing the DGC AvGReg (DGC+). Production of c-di-GMP in the absence of AvGReg (ΔAvin_00420) is also shown. The bars for standard deviations from three independent experiments are shown. Significant differences were analyzed by t test. Statistical significance is indicated. **, P < 0.01; ****, P < 0.0001. (B) Graphic showing the molecular mass distribution of the alginates produced by strains AEIV and DGC+. A representative curve for each strain is shown. A. vinelandii strains were cultivated in Burk’s-sucrose medium for 48 h.
Avin_00420 (GenBank accession number ACO76309, also called AVIN_RS00255) encodes a cytoplasmic DGC that was previously characterized and named AvGReg (32, 33). AvGReg is a 52.8-kDa protein with an N-terminal globin-coupled sensor (GCS) domain and a C-terminal GGDEF transmitter domain, homologous to DosC of Escherichia coli (34). The DGC activity of AvGReg is stimulated by the direct binding of oxygen to its GCS domain, which displays high affinity for this diatomic molecule (33). When the GCS domain does not bind to oxygen, the DGC activity is reduced by 50% (33). The genetic arrangement of the AvGReg-encoding gene suggests that it is transcribed as a monocistron. Furthermore, orthologues of this gene were found in Azotobacter beijerinckii and Azotobacter chroococcum but not in P. aeruginosa.
Overproduction of AvGReg results in increased alginate production and higher alginate molecular mass.
To further test whether artificial elevation of c-di-GMP could affect alginate production and polymerization, we overproduced AvGReg from a constitutive σ70 promoter, resulting in approximately 5-fold and 3-fold increases in c-di-GMP and alginate levels, respectively, compared to the parental AEIV strain (Fig. 4A). These results further support the role of c-di-GMP as a regulator of alginate production. Importantly, the mean molecular mass of the polymer produced by the strain overproducing AvGReg (DGC positive) was 3-fold higher than that of the WT strain (1,697 versus 513 kDa) and also exceeded the mean molecular mass of the reported MucG-deficient mutants (∼1,100 kDa) (Fig. 4B). Analysis of the molecular mass distribution revealed that about 60% of the alginates produced by the DGC-positive strain exhibited a molecular mass in the range of 1,000 to 10,000 kDa, confirming that increased levels c-di-GMP favor the synthesis of longer alginate chains (Fig. 4B).
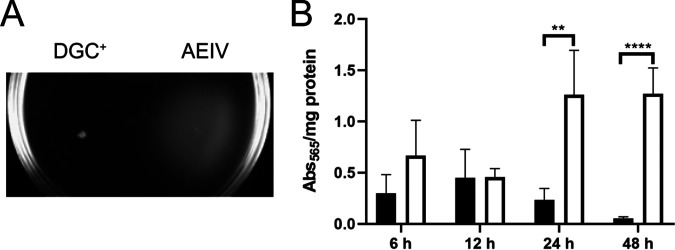
We also explored whether the accumulation of c-di-GMP through overproduction of AvGReg affected other phenotypes regulated by this second messenger. As expected, higher levels of c-di-GMP arrested swimming motility on soft agar plates but enhanced biofilm formation (Fig. 5). At 24 and 48 h, the biofilm formed by the DGC-positive strain increased 3-fold, relative to the WT strain (Fig. 5B).
FIG 5.
Effects of AvGReg overproduction on swimming motility and biofilm formation. (A) Swimming motility of strain AEIV and its derivative overexpressing AvGReg (DGC+) on soft agar after 24 h of incubation. (B) Quantification of biofilm formed in 24-well polystyrene plates by strains AEIV (black) and DGC+ (white) after static incubation at 30°C for the indicated times. Standard deviations from three independent experiments are shown. Significant differences were analyzed by t test. Statistical significance is indicated. **, P < 0.01; ****, P < 0.0001.
The OTR regulates the molecular mass of alginate by modulating the levels of c-di-GMP.
In A. vinelandii, the molecular mass of alginate is regulated by the OTR in the culture medium. When the maximum OTR (OTRmax) is reduced from 5.2 to 2.5 mmol O2/liter/h, the molecular mass of alginate produced by the WT strain AEIV increases from 596 to 969 kDa. This response is not observed in a MucG-deficient strain, since it produces alginates of high molecular mass independently of the OTRmax in the culture medium (Fig. 6A) (25). We hypothesized that changes in the polymer length in response to high or low OTR would be the consequence of different concentrations of c-di-GMP. We found that, in the AEIV strain, the c-di-GMP levels gradually increased when the OTRmax decreased from 5.2 to 1 mmol/liter/h (Fig. 6B). This increase could be the result of reduced activity of the PDE MucG in response to different OTR conditions. To test this hypothesis, we analyzed c-di-GMP levels in the ΔmucG strain grown at the same OTRmax as described for the WT strain. The levels of c-di-GMP in the ΔmucG strain were always higher than those in strain AEIV. At the highest OTRmax (5.2 mmol/liter/h), the amount of the second messenger in the ΔmucG mutant was close to the maximum levels attained in the WT strain (at 1.0 mmol/liter/h), thus explaining the high molecular mass of the alginate produced in the absence of MucG. When OTRmax decreased from 5.2 to 2.5 mmol/liter/h, c-di-GMP levels increased in the ΔmucG strain and were unaffected by a further reduction in OTRmax from 2.5 to 1.0 mmol/liter/h (Fig. 6B). These results suggest that MucG is not the only modulator of c-di-GMP levels in response to the OTR and that further increases in c-di-GMP levels at lower OTRmax in the absence of MucG do not significantly alter the alginate molecular mass. Collectively, our data strongly suggest a key role for the PDE activity of MucG in the control of the c-di-GMP levels that determine the alginate chain length in response to OTRmax changes. Our results also suggest that the overall pool of c-di-GMP is smaller at the highest OTRmax tested, likely because of reduced DGC activity under these conditions.
FIG 6.
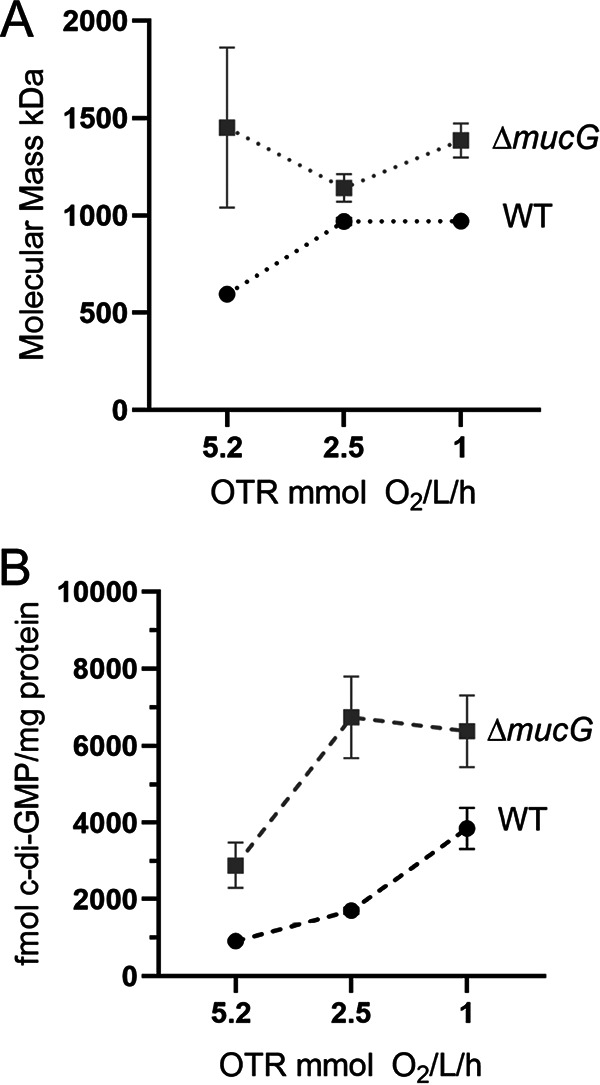
Levels of c-di-GMP vary in response to the aeration conditions of the culture medium. (A) Determination of the molecular mass of the alginates produced by the AEIV strain (WT) and the ΔmucG mutant at the indicated OTR. (B) Quantification of c-di-GMP in the same strains as in panel A. The cells were cultivated in Burk’s-sucrose medium for 48 h. Means and standard deviations from three independent experiments are shown. Statistical analysis of c-di-GMP changes under the different OTRmax conditions evaluated is presented in Fig. S3.
DISCUSSION
The involvement of c-di-GMP signaling in the regulation of alginate biosynthesis has been documented in P. aeruginosa but is less well understood in A. vinelandii. In P. aeruginosa, the DGC involved in the control of alginate biosynthesis was found to be the hybrid GGDEF-EAL protein MucR (23). The mucR gene is regulated by the response regulator AlgR, an important regulator of alginate production in P. aeruginosa (35). The receptor involved in the c-di-GMP-mediated regulation of alginate production in P. aeruginosa was found to be Alg44, a copolymerase that binds c-di-GMP through its PilZ domain (22). Thus, MucR and Alg44 are so far the only known members of the c-di-GMP signaling module responsible for the regulation of alginate polymerization in P. aeruginosa. In a previous report, we showed that in A. vinelandii MucG exerted negative effects on alginate production and alginate molecular mass. MucG has conserved GGDEF and EAL domains; however, the negative effects exerted on alginate biosynthesis and polymerization suggested it may act as a PDE. Our results show that c-di-GMP regulates alginate biosynthesis in A. vinelandii and that MucG is the c-di-GMP-degrading enzyme responsible for limiting alginate production and its molecular mass. The genome of P. aeruginosa does not have an orthologue of mucG, and this observation was an early indication that the c-di-GMP signaling module that controls alginate biosynthesis in A. vinelandii might differ from that previously reported in P. aeruginosa. Furthermore, in P. aeruginosa an interplay between alginate polymerization and its modification was observed, in which the acetylation rate correlated with the molecular mass while the epimerization reduced the polymer chain length (9). In A. vinelandii, the interplay between alginate polymerization and its modification appears to have distinct characteristics. For instance, the increased production and increased molecular mass of alginates in the absence of MucG were not accompanied by changes in the acetylation rate of the polymer, compared to the WT strain (25). Another important difference in the mechanism of alginate modification in P. aeruginosa and A. vinelandii involves the activity of the periplasmic epimerase AlgG, which is crucial in P. aeruginosa but has negligible activity in A. vinelandii (12, 36). The lack of activity of AlgG in A. vinelandii is most likely compensated for by the contribution of a family of extracellular mannuronan C-5 epimerases (37). These important changes in the regulation of the modification events of alginates produced by A. vinelandii and P. aeruginosa might reflect differences in the biological role of this polymer in these two bacteria.
One of the main goals of this work was to determine whether one or multiple DGCs contribute to the regulation of alginate production in A. vinelandii. The genome of this bacterium encodes an orthologue of MucR from P. aeruginosa; however, this protein was dispensable for alginate biosynthesis. Instead, the A. vinelandii AEIV strain uses the oxygen GCS DGC AvGReg as the main c-di-GMP contributor for alginate production; AvGReg is able to bind oxygen directly, which stimulates its DGC activity (33). Its oxygen affinity is very high, one of the highest among the GCSs characterized to date and about 350-fold higher than that reported for DosC of E. coli (0.04 and 14 μM, respectively) (33, 38). A. vinelandii is a nitrogen-fixing, strictly aerobic bacterium; therefore, in order to protect its oxygen-sensitive nitrogenase enzyme, it has evolved adaptations such as a high respiratory rate, which has been proposed to maintain low levels of cytoplasmic oxygen (39). It is unclear whether the oxygen levels that support nitrogen fixation would affect the activity of AvGReg or whether its function might be modulated by ligands that compete with oxygen under certain physiological conditions. The strain lacking AvGReg retains residual alginate production capability (Fig. 3), which is greater than that of the strain overproducing the PDE Avin_50640. This finding suggests that other DGCs may contribute to the activation of alginate biosynthesis.
Very little is known about the signals that modulate alginate production and polymerization. We showed previously that oxygen, and more specifically the OTR, regulates alginate biosynthesis. This is of great importance, since the OTR regulates important physiological traits of A. vinelandii such as nitrogen fixation (40). Here, we showed that the OTR affects c-di-GMP accumulation in a manner that correlates with its regulation of alginate abundance and molecular mass in liquid medium. MucG was the main limiting factor for the production of longer alginate chains in response to changes in OTR; however, other signaling modules appear to further control c-di-GMP accumulation in response to a greater OTR in the absence of MucG (Fig. 6B). Besides the domains for c-di-GMP metabolism, MucG has a HAMP domain and a PAS domain, with the latter predicted to bind the flavin mononucleotide cofactor involved in sensing the intracellular redox state (25). It is possible that differences in the redox state derived from the distinct OTR of the culture medium are sensed by the MucG PAS domain, controlling the activity of the PDE domain. Similar cases have been documented in the past; for instance, the PDE activity of PdeO (formerly YddU) is enhanced by oxygen bound to its PAS domain (34), while the DGC activity of AxDGC2 is stimulated by the oxidized form of its flavin adenine dinucleotide cofactor (41). Also, the PDE activity of RmcA and RbdA depends on their functional PAS domains. The PDE activity is activated under oxidizing conditions in the case of RmcA (42), whereas it is suppressed by the O2-bound PAS domain for RbdA (43), reflecting differences in their physiological roles. Based on these observations, we think it is important to determine whether MucG is a redox sensor and what the role of its sensor domain could be for the function of this enzyme. The activity of AvGReg has been shown to be dependent on oxygen availability; however, the high affinity for oxygen of its globin domain suggests that it might be constitutively active at all physiological levels of oxygen that sustain the growth of A. vinelandii (33). Interestingly, O2 dissociation from AvGReg shows a biphasic behavior in vitro, which has been explained by the existence of two conformational states, i.e., closed and open (33). Displacement of oxygen by competitors such as CO is slower in the closed conformation than in the open complex. It is unclear how changes in the conformation of AvGReg might be regulated, but this could potentially alter signal transduction (33). It is also important to note that AvGReg could be regulated by product inhibition, given its conserved I site, which might also be an important factor that modulates its response (33). Besides its role as an oxygen sensor, the globin domain has been proposed to contribute to NO detoxification, based on some of its in vitro properties (32). The proposed NO-scavenging activity of AvGReg may interfere with its function as a DGC and a modulator of alginate production.
Taken together, our results indicate the existence of a c-di-GMP control module, composed of the DGC AvGReg and the PDE MucG, for the regulation of the amount and chain length of the alginate produced by A. vinelandii under our test conditions (planktonic growth in minimal medium) (Fig. 7). Based on the degree of conservation between Alg44 from P. aeruginosa and that from A. vinelandii, it is presumed that this protein is also the receptor involved in the regulation of alginate polymerization in A. vinelandii. Therefore, it would be important to investigate whether MucG colocalizes or directly interacts with Alg44 and whether its localization affects the allosteric activation of the Alg8-Alg44 polymerase complex. Considering that alginate lyases also affect the alginate chain length, we cannot rule out the possibility that any such enzymes could be regulated by the AvGReg-MucG control module. It is worth pointing out that the observed role for the AvGReg-MucG module is not restricted to the A. vinelandii AEIV strain, as the absence of AvGReg in ATCC 9046 also reduced the production of alginate (see Fig. S2 in the supplemental material). However, this reduction was not as marked as in strain AEIV, implying that other DGCs regulate alginate production in strain ATCC 9046, which could be related to its high polymer levels. In addition, we recently reported that the absence of MucG in the ATCC 9046 genetic background increased the molecular mass of the polymer from 2,170 to 3,112 kDa and tripled the apparent viscosity of alginate solutions when cells were cultivated under microaerophilic conditions (44).
FIG 7.
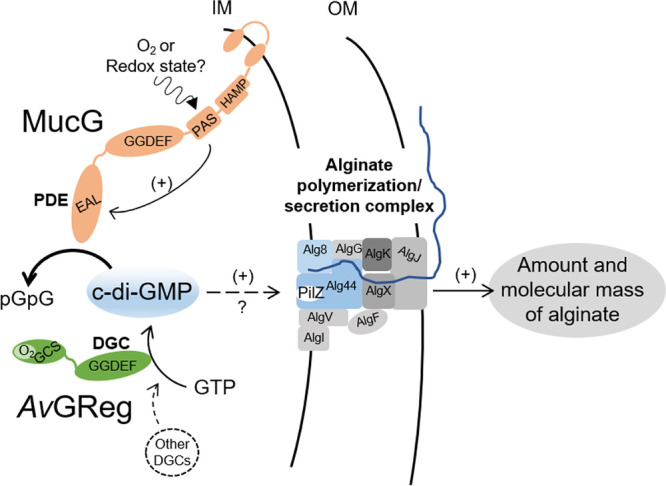
Proposed model for the regulation of alginate production and alginate molecular mass by the AvGReg-MucG control module in strain AEIV of A. vinelandii. The DGC AvGReg and the PDE MucG regulate the intracellular pool of c-di-GMP necessary for the production of alginate, likely through allosteric activation of the polymerase Alg8-Alg44 complex. Increased levels of c-di-GMP favor the synthesis of alginates with higher molecular masses. We propose that the PDE activity of MucG might be stimulated by oxidizing conditions, as detected through its PAS sensor domain, modulating the c-di-GMP pool necessary for alginate production. Other DGCs might contribute mildly, more likely in a nonspecific way, to the pool of c-di-GMP. See text for details. IM, inner membrane; OM, outer membrane.
In an accompanying paper, we demonstrate that in A. vinelandii c-di-GMP also regulates the expression of a family of C-5 epimerases, AlgE1 to AlgE7, involved in the modification of alginate by introducing α-l-guluronic (G) residues into the polymer, improving its gelling properties (31). During encystment, the differentiated A. vinelandii cell enters a dormant stage in which it is protected by two alginate layers containing different proportions of G blocks, which are essential for surviving drought conditions (15, 37). Production of AlgE1 to AlgE7 epimerases required the DGC MucR (31). Therefore, the cyst-like structures formed by the ΔmucR mutant lacked the alginate coat and did not resist desiccation.
Altogether, our data revealed that c-di-GMP regulates not only the amount of the alginate produced but also its physical properties, including the viscosifying power and gelling capacity, by favoring the synthesis of longer alginate molecules with greater contents of G residues. This result has important biotechnological implications, since these two properties are the most exploited within the pharmaceutical and biomedical fields, where alginates of high purity and uniform characteristics are required (1–3). In this context and with the aim of improving the inherent properties of the alginates utilized, they are derivatized through chemical processes that tend to cause depolymerization of the alginate chain (45). The highest molecular mass reported for algal alginates is about 1,000 to 1,200 kDa (46), approximately 3 times lower than the molecular mass of alginates produced by A. vinelandii (about 2,000 to 3,000 kDa). Starting the derivatization process with higher-molecular-mass alginates could improve the chain length of the final polymer, expanding the biotechnological applications.
The biological significance of the synthesis of longer alginate chains in A. vinelandii, yielding a more viscous solution, in response to increasing amounts of c-di-GMP is likely related to biofilm structuring and environmental persistence. This would set MucG and AvGReg as key modulators of bacterial adaptation to oxygen availability in the different environments encountered by A. vinelandii.
MATERIALS AND METHODS
Strains and cultivation conditions.
The bacterial strains and plasmids used in the present work are listed in Table S1 in the supplemental material. A. vinelandii was routinely cultivated in minimum Burk’s nitrogen-free salts medium supplemented with 20 g liter−1 of sucrose (Burk’s-sucrose medium) (47). The composition of the Burk’s medium and the culture conditions have been reported elsewhere (48). Cultures were routinely conducted in 250-ml Erlenmeyer flasks with a filling volume of 50 ml, at 30°C for 48 h at 200 rpm. The calculated OTRmax values used in this study were obtained under conditions in which we varied the volume of the culture medium using 500-ml Erlenmeyer flasks, as reported previously (25). E. coli DH5α was grown in lysogeny broth (LB) at 37°C (49). When needed, the final antibiotic concentrations used for A. vinelandii and E. coli were as follows: tetracycline, 30 and 15 μg ml−1; gentamicin, 1 and 10 μg ml−1; spectinomycin, 100 and 100 μg ml−1; ampicillin, not used and 200 μg ml−1, respectively.
Standard techniques.
DNA isolation and cloning were conducted as described previously (50). The oligonucleotides used for PCR amplifications were designed based on the DJ A. vinelandii genome sequence (39). Sequences of the oligonucleotides used in this study are listed in Table S2. The high-fidelity Phusion DNA polymerase (Thermo Fisher Scientific) was used for all PCR amplifications, which were confirmed by DNA sequencing. DNA sequencing was performed with fluorescent dideoxy terminators using a cycle sequencing method and a 3130xl analyzer (Applied Biosystems).
Real-time qRT-PCR.
The relative levels of mucG mRNA in strains AEIV and ATCC 9046 were determined by qRT-PCR following a protocol previously described (31), using total RNA extracted from cells that had been cultured for 24 and 48 h in Burk`s-sucrose medium, with the primer pair qPCR-mucG-F/qPCR-mucG-R. The results were normalized relative to those obtained for the gyrA gene (Avin_15810).
Bioinformatic analysis.
The genes for annotated proteins of A. vinelandii DJ strain involved in c-di-GMP metabolism were obtained from the NCBI genome (RefSeq accession number NC_012560.1). The SMART (http://smart.embl-heidelberg.de) and Pfam (51) databases were used for the analysis of conserved domains. BLASTp was used for the searching of homologous proteins.
Construction of A. vinelandii mutants.
Construction of the A. vinelandii mutants generated in this study is detailed in Table S1. Mutations of genes encoding enzymes for the metabolism of c-di-GMP were produced by deletion of the genes, followed by the insertion of an antibiotic resistance cassettes. Competent A. vinelandii cells were transformed with linearized plasmids to allow double reciprocal homologous recombination. Transformants were verified by PCR amplification and DNA sequencing of the corresponding locus carrying the desired mutation.
For the MucG D452A E453A variant (designated MucGGGAAF) of the mutant CLAM06, we conducted overlapping PCR. A 224-bp fragment carrying the desired mutations (GAC→GCC and GAA→GCG) was PCR amplified using oligonucleotides GG9-RT-F and GGDEF(GGAAF)-R. The product served as a megaprimer in a second PCR to amplify the N-terminal portion of the mutated gene, using mucG-R as the reverse oligonucleotide. The assembled product was cloned in plasmid pJET1.2/Blunt vector, giving rise to pLA06.
c-di-GMP measurement.
c-di-GMP quantification was conducted following a protocol described previously (52). In brief, 47 ml of culture was centrifuged at 5,752 × g for 7 min. Cell pellets were resuspended in 1 ml of extraction solution (40% acetonitrile, 40% methanol, 0.1% formic acid, and 19.9% H2O) and incubated for 15 min on ice. After this, samples were centrifuged at 17,949 × g for 15 min at 4°C. The supernatant was transferred to a new Eppendorf tube, dried under vacuum, and lyophilized. The samples were resuspended in 100 μl of ultraperformance liquid chromatography (UPLC)-grade water, and 10 μl of this solution was injected into a liquid chromatography-tandem mass spectrometry system. A c-di-GMP standard curve was determined with the following concentrations: 1.9, 3.9, 7.8, 15.6, 31.3, 62.5, and 125 nM. The determinations were conducted by triplicate. The remaining 3 ml of culture was used to quantify protein content by the Lowry method, to normalize the production of c-di-GMP to cell protein levels (53).
Analytical methods.
The specific alginate production was determined in strains grown in Burk’s-sucrose medium for 48 h; 3 ml of culture was centrifuged at 5,752 × g for 7 min, and the supernatant, containing the extracellular alginate, was transferred to a new tube. The cell pellet was resuspended, and the total protein content was determined by the Lowry method (53). The alginate collected was precipitated with 3 volumes of 2-propanol, washed twice with 70% ethanol, dried, and resuspended in 1 ml of distilled water. Alginate was quantitated by spectrophotometric determination of uronic acids with carbazole (54).
Quantification of biofilm formation by A. vinelandii in 24-well microtiter plates was conducted using the crystal violet protocol described previously (55, 56). Twenty-four-well polystyrene microtiter plates were loaded with 200 μl of Burk’s-sucrose medium inoculated with 10% (vol/vol) of a 25-ml culture that had been grown for 18 h in a 125-ml Erlenmeyer flask. We ran a replicate plate that served for estimating cell growth by determining the protein concentration. For all of the determinations, at least three biological replicates were conducted.
Swimming motility was assessed following a protocol reported previously (57). Two microliters of cell suspension from a 16-h culture in Burk’s-sucrose medium was applied to the subsurface of 0.3% agar plates. Visualization of the motility halos was conducted after 24 h of incubation at 30°C.
Determination of the molecular mass of alginate.
The alginate molecular mass was estimated by gel permeation chromatography with a serial set of Ultrahydrogel columns (UG 500 and linear; Waters), using a high-performance liquid chromatography (HPLC) system with a differential refractometer detector (model 2414; Waters). The conditions and the sample preparation details are described elsewhere (25).
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Bahena for computational support and M. Tabche for technical support.
This work was supported by project PAPIIT IN-204818 (Universidad Nacional Autónoma de México [UNAM]) to C.N. and NIH grants GM109259 and AI144395 to C.M.W. C.L.A.-M. received a Ph.D. scholarship from CONACyT to perform his Ph.D. program and from DGAPA, UNAM, for thesis completion.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hay ID, Rehman ZU, Moradali MF, Wang Y, Rehm BHA. 2013. Microbial alginate production, modification and its applications. Microb Biotechnol 6:637–650. doi: 10.1111/1751-7915.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhamecha D, Movsas R, Sano U, Menon JU. 2019. Applications of alginate microspheres in therapeutics delivery and cell culture: past, present and future. Int J Pharm 569:118627. doi: 10.1016/j.ijpharm.2019.118627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee KY, Mooney DJ. 2012. Alginate: properties and biomedical applications. Prog Polym Sci 37:106–126. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. 2005. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J Immunol 175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 5.Lovewell RR, Patankar YR, Berwin B. 2014. Mechanisms of phagocytosis and host clearance of Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol 306:L591–L603. doi: 10.1152/ajplung.00335.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segura D, Núñez C, Espín G. 2020. Azotobacter cysts In eLS. John Wiley & Sons, Chichester, United Kingdom. doi: 10.1002/9780470015902.a0000295.pub3. [DOI] [Google Scholar]
- 7.Urtuvia V, Maturana N, Acevedo F, Peña C, Díaz-Barrera A. 2017. Bacterial alginate production: an overview of its biosynthesis and potential industrial production. World J Microbiol Biotechnol 33:198. doi: 10.1007/s11274-017-2363-x. [DOI] [PubMed] [Google Scholar]
- 8.Rehman ZU, Wang Y, Moradali MF, Hay ID, Rehm BHA. 2013. Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol 79:3264–3272. doi: 10.1128/AEM.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moradali MF, Donati I, Sims IM, Ghods S, Rehm BHA. 2015. Alginate polymerization and modification are linked in Pseudomonas aeruginosa. mBio 6:e00453-15. doi: 10.1128/mBio.00453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakkevig K, Sletta H, Gimmestad M, Aune R, Ertesvåg H, Degnes K, Christensen BE, Ellingsen TE, Valla S. 2005. Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J Bacteriol 187:8375–8384. doi: 10.1128/JB.187.24.8375-8384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ertesvåg H. 2015. Alginate-modifying enzymes: biological roles and biotechnological uses. Front Microbiol 6:523. doi: 10.3389/fmicb.2015.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimmestad M, Steigedal M, ErtesvåG H, Moreno S, Christensen BE, Espín G, Valla S, 2006. Identification and characterization of an Azotobacter vinelandii type I secretion system responsible for export of the AlgE-type mannuronan C-5-epimerases. J Bacteriol 188:5551–5560. doi: 10.1128/JB.00236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galindo E, Peña C, Núñez C, Segura D, Espín G. 2007. Molecular and bioengineering strategies to improve alginate and polydydroxyalkanoate production by Azotobacter vinelandii. Microb Cell Fact 6:7. doi: 10.1186/1475-2859-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trujillo-Roldán M, Moreno S, Segura D, Galindo E, Espín G. 2003. Alginate production by an Azotobacter vinelandii mutant unable to produce alginate lyase. Appl Microbiol Biotechnol 60:733–737. doi: 10.1007/s00253-002-1173-7. [DOI] [PubMed] [Google Scholar]
- 15.Gimmestad M, Ertesvåg H, Heggeset TMB, Aarstad O, Svanem BIG, Valla S. 2009. Characterization of three new Azotobacter vinelandii alginate lyases, one of which is involved in cyst germination. J Bacteriol 191:4845–4853. doi: 10.1128/JB.00455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Mendoza D, Sanjuán J. 2016. Exploiting the commons: cyclic diguanylate regulation of bacterial exopolysaccharide production. Curr Opin Microbiol 30:36–43. doi: 10.1016/j.mib.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Whitney JC, Howell PL. 2013. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol 21:63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schirmer T. 2016. c-di-GMP synthesis: structural aspects of evolution, catalysis and regulation. J Mol Biol 428:3683–3701. doi: 10.1016/j.jmb.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Chou S-H, Galperin MY. 2016. Diversity of cyclic di-GMP-binding proteins and mechanisms. J Bacteriol 198:32–46. doi: 10.1128/JB.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 22.Whitney JC, Whitfield GB, Marmont LS, Yip P, Neculai AM, Lobsanov YD, Robinson H, Ohman DE, Howell PL. 2015. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem 290:12451–12462. doi: 10.1074/jbc.M115.645051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay ID, Remminghorst U, Rehm BHA. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moradali MF, Ghods S, Rehm BHA. 2017. Activation mechanism and cellular localization of membrane-anchored alginate polymerase in Pseudomonas aeruginosa. Appl Environ Microbiol 83:e03499-16. doi: 10.1128/AEM.03499-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahumada-Manuel CL, Guzmán J, Peña C, Quiroz-Rocha E, Espín G, Núñez C. 2017. The signaling protein MucG negatively affects the production and the molecular mass of alginate in Azotobacter vinelandii. Appl Microbiol Biotechnol 101:1521–1534. doi: 10.1007/s00253-016-7931-8. [DOI] [PubMed] [Google Scholar]
- 26.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Liew CW, Wong YH, Tan ST, Poh WH, Manimekalai MSS, Rajan S, Xin L, Liang Z-X, Grüber G, Rice SA, Lescar J. 2018. Insights into biofilm dispersal regulation from the crystal structure of the PAS-GGDEF-EAL region of RbdA from Pseudomonas aeruginosa. J Bacteriol 200:e00515-17. doi: 10.1128/JB.00515-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantoni F, Paiardini A, Brunotti P, D'Angelo C, Cervoni L, Paone A, Cappellacci L, Petrelli R, Ricciutelli M, Leoni L, Rampioni G, Arcovito A, Rinaldo S, Cutruzzolà F, Giardina G. 2018. Insights into the GTP‐dependent allosteric control of c‐di‐GMP hydrolysis from the crystal structure of PA0575 protein from Pseudomonas aeruginosa. FEBS J 285:3815–3834. doi: 10.1111/febs.14634. [DOI] [PubMed] [Google Scholar]
- 29.Bellini D, Caly DL, McCarthy Y, Bumann M, An S-Q, Dow JM, Ryan RP, Walsh MA. 2014. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol 91:26–38. doi: 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores C, Díaz-Barrera Á, Martínez F, Galindo E, Peña C. 2015. Role of oxygen in the polymerization and de‐polymerization of alginate produced by Azotobacter vinelandii. J Chem Technol Biotechnol 90:356–365. doi: 10.1002/jctb.4548. [DOI] [Google Scholar]
- 31.Martínez-Ortiz IC, Ahumada-Manuel CL, Hsueh BY, Guzmán J, Moreno S, Cocotl-Yañez M, Waters CM, Zamorano-Sánchez D, Espín G, Núñez C. 2020. Cyclic di-GMP-mediated regulation of extracellular mannuronan C-5 epimerases is essential for cyst formation in Azotobacter vinelandii. J Bacteriol 202:e00135-20. doi: 10.1128/JB.00135-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thijs L, Vinck E, Bolli A, Trandafir F, Wan X, Hoogewijs D, Coletta M, Fago A, Weber RE, Doorslaer SV, Ascenzi P, Alam M, Moens L, Dewilde S. 2007. Characterization of a globin-coupled oxygen sensor with a gene-regulating function. J Biol Chem 282:37325–37340. doi: 10.1074/jbc.M705541200. [DOI] [PubMed] [Google Scholar]
- 33.Germani F, Nardini M, De Schutter A, Cuypers B, Berghmans H, Van Hauwaert M-L, Bruno S, Mozzarelli A, Moens L, Van Doorslaer S, Bolognesi M, Pesce A, Dewilde S. 2020. Structural and functional characterization of the globin-coupled sensors of Azotobacter vinelandii and Bordetella pertussis. Antioxid Redox Signal 32:378–395. doi: 10.1089/ars.2018.7690. [DOI] [PubMed] [Google Scholar]
- 34.Tuckerman JR, Gonzalez G, Sousa EHS, Wan X, Saito JA, Alam M, Gilles-Gonzalez M-A. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 35.Kong W, Zhao J, Kang H, Zhu M, Zhou T, Deng X, Liang H. 2015. ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res 43:8268–8282. doi: 10.1093/nar/gkv747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steigedal M, Sletta H, Moreno S, Maerk M, Christensen BE, Bjerkan T, Ellingsen TE, Espìn G, Ertesvåg H, Valla S. 2008. The Azotobacter vinelandii AlgE mannuronan C-5-epimerase family is essential for the in vivo control of alginate monomer composition and for functional cyst formation. Environ Microbiol 10:1760–1770. doi: 10.1111/j.1462-2920.2008.01597.x. [DOI] [PubMed] [Google Scholar]
- 37.Hoidal HK, Svanem BIG, Gimmestad M, Valla S. 2000. Mannuronan C-5 epimerases and cellular differentiation of Azotobacter vinelandii. Environ Microbiol 2:27–38. doi: 10.1046/j.1462-2920.2000.00074.x. [DOI] [PubMed] [Google Scholar]
- 38.Kitanishi K, Kobayashi K, Kawamura Y, Ishigami I, Ogura T, Nakajima K, Igarashi J, Tanaka A, Shimizu T. 2010. Important roles of Tyr43 at the putative heme distal side in the oxygen recognition and stability of the Fe(II)-O2 complex of YddV, a globin-coupled heme-based oxygen sensor diguanylate cyclase. Biochemistry 49:10381–10393. doi: 10.1021/bi100733q. [DOI] [PubMed] [Google Scholar]
- 39.Setubal JC, dos Santos P, Goldman BS, Ertesvåg H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, Curatti L, Du Z, Godsy E, Goodner B, Hellner-Burris K, Hernandez JA, Houmiel K, Imperial J, Kennedy C, Larson TJ, Latreille P, Ligon LS, Lu J, Maerk M, Miller NM, Norton S, O'Carroll IP, Paulsen I, Raulfs EC, Roemer R, Rosser J, Segura D, Slater S, Stricklin SL, Studholme DJ, Sun J, Viana CJ, Wallin E, Wang B, Wheeler C, Zhu H, Dean DR, Dixon R, Wood D. 2009. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol 191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noar JD, Bruno-Bárcena JM. 2018. Azotobacter vinelandii: the source of 100 years of discoveries and many more to come. Microbiology (Reading) 164:421–436. doi: 10.1099/mic.0.000643. [DOI] [PubMed] [Google Scholar]
- 41.Qi Y, Rao F, Luo Z, Liang Z-X. 2009. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in Ax DGC2 from Acetobacter xylinum. Biochemistry 48:10275–10285. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- 42.Okegbe C, Fields BL, Cole SJ, Beierschmitt C, Morgan CJ, Price-Whelan A, Stewart RC, Lee VT, Dietrich LEP. 2017. Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc Natl Acad Sci U S A 114:E5236–E5245. doi: 10.1073/pnas.1700264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An S, Wu J, Zhang L-H. 2010. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol 76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García A, Castillo T, Ramos D, Ahumada-Manuel CL, Núñez C, Galindo E, Büchs J, Peña C. 2020. Molecular weight and viscosifying power of alginates produced by mutant strains of Azotobacter vinelandii under microaerophilic conditions. Biotechnol Rep (Amst) 26:e00436. doi: 10.1016/j.btre.2020.e00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawar SN, Edgar KJ. 2012. Alginate derivatization: a review of chemistry, properties and applications. Biomaterials 33:3279–3305. doi: 10.1016/j.biomaterials.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Peña C, Campos N, Galindo E. 1997. Changes in alginate molecular mass distributions, broth viscosity and morphology of Azotobacter vinelandii cultured in shake flasks. Appl Microbiol Biotechnol 48:510–515. doi: 10.1007/s002530051088. [DOI] [Google Scholar]
- 47.Kennedy C, Gamal R, Humphrey R, Ramos J, Brigle K, Dean D. 1986. The nifH, nifM and nifN genes of Azotobacter vinelandii: characterisation by Tn5 mutagenesis and isolation from pLAFR1 gene banks. Mol Gen Genet 205:318–325. doi: 10.1007/BF00430445. [DOI] [Google Scholar]
- 48.Núñez C, Peña C, Kloeckner W, Hernández-Eligio A, Bogachev AV, Moreno S, Guzmán J, Büchs J, Espín G. 2013. Alginate synthesis in Azotobacter vinelandii is increased by reducing the intracellular production of ubiquinone. Appl Microbiol Biotechnol 97:2503–2512. doi: 10.1007/s00253-012-4329-0. [DOI] [PubMed] [Google Scholar]
- 49.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 51.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer ELL, Bateman A. 2006. Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massie JP, Reynolds EL, Koestler BJ, Cong J-P, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci U S A 109:12746–12751. doi: 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]
- 54.Knutson CA, Jeanes A. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem 24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 55.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 56.Huerta JM, Aguilar I, López-Pliego L, Fuentes-Ramírez LE, Castañeda M. 2016. The role of the ncRNA RgsA in the oxidative stress response and biofilm formation in Azotobacter vinelandii. Curr Microbiol 72:671–679. doi: 10.1007/s00284-016-1003-2. [DOI] [PubMed] [Google Scholar]
- 57.León R, Espín G. 2008. flhDC, but not fleQ, regulates flagella biogenesis in Azotobacter vinelandii, and is under AlgU and CydR negative control. Microbiology (Reading) 154:1719–1728. doi: 10.1099/mic.0.2008/017665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.