V. cholerae deploys an arsenal of virulence factors as it attempts to traverse a protective mucus layer and reach the epithelial surface of the distal small intestine. The T6SS used to cull bacterial competition during infection is induced by mucus. We show that this activity may serve an additional purpose by further decreasing motility in the presence of mucin, thereby reducing the probability of speed-dependent, near-perpendicular directional changes. We posit that this encourages cells to maintain course rather than change direction, which may aid those attempting to reach and colonize the epithelial surface.
KEYWORDS: T6SS, Vibrio, motility, mucin
ABSTRACT
Motility is required for many bacterial pathogens to reach and colonize target sites. Vibrio cholerae traverses a thick mucus barrier coating the small intestine to reach the underlying epithelium. We screened a transposon library in motility medium containing mucin to identify factors that influence mucus transit. Lesions in structural genes of the type VI secretion system (T6SS) were among those recovered. Two-dimensional (2D) and 3D single-cell tracking was used to compare the motility behaviors of wild-type cells and a mutant that collectively lacked three essential T6SS structural genes (T6SS−). In the absence of mucin, wild-type and T6SS− cells exhibited similar speeds and run-reverse-flick (RRF) swimming patterns, in which forward-moving cells briefly backtrack before stochastically reorienting (flicking) in a new direction upon resuming forward movement. We show that mucin induced T6SS expression and activity in wild-type bacteria but significantly decreased their swimming speed and flicking, yielding curvilinear or near-surface circular traces for many cells. Conversely, mucin slowed T6SS− cells to a lesser extent, and many continued to flick and produce RRF-like traces. ΔcheY3 cells, which exclusively swim in the forward direction and thus cannot flick, also produced curvilinear traces with or without mucin present and, on occasion, near-surface circular traces in the presence of mucin. The dependence of flicking on swimming speed suggested that mucin-induced T6SS activity further decreased V. cholerae motility and thereby reduced flicking probability during reverse-to-forward transitions. We propose that this encourages cells to continue on their current trajectory rather than reorienting, which may benefit those tracking toward the epithelial surface.
IMPORTANCE V. cholerae deploys an arsenal of virulence factors as it attempts to traverse a protective mucus layer and reach the epithelial surface of the distal small intestine. The T6SS used to cull bacterial competition during infection is induced by mucus. We show that this activity may serve an additional purpose by further decreasing motility in the presence of mucin, thereby reducing the probability of speed-dependent, near-perpendicular directional changes. We posit that this encourages cells to maintain course rather than change direction, which may aid those attempting to reach and colonize the epithelial surface.
INTRODUCTION
Cholera is estimated to cause 3 million to 5 million cases of human infection and over 100,000 deaths globally each year (https://www.cdc.gov/vibrio/index.html). Hallmark symptoms are profuse diarrhea and vomiting. The rapid loss of body fluids (up to 1 liter/h) leads to dehydration and shock that is fatal within hours if left untreated. Infection occurs following the ingestion of Vibrio cholerae in contaminated food or water. Although a high infectious dose of 1011 cells is required to cause infection in healthy individuals, as few as 104 cells can cause disease when stomach acid is buffered. Surviving cells pass through the pyloric sphincter and colonize the distal small intestine (1).
The epithelium of the digestive tract is covered with a viscous mucus layer composed largely of the glycosylated peptide mucin (2), which helps maintain sterility by embedding foreign particles and inhibiting bacterial attack of the underlying epithelium (3). V. cholerae cells penetrate this mucus layer and proliferate on the epithelial surface (1). The toxin-coregulated pilus mediates attachment and microcolony formation on the intestinal epithelium (4), and cholera toxin, a ribosylating enterotoxin, causes the profuse watery diarrhea that is characteristic of cholera (5). During colonization, V. cholerae cells also employ a specialized weapon known as the type VI secretion system (T6SS) to attack and kill neighboring bacterial and eukaryotic cells (6). The T6SS functions similarly to a spear gun (6). It consists of a baseplate complex that attaches to the inner membrane, a transmembrane complex that spans from the inner membrane through the periplasm to the outer membrane, a contractile sheath, and an inner tube tipped with a deliverable toxic payload. The complex is assembled in the cytoplasm on the membrane-associated baseplate and can bridge the width of the cell (7). ATP-dependent contraction of the sheath to nearly half its original length thrusts the inner tube and tip into a target cell. The payload is usually a lethal effector protein(s) that disrupts essential cellular activity such as peptidoglycan biosynthesis, actin polymerization, membrane function, or DNA stability (8–11). If the targeted cell produces the cognate immunity protein, the delivered effector(s) is inactivated and the cell is protected. T6SS-mediated killing reduces microbial competition while promoting nutrient acquisition and adaptation by horizontal gene transfer (12, 13). The T6SS has been shown to be regulated by host-derived factors such as the signaling molecule indole that is present at high concentrations in the intestines (14), bile salts that solubilize dietary fats, and mucins that line the small intestine (15). Mucins and indole activate T6SS-mediated killing, while bile salts repress it.
Chemotaxis is the biased movement of an organism in response to a chemical stimulus (16). Bacteria use transmembrane chemoreceptors and signal transduction systems to monitor the concentration of chemical signals in the immediate vicinity and fine-tune their motility accordingly (17). This signaling network allows bacteria to detect and move toward favorable signals (attractants) such as amino acids, carbohydrates, and other nutrients and away from unfavorable signals (repellents) such as antibiotics and toxins. Most motile bacteria exhibit chemotaxis, and the underlying mechanisms regulating the response are fairly well conserved (18). Chemotaxis is often used by pathogenic bacteria to find suitable colonization sites (19), and colonization of the small intestine by V. cholerae is dependent on its ability to chemotax (20). Mucin is a chemoattractant for V. cholerae (15) and may act as a molecular beacon that primes its colonization of the small intestine (21). Interestingly, disparate roles for motility and chemotaxis in colonization have been reported. Motility is required for colonization of the proximal but not distal small intestine (22). Additionally, motile but nonchemotactic mutants with a counterclockwise (CCW) flagellar rotational bias that generates long forward runs hypercolonize along the entire length of the small intestine, whereas mutants biased toward clockwise (CW) flagellar rotation turn more frequently and are attenuated (23, 24).
Vibrio species are motile by way of a single polar flagellum (25) and move using a three-step run-reverse-flick (RRF) mechanism (26, 27). The direction of rotation of the flagellum determines whether the cell is pushed or pulled. Switching from pushing to pulling results in nearly 180° reversals. Directional changes centered at about 90° (flicks) occur exclusively during reverse-to-forward transitions and result from buckling of the flagellar hook upon resuming forward movement. Because a sufficient compressive load is needed for buckling of the hook to occur, flicking is dependent on the swimming speed of the cell (27). For Vibrio alginolyticus, the speed threshold for flicking is ∼30 μm/s. At lower speeds, a two-step run-reverse (RR) swim pattern dominates (28). The Na+-driven polar flagellar motor and RRF swimming pattern of vibrios are exquisite adaptations for survival in vast and often nutrient-limited aquatic environments, where bacteria must either adjust their metabolism or locate favorable niches such as patches of dissolved organic matter, sinking detritus, and animal hosts (29, 30). Vibrios can attain speeds in excess of 60 μm/s (27, 28, 31–34), and backtracking during RRF motility is proposed to be superior to tumbling in anisotropic aquatic environments by allowing bacteria to rapidly return to an area of higher nutrient concentration when crisscrossing fleeting filamentous nutrient plumes (29). Hence, this mechanism of motility is beneficial for locating and maintaining contact with short-lived nutrient sources before they are dissipated by water currents (30).
Given its crucial role in environmental survival and establishing infection, we sought to identify factors that influence V. cholerae motility. We screened a transposon (Tn) library in motility medium containing mucin for mutants exhibiting altered motility relative to the parental strain. Mutants bearing insertions within components for the T6SS were among those recovered. We constructed a T6SS deletion mutant (ΔMC) that lacked three major T6SS structural components and assessed the impact on bacterial motility. Competition assays and single-cell high-speed two-dimensional (2D) and 3D motility tracking suggested that the T6SS was not active in a minimal medium reflecting estuarine conditions and had little impact on V. cholerae motility behavior. The addition of mucin stimulated T6SS expression and activity in wild-type cells but decreased swimming speed and flicking. Either curvilinear or near-surface circular motility traces were observed for many of them. ΔMC cells were quicker than wild-type cells in the presence of mucin, and many continued to flick and produce RRF-like motility traces, suggesting that the probability of flicking during reverse-to-forward transitions was dependent on swimming speed. Cells lacking cheY3, which cannot move in the reverse direction and consequently cannot flick, also produced curvilinear traces whether or not mucin was present and, occasionally, near-surface circular swimming patterns in the presence of mucin. These results suggested that T6SS activity further decreased V. cholerae swimming motility in the presence of mucin, thereby reducing the probability of flicking during reverse-to-forward transitions. We propose that this may encourage cells to continue on their current course, which would benefit those tracking toward the epithelial surface.
RESULTS
A mucus motility screen identifies T6SS genes.
To identify factors that impact mucus transit, we generated and screened a V. cholerae Tn library for mutants exhibiting altered (increased or decreased) motility relative to the wild type in the presence of mucin. A total of 237 mutants were rescreened to confirm the phenotype, yielding a final collection of 176 mutants. These mutants were pooled, and whole-genome sequencing was used to identify the collective of Tn insertion sites.
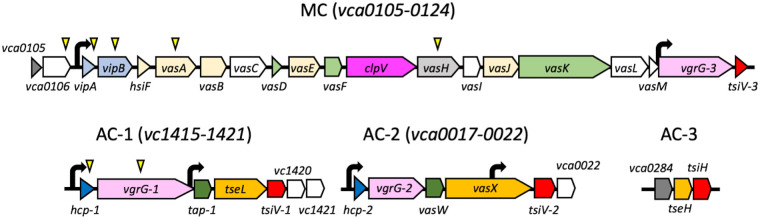
Lesions in a number of genes previously reported to regulate motility were identified. These included insertions in the toxin-coregulated pilus (35), tfoX and tfoY (36), vieS (37), cheA2 (38), flagellar genes (39), and several diguanylate cyclases (40). We turned our attention to insertions that mapped to genes of the T6SS (Fig. 1). VipA and VipB comprise the outer contractile sheath of the structure, and VasA forms part of the baseplate complex (6). Hcp and VgrG-1 form the inner tube and the tip of the spear, respectively. Deletion of vgrG-1 decreases Hcp secretion, a hallmark of T6SS function (41), and VasH is a DNA-binding protein that regulates the expression of T6SS genes, including hcp and vgrG-1 (42). VCA0106 is a putative membrane protein with multiple Sel1-like repeats (SLRs) that is of unknown function; however, SLR proteins typically serve as adaptor proteins for the assembly of macromolecular complexes (43). The recovery of both regulatory and structural mutants of T6SS production suggested that T6SS activity might influence motility in mucin-rich environments.
FIG 1.
Organization of the T6SS loci. The main cluster (MC) and auxiliary clusters (AC-1, AC-2, and AC-3) are shown. Each gene is color-coded according to its predicted role: light blue, outer sheath; dark blue, inner tube; beige, baseplate; light green, inner membrane complex; dark green, adaptor proteins; light pink, spike tip proteins; orange, effector proteins; hot pink, protease; red, immunity proteins; dark gray, cap (PAAR) proteins; light gray, regulator; white, unknown function. Bent black arrows mark promoters, and yellow triangles indicate transposon insertion sites.
The T6SS decreases V. cholerae motility in mucin.
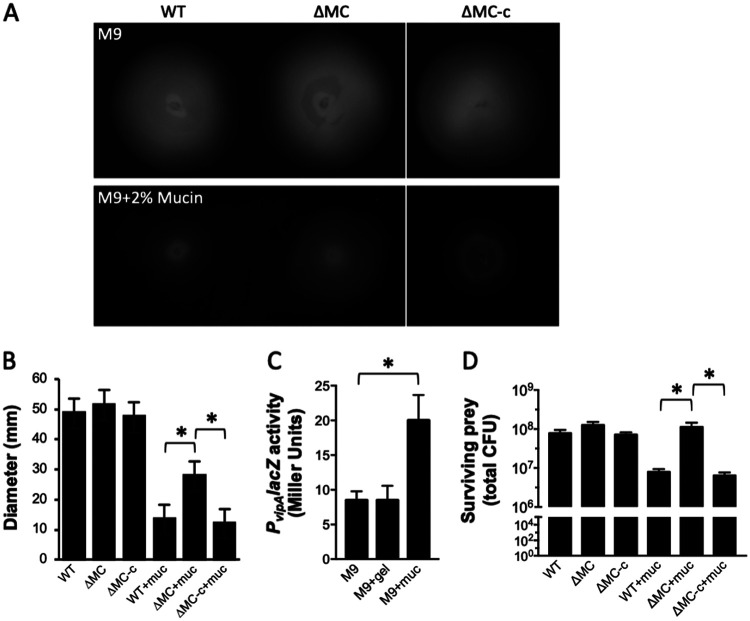
To determine if the T6SS impacted motility, we constructed a T6SS− mutant (ΔMC) that lacked the first three genes (vipA, vipB, and hsiF) of the T6SS main cluster. VipA and VipB form the outer sheath, and HsiF forms part of the baseplate complex. Their collective disruption was anticipated to prevent a functional T6SS from being assembled or fired. Motility by ΔMC and the complemented strain (ΔMC-c) was assessed alongside the parental wild-type strain in M9 motility medium. The motilities of the three strains were similar (Fig. 2A and B). Since it has been demonstrated that V. cholerae pathogenesis correlates with its penetration of intestinal mucin, and host mucins stimulate V. cholerae T6SS-mediated killing (15, 21), the motility of the wild-type and ΔMC strains was also assessed in M9 medium containing mucin (M9-mucin medium). Notably, motility by the ΔMC strain was twice that (P < 0.001) of the wild type but reverted to wild-type levels following complementation. The addition of mucin also increased the expression of a PvipAlacZ reporter 2-fold in wild-type cells (Fig. 2C), suggesting that mucin induced the expression of the T6SS main cluster. To assess this, a killing assay using the wild type, ΔMC, or ΔMC-c as the predator and Escherichia coli as the prey was performed. The number of surviving E. coli cells recovered in M9 medium was the same (∼108 CFU) regardless of the competing predator (Fig. 2D). In the presence of mucin, similar numbers of prey were recovered when competed with ΔMC; however, ∼1 log fewer E. coli cells were recovered when challenged with the wild type or ΔMC-c as the predator. These results suggested that V. cholerae motility was not compromised by the T6SS in the absence of mucin, likely because the system was not active. However, mucin-induced T6SS activity decreased motility.
FIG 2.
T6SS activity decreases motility in the presence of mucin. (A) Motility of wild-type (WT), ΔMC, and complemented (ΔMC-c) strains in M9 motility medium without (top) and with (bottom) 2% mucin. The images were compiled from two identical plates (white line separator). (B) Quantitation of motility of the strains in panel A. (C) PvipA expression in wild-type cells in M9 minimal medium alone or with 2% gelatin (gel) or 2% mucin (muc). (D) Survival of prey E. coli bacteria following incubation with the indicated predator V. cholerae strain in M9 medium alone or with 2% mucin. Plots show the means, and error bars represent the standard deviations. Statistically significant differences between samples (P < 0.001 as determined by the Student t test [unpaired, two-tailed distribution with two-sample, equal variance]) are indicated with an asterisk.
T6SS activity alters single-cell motility behavior in mucin.
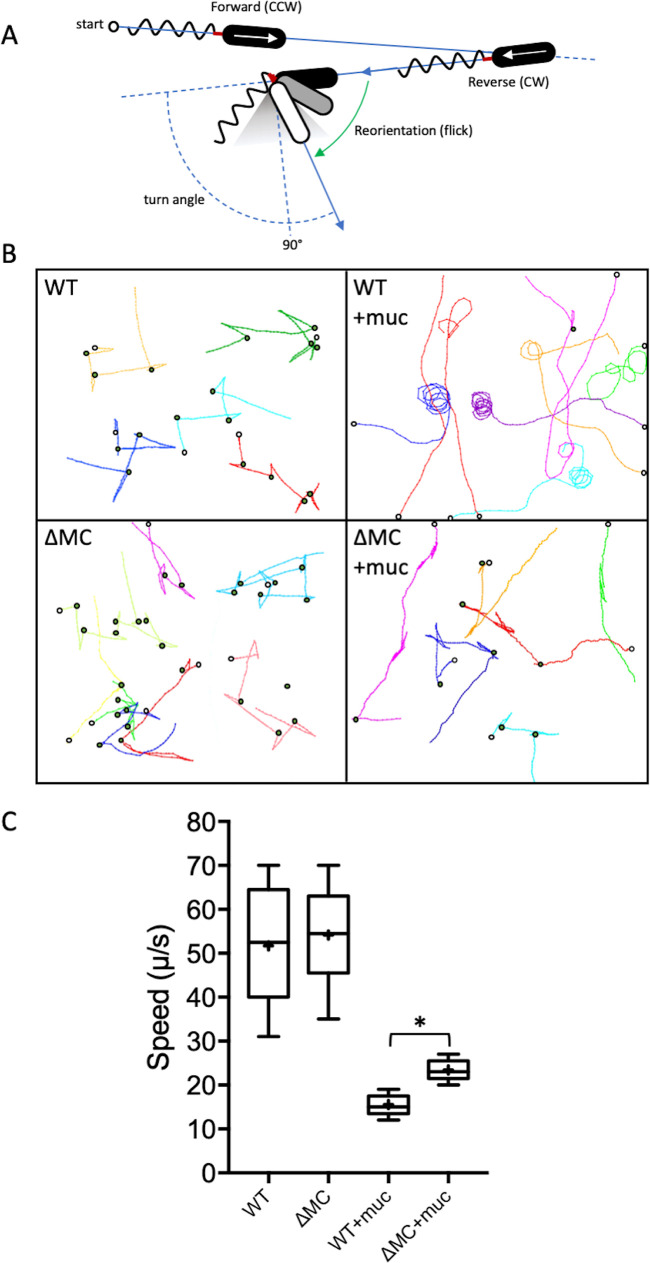
To gain further insight into the effect of T6SS activity on motility, single-cell bacterial trajectories were tracked in 2D (z axis restricted) using high-speed, dark-field microscopy. Since the 2D behavior of the ΔMC-c strain (see Fig. S1 in the supplemental material) was indistinguishable from that of the wild type (Fig. 3B), subsequent analyses focused on the wild-type and ΔMC strains. In M9 medium, wild-type and ΔMC cells exhibited similar RRF patterns of alternating forward and reverse runs interrupted by flicks (Fig. 3B) and had similar speed ranges and medians (53 μm/s and 55 μm/s for the wild type and ΔMC, respectively) (Fig. 3C). The speeds of both wild-type and ΔMC cells deceased in M9-mucin medium, but wild-type cells were slowed to a greater extent (median of 16 μm/s for the wild type compared to 25 μm/s for ΔMC) (Fig. 3C). Curvilinear traces or a circular pattern was observed for most (86%) wild-type cells in mucin, suggesting that flicking when cells switched swimming direction was infrequent at speeds below ∼20 μm/s. The traces of most (87%) ΔMC cells, although largely curvilinear, often included more than one flick and resembled RRF-like patterns, suggesting that flicks were more common during directional changes at speeds of >20 μm/s.
FIG 3.
T6SS activity alters motility behavior in the presence of mucin. (A) V. cholerae swims using the run-reverse-flick mechanism, where the direction of rotation of the polar flagellum (counterclockwise [CCW] or clockwise [CW]) determines whether the cell is pushed or pulled. Switching from pushing to pulling results in nearly 180° reversals, and flicks of 90° ± 30° that reorient the cell (gray-shaded triangle) result from buckling of the flagellar hook (red) upon resuming forward movement. A smaller deflection correlates with a larger flick angle. (B) 2D traces of wild-type and ΔMC cells in M9 medium without or with 2% mucin (+muc). Different single-cell trajectories in x and y are indicated by different colors. Open circles denote the start position for each trace, and green circles indicate flicks. Traces from a single representative experiment are shown. (C) Quantitation of bacterial speed. A box-and-whisker plot shows the minimum, first quartile, median, third quartile, maximum, and mean (marked with +) for the data collected. Data from three stacks of three biological replicates were analyzed for each strain. Statistically significant differences between samples (P < 0.05 as determined by the Student t test [unpaired, two-tailed distribution with two-sample, equal variance]) are indicated with an asterisk.
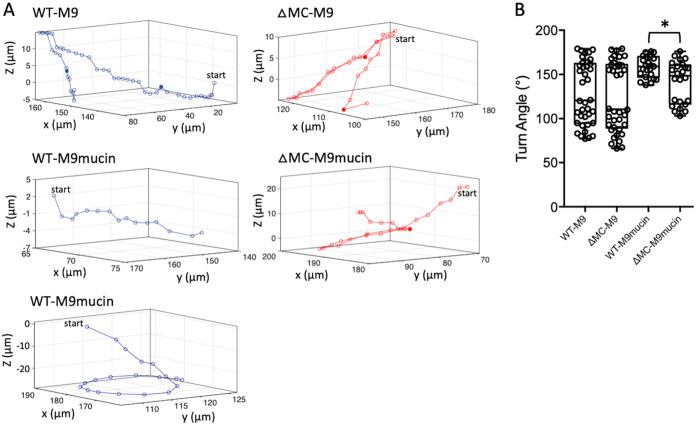
Because 2D motility assays constrain bacteria in a thin sample only a few micrometers thick, trajectory curvature stemming from hydrodynamic interactions between the cell and the chamber surface may be introduced (44). To minimize such interactions, 3D motility traces were captured at micrometer-scale resolution using high-speed, time-lapse phase-contrast microscopy. Tracking of wild-type and ΔMC cells in M9 medium revealed RRF-type swimming motility (Fig. 4A). The range of flick angles for wild-type cells (77° to 178°; median, ∼120°) was greater than that for ΔMC cells (66° to 178°; median, ∼110°), but in both cases, the angles were centered around ∼90° and 163° (Fig. 4B). For both cell types, ∼75% of the angles between reverse and forward vectors fell within the flick range (∼90° ± 30°). In M9-mucin medium, curvilinear (Fig. 4A, middle trace) or circular (bottom trace) motility patterns were observed for most wild-type cells, and flicks were infrequent (only 9% of the angles between reverse and forward vectors were in the flick range). Curvilinear traces were commonly observed for cells in the bulk medium, whereas circular traces, which typically tracked downward, occurred in the vicinity of the coverslip. Conversely, the traces of most ΔMC cells tended to have an RRF-type profile in mucin, and flicks were observed at a higher frequency (23% of the reverse-to-forward vector angles were in the flick range). Although the average turn angle remained high (>130°), angles as low as 98° were detected. A few (<3%) of the traces displayed a circular component. Based on the data, the probability of a flick occurring during a reverse-to-forward transition was roughly the same, about 3/4, for wild-type or ΔMC cells in the absence of mucin. In the presence of mucin, the probability dropped to about 1/10 for wild-type cells and 1/4 for ΔMC cells. Collectively, these results suggested that T6SS activity in the presence of mucin further decreased swimming motility and reduced the probability of a flick occurring during the reverse-to-forward transition.
FIG 4.
T6SS activity reduces near-perpendicular reorientations in the presence of mucin. (A) 3D motility traces of wild-type (WT) and ΔMC cells in M9 medium without or with 2% mucin. The middle and bottom panels for the wild type show the two trajectory types (curvilinear and circular) that were observed in M9-mucin medium. Single representative 3D trajectories are shown for each. Solid points indicate flicks. (B) Quantitation of turn angles. A box-and-whisker plot shows the minimum, first quartile, median, third quartile, and maximum for the data collected. A subset of representative data points is overlaid. Data from three stacks of three biological replicates were analyzed for each strain. Statistically significant differences between samples (P < 0.05 as determined by the Student t test [unpaired, two-tailed distribution with two-sample, equal variance]) are indicated with an asterisk.
Precluding flicks favors curvilinear and circular swimming.
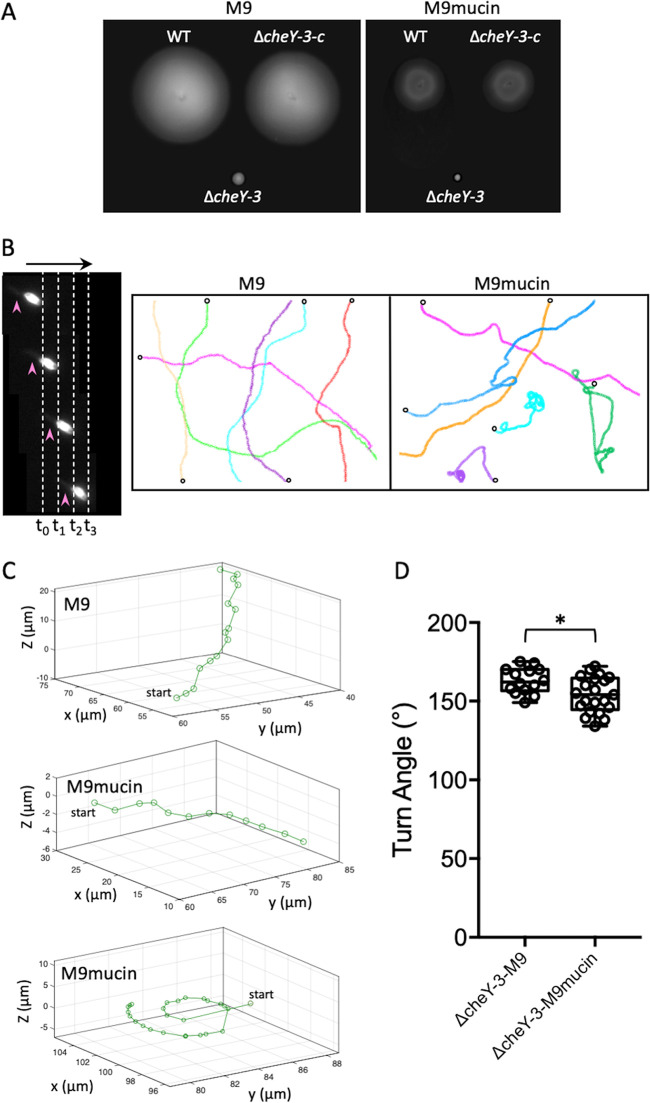
The data suggested that a reduced probability of flicking gave rise to the curvilinear and circular swimming patterns observed for wild-type cells in the presence of mucin. To investigate this further, we monitored the swimming behavior of a cheY-null mutant in medium with and without mucin. In V. cholerae, phosphorylated CheY interacts with the flagellar motor to reverse its rotation (20). The genome codes for five CheY homologs, but only one of these (CheY3) controls flagellar rotation, and its deletion locks cells in the CCW state (45). Since these cells swim exclusively in the forward direction (46), flicking should be nullified. Unlike wild-type cells, the ΔcheY3 strain did not appear to be motile on M9 motility plates, regardless of whether or not mucin was present (Fig. 5A). The ectopic expression of wild-type cheY3 in the mutant restored motility to wild-type levels. However, dark-field microscopy revealed that ΔcheY3 cells were indeed highly mobile in the absence of mucin (Fig. 5B). Cells moved exclusively in the forward direction at up to 60 μm/s. Only long, sweeping curvilinear swimming traces were observed in 2D, and flicks were not detected (Fig. 5B). The swimming speed decreased to ∼21 μm/s in the presence of mucin, and curvilinear traces were observed, along with traces exhibiting significant stretches of circular swimming behavior. Tracking ΔcheY cells in 3D yielded similar overall swim patterns (Fig. 5C), with only curvilinear traces observed in the absence of mucin (top trace). Curvilinear traces and, on occasion, circular traces at the coverslip surface were observed in the presence of mucin (middle and bottom traces, respectively). Only angles of >130° were detected for both patterns under either condition (Fig. 5D). These results supported the notion that the curvilinear and circular swimming patterns of wild-type cells in the presence of mucin were due to a reduced tendency to flick relative to ΔMC cells when transitioning from reverse to forward movement.
FIG 5.
Motility behavior of V. cholerae ΔcheY3 in the absence and presence of mucin. (A) Motility of the wild-type (WT), ΔcheY3, and complemented (ΔcheY3-c) strains in M9 motility medium without (left) and with (right) 2% mucin. (B, left) CCW swimming by ΔcheY3 cells in M9 medium. Four successive dark-field images are shown with time intervals (t0 [0-h time point] to t3) of 0.033 s between consecutive frames (dotted lines), and cells moved up to 2 μm between successive frames (i.e., 60 μm/s). The arrow at the top indicates the swim direction, and the arrowheads mark the polar flagellum, which appears blurred because of its rapid rotation. (Right) 2D traces of ΔcheY3 cells in M9 medium without or with 2% mucin. Different single-cell trajectories in x and y are indicated by different colors. Open circles denote the start position for each trace. Traces from a single representative experiment are shown. (C) Curvilinear 3D motility traces of ΔcheY3 cells in M9 medium without or with 2% mucin. The bottom panel shows the near-surface circular trajectory that was also observed in M9-mucin medium. Single representative 3D trajectories are shown for each. (D) Quantitation of turn angles. A box-and-whisker plot shows the minimum, first quartile, median, third quartile, and maximum for the data collected. A subset of representative data points is overlaid. Data from three biological replicates were analyzed for each strain. Statistically significant differences between samples (P < 0.05 as determined by the Student t test [unpaired, two-tailed distribution with two-sample, equal variance]) are indicated with an asterisk.
DISCUSSION
Despite marked differences between aquatic and human reservoirs, the bactericidal action of the T6SS promotes V. cholerae survival during both inter- and intraspecies competition (13, 47, 48). T6SS activity can be constitutive in some strains or regulated by environmental and host-derived factors in others (15, 49–51). One such signal, mucin, stimulates T6SS activity in clinical isolates (15). Here, we showed that the T6SS activity of a V. cholerae clinical isolate also altered motility in a mucin-rich environment, which we suggest may aid in colonization of the small intestine.
Motility is required for V. cholerae to effectively penetrate intestinal mucus and reach the underlying epithelium of the small intestine (22, 23). Mucin, a chemoattractant for V. cholerae, and intestinal bile, a chemorepellent, are proposed to direct and prime the bacteria for intestinal colonization (15, 21). Only a few bacterial species are known to successfully penetrate the mucus layer and reach the epithelial surface (52). Since encounters between V. cholerae and host microbiota are anticipated to be infrequent as it traverses the mucus layer, it is conceivable that increasing T6SS activity while crossing a viscous 150-μm-thick physical barrier (22, 53) could serve an additional purpose during mucus penetration. Host mucins play a key role in the distribution of V. cholerae during infection. Although V. cholerae can be found along the length of the small intestine, it preferentially colonizes the distal half (24, 54). Motility is dispensable for colonization of the distal niche but is critical for colonization of the proximal region, where the mucus layer is thickest (22) and to which V. cholerae is attracted (55). Bacteria do not have significant inertia due to their small dimensions, and movement is driven largely by the viscous forces of the surrounding fluid (44). For bacteria swimming in aqueous environments, the Reynolds number, which correlates the inertia forces to the viscous forces, is very small (on the order of ∼10−5 to 10−6) (56). Thus, bacteria cannot “coast” and must continually input energy when swimming. The rotation of the helical flagellum and the counterrotation of the cell body act on the surrounding fluid to propel the cell, generating rotational streamlines and laminar flow that influence overall movement (57). Hence, complex surface structures such as the T6SS could potentially affect the trajectory of a swimming cell, particularly in viscous environments, and V. cholerae may assemble up to five T6SS structures per cell (7, 58). Elegant experiments analyzing the dynamics of T6SS production in V. cholerae revealed that the sheath is assembled in the cytoplasm over 20 to 30 s, while sheath contraction takes less than 5 ms (58). Once fired, ClpV-dependent disassembly of the contracted sheath occurs in the attacking cell over 10 to 20 s. The spear tip and shaft are also disassembled, although the time scale over which this happens is not known. Notably, the fate of an ejected spear that fails to contact a neighboring cell is somewhat nebulous. The presence of Hcp in the supernatant suggests that the shaft is released from the cell surface, but it is not known if this happens en masse, if disassembly occurs, or if enzyme input is needed to drive either event. Likewise, the time frame (milliseconds, seconds, or minutes) over which such events might occur is also unknown. It is possible that T6SSs that fire but fail to contact a neighboring cell could remain on the surface sufficiently long as to influence the motility pattern of the cell in viscous environments. The effect could be substantial for fast-moving species such as vibrios that can attain speeds of >60 μm s−1 in aqueous media, particularly when T6SS activity is known to be constitutive or environmentally regulated, as shown for V. cholerae.
The T6SS lesions that we identified mapped to structural genes and a regulator, vasH, that is required for the expression of the auxiliary genes hcp and vgrG-1 (42). Our findings suggest that T6SS activity had a physical impact on swimming motility in the presence of mucin. However, it is possible that the loss of VasH or structural components of the T6SS complex could affect chemotactic behavior that manifests as altered motility, although a chemotactic gradient was not applied in our studies. We posit that mucin-induced T6SS activity altered the motility, rather than the chemotactic, behavior of wild-type cells by further decreasing their swimming speed. Our data also suggested that flicking by V. cholerae during the reverse-to-forward transition was infrequent (<10%) below swimming speeds of ∼20 μm/s. This was in good agreement with a previous report predicting a flick probability of only 8% for V. alginolyticus swimming at this speed (27) and supports the notion that the probability of flicking during reverse-to-forward transitions correlates with swimming speed in Vibrio species. Flicks were infrequent in the curvilinear and circular traces of wild-type cells in the presence of mucin. A shift toward RR motility during transitions at low speed encouraged continued movement along the same trajectory in wild-type cells (curvilinear traces) since each reversal typically resulted in a relatively small change in cell orientation. Moreover, cells bearing a cheY3 deletion, which removed any contribution of flicking to swimming behavior since the cell cannot swim in reverse and flicking occurs exclusively during reverse-to-forward transitions, also produced curvilinear traces. Thus, reduced flicking in the presence of mucin for wild-type cells relative to ΔMC cells likely explains the difference in their motility traces.
Notably, V. cholerae cells that either lack or overexpress cheY3 are nonchemotactic, but the former are forward biased and hypercolonize the small intestine, while the latter are reverse biased and attenuated (22, 24). Similarly, Vibrio coralliilyticus nonchemotactic mutants that were biased toward forward runs were hypervirulent in coral infection assays, while a reverse-biased mutant was avirulent (59). Moreover, the near-surface circular traces of wild-type and ΔcheY3 cells were reminiscent of the orbiting patterns observed for V. cholerae and Vibrio vulnificus cells as they probed a surface for colonization (60, 61). It is tempting to speculate that T6SS activity skews V. cholerae RRF motility toward curvilinear and near-surface circular patterns in mucin-rich environments to encourage cells to continue on their current trajectory rather than reorienting, which may ultimately benefit cells attempting to reach and colonize the epithelial surface.
MATERIALS AND METHODS
Strains and growth conditions.
V. cholerae strain N16961, a clinical O1 El Tor isolate, was used. E. coli S17.1λπ was used for cloning, plasmid maintenance, and conjugation to V. cholerae. A rifampin-resistant variant of E. coli MG1655 was used as the prey in T6SS kill assays. Luria-Bertani (LB) medium was purchased from BD Difco. The following antibiotics and additives were purchased from Sigma and used at the indicated concentrations: ampicillin at 100 μg ml−1, colistin (Col) at 12.5 μg ml−1, chloramphenicol (Cm) at 2 μg ml−1 for V. cholerae and 25 μg ml−1 for E. coli, kanamycin (Kan) at 100 μg ml−1 for V. cholerae and 25 μg ml−1 for E. coli, trimethoprim (Tp) at 10 μg ml−1, isopropyl-β-d-thiogalactopyranoside (IPTG) at 100 μM, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 20 μg/ml. M9 motility medium contained KH2PO4 (22 mM), NaCl (100 mM), NH4Cl (20 mM), Na2HPO4 (50 mM), MgSO4 (2 mM), agar (0.3%, wt/vol), sodium pyruvate (0.4%, wt/vol), and Casamino Acids (0.1%, wt/vol) at pH 7. Mucin from porcine stomach was sterilized as previously described (62). Briefly, purified powder was soaked in 95% ethanol, and the solution was heated at 70°C for 24 h to evaporate the ethanol (62). It was then was added to sterile M9 motility medium (2%, wt/vol) and gently stirred at 50°C for up to 48 h to dissolve the mucin. For assays, strains were grown on selective plates overnight, and an isolated colony was inoculated into freshly poured motility plates.
Transposon mutagenesis.
A V. cholerae transposon mutagenesis library was generated by conjugation with E. coli S17.1λπ that carried the mini-Tn10 delivery vector pNKTXI-SceI (63). Transconjugants were selected on plates containing LB medium with Col and Kan (LB Col Kan plates). A QPixII colony picker (Genetix) was used to transfer a total of 11,520 mutants to 96-well plates. Following growth with shaking at 30°C for 16 h, the QPixII instrument was used to inoculate approximately 2 μl of the culture from each well into 245- by 245-mm plates (Corning) containing 200 ml of M9-mucin motility medium. The plates were incubated for 12 h at 30°C. Mutants exhibiting altered (increased or decreased) motility relative to wild-type controls were manually picked, grown overnight in 96-well plates containing fresh medium, and rescreened in M9-mucin motility medium to confirm the phenotype. Motility mutants from this second round of screening were individually grown in fresh LB Col Kan medium to an optical density at 600 nm (OD600) of 1. Aliquots (50 μl) of each sample were pooled for genomic DNA isolation and whole-genome sequencing at the Center for Genomics and Bioinformatics at Indiana University Bloomington. Geneious (Biomatters) was used to map single-end 150-bp reads to the V. cholerae O1 N16961 reference genome (64) to identify Tn insertion sites.
Construction and complementation of the ΔMC and ΔcheY3 in-frame mutants.
To construct the ΔMC mutant, PCR fragments (see Table S2 in the supplemental material for primer sequences) that included 1 kb upstream to the first 9 bp of vipA (primer pair VchΔvipA-1/VchΔvipA-TpK7-2) and the last 9 bp to 1 kb downstream of hsiF (primer pair VchΔhsiF-TpK7-3/VchΔhsiF-4) were amplified and fused to a central trimethoprim antibiotic resistance cassette (primer pair TmR-F/TmR-R). The primers were designed to include complementary 20-bp overhangs so that the PCR products could be fused using the Gibson assembly kit (New England BioLabs [NEB]) to replace in-frame the vipA-hsiF region (i.e., the first 3 amino acids of VipA and the last 3 amino acids of HsiF are retained) with a trimethoprim marker while leaving the native promoter intact. V. cholerae strains carrying the pMMB-TfoX expression plasmid (65) were induced overnight in LB medium containing ampicillin and IPTG at 30°C. A 10-μl aliquot of cells was added to 500 μl of Instant Ocean at 20 ppt (IO-20) containing IPTG and 25 μl of the Gibson assembly reaction mixture. The transformation mixture was incubated statically overnight at 30°C. The next day, 1 ml of LB medium was added to the tube, and the cells were allowed to outgrow for 3 h before plating on LB Tp plates. Allelic replacement of the target region was confirmed by PCR.
To complement the ΔMC strain, the 2,666-bp region spanning 200 bp upstream of vipA, which included its promoter, to the hsiF stop codon was amplified (primer pair VchvipA-hsiF-F/VchvipA-hsiF-R) and cloned by Gibson assembly into the multiple-cloning site (MCS) of the suicide vector pSW23T (66) that included an internal 400-bp fragment of V. cholerae chromosomal β-galactosidase in its backbone (pSW23T-lacZVc400) (67). The plasmid was conjugated to ΔMC cells for integration into chromosomal lacZVc, and transconjugants were selected on LB Tp Cm X-Gal plates. White colonies were screened by PCR for proper plasmid insertion. The same approach was used to generate and complement the ΔcheY3 strain, except that cheY3 was fused to Ptac and expression was induced with IPTG.
Construction of the PvipAlacZ reporter strain.
To eliminate endogenous V. cholerae β-galactosidase activity, a strain bearing a markerless deletion of the chromosomal lacZVc promoter was created using the pRE112 sucrose counterselection plasmid (68). PCR fragments corresponding to 1 kb upstream (primer pair VchΔPlacZ-1/VchΔPlacZ-2) and downstream (primer pair VchΔPlacZ-3/VchΔPlacZ-4) of the lacZVc promoter region were amplified, fused, and cloned into the MCS of pRE112 by Gibson assembly. Colonies were selected on LB Cm plates, and plasmids containing the correct insert were identified by PCR. Following conjugation to V. cholerae, cointegrates were selected on LB Col Cm plates and then gridded onto plates that lacked Cm but contained 20% sucrose for sacB counterselection. Promoter deletion mutants were confirmed by PCR and were white on LB X-Gal plates.
The V. cholerae vipA promoter (PvipA) and E. coli lacZ (lacZEc) were amplified from the respective genomic DNA (primer pairs VchPvipA-F/VchPvipA-R and EclacZ-F/EclacZ-R, respectively), fused, and inserted by Gibson assembly into the MCS of pSW23T-lacZVc400. The plasmid with the correct insert was identified by PCR and conjugated to V. cholerae for integration into chromosomal lacZVc. Cointegrates were selected on LB Col Cm plates, and proper integration was confirmed by PCR.
β-Galactosidase assays.
Bacteria were grown overnight at 30°C in LB medium. Eighty microliters of the culture was transferred in triplicate into a 96-well flat-bottom microtiter plate, and 120 μl of reaction mix (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 36 mM β-mercaptoethanol, 166 mg ml−1 lysozyme, 1.1 mg/ml o-nitrophenyl-β-d-galactopyranoside [ONPG], and 6.7% PopCulture reagent [Millipore]) was added to each well. Blank wells contained 80 μl LB medium. The plate was quickly transferred to a BioTek Synergy HT1 microplate reader set to 30°C. OD600 and OD420 readings were taken every 60 s for 30 min. The microtiter plate was shaken at 500 rpm (double orbital) between readings to ensure proper mixing and sample lysis. The slope of OD420 readings over time (OD420 per minute) and the initial OD600 reading were used to calculate Miller units as previously described (69), with the following formula: Miller units = (5,000 × OD420 min−1)/OD600. Data from three technical replicates of three biological replicates were collected for all experiments. Plots show the means, and error bars represent the standard deviations.
T6SS kill assay.
Wild-type or ΔMC V. cholerae cells were used as the predator, and rifampin-resistant E. coli MG1655 was used as the prey. Predator and prey strains were streaked onto fresh LB agar plates and grown at 30°C (Vibrio) or 37°C (E. coli) overnight. Isolated colonies were inoculated into fresh medium and grown with shaking overnight at the previous temperature. The strains were washed twice with an equal volume of and resuspended to an OD600 of 1 in M9 or M9-mucin medium. The predator and prey were mixed at a ratio of 10:1, and a 50-μl aliquot of the suspension was spotted on a filter (25 mm, 0.22-μm pore size; Millipore) that was placed on an M9 or M9-mucin plate. The spots were allowed to dry for 10 min, and the plates were then incubated at 37°C for 4 h. The filter was then placed in a 50-ml conical tube containing 5 ml of LB and vigorously vortexed for 30 s to resuspended the bacteria. Serial 10-fold dilutions were spotted on LB rifampin plates and incubated at 37°C overnight before counting the surviving prey (expressed as total CFU).
2D and 3D motility tracking.
V. cholerae was grown overnight in LB Col at 30°C. An aliquot was gently washed once in M9 medium (5 min at 4,000 × g) and then diluted to an OD600 of 0.1 in fresh M9 or M9-mucin medium. A 7-μl aliquot was gently pipetted between a microscope slide and a 22- by 22-mm no. 1.5 coverslip that were separated by a single layer of parafilm on three edges. All four edges of the chamber were quickly sealed with hot VALAP (Vaseline, lanolin, and paraffin), and the sample was imaged immediately. Motility was recorded at 30 Hz by dark-field microscopy on an Olympus IX83 microscope equipped with a Hamamatsu electron microscopy charge-coupled-device (EM-CCD) camera and a 20× extralong-working-distance (ELWD) objective. A stack of at least 150 frames (100-ms exposure time) was recorded for samples in M9 medium and twice that for samples in M9-mucin medium, and the movement of 15 randomly selected motile cells per stack was traced using the MTrackJ plug-in (70) in Fiji (71). Data from three stacks of three biological replicates were analyzed for each strain. At least 300 forward and backward swimming intervals were tracked for determining median and mean speeds (displacement in micrometers between successive frames captured at 0.033-s intervals).
3D tracking data were captured as previously described (32). Briefly, tracking chambers were made by layering three parafilm frames between a microscope slide and a 22- by 22-mm no. 1.5 coverslip. The chamber was sealed by placing it in a 55°C dry oven for 10 min. A culture of bacteria grown overnight was diluted into M9 or M9-mucin medium to an OD600 of 0.1 and flowed into the chamber at one edge by capillary action. The edges of the unit were sealed with hot VALAP, and the samples were immediately imaged on the above-mentioned microscope equipped with an Olympus long-working-distance (LWD) air condenser (numerical aperture [NA], 0.55) and an Olympus LUCPLFLN LWD 40× phase-contrast objective (Ph2; NA, 0.6). The correction collar was set to 1.2. For each movie recording, frames were acquired at a rate of 15 Hz and an exposure time of 5 ms with a Hamamatsu Orca-R2 camera (1,344 by 1,024 pixels). The objective was positioned so that the focus was ∼40 μm above the coverslip. The data were saved as a stack of 16-bit tiff files of 225 images each (twice that for samples in M9-mucin medium). Image stacks were background corrected and aligned to a 3D position reference library in Matlab using a custom-written tracking program to analyze motility behavior (32). Each reorientation event (flicks and reversals) identified by the program was manually confirmed based on the angle and swimming speed between the corresponding vectors for ∼600 trajectories prior to statistical analysis. Trajectories with an average speed of less than 10 μm/s or with a duration of less than 50 frames were ignored. Plots were created using data from three stacks of three biological replicates for each strain. Traces from a single representative experiment are shown.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ankur Dalia for the pMMB-TfoX expression plasmid and trimethoprim resistance gene cassette (TpR-K7) and Katja Taute and Tom Shimizu for providing and helping us to implement their 3D tracking code in Matlab.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Almagro-Moreno S, Pruss K, Taylor RK. 2015. Intestinal colonization dynamics of Vibrio cholerae. PLoS Pathog 11:e1004787. doi: 10.1371/journal.ppat.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelaseyed T, Hansson GC. 2020. Membrane mucins of the intestine at a glance. J Cell Sci 133:jcs240929. doi: 10.1242/jcs.240929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson MEV, Sjövall H, Hansson GC. 2013. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol 10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krebs SJ, Taylor RK. 2011. Protection and attachment of Vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model. J Bacteriol 193:5260–5270. doi: 10.1128/JB.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jobling MG, Gotow LF, Yang Z, Holmes RK. 2015. A mutational analysis of residues in cholera toxin A1 necessary for interaction with its substrate, the stimulatory G protein Gsα. Toxins (Basel) 7:919–935. doi: 10.3390/toxins7030919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, Yildiz FH. 2017. Rules of engagement: the type VI secretion system in Vibrio cholerae. Trends Microbiol 25:267–279. doi: 10.1016/j.tim.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y, Waldor MK, Mekalanos JJ. 2013. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. 2011. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun 79:2941–2949. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jana B, Fridman CM, Bosis E, Salomon D. 2019. A modular effector with a DNase domain and a marker for T6SS substrates. Nat Commun 10:3595. doi: 10.1038/s41467-019-11546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 13.Kostiuk B, Unterweger D, Provenzano D, Pukatzki S. 2017. T6SS intraspecific competition orchestrates Vibrio cholerae genotypic diversity. Int Microbiol 20:130–137. doi: 10.2436/20.1501.01.294. [DOI] [PubMed] [Google Scholar]
- 14.Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. 2009. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J Bacteriol 191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachmann V, Kostiuk B, Unterweger D, Diaz Satizabal L, Ogg S, Pukatzki S. 2015. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl Trop Dis 9:e0004031. doi: 10.1371/journal.pntd.0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 18.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matilla MA, Krell T. 2018. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev 42:fux052. doi: 10.1093/femsre/fux052. [DOI] [PubMed] [Google Scholar]
- 20.Boin MA, Austin MJ, Häse CC. 2004. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett 239:1–8. doi: 10.1016/j.femsle.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Miyashiro T, Tsou A, Hsiao A, Goulian M, Zhu J. 2008. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc Natl Acad Sci U S A 105:9769–9774. doi: 10.1073/pnas.0802241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK. 2014. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SH, Butler SM, Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc Natl Acad Sci U S A 98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci U S A 101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atsumi T, McCarter L, Imae Y. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 26.Xie L, Altindal T, Chattopadhyay S, Wu X-L. 2011. Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc Natl Acad Sci U S A 108:2246–2251. doi: 10.1073/pnas.1011953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Son K, Guasto JS, Stocker R. 2013. Bacteria can exploit a flagellar buckling instability to change direction. Nat Phys 9:494–498. doi: 10.1038/nphys2676. [DOI] [Google Scholar]
- 28.Son K, Menolascina F, Stocker R. 2016. Speed-dependent chemotactic precision in marine bacteria. Proc Natl Acad Sci U S A 113:8624–8629. doi: 10.1073/pnas.1602307113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stocker R, Seymour JR. 2012. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev 76:792–812. doi: 10.1128/MMBR.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. 2008. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci U S A 105:4209–4214. doi: 10.1073/pnas.0709765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie L, Lu C, Wu X-L. 2015. Marine bacterial chemoresponse to a stepwise chemoattractant stimulus. Biophys J 108:766–774. doi: 10.1016/j.bpj.2014.11.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taute KM, Gude S, Tans SJ, Shimizu TS. 2015. High-throughput 3D tracking of bacteria on a standard phase contrast microscope. Nat Commun 6:8776. doi: 10.1038/ncomms9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pu M, Storms E, Chodur DM, Rowe-Magnus DA. 29 July 2019. Calcium-dependent site-switching regulates expression of the atypical iam pilus locus in Vibrio vulnificus. Environ Microbiol doi: 10.1111/1462-2920.14763. [DOI] [PubMed] [Google Scholar]
- 34.Zamorano-Sánchez D, Xian W, Lee CK, Salinas M, Thongsomboon W, Cegelski L, Wong GCL, Yildiz FH. 2019. Functional specialization in Vibrio cholerae diguanylate cyclases: distinct modes of motility suppression and c-di-GMP production. mBio 10:e00670-19. doi: 10.1128/mBio.00670-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harkey CW, Everiss KD, Peterson KM. 1994. The Vibrio cholerae toxin-coregulated-pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect Immun 62:2669–2678. doi: 10.1128/IAI.62.7.2669-2678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzger LC, Stutzmann S, Scrignari T, Van der Henst C, Matthey N, Blokesch M. 2016. Independent regulation of type VI secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep 15:951–958. doi: 10.1016/j.celrep.2016.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Wilson HF, Tamayo R, Tischler AD, Lazinski DW, Camilli A. 2008. The Vibrio cholerae hybrid sensor kinase VieS contributes to motility and biofilm regulation by altering the cyclic diguanylate level. J Bacteriol 190:6439–6447. doi: 10.1128/JB.00541-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gosink KK, Kobayashi R, Kawagishi I, Häse CC. 2002. Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J Bacteriol 184:1767–1771. doi: 10.1128/jb.184.6.1767-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prouty MG, Correa NE, Klose KE. 2001. The novel σ54‐ and σ28‐dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol 39:1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- 40.Hunter JL, Severin GB, Koestler BJ, Waters CM. 2014. The Vibrio cholerae diguanylate cyclase VCA0965 has an AGDEF active site and synthesizes cyclic di-GMP. BMC Microbiol 14:22. doi: 10.1186/1471-2180-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unterweger D, Kostiuk B, Ötjengerdes R, Wilton A, Diaz Satizabal L, Pukatzki S. 2015. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J 34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. 2011. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol 193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mittl PRE, Schneider-Brachert W. 2007. Sel1-like repeat proteins in signal transduction. Cell Signal 19:20–31. doi: 10.1016/j.cellsig.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 44.Lauga E. 2016. Bacterial hydrodynamics. Annu Rev Fluid Mech 48:105–130. doi: 10.1146/annurev-fluid-122414-034606. [DOI] [Google Scholar]
- 45.Hyakutake A, Homma M, Austin MJ, Boin MA, Häse CC, Kawagishi I. 2005. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J Bacteriol 187:8403–8410. doi: 10.1128/JB.187.24.8403-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kojima M, Kubo R, Yakushi T, Homma M, Kawagishi I. 2007. The bidirectional polar and unidirectional lateral flagellar motors of Vibrio alginolyticus are controlled by a single CheY species. Mol Microbiol 64:57–67. doi: 10.1111/j.1365-2958.2007.05623.x. [DOI] [PubMed] [Google Scholar]
- 47.Wong M, Liang X, Smart M, Tang L, Moore R, Ingalls B, Dong TG. 2016. Microbial herd protection mediated by antagonistic interaction in polymicrobial communities. Appl Environ Microbiol 82:6881–6888. doi: 10.1128/AEM.02210-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Y, Ho BT, Mekalanos JJ. 2018. Tracking Vibrio cholerae cell-cell interactions during infection reveals bacterial population dynamics within intestinal microenvironments. Cell Host Microbe 23:274–281.e2. doi: 10.1016/j.chom.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa T, Sabharwal D, Bröms J, Milton DL, Sjöstedt A, Uhlin BE, Wai SN. 2012. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun 80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unterweger D, Kitaoka M, Miyata ST, Bachmann V, Brooks TM, Moloney J, Sosa O, Silva D, Duran-Gonzalez J, Provenzano D, Pukatzki S. 2012. Constitutive type VI secretion system expression gives Vibrio cholerae intra- and interspecific competitive advantages. PLoS One 7:e48320. doi: 10.1371/journal.pone.0048320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernardy EE, Turnsek MA, Wilson SK, Tarr CL, Hammer BK. 2016. Diversity of clinical and environmental isolates of Vibrio cholerae in natural transformation and contact-dependent bacterial killing indicative of type VI secretion system activity. Appl Environ Microbiol 82:2833–2842. doi: 10.1128/AEM.00351-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McGuckin MA, Linden SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 54.Angelichio MJ, Spector J, Waldor MK, Camilli A. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect Immun 67:3733–3739. doi: 10.1128/IAI.67.8.3733-3739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson KM, Gellings PS. 2018. Multiple intraintestinal signals coordinate the regulation of Vibrio cholerae virulence determinants. Pathog Dis 76:ftx126. doi: 10.1093/femspd/ftx126. [DOI] [PubMed] [Google Scholar]
- 56.Purcell EM. 1977. Life at low Reynolds number. Am J Phys 45:3–11. doi: 10.1119/1.10903. [DOI] [Google Scholar]
- 57.Watari N, Larson RG. 2010. The hydrodynamics of a run-and-tumble bacterium propelled by polymorphic helical flagella. Biophys J 98:12–17. doi: 10.1016/j.bpj.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ushijima B, Häse CC. 2018. Influence of chemotaxis and swimming patterns on the virulence of the coral pathogen Vibrio coralliilyticus. J Bacteriol 200:e00791-17. doi: 10.1128/JB.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Utada AS, Bennett RR, Fong JCN, Gibiansky ML, Yildiz FH, Golestanian R, Wong GCL. 2014. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat Commun 5:4913. doi: 10.1038/ncomms5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pu M, Rowe-Magnus DA. 2018. A Tad pilus promotes the establishment and resistance of Vibrio vulnificus biofilms to mechanical clearance. NPJ Biofilms Microbiomes 4:10. doi: 10.1038/s41522-018-0052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeung ATY, Parayno A, Hancock REW. 2012. Mucin promotes rapid surface motility in Pseudomonas aeruginosa. mBio 3:e00073-12. doi: 10.1128/mBio.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakhamchik A, Wilde C, Rowe-Magnus DA. 2007. Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect Immun 75:5550–5558. doi: 10.1128/IAI.00932-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin HY, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dalia AB, McDonough E, Camilli A. 2014. Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A 111:8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babic A, Guérout A-M, Mazel D. 2008. Construction of an improved RP4 (RK2)-based conjugative system. Res Microbiol 159:545–549. doi: 10.1016/j.resmic.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Chodur DM, Rowe-Magnus DA. 2018. Complex control of a genomic island governing biofilm and rugose colony development in Vibrio vulnificus. J Bacteriol 200:e00190-18. doi: 10.1128/JB.00190-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shames SR, Bhavsar AP, Croxen MA, Law RJ, Mak SHC, Deng W, Li Y, Bidshari R, de Hoog CL, Foster LJ, Finlay BB. 2011. The pathogenic Escherichia coli type III secreted protease NleC degrades the host acetyltransferase p300. Cell Microbiol 13:1542–1557. doi: 10.1111/j.1462-5822.2011.01640.x. [DOI] [PubMed] [Google Scholar]
- 69.Schaefer J, Jovanovic G, Kotta-Loizou I, Buck M. 2016. Single-step method for β-galactosidase assays in Escherichia coli using a 96-well microplate reader. Anal Biochem 503:56–57. doi: 10.1016/j.ab.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meijering E, Dzyubachyk O, Smal I. 2012. Methods for cell and particle tracking. Methods Enzymol 504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 71.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.