Vibrio parahaemolyticus possesses at least six osmolyte transporters, which allow the bacterium to adapt to high-salinity conditions. In this study, we identified several additional osmolytes that were utilized by V. parahaemolyticus. We demonstrated that the compound DMG, which is present in the marine environment, was a highly effective osmolyte for Vibrio species. We determined that DMG is transported via BCCT family carriers, which have not been shown previously to take up this compound. BccT1 was a carrier for GB, DMG, and ectoine, and we identified the amino acid residues essential for the coordination of these compounds. The data suggest that for BccT1, GB is more easily accommodated than ectoine in the transporter binding pocket.
KEYWORDS: BCCT, DMG, Vibrio, ectoine, osmolytes
ABSTRACT
Fluctuations in osmolarity are one of the most prevalent stresses to which bacteria must adapt, both hypo- and hyperosmotic conditions. Most bacteria cope with high osmolarity by accumulating compatible solutes (osmolytes) in the cytoplasm to maintain the turgor pressure of the cell. Vibrio parahaemolyticus, a halophile, utilizes at least six compatible solute transporters for the uptake of osmolytes: two ABC family ProU transporters and four betaine-carnitine-choline transporter (BCCT) family transporters. The full range of compatible solutes transported by this species has yet to be determined. Using an osmolyte phenotypic microarray plate for growth analyses, we expanded the known osmolytes used by V. parahaemolyticus to include N,N-dimethylglycine (DMG), among others. Growth pattern analysis of four triple-bccT mutants, possessing only one functional BCCT, indicated that BccT1 (VP1456), BccT2 (VP1723), and BccT3 (VP1905) transported DMG. BccT1 was unusual in that it could take up both compounds with methylated head groups (glycine betaine [GB], choline, and DMG) and cyclic compounds (ectoine and proline). Bioinformatics analysis identified the four coordinating amino acid residues for GB in the BccT1 protein. In silico modeling analysis demonstrated that GB, DMG, and ectoine docked in the same binding pocket in BccT1. Using site-directed mutagenesis, we showed that a strain with all four residues mutated resulted in the loss of uptake of GB, DMG, and ectoine. We showed that three of the four residues were essential for ectoine uptake, whereas only one of the residues was important for GB uptake. Overall, we have demonstrated that DMG is a highly effective compatible solute for Vibrio species and have elucidated the amino acid residues in BccT1 that are important for the coordination of GB, DMG, and ectoine transport.
IMPORTANCE Vibrio parahaemolyticus possesses at least six osmolyte transporters, which allow the bacterium to adapt to high-salinity conditions. In this study, we identified several additional osmolytes that were utilized by V. parahaemolyticus. We demonstrated that the compound DMG, which is present in the marine environment, was a highly effective osmolyte for Vibrio species. We determined that DMG is transported via BCCT family carriers, which have not been shown previously to take up this compound. BccT1 was a carrier for GB, DMG, and ectoine, and we identified the amino acid residues essential for the coordination of these compounds. The data suggest that for BccT1, GB is more easily accommodated than ectoine in the transporter binding pocket.
INTRODUCTION
In order to grow in high-osmolarity environments, bacteria accumulate compounds called compatible solutes (osmolytes) within the cytoplasm of the cell, via either uptake or biosynthesis (1–4). These compounds balance the internal osmolarity with that of the environment and maintain the turgor pressure of the cell (3, 5, 6). Osmolytes also protect proteins, nucleic acids, and other vital molecular machinery by increasing the hydration shell around these molecules (7). Osmolytes fall into several classes of compounds, including sugars (trehalose), polyols (glycerol and mannitol), free amino acids (proline and glutamine), amino acid derivatives (ectoine [ect]), and quaternary amines (glycine betaine [GB], choline, and l-carnitine) (4, 5, 8–12).
Biosynthesis of compatible solutes is energetically costly, and therefore, bacteria encode compatible solute transporters to scavenge available osmolytes from the environment (12–14). Compatible solute transporters include members of the multicomponent ATP binding cassette (ABC) family, such as ProU (proVWX) in Escherichia coli, OpuC in Bacillus subtilis, and OpuC (proVWX) in Pseudomonas syringae (15–19), and the single-component betaine-carnitine-choline transporter (BCCT) family, which are Na+ or H+ dependent. Members of the BCCT family include BetT in E. coli, which transports choline with high affinity, and GB transporters in B. subtilis (OpuD) and Corynebacterium glutamicum (BetP), among many others (20–23).
BCCTs are energized by sodium- or proton-motive force symport and are organized into 12 transmembrane (TM) segments (20, 22, 24, 25). Aromatic residues found in TM4 and TM8 make up the GB binding pocket in BCCT family transporters examined to date. These residues are highly conserved in BCCTs that transport trimethylammonium compounds such as GB, l-carnitine, and γ-butyrobetaine (21, 24, 26, 27). An additional tryptophan residue is present in TM8 just outside the binding pocket and is thought to participate in the coordination of substrates during conformational changes that occur during transport (26).
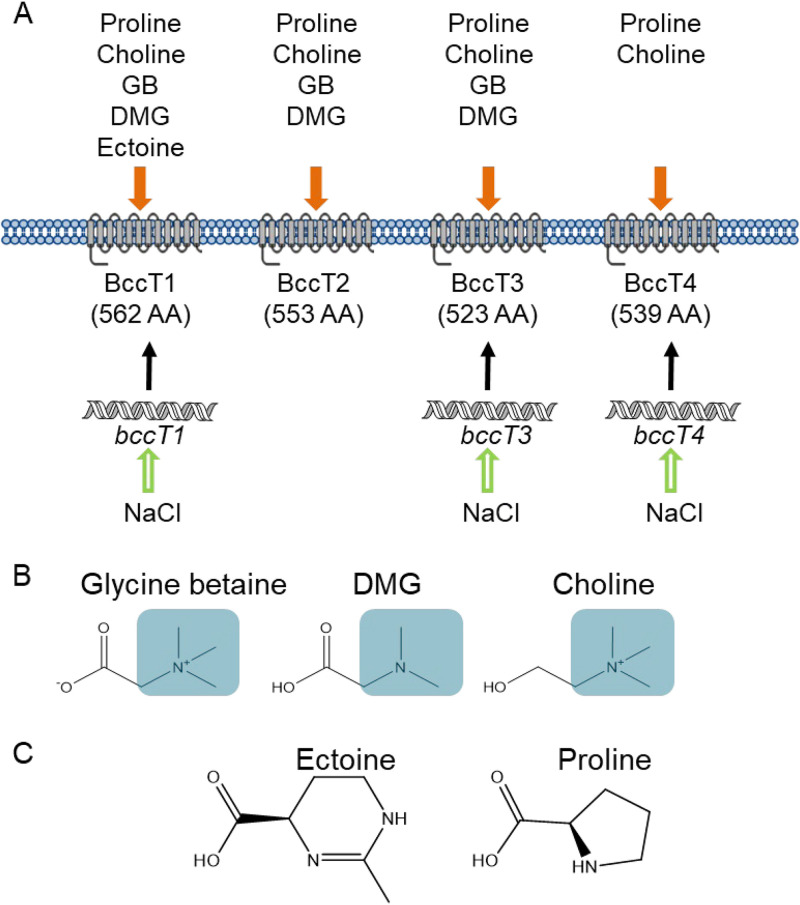
Vibrio parahaemolyticus is a halophilic bacterium present in marine and estuarine environments in association with plankton, fish, and shellfish (28–33). Vibrio parahaemolyticus is the leading bacterial cause of seafood-related gastroenteritis worldwide, frequently associated with consuming raw or undercooked seafood (30, 34). Vibrio parahaemolyticus is a halophile that grows optimally in 3% NaCl (∼500 mM), which is approximately the salinity of seawater, but can grow at salinities of up to 9% NaCl (∼1.5 M) (35). Vibrio parahaemolyticus encounters a range of salinities in tidal habitats and possesses four BCCTs, encoded by bccT1 (VP1456), bccT2 (VP1723), bccT3 (VP1905), and bccT4 (VPA0356), and two ProUs, encoded by proVWX (ProU1) (VP1726 to VP1728) and proXWV (ProU2) (VPA1109 to VPA1111), for the uptake of compatible solutes (36). In addition to compatible solute transporters, V. parahaemolyticus also possesses biosynthesis systems for the compatible solutes ectoine, ectABC-asp_ect (VP1719 to VP1722), and GB, betIBA-proXWV (VPA1112 to VPA1114), whose expression is induced at high salinity (36). It was demonstrated that the expression of these biosynthesis systems in Vibrio species was under tight regulation, controlled by the quorum sensing regulators OpaR and AphA as well as CosR, a global regulator of the osmotic stress response (37–40). Previously, we reported that in V. parahaemolyticus, bccT1, bccT3, bccT4, and both proU operons are responsive to salinity (40, 41). BccT1 had the broadest substrate specificity, transporting GB, choline, proline, and ectoine, while BccT2 and BccT3 transport GB, choline, and proline, and BccT4 transports only choline and proline (Fig. 1) (41). Interestingly, the number of osmolyte transporters present among Vibrio species varies, which suggested differences in the osmolytes utilized and osmotolerance (36). It was shown that Vibrio alginolyticus contained four BCCTs and two ProUs, Vibrio harveyi and Vibrio splendidus possessed six BCCTs and two ProUs, and Vibrio vulnificus possessed a single ProU and a BccT3 homolog, whereas Vibrio cholerae possessed only a BccT3 homolog (36).
FIG 1.
(A) BCCT transporters present in V. parahaemolyticus RIMD2210633 and their known substrates. bccT1, bccT3, and bccT4 are induced by high salinity. (B and C) Structures of BCCT substrates with methylated headgroups highlighted in blue boxes (B) or cyclic compounds (C). AA, amino acids.
In this study, we examined a range of osmolytes utilized by V. parahaemolyticus using an osmolyte phenotypic microarray plate, which identified 14 potential osmolytes. We determined the ability of several Vibrio species to utilize dimethylglycine (DMG), one of the most effective osmolytes that we identified and a compound not previously shown to be an osmolyte in Vibrio. We examined V. parahaemolyticus transport of DMG using several osmolyte transporter mutants and showed that BCCT carriers were required, representing a new transporter family for the uptake of DMG. In V. parahaemolyticus, BccT1 has the broadest substrate uptake ability in terms of the number and diversity of compounds (methylated head groups and cyclic compounds). Our in silico modeling analysis demonstrated that GB, DMG, and ectoine docked in the same binding pocket in BccT1. We investigated via mutagenesis and functional complementation the amino acid residues that are required for the coordination of GB, DMG, and ectoine. This analysis describes for the first time the residues that coordinate DMG and ectoine in a BCCT family transporter.
RESULTS
V. parahaemolyticus can utilize a wide range of compatible solutes.
To determine the range of compatible solutes that can be utilized by V. parahaemolyticus, a Biolog 96-well PM9 osmolyte phenotypic microarray plate was used. Growth analyses were performed using a V. parahaemolyticus ΔectB deletion mutant, which is unable to synthesize ectoine de novo and therefore has a growth defect at high salinity in the absence of exogenous osmolytes (35, 36). The ΔectB mutant was grown in 23 unique osmolytes, 14 of which rescued the growth of the ΔectB mutant (see Fig. S1 in the supplemental material). Previously unrecognized osmolytes for this species included N,N-dimethylglycine (DMG), γ-amino-N-butyric acid (GABA), trimethylamine-N-oxide (TMAO), glutathione, dimethylsulfoniopropionate, morpholinepropanesulfonic acid (MOPS), creatine, N-acetyl l-glutamine, and octopine, in addition to those already known to provide osmoprotection, such as trehalose, β-glutamic acid, GB, ectoine, and l-proline (Fig. S1).
To confirm the data from the phenotypic microarray analysis, we performed growth analyses with wild-type (WT) V. parahaemolyticus RIMD2210633 and the ΔectB mutant in the presence of the compatible solute DMG, GABA, or TMAO in M9 minimal medium supplemented with glucose and 6% NaCl (M9G 6% NaCl) (Fig. 2). In the wild-type strain, in the absence of DMG, there was an ∼4-h lag phase with a growth rate of 0.04 h−1, whereas in the presence of DMG, the lag phase was <1 h, and cells had a higher growth rate (0.06 h−1 in the presence of DMG), indicating that DMG is a very effective compatible solute for V. parahaemolyticus (Fig. 2A). The growth of the ΔectB mutant was rescued by the addition of exogenous DMG, with a lag phase of less than 1 h (Fig. 2A). The growth of the wild-type strain in the presence of GABA had a slightly increased growth rate of 0.045 h−1 (Fig. 2B). The growth of the wild-type strain in the presence of TMAO was not changed, with a growth rate of 0.04 h−1 (Fig. 2C). However, the presence of exogenous GABA or TMAO rescued the growth of the ΔectB mutant, which confirmed that V. parahaemolyticus can utilize these as compatible solutes (Fig. 2). The ΔectB mutant was rescued to a greater extent with exogenous DMG, with a growth rate of 0.07 h−1, than with GABA (0.02 h−1) or TMAO (0.01 h−1) (Fig. 2). In addition, growth analyses of the wild type in M9G 6% NaCl supplemented with either GB or ectoine confirmed that both are effective compatible solutes (Fig. S2) (35).
FIG 2.
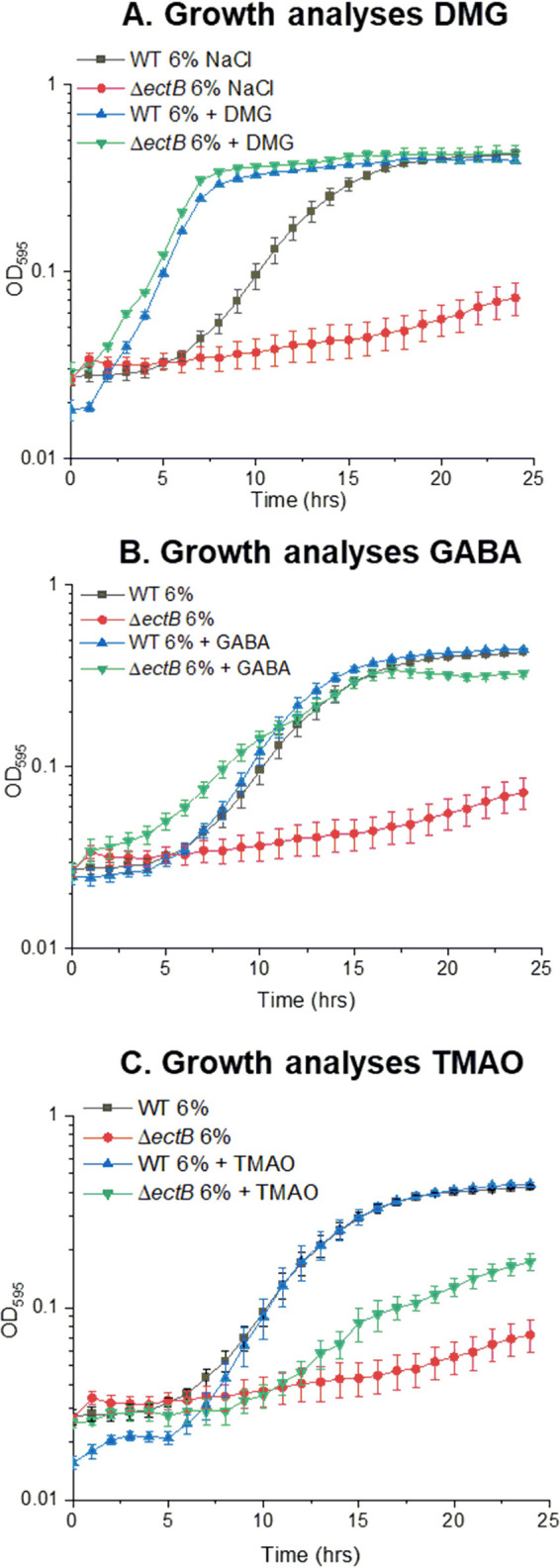
Growth analyses of the V. parahaemolyticus RIMD2210633 wild type (WT) and a ΔectB mutant in M9G 6% NaCl. Growth medium was supplemented with DMG (A), GABA (B), or TMAO (C). The optical density (OD595) was measured every hour for 24 h; the means and standard errors from at least two biological replicates are shown.
DMG is an effective compatible solute for Vibrio species.
To investigate whether DMG can also act as an osmolyte in other Vibrio species, we grew four species representing divergent clades of Vibrio in M9G plus 4% NaCl or 5% NaCl with and without DMG at 37°C for 24 h. Growths of V. harveyi 393, V. cholerae N16961, V. vulnificus YJ016, and V. fluvialis ATCC 33809 were all rescued by exogenous DMG (Fig. S3A to D). In the absence of DMG, V. harveyi grew with a 9-h lag phase, while this strain grew with a 3-h lag phase and an increased growth rate, from 0.04 h−1 to 0.06 h−1, in the presence of DMG (Fig. S3A). In the presence of DMG, the growth rate of V. cholerae was increased from 0.02 h−1 to 0.04 h−1 (Fig. S3B). V. vulnificus is unable to grow in M9G 4% NaCl in the absence of DMG, but growth was rescued in the presence of DMG with a 10-h lag phase and a growth rate of 0.04 h−1 (Fig. S3C). Vibrio fluvialis grown in the presence of DMG also had a reduced lag phase (Fig. S3D). To demonstrate that the changes in the Vibrio species growth patterns were not due to the catabolism of DMG, we grew all strains in M9 medium supplemented with DMG as the sole carbon source. None of the species strains tested grew in the presence of DMG (Fig. S4). Thus, DMG is a bona fide compatible solute for these Vibrio species strains examined.
BCCTs are responsible for the transport of DMG.
We examined whether the BCCTs in V. parahaemolyticus were required for the transport of DMG, and to accomplish this, we used a bccT-null mutant (quadruple ΔbccT1-ΔbccT3-ΔbccT4-ΔbccT2 mutant). We grew the wild type and the bccT-null mutant in M9G 6% NaCl with and without DMG at 37°C for 24 h (Fig. 3). The bccT-null mutant did not exhibit a reduced lag phase or an increased growth rate in the presence of DMG, while the wild-type strain grew with a reduced lag phase of <1 h (Fig. 3), which indicated that a BCCT carrier is required for the transport of DMG. Although V. parahaemolyticus encodes two ABC family compatible solute transporters, ProU1 and ProU2, which are present in the bccT-null mutant, these do not appear to be involved in DMG uptake.
FIG 3.
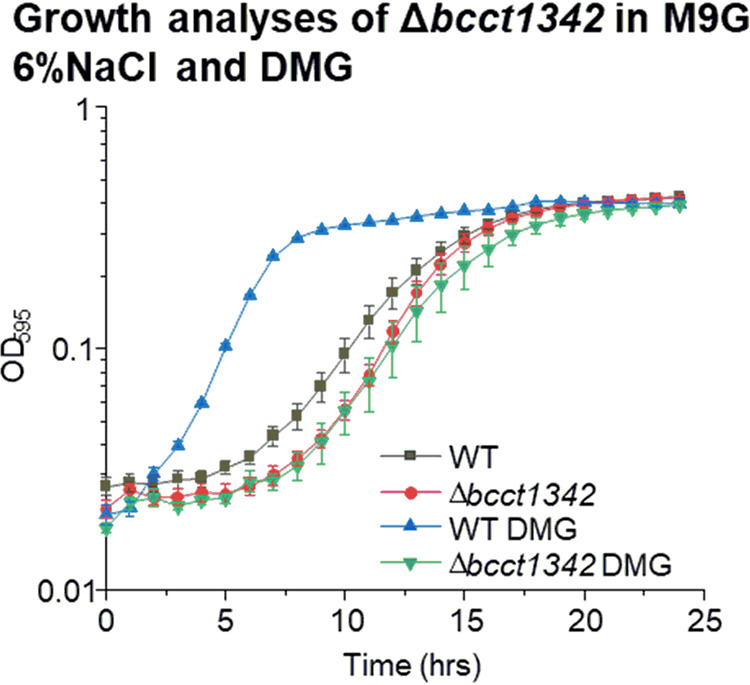
Growth analysis of V. parahaemolyticus RIMD2210633 wild-type and bccT-null (ΔbccT1-ΔbccT3-ΔbccT4-ΔbccT2) mutant strains in M9G 6% NaCl with and without DMG. The optical density (OD595) was measured every hour for 24 h; means and standard errors from at least two biological replicates are displayed.
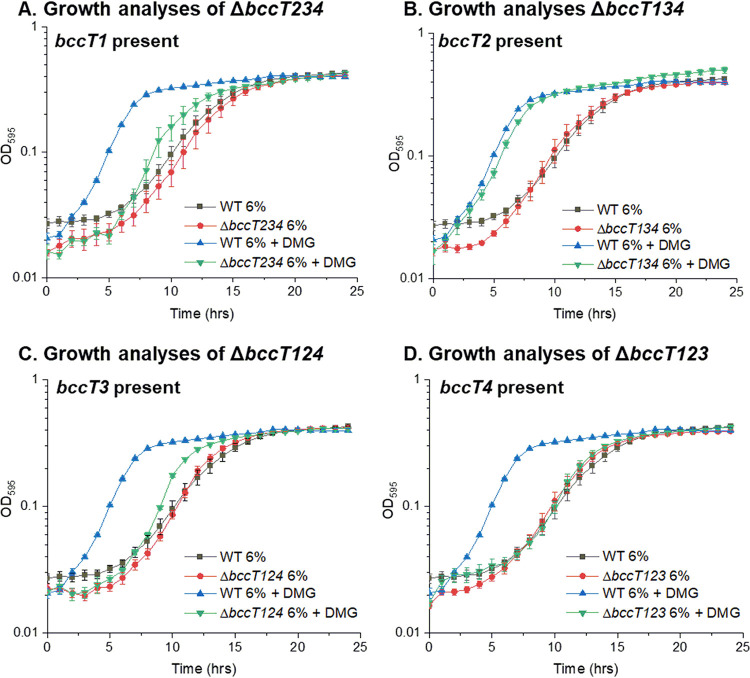
To determine which of the four BCCT family carriers present in V. parahaemolyticus transports DMG, a set of four triple-bccT mutants, each possessing a single functional bccT gene, was utilized in growth assays. The triple ΔbccT2-ΔbccT3-ΔbccT4 mutant, which contains only bccT1, had a slightly reduced lag phase and a slightly higher growth rate through exponential phase in M6G 6% NaCl supplemented with DMG (Fig. 4A). This suggested that BccT1 transported DMG with low efficiency. In contrast, the triple ΔbccT1-ΔbccT3-ΔbccT4 mutant, which contains only bccT2, had a reduction in lag phase similar to that of the wild type (Fig. 4B). This suggested that BccT2 transported DMG as efficiently as the wild-type strain. The triple ΔbccT1-ΔbccT2-ΔbccT4 mutant, which contains only bccT3, grew similarly to the bccT1-only strain, with a marginally reduced lag phase, and grew marginally better through exponential phase in the presence of DMG (Fig. 4C). This suggested that DMG is transported by BccT3 with low efficiency. The triple ΔbccT1-ΔbccT2-ΔbccT3 mutant showed no difference in growth in the absence or presence of DMG, which indicated that BccT4 does not transport DMG into the cell (Fig. 4D).
FIG 4.
Growth analysis of V. parahaemolyticus RIMD2210633 wild-type and triple mutant ΔbccT2-ΔbccT3-ΔbccT4 (A), ΔbccT1-ΔbccT3-ΔbccT4 (B), ΔbccT1-ΔbccT2-ΔbccT4 (C), or ΔbccT1-ΔbccT2-ΔbccT3 (D) strains in M9G 6% NaCl with and without DMG. The optical density (OD595) was measured every hour for 24 h; means and standard errors from at least two biological replicates are displayed.
In order to examine DMG uptake by the BCCT family transporters further, we used E. coli MKH13, a mutant strain that has deletions in betIBA-betT, proU, proP, and putP and cannot grow in M9G 4% NaCl (42). This strain has been utilized successfully in previous studies to assess transporter function in several species (26, 41, 43). We cloned each of the bccT genes into an expression plasmid, used each construct to complement E. coli MKH13, and examined growth in M9G 4% NaCl supplemented with DMG (Fig. 5). Escherichia coli MKH13 strains complemented with bccT1, bccT2, and bccT3 grew in the presence of DMG, confirming that these three are carriers for DMG (Fig. 5). The bccT4 complemented strain and the empty expression plasmid did not grow, which indicated that BccT4 cannot take up DMG (Fig. 5).
FIG 5.
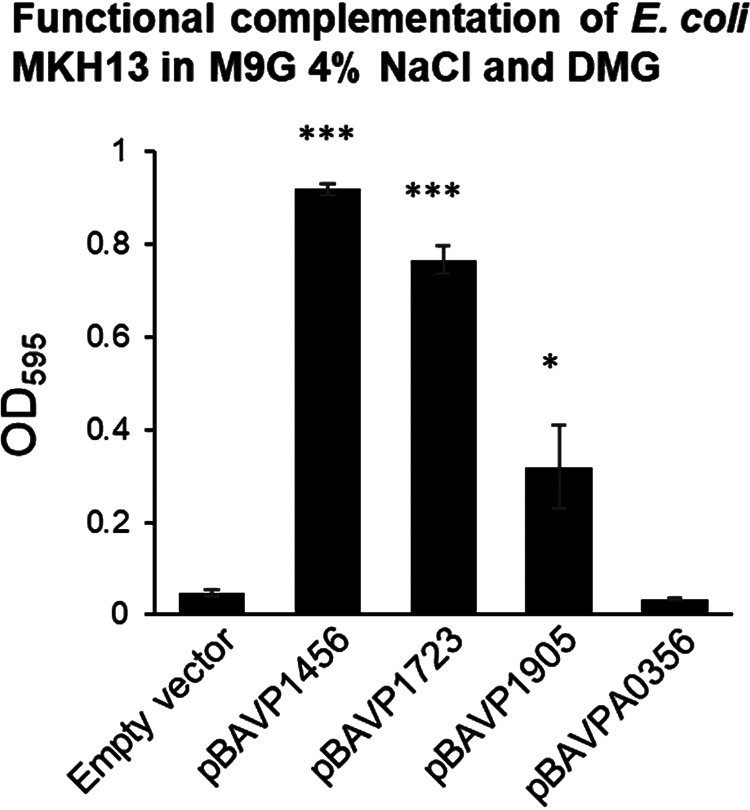
E. coli strain MKH13 complemented with the empty vector, bccT1 (pBAVP1456), bccT2 (pBAVP1723), bccT3 (pBAVP1905), or bccT4 (pBAVPA0356) and grown for 24 h with DMG. The final optical density (OD595) was compared to that of a strain harboring pBAD33. Means and standard errors from at least two biological replicates are shown. Statistics were calculated using Student’s t test (*, P < 0.05; ***, P < 0.001).
BccT1 sequence homology to structurally characterized BCCTs.
From a structure/function standpoint, BccT1 is of particular interest as it can transport GB, DMG, and ectoine, among other substrates (Fig. S5). While GB and DMG have similar structures with methylated head groups, ectoine is a cyclic compound and may require coordination by different amino acid residues in the BCCT transporter (24). Furthermore, BCCT family transporters of ectoine typically do not possess the conserved aromatic residues located in TM4 and TM8 that coordinate GB binding (24). The hydropathy profile of BccT1 was aligned with that of the BCCT transporter BetP from C. glutamicum (CgBetP), whose structure has been studied extensively. We found that BccT1 possessed 12 TM segments (TM1 to TM12) along with N- and C-terminal tail extensions. BccT1 shared matched positions with 89% of residues in CgBetP, which indicated that the structures were highly conserved (Fig. S6). In CgBetP, the residues that form the GB binding pocket are Trp189, Trp194, and Tyr197 in TM4 and one residue located in TM8, Trp374. An additional residue located in TM8 below the binding pocket (Trp377) is thought to be involved in substrate coordination during conformational changes (26). In addition, we aligned the C. glutamicum BCCT family transporters LcoP (CgLcoP) and EctP (CgEctP), which were reported to take up GB and ectoine (44, 45). CgBetP, CgLcoP, and BccT1 possessed amino acids identical to the residues corresponding to TM4 and TM8 described above (Fig. S7).
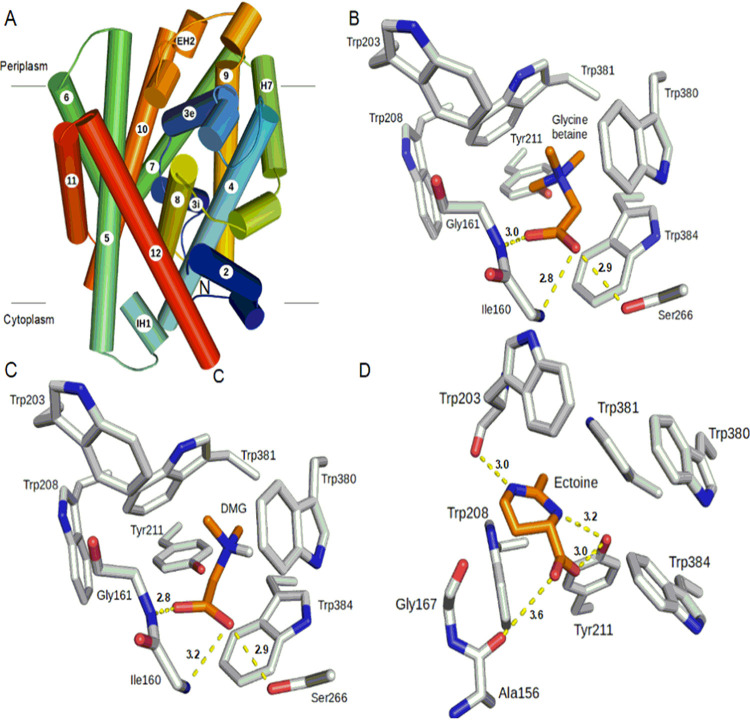
Structural modeling of BccT1.
The structural models of BccT1 consist of 11 transmembrane helices (TM2 to TM12) and a periplasmic helix (H7) (Fig. 6A). The first 108 amino acids of BccT1, including TM1, could not be modeled because of their poor sequence similarity with CgBetP. Nonetheless, the BccT1 models are very similar to the CgBetP structure, with root mean square deviations (RMSDs) of 0.26 Å (399 Cα atoms) and 0.22 Å (391 Cα atoms) for closed-state (CcS) and open substrate-bound-state (CiS) models, respectively. The residues involved in the central binding pocket, cytoplasmic and periplasmic gates, as well as the rest of the substrate pathway are mostly conserved in BccT1. More specifically, a comparison of BccT1 models with the GB-bound structure of CgBetP showed that all the substrate binding residues are conserved and located in TM4 and TM8 (Fig. S7).
FIG 6.
Predicted overall structure of BccT1 and its active-site interactions with different substrates. (A) Side view of the overall structure of BccT1. α-Helices are depicted by cylinders, and the directionality of the polypeptide chain is shown by a color gradient from blue (N terminus) to red (C terminus). Eleven transmembrane helices (TM2 to TM12) and a periplasmic helix (H7) make the core of the structure. Two small helices, IH1 and EH2, connect TM4-TM5 and TM9-TM10, respectively. (B to D) Interactions of the active-site residues of BccT1 with the substrates. Interacting residues of the BccT1 CcS state with docked glycine betaine (B) and DMG (C) as well as those in the CiS state with docked ectoine (D) are represented as sticks. Substrates and BccT1 residues are shown in orange and gray, respectively. Dashed lines show hydrogen-bonding distances between BccT1 residues and various atoms in the substrates. Illustrations were prepared using PyMOL (PyMOL Molecular Graphics System, version 2.0; Schrödinger, LLC).
The closed state (CcS) represents a transition state between outward- and inward-facing open states where the substrate is bound in a central cavity with both substrate entry and exit ports occluded. Unlike open states, the substrate makes optimal interactions with the active-site residues in the CcS state (46); therefore, the CcS model of BccT1 was used for our substrate docking studies. The GB-docked BccT1 model showed a binding free energy change (ΔG) of −4.8 kcal/mol (Table 1). Briefly, the aromatic rings of Trp380, Trp381, and Trp384 form a small hydrophobic pocket that binds GB in this BccT1 model (Table 1 and Fig. 6B). The quaternary amine group of GB binds to this pocket by cation-π and van der Waals interactions. Other aromatic residues, Trp203, Trp208, and Tyr211, of BccT1 also contribute to the hydrophobic interactions with GB. The GB carboxylic group interacts with the peptide backbones of Ile160 and Gly161 as well as the side chain of Ser266 via hydrogen bonding (Fig. 6B).
TABLE 1.
Results of the docking study showing the free energy change upon substrate binding and BccT1 residues mediating interactions with each substrate
| Substrate | Binding free energy (kcal/mol) | Residues involved in hydrogen-bonding interactions | Residues involved in van der Waals interactions |
|---|---|---|---|
| Glycine betaine | −4.8 | Ile160 Gly161, Ser266 | Trp203, Trp208, Tyr211, Trp380, Trp381, Trp384 |
| DMG | −4.4 | Ile160 Gly161, Ser266 | Trp203, Trp208, Tyr211, Trp380, Trp381, Trp384 |
| Ectoine | −6.1 | Trp203, Tyr211 | Trp208, Trp380, Trp381, Trp384 |
Docking analysis of DMG and ectoine binding residues in BccT1.
Next, we sought to determine the DMG and ectoine binding residues in BccT1 by docking. Docking of DMG to the BccT1 CcS model (Fig. 6C) predicted a mode of interaction identical to that of GB with a favorable ΔG of −4.4 kcal/mol (Table 1). This was expected since DMG and GB have very similar chemical structures.
To identify the mode of ectoine binding to BccT1, ectoine was first docked to the active site of the CcS model of BccT1. However, the ΔG of ectoine binding to the CcS model of BccT1 was too low (−2.8 kcal/mol), likely because of the spatially constrained fitting of the large ectoine ring into the smaller hydrophobic pocket comprised of the aromatic rings of Trp380, Trp381, and Trp384. Therefore, we turned our attention to the CiS model of BccT1 ectoine docking, keeping all other docking parameters constant. The substrate binding site in the CiS state differs from that in the CcS state in the relative orientations of the Trp380 and Trp381 side chains remodeling the hydrophobic pocket. More specifically, during the CcS-to-CiS transition, aromatic rings of Trp380 and Trp381 are known to flip 90°, opening up the hydrophobic pocket (46). Interestingly, ectoine docking to the CiS state of BccT1 was accompanied by a significant ΔG of −6.1 kcal/mol (Table 1). The ectoine binding in this BccT1 state is stabilized by hydrogen-bonding interactions between the ectoine carboxyl group and the peptide backbone of Ala157 as well as the side chain of Tyr211 (Fig. 6D). Furthermore, the ectoine ring nitrogens are involved in hydrogen bonding with Tyr211 and the peptide backbone of Trp203. The bound ectoine also shows van der Waals interactions with the aromatic residues Trp203, Trp208, Trp381, and Trp384 (Fig. 6D).
Site-directed mutagenesis of BccT1 uncovers the ectoine binding pocket.
Our structural modeling and docking experiments predicted BccT1 residues that are likely involved in the binding and transport of GB, DMG, and ectoine. Therefore, we selected these residues in BccT1, namely, Trp203, Trp208, Tyr211, and Trp384, for a mutagenesis study. We utilized functional complementation of E. coli MKH13 as a readout for the uptake of GB, DMG, and ectoine by BccT1 with the following single-amino-acid substitutions: Trp203Cys, Trp208Leu, Tyr211Leu, and Trp384Leu. We also tested a BccT1 mutant with all four of these residues replaced by alanine, which is chemically inert and possesses a nonbulky methyl functional group. The replacement of Trp203 resulted in reduced growth of the E. coli MKH13 strain in the presence of GB, while strains harboring the other three single-replacement mutants Trp208Leu, Tyr211Leu, and Trp384Leu grew similarly to a strain expressing WT BccT1 (Fig. 7A). This indicates that Trp203 is important, but not essential, for the uptake of GB by BccT1. In the absence of each of the other three residues 208, 211, and 384, GB may be accommodated by an alternate residue. The replacement of all four residues with alanine resulted in no growth of the E. coli MKH13 strain (Fig. 7A). This indicated that these four residues are involved in the coordination of GB in BccT1.
FIG 7.
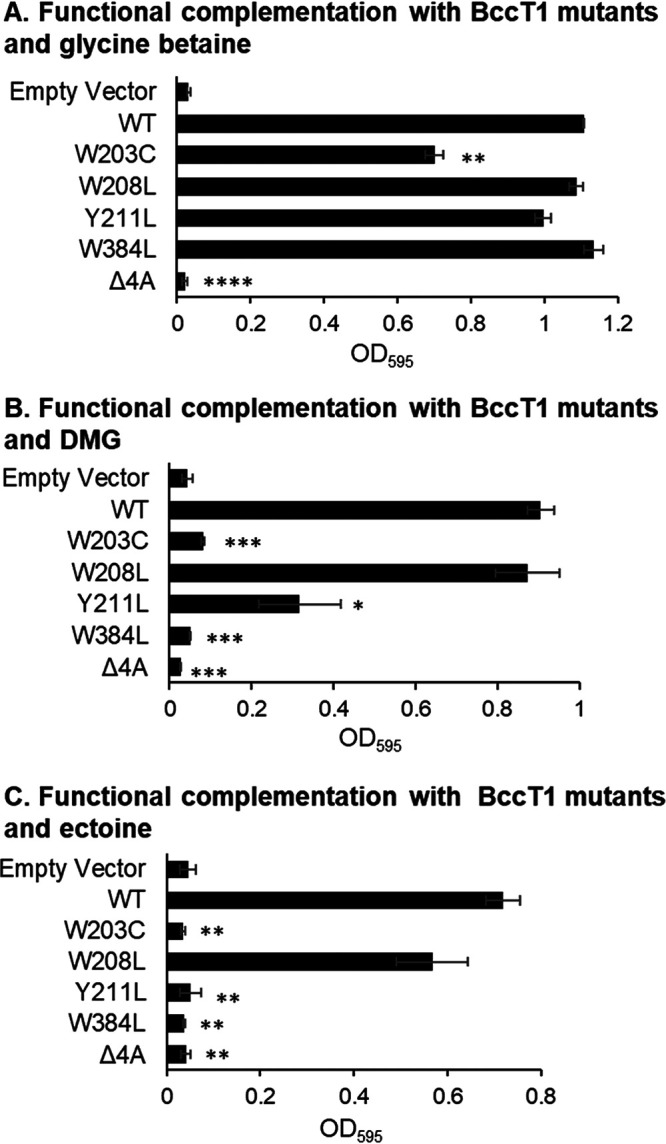
Functional complementation of E. coli MKH13 with BccT1 (WT) and five BccT1 mutants grown in GB (A), DMG (B), or ectoine (C). Strains were grown for 24 h, and the final optical density (OD595) was compared to that of the strain expressing wild-type BccT1 (pBAVP1456). Means and standard errors from at least two biological replicates are shown. Statistics were calculated using Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
The replacement of Trp203 or Trp384 resulted in no uptake of DMG, as strains expressing these mutants did not grow (Fig. 7B). The replacement of Tyr211 resulted in a reduced ability to take up DMG, as evidenced by the growth of this strain to a lower final optical density (OD) (Fig. 7B). The replacement of all four residues resulted in no growth of E. coli MKH13, which indicated that these residues make up the binding pocket for DMG (Fig. 7B).
The replacement of residue Trp203, Tyr211, or Trp384 individually was sufficient to completely abolish the uptake of ectoine by E. coli MKH13, as the strains expressing these mutants were unable to grow (Fig. 7C). However, the replacement of Trp208 did not result in a statistically significant difference in growth from that of a WT BccT1-expressing strain, indicating that Trp208 is not required for the uptake of ectoine (Fig. 7C). The replacement of all four residues with alanine also resulted in the abrogation of ectoine transport (Fig. 7C). Together, these results indicated that DMG and ectoine share a binding pocket with GB. The coordination of DMG requires Trp203 and Trp384, while the coordination of ectoine requires Trp203, Tyr211, and Trp384. These results suggest that the coordination of DMG and ectoine cannot be as easily accommodated by alternate residues as can GB.
Distribution of BccT1.
The BccT1 protein is 562 amino acids long and is present in all sequenced V. parahaemolyticus genomes (>800 genomes). A highly homologous protein is also present in all V. alginolyticus, V. antiquarius, and V. diabolicus strains (95% sequence identity); V. natriegens and V. nereis (90% sequence identity); as well as V. harveyi and V. campbellii sequenced strains (86% sequence identity) (Fig. S8). In general, phylogenetic analysis indicated that BccT1 is conserved within the Harveyi clade, and when present in other clades, it is present in all strains of each species. Overall, within the family Vibrionaceae, BccT1 is present in 35 Vibrio species and in 11 Photobacterium species (Fig. S8). It is of interest to note that in all species, the residues corresponding to Trp203, Trp208, Tyr211, and Trp384, which coordinate substrates, are conserved (data not shown), suggesting an ability to take up a range of osmolytes in these species.
DISCUSSION
Here, we demonstrated that the halophile V. parahaemolyticus can utilize at least 14 compatible solutes, which included DMG, GABA, and TMAO. The transporters BccT1, BccT2, and BccT3 were carriers for DMG, a highly effective osmoprotectant for V. parahaemolyticus, V. vulnificus, V. harveyi, V. cholerae, and V. fluvialis. DMG is the N-dimethyl derivative of glycine, while GB is the N-trimethyl derivative. Some halophilic bacteria were shown to accumulate DMG, an intermediate compound produced during de novo biosynthesis (47–55). DMG was found to be suitable for osmotic adaptation on its own, without modifications by bacteria (47). Additionally, DMG is an intermediate of aerobic GB catabolism and is available in the environment (56–60). None of the Vibrio species strains examined in this study could grow on DMG as a sole carbon and energy source. Several species have been shown to possess genes for the conversion of GB to DMG and the conversion of DMG to sarcosine, which can then be catabolized to glycine (58, 61–66). These genes have not been described within the family Vibrionaceae, and none of the species strains under study contained homologs of these genes.
DMG is known to be transported by major facilitator superfamily (MFS) transporters and ATP binding cassette (ABC) family transporters (56, 67). Here, we demonstrated that the BCCT family of transporters can also take up DMG. BccT1 and BccT2 transport DMG when expressed in a heterologous E. coli background (Fig. 5). However, in the native background, a V. parahaemolyticus strain expressing only BccT2 is fully rescued to wild-type levels by the presence of DMG, while a strain expressing BccT1 is only partially rescued (Fig. 4A and B). This difference may be explained by the fact that bccT2 is not induced by high salinity and has a basal level of transcription in the cell, whereas bccT1 expression is repressed under low-salinity conditions (40, 41). Therefore, in a strain containing only bccT2, expression is constitutively active at low salinity, allowing for more rapid adaptation to high salinity via the uptake of DMG, resulting in a reduced lag phase. In C. glutamicum, the transporter EctP is not induced by external osmolarity, and the EctP transport capacity is maximal at low osmolarity and may act as a rescue system that is available at low osmolarity, ensuring that cells can respond to osmotic stress (45). The same could be true of BccT2 in V. parahaemolyticus, acting as an immediate rescue system to scavenge available osmolytes in the event of osmotic stress.
DMG was not shown previously to be transported by the BCCT family of carriers, and the amino acid residues important for the uptake of DMG by a BCCT are unknown. Our in silico docking study showed that GB was coordinated by residues in the binding pocket of BccT1 (Fig. 6B) identical to those previously reported for CgBetP (26, 27). The coordination of DMG by BccT1 was identical to that of GB (Fig. 6C). Likewise, an ABC transporter, OpuAC, from B. subtilis has been reported to interact very similarly with GB (Protein Data Bank [PDB] ID 2B4L) and a sulfonium analog of DMG, dimethylsulfonioacetate (DMSA) (PDB ID 3CHG) (18, 19). Our mutagenesis studies demonstrated that GB, DMG, and ectoine are coordinated in the same binding pocket of BccT1, but the residues required for coordination are strict for DMG and ectoine, while GB may be accommodated by alternate residues in single-amino-acid mutants (Fig. 7). It was demonstrated previously in an ABC-type transporter that while the replacement of a single aromatic residue in the binding pocket results in decreased affinity of the protein for GB, the substrate is still coordinated with reasonable affinity. Only when any combination of two residues was mutated was transport completely abolished (68). We did not see a reduction in the ability of E. coli MKH13 to grow when only one residue is mutated, and the uptake of GB was completely abolished only when all four residues were mutated. This is likely due to the coordination of GB at alternate positions within the binding pocket that do not affect the overall growth ability of E. coli MKH13 but may affect the affinity of BccT1 for GB. Comparative protein analyses also demonstrated that in other Vibrio species with BccT1 homologs, the residues that coordinate GB, DMG, and ectoine are conserved. There is a very high percent identity shared with BccT1 among BCCT family transporters in these species, and therefore, the ability of these transporters to take up a broad range of substrates is most likely conserved.
All 14 substrates that V. parahaemolyticus was demonstrated to take up are available in the marine environment, including GB, choline, DMG, and ectoine (4, 57, 58, 69). Five of the six compatible solute transporters encoded by V. parahaemolyticus are induced by high salinity and collectively take up a broad range of compatible solutes (40, 41). Although there is redundancy in the compounds taken up by each BCCT, the ability to take up many different compatible solutes likely provides V. parahaemolyticus and other Vibrio species with a fitness advantage in the marine environment that allows them to thrive and grow optimally under high-salt conditions.
Members of the BCCT family are widespread among bacteria, present in both Gram-positive and Gram-negative bacteria as well as Archaea. For example, using the InterPro database (http://www.ebi.ac.uk/interpro/) accession no. IPR000060 for BCCT as a search, 23,000 BCCT proteins fall within the domain Bacteria, 604 BCCT proteins fall within the Archaea, and 78 BCCTs fall within the Eukaryota. Of the 604 Archaea representatives, 593 were from the Stenosarchaea group, of which 549 were within the Halobacteria, suggesting an important function in osmotolerance. Surprisingly, there have been no studies on BCCT function from representatives of Archaea or Eukaryota to date, with the exception of a recent study on the distribution of BCCTs in corals (70).
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
All strains and plasmids used in this study are listed in Table 2. Vibrio parahaemolyticus strains were grown in either lysogeny broth (LB) (Fisher Scientific, Fair Lawn, NJ) with 3% (wt/vol) NaCl (LB3%) or M9 minimal medium (47.8 mM Na2HPO4, 22 mM KH2PO4, 18.7 mM NH4Cl, 8.6 mM NaCl; Sigma-Aldrich) supplemented with 2 mM MgSO4, 0.1 mM CaCl2, and 20 mM glucose as the sole carbon source (M9G) and NaCl as indicated. Dimethylglycine (DMG) was used at a final concentration of 20 mM when supplied as a carbon source. E. coli strains were grown in either LB supplemented with 1% NaCl (LB1%) or M9G supplemented with 1% (wt/vol) NaCl (M9G 1% NaCl). All strains were grown at 37°C with aeration. Chloramphenicol (Cm) was added to the media at 25 μg/ml when necessary.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference(s) or source |
|---|---|---|
| Strains | ||
| V. parahaemolyticus | ||
| RIMD2210633 | O3:K6 clinical isolate; Strr | 87, 88 |
| RIMD2210633 ΔectB | RIMD2210633 ΔectB (VP1721) Strr | 36 |
| SOYBCCT124 | RIMD2210633 ΔVP1456 ΔVP1723 ΔVPA0356 Strr | 41 |
| SOYBCCT123 | RIMD2210633 ΔVP1456 ΔVP1723 ΔVP1905 Strr | 41 |
| SOYBCCT134 | RIMD2210633 ΔVP1456 ΔVP1905 ΔVPA0356 Strr | 41 |
| SOYBCCT234 | RIMD2210633 ΔVP1723 ΔVP1905 ΔVPA0356 Strr | 41 |
| SOYBCCT1342 | RIMD2210633 ΔVP1456 ΔVP1723 ΔVP1905 ΔVPA0356 Strr | 41 |
| V. cholerae N16961 | O1, El Tor strain, Bangladesh, clinical, 1975 | 89 |
| V. vulnificus YJ016 | Clinical isolate | 90 |
| V. fluvialis ATCC 33809 | Synonym, NCTC 11327 or 606; clinical isolate | 91 |
| V. harveyi 393 | Isolated from barramundi in Australia | 92 |
| Escherichia coli | ||
| DH5α λpir | Δlac pir | Thermo Fisher Scientific |
| β2155 λpir | ΔdapA::erm pir for bacterial conjugation | 93 |
| MKH13 | MC4100 (ΔbetTIBA) Δ(putPA)101 Δ(proP)2 Δ(proU) Spr | 42 |
| Plasmids | ||
| pBAD33 | Expression vector; araBAD promoter; Cmr; p15a origin | 71 |
| pBAVP1456 | pBAD33 harboring full-length VP1456 | This study |
| pBAVP1723 | pBAD33 harboring full-length VP1723 | 41 |
| pBAVP1905 | pBAD33 harboring full-length VP1905 | 41 |
| pBAVPA0356 | pBAD33 harboring full-length VPA0356 | This study |
| pBAVP1456W203C | pBAD33 harboring full-length VP1456 with Trp203Cys mutation | This study |
| pBAVP1456W208L | pBAD33 harboring full-length VP1456 with Trp208Lys mutation | This study |
| pBAVP1456Y211L | pBAD33 harboring full-length VP1456 with Tyr211Lys mutation | This study |
| pBAVP1456W377L | pBAD33 harboring full-length VP1456 with Trp377Lys mutation | This study |
| pBAVP1456-3 | pBAD33 harboring full-length VP1456 with Trp203Ala, Trp208Ala, and Tyr211Ala mutations | This study |
| pBAVP1456-4 | pBAVP1456-3 harboring full-length VP1456 with Trp377Ala mutation | This study |
Growth analysis.
Vibrio parahaemolyticus and an in-frame deletion mutant of ectB were grown overnight in M9 minimal medium supplemented with 1% NaCl and 20 mM glucose as the sole carbon source. Cultures were subsequently diluted 1:50 into fresh medium and grown for 5 h. Cultures were pelleted, washed two times with 1× phosphate-buffered saline (PBS) to remove excess salt, and then diluted 1:50 into M9G, and 100 μl was then added to each well of a 96-well Biolog PM9 plate containing different osmolytes and/or salt concentrations (Biolog, Inc., Hayward, CA). The plates were incubated at 37°C with intermittent shaking in a Tecan Sunrise microplate reader, and the optical density at 595 nm (OD595) was measured every hour for 24 h. The area under the curve (AUC) was calculated using Origin 2018 for the wild type (WT) and the ΔectB mutant. Statistics were calculated using Student’s t test; growth in 6% NaCl in the presence of a compatible solute was compared to growth in 6% NaCl with no exogenous compatible solutes.
For growth analysis with individual compatible solutes, wild-type (WT), ΔectB mutant, and quadruple ΔbccT1-ΔbccT3-ΔbccT4-ΔbccT2 mutant (bccT-null) strains were grown overnight in M9G 1% NaCl. Cultures were subsequently diluted 1:50 into fresh medium and grown for 5 h to late exponential phase. Exponential-phase cultures were then diluted 1:40 into 200 μl of M9G 6% NaCl with and without exogenous compatible solutes in a 96-well microplate and grown at 37°C with intermittent shaking for 24 h. The compatible solute N,N-dimethylglycine (DMG), trimethylamine-N-oxide (TMAO), γ-amino-N-butyric acid (GABA), glycine betaine (GB), or ectoine (ect) was added to a final concentration of 500 μM. Growth analysis with each compatible solute was repeated according to the above-described procedure with each of four triple-bccT mutants, ΔbccT2-ΔbccT3-ΔbccT4, ΔbccT1-ΔbccT3-ΔbccT4, ΔbccT1-ΔbccT2-ΔbccT4, and ΔbccT1-ΔbccT2-ΔbccT3.
Growth analyses of V. cholerae N16961, V. harveyi 393, V. vulnificus YJ016, and V. fluvialis ATCC 33809 (synonym, NCTC 11327 or 606) were conducted by growing strains overnight in M9G supplemented with 2% NaCl (M9G 2% NaCl). Strains were diluted 1:40 into 200 μl of M9G 4% NaCl, or M9G 5% NaCl for V. fluvialis, with and without exogenous DMG, in a 96-well microplate and grown at 37°C with intermittent shaking for 24 h. To test DMG as a carbon source, V. cholerae, V. vulnificus, V. fluvialis, and V. parahaemolyticus were grown overnight in LB1%, and V. harveyi was grown in LB2%. Cells were pelleted, washed two times with 1× PBS, and diluted 1:40 into 200 μl of M9 medium with 20 mM DMG as the sole carbon source and 1% NaCl (2% NaCl for V. harveyi). Strains were grown in M9G 1% NaCl (2% NaCl for V. harveyi) as a control. Strains were grown in a 96-well microplate as described above.
Functional complementation of E. coli strain MKH13 with VP1456, VP1723, VP1905, and VPA0356.
Full-length VP1456 (bccT1) or VPA0356 (bccT4) was amplified from the V. parahaemolyticus RIMD2210633 genome using primers listed in Table 3. All primers were purchased from Integrated DNA Technologies (Coralville, IA). The Gibson assembly protocol using NEBuilder HiFi DNA assembly master mix (New England BioLabs, Ipswich, MA) was followed to ligate the VP1456 or VPA0356 fragment with the expression vector pBAD33 (71), which had been linearized with SacI. Regions of complementarity for Gibson assembly are indicated by lowercase type in the primer sequences in Table 3. The resulting expression plasmid, pBAVP1456 or pBAVPA0356, was transformed into E. coli DH5α for propagation. Plasmids were then purified, sequenced, and subsequently transformed into E. coli strain MKH13, which has large deletions that include all compatible solute transporters (putP, proP, and proU) and the choline uptake and GB biosynthesis loci (betT-betIBA) (42). E. coli MKH13 strains containing pBAVP1456, pBAVP1723, pBAVP1905, or pBAVPA0356 were grown overnight in minimal medium supplemented with 1% NaCl and 20 mM glucose (M9G1%) and subsequently diluted 1:40 into M9G supplemented with 4% NaCl (M9G 4% NaCl). E. coli MKH13 strains containing pBAVP1456, pBAVP1723, pBAVP1905, or pBAVPA0356 were grown overnight in M9G 1% NaCl with chloramphenicol and subsequently diluted 1:100 into M9G 4% NaCl with 500 μM the indicated compatible solute and chloramphenicol for plasmid maintenance. The expression of each BccT protein was induced with 0.01% arabinose, and functional complementation was determined by measuring the OD595 after 24 h of growth at 37°C with aeration. Growth was compared to that of an MKH13 strain harboring empty plasmid pBAD33, which cannot grow in M9G 4% NaCl without exogenous compatible solutes. Statistics were calculated using Student’s t test.
TABLE 3.
Primers used in this study
| Use and primer | Sequence (5′–3′)a | Length (bp) |
|---|---|---|
| Expression | ||
| VP1456 Fwd | tgggctagcgaattcgagctTTTGCTGTAAATATGCAATAAAAGTG | 1,740 |
| VP1456 Rev | ggatccccgggtaccgagctTTAGCGGTAAGCGGAAAG | |
| VPA0356 Fwd | tgggctagcgaattcgagctAGCGGCTTTTTGAACATC | 1,671 |
| VPA0356 Rev | ggatccccgggtaccgagctTTAAACCAAACCTTGATCC | |
| Mutagenesis | ||
| BCCT1 TRP203CYS F | CATGTTTCACtgcGGTGTTCACGGTTGGAG | 7,096 |
| BCCT1 TRP203CYS R | GTCGCGCCCATCGCGAGT | |
| BCCT1 TRP208LEU F | TGTTCACGGTctgAGTATTTACGC | 7,096 |
| BCCT1 TRP208LEU R | CCCCAGTGAAACATGGTC | |
| BCCT1 TYR211LEU F | TTGGAGTATTctgGCCCTTGTTGCGTTG | 7,096 |
| BCCT1 TYR211LEU R | CCGTGAACACCCCAGTGA | |
| BCCT1 TRP384LEU F | GTGGGTATCTctgTCTCCGTTTG | 7,096 |
| BCCT1 TRP384LEU R | CAAGCCCAGTAGAACACTG | |
| BCCT1 203208211ALA F | tgcgagtattgcgGCCCTTGTTGCGTTGGCG | 7,096 |
| BCCT1 203208211ALA R | ccgtgaacacccgcGTGAAACATGGTCGCGCC | |
| BCCT1 TRP384ALA F | GTGGGTATCTgcgTCTCCGTTTG | 7,096 |
| BCCT1 TRP384ALA R | CAAGCCCAGTAGAACACTG | |
Regions of complementarity for Gibson assembly are indicated by lowercase type.
Site-directed mutagenesis was performed on pBAVP1456 with a Q5 site-directed mutagenesis kit (New England BioLabs, Ipswich, MA) and primers listed in Table 3. Primers were designed to create nucleotide substitutions resulting in the following amino acid changes: Trp203Cys, Trp203Ala, Trp208Leu, Trp208Ala, Tyr211Leu, Tyr211Ala, Trp384Leu, and Trp384Ala. Site-directed mutagenesis was performed according to the manufacturer’s protocol to create single-amino-acid substitutions Trp203Cys, Trp208Leu, Tyr211Leu, and Trp384Leu. Residues 203, 208, and 211 were mutagenized to encode alanines, and residue 384 was subsequently mutagenized in this plasmid backbone to encode an alanine.
Homology modeling of BccT1 and docking of GB, DMG, and ectoine.
A BLAST search of the BccT1 sequence against the Protein Data Bank (PDB) as the search database showed the highest sequence identity with the 3.2-Å X-ray crystal structure of a GB transporter from C. glutamicum (CgBetP) (PDB ID 4AIN). Homology modeling of BccT1 was done with the SWISS-MODEL server using the CgBetP structure (PDB ID 4AIN) as a template (72). CgBetP is an active trimer correctly represented in its three-dimensional (3D) structure with the three protomers (chains) showing different stages of the substrate transport cycle with an alternating-access mechanism (46). More specifically, chain A of the CgBetP structure represents the closed apo state (Cc), whereas chains B and C represent a closed substrate-bound (CcS) and an open substrate-bound (CiS) state, respectively. Chain B and chain C of CgBetP were used as the templates to generate CcS- and CiS-like homology models of BccT1, respectively. The energy-minimized models of BccT1 were verified using a 3D profile method of Verify3D that evaluates the correlation between the amino acid sequence (one dimensional [1D]) and the model (3D) by comparing it to the other known structures in the database. Residues showing poor stereochemical properties or close contacts in each monomeric model were fixed manually in COOT (73). Models were then subjected to energy minimization using the 3Drefine server to reduce any other structural restraints (74). The quality of the resulting models was finally verified with Verify3D and PROCHECK (75, 76). The CcS and CiS models of BccT1 showed Verify3D scores of 88.6% and 84.0%, respectively, which indicated good quality of the models (see Fig. S9 in the supplemental material). The stereochemical properties of the models were examined by a Ramachandran plot using PROCHECK. Both the CcS and CiS BccT1 models contain 99.2% of all the residues in the allowed region and none in the disallowed region (Fig. S10).
For docking studies, ligand models for GB, DMG, and ectoine were obtained from Chemical Entities of Biological Interest (ChEBI) EMBL (77). Polar hydrogens were added to the ligand structures in PRODRG (78). AutoDock tools v1.5.6 was used to assign the rotatable bonds in the ligands and to add all polar hydrogens in the BccT1 models to prepare them for the docking. The ligand-centered maps for BccT1 models were assigned a grid size of 20 by 20 by 20 Å3. The docking experiments of GB, DMG, and ectoine ligands with the BccT1 receptor model were performed using AutoDock Vina (79). The binding free energies and BccT1 residues making interactions with the ligands are listed in Table 1. The structural illustrations were prepared using PyMOL (PyMOL Molecular Graphics System, version 2.0; Schrödinger, LLC).
Bioinformatics and phylogenetic analyses.
Transmembrane helix probabilities of C. glutamicum BetP (GenBank accession number CAA63771.1) and BccT1 (GenBank accession number Q87PP5.1) were generated using OCTOPUS and aligned via the AlignMe program (http://www.bioinfo.mpg.de/AlignMe) (80–82). The sequences of the V. parahaemolyticus protein BccT1 (GenBank accession number Q87PP5.1), CgBetP (GenBank accession number CAA63771.1), CgEctP (GenBank accession number CAA04760.1), and CgLcoP (GenBank accession number ASW14702.1) were downloaded from the NCBI database and aligned using the ClustalW algorithm (83). Aligned sequences were displayed and annotated using ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) (84).
Phylogenetic analysis was conducted using the BccT1 (VP1456) protein as a seed to identify all homologs within the family Vibrionaceae with completed genome sequences available. Unique protein sequences that had >95% sequence coverage and >70% amino acid identity with BccT1 were obtained from the NCBI database and aligned using the ClustalW algorithm (83). The evolutionary history of BccT1 was inferred by using the maximum likelihood method and the Le_Gascuel_2008 model as determined by best-fit model selection in MEGAX (85, 86). The tree with the highest log likelihood (−10,453.23) is shown. The percentages of trees in which the associated taxa clustered together are shown next to the branches. The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a Jones-Taylor-Thornton (JTT) model and then selecting the topology with the superior log-likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.4143]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 49 amino acid sequences and a total of 525 positions in the final data set.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a National Science Foundation grant (award IOS-1656688) to E.F.B. and a National Institutes of Health grant (award R35GM119504) to V.P. G.J.G. was funded in part by a University of Delaware graduate fellowship award.
We thank members of the Boyd group for constructive feedback on the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Record MT Jr, Courtenay ES, Cayley DS, Guttman HJ. 1998. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem Sci 23:143–148. doi: 10.1016/S0968-0004(98)01196-7. [DOI] [PubMed] [Google Scholar]
- 2.Wood JM. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev 63:230–262. doi: 10.1128/MMBR.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csonka LN. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 53:121–147. doi: 10.1128/MMBR.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.da Costa MS, Santos H, Galinski EA. 1998. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv Biochem Eng Biotechnol 61:117–153. doi: 10.1007/BFb0102291. [DOI] [PubMed] [Google Scholar]
- 5.Galinski EA. 1995. Osmoadaptation in bacteria. Adv Microb Physiol 37:272–328. [PubMed] [Google Scholar]
- 6.Wood JM. 2011. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu Rev Microbiol 65:215–238. doi: 10.1146/annurev-micro-090110-102815. [DOI] [PubMed] [Google Scholar]
- 7.Yancey P. 1994. Compatible and counteracting solutes, p 81–109. In Strange K. (ed), Cellular and molecular physiology of cell volume regulation. CRC Press, Boca Raton, FL. [Google Scholar]
- 8.Galinski EA, Oren A. 1991. Isolation and structure determination of a novel compatible solute from the moderately halophilic purple sulfur bacterium Ectothiorhodospira marismortui. Eur J Biochem 198:593–598. doi: 10.1111/j.1432-1033.1991.tb16055.x. [DOI] [PubMed] [Google Scholar]
- 9.Sleator RD, Hill C. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 10.Roberts MF. 2004. Osmoadaptation and osmoregulation in archaea: update 2004. Front Biosci 9:1999–2019. doi: 10.2741/1366. [DOI] [PubMed] [Google Scholar]
- 11.Roberts MF. 2005. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst 1:5. doi: 10.1186/1746-1448-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 13.Ventosa A, Nieto JJ, Oren A. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544. doi: 10.1128/MMBR.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oren A. 1999. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev 63:334–348. doi: 10.1128/MMBR.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May G, Faatz E, Villarejo M, Bremer E. 1986. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol Gen Genet 205:225–233. doi: 10.1007/BF00430432. [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Beattie GA. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function. J Bacteriol 189:6901–6912. doi: 10.1128/JB.00763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Heide T, Stuart MC, Poolman B. 2001. On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J 20:7022–7032. doi: 10.1093/emboj/20.24.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn C, Sohn-Bösser L, Breed J, Welte W, Schmitt L, Bremer E. 2006. Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J Mol Biol 357:592–606. doi: 10.1016/j.jmb.2005.12.085. [DOI] [PubMed] [Google Scholar]
- 19.Smits SHJ, Höing M, Lecher J, Jebbar M, Schmitt L, Bremer E. 2008. The compatible-solute-binding protein OpuAC from Bacillus subtilis: ligand binding, site-directed mutagenesis, and crystallographic studies. J Bacteriol 190:5663–5671. doi: 10.1128/JB.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, Strom AR. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol 5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 21.Kappes RM, Kempf B, Bremer E. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J Bacteriol 178:5071–5079. doi: 10.1128/jb.178.17.5071-5079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peter H, Burkovski A, Kramer R. 1996. Isolation, characterization, and expression of the Corynebacterium glutamicum betP gene, encoding the transport system for the compatible solute glycine betaine. J Bacteriol 178:5229–5234. doi: 10.1128/jb.178.17.5229-5234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichler K, Bourgis F, Buchet A, Kleber HP, Mandrand-Berthelot MA. 1994. Molecular characterization of the cai operon necessary for carnitine metabolism in Escherichia coli. Mol Microbiol 13:775–786. doi: 10.1111/j.1365-2958.1994.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 24.Ziegler C, Bremer E, Kramer R. 2010. The BCCT family of carriers: from physiology to crystal structure. Mol Microbiol 78:13–34. doi: 10.1111/j.1365-2958.2010.07332.x. [DOI] [PubMed] [Google Scholar]
- 25.Farwick M, Siewe RM, Kramer R. 1995. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J Bacteriol 177:4690–4695. doi: 10.1128/jb.177.16.4690-4695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. 2009. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature 458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 27.Perez C, Koshy C, Ressl S, Nicklisch S, Kramer R, Ziegler C. 2011. Substrate specificity and ion coupling in the Na+/betaine symporter BetP. EMBO J 30:1221–1229. doi: 10.1038/emboj.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiou CS, Hsu SY, Chiu SI, Wang TK, Chao CS. 2000. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J Clin Microbiol 38:4621–4625. doi: 10.1128/JCM.38.12.4621-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph SW, Colwell RR, Kaper JB. 1982. Vibrio parahaemolyticus and related halophilic vibrios. Crit Rev Microbiol 10:77–124. doi: 10.3109/10408418209113506. [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin JB, DePaola A, Bopp CA, Martinek KA, Napolilli NP, Allison CG, Murray SL, Thompson EC, Bird MM, Middaugh JP. 2005. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N Engl J Med 353:1463–1470. doi: 10.1056/NEJMoa051594. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko T, Colwell RR. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J Bacteriol 113:24–32. doi: 10.1128/JB.113.1.24-32.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko T, Colwell RR. 1974. Distribution of Vibrio parahaemolyticus and related organisms in the Atlantic Ocean off South Carolina and Georgia. Appl Microbiol 28:1009–1017. doi: 10.1128/AEM.28.6.1009-1017.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko T, Colwell RR. 1975. Incidence of Vibrio parahaemolyticus in Chesapeake Bay. Appl Microbiol 30:251–257. doi: 10.1128/AEM.30.2.251-257.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J Infect Dis 181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 35.Ongagna-Yhombi SY, Boyd EF. 2013. Biosynthesis of the osmoprotectant ectoine, but not glycine betaine, is critical for survival of osmotically stressed Vibrio parahaemolyticus cells. Appl Environ Microbiol 79:5038–5049. doi: 10.1128/AEM.01008-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naughton LM, Blumerman SL, Carlberg M, Boyd EF. 2009. Osmoadaptation among Vibrio species and unique genomic features and physiological responses of Vibrio parahaemolyticus. Appl Environ Microbiol 75:2802–2810. doi: 10.1128/AEM.01698-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shikuma NJ, Davis KR, Fong JN, Yildiz FH. 2013. The transcriptional regulator, CosR, controls compatible solute biosynthesis and transport, motility and biofilm formation in Vibrio cholerae. Environ Microbiol 15:1387–1399. doi: 10.1111/j.1462-2920.2012.02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Kessel JC, Rutherford ST, Cong JP, Quinodoz S, Healy J, Bassler BL. 2015. Quorum sensing regulates the osmotic stress response in Vibrio harveyi. J Bacteriol 197:73–80. doi: 10.1128/JB.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory GJ, Morreale DP, Carpenter MR, Kalburge SS, Boyd EF. 2019. Quorum sensing regulators AphA and OpaR control expression of the operon responsible for biosynthesis of the compatible solute ectoine. Appl Environ Microbiol 85:e01543-19. doi: 10.1128/AEM.01543-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory GJ, Morreale DP, Boyd EF. 2020. CosR is a global regulator of the osmotic stress response with widespread distribution among bacteria. Appl Environ Microbiol 86:e00120-20. doi: 10.1128/AEM.00120-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ongagna-Yhombi SY, McDonald ND, Boyd EF. 2015. Deciphering the role of multiple betaine-carnitine-choline transporters in the halophile Vibrio parahaemolyticus. Appl Environ Microbiol 81:351–363. doi: 10.1128/AEM.02402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haardt M, Kempf B, Faatz E, Bremer E. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol Gen Genet 246:783–786. doi: 10.1007/BF00290728. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Beattie GA. 2008. Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J Bacteriol 190:2717–2725. doi: 10.1128/JB.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steger R, Weinand M, Kramer R, Morbach S. 2004. LcoP, an osmoregulated betaine/ectoine uptake system from Corynebacterium glutamicum. FEBS Lett 573:155–160. doi: 10.1016/j.febslet.2004.07.067. [DOI] [PubMed] [Google Scholar]
- 45.Weinand M, Kramer R, Morbach S. 2007. Characterization of compatible solute transporter multiplicity in Corynebacterium glutamicum. Appl Microbiol Biotechnol 76:701–708. doi: 10.1007/s00253-007-0938-4. [DOI] [PubMed] [Google Scholar]
- 46.Perez C, Koshy C, Yildiz O, Ziegler C. 2012. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature 490:126–130. doi: 10.1038/nature11403. [DOI] [PubMed] [Google Scholar]
- 47.Menaia J, Duarte J, Boone D. 1993. Osmotic adaptation of moderately halophilic methanogenic Archaeobacteria, and detection of cytosolic N,N-dimethylglycine. Experientia 49:1047–1054. doi: 10.1007/BF01929912. [DOI] [Google Scholar]
- 48.Roberts MF, Lai MC, Gunsalus RP. 1992. Biosynthetic pathways of the osmolytes N epsilon-acetyl-beta-lysine, beta-glutamine, and betaine in Methanohalophilus strain FDF1 suggested by nuclear magnetic resonance analyses. J Bacteriol 174:6688–6693. doi: 10.1128/jb.174.20.6688-6693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai MC, Yang DR, Chuang MJ. 1999. Regulatory factors associated with synthesis of the osmolyte glycine betaine in the halophilic methanoarchaeon Methanohalophilus portucalensis. Appl Environ Microbiol 65:828–833. doi: 10.1128/AEM.65.2.828-833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nyyssölä A, Kerovuo J, Kaukinen P, von Weymarn N, Reinikainen T. 2000. Extreme halophiles synthesize betaine from glycine by methylation. J Biol Chem 275:22196–22201. doi: 10.1074/jbc.M910111199. [DOI] [PubMed] [Google Scholar]
- 51.Nyyssola A, Leisola M. 2001. Actinopolyspora halophila has two separate pathways for betaine synthesis. Arch Microbiol 176:294–300. doi: 10.1007/s002030100325. [DOI] [PubMed] [Google Scholar]
- 52.Nyyssola A, Reinikainen T, Leisola M. 2001. Characterization of glycine sarcosine N-methyltransferase and sarcosine dimethylglycine N-methyltransferase. Appl Environ Microbiol 67:2044–2050. doi: 10.1128/AEM.67.5.2044-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waditee R, Tanaka Y, Aoki K, Hibino T, Jikuya H, Takano J, Takabe T, Takabe T. 2003. Isolation and functional characterization of N-methyltransferases that catalyze betaine synthesis from glycine in a halotolerant photosynthetic organism Aphanothece halophytica. J Biol Chem 278:4932–4942. doi: 10.1074/jbc.M210970200. [DOI] [PubMed] [Google Scholar]
- 54.Lu WD, Chi ZM, Su CD. 2006. Identification of glycine betaine as compatible solute in Synechococcus sp. WH8102 and characterization of its N-methyltransferase genes involved in betaine synthesis. Arch Microbiol 186:495–506. doi: 10.1007/s00203-006-0167-8. [DOI] [PubMed] [Google Scholar]
- 55.Kimura Y, Kawasaki S, Yoshimoto H, Takegawa K. 2010. Glycine betaine biosynthesized from glycine provides an osmolyte for cell growth and spore germination during osmotic stress in Myxococcus xanthus. J Bacteriol 192:1467–1470. doi: 10.1128/JB.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bashir A, Hoffmann T, Smits SH, Bremer E. 2014. Dimethylglycine provides salt and temperature stress protection to Bacillus subtilis. Appl Environ Microbiol 80:2773–2785. doi: 10.1128/AEM.00078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casaite V, Poviloniene S, Meskiene R, Rutkiene R, Meskys R. 2011. Studies of dimethylglycine oxidase isoenzymes in Arthrobacter globiformis cells. Curr Microbiol 62:1267–1273. doi: 10.1007/s00284-010-9852-6. [DOI] [PubMed] [Google Scholar]
- 58.Wargo MJ, Szwergold BS, Hogan DA. 2008. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol 190:2690–2699. doi: 10.1128/JB.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Incharoensakdi A, Waditee R. 2000. Degradation of glycinebetaine by betaine-homocysteine methyltransferase in Aphanothece halophytica: effect of salt downshock and starvation. Curr Microbiol 41:227–231. doi: 10.1007/s002840010125. [DOI] [PubMed] [Google Scholar]
- 60.White RF, Kaplan L, Birnbaum J. 1973. Betaine-homocysteine transmethylase in Pseudomonas denitrificans, a vitamin B12 overproducer. J Bacteriol 113:218–223. doi: 10.1128/JB.113.1.218-223.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hampel K, LaBauve A, Meadows J, Fitzsimmons L, Nock A, Wargo M. 2014. Characterization of the GbdR regulon in Pseudomonas aeruginosa. J Bacteriol 196:7–15. doi: 10.1128/JB.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nock AM, Wargo MJ. 2016. Choline catabolism in Burkholderia thailandensis is regulated by multiple glutamine amidotransferase 1-containing AraC family transcriptional regulators. J Bacteriol 198:2503–2514. doi: 10.1128/JB.00372-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wargo M. 2013. Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa. Appl Environ Microbiol 79:2112–2120. doi: 10.1128/AEM.03565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willsey G, Wargo M. 2016. Sarcosine catabolism in Pseudomonas aeruginosa is transcriptionally regulated by SouR. J Bacteriol 198:301–310. doi: 10.1128/JB.00739-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meskys R, Harris RJ, Casaite V, Basran J, Scrutton NS. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur J Biochem 268:3390–3398. doi: 10.1046/j.1432-1327.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki H, Tamamura R, Yajima S, Kanno M, Suguro M. 2005. Corynebacterium sp. U-96 contains a cluster of genes of enzymes for the catabolism of sarcosine to pyruvate. Biosci Biotechnol Biochem 69:952–956. doi: 10.1271/bbb.69.952. [DOI] [PubMed] [Google Scholar]
- 67.MacMillan SV, Alexander DA, Culham DE, Kunte HJ, Marshall EV, Rochon D, Wood JM. 1999. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim Biophys Acta 1420:30–44. doi: 10.1016/s0005-2736(99)00085-1. [DOI] [PubMed] [Google Scholar]
- 68.Tschapek B, Pittelkow M, Sohn-Bosser L, Holtmann G, Smits SH, Gohlke H, Bremer E, Schmitt L. 2011. Arg149 is involved in switching the low affinity, open state of the binding protein AfProX into its high affinity, closed state. J Mol Biol 411:36–52. doi: 10.1016/j.jmb.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 69.King GM. 1984. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl Environ Microbiol 48:719–725. doi: 10.1128/AEM.48.4.719-725.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ngugi D, Ziegler M, Duarte C, Voolstra C. 2020. Genomic blueprint of glycine betaine metabolism in coral metaorganisms and their contribution to reef nitrogen budgets. iScience 23:101120. doi: 10.1016/j.isci.2020.101120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 74.Bhattacharya D, Nowotny J, Cao R, Cheng J. 2016. 3Drefine: an interactive Web server for efficient protein structure refinement. Nucleic Acids Res 44:W406–W409. doi: 10.1093/nar/gkw336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bowie J, Lüthy R, Eisenberg D. 1991. A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 76.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 77.Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, Turner S, Swainston N, Mendes P, Steinbeck C. 2016. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res 44:D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schüttelkopf A, van Aalten D. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 79.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khafizov K, Staritzbichler R, Stamm M, Forrest L. 2010. A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry 49:10702–10713. doi: 10.1021/bi101256x. [DOI] [PubMed] [Google Scholar]
- 81.Stamm M, Staritzbichler R, Khafizov K, Forrest L. 2013. Alignment of helical membrane protein sequences using AlignMe. PLoS One 8:e57731. doi: 10.1371/journal.pone.0057731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stamm M, Staritzbichler R, Khafizov K, Forrest L. 2014. AlignMe—a membrane protein sequence alignment Web server. Nucleic Acids Res 42:W246–W251. doi: 10.1093/nar/gku291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 87.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 88.Whitaker WB, Parent MA, Naughton LM, Richards GP, Blumerman SL, Boyd EF. 2010. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl Environ Microbiol 76:4720–4729. doi: 10.1128/AEM.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaper J, Lockman H, Baldini M, Levine M. 1984. Recombinant nontoxinogenic Vibrio cholerae strains as attenuated cholera vaccine candidates. Nature 308:655–658. doi: 10.1038/308655a0. [DOI] [PubMed] [Google Scholar]
- 90.Chen C, Wu K, Chang Y, Chang C, Tsai H, Liao T, Liu Y, Chen H, Shen A, Li J, Su T, Shao C, Lee C, Hor L, Tsai S. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res 13:2577–2587. doi: 10.1101/gr.1295503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee J, Shread P, Furniss A, Bryant T. 1981. Taxonomy and description of Vibrio fluvialis sp. nov. (synonym group F vibrios, group EF6). J Appl Bacteriol 50:73–94. doi: 10.1111/j.1365-2672.1981.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 92.Pedersen K, Verdonck L, Austin B, Austin DA, Blanch AR, Grimont PAD, Jofre J, Koblavi S, Larsen JL, Tiainen T, Vigneulle M, Swings J. 1998. Taxonomic evidence that Vibrio carchariae Grimes et al. 1985 is a junior synonym of Vibrio harveyi (Johnson and Shunk 1936) Baumann et al. 1981. Int J Syst Bacteriol 48:749–758. doi: 10.1099/00207713-48-3-749. [DOI] [Google Scholar]
- 93.Dehio C, Meyer M. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol 179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.