Vastly different experimental techniques and conditions have been used to study export in E. coli. We demonstrated that altering experimental conditions can change the step that is observed during study. Investigators should consider specific experimental conditions when comparing data from different laboratories, as well as when comparing data from different experiments within a laboratory. We have shown that each precursor species has inherent properties that determine the translocation rate; thus generalizations from studies of a single species must be made with caution. A summary of advantages and disadvantages in use of nine precursors is presented.
KEYWORDS: protein translocation, protein export, secretion, E. coli, SecA, SecYEG, translocon, single turnover, multiple turnovers, rate-limiting step
ABSTRACT
Precursor proteins are translocated across the cytoplasmic membrane in Escherichia coli by the general secretory, or Sec, pathway. The main components of the pathway are the integral membrane heterotrimeric SecYEG complex and the peripheral membrane ATPase, SecA. In this study, we have applied an in vitro assay using inverted cytoplasmic membrane vesicles to investigate the complex cycle that leads to translocation. We compared the apparent rate constants for nine precursors under two experimental conditions, single turnover and multiple turnovers. For each precursor, the rate constant for a single turnover was higher than for multiple turnovers, indicating that a different step limits the rate under the two conditions. We conclude that the rate-limiting step for a single turnover is an early step in the initial phase of transit through the channel, whereas the rate of multiple turnovers is limited by the resetting of the translocon. The presence of the chaperone SecB during multiple turnovers increased the maximal amplitude translocated for the three precursor species tested, pGBP, pPhoA, and proOmpA, and also increased the apparent rate constants for both pGBP and pPhoA. The rate constant for proOmpA was decreased by the presence of SecB.
IMPORTANCE Vastly different experimental techniques and conditions have been used to study export in E. coli. We demonstrated that altering experimental conditions can change the step that is observed during study. Investigators should consider specific experimental conditions when comparing data from different laboratories, as well as when comparing data from different experiments within a laboratory. We have shown that each precursor species has inherent properties that determine the translocation rate; thus generalizations from studies of a single species must be made with caution. A summary of advantages and disadvantages in use of nine precursors is presented.
INTRODUCTION
The general secretory, or Sec, system in E. coli translocates precursor proteins across the cytoplasmic membrane into the periplasmic space (for a review see reference 1). The channel for passage through the membrane is provided by a highly conserved translocon comprising SecY, SecE, and SecG (SecYEG). The energy for the system is supplied both by proton motive force and by hydrolysis of ATP. The ATP is hydrolyzed by SecA, an essential, peripheral component associated with the integral membrane translocon, SecYEG. Proton motive force is coupled to transport via the heterodimeric complex, SecDF (2), which is associated with SecYEG. The Sec system can export only polypeptides that are devoid of stable tertiary structure (3). SecB, a small cytosolic chaperone, captures precursors before they acquire stable structure and delivers them to SecA (4). SecA is also able to bind precursors directly, as evidenced by the viability of strains of E. coli that lack SecB (5).
Protein export through SecYEG has been studied both in vivo and in vitro over many decades. The approaches vary from conventional biochemical studies to single molecule techniques, such as atomic force microscopy (6, 7) and Förster resonance energy transfer (8). Our recent in vitro work (9) presented a quantification of the parameters of translocation, including the apparent rate constant k, the final amplitude of the precursors translocated, and the efficiency of coupling of ATP hydrolysis to translocation. In all of the activity assays, the translocon SecYEG was limiting. Here, we have extended our studies to a second condition in which SecYEG was in excess and the precursor was limiting.
The observed rate constant for any reaction describes the probability of occurrence of the slowest step (units of min−1), that is, the step that limits the rate under the given experimental conditions. If the conditions are different, the observed step that limits the rate may change. Here, we describe the effects on the observed rate constant of translocation when we change the ratio between translocon and precursor, as well as when we use the mutated translocon SecYEGPrlA4, which has a destabilized SecYEG complex.
RESULTS
In vitro experimental conditions.
In previous work using an in vitro system in which the precursor to be translocated was in excess over the SecYEG translocon (9), we demonstrated that the rate constant was dependent on the precursor species. Here, we expanded our approach by adding the experimental condition in which precursor is limiting and SecYEG is in excess. A key difference between the two conditions is the number of export cycles the translocon could undergo. When the concentration of SecYEG is higher than that of precursor, the majority of active translocons would be used only once due to insufficient precursors for subsequent cycles. However, if precursors were available in sufficient excess over translocons, the active translocons could undergo more than one cycle of translocation. Thus, if the translocon needed to be reset for each cycle, resetting would be a candidate for the rate-limiting step. In addition to examining single and multiple cycles of translocation, we compared translocation through the wild-type SecYEG and the mutationally altered SecYEGPrlA4.
Effects of mutations in SecYEGPrlA4.
The two mutational changes in SecYPrlA4 (10) can affect different aspects of the translocon. The primary effect is the destabilization of the structure. This causes a widening of the channel in SecYEG, which should result in polypeptide chains having a higher probability of passage. However, dependent on the nature of the step affected and the precursor undergoing transfer, other effects of SecYPrlA4 might come into play. Destabilization would cause any changes in conformation of SecYEG to occur more readily. In addition, SecDF dissociates, thereby uncoupling proton motive force from translocation. An often overlooked effect of the mutational changes is that SecA binds more tightly to SecYPrlA4 than it does to the wild-type SecYEG (11).
Determination of the competence of the precursors and the number of cycles.
In order to carry out experiments under the conditions of single and multiple turnovers, it was crucial to know the amount of precursor in each preparation that was competent for translocation, i.e., neither folded nor aggregated. In addition, to be certain that the comparisons between single and multiple turnovers were valid, it was necessary to establish that multiple turnovers had in fact occurred. Figure 1A and B show a single in vitro time course of translocation using vesicles containing the translocon SecYEGPrlA4 as an example of the assay used in this work. In all experiments, the data were fit to a single exponential rise to maximum in order to obtain the values for the apparent rate constant k, and for the maximal amplitude (see the Materials and Methods for a detailed description). All experiments reported here were done at least three times. The maximal amplitude was obtained from the fit of each individual time course. The reported amplitude is the average of the replicates and the error is the standard deviation.
FIG 1.
In vitro translocation of 14C-labeled precursor OmpA. (A) Data quantified from the primary translocation image in panel B were fit to a single exponential rise to maximum, from which values for apparent rate constant and maximal amplitude were obtained (see Materials and Methods for details). (B) Image of radiolabeled proteins sampled from the assay mixture at times indicated in panel A. With the exception of the two samples labeled “0,” which were taken as a measure of precursor input at t = 0 s, the remainder of the samples were subjected to proteolysis by proteinase K. All samples were loaded for SDS-polyacrylamide gel electrophoresis and imaged using a phosphorimager. p, precursor; m, mature; I, intermediates. (C) Time courses of selected precursors. The normalized data points from all replicates for each precursor were globally fit to a single exponential rise to maximum to obtain the rate constants. Lines shown are from the fits. The top two images were single turnover and the bottom two were multiple turnovers.
The fraction of each precursor species that was competent for translocation was determined in the presence of SecB using SecYEG in excess over precursor, so that each polypeptide would have a high probability of engaging a translocon and thus minimizing the time in which the polypeptide might either aggregate or fold and thereby lose competence. Our operational definition of competence of precursors for translocation is taken to be the maximal fraction of the precursor (initially present) that was translocated. Competence was determined in assays using both the wild-type translocon SecYEG as well as the mutationally altered translocon SecYEGPrlA4, which has a destabilized structure that widens the channel (12). Table 1 gives the level of competence determined for each of the nine precursors: (i) four periplasmic proteins, the galactose-binding protein (pGBP), alkaline phosphatase (pPhoA), and slow-folding forms of the maltose-binding protein (pMBPY283D) and the ribose-binding protein (pRBPA248T); (ii) three outer membrane proteins, maltoporin (pLamB), phosphoporin E (pPhoE), and outer membrane protein A (proOmpA); and (iii) two structurally distinct, engineered fragments of proOmpA (see the Materials and Methods for details), proOmpAN176 and properiOmpA. When assessed with wild-type SecYEG, three precursors (proOmpAN176 [0.38], pPhoE [0.35], and pLamB [0.45]) displayed a low fraction of polypeptides that were competent. In their native state, these proteins are entirely β-structures and when unfolded are prone to aggregation (13). In addition, they can associate with the lipid bilayer (14) of the membrane and thereby lose the ability to be translocated. All other polypeptides had levels of competence of 0.6 or greater (Table 1).
TABLE 1.
Fraction of competent precursor
| Precursor | WT IMVa
|
IMVPrlA4a
|
||
|---|---|---|---|---|
| Avg fraction ± SD | n | Avg fraction ± SD | n | |
| proOmpAN176 | 0.38 ± 0.02 | 5 | 0.44 ± 0.07 | 5 |
| pPhoA | 0.77 ± 0.11 | 3 | 0.79 ± 0.04 | 3 |
| proOmpA | 0.80 ± 0.09 | 4 | 0.81 ± 0.05 | 5 |
| pPhoE | 0.35 ± 0.01 | 3 | 0.64 ± 0.07 | 3 |
| pLamB | 0.45 ± 0.05 | 3 | 0.57 ± 0.03 | 3 |
| pGBP | 0.70 ± 0.10 | 3 | 0.84 ± 0.04 | 3 |
| properiOmpA | 0.75 ± 0.06 | 3 | 0.86 ± 0.07 | 3 |
| pMBPY283D | 0.60 ± 0.06 | 5 | 0.57 ± 0.02 | 3 |
| pRBPA248T | 0.67 ± 0.09 | 3 | 0.74 ± 0.13 | 3 |
The average of the maximal amplitudes of the replicates (n) expressed as a fraction of the input and the standard deviation are reported. See the text for definition of competent. IMV, inverted membrane vesicles; WT, wild type; SD, standard deviation.
The level of competence is more properly termed “the apparent level of competence,” since it depends on experimental conditions, as demonstrated by assessing the maximal fraction of precursor translocated using inverted membrane vesicles (IMV) carrying the structurally loosened SecYEGPrlA4. Although most precursors displayed a level of competence with SecYEGPrlA4 similar to that seen with wild-type SecYEG, pPhoE was an exception (Table 1); the fraction of pPhoE translocated was increased 1.8-fold, from 0.35 to 0.64. It is unlikely that the SecYEGPrlA4 translocon actually changed the structural state of the polypeptide to render a higher proportion of the population competent, but rather it is likely that the mutational changes in SecYEGPrlA4, which widen the channel (12), simply allowed the bulkier molecules through with a higher probability.
In the presence of excess precursor, the number of cycles which each precursor underwent (Table 2) was calculated by normalizing the final amplitude expressed in nanomolar to the concentration of accessible, active translocons in the reaction mixture (see Materials and Methods).
TABLE 2.
Cycles of translocation and percentage of competent precursor translocated
| Precursor | WT IMVa
|
IMVPrlA4a
|
||||
|---|---|---|---|---|---|---|
| Avg cycles ± SD | % competent translocated ± SD | n | Avg cycles ± SD | % competent translocated ± SD | n | |
| proOmpAN176 | 1.8 ± 0.0 | 100 ± 2 | 5 | 1.6 ± 0.2 | 77 ± 10 | 3 |
| pPhoA | 3.1 ± 0.5 | 89 ± 20 | 3 | 3.4 ± 0.1 | 91 ± 2 | 3 |
| proOmpA | 3.0 ± 0.4 | 83 ± 15 | 3 | 4.0 ± 0.2 | 107 ± 5 | 3 |
| pPhoE | 0.7 ± 0.2 | 28 ± 2 | 3 | 0.8 ± 0.2 | 27 ± 6 | 4 |
| pLamB | 0.8 ± 0.3 | 30 ± 8 | 3 | 0.7 ± 0.1 | 26 ± 3 | 3 |
| pGBP | 2.6 ± 0.1 | 85 ± 14 | 3 | 3.9 ± 0.3 | 99 ± 8 | 3 |
| properiOmpA | 1.9 ± 0.2 | 54 ± 5 | 4 | 3.0 ± 0.1 | 74 ± 1 | 3 |
| pMBPY283D | 0.9 ± 0.1 | 33 ± 3 | 3 | 3.0 ± 0.5 | 113 ± 19 | 3 |
| pRBPA248T | 1.5 ± 0.2 | 48 ± 5 | 3 | 2.8 ± 0.2 | 81 ± 6 | 3 |
The average of the cycles of the replicates (n) and the standard deviation are reported. See the Materials and Methods for calculation of the number of cycles of translocation. IMV, inverted membrane vesicles; WT, wild type; SD, standard deviation.
Apparent rate constants of translocation.
The apparent rate constant for each of the nine polypeptides was determined with SecYEG in excess (single turnover) and with precursors in excess (multiple turnovers). To minimize the error, the data points for each experimental time course were normalized to the maximal amplitude obtained for the fit of that individual experiment (9). Subsequently, the normalized data points from all replicates were globally fit to obtain the rate constant k (Fig. 1C shows examples of global fits). Figure 2 shows that both with single and with multiple turnovers, the magnitudes of the rate constants are dependent on the precursor species translocated and vary over a 10-fold range. The rate constants determined with SecYEG in excess over the precursor (single turnover) are displayed in the order of the precursor species with the highest to the lowest rate constant (Fig. 2, circle). There is no obvious correlation of the rate constants with the molar mass (given at the bottom of Fig. 2), net charge, or hydrophobicity of the polypeptides. In our previous work (9), to investigate the role of the leader sequence we constructed two forms of proOmpA, called properiOmpA and proOmpAN176, that each carried the same signal peptide as that of native proOmpA followed by one of the two structurally distinct domains of the mature region. Our previous work (9) showed that during multiple turnovers, different rate constants were observed for each of the three species. Here, we extend the work to compare the rate constants during single and multiple turnovers. The full-length proOmpA had a k of 1.7 min−1 during a single turnover and a k of 0.3 min−1 during multiple turnovers. The C-terminal portion, termed properiOmpA, is the final 154 amino acyl residues which, in the periplasmic space, folds into a soluble α-β structure and was among the precursors with the lowest rate constants (0.60 min−1 for a single cycle and 0.18 min−1 for multiple cycles). The species proOmpAN176 comprises the N-terminal 176 amino acyl residues that form a transmembrane eight-stranded β-barrel in the outer membrane. This precursor had the highest rate constant observed, 4.2 min−1 for a single cycle and 2.1 min−1 during multiple cycles. Since the three related species carry the same signal peptide, we conclude that this sequence is not the determinant of the rate constant; rather, the rate constant is determined by the nature of the precursor polypeptide that is undergoing translocation. Furthermore, since the rate-limiting step observed under the two conditions is different, the leader peptide is not a determinant of rate for either step.
FIG 2.

Apparent rate constants with wild-type SecYEG. The apparent rate constants for single turnovers (circles) are compared with those of multiple turnovers (squares). Molar mass for each precursor is given under the x axis. For this and for subsequent figures, apparent rate constants were obtained from global fits of data from three or more replicates (see Materials and Methods for details); an asterisk next to a symbol for a specific precursor indicates that the precursor undergoes only one cycle of translocation. The label on the x axis, pN176, is an abbreviation for proOmpAN176. Errors shown are errors of the global fits; if error bars are not visible, they lie within the symbol.
Comparison of single and multiple turnovers.
Addition of precursor into the assay mixture at a concentration higher than that of the translocon does not always result in more than one cycle (Table 2). One must keep in mind the properties of both the precursor and the translocon. For the multiple-turnover studies reported here, the concentration to be added was corrected for the level of each precursor’s competence (Table 1) to ensure there was an excess that was capable of being translocated. SecB was present in all assays to maximize the amount of competent precursor. In addition, for the assays in which the precursor was in excess, the number of cycles for each precursor was determined (Table 2). The greatest number of cycles observed with the wild-type SecYEG was ∼3 for pPhoA, proOmpA, and pGBP. There was no correlation between the number of cycles a precursor could undergo and the probability it would be translocated, i.e., the rate constant. The rate constants for pPhoA and pPhoE are both ∼0.7 min−1, and yet pPhoA undergoes ∼3 cycles and pPhoE undergoes less than one (Table 2). Even though the precursors were in excess, pLamB and pPhoE each underwent significantly less than one cycle with wild-type SecYEG, as well as with SecYEGPrlA4. These two precursor species are β-structures when folded and when unfolded have a propensity to aggregate (13). It is likely that a time-dependent aggregation occurred during the assay, resulting in a decrease in the level of competence relative to the level at the start of the assay. In the comparisons of “single” (SecYEG in excess) and “multiple” (precursors in excess) turnovers shown in the figures, data for pPhoE and pLamB are marked with asterisks to indicate that in neither experimental condition do the precursors undergo more than one cycle of translocation. Caution should be taken when interpreting these data. Both experimental conditions are in reality “single cycle” with no resetting required and thus one would expect the rate constants to be the same; yet, they were not (Fig. 2). We believe that the reason for these unexpected results lies in the low fraction of the ensembles of pLamB and pPhoE that were competent (Table 1). Addition of a 10-fold higher concentration of pLamB or pPhoE than that used for the comparable single-cycle experiment was required to achieve an effective excess of competent precursor over the SecYEG in the assays. The high concentration of precursor would result in an increased probability for aggregation of folding intermediates. If such aggregated intermediates were not readily able to pass through SecYEG, an unexpectedly low rate constant would result. Details of concentration-dependent aggregation vary with the species of precursor and therefore make it difficult to predict results. Experiments with the precursors pPhoE and pLamB provide examples of the confusion that can arise simply by altering the ratio of precursor and SecYEG if unanticipated changes should occur. Precursors which have a tendency to aggregate should be well characterized before they are used for accurate kinetic or mechanical studies.
The number of cycles that each precursor undergoes appears to be limited by the amount of precursor that is competent for translocation. The fraction of competent precursor that was translocated (Table 2) shows that SecYEGPrlA4 translocated greater than 70% of the competent species for seven precursors, and four of the seven were essentially completely translocated. It appears that transport stops because there is very little competent precursor left in the assay mixture. The number of cycles cannot be increased by addition of more precursor at the start of the assay since that would enhance the probability of aggregation.
The apparent rate constants of translocation through wild-type SecYEG for the precursors that undergo both single and multiple cycles differed by 2- to 6-fold, with the higher rate constant observed for single turnovers (Fig. 2, compare ○ and □, and note that the following discussion does not apply to pPhoE and pLamB, asterisks). Therefore, a different step limits the rate in the two conditions. If the translocon were to undergo more than one turnover of export, a candidate for the rate-limiting step would be the resetting of the channel between cycles. When SecYEG is in excess, only a single turnover is likely and thus resetting would not be observed.
The apparent rate constants of translocation with SecYEGPrlA4 undergoing a single turnover were compared to those for a single turnover with wild-type SecYEG (Fig. 3). As was observed with the wild-type SecYEG, the rate constants were dependent on the mature portion of the precursor species in transit through the destabilized SecYEGPrlA4. Seven of the nine precursors displayed clearly higher rate constants with SecYEGPrlA4 relative to those observed with wild-type SecYEG in excess (Fig. 3, compare red and green). The increase in rate constant likely results from destabilization of SecYEGPrlA4, which would loosen the channel and increase the probability of passage of polypeptides. We cannot be certain that the difference is significant for proOmpAN176 because the data for that protein have such large error. The large error is the result of the very high rate constant for proOmpAN176 (>3 min−1), which means the translocation reaction is ∼60% over in 15 to 20 s. To obtain a reliable rate constant from a fit of the data requires that the initial samples be taken as rapidly as possible. The error in time to sample manually at maximal speed was ∼1 s. Since the magnitude of an exponential rise to maximum is greatest at the start, an error of 1 s results in large differences in amplitude. The precursor proOmpAN176 was among the three precursors which had the highest rate constants and, consequently, the highest errors observed among the precursors. However, the other two precursors pPhoA and proOmpA each displayed a clear, very large difference between the wild-type and the PrlA4 translocon. We favor the interpretation that there is no significant difference between the rate constants observed for proOmpAN176 with the wild-type versus the mutationally altered translocon during a single turnover, implying that whatever limits the rate for this precursor is not affected by the changes in SecYEGPrlA. The dimensions of the wild-type channel might not have restricted proOmpAN176, allowing it to reach its maximal rate of transit. If so, loosening the channel by the changes in SecYEGPrlA4 would have had little or no effect. In the case of precursor properiOmpA, the reason that it shows no increase in rate constant with SecYEGPrlA4 may be the same as described for pOmpAN176. It is also possible that its transit is limited by formation of aggregate structures that are sufficiently large so that even the relaxed channel of SecYEGPrlA4 restricts movement.
FIG 3.

Apparent rate constants for a single turnover. The apparent rate constants observed with wild-type SecYEG (green) compared with SecYEGPrlA4 (red) undergoing a single turnover.
Comparison between wild-type SecYEG and SecYEGPrlA4 (Fig. 4), which underwent transport with precursor in excess, showed that relaxing the channel increased the rate constants for all precursors undergoing multiple cycles of translocation, as well as for pPhoE and pLamB that undergo only one cycle (as indicated by the asterisks). Multiple cycles involve resetting the translocon. Resetting might require changes in conformation which, as pointed out above, would be expected to occur with a higher probability in the structurally destabilized SecYEGPrlA4 relative to that in the wild-type SecYEG. It has been reported that the SecYEG channel is closed in the idle state and must open during translocation (8, 15). Both the closing and the reopening of the channel would involve changes in conformation. However, a single molecule fluorescence technique has been used by Fessl and colleagues (8) to demonstrate that the opening and the closing of the channel each occurred within 10 ms, which is too fast to be observed in our assays. To account for the dependence of the magnitude of the rate constants on the species of precursor, we favor the idea that the step limiting the rate involves the strength of binding of the precursor. This step could occur at the end of a cycle to release the translocated polypeptide and clear the channel. Alternatively, the rate-limiting step might occur at reinitiation of the next cycle to introduce a polypeptide into the translocon through a cascade of transfer from SecB to SecA and on to SecYEG. Speculation on the difference between initiation and reinitiation is found in the Discussion. Either step would involve the strength of binding between the precursor and other proteins in the translocon, which would be specific for each precursor; thus, the rate constant would change with the species of precursor.
FIG 4.

Apparent rate constants for multiple turnovers. The apparent rate constants observed with wild-type SecYEG (cyan) compared with SecYEGPrlA4 (orange) undergoing multiple turnovers.
A direct comparison of single and multiple turnovers with the SecYEGPrlA4 translocon (Fig. 5; note that the data are from Fig. 3 and Fig. 4) shows that, as seen in the comparison of the two conditions with the wild-type SecYEG (Fig. 2), the rate constant is generally higher for a single turnover. The conclusion that we have observed two different steps in the cycle is based on the observation that the rate constants were different under the two conditions. However, apparent inconsistences arise when using SecYEGPrlA4. As seen in Fig. 5, the same rate constant was observed for proOmpAN176 in the two conditions, whereas translocation of proOmpAN176 through wild-type SecYEG showed a large difference in the rate constants, as expected if different steps limit the rate of single and multiple turnovers (Fig. 2). One possible explanation is that changes in SecYEGPrlA4 allowed resetting for translocation of proOmpAN176 to occur at the same or at an even higher rate than the rate for a single cycle, so the same step became rate limiting for both single and multiple turnovers.
FIG 5.

Apparent rate constants with SecYEGPrlA4. The apparent rate constants for single turnovers (circles) compared with those of multiple turnovers (squares). Data are from Fig. 3 and Fig. 4.
As indicated by the asterisks, neither pPhoE nor pLamB underwent more than one turnover in the presence of excess precursor (Table 2) or of excess SecYEG. Since the result in both conditions was less than one cycle, as explained for Fig. 2, one would expect the rate constants to be the same for both conditions. However, as shown in Fig. 2, the rate constants for pPhoE and pLamB were both lower than expected. In contrast, when translocation occurred through the destabilized SecYEGPrlA4 (Fig. 5), pPhoE showed the expected result: rate constants in both conditions were the same. However, in the case of pLamB with precursor in excess, the rate constant was lower than expected. The explanation for the unexpectedly low rate constants in Fig. 2 was the increase of aggregation caused by the high concentration of precursor used. The same degree of aggregation should have occurred in the experiments displayed in Fig. 5, since the concentration used was equally high. The difference, which accounts for the result, is that different translocons were used. If the channel in SecYEGPrlA4 were sufficiently expanded to allow unhindered passage of aggregates of pPhoE formed during the time required for multiple cycles but not those of pLamB, then the same rate constant would be observed for pPhoE in both conditions; however, the rate constant observed for pLamB in excess over the translocon would be unexpectedly low.
Effect of SecB on rate constant of translocation.
All experiments discussed above were done in the presence of SecB. To determine whether SecB has a role in the step that limits the rate, the translocation of each of three precursors (proOmpA, pGBP, and pPhoA) was assessed in both the presence and the absence of SecB for a single turnover (Fig. 6) and for multiple turnovers (Fig. 7). We limited our studies of translocation in the absence of SecB to these three precursors since in our previous study we found that the levels of translocation for pPhoE, pMBPY283D, and pLamB were too low to generate reliable results (9). The dependence on SecB for efficient export in vivo varies with the precursor as well as with growth conditions (1). Export of pGBP to the periplasmic space is highly dependent on SecB, whereas export of proOmpA shows only an enhancement by the presence of SecB. The dependence of pPhoA on SecB in vivo has been reported to vary from highly dependent to no requirement (1). Consistent with this situation in vivo, the rate constant for a single turnover of translocation of proOmpA through the wild-type SecYEG (Fig. 6A) was not changed by the presence of SecB, whereas that of pGBP was increased by 1.8-fold and that of pPhoA by 2.4-fold. When tested with the expanded SecYEGPrlA4 translocon, the presence of SecB increased the rate constant for a single turnover for pGBP by 4.5-fold and by 2.4-fold for pPhoA (Fig. 6B); however, the presence of SecB showed only a 30% increase in rate constant for proOmpA. A simple explanation for the stimulatory effects can be found in the chaperone function of SecB. If a polypeptide were maintained in a state that lacks stable structure, the probability that it would move through the channel without delay is increased.
FIG 6.
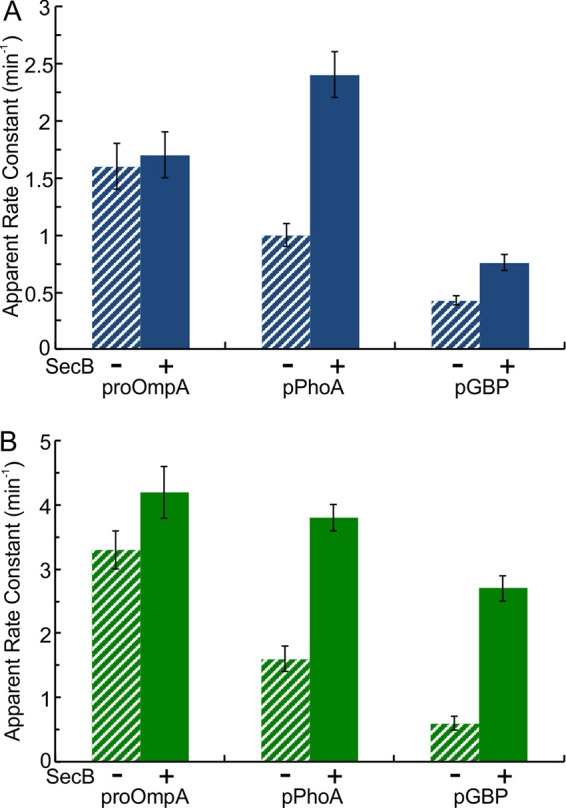
Effect of SecB on apparent rate constants for a single turnover. Translocation was assessed with wild-type SecYEG (A) and SecYEGPrlA4 (B) in the presence (solid; data from Fig. 2 and Fig. 3) or absence (hatched) of SecB.
FIG 7.

Effect of SecB on apparent rate constants for multiple turnovers. Translocation was assessed with wild-type SecYEG (A) and SecYEGPrlA4 (B) in the presence (solid; data from Fig. 2 and Fig. 4) and absence (hatched) of SecB.
During multiple turnovers of translocation, which involves resetting the translocon for reinitiation, the presence of SecB with wild-type SecYEG resulted in an increase of rate constants for pPhoA and for pGBP (Fig. 7A) (2.5-fold and 2.4-fold). The rate constant for pPhoA was enhanced 2.1-fold by the presence of SecB with SecYEGPrlA4 and that of pGBP was enhanced by 3.8-fold (Fig. 7B). These results indicate that, in addition to increasing the rate constant for a single turnover, SecB also plays a role in resetting the translocon for reinitiation between turnovers. Speculation on the role of SecB and the difference between initiation and reinitiation is found in the Discussion.
In contrast to the increase in rate constants seen for both pGBP and pPhoA, the presence of SecB during multiple turnovers reduced the rate constant for proOmpA with both species of SecYEG; it went down by 60% with wild-type SecYEG (Fig. 7A) and by 40% with SecYEGPrlA4 (Fig. 7B). For a precursor bound to SecB, the probability of transfer from SecB to SecA at the translocon would be a function of the relative strengths of binding among the proteins. To ascertain whether the decrease in rate constant for proOmpA might result from tight binding to SecB, we determined the affinities of pGBP, pPhoA, and proOmpA for SecB. The dissociation constants (Kd) for the complexes between SecB and pGBP and SecB and pPhoA were determined by isothermal titration calorimetry to be ∼40 nM (16) and ∼168 nM (Fig. 8A). Titration calorimetry could not be used with proOmpA because of problems with solubility over the time required for a titration in the calorimeter (∼1 to 1.5 h). Therefore, we used competition between pGBP and proOmpA to determine their relative affinities for SecB. Relative to the binding of pGBP to SecB, the binding of proOmpA by SecB was approximately three times tighter (i.e., relative Kd ∼11 nM) (Fig. 8B). Thus, during reinitiation of translocation, if the affinities of the precursors for SecA were similar, pGBP and pPhoA would pass on to the translocon with a higher probability than would proOmpA. This interpretation is consistent with the observed decrease in rate constant for proOmpA in the presence of SecB.
FIG 8.

Affinities of precursors for SecB. (A) Affinity of pPhoA for SecB determined by isothermal titration calorimetry. pPhoA was held at 8°C in the cell at 3.3 μM and was titrated with SecB in the syringe at 54 μM tetramer. Upper, raw data; lower, integrated area of heat as a function of the molar ratio of the reactants. The points are the experimental data and the line is the calculated best fit. The data shown are from one of the three replicates; all three gave a stoichiometry of approximately one pPhoA per SecB tetramer. The average parameters of the three were Kd = 168 ± 51 nM and ΔH = −15 ± 3 kcal/mol. (B) Determination of relative affinities by competition between pGBP-Cy3 and proOmpA for binding to SecB. The following were subjected to size exclusion chromatography: complexes of SecB (4 μM tetramer) and pGBP-Cy3 (4 μM) only (black) or with the addition of competing proOmpA at 1 μM (green), 2 μM (orange), or 3 μM (red), as well as pGBP-Cy3 (4 μM) only (gray). The absorbance of eluent was monitored at 550 nm so the only protein visible was the pGBP labeled with Cy3.
Effect of SecB on final amplitude.
When the precursor is in excess, the maximal amplitude normalized to the concentration of the active translocons is equivalent to the number of cycles of translocation that occurred. Figure 9A shows that the presence of SecB increases the number of cycles for all three precursors. This result is consistent with the occurrence of a time-dependent aggregation of the precursors that would be prevented by binding the precursors to SecB. Destabilization of the channel (Fig. 9B) increased the number of cycles for the precursors both with and without SecB. The magnitude of the increase effected by SecB was less with transit through SecYEGPrlA4 (Fig. 9B) than that observed with transit through the narrower channel in wild-type SecYEG (Fig. 9A); for example, with wild-type SecYEG the enhancement for pGBP was 2.6-fold, versus 1.6-fold with SecYEGPrlA4. This can be explained since the loosened channel would be expected to allow passage of bulkier structures that might form in the absence of SecB, thereby decreasing the effect of SecB.
FIG 9.
Effect of SecB on number of translocation cycles. Translocation undergoing multiple turnovers in the presence (solid) and absence (hatched) of SecB with wild-type SecYEG (A) and SecYEGPrlA4 (B). The total concentration of each precursor in the reaction was 1.3 to 2 μM, and the concentration of accessible, active SecYEG was 0.2 to 0.5 μM. The data in the presence of SecB are from Table 2.
DISCUSSION
Nature of folding intermediates or aggregation depends on the precursor species.
The results of our comparative studies of the precursors with and without SecB in combination with the two forms of SecYEG, the narrow and the loosened channels, allow us to draw conclusions about the types of structures that form upon dilution of the polypeptides from denaturant. Some precursors are completely blocked from passing the channel due to their high propensity for aggregation, while others are hindered but not completely blocked by formation of less bulky species. SecB mediates a kinetic partitioning among the competing pathways open to precursors in order to maintain them in an unstructured state, competent for transport. The affinity between SecB and a precursor, a thermodynamic parameter, can be expressed as the ratio between the kinetic parameters, the rate constant for binding (kon) and the rate constant for dissociation (koff). The kon is diffusion limited (17, 18); therefore, the probability of binding is high. The magnitude of the koff (the probability of dissociation) is much lower and is the main determinate of the strength of binding. An ensemble of unfolded polypeptides, which are substrates for the translocase, undergoes a kinetic partitioning among multiple pathways, which includes folding to a stable structure and formation of aggregates, as well as engaging the translocon. Polypeptides that enter a productive folding pathway populate a dynamic ensemble of intermediates that fluctuate among different states until they reach the final stable tertiary structure. If SecB is present, the precursors remain competent for translocation during a period of time that depends on the rate of rebinding SecB relative to that of entering the competing, irreversible pathways. If the proteins released from SecB have high propensities for rapid aggregation, they have a low probability of being maintained in a competent state; with time they partition to an irreversible, aggregated state.
We have assigned the precursors into four groups based on three parameters for each: (i) the competence for transport, (ii) the extent of translocation, and (iii) the effects of loosening the channel in SecYEGPrlA4 (Table 3). Competence was determined under the condition of an excess of translocon over precursor, which maximizes the number of precursors immediately engaging a translocon. The extent of translocation was determined as the maximal amplitude normalized to the number of active translocons, which is equivalent to the number of cycles. The determination of the extent of translocation required an excess of precursor over translocons.
TABLE 3.
Group assignment of precursors
| Group | Species | Characteristics |
|---|---|---|
| 1 | proOmpA, pPhoA, pGBP | Highest competence, highest percentage translocation of competent precursor |
| 2 | pMBPY283D, pRBPA248T, properiOmpA | Competence increases with destabilized channel, PrlA4 |
| 3 | pLamB, pPhoE | Low competence, aggregates over time |
| 4 | proOmpAN176 | Low competence, but remains competent over time |
Group 1 comprises the precursors that demonstrated the highest level of competence (Table 1), as well as the highest percentage of the competent polypeptides translocated (Table 2). There are three species in group 1: proOmpA, pGBP, and pPhoA. The high degree of competence of proOmpA can be attributed to the fact that the polypeptide carries an internal chaperone within the C-terminal periplasmic region (19). The precursors pPhoA and pGBP each can be maintained in an unfolded state when denaturant is removed under the proper conditions. Precursor pPhoA requires formation of disulfide bonds for stable folding (20); thus, it does not fold if a reducing agent is present in the assay mixture. Since pGBP requires Ca2+ (21) to acquire native structure, folding can be blocked by the presence of the calcium chelator EGTA.
Assignment of proteins to group 2 is based on the behavior of pMBPY283D. The expansion of the channel in SecYEGPrlA4 had a dramatic effect on the translocation of slow-folding pMBPY283D. The initial level of competence was ∼60% as assessed with both the wild-type and the expanded forms of the channel (Table 1). However, the fraction of the competent form that was transferred when the expanded channel was used in place of the wild type went from 30% to 100%, and the number of cycles changed from less than one to three (Table 2). This indicates that pMBPY283D is likely to populate a folding intermediate that is of a size close to that which the channel allows to pass. The ensemble of folding intermediates is in flux among species of various sizes; thus, when a smaller intermediate is formed it would proceed through the channel. Although the effects are less dramatic than that seen for pMBPY283D, two other proteins were assigned to this class based on a similar disproportionate effect of expanding the channel on the fraction competent compared to the percentage of competent that was translocated. When assessed with the expanded channel compared to the wild type, the fraction that was competent increased by only ∼10% both for pRBPA248T and for properiOmpA (Table 1) whereas the percentage of competent precursor that was translocated increased from 50% for both to between 70% and 80% (Table 2). All three proteins in this class have α/β structures and are soluble proteins in the periplasmic space. The binding proteins maltose-binding protein and ribose-binding protein have closely related folds.
Group 3 includes two proteins, pLamB and pPhoE, which show extremely low levels of competence compared to the others. However, the competence of both pPhoE and pLamB is increased significantly when assessed using SecYEGPrlA4 (Table 1), even though the number of cycles is not increased (Table 2). This apparent contradiction can be understood when it is realized that the measure of competence is based on the quantity of the precursor that binds the translocon immediately upon dilution from denaturant. Assuming that the same number of precursors is captured by the wild-type and PrlA4 forms of SecYEG and that the widened channel would pass larger molecules, a higher percentage of the ensemble would be judged to be competent using SecYEGPrlA4. However, the assay for multiple cycles involves an excess of precursor over translocons so that not all precursors can engage a translocon. In addition, several minutes following the initial dilution are required to complete the assay, thus allowing time for further aggregation. The result is a decrease in the number of competent precursors during the time of the assay, thereby affecting the number of cycles that could occur. This behavior is characteristic of precursor species that undergo aggregation during the assay.
The engineered β-barrel proOmpAN176 is currently the only member of group 4. This protein exhibited low competence but unlike the precursors in group 3 did not show any higher competence when examined with SecYEGPrlA4. In addition, there was no evidence of aggregation during the time of the assay, as was observed with the two species assigned to group 3. Our interpretation is that during purification of proOmpAN176, the majority of the polypeptides irreversibly entered large aggregates.
What we have learned about the nature of the individual precursor species should provide researchers with a practical guide in making rational decisions when selecting species for study. It is always desirable to have a high proportion of a population undergo the reaction under study in order to increase the ratio of signal to noise. The proteins in group 1 yield the highest percentage (∼60% to 70%) of translocation of the precursors, calculated as the product of the fraction competent and the fraction of the competent that was translocated. Using a similar calculation, group 2 precursors showed between 20% and 30% translocated and group 3 only 10% to 15%. The polypeptides of the β-barrel proOmpAN176 (group 4) that were competent at the start of the assay were all translocated. Unlike the β-barrels in group 3, which aggregated during the assay, proOmpAN176 seems to be present in two states that are not equilibrating. Further purification to remove the aggregates would decrease the yield, but might at the same time produce a preparation that was 100% competent and completely translocated. This would prove useful in studying the translocation of β-barrels, since the other two β-barrels in this study undergo less than one turnover. In addition, during the assay the other two β-barrels aggregate, indicating they have not reached equilibrium. Therefore, one could not actually remove all aggregates. If the aggregates were removed, the equilibrium would be pulled to repopulate the aggregated state at the cost of the competent state.
An important conclusion based on our previous work (9) and the work presented here, in which we analyzed nine precursor species under two conditions (single turnover and multiple turnovers) in a highly active in vitro system, is that each precursor has specific requirements for efficient translocation through SecYEG. Therefore, one cannot generalize from studies with one or two precursors as examples.
Protein translocation is a complex process, comprising many steps that involve multiple protein complexes, both soluble and integral to the membrane. The in vitro assay that we described here involves the complete ensemble of molecules that are present and is not synchronized; thus, the only step that can be detected is that which is the slowest. We observed different rate constants for single and for multiple cycles, where the simplest interpretation is that different steps limit the rate under the two conditions. Under both conditions, the rate constants observed varied with the species of precursor studied. If passage through the SecYEG pore were rate limiting, in addition to the properties of the polypeptide, the channel itself should be involved in restricting movement. Support for the involvement of the channel is found in the studies of the mutationally altered translocon SecYEGPrlA4. The mutational changes in SecYEGPrlA4 increased the rate constant during a single turnover, consistent with an increase in the probability of the precursor passing through the translocon, while maintaining the dependence of the magnitude of the effect on the nature of the polypeptides in transit. These observations support the conclusion that an aspect of transit through the channel is rate limiting during a single turnover. However, the other effects of the mutational changes must be kept in mind.
We can narrow the possibilities of precisely which step is rate limiting by examining our results in context of a recent study using single molecule fluorescence techniques by Fessl and colleagues (8) that characterized the kinetics of the cycle of protein translocation. The single molecule technique is able not only to resolve the steps in an unsynchronized cycle but also to assign times to each. Fessl and colleagues (8) assigned two kinetic phases to transit through the channel: (i) an early phase that was independent of length of the polypeptide but affected by SecB and (ii) a length-dependent phase. This effect of SecB provides a means of using the results from the single molecule study to assign our rate-limiting step, which was affected by SecB (Fig. 6), to the length-independent phase that occurs early in the initial phase of a single transit.
During this initial phase of transit, SecB is involved in a cascade of transfer of the precursor from SecB to SecA within a complex comprising SecB, precursor, and SecA and subsequent release of SecA carrying the precursor. Upon binding to SecYEG, SecA inserts the two-helix finger in association with the initial portion of the precursor polypeptide into the channel to begin the transit phase. The single cycle of translocation we observed in vitro may not be significant in vivo. Whereas in our experimental design we start with SecYEG in inverted membrane vesicles with SecA removed, in vivo there may not be a significant population of SecYEG without SecA bound. As we suggested in a previous publication (22), SecA might coassemble with SecYEG when the translocon is inserted into the membrane.
The rate constant for the limiting step during multiple turnovers, which we believe involves resetting the channel between cycles, was increased by mutational changes in SecYEGPrlA4 (Fig. 4), as were those for single turnovers. The diverse effects caused by the mutational changes in the PrlA4 form of the translocon (10, 12, 23, 24) can account for the changes we observed in the different steps that limit the rate during single and multiple cycles. The primary effect is the destabilization of the entire SecYEG complex (12). If the steps limiting the rate of resetting the system involve changes in conformation, such changes would occur more readily in the destabilized translocon. The structural changes in SecYEG caused by the mutations not only widen the channel but also increase the stability of the interaction between SecA and SecY (11).
The rate constants of multiple cycles, in addition to those of a single cycle, were affected by SecB (Fig. 7). During multiple cycles, resetting might occur at the beginning of each cycle to prepare the translocase to accept another precursor for reinitiation of the transit phase. The different rate constants for single and multiple cycles indicate that reinitiation differs from initiation of a single cycle. SecA that is associated with SecYEG at the membrane cannot at the same time bind SecB carrying a precursor, because the interactions with SecYEG and those with SecB both occur at the same surface of SecA (25). Therefore, to undergo more than one cycle of transfer may require exchange of free cytosolic SecA for the membrane-bound SecA, which has been activated by SecYEG (26, 27) and was used for the previous cycle. Perhaps the bound SecA remains in a conformation that functions in translocation but not in receiving a precursor from SecB and the insertion of the precursor into the channel. Exchange for SecA in the cytosol would allow interactions between SecA and SecYEG that result in activation of the complex. The idea of an exchange gains support from the demonstration by Morita and colleagues (28) that SecA associated with SecYEG exchanged with cytosolic SecA during translocation of a precursor.
An alternative is that, in vivo, resetting might occur at the end of a cycle to release a translocated polypeptide into the periplasmic space and set the channel to receive another. A deficiency in both our work and that reported by Fessl and colleagues (8) is that in the systems used, SecDF is present at chromosomal levels whereas the plasmid-encoded SecYEG was expressed at 50-fold the chromosomal level. The resulting ratio of SecYEG to SecDF was 200 to 1 (see Materials and Methods for an estimate of the ratio). Therefore, the active role of proton motive force in the release of precursor from SecDF that has been proposed by Tsukazaki and colleagues (29) cannot be addressed. The model put forth by these investigators is well supported by extensive biochemical studies of the function of SecDF, as well as by the crystal structures of the SecDF complex (2, 30–32). This model (29) postulates that a conformational change in SecDF, driven by proton motive force, results in release of the precursor from the translocon to clear it for the next cycle. It is premature to conclude whether or not release of the precursor upon completion of transit is rate limiting in vivo during multiple turnovers of translocation. However, we favor the possibility that release of the precursor from SecDF limits the rate in vivo, and here, in our in vitro studies, rapid spontaneous dissociation occurs that is not rate limiting and thus remains undetected. What we detect in vitro as rate limiting is likely to be resetting of the SecA-SecYEG complex to receive a precursor. In vivo, the step that limits the rate of resetting may occur at the end of the cycle to release the polypeptide from the translocon after termination of translocation. We are developing an in vitro system containing chromosomal levels of SecYEG and SecDF that will allow us to test that possibility.
MATERIALS AND METHODS
Protein purification.
SecA and SecB were purified as described previously (33, 34) and stored in 10 mM HEPES (pH 7.6), 300 mM potassium acetate (KOAc), and 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) at –80°C. All precursors were purified from a temperature-sensitive SecA strain of E. coli harboring a plasmid containing the gene for the precursor of interest (see Table 4 for the plasmids used in this study). The precursors were purified from inclusion bodies by differential solubilization as described in our recent publication (9) (see that publication for details specific to each precursor). The only change in protocol for this work is that some preparations of properiOmpA were stored in 6 M urea, whereas all preparations in the previous work were stored in 4 M urea. We have not seen any difference in results between the two conditions. Three precursors, proOmpA, proOmpAN176 (leader and the aminoacyl residues 1 through 176 of proOmpA), and properiOmpA (leader and periplasmic portion [residues 178 through 325] of proOmpA), contain the same leader sequence as does proOmpA.
TABLE 4.
Plasmids used in this study
| Plasmid | Protein | Accession no.a | PDB no. | Mol wt (kDa) | Mutationsb |
|---|---|---|---|---|---|
| pAL832 | proOmpN176 | 119305 | 1BXW | 21.2 | ompA T95C, I153C, truncated after residue 176 |
| pAL928 | pPhoA | 119853 | 1KHK | 49.4 | phoA C169S, C179S, C287S, C337S, A369C, A427C |
| pAL612 | proOmpA | 119860 | NDc | 37.2 | ompA C290S, G244C |
| pAL950 | pPhoE | 119748 | 1PHO | 38.9 | phoE A249C, Q307C |
| pAL831 | pLamB | 119301 | 1AF6 | 49.9 | lamB C22S, C38S, D329C, N386C |
| pAL725 | pGBP | 119862 | 2FW0 | 35.7 | mglB L244C, D287C |
| pAL800 | properiOmpA | 119306 | 4ERH | 18.5 | ompA 21 residues of the signal sequence followed by residue 178 to the end residue 325; C290S, G244C |
| pAL829 | pMBPY283D | 119697 | 1EZ9 | 43.4 | malE Y283D, G289C, V347C |
| pAL942 | pRBPA248T | 119721 | 2DRI | 31.0 | rbsB A248T, A188C, E246C |
Plasmids have been deposited in Addgene and are available under these accession numbers.
All precursors have 2 cysteines that were introduced for a previous study (22), with the exception of proOmpA, which has one of two native cysteines removed and another added. In addition, pPhoA has the 4 native cysteines removed in order to keep it unfolded, and the precursors of MBP and RBP each carry a mutation that slows folding: malEY283D (37), and rbsA248T (38).
ND, not done.
The concentrations of SecA and SecB were determined spectrophotometrically at 280 nm using the following molar extinction coefficients: SecB tetramer, 47,600 M−1cm−1; SecA dimer, 78,900 M−1cm−1. The concentration of SecY in the cytoplasmic inner membrane vesicles was determined using a quantitative Western blot as described (9). The concentrations of nonradiolabeled precursors (pGBP-Cy3, proOmpA, and pPhoA) were determined using SDS-polyacrylamide electrophoresis and comparing the intensity of the protein bands with those of standards run on the same gel.
Concentration and specific activity of radiolabeled precursors.
Precursors were radiolabeled with [14C]leucine and purified as described by Bariya and Randall (9). The purified 14C-labeled precursors were subjected to analyses of amino acid composition. The submitted sample was hydrolyzed, taken to dryness, and suspended in an accurately measured volume. From the suspended sample, an accurately measured volume was injected onto an amino acid analyzer for composition analysis (AAA Service Laboratory, Inc., Damascus, OR) and another portion was used for determination of radioactivity by scintillation counting. With the result from composition analysis and the known volume injected onto the analyzer, an accurate concentration of the suspended sample was calculated; hence, the specific activity of radiolabeled precursor can be obtained using the concentration and measured radioactivity of the sample.
Preparation of cytoplasmic inner membrane vesicles.
Inverted inner membrane vesicles were prepared from two E. coli strains which harbor plasmids. One plasmid, pEXP2, encodes SecYC329S, C385S, SecE-His tag at the N terminus, and SecG. We have confirmed that the substitution of serine for two native cysteines does not affect translocation activity in vitro (data not shown), and this construct is referred to as wild-type SecYEG. The other plasmid, pEXP26, encodes SecYF286Y, I408N, SecE-His tag, and SecG. This strain is referred to as SecYEGPrlA4. Both strains carry the deletion ΔuncBC to eliminate the proton-translocating ATPase. The cells were grown and the vesicles were prepared as described (9), except the vesicles were not subjected to a urea wash.
Note that neither plasmid encodes SecD and SecF. These two proteins were produced from the genes on the chromosome. A proteomics study (see Table 1 in the supplement of reference 35) determined that in vivo the ratio of SecYEG to SecDF was 4:1. Immunoblots of the inner membrane vesicles used here and of cells that do not carry the plasmid pEXP2 were developed with antiserum to SecY and quantified using a standard curve of purified SecYEG on the same blot (data not shown). Expression from the plasmid produced SecYEG at 50-fold the chromosomal level. Therefore, in our vesicles the ratio of SecYEG to SecDF was 200:1.
In vitro protein translocation.
The reaction mixture of the in vitro translocation assay contained 20 mM HEPES (pH ∼7.6), ∼250 mM KOAc, 5 mM magnesium acetate [Mg(OAc)2], 2 mM dithiothreitol (DTT), 1 mM EGTA, 3.3 mM ATP, 1.7 mM NADH, and an ATP-regenerating system consisting of 7.5 mM phosphocreatine and 37 U/ml creatine phosphokinase. The time for the reaction mixture to come to the assay temperature of 30°C was measured using a thermocouple and was found to be between 15 and 30 s depending on the volume of the reaction mixture. For those precursors with a rate constant of 0.5 min−1 or higher, the reaction mixture without ATP was prewarmed in a 30°C water bath for 25 s before ATP addition to initiate translocation. For assays having SecYEG in excess over precursors, a ratio of active SecYEG to competent precursor between 2 to 10 was used, with SecYEG concentrations kept at ∼1 μM and the SecA dimer between 1 and 2 μM. For assays having precursor in excess over SecYEG, a ratio of competent precursor to active SecYEG between 2 to 4 was used, with SecYEG between 0.4 and 1 μM and the SecA dimer at 1.2 μM. The concentration of SecB when present was equal to that of the precursor.
The reaction mixture was prepared on ice. To serve as a measure of the total amount of precursor present before initiation of translocation (t = 0 min), two samples of 8 μl were removed. The reaction was initiated by transferring the reaction mixture to a water bath at 30°C. At each given time, a sample of 8 μl was taken into tubes held on ice, which contained 6 μl of 42 mM EDTA, 25 mM DTT, and the specific denaturant required for each precursor (see Table 3 in reference 9) to terminate the reaction. Each sample, with the exception of the two taken as a measure of total precursor at t = 0 min, was subjected to proteolysis by addition of proteinase K (4.5 U/ml) and incubation for 15 min on ice, except for proOmpAN176, which required incubation at 40°C for complete degradation. The protease activity was terminated by addition of trichloroacetic acid (final 11%) to precipitate the proteins. The washed precipitate was dissolved in sample buffer for gel electrophoresis containing DTT (20 mM), boiled for 5 min, and subjected to SDS polyacrylamide (12%) gel electrophoresis. All experiments were done three or more times.
Analysis of translocation data.
An example of a single time course of translocation is shown in Fig. 1A. All samples from a given translocation experiment were subjected to SDS polyacrylamide gel electrophoresis. The gels were dried and the intensity of radioactivity in proteins was measured using a Phosphorimager (Fujifilm FLA 3000) in the linear range of its response. The intensities of the radioactive bands were quantified (Image Gauge 4.0) and the percentage of full-length polypeptides protected from proteolysis at each time point was determined by comparing the band intensities with that of the input, i.e., the sample taken from the same reaction mix on ice (t = 0) in duplicate but not subjected to proteinase K treatment. In addition to the full-length species, for some precursors, polypeptides of intermediate length were protected. These intermediates represent polypeptides that were partially translocated at the time the assay was stopped. In such cases, molarities of both full-length and intermediate-length species were included in the calculation of molarity translocated. No intermediate-length species were observed for the three shortest precursors: pRBPA248T, properiOmpA, and proOmpAN176. The amount of precursor protected was plotted as a function of time. Origin software was used to fit data to a single exponential rise to maximum,
| (1) |
where A is the maximal amplitude, k is the apparent rate constant, and t − t0 corrects for the initial time lag. All results shown for the rate constant k are from global fits (Fig. 1C). To minimize the error in the global fits, the data points for each experimental time course were normalized to the maximal amplitude obtained for the fit of that individual experiment (9). The normalized data points from all replicates of the experiment were then globally fit to obtain rate constant k. Errors given are the error of the fit.
Number of cycles of translocation.
Ninety-five percent of the translocons in inverted inner membrane vesicles prepared by a French press are accessible (4) and ∼55% of those accessible were determined to be active as described previously (22, 36). To calculate the number of cycles of translocation for each experiment, the final amplitude (in nanomolar) from the fit of the data to a single exponential rise to maximum was normalized to the concentration of accessible, active translocons in the reaction. The average value of the replicates and its standard deviation for each precursor under the condition for multiple turnovers are reported in Table 2.
Titration calorimetry.
All titrations were carried out using the MicroCal iTC200 titration calorimeter from Malvern Instruments, Inc. (Northampton, MA) and the Origin software supplied with the instrument. The curve-fitting model “One set of sites” with the “Ligand in cell” option as supplied with the software were used to determine Kd, ΔH, and ΔS. The pPhoA was held at 8°C in the cell at 3 or 3.3 μM in 10 mM HEPES-KOH, 0.2 N GnHCl, 280 mM KOAc, 2 mM TCEP, pH 7.6. Wild-type SecB in the same solution at 74, 54, or 50 μM tetramer was titrated in a sequence of 10 injections of 3.6 μl each, spaced at 400 s intervals, after a first injection of 0.2 μl. This first injection was included to expel any air that might be in the tip of the syringe. The heat of the first injection was not included in the analysis. The Kd reported is the average of three determinations.
Labeling of pGBP with the fluorescent dye Cy3.
Purified pGBP with amino acyl residues L244 and D287 replaced by Cys was labeled with Cy3 maleimide (GE Healthcare Life Sciences). pGBP was mixed with Cy3 at a molar ratio of 1 to 0.8 in 10 mM HEPES (pH 7.0), 300 mM KOAc, 1 mM EGTA, and 1N GnHCl and incubated at room temperature for 2 h. Free dye was removed using a Nap10 column (GE Healthcare). The concentration of pGBP-Cy3 was determined as described above in the protein purification section.
Determination of the relative strength of binding to SecB.
Competition between pGBP-Cy3 and proOmpA for binding to SecB was carried out as follows. SecB (4 μM tetramer) was added to pGBP-Cy3 (4 μM) or to a mixture of pGBP-Cy3 (4 μM) and proOmpA at 1, 2, or 3 μM in 10 mM HEPES (pH 7.0), 300 mM KOAc, 2 mM Mg(OAc)2, 1 mM EGTA, and 0.2 N GnHCl. The protein mixtures were subjected to size exclusion chromatography on a TSK G3000 SWXL column (7.8 mm inside diameter [i.d.] by 30 cm) at 7°C with a flow rate of 0.7 ml/min in the same solution. During elution, the absorbance at 280 nm (for total protein) and at 550 nm (specific absorbance for pGBP-Cy3) were recorded and plotted against the volume of eluate. Deconvolution (PeakFit, 4.11 Peaks Auto Fit II) of the profile was used to determine the proportion of pGBP-Cy3 that was bound to SecB and that which was free. This allowed determination of the affinity of the competitor for SecB relative to that of pGBP-Cy3 using the equation R/(A + R), where F is the fraction of pGBP-Cy3 that is free, R is the molar ratio of the competing precursor to the pGBP-Cy3, and A is the ratio of the affinity of pGBP-Cy3 for SecB to the affinity of the competing precursor. Thus, the numbers are higher than one if the competing species binds more weakly than does pGBP-Cy3. In other words, the relative affinity we report corresponds to the ratio of the Kd values.
The design of the competition experiments for determination of relative affinity was validated by a competition between pGBP-Cy3 and unlabeled pGBP for binding to SecB as described above. The ratio of the affinities was determined to be 0.93 (data not shown), thus validating the approach.
ACKNOWLEDGMENTS
We thank Angela Lilly for construction of the plasmids that carry the genes for the precursors used in this work. We are grateful to Ian Collinson for the gift of the plasmid encoding SecYPrlA4.
This work was supported by an endowment from the Hugo Wurdack Trust at the University of Missouri and the National Institutes of Health grant GM29798 (to L.L.R.).
We declare no conflicts of interest with the contents of this article.
REFERENCES
- 1.Crane J, Randall L. 2017. The Sec system: protein export in Escherichia coli. EcoSal Plus 7. doi: 10.1128/ecosalplus.ESP-0002-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, Ito K, Nureki O. 2011. Structure and function of a membrane component SecDF that enhances protein export. Nature 474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Randall LL, Hardy SJS. 1986. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell 46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- 4.Randall LL, Hardy SJS. 2002. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci 59:1617–1623. doi: 10.1007/pl00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumamoto CA, Beckwith J. 1985. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol 163:267–274. doi: 10.1128/JB.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanganna Gari RR, Chattrakun K, Marsh BP, Mao C, Chada N, Randall LL, King GM. 2019. Direct visualization of the E. coli Sec translocase engaging precursor proteins in lipid bilayers. Sci Adv 5:eaav9404. doi: 10.1126/sciadv.aav9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattrakun K, Hoogerheide DP, Mao C, Randall LL, King GM. 2019. Protein translocation activity in surface-supported lipid bilayers. Langmuir 35:12246–12256. doi: 10.1021/acs.langmuir.9b01928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fessl T, Watkins D, Oatley P, Allen WJ, Corey RA, Horne J, Baldwin SA, Radford SE, Collinson I, Tuma R. 2018. Dynamic action of the Sec machinery during initiation, protein translocation and termination. Elife 7:e35112. doi: 10.7554/eLife.35112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bariya P, Randall LL. 2018. Coassembly of SecYEG and SecA fully restores the properties of the native translocon. J Bacteriol 201:e00493-18. doi: 10.1128/JB.00493-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sako T, Iino T. 1988. Distinct mutation sites in prlA suppressor mutant strains of Escherichia coli respond either to suppression of signal peptide mutations or to blockage of staphylokinase processing. J Bacteriol 170:5389–5391. doi: 10.1128/jb.170.11.5389-5391.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Wolk JP, Fekkes P, Boorsma A, Huie JL, Silhavy TJ, Driessen AJ. 1998. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J 17:3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duong F, Wickner W. 1999. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J 18:3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming KG. 2015. A combined kinetic push and thermodynamic pull as driving forces for outer membrane protein sorting and folding in bacteria. Philos Trans R Soc B 370:20150026. doi: 10.1098/rstb.2015.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinschmidt JH. 2015. Folding of beta-barrel membrane proteins in lipid bilayers—unassisted and assisted folding and insertion. Biochim Biophys Acta 1848:1927–1943. doi: 10.1016/j.bbamem.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Knyazev DG, Winter L, Bauer BW, Siligan C, Pohl P. 2014. Ion conductivity of the bacterial translocation channel SecYEG engaged in translocation. J Biol Chem 289:24611–24616. doi: 10.1074/jbc.M114.588491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randall LL, Topping TB, Suciu D, Hardy SJS. 1998. Calorimetric analyses of the interaction between SecB and its ligands. Protein Sci 7:1195–1200. doi: 10.1002/pro.5560070514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randall LL, Hardy SJS. 1995. High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends Biochem Sci 20:65–69. doi: 10.1016/S0968-0004(00)88959-8. [DOI] [PubMed] [Google Scholar]
- 18.Fekkes P, den Blaauwen T, Driessen AJ. 1995. Diffusion-limited interaction between unfolded polypeptides and the Escherichia coli chaperone SecB. Biochemistry 34:10078–10085. doi: 10.1021/bi00031a032. [DOI] [PubMed] [Google Scholar]
- 19.Danoff EJ, Fleming KG. 2011. The soluble, periplasmic domain of OmpA folds as an independent unit and displays chaperone activity by reducing the self-association propensity of the unfolded OmpA transmembrane β-barrel. Biophys Chem 159:194–204. doi: 10.1016/j.bpc.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamitani S, Akiyama Y, Ito K. 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J 11:57–62. doi: 10.1002/j.1460-2075.1992.tb05027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topping TB, Randall LL. 1997. Chaperone SecB from Escherichia coli mediates kinetic partitioning via a dynamic equilibrium with its ligands. J Biol Chem 272:19314–19318. doi: 10.1074/jbc.272.31.19314. [DOI] [PubMed] [Google Scholar]
- 22.Mao C, Cheadle CE, Hardy SJS, Lilly AA, Suo Y, Gari RRS, King GM, Randall LL. 2013. Stoichiometry of SecYEG in the active translocase of Escherichia coli varies with precursor species. Proc Natl Acad Sci U S A 110:11815–11820. doi: 10.1073/pnas.1303289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collinson I, Breyton C, Duong F, Tziatzios C, Schubert D, Or E, Rapoport T, Kuhlbrandt W. 2001. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J 20:2462–2471. doi: 10.1093/emboj/20.10.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veenendaal AK, van der Does C, Driessen AJ. 2004. The protein-conducting channel SecYEG. Biochim Biophys Acta 1694:81–95. doi: 10.1016/j.bbamcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. 2008. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol 382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. 2008. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmer J, Nam Y, Rapoport TA. 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morita K, Tokuda H, Nishiyama K. 2012. Multiple SecA molecules drive protein translocation across a single translocon with SecG inversion. J Biol Chem 287:455–464. doi: 10.1074/jbc.M111.301754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsukazaki T. 2018. Structure-based working model of SecDF, a proton-driven bacterial protein translocation factor. FEMS Microbiol Lett 365:fny112. doi: 10.1093/femsle/fny112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furukawa A, Yoshikaie K, Mori T, Mori H, Morimoto YV, Sugano Y, Iwaki S, Minamino T, Sugita Y, Tanaka Y, Tsukazaki T. 2017. Tunnel formation inferred from the I-form structures of the proton-driven protein secretion motor SecDF. Cell Rep 19:895–901. doi: 10.1016/j.celrep.2017.04.030. [DOI] [PubMed] [Google Scholar]
- 31.Mio K, Tsukazaki T, Mori H, Kawata M, Moriya T, Sasaki Y, Ishitani R, Ito K, Nureki O, Sato C. 2014. Conformational variation of the translocon enhancing chaperone SecDF. J Struct Funct Genomics 15:107–115. doi: 10.1007/s10969-013-9168-4. [DOI] [PubMed] [Google Scholar]
- 32.Furukawa A, Nakayama S, Yoshikaie K, Tanaka Y, Tsukazaki T. 2018. Remote coupled drastic beta-barrel to beta-sheet transition of the protein translocation motor. Structure 26:485–489. doi: 10.1016/j.str.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJS. 2005. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J Mol Biol 348:479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 34.Randall LL, Topping TB, Smith VF, Diamond DL, Hardy SJS. 1998. SecB: a chaperone from Escherichia coli. Methods Enzymol 290:444–459. doi: 10.1016/s0076-6879(98)90037-4. [DOI] [PubMed] [Google Scholar]
- 35.Li G-W, Burkhardt D, Gross C, Weissman Jonathan S. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Futai M. 1974. Orientation of membrane vesicles from Escherichia coli prepared by different procedures. J Membr Biol 15:15–28. doi: 10.1007/BF01870079. [DOI] [PubMed] [Google Scholar]
- 37.Liu GP, Topping TB, Cover WH, Randall LL. 1988. Retardation of folding as a possible means of suppression of a mutation in the leader sequence of an exported protein. J Biol Chem 263:14790–14793. [PubMed] [Google Scholar]
- 38.Kim J, Lee Y, Kim C, Park C. 1992. Involvement of SecB, a chaperone, in the export of ribose-binding protein. J Bacteriol 174:5219–5227. doi: 10.1128/jb.174.16.5219-5227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]




