Abstract
BACKGROUND
Serological assays are of critical importance to investigate correlates of response and protection in coronavirus disease 2019 (COVID-19), to define previous exposure to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in populations, and to verify the development of an adaptive immune response in infected individuals.
METHODS
We studied 509 patients confirmed to have COVID-19 from the San Raffaele Hospital of Milan and 480 samples of prepandemic organ donor sera collected in 2010–2012. Using fluid-phase luciferase immune precipitation (LIPS) assays, we characterized IgG, IgM, and IgA antibodies to the spike receptor binding domain (RBD), S1+S2, nucleocapsid, and ORF6 to ORF10 of SARS-CoV-2, to the HCoV-OC43 and HCoV-HKU1 betacoronaviruses spike S2, and the H1N1Ca2009 flu virus hemagglutinin. Sequential samples at 1 and 3 months after hospital discharge were also tested for SARS-CoV-2 RBD antibodies in 95 patients.
RESULTS
Antibodies developed rapidly against multiple SARS-CoV-2 antigens in 95% of patients by 4 weeks after symptom onset and IgG to the RBD increased until the third month of follow-up. We observed a major synchronous expansion of antibodies to the HCoV-OC43 and HCoV-HKU1 spike S2. A likely coinfection with influenza was neither linked to a more severe presentation of the disease nor to a worse outcome. Of the measured antibody responses, positivity for IgG against the SARS-CoV-2 spike RBD was predictive of survival.
CONCLUSION
The measurement of antibodies to selected epitopes of SARS-CoV-2 antigens can offer a more accurate assessment of the humoral response in patients and its impact on survival. The presence of partially cross-reactive antibodies with other betacoronaviruses is likely to impact on serological assay specificity and interpretation.
TRIAL REGISTRATION
COVID-19 Patients Characterization, Biobank, Treatment Response and Outcome Predictor (COVID-BioB). ClinicalTrials.gov identifier: NCT04318366.
FUNDING
IRCCS Ospedale San Raffaele and Università Vita Salute San Raffaele.
Keywords: COVID-19, Immunology
Keywords: Adaptive immunity, Immunoglobulins, Influenza
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread rapidly worldwide since it was confirmed as the causative agent of coronavirus disease 2019 (COVID-19) (1). Accumulating evidence highlights the development of antibodies to the virus in patients with COVID-19 (2–10). Accordingly, serological assays are of critical importance to investigate correlates of response and protection, to define previous exposure to SARS-CoV-2 in populations, and to verify the development of an adaptive immune response in infected (and in the future also vaccinated) individuals (11).
To date, many commercial companies and research institutes have developed serological assays to detect SARS-CoV-2 antibodies in a patient’s serum or plasma (12–14). The performances of the assays were assessed mostly in small cohorts (14). Usually, these assays detect binding to single coronavirus antigens, mainly the SARS-CoV-2 spike protein, which is exposed on the virion surface, and the nucleocapsid protein, a major virus structural component. The receptor binding domain (RBD), located within the S1 portion of the spike protein, has also attracted particular interest because of its crucial role in cell entry (15, 16).
Although early studies prove that seroconversion is detectable in infected individuals after symptom onset, the complexity of the humoral response in COVID-19 is not fully elucidated and the relevance of the SARS-CoV-2 antibody response for long-term clinical outcome or viral clearance is still lacking. Similarly, the relationship between preexisting humoral responses against other endemic coronaviruses (e.g., HCoV-OC43 or HKU1) or other seasonal respiratory viruses (e.g., influenza) and the outcome of COVID-19 is still unclear. In particular, it is still unconfirmed whether infection with endemic coronaviruses produces antibodies cross-reactive with SARS-CoV-2 antigens (17, 18), as previously observed for SARS-1 and MERS (19–22), and whether this cross-reactivity has any impact on disease severity. Moreover, recent exposure to influenza virus in patients and the presence of a flu protective humoral response during SARS-CoV-2 infection needs to be defined, since these may have implications on susceptibility to SARS-CoV-2 infection and disease severity, as suggested by the described significant upregulation of ACE2 mRNA expression in alveolar epithelial cells after influenza A virus infection (23).
Using newly developed, highly specific, and sensitive measurement of antibodies by fluid-phase luciferase immunoprecipitation system (LIPS) assays, we conducted an extended analysis of the antibody response in a large cohort of patients with COVID-19 admitted to the emergency or clinical departments of the San Raffaele Hospital in Milan, Italy, between February 25 and April 19, 2020, at the peak of the local pandemic. In 509 patients with COVID-19 infection and prospectively followed for clinical outcome, we characterized the IgG, IgM, and IgA response to multiple antigens of SARS-CoV-2 and of beta coronavirus HCoV-OC43 and HKU1, as well as to influenza hemagglutinin (HA).
Results
Study cohorts.
From February 25 to April 19, 2020, 1031 consecutive adult patients with suspected COVID-19 pneumonia were admitted to the emergency or clinical departments at the Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) San Raffaele Hospital (24, 25). Serum samples obtained at a time close to diagnosis were available for 582 patients. A confirmed infection (defined as a SARS-CoV-2–positive real-time reverse-transcriptase polymerase chain reaction [RT-PCR] from a nasal/throat swab and/or signs, symptoms, and radiological findings suggestive of COVID-19 pneumonia) was present in 509 of 582 (87.4%) cases (Figure 1 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI142804DS1). The median age of these patients was 63 years (range 54–75 years); sex was 66.2% male and 43.8% female; ethnicity was 84.1% White, 10.0% Hispanic or Latino, 3.1% Black, and 2.8% Asian. With the exception of 29 patients, the date of symptom onset was identified. The median time from symptoms to admission was 7 days (range 4.5–10 days). The median time from symptoms to blood sampling was 10 days (range 7–16 days). As of May 25, 2020, median time from symptoms to last follow up was 59 days (95% CI: 58–60 days). The median time from symptoms to swab negativization was 40 days (95% CI: 38–42 days). A total of 452 patients (88.8%) were hospitalized and 79 were admitted to the ICU with a median time from hospitalization of 1 day (0–5.25 days). Out of 451 hospitalized patients, 93 died (20.6%) (30 of whom died after entering the ICU). Twenty-three (5.1%) patients were still hospitalized as of May 25, 2020, and 335 (74.3%) had been discharged. The median length of stay from hospitalization to discharge was 14 days (95% CI: 13–15 days), while the median time from hospitalization to death was 12 days (range 6–21 days). For patients who were still hospitalized, the median time from hospitalization to last follow up was 55 days (range 48–64 days). In addition, we included 8 paucisymptomatic subjects with confirmed SARS-CoV-2 infection but with mild symptoms, no history of hospital admission, a median age of 54 years (range 39–62 years), of which 4 (50%) were males, and 480 sera samples from organ donors collected from 2010 to 2012 as control samples (Supplemental Table 1).
Figure 1. Flow diagram of patients characterized in the study.
All patients included in the study had a confirmed SARS-CoV-2 infection as defined by a positive RT-PCR from a nasal/throat swab and/or signs, symptoms, and radiological findings suggestive of COVID-19 pneumonia.
Recombinant antigen production and evaluation of antibody binding.
We explored the IgG binding in LIPS to a panel of recombinant antigens (Supplemental Figure 1) in COVID-19 using patient sera collected at least 3 weeks after the disease onset and control sera collected between 2010 and 2012. The ROC-AUC analysis showed the absence of COVID-19–specific IgG antibodies against ORF7a, ORF8, and ORF10 proteins. A potentially antigen-specific reactivity was present against ORF6 and ORF9b proteins in a minor fraction of patients (Supplemental Figure 2). Regarding the reactivity against major SARS-CoV-2 antigens, an alternative version of the spike glycoprotein S1 RBD recombinant antigen (SARS-CoV-2 RBD) exhibited large differences in antigenicity (Supplemental Figure 3). Upon selection of the construct with best assay performance, we could detect strong and SARS-CoV-2–specific antibody binding in LIPS against an N-terminally luciferase tagged monomeric RBD construct (AUC = 1), a trimeric SARS-CoV-2 S1+S2 spike glycoprotein (SARS-CoV-2 S1+S2) (AUC = 1), and a monomeric nucleocapsid protein (SARS-CoV-2 NP) (AUC = 1) (Supplemental Figures 3–5).
Antibody responses to SARS-CoV-2 spike RBD stratified by time from symptom onset and Ig class.
We analyzed by LIPS 480 patients with COVID-19 in which available information allowed the stratification according to time from symptom onset (weeks 1, 2, 3, ≥ 4) and 95 follow-up samples after hospital discharge (Supplemental Table 2).
SARS-CoV-2 RBD IgM antibodies increased in prevalence from week 1 (43.4%) to week 3 after symptom onset (87.9%) followed by a small decline thereafter (Figure 2 and Figure 3). In prepandemic controls, the SARS-CoV-2 RBD-IgM LIPS showed a specificity of 98.5% (7/480). In antibody-positive patients, the IgM level peaked at week 2 and was followed by a decline thereafter (P < 0.0217, Supplemental Table 2).
Figure 2. Kinetics of SARS-CoV-2 spike RBD antibody development.
Antibody levels in COVID-19 patient (n = 575) and control (n = 480) sera were stratified by the symptom duration (weeks 1, 2, 3, ≥4) at serum sampling and by IgM, IgA, IgG immunoglobulin class (A–C, respectively). For each assay and time point are shown the percentage and count of antibody positive sera, the arbitrary units measured in each sample (circles), their probability density estimate (with the half violin plots upscaled to maximum width for better visualization), a box plot showing median, IQR, and whiskers extending to 1.96 times the median. The dashed vertical lines correspond to the threshold for positivity. Fill colors correspond to an antibody-positive (magenta) or -negative (light blue) score. A schematic depiction of the recombinant antigen is shown on top.
Figure 3. Assay performance of the SARS-CoV-2 spike RBD LIPS in COVID-19.
Antibody levels in COVID-19 patient (n = 575) and control (n = 480) sera were stratified by symptom duration (weeks 1, 2, 3, ≥4) at serum sampling and IgM, IgA, IgG immunoglobulin class. Left panels: ROC curve analysis of LIPS assays measuring either IgM, IgA, or IgG at 1 week to 4 weeks or later after symptom onset. Shown are the total ROC-AUC and the pAUC95. Middle panels: Venn diagrams of spike RBD antibody-positive or -negative score combinations (shown as count and percentages) for different immunoglobulin classes at the same time points. Right panels: ROC-AUC, pAUC95, sensitivity, specificity, positive, and negative predictive values of an algorithm combining results from IgG and IgM immunoglobulin class–specific LIPS assays at the same time points.
SARS-CoV-2 RBD IgA antibodies increased in prevalence from week 1 (34.3%) to week 4 or later (87.4%) (Figure 2 and Figure 3). The RBD-IgA test specificity was 94.8% (25/480). The RBD IgA level increased modestly from week 1 to week 3 followed by a marginal decline (P = 0.0002).
SARS-CoV-2 RBD IgG antibodies increased in prevalence from week 1 (25.9%) to week 4 or later (96.5%) after symptom onset. The specificity of the RBD IgG LIPS test was 100% (0/480) (Figure 2 and Figure 3). In antibody-positive cases, the IgG level increased over time from week 1 to week 4 (P < 10–12).
Positivity for RBD antibodies of more than one class progressively expanded over time, with patients simultaneously positive for 2 or 3 Ig classes increasing from 30% at week 1 to 95% at week 4 (Figure 3). In the first week, antibody-positive subjects had immunoglobulins of all 3 classes in 30% of cases (24/80), of 2 classes in 25% (20/80), and of a single class in 45% (36/80) that were predominantly IgM or IgA (Figure 3).
In patients, a partial correlation was present between SARS-CoV-2 RBD antibody levels of different Ig class that peaked in samples collected 2 weeks after symptom onset (R2: 0.474–0.694) and decreased thereafter (Supplemental Figure 6).
SARS-CoV-2 RBD antibody levels after hospital discharge.
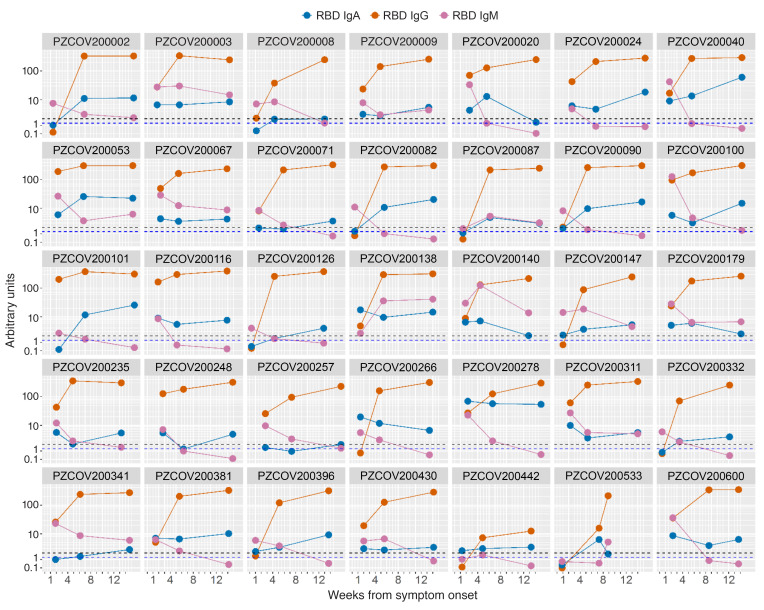
We collected sera from 95 patients at the 1 month visit after hospital discharge. All samples tested positive for SARS-CoV-2 RBD IgG antibodies. For a subset of 35 patients, samples were available from the first hospital admission and from visits 1 month and 3 months after discharge. In all patients, SARS-CoV-2 RBD IgG antibodies continued to rise until month 3, with levels converging toward the upper limit of the assay range (P < 10–6) (Figure 4 and Supplemental Table 3).
Figure 4. SARS-CoV-2 spike RBD antibody titer after hospital discharge.
The line plots show the titer of IgG (orange), IgM (purple), and IgA (blue) according to time from development of symptom onset in sequential samples from the same patients (n = 35). Samples were collected at baseline and at follow-up visits 1 and 3 month after hospital discharge. The dashed lines indicate the cutoff of the IgG and IgM (black) and IgA (blue) assays.
The SARS-CoV-2 RBD IgM and IgA levels showed a more variable profile during follow-up. IgM levels decreased in most patients and seroconverted to negative in 60% (21/35) at month 3. IgA antibodies showed a trend toward a reduction in patients with a longer disease duration at baseline that did not reach statistical significance (Figure 4 and Supplemental Table 3).
Antibody responses to SARS-CoV-2 S1+S2 stratified by time from symptom onset and Ig class.
SARS-CoV-2 S1+S2 IgM antibody prevalence increased in patients with COVID-19 from week 1 (62.2%) through week 3 (91.9%) followed by a decline. IgM levels peaked in week 2 and declined thereafter (P < 10–5) (Supplemental Figure 7 and Supplemental Table 2).
IgA antibody prevalence ranged from 62.9% in week 1 to 95.8% in week 4 or later with levels increasing modestly from up to week 3 followed by a marginal decline (P = 0.007) (Supplemental Figure 7 and Supplemental Table 2).
IgG prevalence progressively increased from week 1 (34.3%) to week 4 or later (97.2%) accompanied by a continuous rise in antibody levels (P < 10–16) (Supplemental Figure 7 and Supplemental Table 2).
SARS-CoV-2 S1+S2 assays showed an overall lower specificity compared with the SARS-CoV-2 RBD LIPS, in particular for the IgM and IgA tests (IgM: 76.4%; IgA: 87.1%; IgG: 98.8%).
Positivity for SARS-CoV-2S1+S2 antibodies of more than one class progressively expanded over time, with patients being simultaneously positive for 2 or 3 classes increasing from 51% in week 1 to 95% in week 4 (Supplemental Figure 8). The correlation of S1+S2 antibody levels of different Ig class was partial and peaked in samples collected 2 weeks after symptom onset (R2 from 0.626–0.671) (Supplemental Figure 9).
Overall, we observed a correlation of SARS-CoV-2 S1+S2 and RBD antibodies of all antibody classes. This was highest for IgG immunoglobulins (R2 = 0.866) followed by that of IgM and IgA antibodies (R2 = 0.714 and 0.692, respectively) (Supplemental Figure 10).
Antibody response to SARS-CoV-2 NP stratified by time from symptom onset.
The SARS-CoV-2 NP IgG prevalence was 53.1% in week 1 and peaked at 93.7% at week 4, while in prepandemic controls the LIPS test had a specificity of 95% (26/480). The SARS-CoV-2 NP IgG levels increased in patients with COVID-19 throughout the study from week 1 to week 4 (P < 10–12) (Supplemental Figure 11 and Supplemental Table 2).
Diagnostic performance of SARS-CoV-2 antibody assay and their combination in COVID-19.
After patient stratification according to time from symptom onset, we evaluated the diagnostic performance of each LIPS assay by determining the ROC-AUC, the partial ROC-AUC after imposing a 95% specificity (pAUC95), sensitivity, specificity, positive, and negative predictive values (Figure 3 and Supplemental Figures 8, 12, 13).
In addition, we evaluated alternative combinations of independent tests to select the best-performing algorithm to identify early seroconversion in patients with COVID-19. The combination of the SARS-CoV-2 RBD IgM and IgG LIPS showed the best positive predictive value at both week 1 (91%) and week 2 (96%) from symptom onset (Figure 3 and Supplemental Figure 14), a sensitivity of 48% in week 1, 83% in week 2, and a 99% specificity.
Antibody responses to SARS-CoV-2 and clinical outcome.
Patients were classified into 6 groups based on increasing disease severity: discharged without hospitalization (57/509, 11.2%); hospitalized 7 days or fewer (78/509, 15.3%); hospitalized more than 7 days (232/509, 45.6%); hospitalized and admitted to ICU and survived (49/509, 9.6%); hospitalized and died after admission to the ICU (30/509, 5.9%); hospitalized and died without admission to the ICU (63/509, 12.4%). The baseline characteristics, laboratory testing, and antibody prevalence at blood sampling of the patients with COVID-19 according to disease severity are summarized in Supplemental Table 4.
Going from best to worst outcome we saw an increase in age, prevalence of comorbidities, and male sex. Furthermore, among laboratory values determined at or in proximity to the time of sampling, the disease severity was associated with an increase in the neutrophils/white blood cells count and biomarkers of inflammation (CRP, IL-6, ferritin, platelet count), tissue damage (LDH, AST, ALT), coagulatory cascade activation (D-Dimer), and a decrease of hemoglobin values and lymphocyte count.
We then performed a time-dependent covariate Cox regression analysis of antibody responses (adjusted for sex and age and stratified for symptom duration at the time of sampling) on the time to death, time to ICU, and time to swab negativization (Figure 5 and Supplemental Table 5). The development of SARS-CoV-2 RBD IgG antibodies was associated with improved patient survival in regression analysis with a hazard ratio (HR) for time to death of 0.45 (95% CI 0.26–0.78, P = 0.0141). A trend was also present for the development of IgG to the SARS-CoV-2 S1+S2 (HR and 95% CI for time to death 0.6, 0.35–1.02, P = 0.057). Neither the response to the nucleocapsid protein nor the responses to the same antigens of other immunoglobulin classes were linked to survival. Similarly, when SARS-CoV-2 RBD–specific IgG and other antibodies or antibody classes responses were combined the association with the survival did not improve.
Figure 5. Hazard ratios for death, ICU admission, and nasopharyngeal swab SARS-CoV-2 viral RNA RT-PCR negativization in patients with COVID-19.
The forest plots show the corresponding hazard ratios for each variable at the time of antibody sampling. The univariable Cox regression analysis was adjusted for sex and age and stratified for the duration of symptoms at serum sampling. Antibody positivity was considered as a time-dependent covariate. Dots represent the HR, filled dots indicate P < 0.05.
A multivariable analysis using 2 different models confirmed the positivity for SARS-CoV-2 RBD IgGs (HR and 95% CI: 0.47 [0.25–0.87]; P = 0.016 and 0.42 [0.19–0.97]; P = 0.041, respectively) as an independent predictor of patient survival (Figure 6A). None of the evaluated antibody responses were associated with ICU admission. A trend for a positive association between the development of SARS-CoV-2 S1/S2 IgA antibodies and the time to swab negativization was observed in the univariable analysis (HR and 95% CI: 1.37 [0.97–1.93]; P = 0.074) that was confirmed as statistically significant in the multivariable analysis (HR and 95% CI: 1.74 [1.1–2.7]; P = 0.017) (Figure 6B).
Figure 6. Multivariable hazard ratios for anti–SARS-CoV2 spike antibodies and time to death or to swab RT-PCR negativization in patients with COVID-19.
(A) Forest plots of hazard ratios for time to death obtained with 2 models of multivariable Cox regression analysis using SARS-CoV-2 RBD IgG-positive score and the shown variables measured at the time of antibody sampling. (B) Forest plot of the hazard ratio of a multivariable model for time to nasopharyngeal swab SARS-CoV-2 RNA RT-PCR negativization based on SARS-CoV-2 S1+S2 IgA-positive score and the shown variables. A higher hazard ratio corresponds to a decreased time to swab negativization. The multivariable Cox regression analysis was adjusted for sex and age and stratified for the duration of symptoms at serum sampling. Antibody positivity was considered as a time-dependent covariate. Dots represent the HR, filled dots indicate P < 0.05.
Antibody responses to HA flu in COVID-19.
We measured IgM and IgG antibodies against the HA1 antigen of the H1N1 Ca2009 virus in the COVID-19 patient and prepandemic control cohorts. The results showed that an average of 22.6% of patients with COVID-19 were HA1 IgM antibody positive in March and April as compared with 8.9% of prepandemic controls collected in the same months of years 2010 to 2012 (Supplemental Figure 15A and Supplemental Table 6). No differences were observed across patients in HA IgM prevalence after stratification according to disease severity (P = 0.475) (Supplemental Figure 15B). In light of the ubiquitous presence of HA IgG antibodies, we titrated the sera and stratified the levels into terciles. We observed no differences in high titer HA IgG (defined as those falling in the upper tercile) prevalence across patients with different disease severity (P = 0.919) (Supplemental Figure 15C). Flu antibodies were not correlated with those against SARS-CoV-2 antigens, as indicated by both regression and PCA analyses (Supplemental Figure 16).
Antibody responses to HCoV-OC43 and HCoV-HKU1 antigens in COVID-19.
In both COVID-19 patients and prepandemic controls, IgG antibody binding to the HCoV-OC43 spike RBD was low, did not correlate with SARS-CoV-2 RBD IgG antibodies, and increased modestly from week 1 to week 3 after symptom onset (ANOVA P < 0.001) (Supplemental Figure 17 and Supplemental Table 2).
IgG antibody binding to the S2 antigen of both HCoV-OC43 and HCoV-HKU1 was instead strong and generalized in both case and control sera. We confirmed the specificity of this binding using monoclonal antibodies to irrelevant antigens (insulin and IA-2), that showed a signal overlapping with the assay background (Supplemental Figure 18). After serum titration and stratification of the S2 antibody reactivities into terciles, we observed that high titer IgG antibodies to the HCoV-OC43 or the HCoV-HKU1 S2 antigens progressively increased in prevalence and level in patients from week 1 to week 4 or later (P < 10–15 for both antigens) (Figure 7, A and B; and Supplemental Table 2).
Figure 7. HCoV-OC43 and HUK1 S2 IgG antibodies in patients with COVID-19.
(A, B) Kinetics of HCoV-OC43 and HKU1 S2 IgG expansion in COVID-19 (n = 575) and control (n = 480) sera stratified by the duration of symptoms at serum sampling. For each sample are shown the measured arbitrary units (circles), the probability density estimate (with the half violin plot upscaled to maximum width for better visualization), box plot displaying median, IQR, and whiskers extending to 1.96 times the IQR. Fill colors correspond to AU greater than 66th (light blue), greater than 33rd (purple), or less than 33rd (orange) percentile in patients with COVID-19. Shown are the percentages and count of subjects with AU greater than the 66th percentile. (C) Correlation of SARS-CoV-2, HCoV-OC43, and HCoV-HKU1 spike IgG in symptomatic COVID-19 sera. Shown are the linear regression (black lines) of log-transformed AU (circles), its 95% CI (gray areas), and its coefficients. (D) Dumbbell plot of IgG binding reduction in a selection of symptomatic and paucisymptomatic patients with COVID-19. LIPS using the indicated HCoV-OC43 and SARS-CoV-2 antigens were performed with (orange fill) or without (light blue fill) competition with untagged SARS-CoV-2 S1+S2 protein.
In patients, a partial but clear correlation between HCoV-OC43 and HCoV-HKU1 S2 IgG antibodies was present that extended also to SARS-CoV-2 S1+S2 IgG antibodies (R2 = 0.616–0.786) (Figure 7C).
We then conducted an inhibition of antibody-binding experiment using the SARS-CoV-2 S1+S2 antigen as competitor and a selection of sera from symptomatic patients with COVID-19 and paucisymptomatic subjects with a proven SARS-CoV-2 infection and high titer SARS-CoV-2 spike antibodies (Figure 7D). All sera were diluted to bring the antibody binding within the linear range of the assay. The results showed that IgG binding to the cognate antigen sNLuc-SARS-CoV-2 S1+S2 was reduced up to 90% in all symptomatic and paucisymptomatic COVID-19 sera (median percentage of inhibition of binding: 86% and 81%, respectively). The IgG binding to the HCoV-OC43 S2 antigen was instead only partially competed by SARS-CoV-2 S1+S2 in symptomatic (5/6, median binding reduction: 9%, IQR 3%–16%) and paucisymptomatic COVID-19 (8/9, median binding reduction: 36%, IQR 6%–49%).
The prevalence of serum samples with high levels of HCoV-OC43 S2 IgA increased from week 1 through week 3 followed by a modest decline (Supplemental Figure 19C). The S2 IgA levels in COVID-19 increased over time from week 1 to week 3 after symptom onset (P < 10–5) but were only modestly greater than in prepandemic controls (Supplemental Figure 19A). No correlation was observed between HCoV-OC43 S2 and SARS-CoV-2 IgA levels (Supplemental Figure 19B).
Discussion
The dynamics of the antibody response against SARS-CoV-2 is currently under intense investigation, as antibodies are considered a potent tool to complement RT-PCR–based diagnostics and to perform an immune response tracking that is in high demand for the development of improved preventive and therapeutic approaches (26). It is thus not surprising that numerous reports have focused on the measurement and description of antibodies in COVID-19, but the design, size and implementation of most of these studies, adopting a variety of technical approaches, do not provide solid qualitative and quantitative information on the SARS-CoV-2 humoral response (14).
For this study, we leveraged our prior expertise (27–31) to develop novel LIPS liquid phase immunoassays, a format that has already been applied to the study of antibody responses to pathogens (32–34). LIPS assays have also found extensive application in autoimmunity, where they demonstrated the ability to measure with high sensitivity and specificity autoantibody responses, often varying by orders of magnitude across patients, and to detect antibody binding to conformational epitopes on antigens undergoing posttranslational modifications and with complex tertiary structures (28, 31, 35, 36). To our knowledge, at least 3 recent reports presented data on antibodies in COVID-19 measured by LIPS in small cohorts of patients and controls using different SARS-CoV-2 antigens (37–40). We analyzed hundreds of COVID-19 and prepandemic sera samples using independent measurements by LIPS of IgG, IgM, and IgA to the SARS-CoV-2 spike, either whole or just its RBD, and nucleocapsid proteins. Our observations showed a progressive expansion of SARS-CoV-2 antibodies in terms of targeted antigens/epitopes and of immunoglobulin class diversification in the course of the disease, already detectable during the first week after symptom onset.
Our data indicate that some of the previously reported LIPS measurements in COVID-19 should be interpreted with caution. We were not able to confirm the existence of disease-specific antibodies to several SARS-CoV-2 ORFs and, similar to previous findings (41), we found important differences in assay performance dependent on the type of recombinant antigen used in the immunoassay. This was particularly true in the case of the recombinant SARS-CoV-2 spike RBD antigen, where the identification and selection of the best antigen configuration was instrumental in conferring to the corresponding LIPS both remarkably high sensitivity and specificity.
Thanks to the availability of large cohorts of clinically well-characterized cases and controls, we were able to address some knowledge gaps (14). First, our data indicate that strategies aimed at the earliest and most sensitive detection of SARS-CoV-2 antibodies in subjects at risk of COVID-19 disease should be based on high specificity tests that discriminate reactivities to multiple antigens and/or immunoglobulin classes. For instance, in our study the distinct measurement of both IgM and IgG antibodies against the SARS-CoV-2 spike RBD led to a positive predictive value of the combined test comparable if not superior to that of nasopharyngeal swabs in patients during the first week after symptom onset (42). Second, the less-than-perfect specificity we observed for several assays measuring antibodies to some SARS-CoV-2 antigens suggests that more stringent criteria are needed for confirming the positivity for SARS-CoV-2 antibodies. In particular, the independent measurement in the same sample of responses against more than one antigen might lead to improved discrimination of bona fide exposure to SARS-CoV-2 and potential COVID-19 disease risk. Third, of the measured SARS-CoV-2 antibodies, the IgG response against the spike RBD domain was associated with improved patient survival independently of other factors such as sex or age, supporting the concept that these antibodies are a major contributor to the protective effect of humoral immunity in COVID-19 (43). Also observed was a less pronounced positive association of whole SARS-CoV-2 spike IgA with a faster clearance of SARS-CoV-2 virus from the upper respiratory tract, as indicated by the reduced time to nasopharyngeal swab negativization in IgA-positive patients. Fourth, unlike in some other reports (44, 45) but consistent with recently described findings, in our COVID-19 cohort the prevalence and levels of RBD IgG antibodies in cured or paucisymptomatic patients showed a further increase for at least 3 months after hospital discharge.
Furthermore, we investigated the antibody response to other respiratory tract viruses: the H1N1Ca2009 flu virus and the HCoV-OC43 and HKU1 betacoronaviruses. Regarding flu antibodies, neither the presence of IgM antibodies nor the titer of the IgG response was associated with survival or disease severity in patients with COVID-19. The analysis of clinical data suggests that the likely coinfection with influenza was neither linked to a more severe presentation of the disease nor to a worse outcome.
More interestingly, in patients with COVID-19 we observed a major expansion of the antibody response against the HCoV-OC43 and HCoV-HKU1 S2 antigen, a spike protein component that shares at least 40% amino-acid identity and up to 63% conserved residues with its SARS-CoV-2 homologue. This was not a generalized case, since antibodies to a region of the HCoV-OC43 spike 1 carboxy terminal domain, corresponding roughly to the location of the SARS-CoV-2 RBD albeit with little homology, were instead rare in both patient and control sera.
While HCoV-OC43 or HCoV-HKU1 S2 IgG antibodies were found essentially in all prepandemic controls, their titer was larger in most patients with COVID-19 and progressively increased according to disease duration, mirroring the course of the SARS-CoV-2 humoral response. However, the HCoV-OC43 and HCoV-HKU1 S2 IgG titers were only partially correlated with the levels of SARS-CoV-2 spike antibodies, and an antibody binding inhibition experiment, while confirming the existence of cross-reactive antibodies, showed a reduced ability of the SARS-CoV-2 antigen to compete antibodies to HCoV-OC43 in LIPS.
Intriguingly, the observed heterogeneity across sera in the competition experiment was suggestive of an increased prevalence of cross-reactive HCoV-OC43 antibodies (i.e., those whose binding could be at least partially competed with SARS-CoV-2 antigen) in the small set of paucisymptomatic individuals with confirmed SARS-CoV-2 infection compared with symptomatic patients with COVID-19. These data are consistent with previously published findings in SARS-CoV-1–infected patients (46) and recent reports describing immune responses to SARS-CoV-2 already present before the pandemic (47–50). They also suggest that immunological phenomena often referred to as antigenic sin (51), i.e., the expansion of a preexisting memory response against partially homologous antigens of a related pathogen, might play a role in the progression from SARS-CoV-2 infection to full blown COVID-19 disease.
Our study encompasses some obvious limitations: the data we generated in this study derive mostly from samples collected from patients with COVID-19 with important symptoms, i.e., prompting hospitalization or at least a visit to the emergency room, and at the time of the pandemic peak, i.e., before the dramatic reduction of new and symptomatic cases observed in our country. For this reason, we should be cautious in generalizing our findings to subjects with an asymptomatic or paucisymptomatic SARS-CoV-2 infection. Moreover, we still need to determine whether certain antigen-specific antibodies assayed by LIPS may be surrogate markers for the presence of neutralizing antibodies.
Nevertheless, we believe that several useful lessons can be derived from our study that might find potential application to the current and future clinical and population settings. Our results stress the need to improve the practice of SARS-CoV-2 antibody measurement. Ideally, the establishment of a blinded serum exchange and unbiased evaluation program might better inform the decision process by public authorities regarding screening strategies for SARS-CoV-2 infection. In addition, our data indicate that the selection of assays aimed at implementing screening strategies must take into account the potential detection of cross-reactive antibodies in some assays. Finally, our study indicates that to anticipate the impact of SARS-CoV-2 antibodies either induced by the virus or by vaccination on disease protection, viral neutralization and disease course will require a deeper knowledge of the interaction with preexisting humoral responses to other betacoronaviruses.
Methods
Study population and data sources.
The study population consisted of 509 adult patients (≥ 18 years) who matched the criteria for a confirmed SARS-CoV-2 infection admitted between February 25 and April 19, 2020, to the emergency or clinical departments at the IRCCS San Raffaele Hospital, a 1350-bed tertiary care hospital in Milan, Italy. A confirmed infection case was defined as a SARS-CoV-2–positive RT-PCR from a nasal/throat swab and/or symptoms and radiological findings suggestive of COVID-19 pneumonia. This series included patients for whom a serum sample was stored in the San Raffaele institutional COVID-19 clinical-biological biobank (COVID-BioB). In addition, we included in our study sera collected from patients returning to our center for follow-up visits at 1 month and 3 months after hospital discharge, sera from 8 paucisymptomatic subjects with confirmed SARS-CoV-2 infection but with mild symptoms and no history of hospital admission, and sera from 480 organ donors collected from 2010 to 2012 as controls.
The clinical data were collected from medical chart review or directly by patient interview, cross-checked for accuracy by data managers and clinicians, and entered in a dedicated electronic case record form (eCRF) specifically developed on site for the COVID-BioB study. Routine blood tests included complete blood count with differential, renal and liver function tests, C-reactive protein (CRP), lactate dehydrogenase (LDH), serum ferritin, D-Dimer, and IL-6. Subject characteristics are reported in Supplemental Table 1, A and B.
Cloning of recombinant nanoluciferase-tagged viral antigens.
We produced several recombinant monomeric or multimeric SARS-CoV-2 proteins tagged with a Nanoluciferase reporter (Promega)—the whole spike glycoprotein S1+S2, spike glycoprotein RBD, nucleocapsid protein (NP), and a panel of ORFs (ORF6, ORF7, ORF8, ORF9, ORF10) (Supplemental Figure 1). In addition, we produced the spike RBD and spike S2 proteins of the HCoV-OC43 and HCoV-HKU1 betacoronaviruses and the hemagglutinin HA1 protein of the 2009 H1N1 pandemic flu virus.
Modified coding sequences were designed and obtained as synthetic genes (Eurofins Genomics, Eurofins Scientific Group) to facilitate the subcloning into modified pCMV-TnT (Promega) vectors containing standard (NLuc) or secretory Nanoluciferase (sNLuc) reporters. A sNLuc tagged SARS-CoV-2 whole spike antigen was instead obtained as a derivative from a previously described trimeric S1+S2 spike protein (52) (plasmid donated by Florian Krammer, Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, New York, USA).
To achieve optimal expression and improve antibody binding, recombinant antigens were produced in alternative versions that differed for the included span of the coding sequence, the use of a secretory or not-secretory nanoluciferase reporter, and its placement relative to the antigen (N- or C-terminal). To induce secretion upon expression, recombinant antigens included the IL-6 signal peptide while a C-terminal T4 foldon trimerization domain was added to achieve trimerization.
Expression of recombinant antigens.
Recombinant nanoluciferase-tagged antigens were expressed by transient transfection of their corresponding plasmid into Expi293F cells (Expi293 Expression System, Thermo Fisher Scientific Life Technologies) according to the manufacturer’s instructions. Recombinant proteins were harvested after 48 hours either from the supernatant or upon lysis of the cell pellet with 0.1% dodecyl maltoside (DDM) in PBS. The recombinant antigens were then aliquoted and stored frozen at –80°C.
High expression levels of secreted proteins were obtained for several antigens, e.g., in the case of the monomeric sNLuc-tagged SARS-CoV-2 spike RBD the luciferase activity yield from a single well of a 6-well-plate transfection was 1012 LU/μL supernatant with a total antigen production sufficient to test more than 450,000 sera by LIPS.
LIPS assay.
For the LIPS assay, the antigen of interest was thawed, diluted in 20 mM Tris Buffer, 150 mM NaCl, 0.5% Tween-20, pH 7.4 (TBST) buffer, filtered with a Durapore PVDF 0.45-μm Millex-HV syringe filter (Millipore) and adjusted to achieve a luciferase activity corresponding to a final concentration of 4 × 106 LU/25 μL.
For antibody measurement, 1 μL (for IgG or IgA measurements) or 5 μL (for IgM measurement) of serum was then seeded into the well of a 96-deep-well plate (Beckman Coulter) added with 25 μL of the diluted antigen preparation, and incubated for 2 hours at room temperature.
For IgG antibody measurement, immunocomplexes were then captured with 5 μL of a 50% weight/volume blocked (53) rProtein A slurry (GE Healthcare Europe GmbH) for 1 hour at 4°C with shaking. Plates were washed 5 times by sequentially dispensing 750 μL/well of TBST, followed by centrifugation at 500g for 3 minutes at 4°C and removal of the wash buffer using a micro-plate washer/dispenser (BioTek Instruments). For IgM or IgA antibody measurements, rProtein A was replaced with 5 μL goat anti–human IgM- or anti–human IgA agarose (Merck Life Sciences), respectively.
After washing, the resin pellets were transferred to an OptiPlate 96-well plate (PerkinElmer) and upon the addition of 40 μL/well of Nano-Glo substrate (Promega) the recovered luciferase activity was measured over 2 seconds per well in a Berthold Centro XS3 luminometer (Berthold Technologies GmbH & Co. KG). Raw data were converted to arbitrary units (AU) using either a local positive serum as index or serial dilution of a SARS-CoV-2 spike protein antibody–positive serum (a gift of Ezio Bonifacio, DFG Center for Regenerative Therapies Dresden, Technische Universität Dresden, Dresden, Germany) and showed a median interassay coefficient of variation of 11.6% (range: 6.4%–19.3%). For antibody titrations, the sera that bound recombinant antigens above the linear range of the assay were serially diluted (1:10, 1:100, 1:1000) in TBST, retested until binding fell into the linear range, and the calculated AU corrected by multiplying for the corresponding dilution factor.
Thresholds for antibody positivity were established upon a QQ plot analysis by selecting AU values at which the distribution of calculated arbitrary units deviated from normality. For ubiquitously present antibody responses like those against the 2009 pandemic flu HA and the HCoV-OC43 and HCoV-HKU1 S2 spike proteins, subjects were binned into terciles.
Antibody binding displacement experiments were performed by adding to replicate test reactions 100 ng untagged affinity purified RBD protein, expressed by transient transfection in Expi293F cells.
Statistics.
Median values with IQRs were used to describe continuous variables while frequencies in percentages and counts were used for categorical variables. Patient-related variables were compared using the χ2 or Fisher’s exact test for categorical variables, and the Wilcoxon rank sum for continuous variables. Imputation for missing data was not performed. Sensitivity and specificity for each assay were calculated as the percentage of case sera reported as antibody positive and as the percentage of control sera reported as negative, respectively. Positive and negative predictive values were calculated using the epiR R package. Assay performance in discriminating health from disease was analyzed using the area under the receiver operator characteristics curve (ROC-AUC) and the partial ROC-AUC at 95% specificity (pAUC95), to exclude diagnostically irrelevant ROC-AUC regions corresponding to very low assay specificity (36, 54). The ROC-AUCs of combination of markers were calculated by performing an initial binary logistic regression for the biomarkers to be combined and then using the obtained probabilities as the test variable to build a ROC curve. Comparisons of antibody levels across groups were performed using ANOVA with Tukey’s HSD post hoc test or the Wilcoxon signed-rank test as appropriate. The correlation between LIPS antibody levels was assessed using linear regression upon log transformation of the data. The time-to-events was calculated from the date of symptom onset to the date of the event, or to the date of last available visit, whichever occurred first. We calculated univariable and multivariable Cox proportional hazards models to evaluate the association between patient characteristics and laboratory findings with time to death, time to intensive care unit (ICU) admission or time to swab negativization (as defined by 2 consecutive negative RT-PCR results). The effect estimates were reported as HRs with the corresponding 95% CI, estimated according to the Wald approximation. All analysis of survival and association were stratified according to time from symptoms to blood sampling (days ≤ 7; days 8–14; days 15–21; days > 21) and antibody positivity was considered as a time-varying covariate. The Cox regression analyses were estimated according to antibodies’ positivity on validated reference cutoffs (IgG, IgM, and IgA vs. SARS-CoV-2 RBD, S1/S2, nucleocapsid protein; IgG vs. HCoV-OC43 RBD; IgM vs. A/H1N1 [flu] HA) or on the overall tercile values (IgG vs. A/H1N1 [flu] HA; IgG vs. HCoV-OC43 S2; IgG vs. HCoV-HKU1 S2). P values less than 0.05 were considered significant. Two-tailed P values not adjusted for multiple testing are reported for Cox analyses with P value less than 0.05 considered to indicate statistical significance, with the exception of antibody comparisons, in which due to the high degree of correlation across antibody responses the P value was corrected for the number of principal components analysis (PCA) that explained most of the variance in the antibody data. All confidence intervals were 2-sided. Statistical analyses were performed with the SPSS 24 (SPSS Inc./IBM) and the R software version 3.4.0 (R Core Team (2017) (https://www.R-project.org/).
Study approval.
This study was approved by the IRCCS Ospedale San Raffaele IRB, protocol number 34/int/2020. Written informed consent was obtained from patients or their guardians according to the IRB guidelines.
Author contributions
MS, EB, and CB performed the cloning and expression of recombinant proteins and the LIPS assays used throughout the study. IM contributed to the data analysis. AP, CT, and PRQ contributed to the clinical data collection and analysis. AC, GS, AZ, FC, LP, and VL contributed to the study design, data analysis, and writing of the manuscript.
Supplementary Material
Acknowledgments
We wish to thank Ezio Bonifacio, Anette Ziegler, William Hagopian, Alistair Williams, and Ake Lernmark for useful discussions. We also wish to acknowledge the contribution of Alexander Lindt to the validation of the flu HA LIPS. We are indebted to Florian Krammer for his generous sharing of the SARS-CoV-2 spike S1+S2 coding sequence. This study was supported by the IRCCS Ospedale San Raffaele and Università Vita Salute San Raffaele grant Program Project COVID-19 OSR-UniSR
Version 1. 09/29/2020
In-Press Preview
Version 2. 11/09/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6366–6378.https://doi.org/10.1172/JCI142804.
See the related Commentary at What are protective antibody responses to pandemic SARS-CoV-2?.
Contributor Information
Massimiliano Secchi, Email: secchi.massimiliano@hsr.it.
Elena Bazzigaluppi, Email: bazzigaluppi.elena@hsr.it.
Cristina Brigatti, Email: brigatti.cristina@hsr.it.
Ilaria Marzinotto, Email: marzinotto.ilaria@hsr.it.
Cristina Tresoldi, Email: tresoldi.cristina@hsr.it.
Patrizia Rovere-Querini, Email: rovere.patrizia@hsr.it.
Andrea Poli, Email: poli.andrea@hsr.it.
Antonella Castagna, Email: castagna.antonella@hsr.it.
Gabriella Scarlatti, Email: scarlatti.gabriella@hsr.it.
Alberto Zangrillo, Email: zangrillo.alberto@hsr.it.
Fabio Ciceri, Email: ciceri.fabio@hsr.it.
Lorenzo Piemonti, Email: piemonti.lorenzo@hsr.it.
Vito Lampasona, Email: lampasona.vito@hsr.it.
References
- 1.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao J, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. doi: 10.1093/cid/ciaa344. [published online May 28, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo L, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okba NMA, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerging Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To KK, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long QX, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 7.Lou B, et al. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur Respir J. 2020;56(2):2000763. doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao AT, Gao C, Zhang S. Profile of specific antibodies to SARS-CoV-2: The first report. J Infect. 2020;81(1):147–178. doi: 10.1016/j.jinf.2020.03.012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6):e00461-20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter AK, Hegde ST. The important role of serology for COVID-19 control. Lancet Infect Dis. 2020;20(7):758–759. doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theel ES, Harring J, Hilgart H, Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(8):e01243-20. doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J Clin Virol. 2020;129:104480. doi: 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deeks JJ, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;6:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. doi: 10.1101/2020.06.22.20137695. Hicks J, et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal betacoronaviruses. medRxiv . Published June 23, 2020. Accessed October 1, 2020. [DOI] [PMC free article] [PubMed]
- 18. doi: 10.1101/2020.03.24.006544. Khan S, et al. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. bioRxiv . Published March 25, 2020. Accessed October 1 , 2020. [DOI]
- 19.Che XY, et al. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191(12):2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan KH, et al. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin Diagn Lab Immunol. 2005;12(11):1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patrick DM, et al. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can J Infect Dis Med Microbiol. 2006;17(6):330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okba NMA, et al. Sensitive and specific detection of low-level antibody responses in mild Middle East respiratory syndrome coronavirus infections. Emerging Infect Dis. 2019;25(10):1868–1877. doi: 10.3201/eid2510.190051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui KPY, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020;8(7):687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciceri F, et al. Early predictors of clinical outcomes of COVID-19 outbreak in Milan, Italy. Clin Immunol. 2020;217:108509. doi: 10.1016/j.clim.2020.108509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zangrillo A, et al. Fast reshaping of intensive care unit facilities in a large metropolitan hospital in Milan, Italy: facing the COVID-19 pandemic emergency. Crit Care Resusc. 2020;22(2):91–94. doi: 10.51893/2020.2.pov1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siracusano G, Pastori C, Lopalco L. Humoral immune responses in COVID-19 patients: a window on the state of the art. Front Immunol. 2020;11:1049. doi: 10.3389/fimmu.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lampasona V, et al. Autoantibodies to harmonin and villin are diagnostic markers in children with IPEX syndrome. PLoS One. 2013;8(11):e78664. doi: 10.1371/journal.pone.0078664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaughlin KA, et al. Identification of tetraspanin-7 as a target of autoantibodies in type 1 diabetes. Diabetes. 2016;65(6):1690–1698. doi: 10.2337/db15-1058. [DOI] [PubMed] [Google Scholar]
- 29.Lahner E, et al. Luminescent immunoprecipitation system (LIPS) for detection of autoantibodies against ATP4A and ATP4B subunits of gastric proton pump H+,K+-ATPase in atrophic body gastritis patients. Clin Transl Gastroenterol. 2017;8(1):e215. doi: 10.1038/ctg.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzinotto I, et al. Autoantibody binding in liquid phase to IL-2 in human sera is not type 1 diabetes specific. Diabetologia. 2017;60(9):1834–1835. doi: 10.1007/s00125-017-4376-2. [DOI] [PubMed] [Google Scholar]
- 31.Liberati D, et al. A novel LIPS assay for insulin autoantibodies. Acta Diabetol. 2018;55(3):263–270. doi: 10.1007/s00592-017-1082-y. [DOI] [PubMed] [Google Scholar]
- 32.Burbelo PD, et al. Serological diagnosis of human herpes simplex virus type 1 and 2 infections by luciferase immunoprecipitation system assay. Clin Vaccine Immunol. 2009;16(3):366–371. doi: 10.1128/CVI.00350-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burbelo PD, et al. Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PLoS One. 2011;6(8):e22576. doi: 10.1371/journal.pone.0022576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zubair A, Burbelo PD, Vincent LG, Iadarola MJ, Smith PD, Morgan NY. Microfluidic LIPS for serum antibody detection: demonstration of a rapid test for HSV-2 infection. Biomed Microdevices. 2011;13(6):1053–1062. doi: 10.1007/s10544-011-9575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fishman D, et al. Autoantibody repertoire in APECED patients targets two distinct subgroups of proteins. Front Immunol. 2017;8:976. doi: 10.3389/fimmu.2017.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lampasona V, et al. Islet Autoantibody Standardization Program 2018 Workshop: interlaboratory comparison of glutamic acid decarboxylase autoantibody assay performance. Clin Chem. 2019;65(9):1141–1152. doi: 10.1373/clinchem.2019.304196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haljasmägi L, et al. LIPS method for the detection of SARS-CoV-2 antibodies to spike and nucleocapsid proteins. Eur J Immunol. 2020;50(8):1234–1236. doi: 10.1002/eji.202048715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grzelak L, et al. SARS-CoV-2 serological analysis of COVID-19 hospitalized patients, pauci-symptomatic individuals and blood donors. Sci Transl Med. 2020;12(559):eabc3103. doi: 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burbelo PD, et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis. 2020;222(2):206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. doi: 10.1101/2020.04.30.20085670. Hachim A, et al. Beyond the spike: identification of viral targets of the antibody response to SARS-CoV-2 in COVID-19 patients. medRxiv . Published May 2, 2020. Accessed October 1, 2020. [DOI]
- 41.Woo PCY, et al. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid enzyme-linked immunosorbent assay due to HCoV-OC43 and HCoV-229E rectified by western blotting with recombinant SARS-CoV spike polypeptide. J Clin Microbiol. 2004;42(12):5885–5888. doi: 10.1128/JCM.42.12.5885-5888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Wernike K, Keller M, Conraths FJ, Mettenleiter TC, Groschup MH, Beer M. Pitfalls in SARS-CoV-2 PCR diagnostics. Transbound Emerg Dis. doi: 10.1111/tbed.13684. doi: 10.1111/tbed.13684. [published online June 14, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brouwer PJM, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long QX, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 45. doi: 10.1101/2020.07.09.20148429. Seow J, et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv . Published July 11, 2020. Accessed October 1, 2020. [DOI]
- 46.Chan KH, et al. Cross-reactive antibodies in convalescent SARS patients’ sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect. 2013;67(2):130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Díez JM, et al. Cross-neutralization activity against SARS-CoV-2 is present in currently available intravenous immunoglobulins. Immunotherapy. doi: 10.2217/imt-2020-0220. [published online September 9, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun J, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. doi: 10.1038/s41586-020-2598-9. [published online July 29, 2020]. [DOI] [PubMed] [Google Scholar]
- 50. doi: 10.1101/2020.07.20.213298. Nguyen-Contant P, et al. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. bioRxiv . Published July 21, 2020. Accessed October 1, 2020. [DOI] [PMC free article] [PubMed]
- 51.Vatti A, Monsalve DM, Pacheco Y, Chang C, Anaya JM, Gershwin ME. Original antigenic sin: A comprehensive review. J Autoimmun. 2017;83:12–21. doi: 10.1016/j.jaut.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Amanat F, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams AJ, Norcross AJ, Chandler KA, Bingley PJ. Non-specific binding to protein A Sepharose and protein G Sepharose in insulin autoantibody assays may be reduced by pre-treatment with glycine or ethanolamine. J Immunol Methods. 2006;314(1–2):170–173. doi: 10.1016/j.jim.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Ma H, Bandos AI, Gur D. On the use of partial area under the ROC curve for comparison of two diagnostic tests. Biom J. 2015;57(2):304–320. doi: 10.1002/bimj.201400023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.