Abstract
SARS-CoV-2 is responsible for the development of coronavirus disease 2019 (COVID-19) in infected individuals, who can either exhibit mild symptoms or progress toward a life-threatening acute respiratory distress syndrome (ARDS). Exacerbated inflammation and dysregulated immune responses involving T and myeloid cells occur in COVID-19 patients with severe clinical progression. However, the differential contribution of specific subsets of dendritic cells and monocytes to ARDS is still poorly understood. In addition, the role of CD8+ T cells present in the lung of COVID-19 patients and relevant for viral control has not been characterized. Here, we have studied the frequencies and activation profiles of dendritic cells and monocytes present in the blood and lung of COVID-19 patients with different clinical severity in comparison with healthy individuals. Furthermore, these subpopulations and their association with antiviral effector CD8+ T cell subsets were also characterized in lung infiltrates from critical COVID-19 patients. Our results indicate that inflammatory transitional and nonclassical monocytes and CD1c+ conventional dendritic cells preferentially migrate from blood to lungs in patients with severe COVID-19. Thus, this study increases the knowledge of specific myeloid subsets involved in the pathogenesis of COVID-19 disease and could be useful for the design of therapeutic strategies for fighting SARS-CoV-2 infection.
Keywords: COVID-19, Immunology
Keywords: Dendritic cells, Monocytes, T cells
Introduction
The new SARS-CoV-2 virus causing coronavirus disease 2019 (COVID-19) has unleashed the current pandemic. Individuals infected with this pathogen can either remain asymptomatic or progress from mild to severe clinical conditions that include acute respiratory distress syndrome (ARDS) and death. Analytical parameters, such as high concentrations of IL-6 and acute-phase reactants in plasma, correlate with the development of severe symptoms in COVID-19 patients (1, 2), suggesting dysregulated immune responses in these individuals. Indeed, a cytokine storm–related syndrome has been proposed as a trigger of ARDS, (3, 4) and in accordance, treatments to control inflammatory cytokine signaling are being used to reduce mortality of COVID-19 patients (5, 6). However, it is unknown whether specific subsets of innate and adaptive immune cells could be differentially contributing to a dysregulated immune response underlying the development of ARDS in COVID-19. The most recent studies indicate that immune exhaustion of effector T lymphocytes and altered humoral responses (7–9), combined with alterations in myeloid cells such as monocytes (Mo) (10–12), might be related to increased inflammation and contribute to disease progression in SARS-CoV-2–infected patients. Dendritic cells (DCs) are a heterogeneous lineage of antigen-presenting cells (APCs) that include different subsets of CD123hi plasmacytoid DCs (pDCs) and conventional DCs (cDCs, CD1c+ and CD141+). These innate cells are critical for activation of adaptive CD4+ and CD8+ T cell responses and are key players in the immune responses to viral and bacterial infections (13–17). A second important player in the immune response against pathogens are Mo, which can be subdivided into immature classical (CD14++CD16–, C) and more differentiated inflammatory transitional (CD14+CD16+, T) and nonclassical (CD14–CD16++, NC) subsets. Altered homeostasis of these specific populations has been linked to chronic inflammation and autoimmunity (18–20). Therefore, it is critical to assess in depth myeloid cell populations in COVID-19 to generate new knowledge that may contribute to the development of effective specific treatments. Additionally, it is important to better understand adaptive antiviral responses potentially present in COVID-19 patients and how they might be associated with altered myeloid cells. Recent studies suggest that exhausted CD4+ T cell responses might be dysregulated in COVID-19 patients (21), but less is known about antiviral CD8+ T cells, which could be important for virus control. Assessment of a small number of COVID-19 patients suggests that CD8+ T cells infiltrate lungs during the course of the disease, but little information is available about which specific effector subsets are recruited (9). Therefore, an integrative view of the myeloid and lymphoid populations altered in peripheral blood and lung tissue of the patients and their association with the severity of COVID-19 is clearly needed. In the present study, we contribute to this aim by analyzing the frequencies and activation profiles of different DC and Mo subsets present in the blood of COVID-19 patients with different levels of clinical severity and by identifying cell subsets associated with protection or disease progression. Moreover, we have characterized inflammatory Mo and DC subsets as well as effector CD8+ T lymphocytes that are specifically recruited to the lung during progressive ARDS.
Results
Inflammatory patterns and severity define subgroups of COVID-19 patients.
Our aim is to evaluate the impact of SARS-CoV-2 infection in circulating myeloid populations. To this end, we followed a cross-sectional study strategy (see STROBE flow chart, Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI140335DS1) and collected peripheral blood samples from a total of n = 64 individuals with COVID-19 (median age 61 years, minimum-maximum, 22–89 years; 57% male, 43% female; 95% receiving treatment) who were recruited after a median of 3 days (range, 0–25 days) upon admission (Supplemental Table 1). For comparison purposes, 22 non–COVID-19 donors were included in the study (Supplemental Table 1). To correlate particular profiles of frequencies and activation of peripheral blood inflammatory cells and the severity of the disease, we stratified our cohort of COVID-19 patients into 3 groups of those showing mild (G1), severe (G2), and critical (G3) disease, following recently described criteria (22). Their demographic and clinical characteristics are summarized in Supplemental Table 1 and Supplemental Table 2. As shown in Supplemental Figure 1B and in agreement with previous studies (2), we observed a significant and progressive decrease of partial pressure of oxygen (PaO2) to fraction of inspired oxygen (FiO2) ratio (PaFiO2) (P = 0.0008 for G1 vs. G3) and an increase of plasma IL-6 (P = 0.001 for G1 vs. G3) and other inflammatory parameters, such as procalcitonin (PCT) (P = 0.005 G1 vs. G3) and C reactive protein (CRP) (P = 0.0022 G1 vs. G3), that was correlated with COVID-19 severity (Supplemental Figure 1B). In contrast, IFN-α was almost undetectable in the plasma from most COVID-19 patients, with the exception of a few individuals from the mild G1 group, suggesting that type I IFNs are not associated with increased disease severity (Supplemental Figure 1B). Finally, critical G3 patients were characterized by a significantly longer hospitalization time than severe G2 (P = 0.0313) and mild G1 (P = 0.0005) individuals (Supplemental Figure 1C and Supplemental Table 1). However, the total duration of symptoms until sample collection was not significantly different between critical G3 and severe G2 patients and only weakly significantly increased when compared with mild G1 individuals (P = 0.0428) (Supplemental Figure 1C).
Differential redistribution of myeloid cell subsets in the blood of COVID-19 patients is associated with severity.
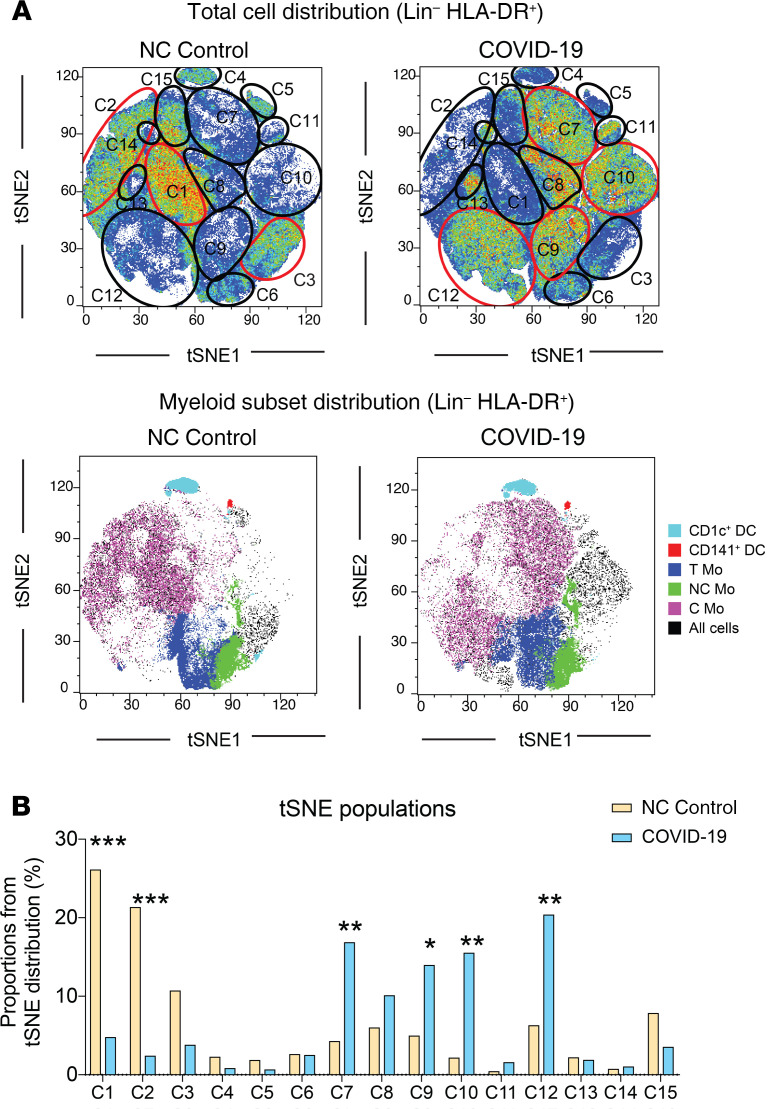
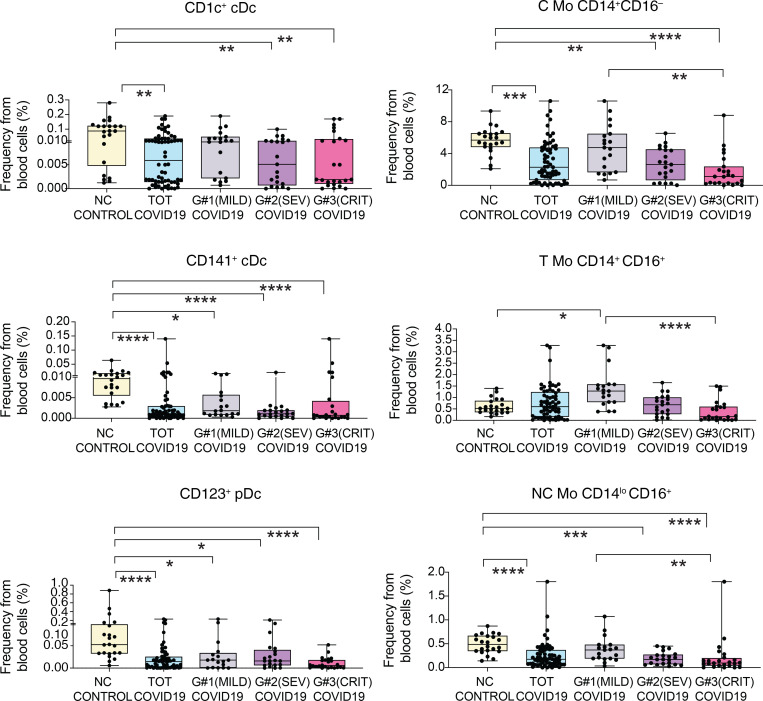
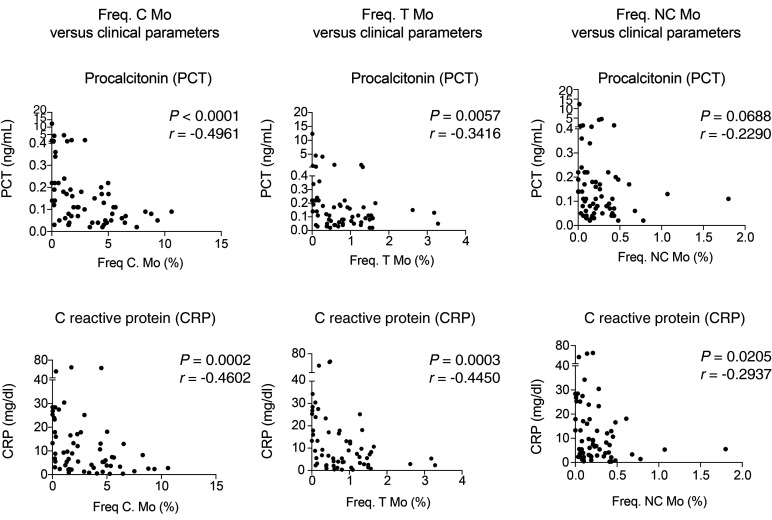
To evaluate the impact of SARS-CoV-2 infection in circulating myeloid populations, we next examined whether specific myeloid subsets could be differentially altered in the 3 groups of COVID-19 patients and associated with disease progression. Using a multicolor flow cytometry panel, we determined the frequencies of the following circulating myeloid cell subsets: C Mo, T Mo, NC Mo, granulocytes, CD1c+ and CD141+cDCs, and CD123hi pDCs (for gating strategy, see Supplemental Figure 2A). Using a t-SNE tool, we were able to observe differences in mononuclear myeloid cell distribution based on the abundance of cell populations between COVID-19 and healthy control individuals (Figure 1A). A more detailed analysis of t-SNE subpopulations suggested that these changes were due to alterations on specific myeloid subsets in COVID-19 individuals (Figure 1A). Indeed, the t-SNE populations that were more significantly altered in COVID-19 patients were highly enriched in different Mo subsets (Figure 1, A and B). An individual analysis of each myeloid subset in blood showed a significant reduction of almost all circulating myeloid cell subsets when considering all COVID-19 patients (Figure 2). Interestingly, certain myeloid populations were more significantly affected in the different patient subgroups. In particular, CD123hi pDCs and CD141+ cDCs were significantly diminished in the 3 patient groups, suggesting a disease- rather than severity-related impact on these cells (Figure 2). On the other hand, the decrease of CD1c+ cDCs and C and NC Mo was more pronounced in severe G2 and critical G3 COVID-19 patients and therefore indicating an association with severity (Figure 2). In contrast, T Mo followed a completely different pattern, since they were significantly increased in mild G1 COVID-19 patients compared with healthy controls, but markedly decreased in critical G3 COVID-19 patients (Figure 2). Moreover, T Mo/CD1c+ cDC ratios tended to be higher in G1 and G2 patients with less severe disease progression. In addition, large CD14–CD16hiHLADR– granulocytes were increased in G1 and G2, but not G3 COVID-19 subgroups; and higher ratios of these cells relative to CD1c+ DCs were specifically increased in severe G2 patients (Supplemental Figure 2, B and C). Importantly, similar differences were observed when absolute numbers of most myeloid subsets were analyzed (Supplemental Figure 3A). Moreover, although a large proportion of critical G3 patients presented microbial superinfection in the lung at the time of sample collection (see Methods and Supplemental Table 2), we observed similar significant decreases of most populations between these individuals and those who did not present superinfection (Supplemental Figure 3B). In addition, although G3 patients were hospitalized for a longer time before sample collection than the other 2 COVID-19 subgroups, no significant variations were observed in frequencies of circulating populations within G3 patients stratified according to sample collection prior to or after 6 days from hospital admission (Supplemental Figure 4). Alterations in frequencies of specific myeloid populations have been related to clinical parameters associated with COVID-19 severity (2). Our analyses revealed that higher frequencies of circulating C, T, and NC Mo and CD141+ cDCs (Supplemental Figure 5, B and C) were weakly correlated with higher levels of blood oxygenation. Importantly, depletion of C and T Mo from the blood correlated most significantly with higher plasma levels of inflammatory markers, such as PCT, CRP, and in the case of C and T Mo, to some extent with higher IL-6 (Figure 3 and Supplemental Figure 5A). Similar trends were observed for cDCs (Supplemental Figure 5, A and B), while no significant association was observed for granulocytes (Supplemental Figure 5, A and B). On the other hand, depletion of inflammatory NC Mo in COVID-19 did not associate with IL-6 plasma levels (Supplemental Figure 5A). Together, these data indicate that frequencies of C, T, and NC Mo and CD1c+ cDCs are preferentially reduced in the blood during severe disease progression and that these populations are differentially correlated with inflammatory markers.
Figure 1. t-SNE visualization of cell subset distribution in the blood of COVID-19 patients.
(A) t-SNE analysis of myeloid cells from a total of 49 samples (34 COVID-19 patients and 15 non–COVID-19 [NC] controls) gated after exclusion of lineage-positive cells and excluding granulocytes. Upper dot plots on the left show combined density of cell clusters in both patient groups. Lower dot plots on the right display highlighted distribution of each indicated myeloid cell population. Cell populations present in both t-SNE plots are highlighted with a number. Those populations changing in the 2 patient groups are highlighted in red. (B) Quantification of numbered cell populations identified by t-SNE in A from the blood of COVID-19 and NC controls. Statistical significance was calculated using χ2 test and FDR multiple-comparison correction. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 2. Analysis of different myeloid subsets in the blood of COVID-19 patients with different clinical severity.
Box-and-whisker plots representing proportions of indicated myeloid cell populations present in the blood of non–COVID-19 (n = 22) control individuals versus either total COVID-19 patients included in the study or patients stratified into groups according to mild (G1; n = 19), severe (SEV, G2; n = 21), and critical (CRIT, G3; n = 24) clinical status as shown in Supplemental Table 1. Median of values is shown. Error bars represent maximum and minimum values. Statistical significance of differences between patient groups was calculated using a Kruskal-Wallis test followed by Dunn′s post hoc test for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Figure 3. Analysis of associations between Mo subset frequencies and clinical parameters from COVID-19 patients.
Spearman’s correlations between frequencies of C (left), T (center), and NC (right) Mo and values of PCT (upper plots) and CRP (lower plots) detected in the blood of all COVID-19 patients included in the study. P and R values are shown in the upper right corner on each plot.
Activation profiles in circulating Mo versus DCs from mild, severe, and critical COVID-19 patients.
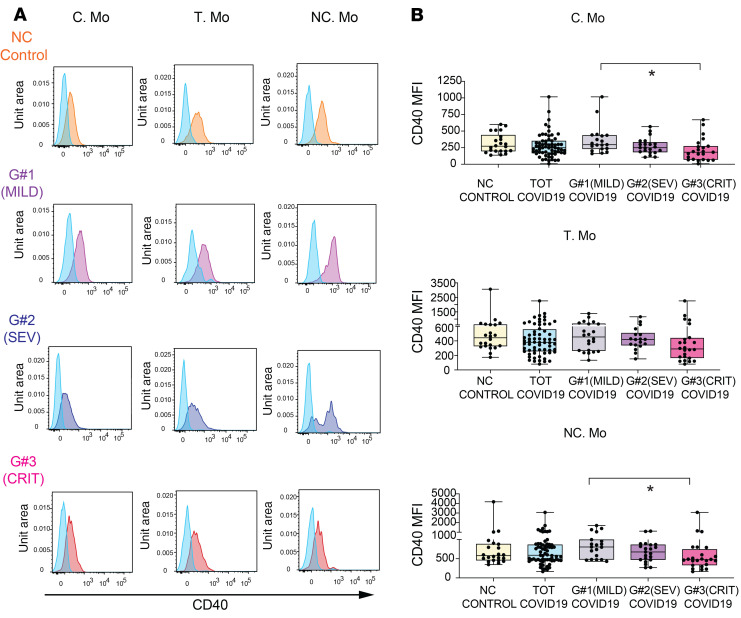
We next sought to determine whether the activation state of DC and Mo subsets was different in the subgroups of COVID-19 patients. Our previous t-SNE analysis involved redistribution of cell populations considering both frequencies and expression of the maturation marker CD40, which has been linked to cellular activation and the expression of IL-6 cytokine (23, 24). As shown in Supplemental Figure 6A, t-SNE distribution of CD40 expression was restricted to specific myeloid cell populations, while other cells seemed to have downregulated this molecule on COVID-19 patients. These patterns were particularly evident for t-SNE from severe G2 and critical G3 patients, which were characterized by the depletion of cells expressing CD40 (Supplemental Figure 6A). In contrast, cells from mild G1 patients seemed to show similar or even upregulated expression of CD40 (Supplemental Figure 6A). When CD40 expression levels were analyzed within each myeloid cell subset in total and stratified COVID-19 patients, we observed again differences associated with disease severity in some populations (Figure 4, A and B; and Supplemental Figure 6, B and C). Interestingly, CD40 expression tended to be lower in all Mo subsets from G3 patients, but most significantly in C and NC Mo (Figure 4B). In contrast, no significant changes in CD40 expression were observed for CD1c+ and CD141+ cDCs, while a mild downregulation of CD40 occurred on pDCs (Supplemental Figure 6, B and C). Remarkably, we observed no significant or weak associations of CD40 expression in cDC or Mo subsets with plasma IL-6 and PCT levels or the frequency of these subsets in the blood (Supplemental Figure 6, D–F). Therefore, these data highlight a differential relationship of innate activation between T and NC Mo versus cDC in COVID-19 patients.
Figure 4. Activation profiles of myeloid cells from the blood of COVID-19 patients and association with clinical parameters.
(A) Flow cytometry dot plots showing CD40 expression on gated C, T, and NC Mo from representative non–COVID-19 controls and mild (G1), severe (G2), and critical (G3) COVID-19 patients. Fluorescence minus one (FMO) controls (blue) for each cell subset are included for comparison purposes. (B) Box-and-whisker plots representing CD40 MFI on the indicated myeloid cell populations present in the blood of healthy individuals versus either total COVID-19 patients included in the study or patients stratified into groups according to mild (G1; n = 19), severe (G2; n = 21),and critical (G3; n = 24) clinical characteristics specified in Supplemental Table 1. Median is highlighted. Error bars represent maximum and minimum values. Statistical differences between patient groups were calculated using a Kruskal-Wallis test followed by Dunn′s post hoc test for multiple comparisons. TOT, total. *P < 0.05.
Inflammatory T and NC Mo are enriched and activated in bronchoscopy infiltrates from COVID-19 patients with ARDS.
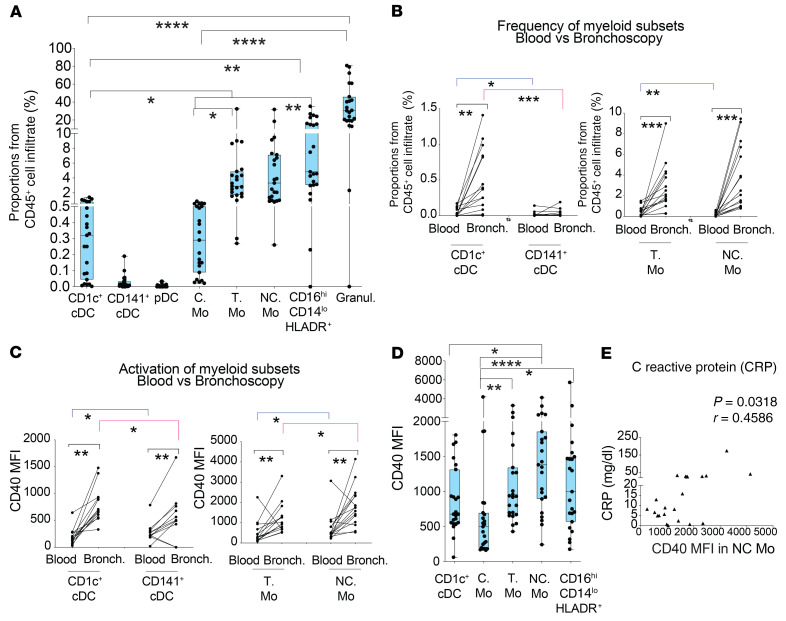
Based on these data, we postulated that activated Mo and DC subsets might be actively recruited to the lung in COVID-19 patients developing severe disease and ARDS. Most studies analyzing inflammatory cell populations in the lung have used bronchoalveolar lavage (BAL) samples, since they are most representative of deep alveolar structure (25–27). However, clinical features of COVID-19 patients requiring artificial respiratory support make it difficult to collect enough BAL samples to assess significant differences in immune cell subsets. Hence, we assessed the proportions of CD45+ leukocytes (Supplemental Figure 7A) and their subsets (see strategy of flow cytometry gating in Supplemental Figure 7B) in dense secretions obtained from bronchoscopy samples routinely obtained to allow ventilation in critical patients in the intensive care unit (ICU). Proportions of hematopoietic cell infiltrate from bronchoscopy samples were not significantly associated with differences in clinical parameters in patients (Supplemental Figure 7A). Remarkably, cell distribution patterns in bronchoscopy samples were very similar to those observed in the 2 cases in which a BAL sample could also be obtained in parallel from the same individual (Supplemental Figure 7, B and C). Using this approach, a significant enrichment of granulocytes (which represented the vast majority of the leukocytes in those samples; Figure 5A and Supplemental Figure 7, B and C), CD16hiCD14lo/– HLA-DR+ cells (Figure 5A and Supplemental Figure 7, B and C), and inflammatory T and NC Mo was observed (Figure 5A). In contrast, C Mo and CD1c+ DCs were also enriched, but made up a very low percentage of the bronchoscopy samples. No pDCs or CD141+ cDCs were found in bronchoscopy infiltrates (Figure 5A). In addition, analysis of blood simultaneously obtained with bronchoscopy lung samples from the same patients (n = 15) demonstrated that T and NC Mo and CD1c+ cDCs were specifically enriched in the lung (Figure 5B). Similar enrichment patterns were observed with granulocytes (Supplemental Figure 7F). Importantly, myeloid cells infiltrated in the lung expressed higher levels of CD40 than their circulating counterparts (Figure 5C). In the lung, we observed increasing expression of CD40 on C, T, and NC Mo. The latter represented the myeloid cell subset expressing the highest levels of CD40 compared with CD1c+ cDCs as well (Figure 5D). In addition, levels of CD40 on NC Mo correlated with higher CRP within this group of critical COVID-19 patients (Figure 5E). Although all bronchoscopy samples were collected from critical G3 patients that presented microbial superinfection either in the lung, blood, urine, or both blood and urine (Supplemental Table 2), we did not observe significant differences in the distribution of myeloid cells in the lung after subclassifying patients according to either site of microbial superinfection or time between hospital admission and sample collection (Supplemental Figure 7, D and E). However, levels of activation on NC Mo were significantly more increased in patients presenting superinfection in the lung (Supplemental Figure 7D). Together, these data indicate preferential enrichment inflammatory Mo in the lung and contrasting maturation profiles between infiltrated NC Mo compared with T Mo and CD1c+ cDCs.
Figure 5. Characterization of myeloid cell subsets present in bronchoscopy infiltrates from COVID-19 patients with ARDS.
(A) Box-and-whiskers plots showing percentages of the indicated cell populations in the hematopoietic CD45+ infiltrate present in bronchoscopy mucus samples from severe COVID-19 patients (n = 23) presenting ARDS and receiving IMV at ICU. Error bars represent maximum and minim values. Statistical differences between proportions of cell populations within the same infiltrates were calculated using Friedman’s test for multiple comparisons. *P < 0.05; **P < 0.01; ****P < 0.0001. (B and C) Frequencies (B) and CD40 MFI (C) of CD1c+ and CD141+ cDCs (left) and T and NC Mo (right) in paired blood and bronchoscopy samples from COVID-19 patients presenting with ARDS (n = 15). Statistical significance of differences in frequencies between paired blood vs. bronchoscopy samples (black) or between different cell subsets within either blood (blue) or bronchoscopy infiltrates (pink) was calculated using a 2-tailed matched pairs Wilcoxon’s test. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Box-and-whiskers plots representing comparison of CD40 MFI on the indicated myeloid cell populations present in the bronchoscopy infiltrates of total critical G3 COVID-19 patients. Error bars represent maximum and minimum values. Statistical significance of differences was calculated using Friedman’s test for multiple comparisons. *P < 0.05; **P < 0.01; ****P < 0.0001. (E) Spearman’s correlations between CRP levels in plasma and CD40 MFI on NC Mo present in the bronchoscopy infiltrates of severe COVID-19 patients. Spearman’s P and R values are shown in the upper right corner of the plot.
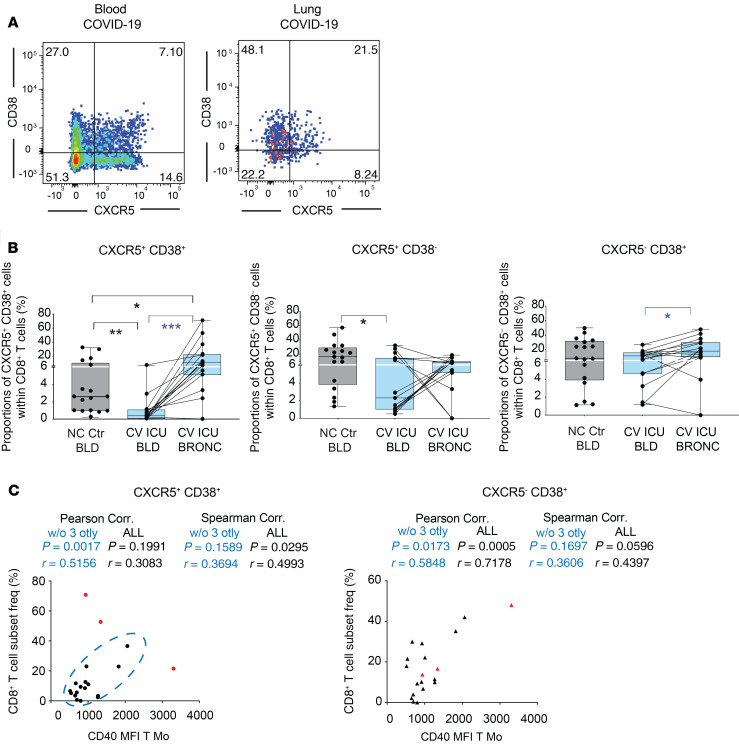
Effector CD8+ T cell populations in the lung of COVID-19 patients: association with inflammatory Mo subsets and CD1c+ DC.
To better understand the contribution of CD1c+ and inflammatory Mo to either disease progression or protective antiviral immunity in COVID-19 patients, we studied the presence of infiltrating effector T cells in bronchoscopy samples. We did not observe any significant increase of T cells or changes in the ratios of CD4+CD8+ T cells in the lung as compared with blood (Supplemental Figure 8A). Expression of CD38 and CXCR5 in CD8+ T cells has been previously linked to effector function and immune exhaustion during viral infections (Figure 6A) (28, 29). Next, we analyzed expression of these activation markers in circulating CD8+ T cells, finding a significant enrichment of activated CD38+CD8+ T cells, more significantly those that coexpressed CXCR5 in bronchoscopies from critical COVID-19 patients requiring invasive mechanical ventilation (IMV) (Figure 6B and Supplemental Figure 8B). Of note, CD38+CXCR5+CD8+ T cells were the only subset that was significantly altered compared with both paired blood in COVID-19 patients and blood from non–COVID-19 individuals (Figure 6B). Finally, we assessed whether frequencies or activation status of inflammatory Mo and CD1c+ cDCs could be associated with any of these effector CD8+ T cell subsets. Interestingly, we observed that higher ratios of inflammatory T and NC Mo/CD1c+ cDCs were negatively associated with proportions of CXCR5+CD38+CD8+ T cells, suggesting a detrimental role of more preferential infiltration of inflammatory Mo versus DCs during disease progression (Supplemental Figure 8C). In addition, levels of CD40 in T Mo, but no other myeloid cells in the lung, were positively associated with proportions of infiltrated CD38+ (both CXCR5+ and CXCR5–) CD8+ T cells (Figure 6C and Supplemental Figure 8D).
Figure 6. Association between effector CD8+ T cell and inflammatory myeloid cells present in bronchoscopy from COVID-19 patients with ARDS.
(A) Representative flow cytometry analysis of CD38 versus CXCR5 expression on gated CD8+ T cells present in the blood (left) and paired bronchoscopy infiltrate (right) from COVID-19 patients with ARDS. Numbers on quadrants represent percentages of positive cells. (B) Box-and-whiskers plots representing analysis of frequencies of CXCR5+CD38+ (left), CXCR5+CD38– (center), and CXCR5–CD38+ (right) CD8+ T cells present in paired blood and bronchoscopy samples from n = 15 COVID-19 patients presenting with ARDS. Frequencies of these CD8+ T cell subsets on the blood of n = 17 non–COVID-19 controls were included for reference. Error bars represent maximum and minimum values. Statistical significance of differences in frequencies between paired blood vs. bronchoscopy samples (blue) or comparison with healthy controls (black) was calculated using a 2-tailed matched pairs Wilcoxon’s and Mann-Whitney U test with Bonferroni’s multiple comparison correction, respectively. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Spearman’s and Pearson’s correlations between proportions of the indicated effector CD8+ T cells subset and CD40 MFI on T Mo. Spearman’s and Pearson’s P and R values are shown in the upper right corner on each correlation plot.
Discussion
In this study, associations of specific Mo and DC subsets with disease progression in COVID-19 patients are established. Our results indicate that increased proportions of T Mo in the blood could be used as a marker of viral control or mild clinical conditions in infected patients. In contrast, dramatic decreases of T and NC Mo and CD1c+ DCs are associated with severe clinical outcomes in COVID-19 patients. In addition, our study shows that reduction of frequency of T and NC Mo in the blood is associated with increased inflammatory markers, such as PCT, CRP, and weakly, with IL-6, while no link with type I IFN was observed. This reduction is also associated with the selective recruitment to the lung of these populations and CD1c+ cDCs during the development of ARDS. Interestingly, remaining circulating Mo tended to express lower levels of CD40, which could reflect either impaired maturation or selective migration of activated cells to the lung or other anatomical locations. On the other hand, increased activation of NC Mo and defective maturation of T Mo might be associated with uncontrolled inflammation in the lung and the activation of effector CD8+ T cells. Moreover, our data indicate that frequency of granulocytes is reduced in the blood from critical patients, while numbers are not significantly affected, in contrast with other studies (30). However, these cells are highly enriched in the lungs of individuals undergoing critical COVID-19. In addition, CD16hiCD14lo/– HLA-DR+ cells are also enriched in the lungs from critical COVID-19 patients and might represent macrophages that are differentiated in situ from inflammatory NC Mo that could also contribute to the disease. Therefore, our data highlight differential involvement of myeloid cell subsets in the pathogenesis of COVID-19 disease and immunity against SARS-CoV-2 virus. Some of these patterns are similar to those described in pneumonia from different pathological contexts (31, 32). Although these data provide cellular insights into the pathogenesis of severe COVID-19, future studies will need to focus on mechanisms responsible for these altered cellular patterns. In this regard, recent single-cell transcriptomic analyses on inflammatory infiltrates in the lung of COVID-19 patients suggest the altered expression of genes coding for proinflammatory cytokines and/or IFN-associated and IFN-stimulated proteins that have been previously associated with the activation of antibacterial innate pathways (10).
Our data indicate that a large proportion of individuals with COVID-19 from the critical G3 group developing ARDS and requiring IMV were superinfected with bacteria or candida at the time of sample collection. While these superinfections were mostly detected after the initiation of IMV, the majority of critical G3 COVID-19 patients exhibited high levels of PCT upon admission, a parameter commonly associated with bacterial infection (33) and also an indicator of inflammation and severe COVID-19 prognosis (2). In addition, although the majority of myeloid cell populations analyzed were similarly decreased in the blood of G3 patients independently of whether they presented superinfection in the lung or not, T Mo seemed to be more markedly affected specifically in patients with superinfection in the lung. Also, those G3 patients positive for microbial superinfection in blood and urine seemed to behave differently from those positive in the lung. Therefore, it is possible that COVID-19 patients might be more susceptible to microbial superinfection early or during ICU treatment and this could contribute to changes in inflammation, migration, and homeostasis of myeloid cells from COVID-19 patients specifically undergoing severe disease progression. If this were the case, the simultaneous activation of antiviral and antibacterial innate recognition pathways could be a potential mechanism that might contribute to uncontrolled inflammation and immune exhaustion in these individuals, similarly to what has been described in HIV+ individuals coinfected with mycobacterium tuberculosis and developing immune reconstitution inflammatory syndrome (IRIS) (34). In this regard, type I IFN responses have been described as inhibiting expression of CD40 (35); however,we did not find higher levels of type I IFN in the blood of critical G3 patients. In fact, some levels of type I IFN were detected in mild COVID-19 patients, suggesting a potential protective role of this type of response, as proposed by a recent study (36). Therefore, further research is required to specifically address this issue.
Information about critical cell subsets participating in the pathogenesis of COVID-19, significant sample size, and the availability of healthy non–COVID-19 controls as a reference group are strengths of our research. However, a number of limitations need to be discussed. This is a cross-sectional study design comparing severe COVID-19 patients with 2 sets of controls: mild/severe COVID-19 patients and non–COVID-19 healthy controls. In addition, a limitation of our study is the different time periods of sample collection after hospitalization in critical G3 patients compared with severe and mild patients. Although we did not observe significant differences in myeloid cell distribution between patients that had been hospitalized for fewer or more than 6 days or significant differences in symptom duration between severe and critical patients, we cannot formally rule out that some of the characteristics of this patient subgroup might be due to the timing after infection and their inability to control the virus. Obviously, it is difficult to obtain time-matched samples for the other 2 subgroups, since their clinical progression is different from that G3 patients. Because of this cross-sectional nature, any assessment of progression will require a confirmation in prospective follow-up studies. In addition, our study provides data on bronchoscopy infiltrates rather than BAL, given the fragile status of COVID-19 patients with ARDS. Although bronchoscopies might not necessarily reflect the characteristics of infiltrates present in terminal bronchioles and alveoli, our data from 2 patients indicate that the composition of myeloid cells in the infiltrates obtained by these methods are comparable. On the other hand, our study did not directly address the protective versus detrimental function of DCs during COVID-19 infection. In this sense, CD141+ cDCs are important for the priming of antiviral CD8+ T cell responses (15, 37) and are severely depleted from the blood in all COVID-19 patients regardless of their clinical status, but they are almost completely undetectable in bronchoscopy infiltrates from individuals developing severe ARDS. These data might support a defect in the priming, maintenance, or exhaustion of virus-specific CD8+ T cells in the lungs. In addition, our data suggest a certain level of association of activation of T Mo and proportions of a new subset of CXCR5+CD38+CD8+ T cells that we have identified in the lung. CXCR5+CD8+ T cells have been associated with control of respiratory and nonrespiratory viruses (29, 38, 39). In contrast, CD38+CD8+ T cells can act as an immunosuppressive subset (40). However, the functional nature of this population of CXCR5+CD38+CD8+ T cells remains to be tested and these issues must be investigated in depth with a larger patient cohort and specific functional assays. In addition, CD1c+ cDCs are known to support CD4+ T cell responses and stimulate follicular helper T cells required for effective humoral antiviral adaptive immunity (14). While it has been proposed that protection against SARS-CoV-2 might also be at least partially mediated by specific Abs (41), the role of CD1c+ DCs inducing Tfh responses in these patients has not been addressed in our study and requires further research.
To conclude, our study unveils immune cellular networks associated with control or progression of COVID-19 disease, which might be useful for early diagnosis and the design of new more targeted and individualized treatments. Eventually, further research on these mechanisms may lead to the development of new therapeutic or preventive strategies.
Methods
Study patient cohort and samples.
A total of 64 patients diagnosed with COVID-19 after testing positive for SARS-CoV-2 RNA by quantitative PCR (qPCR) were included in the study. As specified in Supplemental Table 1, 95% of patients initiated different treatments upon hospital admission. Our cohort of COVID-19 patients was stratified into 3 groups of mild (G1), severe (G2), and critical (G3) prognosis based on respiratory frequency (RF), blood oxygen saturation (StO2), PaFiO2, and respiratory failure values as well as by following recently described criteria (22). Plasma levels of INF-α and IL-6 were also determined by ELISA (Human IFN Alpha Multi-Subtype Serum ELISA Kit, pbl ASSAY Science; Human IL-6 Quantikine High Sensitivity Enzyme-Immune Assay; R&D Systems). Blood samples were obtained for all participants and directly used for analysis without prior Ficoll gradient centrifugation to avoid the depletion of polymorphonuclear cells.
Of note, critical G3 patients from whom blood (n = 24) and/or bronchoscopy samples (n = 23) were collected displayed 80.95% and 100% of microbial superinfection at the time of sample collection, respectively. The majority of superinfections in these patients were observed 6 days after ICU admission and IMV support (88.24% from blood and 100% from bronchoscopy samples) and were caused by either bacteria alone (66.66% from blood and 65% from bronchoscopy samples), fungi alone (26.66% from blood and 30% from bronchoscopy samples), or both types of pathogens (6.66% from blood and 5% from bronchoscopy samples) (Supplemental Table 2).
Flexible bronchoscopy procedures were conducted in all ICU patients under sedation and muscle relaxation. The main indication for performing this technique was due to the poor respiratory and clinical situation of the patients. This technique enables the visualization of the lumen and mucosa of the trachea and also of the proximal and distal airways. Flexible bronchoscopy is indicated to evaluate pneumonia, to asses infiltrates of unclear etiology, to aspirate secretions, and to prevent development of atelectasis and pneumonia associated with invasive ventilation. Furthermore, in patients suspected of having pneumonia or infections, microbiological specimens can be collected with bronchial washings. Bronchial washings consist of aspirating bronchial secretions directly or after instilling physiological serum. We did not employ local anesthetic to avoid sample alterations. BAL can also be performed in pneumonia patients. Nevertheless, this technique entails a high risk for patients with severe respiratory failure. This is true specially for patients with high oxygen concentration requirements, such as COVID pneumonia patients. Therefore, we performed BAL solely in 2 cases (42). The technique used in the study was based on an initial bronchial aspiration that was discarded and a subsequent aspiration to collect sample from distal bronchial portions; after that, a bronchial washing sample was obtained in the distal bronchial portions. In situations where the clinical and ventilatory situation allowed it, BAL was performed, which was obtained after nailing the bronchoscope in a subsegmental bronchus with the instillation of 3 aliquots of 20 cc of 0.9% saline, with recovery of more than 50% of the instilled fluid. Analysis of the BAL samples showed results similar to those obtained in bronchial lavage. Finally, the hematopoietic lung infiltrates were obtained from bronchoscopy samples diluted in 0.9% sodium chloride at a 1: 5 ratio and after 3 sequential centrifugations at 830 g over 10 minutes.
Flow cytometry reagents and data analysis.
For phenotypical studies of cell populations present in peripheral whole blood, cells were incubated with a panel of different combinations of the following mAbs: anti-human CD3-Pacific blue and PerCP-Cy5.5, CD19PerCP-Cy5.5, CD56 PerCP-Cy5.5, CD8-APC-Cy7, CXCR5-PE, CD14-PE, CD16-Pacific blue, CD40-FITC, HLA-DR-APC-Cy7 and HLA-DR-APC, CD11c-Pacific blue, CD1c-PE-Cy7, CD141-APC (BioLegend) and CD38-FITC, CD123-APC, CD20-PerCP_Cy5.5, and HLA-DR-APC (BD) for 30 minutes. All details for commercial Abs are summarized in Supplemental Table 3. Subsequently, whole blood cells were treated with BD FACS Lysing Solution for 15 minutes, centrifuged at 830 g, and finally resuspended in 1× PBS (Lonza) and analyzed using a BD FACSCanto II Flow Cytometer (BD). The gating strategy for myeloid cells is shown in Supplemental Figure 2. CD1c+ and CD141+ cDC subsets were identified as lineage-negative (CD3–CD19–CD20–CD56–), CD14- HLA–DR+ cells that expressed either CD1c and CD141 (panel I). pDCs were defined as Lin–HLA-DR+CD11c–CD123hi cells (panel II). CD14hiCD16– C, CD14int CD16+ T, and CD14loCD16hi NC Mo were defined on the basis of CD14 and CD16 expression levels. Granulocytes were identified as large CD14lo/–CD16hi HLADR– cells. Maturation status of myeloid cells was defined based on CD40 MFI. In the case of bronchoscopy samples from critical COVID-19 patients, we added anti-CD45–Pacific Orange mAbs (BioLegend) to the previously described Ab cocktails and performed an additional panel to analyze frequencies and distribution of CD3+CD8+ and CD4+ T cell subsets defined by expression of CD38 and CXCR5 or HLA-DR, respectively. Individual and multiparametric analysis of flow cytometry data was performed using FlowJo software (Tree Star) and t-SNE.
Statistics.
Quantitative variables were represented as median and interquartile range. Statistical differences between different cell populations or between patient cohorts was calculated using a nonparametric 2-tailed Mann-Whitney U test or using a Kruskal-Wallis test, Friedman test, or Bonferroni’s for multiple comparisons, as appropriate. Significant differences of paired analyses were calculated using a 2-tailed Wilcoxon’s matched pairs test and a Bonferroni’s multiple comparison correction when possible. Association between cellular and clinical parameters was calculated using nonparametric Spearman’s correlations. Statistical significance was set at P < 0.05. Statistical analyses were performed using the GraphPad Prism 8 software.
Study approval.
This study was approved by the Research Ethics Committee of Hospital Universitario de La Princesa in the context of REINMUN-COVID and EDEPIMIC projects, and it was carried out following the ethical principles established in the Declaration of Helsinki. All included patients (or their representatives) were informed about the study and gave written, informed consent.
Author contributions
EMG, JA, JBS, FSM, CMC, ISC, and PL developed the research idea and study concept, designed the study, and wrote the manuscript. EMG, JA, CMC, AA, and JBS supervised the study. ISC and PL designed and conducted most experiments and contributed equally to the study. JA, JBS, and PL provided peripheral blood and bronchoscopy samples from study patient cohorts. HDLF and ISC performed the analysis of plasma levels of IFN-α by ELISA. All other authors participated in patient sample processing and the creation of a clinical database used for the study.
Supplementary Material
Acknowledgments
EMG was supported by the Comunidad de Madrid Talento program (2017-T1/BMD-5396), the Ramón y Cajal program (RYC2018-024374-I), the MINECO RETOS program (RTI2018-097485-A-I00), and the NIH R21 program (R21AI140930). EMG and JBS have applied for funding for the EDEPIMIC study of the COVID-19 pandemic. Grants PREDINMUN-COVID from Fondo SUPERA COVID19, SAF2017-82886-R to FSM from the Ministerio de Economía y Competitividad, and HR17-00016 grant from La Caixa Banking Foundation to FSM supported the study. AA was supported by Fondo de Investigaciones Sanitarias (FIS) PI19/00549. CMC was supported by FIS PI18/01163. This work has also been cofinanced by the Community of Madrid through the COVID 2019 aid/funds. We would like to thank the REINMUN-COVID and EDEPIMIC groups (see Supplemental Acknowledgments for consortium details).
Version 1. 08/12/2020
In-Press Preview
Version 2. 10/26/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Funding Statement
Supported EMG
Supported EMG
Supported EMG
Supported EMG
Supported FS-M
Supported FS-M
Supported FS-M
Supported AA
Supported CMC
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6290–6300.https://doi.org/10.1172/JCI140335.
See the related Commentary at COVID-19 and myeloid cells: complex interplay correlates with lung severity.
Contributor Information
Ildefonso Sánchez-Cerrillo, Email: ildefonso_92@hotmail.com.
Beatriz Aldave, Email: beatrizaldave@gmail.com.
Santiago Sánchez-Alonso, Email: sanchezalonsosantiago@gmail.com.
Ana Sánchez-Azofra, Email: sanchezazofra@gmail.com.
Ana Marcos-Jiménez, Email: anamarcosjim@gmail.com.
Elena Ávalos, Email: elena.avapu@gmail.com.
Ana Alcaraz-Serna, Email: ana.alcaraz.serna@gmail.com.
Ignacio de los Santos, Email: isantosg@hotmail.com.
Tamara Mateu-Albero, Email: tamaltma@gmail.com.
Laura Esparcia, Email: lau.esparpi@gmail.com.
Celia López-Sanz, Email: celia1096@hotmail.com.
Pedro Martínez-Fleta, Email: martinezq93@gmail.com.
Ligia Gabrie, Email: lgabrie.652@gmail.com.
Luciana del Campo Guerola, Email: luciana.del.campo.g@gmail.com.
Hortensia de la Fuente, Email: hortensia.fuente@salud.madrid.org.
Isidoro González-Álvaro, Email: isidoro.ga@ser.es.
Arantzazu Alfranca, Email: aalfranca@gmail.com.
Francisco Sánchez-Madrid, Email: fsanchez.hlpr@salud.madrid.org.
Cecilia Muñoz-Calleja, Email: cmunoz.hlpr@salud.madrid.org.
Joan B. Soriano, Email: jbsoriano2@gmail.com.
Julio Ancochea, Email: juli119@separ.es.
Enrique Martín-Gayo, Email: enrique.martin@uam.es.
References
- 1.Gao Y, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92(7):791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan WJ, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thevarajan I, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 10.Xiong Y, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giamarellos-Bourboulis EJ, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen W, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Keeffe M, Mok WH, Radford KJ. Human dendritic cell subsets and function in health and disease. Cell Mol Life Sci. 2015;72(22):4309–4325. doi: 10.1007/s00018-015-2005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin-Gayo E, et al. Immunological fingerprints of controllers developing neutralizing HIV-1 antibodies. Cell Rep. 2020;30(4):984–996.e4. doi: 10.1016/j.celrep.2019.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jongbloed SL, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207(6):1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson DA, Murphy KM, Briseño CG. Development, diversity, and function of dendritic cells in mouse and human. Cold Spring Harb Perspect Biol. 2018;10(11):a028613. doi: 10.1101/cshperspect.a028613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancel JC, Crozat K, Dalod M, Mattiuz R. Are conventional type 1 dendritic cells critical for protective antitumor immunity and how? Front Immunol. 2019;10:9. doi: 10.3389/fimmu.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narasimhan PB, Marcovecchio P, Hamers AAJ, Hedrick CC. Nonclassical monocytes in health and disease. Annu Rev Immunol. 2019;37:439–456. doi: 10.1146/annurev-immunol-042617-053119. [DOI] [PubMed] [Google Scholar]
- 19.Wolf AA, Yáñez A, Barman PK, Goodridge HS. The ontogeny of monocyte subsets. Front Immunol. 2019;10:1642. doi: 10.3389/fimmu.2019.01642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity. 2018;49(4):595–613. doi: 10.1016/j.immuni.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Zheng HY, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 23.Perona-Wright G, et al. A pivotal role for CD40-mediated IL-6 production by dendritic cells during IL-17 induction in vivo. J Immunol. 2009;182(5):2808–2815. doi: 10.4049/jimmunol.0803553. [DOI] [PubMed] [Google Scholar]
- 24.Yanagawa Y, Onoé K. Distinct regulation of CD40-mediated interleukin-6 and interleukin-12 productions via mitogen-activated protein kinase and nuclear factor kappaB-inducing kinase in mature dendritic cells. Immunology. 2006;117(4):526–535. doi: 10.1111/j.1365-2567.2006.02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon SB, Irving GR, Lawson RA, Lee ME, Read RC. Intracellular trafficking and killing of Streptococcus pneumoniae by human alveolar macrophages are influenced by opsonins. Infect Immun. 2000;68(4):2286–2293. doi: 10.1128/IAI.68.4.2286-2293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon SB, et al. Pulmonary immunoglobulin responses to Streptococcus pneumoniae are altered but not reduced in human immunodeficiency virus-infected Malawian adults. J Infect Dis. 2003;188(5):666–670. doi: 10.1086/377480. [DOI] [PubMed] [Google Scholar]
- 27.Jambo KC, et al. Bronchoalveolar CD4+ T cell responses to respiratory antigens are impaired in HIV-infected adults. Thorax. 2011;66(5):375–382. doi: 10.1136/thx.2010.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann M, et al. Exhaustion of activated CD8 T cells predicts disease progression in primary HIV-1 infection. PLoS Pathog. 2016;12(7):e1005661. doi: 10.1371/journal.ppat.1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He R, et al. Follicular CXCR5-expressing CD8(+) T cells curtail chronic viral infection. Nature. 2016;537(7620):412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, et al. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Front Mol Biosci. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, et al. Advanced role of neutrophils in common respiratory diseases. J Immunol Res. 2017;2017:6710278. doi: 10.1155/2017/6710278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florentin J, et al. Inflammatory macrophage expansion in pulmonary hypertension depends upon mobilization of blood-borne monocytes. J Immunol. 2018;200(10):3612–3625. doi: 10.4049/jimmunol.1701287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massaro KS, Costa SF, Leone C, Chamone DA. Procalcitonin (PCT) and C-reactive protein (CRP) as severe systemic infection markers in febrile neutropenic adults. BMC Infect Dis. 2007;7:137. doi: 10.1186/1471-2334-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cevaal PM, Bekker LG, Hermans S. TB-IRIS pathogenesis and new strategies for intervention: Insights from related inflammatory disorders. Tuberculosis (Edinb) 2019;118:101863. doi: 10.1016/j.tube.2019.101863. [DOI] [PubMed] [Google Scholar]
- 35.Qin H, Wilson CA, Lee SJ, Benveniste EN. IFN-beta-induced SOCS-1 negatively regulates CD40 gene expression in macrophages and microglia. FASEB J. 2006;20(7):985–987. doi: 10.1096/fj.05-5493fje. [DOI] [PubMed] [Google Scholar]
- 36. Lokugamage KG, Hage A, Schindewolf C, Rajsbaum R, Menachery VD. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv. https://doi.org/10.1101/2020.03.07.982264. Published April 9, 2020. Accessed September 11, 2020.
- 37.Haniffa M, et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37(1):60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, et al. CXCL13-mediated recruitment of intrahepatic CXCR5+CD8+ T cells favors viral control in chronic HBV infection. J Hepatol. 2020;72(3):420–430. doi: 10.1016/j.jhep.2019.09.031. [DOI] [PubMed] [Google Scholar]
- 39.Qiu L, Wang H, Yu Q, Liu J, Chen S, Zhao Z. Protective role of follicular CXCR5+CD8+ T cells against dengue virus 2 infection. Int J Infect Dis. 2019;83:12–19. doi: 10.1016/j.ijid.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bahri R, Bollinger A, Bollinger T, Orinska Z, Bulfone-Paus S. Ectonucleotidase CD38 demarcates regulatory, memory-like CD8+ T cells with IFN-γ-mediated suppressor activities. PLoS One. 2012;7(9):e45234. doi: 10.1371/journal.pone.0045234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padoan A, et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin Chim Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du Rand IA, et al. British Thoracic Society guideline for diagnostic flexible bronchoscopy in adults: accredited by NICE. Thorax. 2013;68 Suppl 1:i1–i44. doi: 10.1136/thoraxjnl-2013-203618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.