Abstract
Gain-of-function mutations in with no lysine (K) 1 (WNK1) and WNK4 genes are responsible for familial hyperkalemic hypertension (FHHt), a rare, inherited disorder characterized by arterial hypertension and hyperkalemia with metabolic acidosis. More recently, FHHt-causing mutations in the Kelch-like 3–Cullin 3 (KLHL3-CUL3) E3 ubiquitin ligase complex have shed light on the importance of WNK’s cellular degradation on renal ion transport. Using full exome sequencing for a 4-generation family and then targeted sequencing in other suspected cases, we have identified new missense variants in the WNK1 gene clustering in the short conserved acidic motif known to interact with the KLHL3-CUL3 ubiquitin complex. Affected subjects had an early onset of a hyperkalemic hyperchloremic phenotype, but normal blood pressure values”Functional experiments in Xenopus laevis oocytes and HEK293T cells demonstrated that these mutations strongly decrease the ubiquitination of the kidney-specific isoform KS-WNK1 by the KLHL3-CUL3 complex rather than the long ubiquitous catalytically active L-WNK1 isoform. A corresponding CRISPR/Cas9 engineered mouse model recapitulated both the clinical and biological phenotypes. Renal investigations showed increased activation of the Ste20 proline alanine–rich kinase–Na+-Cl– cotransporter (SPAK-NCC) phosphorylation cascade, associated with impaired ROMK apical expression in the distal part of the renal tubule. Together, these new WNK1 genetic variants highlight the importance of the KS-WNK1 isoform abundance on potassium homeostasis.
Keywords: Genetics, Nephrology
Keywords: Epithelial transport of ions and water, Genetic diseases, Protein kinases
Introduction
Familial hyperkalemic hypertension (FHHt), also known as Gordon syndrome and pseudohypoaldosteronism type 2, is a rare disease associated with net positive Na+ balance and renal K+ retention resulting in hypertension, hyperkalemia, and hyperchloremic metabolic acidosis (1). Mutations causing FHHt were first identified in the WNK1 and WNK4 genes that encode 2 members of the with no lysine (K) (WNK) serine-threonine kinase family (2). More recently, we and others have identified disease-causing mutations in the Kelch-like 3 (KLHL3) and Cullin 3 (CUL3) proteins belonging to a ubiquitin-protein ligase complex (3, 4). This complex has been shown in vitro to interact with WNK1 and WNK4, induce their ubiquitination, and regulate their protein levels through proteasomal degradation (5–8). Ohta and collaborators mapped the interaction site in WNK1 to a region containing a short acidic motif, which is highly conserved among the members of the WNK family (5). Interestingly, most of the FHHt-causing WNK4 variants cluster in this motif and are charge changing. WNK4 proteins carrying the same mutations fail to interact with the KLHL3 adaptor protein in vitro (5–7). This causes increased WNK4 abundance with secondary activation of the Na+-Cl– cotransporter (NCC) and development of FHHt (6, 7).
The genetic heterogeneity of FHHt is reflected in its phenotypic heterogeneity, ranging from severe cases presenting in childhood to mild and sometimes asymptomatic cases presenting in late adulthood (9, 10). A similar spectrum exists for the more recent causal genes KLHL3 and CUL3, with CUL3 mutations causing the most severe hypertension and electrolyte abnormalities (3). A milder phenotype was suggested for the WNK1-FHHt cases that could be partly explained by a different mutational mechanism. Indeed, the WNK1 gene gives rise to 2 main isoforms: a long ubiquitously expressed and catalytically active isoform (L-WNK1) and a short kidney-specific isoform (KS-WNK1) that is only expressed in the distal nephron (11, 12). FHHt-causing WNK1 mutations were found to be large deletions located in intron 1, resulting in an increased expression of L-WNK1 in the distal convoluted tubule (DCT) and connecting tubule (CNT) (13).
We have now identified cases and families with inherited hyperkalemic hyperchloremic acidosis carrying missense variants in the WNK1 gene that all cluster in the conserved acidic motif and are similar to those found previously in WNK4. Affected subjects had a clear electrolyte phenotype, but no significant blood pressure (BP) increase, especially in comparison with those who had similar WNK4 mutations. Their functionality was tested in Xenopus laevis oocytes and HEK293T cells, showing that they mainly affect the regulation of KS-WNK1 by the KLHL3-CUL3 ubiquitin complex. Using the CRISPR/Cas 9 genome-editing system, a mouse line was created that replicated the human phenotype and that was used to analyze the consequences of mutations at the WNK1 acidic motif on renal transport.
Results
A missense variant in the WNK1 acidic motif in the first FHHt kindred.
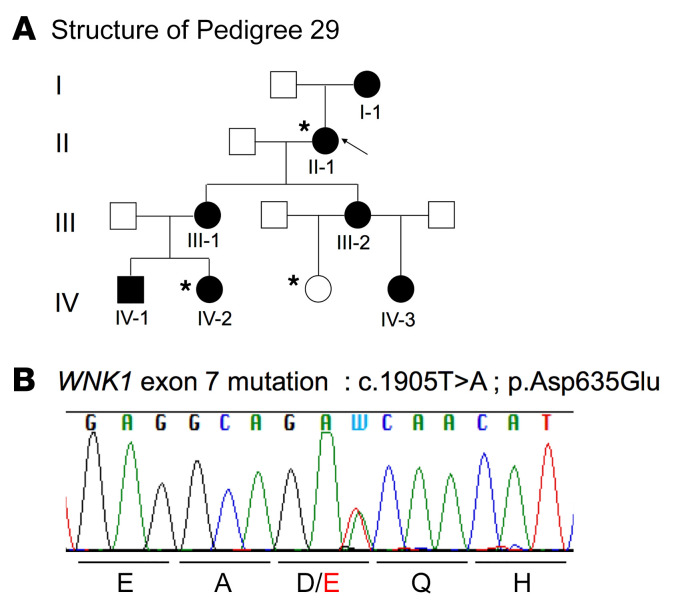
We took advantage of one 4-generation family to combine linkage analysis and whole exome sequencing (WES). This family included 7 affected individuals (all with metabolic acidosis and hyperkalemia, but with normal BP) and 6 unaffected individuals (Figure 1A and Supplemental Table 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI94171DS1). Using a SNP-based linkage approach, 7 suggestive linkage regions (maximum logarithm of odds [LOD] score = 1.8 in all linked regions) were identified in this panel (Supplemental Table 1B). In total, linked regions spanned 69 Mb and included 829 protein-coding genes. After filtering, WES performed in 1 unaffected and 2 affected individuals identified 71 possible disease-causing coding variants (Supplemental Table 1C). Four missense variants mapped to the linkage regions and were predicted in silico to be damaging. These variants were located in solute carrier family 30 member 7, a zinc transporter (SLC30A7), kinesin family member 11 (KIF11), tectonic family member 3 (TCTN3), and WNK1. The WNK1 variant (c.1905T>A; P.Asp635Glu) (Figure 1B) was located in exon 7 (ex7), which encodes the conserved acidic motif, previously shown to mediate the interaction with the substrate adaptor KLHL3 (14).
Figure 1. Missense variant in the WNK1 acidic motif in the FHHt pedigree 29.
(A) Kindred 29: family affected by FHHt composed of 7 affected (black) and 6 unaffected (white) members. Arrow indicates the index case. Asterisks indicate exome-sequenced individuals. (B) Electrophoregram obtained by Sanger sequencing showing the double-peak A/T corresponding to the WNK1 heterozygous mutation (c.1905T>A; P.Asp635Glu).
Additional variants in the WNK1 acidic motif in other cases and kindreds.
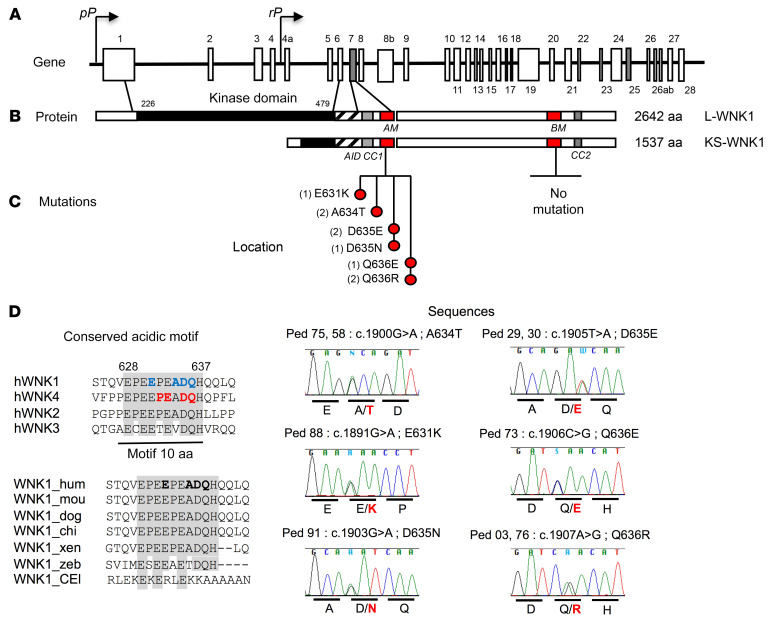
FHHt-causing WNK4 mutations are located in ex7 and ex17, encoding highly conserved acid and base motifs, respectively. Thus, we screened the homologous motifs of WNK1 encoded by ex7 and ex25, respectively, in 26 unrelated affected cases previously found as negative for the classical mutations in WNK4, KLHL3, CUL3, or the intron 1 deletion in WNK1. Direct sequencing identified 5 additional nonsynonymous heterozygous variants in ex7 in 8 unrelated subjects (Figure 2, A–C). The in silico pathogenicity of these variants is described in Supplemental Table 2. All were located within the acidic motif, between positions 631 and 636 of the L-WNK1 protein, and were predicted to be pathogenic. Four of the 6 missense variants were charge changing (E631K, D635N, Q636R, Q636E); 2 affected residues (D635, Q636) were also found mutated in the homologous acidic motif of WNK4 (D564 and Q565, Figure 2, C and D)
Figure 2. Acidic motif WNK1 mutations.
(A) Schematic representation of the WNK1 gene. The coordinates of the different exons are indicated above or below the structure with ex7 and ex25 (gray), which code for the conserved acidic and basic motifs. The proximal promoter (pP) drives the expression of the long ubiquitous kinase active isoform (L-WNK1), whereas the renal promoter (rP) drives the expression of the kinase defective kidney-specific isoform (KS-WNK1). (B) Schematic linear structure of LWNK1 and KS-WNK1. The kinase domain is represented in full black, the autoinhibitory domain (AID) is striped, coiled-coil domain 1 (CC1) and CC2 are represented in gray, and the conserved acidic motif (AM) and basic motif (BM) are represented in red. (C) Location and sequences of the mutated residues clustering in the acidic motif. The brackets indicate the number of unrelated affected subjects for each mutation. On the lower right are shown the Sanger sequencing electrophoregrams showing the various missense WNK1 mutations. (D) Conservation of the acidic motif showing residues mutated in FHHt among human WNK family members. The previously described WNK4 mutations are indicated in red; those at WNK1 and identified in this study in blue. All are located at completely conserved residues. The bottom part shows the conservation of the WNK1 acidic motif across species. The mutated residues are indicated in bold.
Clinical and biochemical characteristics: hydrochlorothiazide-sensitive hyperkalemic acidosis without hypertension.
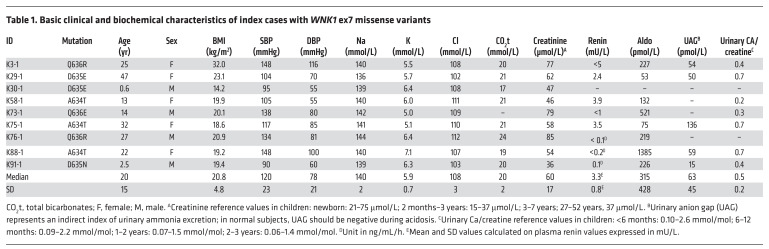
Detailed clinical and biological characteristics of index cases are given in Table 1. The circumstances of discovery and the clinical symptoms of these index patients are detailed in Supplemental Table 3. In most of the cases, patients were asymptomatic and showed with no signs of hyperkalemia on an electrocardiogram. All displayed the electrolyte anomalies typical of FHHt, including marked hyperkalemia (median, 5.9 mmol/L; IQR, 5.3–6.3), hyperchloremia (median, 108 mmol/L; IQR, 106–110), and metabolic acidosis (total CO2, 20 mmol/L; IQR, 19–21) despite a normal glomerular filtration rate (GFR) (median creatinine, 58 μmol/L; IQR, 47–74). For the 7 index cases with prospective reliable clinical data, hyperkalemia and hyperchloremia were rapidly corrected with low doses of hydrochlorothiazide (HCTZ) (6.25 to 25 mg/d; Supplemental Figure 1). In comparison, an average drop in potassium of only 0.7 mmol/L was observed in normal, healthy subjects administered a much higher dose of HCTZ (50 mg for 3 weeks) (15). Compared with reference values (16), we also observed a tendency for significant hypercalciuria in infancy and adulthood (Supplemental Figure 2).
Table 1. Basic clinical and biochemical characteristics of index cases with WNK1 ex7 missense variants.
Surprisingly, most of the index cases had casual BP values in the normal range, except a 25-year old woman with associated obesity (K3-1, BMI, 32 kg/m2) and a 22-year old woman (K88-1) without other cardiovascular risk factors, but who had short stature and mild mental retardation (Table 1). This tendency for normal BP values was confirmed in the first-degree relatives harboring the familial mutations, since only 2/10 were mildly hypertensive (Supplemental Table 4). Overall, the 20 mutated individuals with measured BP (23 ± 18 years) belonging to 9 affected families had normal systolic BP (SBP) (116 ± 20 mmHg) and diastolic BP (DBP) (73 ± 21 mmHg). Low or suppressed plasma renin levels were observed with normal to slightly elevated plasma aldosterone concentrations (Table 1 and Supplemental Table 4), as has been observed with other FHHt genotypes.
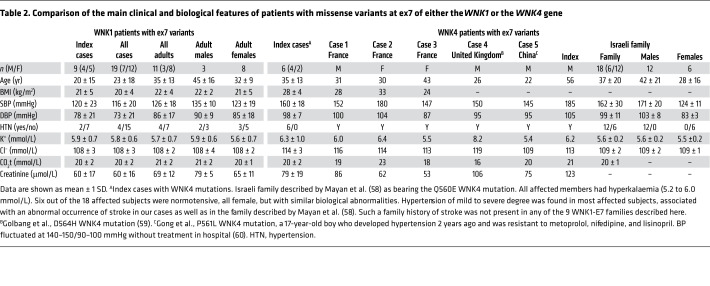
We then compared the clinical phenotypes of FHHt patients with WNK1 ex7 missense mutations with those who had WNK4 missense variants affecting the same acidic motif collected in our center and in the literature (Table 2). Interestingly, WNK4-related patients consistently had the strongest BP and electrolyte phenotype, whatever the subclassification (index cases, adults or males only); BP was much higher in patients with WNK4 mutations (SBP and DBP averaged ~20 mmHg and ~10 mmHg higher) compared with that in patients with WNK1 ex7 mutations.
Table 2. Comparison of the main clinical and biological features of patients with missense variants at ex7 of either the WNK1 or the WNK4 gene.
The WNK1 ex7 mutations selectively abolish the ubiquitination and degradation of the KS-WNK1 isoform in vitro.
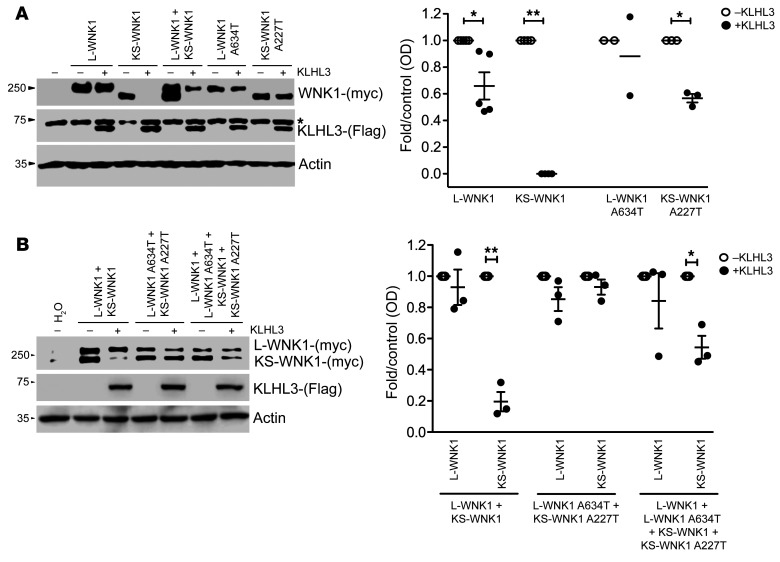
The acidic motif common to WNK1 and WNK4 has been reported as a binding site for the CUL3 and KLHL3 E3-ubiquitin ligase complex (14). Therefore, we tested the effect of KLHL3 on the abundance of WT and mutant L- and KS-WNK1 isoforms (Figure 3). The expression of KLHL3 in X. laevis oocytes resulted in a small (about 30%), but significant, decrease in the amount of L-WNK1 and almost completely abolished KS-WNK1. We then analyzed the recurrent WNK1 A634T variant, found in the K58 and K75 kindreds. No significant change in L-WNK1 abundance was observed between the WT and mutant isoforms, whereas the KS-WNK1-A227T mutant was highly expressed in comparison with the WT KS-WNK1 isoform, suggesting that the mutation abrogates the degradation of KS-WNK1 (Figure 3A). We also tested the other observed WNK1 missense variants of the acidic motif using the same experimental system (Supplemental Figure 3A). All were protective with regard to KLHL3/CUL3-mediated degradation.
Figure 3. Differential effect of the WNK1 Ex7 G1900A (A634T-L-WNK1; A227T-KS-WNK1) variant on the interaction between KLHL3 and L-WNK1 or KS-WNK1 isoforms.
(A) Representative immunoblot of proteins extracted from X. laevis oocytes (left panel) that were injected with WT or mutant L-WNK1 or KS-WNK1 in the absence or presence of KLHL3 cRNA, as stated. The upper blot shows c-myc–positive bands corresponding to L-WNK1 and KS-WNK1. The middle blot shows unspecific upper band present in all lanes (*), including water-injected oocytes, and a lower band corresponding to KLHL3 only present in KLHL3-injected oocytes. The lower blot shows actin. Densitometry of several (n = 3) blots in which the effect of KLHL3 was tested in L-WNK1 or KS-WNK1 WT or mutants separately (right panel). In the absence of KLHL3, mean values were arbitrarily set to 1.0, and in the presence of KLHL3, values were normalized accordingly. *P < 0.05; **P < 0.01, unpaired Student’s t test. (B) Representative immunoblot of proteins extracted from X. laevis oocytes (left panel) that were injected with mixture of WT or mutant L-WNK1 and KS-WNK1 in the absence or presence of KLHL3 cRNA in order to analyze the consequences of L-WNK1 and KS-WNK1 coexpression. Immunoblot (left panel) and densitometry analysis of several blots (right panel) are as in Figure 3A.
Since both WNK1 isoforms are physiologically coexpressed in the distal nephron (12), we sought to characterize how KLHL3/CUL3-mediated degradation affects KS- and L-WNK1 abundance in coexpression experiments (Figure 3B). On KLHL3 expression, a 70%–80% decrease in WT KS-WNK1 abundance was observed, while there was no change in L-WNK1. This major difference was not observed after transfection with the mutated isoforms. The coexpression of WT and mutant isoforms led to an intermediate decrease in KS-WNK1 abundance, suggesting that only the WT KS-WNK1 was degraded. Overall, these results suggest that human mutations of the WNK1 acidic motif selectively increase KS-WNK1 abundance in the distal nephron where KLHL3 is exclusively expressed.
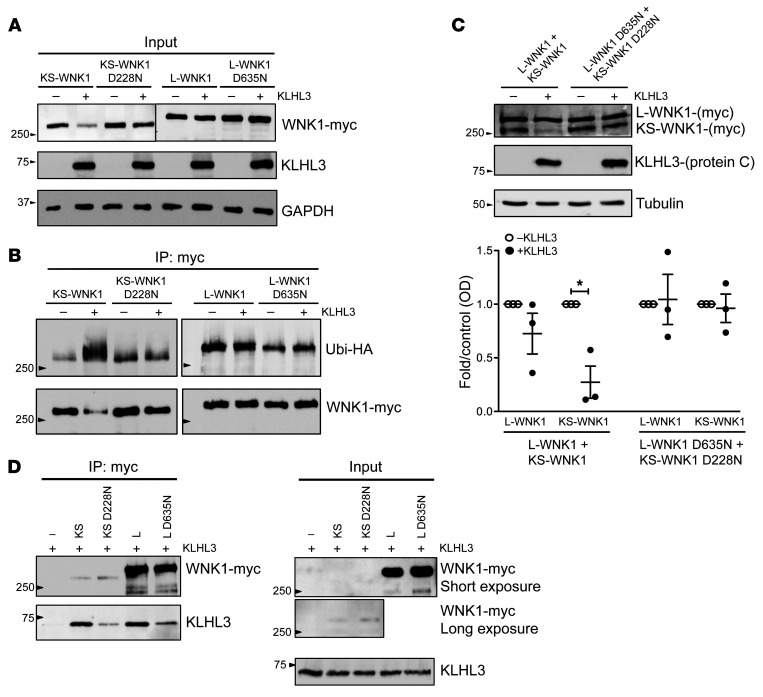
We also tested the effect of KLHL3 on the WT and mutant WNK1 isoforms in human tissue culture HEK293T cells. Since we found that this cell line expresses high levels of CUL3, but undetectable levels of KLHL3, we first established a stable tetracycline-inducible cell line expressing KLHL3 (see Supplemental Methods). The induction of KLHL3 resulted in a dramatic decrease in the level of transfected KS-WNK1, but not L-WNK1 (Figure 4A). The introduction of the D635N mutation (D228N for the KS-WNK1 isoform) prevented this decrease. Interestingly, all other mutations were also protective with regard to KLHL3/CUL3-mediated degradation, with the exception of KS-WNK1 D228E (Supplemental Figure 3B). The substitution of an aspartic acid (D) by a glutamic acid (E) was not charge changing, which might explain the milder phenotype. The coexpression of L- and KS-WNK1 in HEK293T cells confirm the observations made in Xenopus oocytes (Figure 4B). Next, we immunoprecipitated L- and KS-WNK1 and assayed their ubiquitination status in denaturing conditions to exclude any ubiquitination signal linked to WNK1 interactors. KLHL3 induction resulted in heavy KS-WNK1 ubiquitination, whereas it had little or no effect on L-WNK1 or D635N (D228N) mutants (Figure 4C). KS-WNK1 ubiquitination by the KLHL3/CUL3 complex suggests that the E3 ligase adaptor (KLHL3) and the substrate (WNK1) interact. Indeed, both L-WNK1 and KS-WNK1 coimmunoprecipitated KLHL3 in native conditions. However, KS-WNK1 did so with a much higher efficiency (Figure 4D). As expected, the D635N mutation in the acidic domain (D228N in KS-WNK1) reduced the interaction with KLHL3. Taken together, our in vitro studies strongly suggest that KS-WNK1 is the preferential WNK1 isoform target of the KLHL3/CUL3 ubiquitin complex and that the mutations identified in WNK1 ex7 prevent this ubiquitination.
Figure 4. KLHL3 interaction with WNK1 isoforms in HEK293T cells: KLHL3 ubiquitinates KS-WNK1 and significantly reduces its protein levels.
(A) Flp-In T-Rex 293 cells stably and inducibly expressing (His)6-protein C-Flag-hKLHL3 were transfected with myc-tagged L-WNK1 (WT or D635N mutant) or KS-WNK1 (WT or D228N mutant), as indicated. At 34 hours after transfection, cells were induced with tetracycline. Fourteen hours later (48 hours after transfection), cells were harvested and lysed in denaturing conditions. Cell lysates were subjected to immunoblot analysis with the indicated antibodies. Data shown are representative of 3 independent experiments. (B) Flp-In T-Rex 293 cells stably and inducibly expressing (His)6-protein C-Flag-hKLHL3 were transfected with ubiquitin-HA and myc-tagged L-WNK1, L-WNK1 D635N, KS-WNK1 or KS-WNK1 D228N, as indicated. At 34 hours after transfection, cells were induced with tetracycline. Fourteen hours later (48 hours after transfection), cells were harvested and lysed in denaturing conditions. Upper panel: Myc-tagged WNK1 isoforms were immunoprecipitated with anti-myc antibody (9B11, Cell Signaling Technology); immunoprecipitates were analyzed by immunoblotting with anti-HA antibody (3724S; Cell Signaling Technology). Nitrocellulose membranes were stripped and reblotted with anti-myc antibody. Immunoblot of cell lysates is represented in D (input). Data shown are representative of 3 independent experiments. (C) Cells were transfected with myc-tagged L-WNK1 (WT or D635N mutant) and KS-WNK1 (WT or D228N mutant), as indicated and in conditions similar to those in A. Cell lysates were subjected to immunoblot analysis with the indicated antibodies. Densitometric analysis was performed using FUJI FILM Multi-Gauge software. Results are shown as mean ± SEM. *P < 0.05 compared with control, unpaired Student’s t test. n = 3. (D) Flp-In T-Rex 293 cells stably and inducibly expressing (His)6-protein C-Flag-hKLHL3 were transfected with myc-tagged L-WNK1, L-WNK1 D635N, KS-WNK1, or KS-WNK1 D228N, as indicated. At 43 hours after transfection, cells were induced with tetracycline and simultaneously treated with MG132 for 5 hours. At 48 hours after transfection, cells were harvested and lysed in native conditions. Left panel: cell lysates were immunoprecipitated with anti-myc antibody, and immunoprecipitates were analyzed by immunoblotting with anti-protein C and anti-myc antibodies. Right panel: cell lysates (input) were subjected to immunoblot analysis with anti-myc and anti–protein C antibodies (HPC4, Roche) to check for even expression of KLHL3. Data shown are representative of 3 independent experiments.
A mouse model bearing a mutation at the WNK1 acidic motif confirms the absence of arterial hypertension despite a typical electrolyte phenotype.
To understand why WNK1 ex7 missense mutations lead to a hyperkalemic metabolic acidosis with normal BP in most of the affected patients, we sought to generate a mouse model bearing the D635E mutation at the WNK1 acidic motif using CRISPR/Cas9 genome-editing technology (Supplemental Figure 4A). The knockin of the point mutation was not found in 300 microinjected oocytes, but an in-phase suppression of a GAA triplet leading to the deletion of the first amino acid of the WNK1 acidic motif (delE631) was observed in 7/300 injected oocytes (Supplemental Figure 4B). In vitro, this mutant had an effect similar to that of the reference D635N variant, abrogating the KLHL3-induced decrease in KS-WNK1 abundance (Supplemental Figure 4C). The analysis of the BP and plasma electrolyte concentration confirmed that the heterozygous Wnk1+/delE631 mice represent an adequate model in which to study the in vivo consequences of the WNK1 mutations identified in our patients.
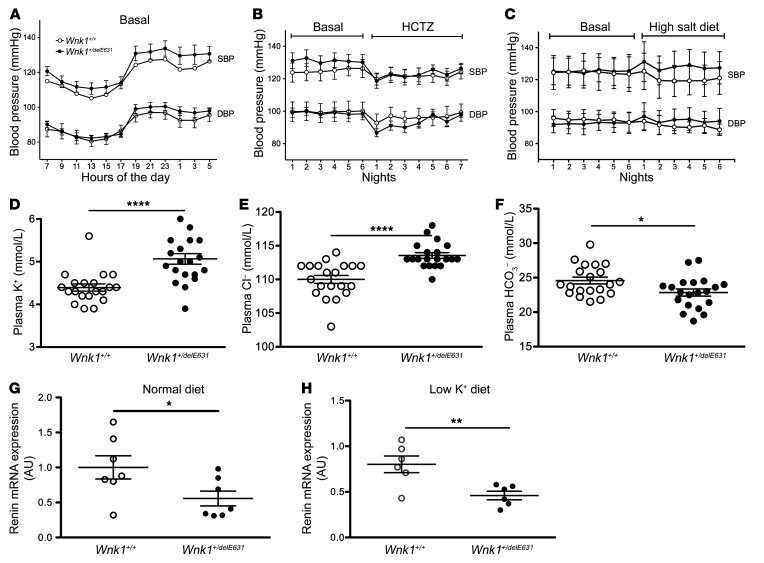
Using telemetry, we found that basal SBP and DBP were similar in heterozygous Wnk1+/delE631 mice and WT littermates (Figure 5A) as well as on HCTZ (Figure 5B). Tail-cuff measured SBP on a larger set of adult males also revealed the absence of hypertension (113 ± 5 mmHg, n = 20 Wnk1+/delE631 vs. 108 ± 5 mmHg, n = 20 Wnk1+/+, NS). The homozygous Wnk1delE631/delE631 mice were perfectly viable, with no departure from the expected Mendelian proportions when heterozygous mice were crossed. The biological phenotype of these homozygous mice did not differ from that the heterozygous mice, with the exception of SBP, which was significantly higher than in heterozygous or WT littermates when measured by tail cuff (Supplemental Table 5). We focused our studies on the heterozygous model, since it corresponds to the human autosomal dominant pathology.
Figure 5. Normal BP on both normal and high-salt diet in Wnk1+/delE631 mice.
(A–C) SBP and DBP profiles over 24 hours of SBP and DBP under a12-hour day/12-hour night schedule in Wnk1+/+ (n = 6) and Wnk1+/delE631 (n = 7) mice examined with a telemetric system under basal conditions (A). Night SBP and DBP of the same mice before (6 nights = basal) or during (7 nights) oral administration of HCTZ (240 mg/kg/d) (B). Night SBP and DBP of another group of mice (n = 4 Wnk1+/+, n = 4 Wnk1+/delE631) before (6 nights = basal) or during (6 nights) the administration of high (3%) Na+ diet (C). (D–F) Biological characteristics. In the mutant mice, significant hyperkalemia (5.1 ± 0.5 vs. 4.3 ± 0.2 mmol/L, P < 0.0001; n = 20) (D); hyperchloremia (114 ± 2 vs. 110 ± 3 mmol/L, P < 0.0001; n = 20) (E); and metabolic acidosis (HCO3–, 22.8 ± 2.1 vs. 24.6 ± 2.5 mmol/L, P < 0.05; n = 20) (F) were observed together with normal creatinine values (not shown). Data are represented as mean ± SEM. Statistical comparisons were made using unpaired Student’s t test. (G and H) Renin expression. Levels of renin mRNA were measured by RT-qPCR in the kidney cortex of Wnk1+/+ (n = 7) and Wnk1+/delE631 (n = 7) mice in baseline conditions (G) or of Wnk1+/+ (n = 6) and Wnk1+/delE631 (n = 6) mice fed a low (0%) K+ diet (H). Results (mean ± SEM) are expressed in arbitrary units relative to the expression of ubc. The expression level in Wnk1+/+ mice under basal conditions was arbitrarily set to 1. *P< 0.05; **P < 0.01; ****P < 0.0001, unpaired Student’s t test.
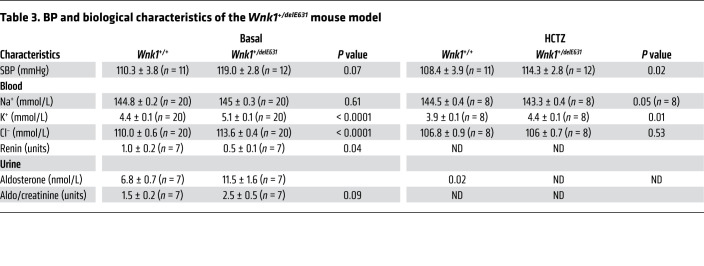
Wnk1+/delE631 mice displayed hyperkalemia (5.1 ± 0.5 mmol/L) and hyperchloremia (114 ± 2 mmol/L) (Figure 5, D–F) that were corrected by 3-day HCTZ oral administration (Table 3). Urinary aldosterone excretion was increased 2-fold, whereas renin expression was decreased by 45% in Wnk1+/delE631 mice (Figure 5G, Table 3 and Supplemental Table 6). This difference remained significant after a 7-day low K+ diet and normalization of kalemia (Figure 5H), suggesting that the lower renin was probably the consequence of a mild volume expansion rather than the hyperkalemic state (17). To better appreciate the absence of hypertension in this model, we performed BP telemetry measurements in Wnk1+/delE631 and Wnk1+/+ male littermates (6 to 7 months old) on a 1-week normal standard diet (0.3% NaCl) followed by a 1-week high NaCl (3%) diet (Figure 5C). The results confirm that there were no differences in basal BP between the 2 groups (SBP, 124.5 ± 10.4 in Wnk1+/+ vs. 125.2 ± 8.4 mmHg in Wnk1+/delE631) as well as no significant BP increase on a high-sodium diet (SBP: 120.6 ± 10.1 in Wnk1+/+ vs. 128.1 ± 10.1 mmHg in Wnk1+/delE631; Supplemental Table 6).
Table 3. BP and biological characteristics of the Wnk1+/delE631 mouse model.
Increased activation of the SPAK-NCC phosphorylation cascade.
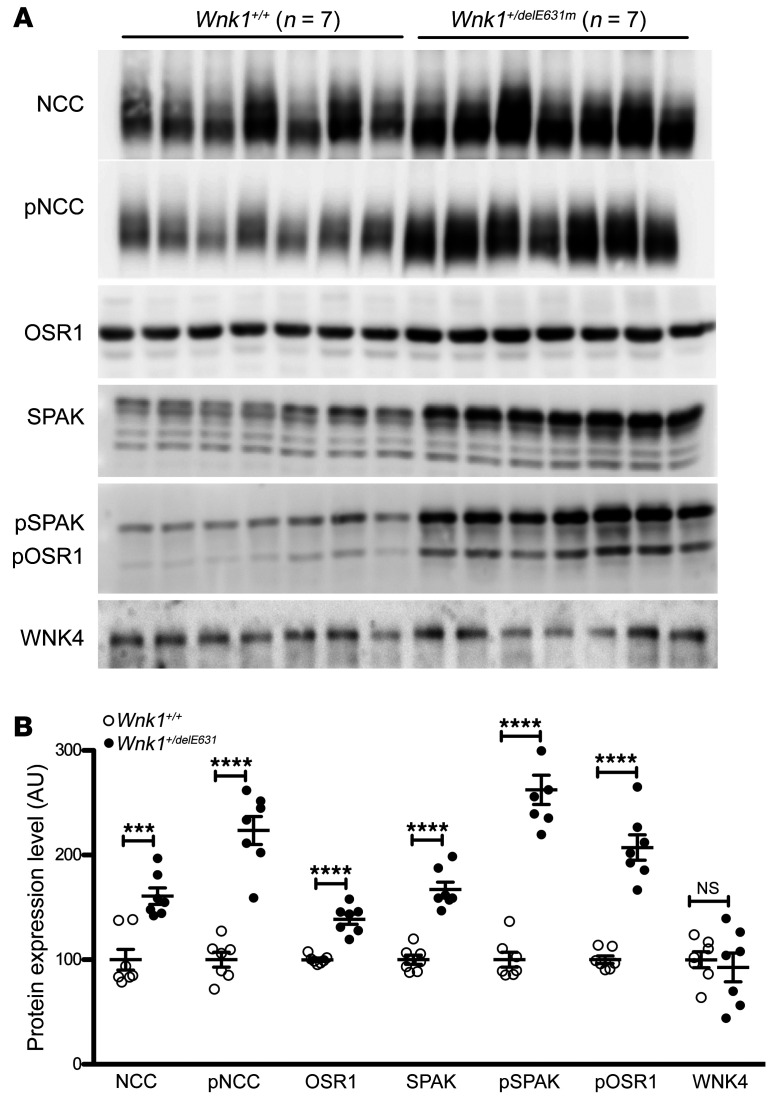
The extreme sensitivity of FHHt patients to thiazide diuretics initially suggested that the syndrome resulted from an increased activity of the Na+-Cl– cotransporter NCC. This hypothesis was confirmed by the demonstration that NCC abundance and phosphorylation are increased in all FHHt mouse hypertensive models (13, 18–20). Despite the absence of arterial hypertension, we observed the same phenomenon in the renal cortex of Wnk1+/delE631 mice compared with Wnk1+/+ littermates (Figure 6). The Ste20 proline alanine rich kinase (SPAK) kinase is essential for the expression and phosphorylation of NCC in vivo (20, 21). Immunoblotting on whole-kidney cortex homogenates showed that SPAK phosphorylation was greater in Wnk1+/delE631 mice compared with Wnk1+/+ (Figure 6). We also observed an increased abundance of the full-length SPAK isoform, which is the one predominantly expressed in the DCT (22), as well as a significant (P < 0.0001) increase in oxidative stress–responsive kinase 1 (OSR1). WNK4 abundance was not significantly changed in the kidney of Wnk1+/delE631 mice (Figure 6). However, due to the lack of a suitable antibody, we could not measure the phosphorylation level of the T-loop serine, which reflects the activation state of the kinase (23).
Figure 6. Activation of the SPAK-NCC phosphorylation cascade in Wnk1+/delE631 mice.
(A) Representative immunoblots with the indicated antibodies performed on the membrane-enriched fractions (NCC and pNCC) or total homogenates of the renal cortex of mice of each genotype. (B) Densitometric analysis. NCC, SPAK, and OSR1 abundance and phosphorylation are increased in Wnk1+/delE631 mice compared with Wnk1+/+ mice. WNK4 expression is similar between the 2 groups of mice. The expression level in Wnk1+/+ mice was arbitrarily set to 100. Data are represented as mean ± SEM. ***P < 0.001 ; **** P < 0.0001, unpaired Student’s t test.
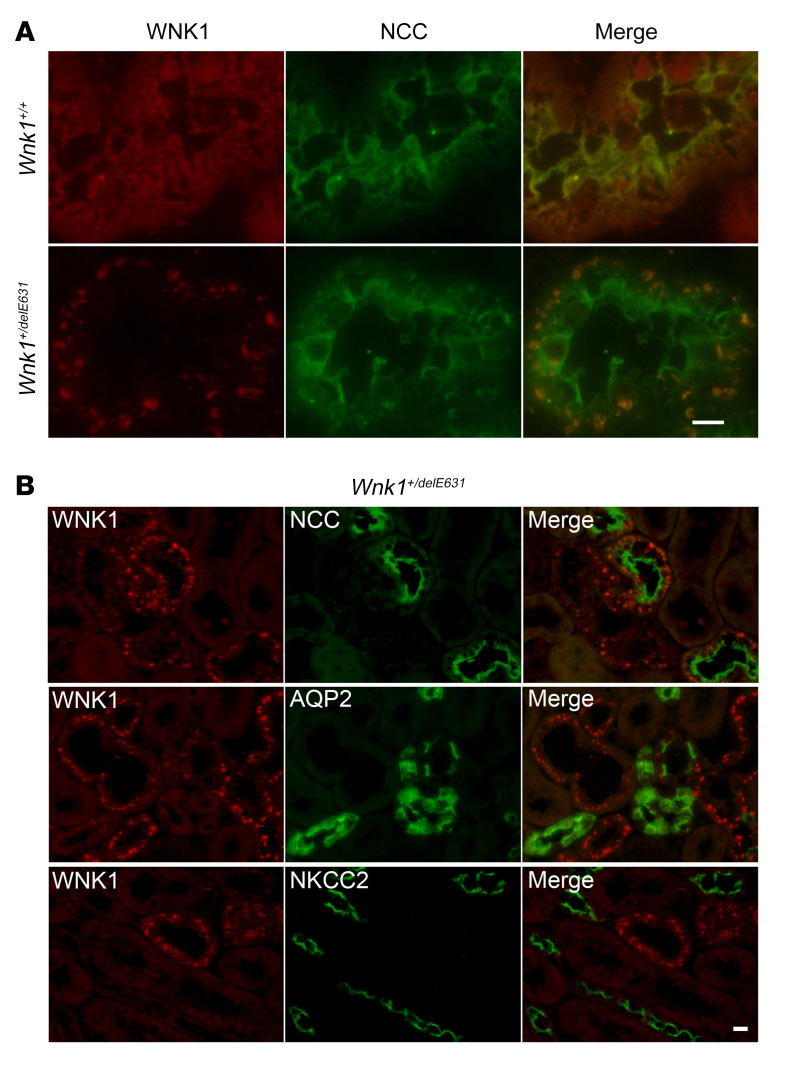
To further characterize the activation of the WNK1-SPAK/OSR1 cascade in the kidney of Wnk1+/delE631 mice, we performed immunofluorescence experiments with an antibody recognizing both L- and KS-WNK1 isoforms (a KS-WNK1–specific antibody was not available). While the DCT of Wnk1+/+ mice contained small WNK1-positive puncta, large WNK1-positive structures, resembling the previously described WNK bodies, were observed in Wnk1+/delE631 NCC-positive DCTs (Figure 7). These bodies were also found in some AQP2-positive, but not in NKCC2-positive, tubules. Previous studies demonstrated that KS-WNK1 is required for the formation of these WNK1 bodies, which are dynamic membraneless structures (24). Containing the components of the WNK/SPAK pathway, they are usually not present at baseline conditions, but form under conditions of low potassium intake when WNKs become activated (25). The fact that we observed them at baseline in Wnk1+/delE631 kidneys provides further support that the WNK1/SPAK cascade is activated in Wnk1+/delE631 mice.
Figure 7. The DCTs of Wnk1+/delE631 mice contain large WNK bodies.
(A) Immunofluorescence experiments using WNK1 and NCC antibodies. While only small puncta are observed in Wnk1+/+ DCT, large WNK1-positive structures are found in Wnk1+/delE631 DCT, resembling the previously described WNK bodies. Scale bar: 10 μm. (B) Costaining with NCC, AQP2, and NKCC2 revealed that these bodies are localized in the DCT and CNT/CCD, but not in Henle’s loop. Scale bar: 20 μm.
Potassium secretory capacity is diminished without change of ENaC expression in Wnk1+/delE631 mice.
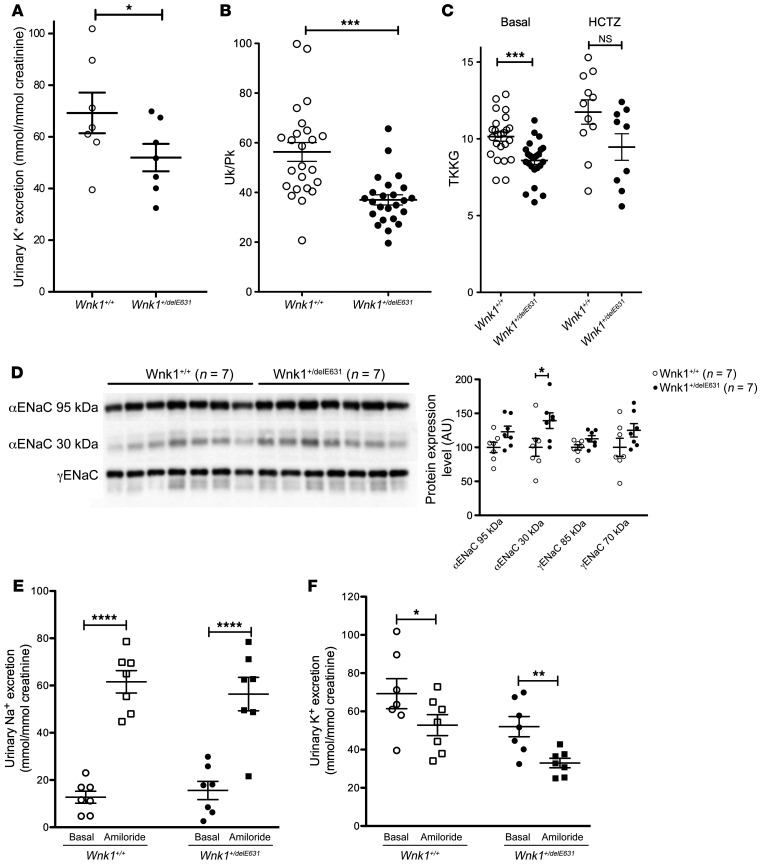
Basal urinary K+ excretion and urinary/plasma ratio of K+ concentration were lower in Wnk1+/delE631 mice, indicating a defect in urinary potassium excretion (Figure 8, A and B). We calculated the transtubular potassium gradient (TTKG) to estimate the potassium secretory capacity of the aldosterone sensitive distal nephron (ASDN), connective tubule and collecting duct (26). Wnk1+/delE631 mice exhibited significantly lower TTKG (8.6 ± 0.3, n = 24) than Wnk1+/+ mice (10.1 ± 0.3, n = 24, P = 0.0003), despite higher aldosterone levels, consistent with a potassium secretory defect (Figure 8C). As observed in patients, HCTZ rapidly corrected the hyperkalemia in the Wnk1+/delE631 mice (see above) and partially abolished the TTKG difference between the Wnk1+/+ and Wnk1+/delE631 mice (Figure 8C), suggesting the defect develops as a consequence of increased Na+ reabsorption by NCC and reduced sodium delivery to ASDN. Metolazone, a thiazide diuretic suggested as having less carbonic anhydrase inhibitory effect, did not correct the phenotype when administered intraperitoneally at a dose of 50 μg/kg/d over 4 days (Supplemental Figure 5).
Figure 8. Abnormal K+ handling in Wnk1+/delE631 mice.
(A–C) Decreased UK+ excretion, Uk/Pk ratio, and TTKG. (A) Urinary K+ excretion was lower in Wnk1+/delE631 (n = 7, 52 ± 5.3 mmol/mmol creatinine) than in Wnk1+/+ mice (n = 7, 69.3 ± 7.9 mmol/mmol creatinine). *P < 0.05, unpaired Student’s t test. (B) Basal urinary/plasma ratio of K+ concentration was lower in Wnk1+/delE631 (n = 24, 37.2 ± 2.1) than in Wnk1+/+ mice (n = 24, 56.3 ± 3.8). ***P < 0.0001, unpaired Student’s t test. (C) TTKG was significantly lower in Wnk1+/delE631 (n = 24, 8.6 ± 0.3) than in Wnk1+/+ mice (n = 24, 10.1 ± 0.3). ***P = 0.0003. Following 4-day HCTZ oral (240 mg/kg/d) administration, the difference in TTKG between Wnk1+/+ and Wnk1+/delE631 remained the same (9.5 ±0.9 versus 11.8 ±0.8, respectively) although no more significant (P = 0.068), likely because of the smaller number of animals studied (n = 9 and n = 11, respectively). Statistical comparisons were made using unpaired t tests. (D) ENaC expression. Representative immunoblots with the indicated antibodies performed on the membrane-enriched fractions of the renal cortex of mice of each genotype. Densitometric analysis. The abundance of the cleaved form of the α-subunit of ENaC was significantly increased in Wnk1+/delE631 mice compared with Wnk1+/+ mice. The expression level in Wnk1+/+ mice was arbitrarily set to 100. Data are represented as mean ± SEM.*P < 0.05, unpaired Student’s t test. (E and F) Natriuretic and kaliuretic response to amiloride. Urinary Na+ (E) and K+ (F) excretion in response to amiloride injection. Wnk1+/+ and Wnk1+/delE631 males (n = 7 in each group) were housed in metabolic cages and received 1 injection of vehicle or amiloride on 2 consecutive days. Urine was collected 6 hours after injection. Data are represented as mean ± SEM. *P < 0.05; **P< 0.01; ****P < 0.0001 versus vehicle, unpaired Student’s t test.
Decreased activity of the epithelial sodium (Na) Channel ENaC and the renal outer medullary potassium (K) channel ROMK are thought to contribute to the hyperkalemia in other forms of FHHt (27). Abundance of ENaC α and γ subunits in the membrane-enriched fraction of the renal cortex was assessed as a surrogate for channel function (28, 29). No significant increases in the abundance in full-length or cleaved forms of the ENaC subunits were observed in Wnk1+/delE631 mice compared with Wnk1+/+, except for the short fragment of α subunit (Figure 8D). We then acutely treated Wnk1+/delE631 and Wnk1+/+ mice with amiloride, the specific pharmacological inhibitor of ENaC. Six hours after the amiloride injection, the natriuresis increased very significantly and similarly in both groups that had similar Na and K intake (Figure 8E). Urinary K+ excretion also decreased similarly in Wnk1+/+ and in Wnk1+/delE631 (Figure 8F), the difference in postamiloride absolute values reflecting the difference in basal values. Together, these results make it seem unlikely that alterations in ENaC contribute to the potassium secretory defect.
Altered regulation of ROMK in Wnk1+/delE631 mice.
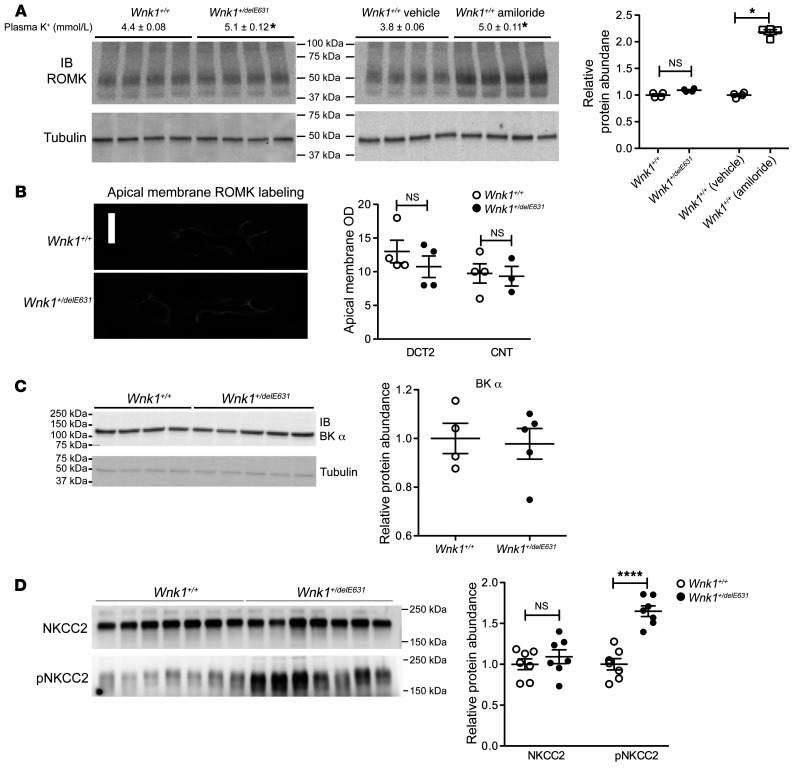
We then analyzed ROMK expression in the kidney cortex and at the apical pole of the distal part of the DCT (DCT2) and CNT cells, where the channel is most significantly upregulated in response to dietary potassium loading (30). At the basal level, Western blots showed that ROMK protein expression was not increased in Wnk1+/delE631 mice compared with Wnk1+/+ littermates despite significant hyperkalemia (5.1 ± 0.1 vs. 4.4 ± 0.1). In contrast, a significant increase in ROMK abundance was observed in WT mice when a similar rise in plasma potassium was induced by amiloride administration (Figure 9A). Immunofluorescence experiments revealed that apical ROMK expression was not modified in either the DCT2 or CNT of the hyperkalemic Wnk1+/delE631 mice compared with normokalemic Wnk1+/+ mice (Figure 9B). This lack of ROMK increase in the distal nephron is considered abnormal in these hyperkalemic Wnk1+/delE631 mice.
Figure 9. Expression of ROMK, BK channel, and NKCC2.
(A) ROMK protein abundance. No change in cortical ROMK expression was observed in Wnk1+/delE631 (n = 4) vs. littermate Wnk1+/+ (n = 4) mice, despite the latter group having significantly higher plasma K+ levels (4.4 ± 0.08 mM, n = 20 vs. 5.1 ± 0.12 mM, n = 20). Conversely, a more than 2-fold increase was observed in Wnk1+/+ mice treated by amiloride (25 mg/kg/d for 4 days) achieving a similar rise in plasma potassium (Wnk1+/+ vehicle: 3.8 ± 0.06 mM, n = 4 vs. Wnk1+/+ amiloride: 5.0 ± 0.11 mM). Quantification of cortical ROMK expression (n = 4 per group). *P < 0.05, t test. (B) ROMK immunofluorescence in the distal tubule. Left panel: immunolocalization of ROMK in the DCT2 of Wnk1+/+ and Wnk1+/delE631 mice. Right panel: analysis of membrane labeling intensity showed no change in ROMK apical expression in the DCT2 and CNT of Wnk1+/delE631 mice (n = 4 animals per genotype). Scale bar: 10 microns. (C) Basal BK α channel protein abundance in Wnk1+/delE631 mice and Wnk1+/+ littermates. BK α immunoblots (left panel) and quantification (right panel) demonstrate that cortical BK α expression is unchanged between Wnk1+/+ (n = 4) and Wnk1+/delE631 mice (n = 5). (D) Basal NKCC2 and P-NKCC2 protein abundance in Wnk1+/delE631 mice and Wnk1+/+ littermates. (n = 7 per group). NKCC2 and pNKCC2 immunoblots (left panel) and quantification (right panel) demonstrate that NKCC expression is unchanged between Wnk1+/+ (n = 7) and Wnk1+/delE631 mice (n = 7), but pNKCC2 is significantly increased in Wnk1+/delE631 mice compared with Wnk1+/+. ****P < 0.0001, unpaired Student’s t test.
We also studied the large-conductance Ca++-activated K+ (BK) channel, the expression of which was unchanged. Likewise, the protein abundance of the Na-K-2Cl cotransporter (NKCC2) (Figure 9, C and D), which has been shown to be regulated in vivo by KS-WNK1 (31), was unchanged. In addition, a significant increase in NKCC2 phosphorylation was observed (Figure 9D) in accordance with the observed increase in SPAK and OSR1, the main kinases involved in the phosphorylation of NKCC2 (32), and opposite of the expected inhibitory effect KS-WNK1. We did not detect an increase in KS-WNK1 in NKCC2-positive tubules (Figure 7), suggesting that other regulatory mechanisms are at play (33).
Discussion
This is the first report, to our knowledge, of missense mutations in the WNK1 gene leading to an autosomal dominant tubulopathy. The syndrome is striking in that it resembles more an isolated form of hyperkalemic renal distal tubular acidosis than a classic FHHt. Our cases are very similar to the 3 published cases of Spitzer-Weinstein syndrome, characterized by normotension and early onset hyperkalemic tubular acidosis and sensitive to HCTZ (34–36). The mutations are all located in the acidic motif, which is highly conserved across members of the WNK family and pivotal for their recruitment by KLHL proteins for ubiquitination by the CUL3-based E3 ligase ubiquitin complex (5, 37). Our in vitro experiments suggest that WNK1 mutations result in a differential regulation of the 2 major WNK1 isoforms, with an increase in abundance of KS-WNK1 being preferentially affected compared with the kinase-active L-WNK1.
Since the discovery that mutations in KLHL3 and CUL3 cause FHHt (3, 38), several studies have demonstrated that the WNK kinases are substrates for the KLHL3-CUL3 E3 ligase complex (37). Ohta et al. mapped the WNK interaction site to a region containing a motif of 10 amino acids, EPEEPEADQH, called the acidic motif because of the predominance of negatively charged residues (5). Most of the FHHt-causing mutations in WNK4 cluster in this highly conserved motif (2). The analysis of the Kelch domain of KLHL3 crystal structure in complex with the WNK4 acidic motif revealed close polar contacts between several residues at this motif and other conserved residues at the surface of the Kelch domain β-propeller (14). In particular, the D635 residue of L-WNK1 (equivalent to D564 in WNK4) establishes ionic interactions with R528 of KLHL3, both residues being mutated in FHHt. Mutations in this degron motif abolish the interaction of WNK4 with the ubiquitin-ligase complex, thereby preventing their ubiquitination and proteasomal degradation. Accordingly, WNK4 protein abundance increases in the kidney of WNK4+/D561A mice, which carried 1 of the FHHt mutations (7, 39).
We found that KS-WNK1 abundance is much more affected by the acidic motif mutations than L-WNK1. While the expression of KLHL3 only slightly reduced L-WNK1 abundance, the expression of KS-WNK1 was drastically decreased in both X. laevis oocytes and mammalian cells (HEK293T). We also found that KS-WNK1 is more ubiquitinated than L-WNK1 when KLHL3 is overexpressed. Each of the mutations in the acidic motif abolished the degradation of KS-WNK1, with the exception of KS-WNK1 D228E. Ohta et al. previously demonstrated that L-WNK1 interacts with KLHL3 and that this interaction is prevented by mutations in either KLHL3 or the acidic motif of L-WNK1 (5). However, the effect of KLHL3 overexpression on KS-WNK1 abundance and ubiquitination was not tested. Therefore, the potential difference in sensitivity of WNK1 isoforms to KLH3-induced degradation was not analyzed. The acidic motif is present in both L- and KS-WNK1 isoforms. Thus, one might have expected that the missense mutations would have the same consequence on both isoforms. However, the 2 proteins have a different 3D structure, the conformation of which is probably regulated by different factors. Piala et al. showed that L-WNK1 is maintained in an inactive conformation by the binding of chloride to the catalytic site (39). KS-WNK1 is insensitive to this regulation because it lacks the major part of the kinase domain. Under physiological conditions, KS-WNK1 mRNA is markedly more abundant than L-WNK1 in the distal nephron (13, 40), but data on KS-WNK1 protein abundance are still lacking, since there are no specific antibodies against this isoform. Our results obtained in both Xenopus oocytes and HEK293T cells suggest that KS-WNK1 protein could be physiologically degraded in the distal nephron by ubiquitination through the KLHL3-CUL3 E3 ligase complex.
The present study suggests that mutations in the WNK1 acidic motif drive altered potassium metabolism primarily by increasing KS-WNK1 abundance. The most parsimonious explanation for the phenotype is that the selective increase in KS-WNK1 abundance directly stimulates NCC activity and abrogates potassium-dependent upregulation of ROMK. The explanation is consistent with recent observations indicating that KS-WNK1 is critical for the coalescence of WNK-SPAK signaling molecules into large membraneless structures in the DCT, called WNK bodies (24), which appear to be key intermediates in WNK-SPAK phosphorylation of NCC, particularly during dietary potassium restriction (41). Because binding of KS-WNK1 to WNK4 increases the phosphorylation of the WNK4 T-loop (23) and the WNK bodies become larger in DCT of Wnk1+/delE631 mice, we speculate that increased abundance of KS-WNK1 in Wnk1+/delE631 mice drives phosphorylation of NCC through activation of the WNK4-SPAK cascade in WNK bodies. As evidenced by the rapid correction of hyperkalemia in Wnk1+/delE631 mice and patients with HCTZ, the aberrant activation of NCC most likely contributes to urinary potassium retention by reducing sodium delivery to ASDN.
The fact that an increased KS-WNK1 abundance stimulates NCC expression and phosphorylation in the Wnk1+/delE631 mice might be considered surprising, given that the same phenomenon was observed in mice carrying an inactivation of KS-WNK1 (KS-Wnk1–/– mice; ref. 42). However, given the information raised in the last 10 years, we now believe that increased NCC activity in KS-Wnk1–/– mice is not directly caused by KS-WNK1 inactivation, but is rather the consequence of the potassium-losing phenotype. The latter could be caused by the increased ROMK expression observed in the DCT2/CNT of these mice and associated with a decrease in aldosterone secretion in the absence of decreased renin expression (our unpublished observations). This new interpretation is supported by the fact that all the other mouse models in which NCC is stimulated display hypertension and hyperkalemia (Wnk4-PHAII transgenic mice ref. 19; WNK4D561A/+ knockin mice, ref. 18; KLHL3–/– mice, ref. 43, KLHL3R582/+ mice, ref. 44, and constitutively active SPAK mutant mice, ref. 45). In addition, our original interpretation of the phenotype resulting from KS-WNK1 inactivation (42) was guided by the early oocyte studies, which suggested that this WNK1 isoform exerted a dominant negative effect on L-WNK1 (46). However, we have since shown that an unfortunate mutation in the L- and KS-WNK1 cDNA used in these oocyte studies caused spurious loss of function (47). A newer study in oocytes, using the corrected cDNA, now indicates that KS-WNK1 in an activator of WNK4 (48).
Based on our observation that Wnk1+/delE631 mice fail to properly upregulate ROMK in the setting of hyperkalemia, it seems likely that KS-WNK1 inhibits pathways that normally upregulate ROMK in the DCT2/CNT and this contributes to the potassium-retention phenotype. This conclusion is consistent with observations in mice that KS-WNK1 gene ablation causes apical membrane expression of ROMK to increase in the DCT2 and CNT (42), but does not easily translate into an understanding of how KS-WNK1 physiologically regulates ROMK. However, recent patch-clamp studies in mice demonstrated that KS-WNK1 is necessary for the physiological stimulation of ROMK currents in response to a dietary potassium load (49), a condition known to stimulate KS-WNK1 transcription (40). We speculate that KS-WNK1 may affect ROMK differently depending on the circumstance and where it is expressed. In Wnk1+/delE631 mice and patients, overexpressed KS-WNK1 in the DCT may indirectly inhibit ROMK in the CNT through distal nephron remodeling processes, as have been observed in a DCT-specific mouse model of FHHt (45). But when KS-WNK1 is physiologically activated in the CNT, its direct positive effects on ROMK would be completely opposite. Obviously, a test of these ideas will require further studies, perhaps with nephron-specific KS-WNK1 knockout and overexpression models.
The absence of hypertension in the patients and Wnk1+/delE631 mice is puzzling, given the increase in NCC phosphorylation. In that regard, it was interesting to compare the phenotypes of our FHHt patients depending on the WNK1 or WNK4 gene affected with similar genetic variants at the acidic motif of the kinase. Despite a relatively small number of affected subjects and difference in median age of the 2 groups (39.5 vs. 23.0 years), it was clear that WNK4 patients had a much higher BP than those with similar mutations at the WNK1 gene (median 163/102 mmHg vs. 111/75 mmHg, respectively). In contrast, these 2 groups had a similar biological profile, characterized by marked hyperkalemia, hyperchloremia, and metabolic acidosis. In addition, the Wnk1+/delE631 mice are normotensive, even on a high–sodium chloride diet. Only homozygous mice displayed a higher basal SBP (Supplemental Table 5). These in vivo observations highly suggest a differential effect on NaCl reabsorption between the 2 genes, independent of NCC stimulation. Interestingly, renin secretion was low in the patients with WNK1 mutations at the acidic motif and renin transcription was reduced in Wnk1+/delE631 mice, as expected from the negative feedback loop of increased intravascular volume. However, ENaC expression was unchanged and natriuresis sensitivity to acute amiloride was not affected in mutant mice, suggesting that the hypertensive phenotype in FHHt may require increased ENaC activity. Finally, BP remained lower in Wnk1+/delE631 mice fed a low K+ diet compared with WT littermates, demonstrating that the hyperkalemia was not responsible for the renin inhibition. Taken together, these results suggest that Wnk1+/delE631 mice and the patients display a mild hypervolemic state and that the full understanding of the mechanisms underlying the absence of hypertension remain to be identified.
In conclusion, the discovery of mutations within the WNK1 acidic motif and their in vitro and in vivo characterization uncover what we believe are new features of the KS-WNK1 isoform. We found that KS-WNK1 acts as a positive modulator of NCC that is preferentially degraded at the protein level by the KLHL3-CUL3 ubiquitin complex compared with the L-WNK1 isoform. In the pathological conditions observed here, mutations in the acidic motif prevent KLHL3-CUL3 interaction, leading to increased KS-WNK1 abundance, aberrant activation of NCC, and hyperkalemia without hypertension.
Methods
Human genetic studies
Linkage analysis.
Linkage in Ped29 was analyzed using markers generated by the 250K Affymetrix array, as previously performed (38). We excluded nonpolymorphic and low-frequency (minor allele frequency [MAF] < 0.1) SNPs. To decrease the SNPs list to a manageable set of 49,187 SNPs for linkage analysis, we applied a filter using the Bayesian robust linear model with Mahalanobis distance classifier (BRLMM) and only analyzed SNPs with BRLMM = 0.15. We applied the 1 Mb to 1 centimorgan conversion before analysis as recommended (50). Checks for Mendelian errors and parametric linkage analyses were computed by MERLIN (51) under a rare dominant model with full penetrance (100%) and a disease allele frequency of 0.0005.
WES.
IntegraGen performed library preparation, capture, sequencing, and variant detection and annotation. Exons of genomic DNA samples were captured using Agilent in-solution enrichment methodology with their biotinylated oligonucleotides probes library, followed by paired-end 75 bp massively parallel sequencing on Illumina HiSeq 2000 (see ref. 52 for detailed explanation of the process). The bioinfomatics analysis of deep-sequencing data was based on the Illumina pipeline (CASAVA 1.8).
Direct Sanger sequencing
PCR amplification and Sanger sequencing from genomic DNA was performed using standard methods. The sequencing of the WNK1 ex7 and ex25 was performed using the primers listed in Supplemental Table 7A.
In vitro functional characterization
Mutant proteins in X. laevis oocytes.
Vectors used for expression in Xenopus oocytes and HEK293 cells are described in the Supplemental Methods. cRNA for Xenopus oocyte injections were prepared by in vitro transcription of the corresponding human clones after plasmid linearization using the T7 RNA polymerase m MESSAGE kit (Ambion).
We used X. laevis oocytes as an expression system. Oocyte preparation protocol has been described before (53). In brief, oocytes were obtained from female frogs under anesthesia and the follicular layer was removed. After 24 hours, oocytes were injected with 50 nL of H2O alone or containing 0.2 μg/μl of each clone cRNA. After 48 hours of incubation, protein extracts were obtained using lysis buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EGTA, 1 mM EDTA, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1% (wt/vol) Nonidet P-40, 0.27 M sucrose, 0.1% (vol/vol) 2-mercaptoethanol, and protease inhibitors (Complete Tablets; Roche). Protein extracts equivalent to 1 oocyte (60 μg) were resolved using SDS-PAGE and transferred to PVDF membranes for Western blot analysis. The antibodies used were commercial c-myc (catalog 11814150001, Roche/MilliporeSigma) and Flag antibody (M2, catalog A6592; MilliporeSigma) at 1:1000 and 1:5000 as well as commercial anti–β-actin (catalog sc47778; Santa-Cruz Biotechnology Inc.) at 1:2500. Immobilized antigens were detected by chemiluminesence using the Luminata Forte Western HRP substrate (Merck Millipore).
Mutant proteins in HEK293T cells
Cell culture and transfection.
Flp-InTM T-RExTM 293 cells (Invitrogen) were stably transfected with pCDNA5/FRT/(His)6-protein C-Flag-hKLHL3 WT or R528H vector following the manufacturer’s instructions. Conditions of culture and induction are described in Supplemental Methods. For expression of WNK1-myc constructs (pcDNA vectors described in Supplemental Methods) and ubiquitin-HA, cells were transiently transfected using Effectene (QIAGEN) following the manufacturer’s instructions.
Immunoprecipitation.
Cells were harvested 48 hours after transfection, washed in cold PBS, and frozen in liquid nitrogen. Lysis/immunoprecipitation of cell pellets in denaturing or native conditions were performed according to classical procedures described in Supplemental Methods.
Immunoblotting.
Lysates and immunoprecipitates were analyzed by SDS-PAGE (6% or 8% gels), transferred to nitrocellulose membrane, and immunoblotted with primary antibodies including anti-HA (Cell Signaling Technology), anti-myc (clone 9B11, Cell Signaling Technology), anti-protein C (clone hpc4, Roche), rabbit anti-GAPDH (catalog ab37168; Abcam), and goat anti-actin (catalog sc47778; Santa Cruz Biotechnology Inc.). Thereafter, the membranes were incubated with a horseradish peroxidase–conjugated secondary antibody (1:5000 dilution). Images were obtained with chemiluminescence (Pierce ECL Western Blotting Substrate) using a luminescent image analyzer (LAS-4000 mini, Fujifilm) and quantified with MultiGauge software.
Mouse experimental studies
Crispr/Cas9 engineered mice.
In order to produce the c.1905T>A, P.D635E mutation at ex7 of the mouse Wnk1 gene using the CRISPR/Cas9 system, we chose three 20 mer sgRNAs (Supplemental Table 7C), using the CRISPR Design algorithm (http://zlab.bio/guide-design-resources). sgRNAs were produced with Cas9 SmartNuclease RNA System (SBI Ozyme), and quality was controlled by the Experion Automated Electrophoresis System (Bio-Rad).
One-cell zygotes (3 series of 100 oocytes) were coinjected with CAS9WT protein (New England Biolabs), 1 of the 3 sgRNAs, and a single-strand 160 mer nucleotide (ssODN) carrying the acidic motif mutation and 80 bases of flanking sequence on each side of the cut (ref. 54 and Supplemental Table 7C). This ssODN has been designed to achieve high HDR efficiency; sgRNA/CAS9 system efficiency was validated in vitro by PCR digestion. About 10% of injected zygotes (30 blastocysts) were amplified by PCR (primers in Supplemental Table 7C) and analyzed by high-resolution melting (HRM), droplet digital PCR (ddPCR) (Bio-Rad), and sequencing to identify indel or mutations. After birth, mouse biopsies were analyzed by sequencing.
Radiotelemetry monitoring of BP and heart rate.
The study was performed on 14 C57BL6J male mice divided in 2 subgroups (7 control Wnk1+/+, 7 Wnk1+/delE631 mice). Mice were implanted with BP and heart-rate measuring telemetric probes (DSI). The precise monitoring of the animals is described in Supplemental Methods.
Basal conditions.
Animals were housed in metabolic cages and fed a standard diet (0.3% NaCl) with free access to tap water. After a 3-day adaptation period, urine was collected daily for electrolyte measurements for 2 days.
Diets.
The standard diet contained 0.3% NaCl and 1% KCl; low K diet contained 0.3% NaCl and 0% KCl; high-salt diet contained 3% NaCl and 1% KCl.
Amiloride.
After a 3-day adaptation period, physiological saline was injected intraperitoneally at 10:00 am and urine was collected 6 hours after the injection. The same procedure was repeated the day after with an injection of amiloride (1.45 mg/kg body weight) dissolved in physiological saline. For the analysis of the variations in physiological ROMK expression response to blood potassium increase as well as for the measurement of TTKG, amiloride was administered intraperitoneally at the same dose over 4 days.
HCTZ.
Animals were fed with a specific diet supplemented with HCTZ (240 mg/kg/d, oral) over 4 to 7 days such as done in previous studies (13, 19) for analysis of BP response and analysis of TTKG variations.
Metolazone.
Metolazone (50 μg/kg/d) was administered intraperitoneally over 4 days.
Blood and urine measurements.
Plasma electrolytes were measured using an i-STAT system (Abbott) and EC-8+ cartridge test. Blood samples were collected under isoflurane anesthesia by retroorbital puncture. Creatinine was determined using an Olympus AU400 analyzer. Urinary aldosterone was measured by competitive chemiluminescent immunoassay (LIAISON Aldosterone Kit, Diasorin). TTKG was calculated as follows (26): TTKG = ([K+]urine/[K+]blood) × (osmblood/osmurine), as has been used widely before in animal model studies to estimate the potassium secretory capacity of the distal nephron. Although urea recycling has been argued as making TTKG greater than the actual value in vivo (55), the error is small (~10%) because the urea reabsorption is offset by passive potassium reabsorption in the inner medullary collecting duct (56). This is in agreement with experimental measurements that have validated the TTKG calculation as a reasonable estimate of potassium secretory capacity in rodents (57). Although we cannot rule out that urea recycling or passive potassium reabsorption is affected by the WNK1 ex7 mutation, it seems unlikely. Nevertheless, we acknowledge this as a limitation.
RNA extraction and RT-qPCR for renin expression.
The extraction of total RNA from mouse kidneys, reverse transcription and quantitative PCR (RT-qPCR) were performed as described in ref. 12. Primer sequences are indicated in Supplemental Table 7D. Ubiquitin c (ubc) was used a reference gene, and comparative quantification of the gene of interest between the 2 genotypes was obtained using the 2−ΔΔCt method.
Mouse kidney immunoblotting.
At the end of the experimental period, animals were sacrificed with ketamine and xylazine (0.1 and 0.01 mg/g of body weight, respectively). Renal cortex or total renal samples were homogenized, extracted, and submitted to SDS-PAGE electrophoresis, and immunoblotting was performed as detailed in Supplemental Methods.
WNK1 and ROMK1 immunolocalization in the cortical nephron.
WNK1 and ROMK1 immunofluorescence experiments were performed on kidneys embedded in paraffin. All technical details are given in Supplemental Methods.
Statistics
Human studies.
Data were analyzed using Fisher’s exact and t tests. All tests were 2 sided. P values of less than 0.05 were considered significant.
Mouse studies.
When analyzing 2 groups of mice, we used an unpaired Student’s t test. One-way ANOVA followed by Holm-Šidák’s multiple comparisons test was used to analyze more than 2 groups of 5 or more while a Kruskal-Wallis test followed by a Mann-Whitney U test was used to analyze more than 2 groups of 5 or less. Data are given as mean ± SEM. Differences between groups were considered significant at P < 0.05.
Study approval
Affected individuals were recruited at the Department of Genetics of the Hôpital Européen Georges Pompidou as well as at other Departments of Nephrology located in France, Italy, and the United Kingdom. Genetic testing and research were performed according to the French ethic laws published in 2001 (articles L.1110-4 al 1 CSP and R 1131-14 CSP). Informed, written consent was obtained from all study participants. All mouse studies were conducted on 3- to 5-month-old male mice and performed in accordance with the European Communities Council Directive. The project was approved by the French Ministry of Research (no. 02650.02). Work with X. laevis was approved and performed following the guidelines set by the Institutional Animal Care Committee of our institution. The Xenopus oocytes studies were approved by the IACUC from the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán in Mexico City.
Author contributions
HLDP and NT conducted the human genetic studies, IK, WAK, CR, TM, and MCC performed the in vitro cellular experiments from mouse models. ERA, SV, OS, and GG performed the Xenopus oocytes studies. RC, RG, and PAW performed the ROMK1 experiments. ILF, MDC, SB, TM, CR, and JH conducted in vivo experiments from mouse studies. MH, SD, XG, KO, PM, GR, IT, RU, and RVP acquired and analyzed clinical human data. PAW, GG, EC, JH, and XJ designed the research studies, assembled the figures and tables, and wrote the manuscript. XJ coordinated the entire study.
Supplementary Material
Acknowledgments
We thank all patients and families for their participation in the study. We acknowledge the contributions of additional clinicians for the clinical and biochemical characterization of FHHt patients, especially Béatrice Fiquet and Geneviève Beaurain (Paris, France), Patrick Giraud (Montauban, France), and Silvia Monticone (Torino, Italy). We also thank Valérie Boccio for her contribution to the genetic analysis of the WNK1 gene. This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM); the Fondation pour la Recherche pour l’Hypertension Artérielle (to XJ and JH); the Transatlantic Network on Hypertension funded by the Fondation Leducq (to XJ, GG, JH, and OS); the Transatlantic Network on Potassium and Hypertension funded by the Fondation Leducq (to PAW and OS); and the Agence Nationale pour la Recherche (ANR) (grant to JH and GG). This study also received funding from the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305608 (EURenOmics). IK was supported by a grant from Institut de Recherche Servier, and PM is supported by a grant from MIUR (ex-60% 2016 and 2017). This study was also supported by grants no. A1-S-8290 from the Mexican Council of Science and Technology (CONACYT) and no. IN201519 from PAPIIT-DGAPA (to GG) and collaborative funding from CONACYT–Agence Nationale pour la Recherche (Conacyt 188712-ANR-12-ISVS1-0001-01 to GG and JH). OS and SV were supported by Swiss National Science Foundation grant 310030-141013 and the NCCR-Kidney.ch (Swiss National Science Foundation). PAW was supported by grants from the NIH (DK054231 and DK093501).
Version 1. 08/13/2020
In-Press Preview
Version 2. 10/26/2020
Electronic publication
Version 3. 10/28/2020
Corrected affiliation number 5
Version 4. 12/01/2020
Print issue publication
Funding Statement
GRANT NUMBER : FP7 / 2007-2017
Grants No. 165815 from the Mexican Council of Science and Technology, IN200513 from PAPIIT-DGAPA
CONACYT - Agence Nationale pour la Recherche (Conacyt 188712-ANR-12-ISVS1-0001-01)
Footnotes
Conflict of interest: RU is currently working in early clinical development and early cardiovascular, renal, and metabolism studies at R&D BioPharmaceuticals and AstraZeneca.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6379–6394.https://doi.org/10.1172/JCI94171.
Contributor Information
Ilektra Kouranti, Email: ilektra.kouranti@gmail.com.
Irmine Loisel-Ferreira, Email: irmine.ferreira@inserm.fr.
Maria Chavez-Canales, Email: maria.chc@gmail.com.
Sarah Vacle, Email: sarah.vacle@unil.ch.
Tiffany Migeon, Email: tiffany.migeon@sorbonne-universite.fr.
Richard Coleman, Email: rcolem15@jhmi.edu.
Marcio Do Cruzeiro, Email: marcio.do-cruzeiro@inserm.fr.
Marguerite Hureaux, Email: marguerite.hureaux@inserm.fr.
Nirubiah Thurairajasingam, Email: nirubiah.thurairajasingam@gmail.com.
Stéphane Decramer, Email: decramer.s@chu-toulouse.fr.
Xavier Girerd, Email: xavier.girerd@aphp.fr.
Paolo Mulatero, Email: paolo.mulatero@libero.it.
Ivan Tack, Email: tack.i@chu-toulouse.fr.
Rosa Vargas-Poussou, Email: rosa.vargas@aphp.fr.
Olivier Staub, Email: Olivier.staub@unil.ch.
Paul A. Welling, Email: pwellin1@jhmi.edu.
Gerardo Gamba, Email: gamba@biomedicas.unam.mx.
Eric Clauser, Email: Eric.clauser@inserm.fr.
Juliette Hadchouel, Email: juliette.hadchouel@inserm.fr.
Xavier Jeunemaitre, Email: xavier.jeunemaitre@inserm.fr.
References
- 1. Gordon RD, et al. In: Brenner JHLaBM ed. Hypertension: Pathology, Diagnosis and Management. Raven Press Ltd; 1995:2111–2113. [Google Scholar]
- 2.Wilson FH, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293(5532):1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 3.Boyden LM, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482(7383):98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis-Dit-Picard H, Hadchouel J, Jeunemaitre X. [KLHL3 and CULLIN-3: new genes involved in familial hypertension] Med Sci (Paris) 2012;28(8–9):703–706. doi: 10.1051/medsci/2012288010. [DOI] [PubMed] [Google Scholar]
- 5.Ohta A, et al. The CUL3-KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J. 2013;451(1):111–122. doi: 10.1042/BJ20121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP. Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci U S A. 2013;110(19):7838–7843. doi: 10.1073/pnas.1304592110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakabayashi M, et al. Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep. 2013;3(3):858–868. doi: 10.1016/j.celrep.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Wu G, Peng JB. Disease-causing mutations in KLHL3 impair its effect on WNK4 degradation. FEBS Lett. 2013;587(12):1717–1722. doi: 10.1016/j.febslet.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Achard JM, Disse-Nicodeme S, Fiquet-Kempf B, Jeunemaitre X. Phenotypic and genetic heterogeneity of familial hyperkalaemic hypertension (Gordon syndrome) Clin Exp Pharmacol Physiol. 2001;28(12):1048–1052. doi: 10.1046/j.1440-1681.2001.03575.x. [DOI] [PubMed] [Google Scholar]
- 10.Hadchouel J, Delaloy C, Faure S, Achard JM, Jeunemaitre X. Familial hyperkalemic hypertension. J Am Soc Nephrol. 2006;17(1):208–217. doi: 10.1681/ASN.2005030314. [DOI] [PubMed] [Google Scholar]
- 11.Delaloy C, et al. Multiple promoters in the WNK1 gene: one controls expression of a kidney-specific kinase-defective isoform. Mol Cell Biol. 2003;23(24):9208–9221. doi: 10.1128/MCB.23.24.9208-9221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal-Petiot E, et al. A new methodology for quantification of alternatively spliced exons reveals a highly tissue-specific expression pattern of WNK1 isoforms. PLoS One. 2012;7(5):e37751. doi: 10.1371/journal.pone.0037751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidal-Petiot E, et al. WNK1-related familial hyperkalemic hypertension results from an increased expression of L-WNK1 specifically in the distal nephron. Proc Natl Acad Sci U S A. 2013;110(35):14366–14371. doi: 10.1073/pnas.1304230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher FR, Sorrell FJ, Alessi DR, Bullock AN, Kurz T. Structural and biochemical characterization of the KLHL3-WNK kinase interaction important in blood pressure regulation. Biochem J. 2014;460(2):237–246. doi: 10.1042/BJ20140153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuermser LA, Reilly C, Poindexter JR, Sakhaee K, Pak CY. Potassium-magnesium citrate versus potassium chloride in thiazide-induced hypokalemia. Kidney Int. 2000;57(2):607–612. doi: 10.1046/j.1523-1755.2000.00881.x. [DOI] [PubMed] [Google Scholar]
- 16.Matos V, van Melle G, Boulat O, Markert M, Bachmann C, Guignard JP. Urinary phosphate/creatinine, calcium/creatinine, and magnesium/creatinine ratios in a healthy pediatric population. J Pediatr. 1997;131(2):252–257. doi: 10.1016/S0022-3476(97)70162-8. [DOI] [PubMed] [Google Scholar]
- 17.Sealey JE, Clark I, Bull MB, Laragh JH. Potassium balance and the control of renin secretion. J Clin Invest. 1970;49(11):2119–2127. doi: 10.1172/JCI106429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang SS, et al. Molecular pathogenesis of pseudohypoaldosteronism type II: generation and analysis of a Wnk4(D561A/+) knockin mouse model. Cell Metab. 2007;5(5):331–344. doi: 10.1016/j.cmet.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Lalioti MD, et al. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat Genet. 2006;38(10):1124–1132. doi: 10.1038/ng1877. [DOI] [PubMed] [Google Scholar]
- 20.Rafiqi FH, et al. Role of the WNK-activated SPAK kinase in regulating blood pressure. EMBO Mol Med. 2010;2(2):63–75. doi: 10.1002/emmm.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang SS, et al. SPAK-knockout mice manifest Gitelman syndrome and impaired vasoconstriction. J Am Soc Nephrol. 2010;21(11):1868–1877. doi: 10.1681/ASN.2009121295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick JA, Ellison DH. The WNKs: atypical protein kinases with pleiotropic actions. Physiol Rev. 2011;91(1):177–219. doi: 10.1152/physrev.00017.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rafael C, Soukaseum C, Baudrie V, Frere P, Hadchouel J. Consequences of SPAK inactivation on hyperkalemic hypertension caused by WNK1 mutations: evidence for differential roles of WNK1 and WNK4. Sci Rep. 2018;8(1):3249. doi: 10.1038/s41598-018-21405-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd-Shiwarski CR, et al. Potassium-regulated distal tubule WNK bodies are kidney-specific WNK1 dependent. Mol Biol Cell. 2018;29(4):499–509. doi: 10.1091/mbc.E17-08-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson MN, Schneider W, Mutig K, Ellison DH, Kettritz R, Bachmann S. Patients with hypokalemia develop WNK bodies in the distal convoluted tubule of the kidney. Am J Physiol Renal Physiol. 2019;316(2):F292–F300. doi: 10.1152/ajprenal.00464.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi MJ, Ziyadeh FN. The utility of the transtubular potassium gradient in the evaluation of hyperkalemia. J Am Soc Nephrol. 2008;19(3):424–426. doi: 10.1681/ASN.2007091017. [DOI] [PubMed] [Google Scholar]
- 27.Richardson C, et al. Regulation of the NKCC2 ion cotransporter by SPAK-OSR1-dependent and -independent pathways. J Cell Sci. 2011;124(Pt 5):789–800. doi: 10.1242/jcs.077230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughey RP, et al. Maturation of the epithelial Na+ channel involves proteolytic processing of the alpha- and gamma-subunits. J Biol Chem. 2003;278(39):37073–37082. doi: 10.1074/jbc.M307003200. [DOI] [PubMed] [Google Scholar]
- 29.Masilamani S, Kim GH, Mitchell C, Wade JB, Knepper MA. Aldosterone-mediated regulation of ENaC alpha, beta, and gamma subunit proteins in rat kidney. J Clin Invest. 1999;104(7):R19–R23. doi: 10.1172/JCI7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wade JB, et al. WNK1 kinase isoform switch regulates renal potassium excretion. Proc Natl Acad Sci U S A. 2006;103(22):8558–8563. doi: 10.1073/pnas.0603109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Xie J, Wu T, Truong T, Auchus RJ, Huang CL. Downregulation of NCC and NKCC2 cotransporters by kidney-specific WNK1 revealed by gene disruption and transgenic mouse models. Hum Mol Genet. 2011;20(5):855–866. doi: 10.1093/hmg/ddq525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castrop H, Schiessl IM. Physiology and pathophysiology of the renal Na-K-2Cl cotransporter (NKCC2) Am J Physiol Renal Physiol. 2014;307(9):F991–F1002. doi: 10.1152/ajprenal.00432.2014. [DOI] [PubMed] [Google Scholar]
- 33.Cheng CJ, Truong T, Baum M, Huang CL. Kidney-specific WNK1 inhibits sodium reabsorption in the cortical thick ascending limb. Am J Physiol Renal Physiol. 2012;303(5):F667–F673. doi: 10.1152/ajprenal.00290.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolis BL, Lifschitz MD. The Spitzer-Weinstein syndrome: one form of type IV renal tubular acidosis and its response to hydrochlorothiazide. Am J Kidney Dis. 1986;7(3):241–244. doi: 10.1016/S0272-6386(86)80011-7. [DOI] [PubMed] [Google Scholar]
- 35.Spitzer A, Edelmann CM, Jr, Goldberg LD, Henneman PH. Short stature, hyperkalemia and acidosis: A defect in renal transport of potassium. Kidney Int. 1973;3(4):251–257. doi: 10.1038/ki.1973.38. [DOI] [PubMed] [Google Scholar]
- 36.Weng CH, Cheng JW, Hung CC, Wu MS, Yang CW, Chang CT. A young female with Spitzer-Weinstein syndrome diagnosed by thiazide test. Ren Fail. 2007;29(2):239–241. doi: 10.1080/08860220601100510. [DOI] [PubMed] [Google Scholar]
- 37.Uchida S, Chiga M, Sohara E, Rai T, Sasaki S. Does a β2-adrenergic receptor-WNK4-Na-Cl co-transporter signal cascade exist in the in vivo kidney? Nat Med. 2012;18(9):1324–1325. doi: 10.1038/nm.2809. [DOI] [PubMed] [Google Scholar]
- 38.Louis-Dit-Picard H, et al. KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet. 2012;44(4):456–460. doi: 10.1038/ng.2218. [DOI] [PubMed] [Google Scholar]
- 39.Piala AT, Moon TM, Akella R, He H, Cobb MH, Goldsmith EJ. Chloride sensing by WNK1 involves inhibition of autophosphorylation. Sci Signal. 2014;7(324):ra41. doi: 10.1126/scisignal.2005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Reilly M, et al. Dietary electrolyte-driven responses in the renal WNK kinase pathway in vivo. J Am Soc Nephrol. 2006;17(9):2402–2413. doi: 10.1681/ASN.2005111197. [DOI] [PubMed] [Google Scholar]
- 41.Thomson MN, et al. WNK bodies cluster WNK4 and SPAK/OSR1 to promote NCC activation in hypokalemia. Am J Physiol Renal Physiol. 2020;318(1):F216–F28. doi: 10.1152/ajprenal.00232.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadchouel J, et al. Decreased ENaC expression compensates the increased NCC activity following inactivation of the kidney-specific isoform of WNK1 and prevents hypertension. Proc Natl Acad Sci U S A. 2010;107(42):18109–18114. doi: 10.1073/pnas.1006128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki E, et al. KLHL3 knockout mice reveal the physiological role of KLHL3 and the pathophysiology of pseudohypoaldosteronism type II caused by mutant KLHL3. Mol Cell Biol. 2017;37(7):e00508–e00516. doi: 10.1128/MCB.00508-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Susa K, et al. Impaired degradation of WNK1 and WNK4 kinases causes PHAII in mutant KLHL3 knock-in mice. Hum Mol Genet. 2014;23(19):5052–5060. doi: 10.1093/hmg/ddu217. [DOI] [PubMed] [Google Scholar]
- 45.Grimm PR, Coleman R, Delpire E, Welling PA. Constitutively active SPAK causes hyperkalemia by activating NCC and remodeling distal tubules. J Am Soc Nephrol. 2017;28(9):2597–2606. doi: 10.1681/ASN.2016090948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanya AR, Yang CL, Zhu X, Ellison DH. Dominant-negative regulation of WNK1 by its kidney-specific kinase-defective isoform. Am J Physiol Renal Physiol. 2006;290(3):F619–24. doi: 10.1152/ajprenal.00280.2005. [DOI] [PubMed] [Google Scholar]
- 47.Chavez-Canales M, et al. WNK-SPAK-NCC cascade revisited: WNK1 stimulates the activity of the Na-Cl cotransporter via SPAK, an effect antagonized by WNK4. Hypertension. 2014;64(5):1047–1053. doi: 10.1161/HYPERTENSIONAHA.114.04036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Argaiz ER, Chavez-Canales M, Ostrosky-Frid M, Rodriguez-Gama A, Vazquez N, Gonzalez-Rodriguez X, et al. Kidney-specific WNK1 isoform (KS-WNK1) is a potent activator of WNK4 and NCC. Am J Physiol Renal Physiol. 2018;315(3):F734–F745. doi: 10.1152/ajprenal.00145.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu P, Gao ZX, Su XT, Ellison DH, Hadchouel J, Teulon J, et al. Role of WNK4 and kidney-specific WNK1 in mediating the effect of high dietary K+ intake on ROMK channel in the distal convoluted tubule. Am J Physiol Renal Physiol. 2018;315(2):F223–F230. doi: 10.1152/ajprenal.00050.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulgen A, Li W. Comparing single-nucleotide polymorphism marker-based and microsatellite marker-based linkage analyses. BMC Genet. 2005;6(Suppl 1):S13. doi: 10.1186/1471-2156-6-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 52.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monroy A, Plata C, Hebert SC, Gamba G. Characterization of the thiazide-sensitive Na(+)-Cl(-) cotransporter: a new model for ions and diuretics interaction. Am J Physiol Renal Physiol. 2000;279(1):F161–F169. doi: 10.1152/ajprenal.2000.279.1.F161. [DOI] [PubMed] [Google Scholar]
- 54.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8(9):753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamel KS, Halperin ML. Intrarenal urea recycling leads to a higher rate of renal excretion of potassium: an hypothesis with clinical implications. Curr Opin Nephrol Hypertens. 2011;20(5):547–554. doi: 10.1097/MNH.0b013e328349b8f9. [DOI] [PubMed] [Google Scholar]
- 56.Wen D, et al. Relation between BK-α/β4-mediated potassium secretion and ENaC-mediated sodium reabsorption. Kidney Int. 2014;86(1):139–145. doi: 10.1038/ki.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West ML, et al. Development of a test to evaluate the transtubular potassium concentration gradient in the cortical collecting duct in vivo. Miner Electrolyte Metab. 1986;12(4):226–233. [PubMed] [Google Scholar]
- 58.Mayan H, et al. Hypercalciuria in familial hyperkalemia and hypertension accompanies hyperkalemia and precedes hypertension: description of a large family with the Q565E WNK4 mutation. J Clin Endocrinol Metab. 2004;89(8):4025–4030. doi: 10.1210/jc.2004-0037. [DOI] [PubMed] [Google Scholar]
- 59.Golbang AP, et al. A new kindred with pseudohypoaldosteronism type II and a novel mutation (564D>H) in the acidic motif of the WNK4 gene. Hypertension. 2005;46(2):295–300. doi: 10.1161/01.HYP.0000174326.96918.d6. [DOI] [PubMed] [Google Scholar]
- 60.Gong H, et al. A patient with pseudohypoaldosteronism type II caused by a novel mutation in WNK4 gene. Endocrine. 2008;33(3):230–234. doi: 10.1007/s12020-008-9084-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.