Abstract
The transcription factor IFN regulatory factor 5 (IRF5) is a central mediator of innate and adaptive immunity. Genetic variations within IRF5 are associated with a risk of systemic lupus erythematosus (SLE), and mice lacking Irf5 are protected from lupus onset and severity, but how IRF5 functions in the context of SLE disease progression remains unclear. Using the NZB/W F1 model of murine lupus, we show that murine IRF5 becomes hyperactivated before clinical onset. In patients with SLE, IRF5 hyperactivation correlated with dsDNA titers. To test whether IRF5 hyperactivation is a targetable function, we developed inhibitors that are cell permeable, nontoxic, and selectively bind to the inactive IRF5 monomer. Preclinical treatment of NZB/W F1 mice with an inhibitor attenuated lupus pathology by reducing serum antinuclear autoantibodies, dsDNA titers, and the number of circulating plasma cells, which alleviated kidney pathology and improved survival. Clinical treatment of MRL/lpr and pristane-induced lupus mice with an inhibitor led to significant reductions in dsDNA levels and improved survival. In ex vivo human studies, the inhibitor blocked SLE serum–induced IRF5 activation and reversed basal IRF5 hyperactivation in SLE immune cells. We believe this study provides the first in vivo clinical support for treating patients with SLE with an IRF5 inhibitor.
Keywords: Autoimmunity, Immunology
Keywords: Autoimmune diseases
Introduction
Systemic lupus erythematosus (SLE) is a debilitating systemic autoimmune disease characterized by elevated levels of circulating antinuclear autoantibodies (ANAs) and severe immune dysregulation. Immune dysregulation may be conferred by genetic susceptibility and/or environmental triggers. In the past 50 years, only 1 new drug has been approved for the treatment of SLE, the monoclonal antibody belimumab; however, global immunosuppression to control disease activity remains the standard of care. Thus, extensive efforts are underway to develop drugs against targets involved in disease progression. One such new target is IFN regulatory factor 5 (IRF5), a member of the IRF family of transcription factors. IRF5 was originally identified as a regulator of type I IFNs (IFNs) and IFN-stimulated genes (ISGs) in response to virus infection (1–3). Subsequent studies revealed important roles for IRF5 in innate and adaptive immunity, macrophage polarization, cell growth regulation, and apoptosis (4, 5). IRF5 was later identified as an autoimmune susceptibility gene. IRF5 polymorphisms associate with autoimmune and inflammatory conditions, including inflammatory bowel disease, primary biliary cirrhosis, rheumatoid arthritis, SLE, and systemic sclerosis (6–11). The most well studied is the role of IRF5 in SLE pathogenesis, and a common characteristic among patients with SLE is increased expression of inflammatory cytokines and type I IFNs that contribute to sustained and persistent autoimmunity (12–17). IRF5 expression is significantly elevated in PBMCs from SLE patients with SLE compared with PBMCs from age-matched healthy donors (18), and IRF5 was found to be constitutively activated, i.e., nuclear localized, in SLE monocytes (19). These findings, which implicate IRF5 dysfunction in SLE pathogenesis, are supported by multiple models of murine lupus showing that mice lacking Irf5 (Irf5–/–) are protected from disease onset and severity (11, 20–26). Equally important and relevant to the therapeutic potential of IRF5 is the finding that lupus disease is abrogated in Irf5+/– mice, indicating that a reduction in IRF5 expression and/or activity by only half is sufficient for therapeutic effect (21, 24).
Although the mechanism or mechanisms by which IRF5 contributes to disease pathogenesis remain unclear, much of the data point to its role in regulating the expression of proinflammatory cytokines, including IFN-α, IL-6, TNF-α, and IL-12, as well as pathogenic autoantibody production (3, 5, 11, 21–28). Dysregulation of many of these cytokines is associated with disease pathogenesis, and IRF5 is predominantly expressed in immune cells (monocytes, DCs, and B cells) responsible for their production (29). In an unstimulated cell, IRF5 is localized in the cytoplasm as an inactive monomer (30). While in the inactive conformation, the C-terminal autoinhibitory domain (AID) of IRF5 is thought to either mask the N-terminal DNA-binding domain (DBD) and/or the C-terminal protein interaction domain (IAD) that is required for homo- or heterodimerization (30, 31). Upon activation by posttranslational modification events downstream of TLRs, DNA damage, or other antigenic signaling cascades, IRF5 undergoes a conformational change that exposes the IAD for dimerization and nuclear localization signals (NLSs) for translocation (1, 30–32). Although a significant body of in vitro work suggests that this conformational shift is dependent on phosphorylation of C-terminal serine (Ser) residues by activating kinases (33–35), nuclear translocation remains the essential regulatory step that mediates IRF5 transcriptional activity (1, 30).
Identification of IRF5 as a global risk factor for autoimmune and inflammatory diseases (5, 11, 20, 36–38), coupled with its increased activation in the blood of patients with SLE, indicates that IRF5 is an attractive target for therapeutic inhibition. While C-terminal phosphorylation and dimerization represent steps amenable to inhibition (39), neither has been definitively shown to be an absolute requirement for nuclear translocation (35). An alternate approach to inhibiting IRF5 stems from the finding that either N- or C-terminal regions of IRFs can act as dominant-negative (DN) mutants to block transactivation ability (2, 29, 40–44). Though the mechanism or mechanisms by which DN mutants inhibit IRFs remain unclear, their activity suggests that IRF peptide mimetics may be an effective approach for blocking function. We detail here the ex vivo characterization of IRF5 peptide mimetics in healthy and SLE immune cells and the in vivo characterization in the NZB/W F1, MRL/lpr, and pristane-induced models of murine lupus.
Results
IRF5 hyperactivation in patients with SLE associates with clinical disease activity.
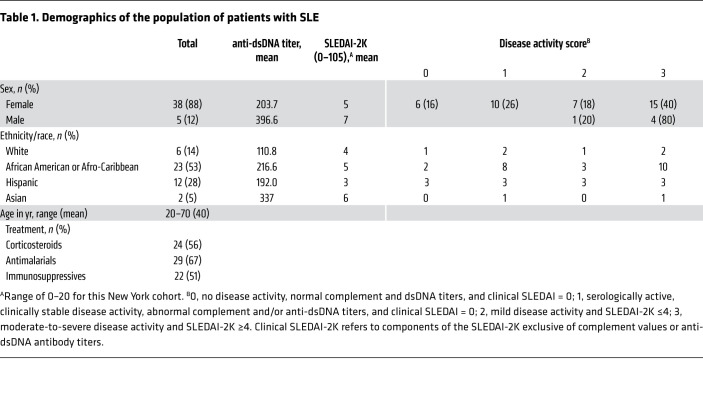
We previously reported elevated IRF5 activation, which we refer to here as IRF5 hyperactivation, in SLE monocytes from a cohort of patients with SLE from Sweden (18). We have extended these original findings in 2 additional independent cohorts of age- and sex-matched patients with SLE and healthy donors from University Hospital in Newark, New Jersey, and Northwell Health in Long Island, New York (Table 1). In agreement with previous work, we detected a significant increase in basal IRF5 hyperactivation in SLE monocytes (CD45+CD14+) as compared with healthy donor monocytes (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI120288DS1). We examined SLE B cells (CD45+CD19+) and found a similarly significant increase (Figure 1B). We attempted to measure IRF5 activation in SLE plasmacytoid DCs (pDCs) but were unable to acquire sufficient cellular events for robust statistical analysis (45). We next recruited patients in different stages of clinical disease activity, which led to the finding of a stage-dependent increase in IRF5 hyperactivation within monocytes and B cells (Figure 1, C and D). Disease activity scoring is defined in Table 1 and in Methods. We further stratified patient data according to the SLE disease activity index (SLEDAI) and dsDNA titers to determine whether IRF5 hyperactivation is associated with either clinical phenotype, as neither alone defines clinically active disease. We found that basal levels of IRF5 activation were significantly higher in monocytes from patients with SLE who had a SLEDAI of 4 or higher compared with those with a SLEDAI of zero, and found no significant difference when we compared with patients with a SLEDAI of 4 or less (Figure 1E). Similarly, we found that IRF5 hyperactivation was significantly elevated in B cells from SLE patients with a SLEDAI of 4 or higher. Although we detected a positive association between IRF5 hyperactivation and increased SLEDAI score in both cell types, neither showed a significant correlation (B cells, r2 = 0.03, P = 0.32; monocytes, r2 = 0.08, P = 0.07; Supplemental Figure 2, A and B). Instead, we found a significant correlation between IRF5 hyperactivation in SLE B cells or monocytes and dsDNA titers (Figure 1, F–H). In addition, IRF5 hyperactivation in SLE B cells and monocytes was significantly correlated (Figure 1I). Given that IRF5 expression and activation have been previously implicated in a type I IFN gene signature in patients with SLE (5, 11, 19, 20, 46), we examined whether IRF5 hyperactivation in either cell type correlated with serum IFN-α levels. Somewhat to our surprise, we detected a significant positive correlation between IRF5 activation in SLE B cells and IFN-α levels but not in SLE monocytes (Figure 1, J and K). No significant correlation was found between dsDNA and SLEDAI or between dsDNA or SLEDAI and IFN-α levels (Supplemental Figure 2, C–E). Together, the data support the idea that IRF5 hyperactivation may be a systemic marker of disease activity and severity.
Table 1. Demographics of the population of patients with SLE.
Figure 1. IRF5 is hyperactivated in immune cells from patients with SLE and in NZB/W F1 lupus-prone mice.
IRF5 activation was assessed by nuclear localization in CD45+CD14+ monocytes (Mo) (A) and CD45+CD19+ B cells (B) from healthy donors and patients with SLE in the New Jersey cohort using imaging flow cytometry. Data represent the percentage of IRF5 nuclear translocation; circles represent independent donors. (C and D) IRF5 localization determined in monocytes (C) and B cells (D) from healthy donors and patients with SLE in the New York cohort with clinically inactive (score = 0/1) or active (score = 2/3) disease. (E and F) Percentage of IRF5 nuclear translocation in monocytes and B cells from patients with SLE stratified by SLEDAI (E) and dsDNA antibody titers (F). (G–K) Correlation between the percentage of IRF5 translocation in B cells or monocytes and dsDNA titers (G and H) or serum IFN-α levels (J and K) by linear regression analysis. (L and M) IRF5 nuclear translocation in CD11b+ monocytes from cohort 1 (L) and cohort 2 (M) consisting of aging female NZB/W F1 and BALB/c mice. Black circles, NZB/W F1 mice; white circles, BALB/c. n = 3 mice/group/cohort. (N) Inhibition of IRF5 activation (10–21 weeks old) by N5-1 in CD11b+ monocytes. (O and P) Same as in L and M, except in B220+ B cells from cohort 1 (O) and cohort 2 (P). (Q) Same as in N, except inhibition of IRF5 activation is shown in B220+ B cells. (R and S) IRF5 translocation in CD3+CD4+ T cells (R) and CD3+CD8+ T cells (S) from aging female NZB/W F1 and BALB/c mice. n = 6 mice/group. Data represent the mean ± SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001, by 2-way ANOVA with Bonferroni’s post hoc test.
IRF5 is hyperactivated in monocytes and B cells from NZB/W F1 mice.
Irf5–/– mice have been examined in numerous models of murine lupus, with all reports showing that loss of Irf5 protects mice from disease onset and severity (21–26, 47). What is lacking from these studies, though, is an understanding of how IRF5 drives lupus pathogenesis, which is relevant to human SLE. Here, we used the NZB/W F1 model of murine lupus to characterize changes in IRF5 activation by imaging flow cytometry in immune cell subsets before and during clinical onset (Supplemental Figure 3A). NZB/W F1 is a classic, spontaneous model that develops severe lupus-like phenotypes comparable to that of patients with SLE (48). Disease onset and severity can be heterogeneous among mice, yet clinical onset generally occurs approximately at approximately 19–21 weeks of age, when proteinuria levels begin to increase and dsDNA antibodies are detectable. Somewhat surprisingly, we detected dramatic increases in basal IRF5 activation in CD11b+ monocytes and B220+ B cells during early clinical onset, as early as approximately 10 weeks of age (Figure 1, L–Q). This increase was not detected in CD4+ or CD8+ T cells (Figure 1, R and S), nor was it detected in any immune cell subset from age- and sex-matched WT BALB/c or C57BL/6 mice (Figure 1, L–S, and data not shown). Further, IRF5 expression remained relatively unchanged over the course of the disease (Supplemental Figure 3, B and C). These data support the idea that IRF5 hyperactivation may be a driver of murine lupus onset in NZB/W F1 mice.
Design of peptide mimetics that specifically bind to IRF5.
Given the distinct findings of IRF5 hyperactivation in immune cells from patients with SLE and NZB/W F1 lupus mice, we designed a series of inhibitors that would potentially bind to and inhibit IRF5 activation. We used data from IRF crystal structures and IRF5 DN mutants (2, 29, 31, 40) to generate a series of peptide mimetics that corresponded to the N-terminus of IRF5 and might stabilize or maintain the inactive IRF5 monomer, thus inhibiting IRF5 nuclear translocation. Since a crystal structure containing the IRF5 N-terminus has yet to be resolved (31), and the DBD of IRFs is highly homologous, we used coordinates from the resolved IRF3 DBD to build an N-terminal homology model of IRF5 (49). This model was used to predict amino acid sequences with different characteristics that may lead to interaction with the IRF5 C-terminus. Sequence predictions were based on a solvent-accessible surface, charge, and hydrophobicity (Figure 2, A and B, and Supplemental Table 1). In order for the peptides to transduce the cell membrane, we combined the IRF5 sequences with a protein transduction domain (PTD). The PTD has been previously shown to facilitate cell permeability of small peptides (50).
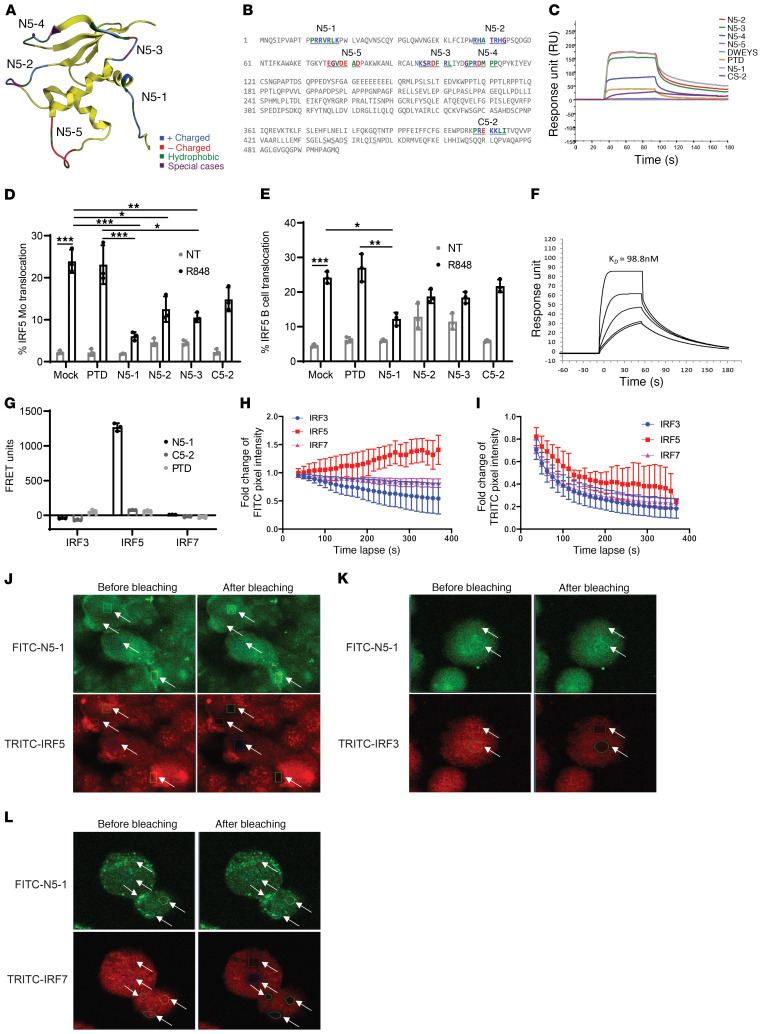
Figure 2. Design of IRF5 peptide mimetics.
(A) Homology model of the IRF5 DBD with location of N-terminal peptides and amino acid characteristics. (B) Position of N- and C-terminal peptides highlighted within the full-length IRF5 V5 sequence. The color code is based on the amino acid characteristics defined in A. (C) Biacore T200 SPR analysis of peptide mimetics. Data are representative of 4 independent experimental replicates per peptide. (D and E) IRF5 nuclear translocation quantified in healthy donor PBMCs preincubated with 10 μM peptide for 1 hour and stimulated with 500 ng/mL R848 for 2 hours using imaging flow cytometry. Plots show quantification in CD45+CD14+ monocytes (D) and CD45+CD19+ B cells (E). n = 3 independent samples from healthy donors. (F) Kinetics analysis of N5-1 peptide binding to IRF5 by SPR. Data are representative of 4 independent experimental replicates. (G) Purified human monocytes were preincubated with 2.5 μM FITC-PTD, –N5-1, or –C5-2 for 1 hour followed by permeabilization and staining for intracellular IRF3, IRF5, or IRF7 with TRITC-conjugated antibodies. FRET units were calculated from fluorescence emissions (see Supplemental Methods). n = 3 independent samples from healthy donors. (H–L) In vivo monitoring of the interaction between FITC–N5-1 and endogenous IRF3, IRF5, or IRF7 in THP1 cells by acceptor photobleaching FRET microscopy. (H and I) Fold change in donor pixel intensity was monitored in the photobleached regions (J–L) and plotted over time. Photobleached regions are indicated by white arrows. Images were acquired before and after acceptor photobleaching. Representative images of FITC–N5-1 and TRITC-IRF5 (J), TRITC-IRF3 (K), and TRITC-IRF7 (L) are shown (original magnification, ×60). Data are representative of 3 independent biological replicate experiments performed in triplicate. Data represent the mean ± SD. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, by 1-way ANOVA.
We tested peptides (1 μM) for their ability to directly interact with the human full-length recombinant IRF5 variant 5 (isoform V5) by surface plasmon resonance (SPR) analysis. The DWEYS peptide served as a nontargeted control and PTD as a control for the cell-permeable sequence. DWEYS showed no affinity for IRF5, and PTD had minimal binding affinity (Figure 2C). N5-1 and N5-2 showed the strongest affinity for IRF5, with N5-3 binding to a slightly lesser extent; N5-5 showed no affinity for IRF5 (binding affinity for IRF5: N5-1 ≥N5-2 >N5-3 >N5-4 >PTD >N5-5). A shared similarity between N5-1 and N5-2 is their relatively stronger positive charge compared with the others (Supplemental Table 1).
Human IRF5 contains 2 NLSs, 1 in the N-terminus and 1 in the C-terminus (30). N5-1 corresponds to the N-terminal NLS (PRRVRLK). To determine whether any NLS is capable of binding to IRF5, we generated C5-2, corresponding to the C-terminal NLS (PREKKLI), and examined binding by SPR. C5-2 and PTD bound with similarly low affinities (Figure 2C). We have thus identified first-generation peptide mimetics (N5-1, N5-2, and N5-3) that directly bind to the full-length inactive IRF5 monomer. The observed difference in function between N5-1 and C5-2 supports the idea that the NLS is not the driver of inhibitor activity and, instead, that peptide mimetics showing the strongest binding affinity for IRF5 (N5-1, N5-2) are those that are positively charged and relatively surface accessible.
Peptide mimetics inhibit TLR7-induced IRF5 nuclear translocation.
We next sought to determine whether in vitro binding data would translate into IRF5 cellular inhibition. IRF5 is a key downstream mediator of TLR7-induced cytokine expression, and TLR7 signaling has been implicated in SLE pathogenesis (2, 51–54). We examined the ability of IRF5 peptide mimetics to inhibit IRF5 nuclear translocation following stimulation of PBMCs with R848. We focused on peptides that showed binding to IRF5 by SPR and included PTD and C5-2 as negative controls. For the initial screening, isolated PBMCs from healthy donors were preincubated in the presence of mock (PBS) or 10 μM PTD, N5-1, N5-2, N5-3, or C5-2 inhibitor for 1 hour followed by stimulation with 500 ng/mL R848 for 2 hours. Cells were surface stained with anti-CD14 (monocytes) and anti-CD19 (B cells) antibodies and then permeabilized and stained for intracellular IRF5 and DRAQ5. R848 induced significant IRF5 nuclear translocation in mock-incubated monocytes (2) (Figure 2D) and B cells (Figure 2E). Although preincubation with PTD had no significant effect on R848-induced IRF5 nuclear translocation in either cell type, N5-1, N5-2, and N5-3 induced a significant reduction in R848-induced nuclear translocation in monocytes (Figure 2D). In B cells, only N5-1 significantly reduced R848-induced IRF5 nuclear translocation (Figure 2E). Surprisingly, preincubation with C5-2 showed some reduction in nuclear localization of IRF5, even though there was low binding affinity (Figure 2C); inhibition failed to achieve statistical significance. In unstimulated cells, it is noteworthy that N5-1 and C5-2 had no effect on baseline levels of nucleus-localized IRF5 (gray bars), whereas N5-2 and N5-3 resulted in increased basal IRF5 nuclear translocation (Figure 2, D and E). Since N5-1 conferred potent inhibition in both cell types, we determined the equilibrium dissociation constant (KD) for N5-1 binding to IRF5 (Figure 2F). A KD of 98.8 nM was calculated, confirming a strong binding affinity between N5-1 and the inactive full-length IRF5 monomer, which is in agreement with our functional data (Figure 2, D and E). To further confirm the SPR data and analyze inhibitor specificity, we developed an in-cell fluorescence resonance energy transfer (FRET) assay to measure binding of inhibitors to endogenous IRF5, as well as other IRF family members — IRF3 and IRF7 — with similar structural and functional domains. We obtained a positive FRET signal only for N5-1 binding to endogenous IRF5, but not IRF3 or IRF7, in human primary monocytes (Figure 2G). We also tested whether PTD that is positively charged like N5-1 and C5-2 that has a neutral charge could bind to the IRFs. We were unable to detect binding of PTD or C5-2 to any IRF, confirming that N5-1 specificity is more related to the peptide sequence than the positive charge. Last, we performed an acceptor photobleaching FRET assay as a secondary, independent method of confirmation and found that N5-1 specifically bound to IRF5, and not to IRF3 or IRF7 (Figure 2, H–L, and Supplemental Figure 4, A–C). Together, these findings confirm the specificity of N5-1 for IRF5.
N5-1 binds to and stabilizes the inactive IRF5 monomer.
To gain insight into N5-1 binding to IRF5 at the atomic level, we performed molecular modeling studies. We reasoned that N5-1 must interact with monomeric IRF5 at the IAD in the cytoplasm of a cell rather than the DBD, since data from both SPR analysis and in-cell FRET assays indicated direct binding of N5-1 to the inactive full-length IRF5 monomer. To test this, we generated a homology model of an inactive IRF5 C-terminal domain using the monomeric autoinhibited IRF3 C-terminal domain (Protein Data Bank [PDB]: 1QWT) (49) as a template. The model showed good overall alignment with the α-carbon backbone of the template, with a root mean square difference (RMSD) of less than 0.7Å. N5-1 was then docked to IRF5 and ranked by molecular mechanics generalized Born surface area (MM-GBSA) binding free energy (ΔGbinding) (Supplemental Table 2 and Supplemental Figure 4D). The top-ranked N5-1 peptide–docked pose with a ΔGbinding value of –111.087 kcal/mol is shown in Figure 3A. Meanwhile, C5-2 was docked to the same model as a reference, and no pose was predicted. These data provide further support for the select binding of N5-1 to IRF5.
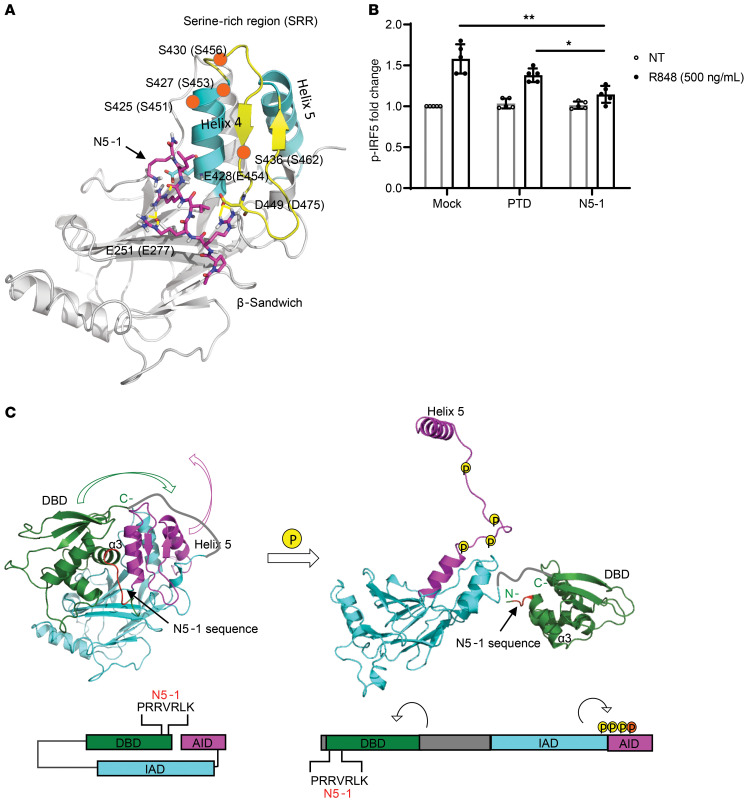
Figure 3. N5-1 is predicted to bind to the C-terminal IAD of an inactive IRF5 monomer and inhibit phosphorylation of Ser462.
(A) Schematic diagram of N5-1 (pink) binding to the C-terminal IAD of IRF5 from peptide docking using the Schrodinger suite (see Supplemental Methods). N5-1 stabilizes the nonphosphorylated, inactive IRF5 monomer. Serine phosphorylation sites are shown by orange circles. (B) PBMCs were preincubated with 10 μM inhibitor for 1 hour and stimulated with R848. p-IRF5 phosphorylation at Ser462 was detected by flow cytometry following gating on CD14+ monocytes. The fold change in p-IRF5 relative to unstimulated mock samples is shown. n = 5 independent samples from healthy donors. Data represent the mean ± SD. *P ≤ 0.05 and **P ≤ 0.01, by 1-way ANOVA. (C) On the basis of the binding of N5-1 to full-length inactive IRF5, we propose that the DBD masks the IAD of IRF5 and that the AID masks the C-terminal phosphorylation sites, thus stabilizing a closed, unphosphorylated conformation of the IRF5 monomer (left panel). In this conformation, the DBD α3 helix, which contains all the conserved residues and is responsible for protein-DNA contacts, is shielded. Upon phosphorylation, the AID unfolds, which unmasks the C-terminal phosphorylation sites and frees helix 5 for dimerization (right panel). The DBD will also be released from this folded, inactive position and exposed to DNA for binding. The colors correspond to the specified regions of IRF5 in the crystal structure (above) and the stick model (below). The DBD is indicated in green, the IAD in blue, and the AID in purple. The N5-1 sequence is shown in red in both models.
We identified 3 arginine residues within N5-1 that formed stable hydrogen bonds and salt bridges with the acidic amino acids Asp449, Glu251, and Glu428 on IRF5 (Figure 3A). These amino acids are located on the AID folded loop, the AID helix bundle 4, and the β-sandwich region on the IAD, respectively. Therefore, N5-1 binds to IRF5 and anchors the flexible AID loop onto the IAD. Since activation of IRF5 requires phosphorylation on the Ser-rich region (SRR) to generate the charge repulsion force to destabilize the inactive folded conformation of the AID, we proposed that stabilization of the AID loop in a folded conformation near helix 4 would mask C-terminal IRF5 phosphorylation sites (30, 31, 33–35). We and others previously identified key C-terminal Ser residues critical for IRF5 activation, yet antibodies to detect these residues are unavailable (30, 31, 35). Subsequent studies identified IKK-β as a kinase that phosphorylates human IRF5 at Ser462 in response to TLR stimulation (33, 34). In an effort to develop experimental data that would prove or disprove this binding model, we examined IRF5 phosphorylation at Ser462 (p-IRF5). PBMCs were stimulated with R848, and IRF5 phosphorylation was detected with p-IRF5 antibodies (34). We detected elevated p-IRF5 levels in R848-stimulated cells and observed that preincubation with N5-1 significantly reduced p-IRF5 to mock levels (Figure 3B). These data provide the initial mechanistic support for the idea that N5-1 binds to a region that stabilizes the inactive IRF5 monomer in a conformation that inhibits phosphorylation at Ser462.
Since the N5-1 sequence was extracted from the IRF5 DBD, we projected that the DBD folds onto the C-terminal IAD (Figure 3C). Molecular modeling indicates that this occurs without much steric hindrance. Thus, we propose that in unstimulated cells, the N-terminal DBD is folded over the C-terminal IAD in the full-length, inactive IRF5 monomer (Figure 3C, left). The N5-1 sequence within the DBD offers electrostatic interactions to stabilize the folded conformation of the AID loop, which masks the critical phosphorylation sites. Meanwhile, the DBD α3 helix, which contains all of the conserved residues and is responsible for protein-DNA contacts (55, 56), is shielded in this position. Upon phosphorylation, the large charge repulsion force on the SRR causes the dramatic conformational change to unfold the AID loop, which frees helix 5 for dimerization (Figure 3C, right). It has been observed in the IRF3 crystal structure that the N-terminal region of the IAD undergoes large conformational changes during activation (55, 57). Since the DBD links to the N-terminus of the IAD, we predict that the DBD can be released from this folded, inactive position and be exposed to DNA for binding after the conformational change (56) (Figure 3C).
IRF5 peptide mimetics readily enter cells and have low associated toxicity.
Next, we synthesized peptides conjugated to FITC (Supplemental Table 1) to measure cellular uptake by flow cytometry. Isolated PBMCs were treated with FITC-conjugated PTD, N5-1, or C5-2 for 1 hour and surface stained to identify monocytes and B cells. Representative histograms in Figure 4A show increased FITC intensity from PTD and N5-1 in monocytes and B cells. We obtained similar results for C5-2 (data not shown). Summarized data from multiple independent donors over a dose range revealed the preferential uptake of N5-1 into monocytes rather than B cells (Figure 4B). However, unlike monocytes that showed similar uptake of N5-1 over the range, uptake into B cells was dose dependent (Figure 4B). To confirm that FITC-conjugated inhibitors enter cells and do not simply bind to the surface upon preincubation, we examined cell uptake using imaging flow cytometry. At 10 μM, we found that multiple cell types — monocytes, B cells and pDCs — took up the inhibitors efficiently, as determined by costaining with nuclear DRAQ5 (Figure 4C). We assessed inhibitor toxicity by staining PBMCs with propidium iodide or trypan blue after treatment and found minimal toxicity (Supplemental Figure 5, A and B). IRF5 has been previously shown to regulate the cell cycle and apoptosis (58). We examined cell-cycle progression as a potential off-target effect of the inhibitors and found no significant difference (Supplemental Figure 5C). No significant change in cellular apoptosis was detected by annexin V and SYTOX costaining (Supplemental Figure 5D). Together, these data confirm that IRF5 inhibitors enter the cell, are nontoxic, and have limited off-target effects. They also suggest that cell-type specificity may be achieved through varied inhibitor dosing.
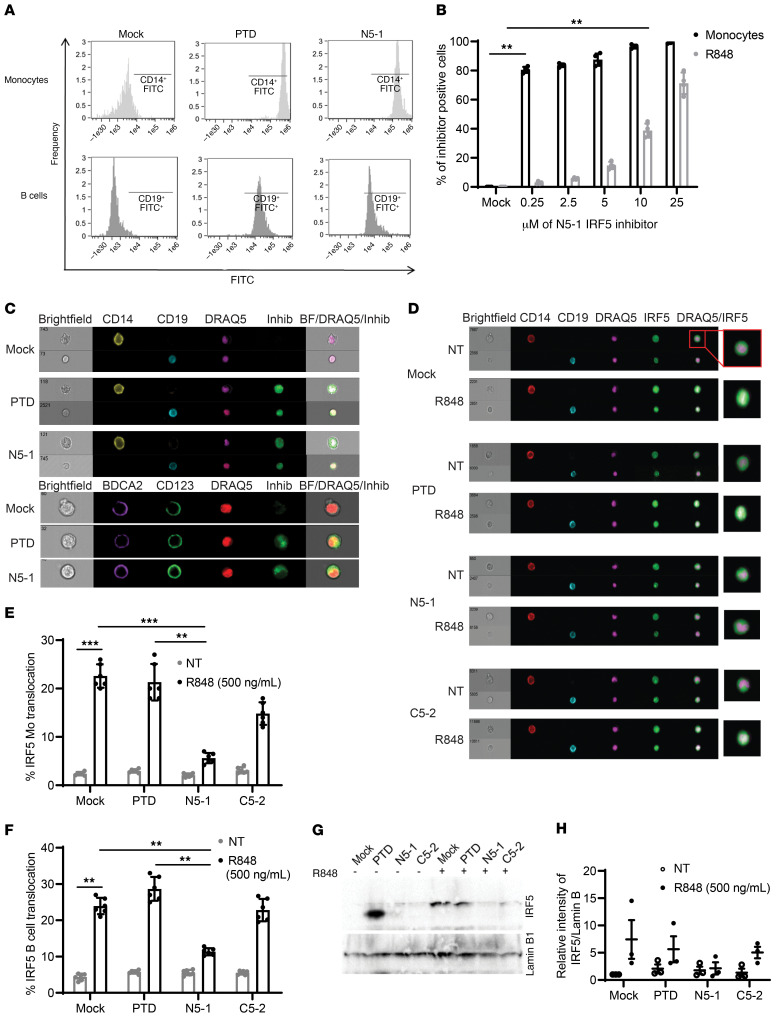
Figure 4. IRF5 peptide inhibitors readily enter primary immune cells to inhibit R848-induced IRF5 nuclear translocation.
(A) Representative flow cytometry histograms showing uptake of 10 μM FITC-conjugated PTD or N5-1 after incubation of human PBMCs with an inhibitor for 1 hour. For inhibitor uptake, an FITC intensity of greater than 104 in CD14+ monocytes (light gray) and 103 in CD19+ B cells (dark gray) was considered positive. (B) Percentage of total monocytes and B cells positive for FITC-conjugated N5-1. n = 4 independent samples from healthy donors. (C) Representative images of cellular uptake of 10 μM FITC-conjugated PTD or N5-1 in monocytes (top row), B cells (bottom row), and pDCs (bottom panel). Inhib, inhibitor. (D) Representative images of IRF5 cellular localization in monocytes (CD14) and B cells (CD19) after preincubation of PBMCs with 10 μM mock, PTD, N5-1, or C5-2 inhibitors followed by stimulation with 500 ng/mL R848 for 2 hours. (E and F) Quantification of IRF5 nuclear translocation in monocytes (E) and B cells (F) was done by imaging flow cytometry. n = 6 independent samples from healthy donors. (G) Representative Western blot of nuclear extracts from primary human monocytes following treatment with 2.5 μM mock, PTD, N5-1, or C5-2 inhibitors and stimulation with 500 ng/mL R848 for 2 hours. (H) Quantification of nuclear IRF5 from G relative to lamin B1. n = 3 independent samples from healthy donors. Data are representative of 3 or more independent experimental replicates. Data represent the meant ± SEM. **P ≤ 0.01 and ***P ≤ 0.001, by 1-way ANOVA.
N5-1 is a potent inhibitor of TLR7-induced IRF5 nuclear translocation in monocytes and B cells.
To confirm a block in IRF5 nuclear translocation by the inhibitors, we performed imaging flow cytometry and cell fractionation. PBMCs from healthy donors were pretreated with 10 μM PTD, N5-1, or C5-2 before R848 stimulation. Representative images of mock preincubated and unstimulated (nontreated [NT]) PBMCs show cytoplasmic IRF5 staining as a green halo around the DRAQ5-positive nucleus in both monocytes and B cells (Figure 4D). Upon stimulation, IRF5 translocated to the nucleus, which is shown by the yellow nuclear costain. Preincubation with inhibitors followed by stimulation resulted in a significant reduction of nuclear IRF5 by N5-1 (Figure 4, E and F). Additionally, cell fractionation was performed on isolated primary monocytes that were pretreated with 2.5 μM inhibitor and then stimulated with R848 for 2 hours. Western blot analysis of nuclear lysates confirmed the imaging flow data showing a marked decrease in R848-induced IRF5 nuclear translocation by N5-1 (Figure 4, G and H; see the complete, unedited blot for panel G in the supplemental material). We next quantified IRF5 expression in monocytes from imaging flow data to determine whether inhibitors altered IRF5 expression. Although no change in basal IRF5 protein expression was detected in NT monocytes preincubated with inhibitor, the observed upregulation of IRF5 by R848 was not seen in cells preincubated with N5-1 (Supplemental Figure 6A).
N5-1 selectively inhibits IRF5-mediated inflammatory cytokine expression.
Following IRF5 phosphorylation, homodimerization, and nuclear translocation, IRF5 binds to the promoters of target genes and regulates their expression (1, 3, 59). In SLE, it is thought that increased levels of proinflammatory cytokines contribute to systemic inflammation (12, 13). We thus determined whether inhibition of IRF5 nuclear translocation would reduce inflammatory cytokine expression. PBMCs were pretreated with 10 μM inhibitor and stimulated with R848 for 2 hours, and total RNA was isolated to determine IL6 and IFNA expression by quantitative reverse transcriptase PCR (qRT-PCR). Expression of both cytokines was significantly reduced by N5-1 (Supplemental Figure 6, B and C). We observed no significant difference with PTD or C5-2. Since other transcription factors, such as NF-κB and IRF7, undergo nuclear translocation in response to TLR signaling that results in similar levels of proinflammatory cytokine expression (60, 61), we examined the effect of N5-1 on R848-induced NF-κB nuclear translocation in monocytes and CpG-A–induced IRF7 nuclear translocation in pDCs. Importantly, the kinetics of NF-κB nuclear translocation are distinct from the kinetics of IRF5. In R848-stimulated monocytes, NF-κB nuclear translocation was more rapid and was not detected at 2 hours. Instead, the effect of N5-1 on NF-κB nuclear translocation in PBMCs was examined after preincubation with an inhibitor and a 30-minute post-R848 stimulation. As expected, R848 induced significant accumulation of nuclear NF-κB (Supplemental Figure 6D), whereas inhibitors had no significant effect on basal or R848-induced nuclear NF-κB levels. We obtained similar results for IRF7 in pDCs after stimulation with CpG-A for 2 hours (Supplemental Figure 6E). These data support the idea that N5-1 reduces proinflammatory cytokine expression through the select inhibition of IRF5 activation.
N5-1 protects NZB/W F1 mice from spontaneous onset of lupus.
To determine whether IRF5 hyperactivation is a driver of lupus onset and severity in NZB/W F1 mice, we tested to see if N5-1 could inhibit murine IRF5 nuclear translocation. RAW264.7 macrophages were preincubated with N5-1 and stimulated with LPS or R848 for 2 hours, followed by isolation of nuclear extracts for Western blot analysis. Similar to our findings in human monocytes (Figure 4, G and H), N5-1 resulted in a significant dose-dependent reduction in nuclear translocation of IRF5 (Figure 5, A and B, and Supplemental Figure 7; see the complete, unedited blot for Figure 5A in the supplemental material). We next examined the ability of FITC-conjugated inhibitors to be taken up in vivo. NZB/W F1 mice were injected with inhibitor, and uptake was monitored by imaging flow cytometry over a 2-hour period. Similar to our findings in human PBMCs (Figure 4, B and C), we detected cell-type–specific differences, yet all cells showed uptake of the inhibitors (Supplemental Figure 8). Since it is well established that Irf5–/– mice have impaired production of IL-6 (3, 47), we used this model to test the ability of N5-1 to inhibit IL-6 production in vivo. To confirm in vivo specificity, we compared IL-6 production in Irf5+/+, Irf5+/–, and Irf5–/– littermate mice treated with N5-1 and injected with R848. As expected, we found that R848 induced IL-6 production in Irf5+/+ mice, while it attenuated IL-6 production in Irf5+/– mice and significantly reduced IL-6 production in Irf5–/– mice (3, 62) (Figure 5C). N5-1–treated Irf5+/+ mice mimicked the level of IL-6 produced in R848-injected Irf5–/– mice, and no further effect of N5-1 was seen in Irf5–/– mice. Last, Irf5+/+ mice treated with PTD or C5-2 had no significant effect on R848-induced IL-6 production. Altogether, these data support the specific inhibition of murine IRF5 function in vivo by N5-1.
Figure 5. N5-1 protects NZB/W mice from spontaneous onset of lupus.
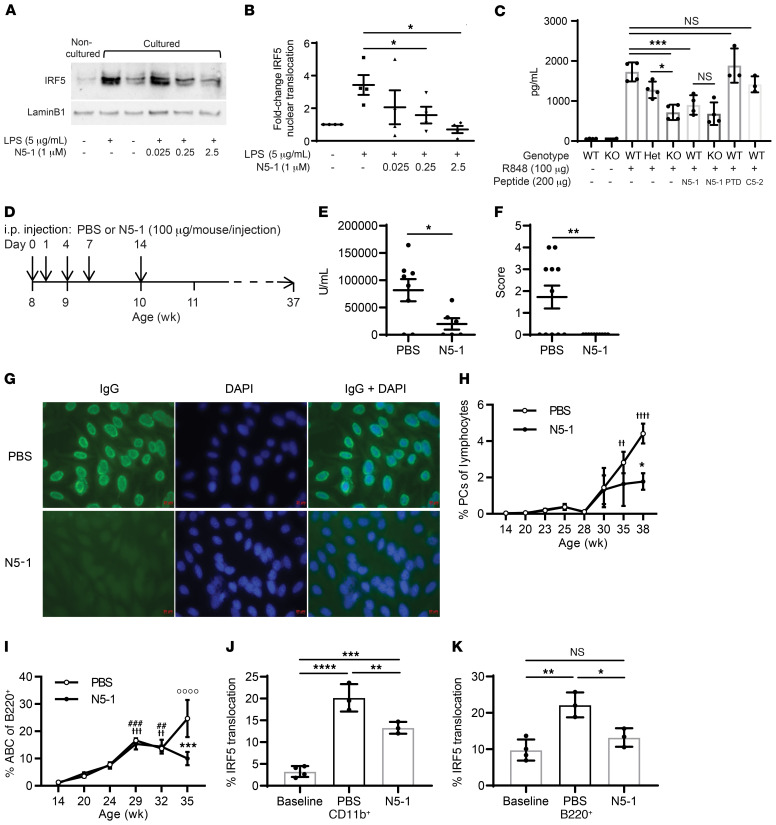
(A) Representative Western blot of nuclear extracts from RAW264.7 macrophages pretreated for 1 hour with N5-1 followed by LPS for 2 hours. Noncultured cells, 0 hours before pretreatment; cultured cells, 3 hours after treatment. (B) Quantification of nuclear IRF5 in A relative to lamin B1 from 3 independent replicates. Statistical significance was determined by 1-way ANOVA. (C) In vivo inhibition of IL-6 secretion by N5-1 in WT (Irf5+/+), heterozygous (Het) (Irf5+/–), and KO (Irf5–/–) mice. Sera were harvested 1.5 hours after R848 administration. n = 3–4 mice/group. Statistical significance was determined by 1-way ANOVA. (D) N5-1 dosing strategy for NZB/W F1 mice. (E) Anti-dsDNA Ig titers (1:500 serum dilution) in mice at 20 weeks of age. (F) ANA immunofluorescence scoring for sera from 11 PBS- and 10 N5-1–treated mice. 0, negative signal; 4, strongest signal. Statistical significance was determined by Mann-Whitney U test. (G) Representative ANA images from 27-week-old treated mice (×20 objective and ×10 eyepiece). (H and I) Percentage of circulating IgD–B220CD138+ PCs (H) and B220+CD11c+CD11b+ ABCs (I). n = 4 mice/time point. Statistical significance was determined by 2-way ANOVA and Bonferroni’s multiple-comparison test. (H) F(7,35) = 10.27, P < 0.0001, age; F(1,35) = 4.125, P = 0.049, treatment; F(7,35) = 1.627, P = 0.1603, interaction. *P = 0.0133 vs. PBS, week 38; ††P < 0.0081 and ††††P < 0.0001 vs. PBS, week 14. (I) F(5,32) = 20.63, P < 0.0001, age; F(1,32) = 4.402, P = 0.0439, treatment; F(5,32) = 4.146, P = 0.0051, interaction. ***P = 0.0001 versus PBS, week 35; ††P = 0.0033, †††P = 0.0002 and ††††P < 0.0001 vs. PBS, week 14; ##P = 0.0005 and ###P = 0.0029 vs. N5-1, week 14. (J and K) Inhibition by N5-1 of IRF5 activation in cohort 2 (14–21 weeks old). CD11b+ monocytes (J) and B220+ (K) B cells. n = 4 mice/group. Statistical significance was determined by 1-way ANOVA. Data represent the mean ± SEM. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
On the basis of the observed peak of IRF5 hyperactivation in NZB/W F1 mice (Figure 1, N and Q), we developed a pilot dosing regimen to test the effects of N5-1 on lupus disease onset in female mice (Figure 5D). Mice received 5 equal doses of 100 μg N5-1 or an equal volume of vehicle control from 8–10 weeks of age, and proteinuria was measured weekly to track disease onset. At 20 weeks of age, proteinuria levels began to significantly drop in the N5-1–treated mice (Supplemental Figure 9A), and serum dsDNA autoantibodies were significantly reduced (Figure 5E). Measurement of serum antinuclear IgG antibodies by HEp-2 antinuclear antibody (ANA) assay at 27 weeks of age revealed a significant reduction by N5-1 (Figure 5, F and G). Given the recent studies implicating IRF5 in human plasma cell (PC) differentiation (63), accumulation of age-or autoimmune-associated B cells (ABCs) (27), and antibody secretion, we examined cells in the blood of PBS- and N5-1–treated mice. As expected, the percentage of circulating PCs and ABCs increased with disease severity (64) (Figure 5, H and I), even though the total number of B220+ B cells remained fairly unchanged (Supplemental Figure 9B). Although the percentage of PCs increased significantly during later stages of disease development in PBS-treated mice, we observed no significant change in N5-1–treated mice over the course of the disease; however, N5-1 significantly reduced the accumulation of PCs beyond 35 weeks of age (Figure 5H and Supplemental Figure 9C). Conversely, the percentage of circulating ABCs was significantly increased in both PBS- and N5-1–treated mice, however, a significant N5-1–induced reduction in ABCs was detected at 35 weeks of age (Figure 5I and Supplemental Figure 9C).
N5-1 attenuates IRF5 hyperactivation in NZB/W F1 mice.
Next, we monitored the kinetics of IRF5 hyperactivation in monocytes and B cells from N5-1–treated NZB/W F1 mice. At the observed peak in IRF5 hyperactivation, between approximately 10 and 19 weeks of age for both monocytes and B cells (Figure 1, N and Q), we detected a significant reduction in IRF5 nuclear translocation in N5-1–treated mice (Figure 5, J and K). No effect on IRF5 expression was found (Supplemental Figure 9, D and E). These data confirm the in vivo efficacy of N5-1 in reducing IRF5 hyperactivation.
Reduced kidney pathology and increased overall survival.
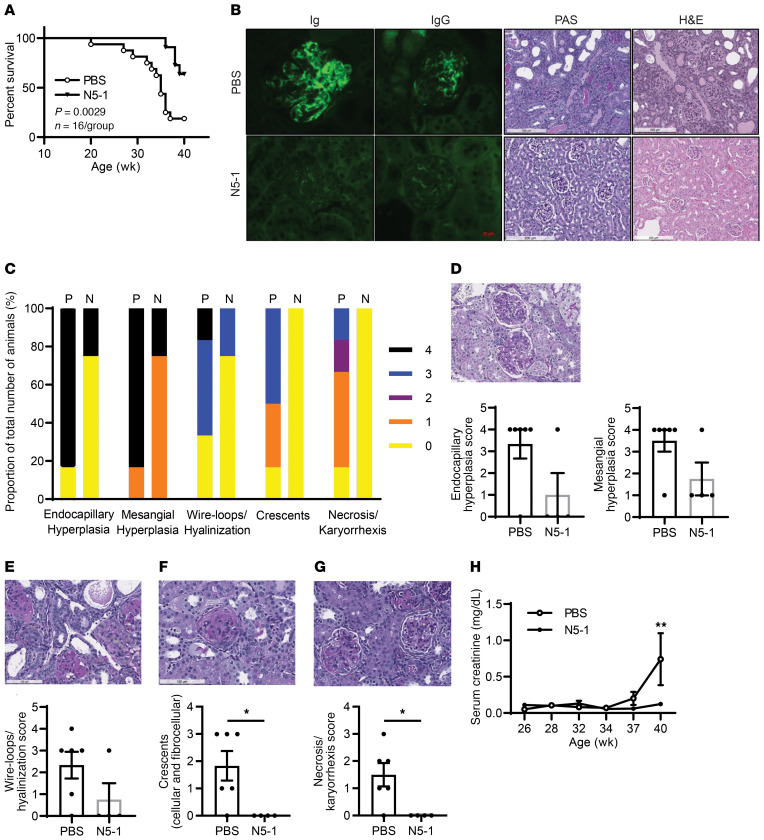
Survival of a cohort of NZB/W F1 mice was monitored until 40 weeks of age, revealing significant protection of N5-1–treated mice from lupus-induced mortality (Figure 6A). Histologic analysis of kidneys revealed amelioration of several parameters of renal injury in N5-1–treated mice, including expansion of the mesangial matrix, presence of hyaline deposits, decreased capillary loops, presence of cellular/fibrocellular crescents, tubular necrosis, and deposition of immunocomplexes (Figure 6, B–G). In N5-1–treated mice, albumin/creatinine ratios began trending downward after 27 weeks (Supplemental Figure 9F), and serum creatinine levels were significantly reduced by 40 weeks of age (Figure 6H). We observed no significant change in body weight between the groups (Supplemental Figure 9G). Last, we monitored IFN-α levels over the course of the disease and found that low levels were detectable around 17 weeks of age (65–68), which occurred after or concurrently with IRF5 activation (Supplemental Figure 9H). Unfortunately, IFN-α levels were too low to detect significant differences between groups (data not shown). These data show the in vivo utility of N5-1 in protecting NZB/W F1 mice from spontaneous onset of lupus and mortality.
Figure 6. N5-1 reduces kidney pathology and increases overall survival.
(A) Kaplan-Meier survival curves. Differences were determined by Gehan-Breslow-Wilcoxon test. n = 11 mice/group. (B) Representative microscopic images of kidney sections; fluorescence deposition of Ig and IgG (×40 magnification), periodic acid–Schiff (PAS) staining, and H&E staining (×10 magnification). (C) Summarized scoring of renal inflammation and damage shown in D–G from 6 PBS-treated mice and 4 N5-1–treated mice. (D–G) Microscopic images of kidney sections assessed by PAS staining (original magnification, ×20) showing images of endocapillary and mesangial hyperplasia (D), wire-loops/hyalinization (E), crescents (F), and necrosis/karyorrhexis (G). Scoring for 100 glomeruli per case is shown. (H) Serum creatinine levels were plotted over the course of the disease. n = 8 mice/group. *P ≤ 0.05, by Mann-Whitney U test.
N5-1 provides therapeutic efficacy in NZB/W F1, MRL/lpr, and pristane-induced lupus mice.
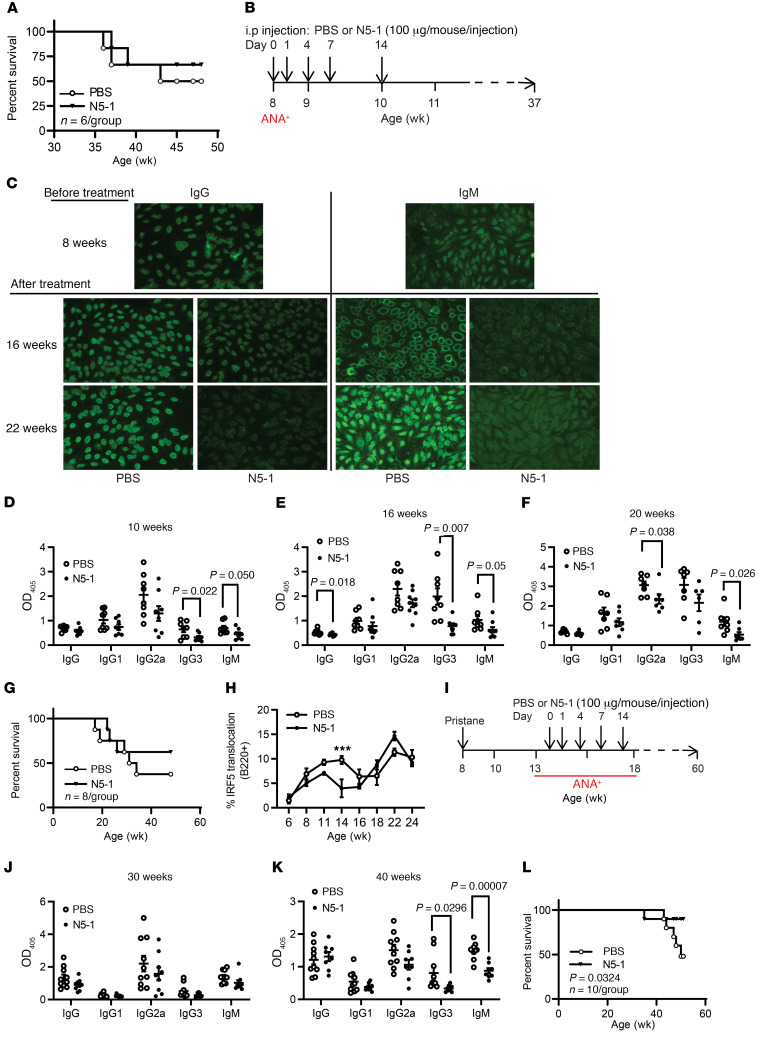
We next examined whether N5-1 would increase survival of NZB/W F1 mice in a therapeutic efficacy model in which mice already had clinically elevated dsDNA titers, positive ANAs, elevated proteinuria, and kidney disease (https://www.jax.org/jax-mice-and-services/in-vivo-pharmacology/immunology-services/autoimmune-diseases/lupus-studies). We initiated the 2-week N5-1 dosing regimen in a cohort of 27-week-old NZB/W F1 mice. At this later stage of clinical disease, we detected a modest increase in survival that did not reach statistical significance (Figure 7A). Given the slow onset of lupus in NZB/W F1 mice, we switched to MRL/lpr and pristane-induced lupus mice to further examine N5-1 clinical efficacy. Dosing in MRL/lpr mice was initiated at 8 weeks of age and after confirmation of ANA IgG positivity (Figure 7, B and C). We monitored ANAs, anti-dsDNA antibody titers, proteinuria, IRF5 cellular activation, and survival. As early as 10 weeks of age, we detected significant reductions in dsDNA titers in the N5-1–treated mice that were maintained over the course of disease and contributed to increased survival; significant reductions in proteinuria occurred at 30 weeks (Figure 7, C–G, and Supplemental Figure 10A). The observed reduction in dsDNA titers corresponded to a significant, concomitant reduction in B220+ IRF5 activation (Figure 7H). Unlike NZB/W F1 mice, however, which showed an early single peak in IRF5 activation (Figure 1, L–Q), we detected at least 2 peaks in IRF5 B cell and monocyte activation that continued to increase with age and disease severity (Figure 7H and Supplemental Figure 11A). We detected significant, albeit small, reductions in IRF5 activation in monocytes and CD8+ T cells from N5-1–treated mice (Supplemental Figure 11, A–C). These data suggest that IRF5 activity in MRL/lpr mice more closely mirrors that seen in SLE patients, where increased IRF5 activation is associated with clinical disease activity and dsDNA titers (Figure 1, C and D and F–H).
Figure 7. Therapeutic efficacy of N5-1 in ANA-positive NZB/W F1, MRL/lpr, and pristane-induced lupus mice.
(A) Kaplan-Meier survival curves of NZB/W F1 mice treated at 27 weeks of age. Differences were determined by Gehan-Breslow-Wilcoxon test. n = 6 mice/group. (B) N5-1 dosing strategy for MRL/lpr mice. (C) Representative ANA images from 8-week-old pretreated mice and 16- and 22-week-old treated mice (×20 objective and ×10 eyepiece). (D–F) Anti–dsDNA IgG isotype titers (1:500 serum dilution) were measured at 10, 16, and 20 weeks of age. (G) Kaplan-Meier survival curves of treated MRL/lpr mice. Differences determined by Gehan-Breslow-Wilcoxon test. n = 8 mice/group. (H) Analysis of IRF5 nuclear translocation in B220+ B cells from PBS- and N5-1–treated MRL/lpr mice. n = 8 mice/group. (I) N5-1 dosing strategy for pristane-injected mice. (J and K) Anti–dsDNA IgG isotype titers (1:500 serum dilution) were determined at 30 (J) and 40 (K) weeks of age. (L) Kaplan-Meier survival curves of pristane-induced BALB/c mice. Differences were determined by Gehan-Breslow-Wilcoxon test. n = 10 mice/group. (D–F, H, J, and K) ***P ≤ 0.0001, by Mann-Whitney U test.
To further confirm the clinical utility of N5-1, we injected BALB/c mice with pristane (21, 47) and treated them with N5-1 after ANA detection (Figure 7I). Similar to NZB/W F1 and MRL/lpr mice, we detected significant reductions in dsDNA titers that corresponded to a significant increase in overall survival (Figure 7, J–L, and Supplemental Figure 10B). Altogether, these data show that, independent of the mechanism of lupus onset (NZB/W F1, MRL/lpr, pristane-induced), N5-1 provided significant clinical benefit at later stages of disease development, which holds tremendous promise for patients with SLE.
N5-1 inhibits SLE serum–induced IRF5 activation and reverses IRF5 hyperactivation in SLE immune cells.
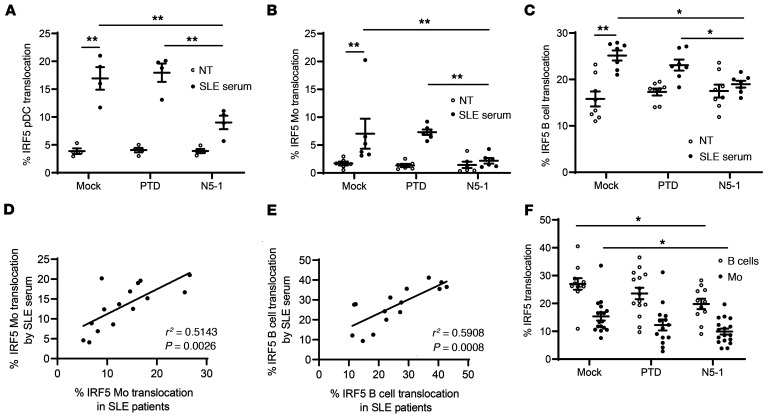
Here, we report for the first time to our knowledge that IRF5 is hyperactivated in immune cells from NZB/W F1– and MRL/lpr lupus–prone mice before and during clinical onset, respectively. Patients with SLE present with elevated basal IRF5 activation that is further increased during active flares (Figure 1, A–D). Since the mechanism or mechanisms by which IRF5 becomes activated in patients with SLE and murine models of lupus are not yet known and are likely mediated by multiple triggers and pathways (19, 30–35), we evaluated the effects of N5-1 ex vivo in response to SLE serum, which is a more complex and disease-relevant trigger of IRF5 activation than are pure TLR ligands (19). Healthy donor PBMCs were preincubated with an inhibitor and stimulated with SLE sera for 2 hours, and then IRF5 activation was assessed (19). N5-1 induced a significant reduction in SLE serum–induced IRF5 activation in pDCs, monocytes, and B cells (Figure 8, A–C), supporting the idea that N5-1 works in the context of a human SLE–like environment. We next asked whether SLE serum–induced IRF5 activation in healthy donors correlated with IRF5 activation in SLE monocytes and B cells from matched patients. Indeed, we found a significant correlation between ex vivo and in vivo IRF5 activation (Figure 8, D and E). Last, we examined whether N5-1 could reverse IRF5 hyperactivation in monocytes and B cells from patients with active SLE. To our surprise, treatment of SLE PBMCs with N5-1 for 1 hour led to a significant reduction in basal IRF5 hyperactivation (Figure 8F). These data support the use of N5-1 to treat patients with SLE at different stages of disease development.
Figure 8. N5-1 inhibits SLE serum–induced IRF5 activation and reverses IRF5 hyperactivation in SLE immune cells.
(A) Healthy donor PBMCs (n = 6) were preincubated with an inhibitor (10 μM) followed by stimulation with 2% SLE serum for 2 hours. The percentage of IRF5 nuclear translocation is shown in pDCs (A), monocytes (B), and B cells (C) from imaging flow cytometry. (D and E) Correlation between the percentage of IRF5 translocation in SLE serum–stimulated monocytes (D) or B cells (E) and in vivo IRF5 activation in matched SLE monocytes or B cells, respectively, by linear regression analysis. (F) SLE PBMCs were mock or inhibitor treated (10 μM) for 1 hour, and IRF5 activation in monocytes and B cells was quantified by imaging flow cytometry. The percentage of IRF5 nuclear translocation is shown. Data represent the mean ± SEM. Differences between groups were determined by 2-way ANOVA with Bonferroni’s multiple-comparison test. *P ≤ 0.05 and **P ≤ 0.01.
Discussion
IRF5 genetic variants that associate with SLE risk were originally identified in 2005 (6) and only recently are studies beginning to shed light on how IRF5 genetic risk contributes to SLE pathogenesis (20, 28, 69, 70). While these studies led to a multitude of studies in murine lupus models lacking Irf5, together, they support genetic and nongenetic roles for IRF5 in lupus disease development (21–26, 71). Similar to our recent findings in healthy donor IRF5 risk carriers (69), we demonstrate here that dysregulated IRF5 activity, rather than expression, is a driver of SLE disease onset and severity. In NZB/W F1 mice, the observed increase in basal IRF5 hyperactivation occurred in both monocytes and B cells, but not T cells, and preceded clinical disease onset. However, in MRL/lpr mice, IRF5 activation coincided with or occurred after clinical disease onset and was detected in all cell types examined (Figure 7H, and Supplemental Figure 11, A–C). The observed differences in the kinetics of IRF5 activation in these 2 models of spontaneous murine lupus point to distinct mechanisms of lupus onset, yet implicate IRF5 immune cell hyperactivation in both (72). In human SLE, we found that IRF5 hyperactivation in monocytes and B cells was associated with disease activity and correlated with dsDNA titers.
Baseline increases in murine IRF5 activation were first reported in DCs from Lyn-deficient mice with lupus (73). Lyn is a Src family kinase that functions in multiple aspects of immune signaling as both a positive and negative regulator (74, 75). Lyn was identified as a negative regulator of IRF5 posttranslational modification via direct binding to IRF5 (73). Given that basal IRF5 hyperactivation has been detected in multiple cohorts of patients with lupus and, more recently, in healthy donor IRF5 genetic risk carriers (69), and given our finding that IRF5 hyperactivation in NZB/W F1 and MRL/lpr lupus mice was not constitutive over the course of the disease, it is unlikely that alterations in Lyn are driving IRF5 hyperactivation. Further, since SLE serum recapitulated the IRF5 activation detected in matched SLE patients, this points to a circulating trigger or triggers that induce IRF5 activation rather than a loss in negative control regulators (Figure 8, D and E) (19). Last, given the heterogeneity of SLE and the differential kinetics and cell-type specificity of IRF5 activation in NZB/W F1 and MRL/lpr mice, it is unlikely that there is a single trigger or pathway leading to IRF5 activation. Indeed, we recently reported that the kinetics of Ser462 IRF5 phosphorylation and nuclear translocation were distinct depending on the stimulus (39). Unfortunately, this phospho-antibody does not detect endogenous IRF5 by Western blotting (34), nor were we successful in detecting murine IRF5. Thus, mapping posttranslational events on IRF5 in these different model systems would provide valuable insight into the mechanisms of activation.
Signaling pathways have emerged as key targets for the development of small-molecule inhibitors, with the primary targets being protein kinases and phosphatases (76, 77). A caveat to this type of therapeutic targeting is that it requires a priori knowledge of the signalling molecules leading to activation. Additionally, kinase inhibitors are often not specific to 1 kinase, 1 signaling pathway, or 1 downstream target protein. In the case of IRF5, it is well documented that IRF5 becomes activated in a cell-type– and stimuli-dependent manner (1, 29, 33–35, 78–80). Regulation of cytokine production by IRF5 requires nuclear translocation and transcriptional modulation of target genes. Previous work suggested a requirement for ubiquitination and/or acetylation before phosphorylation and homo- or heterodimerization, which may or may not lead to nuclear translocation (35, 81, 82). Further, IRF5 phosphorylation occurs at multiple sites that are dependent on the pathway of activation (30–35). Thus, in order to bypass the ambiguity of posttranslational modifications and dimerization, we developed peptide mimetics that directly bind to and inhibit IRF5 activation independently of the initiating pathway.
In support of this rationale, we recently characterized another family of cell-permeable peptides that inhibit IRF5 dimerization (39). Unfortunately, these inhibitors were not stable for in vivo analysis, nor did they similarly inhibit both human and murine IRF5. Targeting IRF5 inhibition in a cell-type–specific manner may be feasible, as we observed cell-type specificity for N5-1, N5-2, and N5-3 that may be dose dependent and due to distinct physicochemical properties of each inhibitor (Figures 2 and 4). Although N5-2 and N5-3 were capable of inhibiting R848-induced IRF5 nuclear translocation in monocytes, they had little effect on IRF5 in B cells. This may be due to differential binding of N5-2 and N5-3 to IRF5 or differential uptake. Given the high binding affinity of N5-2 and N5-3 for IRF5, the data warrant further investigation of these inhibitors. The PTD enables cell permeability, yet endocytosis and macropinocytosis play a role in peptide uptake (50). Thus, differences in both endocytosis and pinocytosis between cell types may account for differences in uptake (Supplemental Figure 8). Additional studies will be required to discern the cell-type–specific effects of the inhibitors.
Among the most significant findings from the N5-1 studies was the long-term protection from spontaneous lupus onset and severity in NZB/W F1 mice treated for only 2 weeks. Sustained effects were observed in the mice out to 40 weeks of age. These data suggest that N5-1 is stable over long periods of time in vivo and/or that IRF5 hyperactivation is an early, targetable driver of lupus onset. The latter is more likely, given the expected shorter half-life of peptide inhibitors; however, we were able to detect a significant reversal in IRF5 hyperactivation in mice between 14 and 21 weeks of age (Figure 5, J and K). Another striking finding was that N5-1 proved clinically efficacious in NZB/W F1, MRL/lpr, and pristane-induced lupus mice when given after disease onset (Figure 7). In NZB/W F1 mice, the in vivo inhibition of IRF5 hyperactivation by N5-1 resulted in the reversal of several key pathogenic phenotypes that are associated with lupus severity, including serum ANA positivity, elevated dsDNA titers, expansion of circulating PCs and ABCs, and renal injury. Importantly, similar observations of reduced ANA and dsDNA titers along with increased survival were made in MRL/lpr and pristane-induced lupus mice after clinical treatment. The finding of decreased ANA and dsDNA titers in N5-1–treated NZB/W F1 mice before detection of a significant decrease in PCs and ABCs (Figure 5, E–I) is interesting and reminiscent of findings with IRF5 knockdown in human primary B cells that showed a larger decrease in secreted IgG isotypes than was detected in intracellular isotypes (63). Thus, in addition to playing a role in PC differentiation, these data provide added support for the notion that IRF5 may regulate antibody secretion (63). Our finding of in vivo IL-6 inhibition by N5-1 was also notable, as it mimicked the IL-6 levels seen in R848-injected Irf5–/– mice. Further, N5-1 exerted no additional effect on IL-6 production in Irf5–/– mice, confirming the specificity of N5-1 for IRF5. Treatment of healthy donor blood ex vivo with N5-1 confirmed the reduction in IRF5-mediated proinflammatory cytokines (IL6 and IFNA). Interestingly, N5-1 had no impact on TLR signaling itself, as TLR7 and TLR9 induced normal NF-κB and IRF7 nuclear translocation (Supplemental Figure 6, D and E). Altogether, these data show that IRF5 represents a particularly valuable, dual-function therapeutic target to treat autoimmune and inflammatory diseases. To date, this is the first report to our knowledge of a selective IRF5 inhibitor that directly binds to IRF5 to inhibit nuclear translocation and has in vivo clinical efficacy in murine models of lupus (39, 83, 84).
Methods
Details on methods are provided in the Supplemental Methods.
Statistics.
A 2-tailed Student’s t test was used for comparisons between 2 samples with normal distribution. Prior to testing, graph kurtosis was analyzed to ensure normal distribution. For comparisons of 1 factor across multiple groups, 1-way ANOVA was performed followed by Bonferroni’s multiple-comparison test. For comparisons of multiple factors over multiple groups, 2-way ANOVA was performed followed by Bonferroni’s multiple-comparison test. Correlation analysis was performed by linear regression using Pearson’s correlation coefficient. Proteinuria levels and body weight were compared by multiple t test, and FDR values were calculated using the Benjamini-Hochberg method to consider false-positive associations (threshold of 0.05). The Mann-Whitney U test was used for ANA-HEp2 and pathologic scoring. Survival curves were derived using the Kaplan-Meier method. GraphPad Prism 6 (GraphPad Software) was used for statistical analysis and graphing. A P value of less than 0.05 was considered significant. All data for SLE patients and mice are presented as the mean ± SEM; all other data are presented as the mean ± SD.
Study approval.
All human work was conducted in accordance with the IRBs of Rutgers Biomedical and Health Sciences and the Feinstein Institute for Medical Research. All animal care and experimental procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011)) and approved by the IACUC of the Feinstein Institute for Medical Research. Informed consent was obtained from all healthy donors and patients with SLE.
Author contributions
BJB and S. Song designed experiments. S. Song, SD, VN, MH, DL, TS, and NT performed in vitro experiments. CDT and KK performed FRET assays. S. Song, SC, and ML performed in vivo experiments. S. Sun performed modeling and docking analyses. SD, S. Song, S. Sun, BJB and YAA wrote the manuscript. EC, MM, and CA provided samples from patients with SLE. WLC performed pathologic analysis of blinded kidney samples. S. Song is designated the first co-author, as she performed all long-term in vivo experiments and completed the project; SD is designated the second co-author, as he began the project and performed all initial inhibitor studies in human cells. All authors participated in data analysis and preparation of the manuscript.
Supplementary Material
Acknowledgments
We thank EMD Serono for human p-IRF5 antibodies provided by MRC-PPU Reagents (https://mrcppureagents.dundee.ac.uk) and L. Persaud, S. Kandasami, and A. Shaw for obtaining consent from the patients. We thank George M. O’Brien from the Kidney Center at Yale for urine and sera analyses (NIH P30 DK079310). This work was supported by grants from the Lupus Research Alliance, the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) (AR065959-01), Flight Attendant Medical Research Institute (FAMRI) (grant 123059), the US Department of Defense (DoD) Congressionally Directed Medical Research Programs (CDMRP) Lupus Research Program (LRI170107, to BJB), the New Jersey Commission on Cancer Research (to SD), and the NIH (S10 RR033072-01, to YAA).
Version 1. 09/08/2020
In-Press Preview
Version 2. 11/16/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Footnotes
SD’s present address is: Department of Immunology and Inflammation, Pfizer Inc., Cambridge, Massachusetts, USA.
Conflict of interest: BJB is an inventor on a US patent application (US20200071370A1, Cell-penetrating peptides that inhibit IRF5 nuclear localization), assigned to Rutgers. BJB and S. Sun are inventors on a US provisional patent application (62/844,894, Inhibition of IRF5 protects from lupus onset and severity), assigned to Feinstein Institutes.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6700–6717.https://doi.org/10.1172/JCI120288.
Contributor Information
Su Song, Email: ssong2@northwell.edu.
Saurav De, Email: saurav.de@pfizer.com.
Victoria Nelson, Email: victoria.nelson.525@outlook.com.
Samin Chopra, Email: schopra3@northwell.edu.
Margaret LaPan, Email: mlapan@northwell.edu.
Kyle Kampta, Email: kylekampta@gmail.com.
Shan Sun, Email: ssun@tritdi.org.
Mingzhu He, Email: mhe@nshs.edu.
Cherrie D. Thompson, Email: cherriesherman.phd@gmail.com.
Dan Li, Email: dli5@northwell.edu.
Tiffany Shih, Email: tiffshih168@gmail.com.
Natalie Tan, Email: natalie_p_tan@college.harvard.edu.
Yousef Al-Abed, Email: yalabed@nshs.edu.
Eugenio Capitle, Email: capitlem@njms.rutgers.edu.
Cynthia Aranow, Email: caranow@nshs.edu.
Meggan Mackay, Email: mmackay@northwell.edu.
William L. Clapp, Email: clapp@pathology.ufl.edu.
Betsy J. Barnes, Email: bbarnes1@northwell.edu.
References
- 1.Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276(26):23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 2.Schoenemeyer A, et al. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280(17):17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 3.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434(7030):243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 4.Kaur A, Lee LH, Chow SC, Fang CM. IRF5-mediated immune responses and its implications in immunological disorders. Int Rev Immunol. 2018;37(5):229–248. doi: 10.1080/08830185.2018.1469629. [DOI] [PubMed] [Google Scholar]
- 5.Ban T, Sato GR, Tamura T. Regulation and role of the transcription factor IRF5 in innate immune responses and systemic lupus erythematosus. Int Immunol. 2018;30(11):529–536. doi: 10.1093/intimm/dxy032. [DOI] [PubMed] [Google Scholar]
- 6.Sigurdsson S, et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet. 2005;76(3):528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirschfield GM, et al. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet. 2010;42(8):655–657. doi: 10.1038/ng.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawidowicz K, et al. The interferon regulatory factor 5 gene confers susceptibility to rheumatoid arthritis and influences its erosive phenotype. Ann Rheum Dis. 2011;70(1):117–121. doi: 10.1136/ard.2010.129171. [DOI] [PubMed] [Google Scholar]
- 9.Gathungu G, Zhang CK, Zhang W, Cho JH. A two-marker haplotype in the IRF5 gene is associated with inflammatory bowel disease in a North American cohort. Genes Immun. 2012;13(4):351–355. doi: 10.1038/gene.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saigusa R, et al. Multifaceted contribution of the TLR4-activated IRF5 transcription factor in systemic sclerosis. Proc Natl Acad Sci U S A. 2015;112(49):15136–15141. doi: 10.1073/pnas.1520997112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matta B, Song S, Li D, Barnes BJ. Interferon regulatory factor signaling in autoimmune disease. Cytokine. 2017;98:15–26. doi: 10.1016/j.cyto.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25(3):383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Tackey E, Lipsky PE, Illei GG. Rationale for interleukin-6 blockade in systemic lupus erythematosus. Lupus. 2004;13(5):339–343. doi: 10.1191/0961203304lu1023oa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banchereau R, et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165(3):551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li QZ, et al. Interferon signature gene expression is correlated with autoantibody profiles in patients with incomplete lupus syndromes. Clin Exp Immunol. 2010;159(3):281–291. doi: 10.1111/j.1365-2249.2009.04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rönnblom L, Eloranta ML, Alm GV. The type I interferon system in systemic lupus erythematosus. Arthritis Rheum. 2006;54(2):408–420. doi: 10.1002/art.21571. [DOI] [PubMed] [Google Scholar]
- 17.Ripley BJ, Goncalves B, Isenberg DA, Latchman DS, Rahman A. Raised levels of interleukin 6 in systemic lupus erythematosus correlate with anaemia. Ann Rheum Dis. 2005;64(6):849–853. doi: 10.1136/ard.2004.022681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng D, et al. Genetic variants and disease-associated factors contribute to enhanced interferon regulatory factor 5 expression in blood cells of patients with systemic lupus erythematosus. Arthritis Rheum. 2010;62(2):562–573. doi: 10.1002/art.27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone RC, et al. Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arthritis Rheum. 2012;64(3):788–798. doi: 10.1002/art.33395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes BJ. Genetic versus non-genetic drivers of SLE: Implications of IRF5 dysregulation in both roads leading to SLE. Curr Rheumatol Rep. 2019;21(1):2. doi: 10.1007/s11926-019-0803-3. [DOI] [PubMed] [Google Scholar]
- 21.Feng D, Yang L, Bi X, Stone RC, Patel P, Barnes BJ. Irf5-deficient mice are protected from pristane-induced lupus via increased Th2 cytokines and altered IgG class switching. Eur J Immunol. 2012;42(6):1477–1487. doi: 10.1002/eji.201141642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, et al. Pleiotropic IFN-dependent and -independent effects of IRF5 on the pathogenesis of experimental lupus. J Immunol. 2012;188(8):4113–4121. doi: 10.4049/jimmunol.1103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tada Y, et al. Interferon regulatory factor 5 is critical for the development of lupus in MRL/lpr mice. Arthritis Rheum. 2011;63(3):738–748. doi: 10.1002/art.30183. [DOI] [PubMed] [Google Scholar]
- 24.Richez C, et al. IFN regulatory factor 5 is required for disease development in the FcgammaRIIB–/–Yaa and FcgammaRIIB–/– mouse models of systemic lupus erythematosus. J Immunol. 2010;184(2):796–806. doi: 10.4049/jimmunol.0901748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watkins AA, et al. IRF5 deficiency ameliorates lupus but promotes atherosclerosis and metabolic dysfunction in a mouse model of lupus-associated atherosclerosis. J Immunol. 2015;194(4):1467–1479. doi: 10.4049/jimmunol.1402807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savitsky DA, Yanai H, Tamura T, Taniguchi T, Honda K. Contribution of IRF5 in B cells to the development of murine SLE-like disease through its transcriptional control of the IgG2a locus. Proc Natl Acad Sci U S A. 2010;107(22):10154–10159. doi: 10.1073/pnas.1005599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manni M, et al. Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat Immunol. 2018;19(4):407–419. doi: 10.1038/s41590-018-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedl M, Abraham C. IRF5 risk polymorphisms contribute to interindividual variance in pattern recognition receptor-mediated cytokine secretion in human monocyte-derived cells. J Immunol. 2012;188(11):5348–5356. doi: 10.4049/jimmunol.1103319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancl ME, et al. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J Biol Chem. 2005;280(22):21078–21090. doi: 10.1074/jbc.M500543200. [DOI] [PubMed] [Google Scholar]
- 30.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol. 2002;22(16):5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, et al. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15(11):1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin R, Yang L, Arguello M, Penafuerte C, Hiscott J. A CRM1-dependent nuclear export pathway is involved in the regulation of IRF-5 subcellular localization. J Biol Chem. 2005;280(4):3088–3095. doi: 10.1074/jbc.M408452200. [DOI] [PubMed] [Google Scholar]
- 33.Ren J, Chen X, Chen ZJ. IKKβ is an IRF5 kinase that instigates inflammation. Proc Natl Acad Sci U S A. 2014;111(49):17438–17443. doi: 10.1073/pnas.1418516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Pelaez M, Lamont DJ, Peggie M, Shpiro N, Gray NS, Cohen P. Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci USA. 2014;111(49):17432–17437. doi: 10.1073/pnas.1418399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang Foreman HC, Van Scoy S, Cheng TF, Reich NC. Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS ONE. 2012;7(3):e33098. doi: 10.1371/journal.pone.0033098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alzaid F, et al. IRF5 governs liver macrophage activation that promotes hepatic fibrosis in mice and humans. JCI Insight. 2016;1(20):e88689. doi: 10.1172/jci.insight.88689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun K, et al. IRF5 regulates lung macrophages M2 polarization during severe acute pancreatitis in vitro. World J Gastroenterol. 2016;22(42):9368–9377. doi: 10.3748/wjg.v22.i42.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrne AJ, et al. A critical role for IRF5 in regulating allergic airway inflammation. Mucosal Immunol. 2017;10(3):716–726. doi: 10.1038/mi.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banga J, et al. Inhibition of IRF5 cellular activity with cell-penetrating peptides that target homodimerization. Sci Adv. 2020;6(20):eaay1057. doi: 10.1126/sciadv.aay1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin HJ, Lee JM, Walls D, Hayward SD. Manipulation of the toll-like receptor 7 signaling pathway by Epstein-Barr virus. J Virol. 2007;81(18):9748–9758. doi: 10.1128/JVI.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L, et al. Functional analysis of a dominant negative mutation of interferon regulatory factor 5. PLoS ONE. 2009;4(5):e5500. doi: 10.1371/journal.pone.0005500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren Z, et al. Overexpression of the dominant-negative form of interferon regulatory factor 1 in oligodendrocytes protects against experimental autoimmune encephalomyelitis. J Neurosci. 2011;31(23):8329–8341. doi: 10.1523/JNEUROSCI.1028-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim TY, et al. Oncogenic potential of a dominant negative mutant of interferon regulatory factor 3. J Biol Chem. 2003;278(17):15272–15278. doi: 10.1074/jbc.M205792200. [DOI] [PubMed] [Google Scholar]
- 44.Ning S, Huye LE, Pagano JS. Regulation of the transcriptional activity of the IRF7 promoter by a pathway independent of interferon signaling. J Biol Chem. 2005;280(13):12262–12270. doi: 10.1074/jbc.M404260200. [DOI] [PubMed] [Google Scholar]
- 45.Liao X, Reihl AM, Luo XM. Breakdown of immune tolerance in systemic lupus erythematosus by dendritic cells. J Immunol Res. 2016;2016:6269157. doi: 10.1155/2016/6269157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58(8):2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Feng D, Bi X, Stone RC, Barnes BJ. Monocytes from Irf5–/– mice have an intrinsic defect in their response to pristane-induced lupus. J Immunol. 2012;189(7):3741–3750. doi: 10.4049/jimmunol.1201162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theofilopoulos AN, Dixon FJ. Murine models of systemic lupus erythematosus. Adv Immunol. 1985;37:269–390. doi: 10.1016/S0065-2776(08)60342-9. [DOI] [PubMed] [Google Scholar]
- 49.Takahasi K, et al. X-ray crystal structure of IRF-3 and its functional implications. Nat Struct Biol. 2003;10(11):922–927. doi: 10.1038/nsb1001. [DOI] [PubMed] [Google Scholar]
- 50.Lin YZ, Yao SY, Veach RA, Torgerson TR, Hawiger J. Inhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J Biol Chem. 1995;270(24):14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- 51.Komatsuda A, et al. Up-regulated expression of Toll-like receptors mRNAs in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin Exp Immunol. 2008;152(3):482–487. doi: 10.1111/j.1365-2249.2008.03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyn-Cook BD, et al. Increased expression of Toll-like receptors (TLRs) 7 and 9 and other cytokines in systemic lupus erythematosus (SLE) patients: ethnic differences and potential new targets for therapeutic drugs. Mol Immunol. 2014;61(1):38–43. doi: 10.1016/j.molimm.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Subramanian S, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103(26):9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312(5780):1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 55.Qin BY, et al. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol. 2003;10(11):913–921. doi: 10.1038/nsb1002. [DOI] [PubMed] [Google Scholar]
- 56.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129(6):1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin BY, et al. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13(9):1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 58.Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 2003;63(19):6424–6431. [PubMed] [Google Scholar]
- 59.Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem. 2004;279(43):45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- 60.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 61.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 62.Purtha WE, Swiecki M, Colonna M, Diamond MS, Bhattacharya D. Spontaneous mutation of the Dock2 gene in Irf5–/– mice complicates interpretation of type I interferon production and antibody responses. Proc Natl Acad Sci U S A. 2012;109(15):E898–E904. doi: 10.1073/pnas.1118155109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De S, et al. B cell-intrinsic role for IRF5 in TLR9/BCR-induced human b cell activation, proliferation, and plasmablast differentiation. Front Immunol. 2017;8:1938. doi: 10.3389/fimmu.2017.01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubtsov AV, et al. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c+ B-cell population is important for the development of autoimmunity. Blood. 2011;118(5):1305–1315. doi: 10.1182/blood-2011-01-331462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhuang H, Szeto C, Han S, Yang L, Reeves WH. Animal models of interferon signature positive lupus. Front Immunol. 2015;6:291. doi: 10.3389/fimmu.2015.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Z, Davidson A. IFNα inducible models of murine SLE. Front Immunol. 2013;4:306. doi: 10.3389/fimmu.2013.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leng L, et al. A small-molecule macrophage migration inhibitory factor antagonist protects against glomerulonephritis in lupus-prone NZB/NZW F1 and MRL/lpr mice. J Immunol. 2011;186(1):527–538. doi: 10.4049/jimmunol.1001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathian A, Weinberg A, Gallegos M, Banchereau J, Koutouzov S. IFN-alpha induces early lethal lupus in preautoimmune (New Zealand Black x New Zealand White) F1 but not in BALB/c mice. J Immunol. 2005;174(5):2499–2506. doi: 10.4049/jimmunol.174.5.2499. [DOI] [PubMed] [Google Scholar]
- 69.Li D, et al. IRF5 genetic risk variants drive myeloid-specific IRF5 hyperactivation and presymptomatic SLE. JCI Insight. 2020;5(2):e124020. doi: 10.1172/jci.insight.124020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calise J, Marquez Renteria S, Gregersen PK, Diamond B. Lineage-specific functionality of an interferon regulatory factor 5 lupus risk haplotype: lack of b cell intrinsic effects. Front Immunol. 2018;9:996. doi: 10.3389/fimmu.2018.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnes BJ. Genetic versus non-genetic drivers of SLE: implications of IRF5 dysregulation in both roads leading to SLE. Curr Rheumatol Rep. 2019;21(1):2. doi: 10.1007/s11926-019-0803-3. [DOI] [PubMed] [Google Scholar]
- 72.Richard ML, Gilkeson G. Mouse models of lupus: what they tell us and what they don’t. Lupus Sci Med. 2018;5(1):e000199. doi: 10.1136/lupus-2016-000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ban T, et al. Lyn kinase suppresses the transcriptional activity of IRF5 in the TLR-MyD88 pathway to restrain the development of autoimmunity. Immunity. 2016;45(2):319–332. doi: 10.1016/j.immuni.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 74.Scapini P, Pereira S, Zhang H, Lowell CA. Multiple roles of Lyn kinase in myeloid cell signaling and function. Immunol Rev. 2009;228(1):23–40. doi: 10.1111/j.1600-065X.2008.00758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu Y, Harder KW, Huntington ND, Hibbs ML, Tarlinton DM. Lyn tyrosine kinase: accentuating the positive and the negative. Immunity. 2005;22(1):9–18. doi: 10.1016/j.immuni.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 76.Cohen P. Protein kinases — the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1(4):309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 77.Ott PA, Adams S. Small-molecule protein kinase inhibitors and their effects on the immune system: implications for cancer treatment. Immunotherapy. 2011;3(2):213–227. doi: 10.2217/imt.10.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cushing L, et al. IRAK4 kinase activity controls Toll-like receptor-induced inflammation through the transcription factor IRF5 in primary human monocytes. J Biol Chem. 2017;292(45):18689–18698. doi: 10.1074/jbc.M117.796912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu G, Mancl ME, Barnes BJ. Signaling through IFN regulatory factor-5 sensitizes p53-deficient tumors to DNA damage-induced apoptosis and cell death. Cancer Res. 2005;65(16):7403–7412. doi: 10.1158/0008-5472.CAN-05-0583. [DOI] [PubMed] [Google Scholar]
- 80.Hu G, Barnes BJ. IRF-5 is a mediator of the death receptor-induced apoptotic signaling pathway. J Biol Chem. 2009;284(5):2767–2777. doi: 10.1074/jbc.M804744200. [DOI] [PubMed] [Google Scholar]
- 81.Balkhi MY, Fitzgerald KA, Pitha PM. Functional regulation of MyD88-activated interferon regulatory factor 5 by K63-linked polyubiquitination. Mol Cell Biol. 2008;28(24):7296–7308. doi: 10.1128/MCB.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng D, et al. Differential requirement of histone acetylase and deacetylase activities for IRF5-mediated proinflammatory cytokine expression. J Immunol. 2010;185(10):6003–6012. doi: 10.4049/jimmunol.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu H, et al. 4F decreases IRF5 expression and activation in hearts of tight skin mice. PLoS ONE. 2012;7(12):e52046. doi: 10.1371/journal.pone.0052046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weihrauch D, et al. An IRF5 decoy peptide reduces myocardial inflammation and fibrosis and improves endothelial cell function in tight-skin mice. PLoS ONE. 2016;11(4):e0151999. doi: 10.1371/journal.pone.0151999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.