Abstract
Germ cell tumors (GCTs) are the most common cancer in men between the ages of 15 and 40. Although most patients are cured, those with disease arising in the mediastinum have distinctly poor outcomes. One in every 17 patients with primary mediastinal nonseminomatous GCTs develop an incurable hematologic malignancy and prior data intriguingly suggest a clonal relationship exists between hematologic malignancies and GCTs in these cases. To date, however, the precise clonal relationship between GCTs and the diverse additional somatic malignancies arising in such individuals have not been determined. Here, we traced the clonal evolution and characterized the genetic features of each neoplasm from a cohort of 15 patients with GCTs and associated hematologic malignancies. We discovered that GCTs and hematologic malignancies developing in such individuals evolved from a common shared precursor, nearly all of which harbored allelically imbalanced p53 and/or RAS pathway mutations. Hematologic malignancies arising in this setting genetically resembled mediastinal GCTs rather than de novo myeloid neoplasms. Our findings argue that this scenario represents a unique clinical syndrome, distinct from de novo GCTs or hematologic malignancies, initiated by an ancestral precursor that gives rise to the parallel evolution of GCTs and blood cancers in these patients.
Keywords: Genetics, Hematology
Keywords: Cancer, Leukemias
Introduction
Germ cell tumors (GCTs) are a model of curable cancer, as most patients with metastatic GCTs are successfully treated with cisplatin-based chemotherapy (1). However, up to 30% of patients with advanced GCTs develop cisplatin-resistant disease, which requires intensive salvage treatment and has a 50% risk of cancer-related death (2). In particular, primary mediastinal nonseminomatous GCTs (PM NSGCTs) have an adverse prognosis and a high frequency of cisplatin resistance (3). Moreover, diverse forms of somatic type malignancies including sarcomas, carcinomas, and a variety of hematologic malignancies can arise in patients with mediastinal GCTs (4, 5). For example, 1 in every 17 patients with PM NSGCT will develop a hematologic malignancy (5, 6), a clinical presentation associated with a dismal median survival of less than 6 months.
Although initially presumed to represent a therapy-induced secondary complication of chemotherapy, a shared clonal origin for GCTs and hematologic malignancies arising in the same individual has been suggested by the occasional shared presence of isochromosome 12p [i(12p)], a clonal marker common in GCTs but absent in hematologic cancers (7–10). Although the identification of i(12p) in hematologic malignancies arising in GCT patients suggests a clonal relationship between the 2 disorders (7–10), the evolutionary dynamics and comprehensive characterization of the genomic features of this syndrome have thus far been lacking. Given the totipotent nature of GCTs, it has been postulated that hematologic malignancies arising in this setting are derived directly from cells within the fully formed GCT that have the capacity to differentiate into hematopoietic tissue (11, 12). However, a competing hypothesis is that a progenitor with the capacity to differentiate into germ cell and hematopoietic lineages harbors the initiating genetic alterations and drives development of both malignancies (10) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI139682DS1). To trace the clonal origins of these multiple malignancies within individual patients and understand their comprehensive genomic features, we performed broad mutational and copy number analyses of GCTs, the hematologic malignancies developing in these individuals, and patient-matched normal DNA from a cohort of 15 patients.
Results
We identified 15 individuals diagnosed with GCT and co-incident hematologic malignancies over a 21-year period (1998–2019; Figure 1A and Supplemental Table 1; Methods). Despite allowing the collection of GCTs from any gender and histologic subtype, all patients developing GCTs and concurrent blood cancers were males between the age of 18 and 33 (median age 25) with PM NSGCTs. The most common GCT histology was a teratoma component (present in 73% and as the predominant histology in 60% of cases) mixed with nonseminomatous components, most commonly a yolk sac tumor component (present in 53%; more details in Supplemental Table 1).
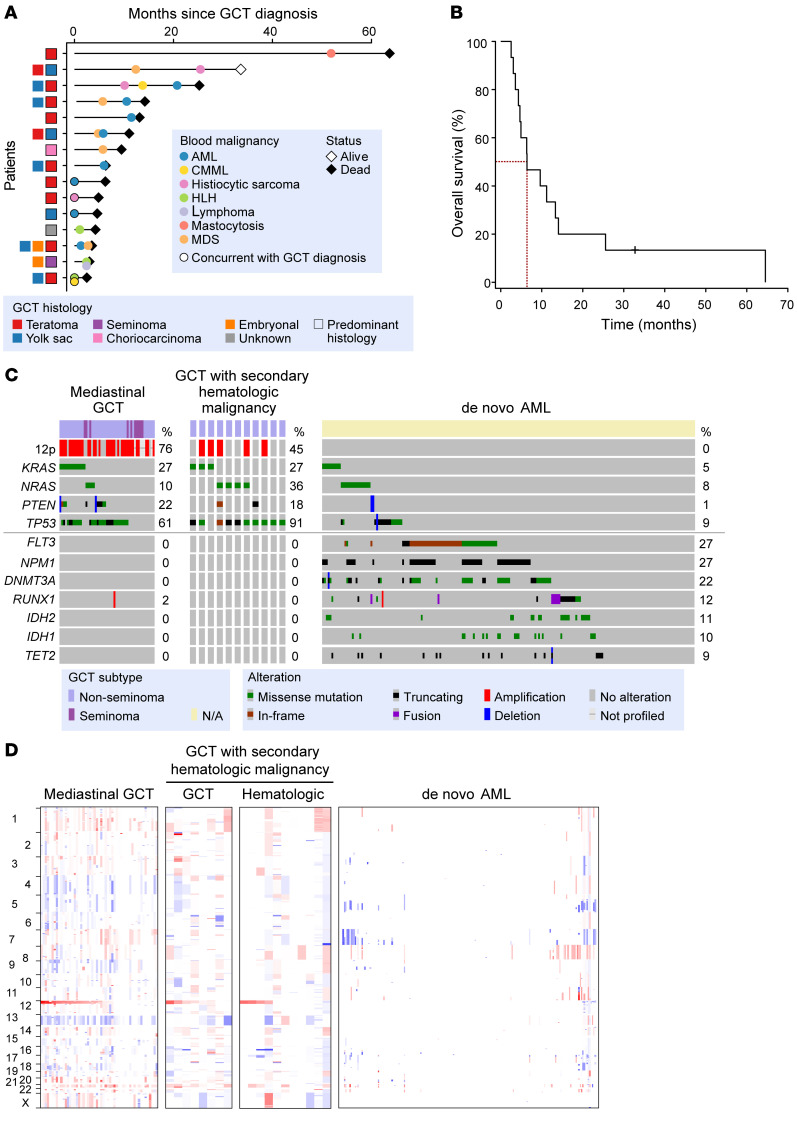
Figure 1. Genetic and clinical characteristics of patients with germ cell tumors (GCTs) and concomitant hematologic malignancies.
(A) Timeline of diagnoses and histologic characteristics of GCT and blood cancers from all 15 patients. Each patient is shown in a row on the x axis and timeline of diagnosis is shown on the y axis. AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; HLH, hemophagocytic lymphohistiocytosis; MDS, myelodysplastic syndrome. (B) Kaplan-Meier curve of the patients from A. Survival is shown as time from diagnosis of GCT. Median survival is 6.3 months (95% CI 4.6–25.2 months). (C) Shown are the most prevalent genetic alterations in patients with mediastinal GCTs and no secondary malignancy diagnosis (left; n = 51), mediastinal GCT with hematologic malignancy (middle; n = 11; composite for GCT and hematologic malignancy samples shown), or de novo AML (n = 200, from the AML TCGA; ref. 13). (D) DNA copy number alterations (CNAs) in the same cohorts of patients from C (genomic gains and losses are red and blue, respectively). GCT and hematologic malignancy samples displayed separately for CNAs (GCT = 8, HM = 9 patients [11 samples]).
Overall, the median time between GCT and blood cancer diagnoses was only 4.8 months, while 4 patients concurrently developed their GCT and hematologic cancer before any anticancer therapy (Figure 1A). The most common hematologic malignancy was acute myeloid leukemia (AML) in 53% of patients, but myelodysplastic syndromes (MDS), histiocytic sarcoma, and chronic myelomonocytic leukemia (CMML) were also diagnosed in 2 or more patients each. Nearly half of the AMLs seen in this setting were acute megakaryoblastic leukemias (M7 AML). However, one-third of the cohort developed multiple distinct clinical subtypes of hematologic malignancies, often in a temporally distinct manner (Figure 1A and Supplemental Table 1).
Consistent with prior reports documenting the poor survival of patients who develop both GCTs and hematologic malignancies (7–10), the median survival of this cohort was 6.3 months (95% confidence interval [CI] 4.6–25.2 months) (Figure 1B). Only one patient from the cohort is alive to date (and the cause of death for each patient is noted in Supplemental Table 1). In order to understand the genetic features of the GCTs and hematologic malignancies arising in this aggressive clinical setting, in 11 patients with available tissue, we performed targeted and/or whole-exome sequencing (WES) of tumor DNA from the GCT, hematologic malignancy (blood, marrow, or biopsy of affected solid tissue), and patient-matched normal controls using DNA from fingernails. The genetic features of each tumor in these patients were then compared with those of a cohort of 51 patients with mediastinal GCTs (including 10 with seminoma and 41 with nonseminoma histology) who did not develop additional malignancies after long-term clinical follow-up as well as 200 de novo AML patients of a similar age range (from the AML TCGA data set; ref. 13) (Figure 1, C and D, and Supplemental Figure 2).
Blood cancers developing in the setting of PM NSGCTs were far more genetically similar to PM NSGCTs overall than to de novo AML. For example, i(12p) was commonly seen in mediastinal GCTs and hematologic malignancies in our cohort, whereas i(12p) was never observed in de novo AML (Figure 1, C and D, and Supplemental Table 2). Although a complex karyotype with multiple chromosome abnormalities was evident in the majority of hematologic malignancies developing in the setting of GCTs, i(12p) could be identified as an isolated clone by conventional cytogenetics in several of these individuals, suggesting i(12p) as an early event in these cases (Supplemental Figure 3 and Supplemental Table 3). In addition to i(12p), shared mutations in TP53 were present in 91% of patients with mediastinal GCTs with hematologic malignancies versus 61% of patients with mediastinal GCTs lacking hematologic malignancies and in only 9% of patients with de novo AML (Figure 1C). Given recent data on the association of p53 pathway alterations and cisplatin resistance in GCTs (14) and the known adverse prognostic effect of TP53 alterations in myeloid neoplasms (15–17), the nearly obligate presence of TP53 mutations in GCTs and their associated secondary blood cancers provides one basis for the short survival of these patients. Activating mutations in KRAS and NRAS accounted for 63% and 37% of mediastinal GCTs with hematologic malignancies and without accompanying hematologic malignancies, respectively, but were present in only 13% of de novo AML. In contrast to shared genetic similarities in i(12p), TP53 alterations, and mutations activating RAS/PI3K signaling in GCTs and their associated blood cancers, blood cancers arising in association with GCTs lacked mutations in the most commonly mutated genes in AML, MDS, or clonal hematopoiesis (such as FLT3, DNMT3A, TET2, and NPM1 mutations; Figure 1, C and D). These findings suggest that the hematologic malignancies developing in the setting of GCTs are clonally related to mediastinal GCTs and have a pathogenesis distinct from other hematopoietic malignancies. This represents a possible basis for their treatment refractoriness, as these blood cancers are molecularly distinct from the hematologic malignancies they histologically mimic.
We next sought to trace the evolutionary origin of each distinct cancer type developing in these patients by using the somatic mutational and DNA copy number data from WES in 5 patients (Figure 2, Figure 3C, and Figure 4F). In every such patient, we identified divergent genetic evolution of the GCT and blood cancer, with putative driver alterations unique to each malignancy (sometimes in the same gene). In contrast to prior data that secondary malignancies developing in the setting of GCTs represent malignant transformation of differentiated GCT components such as teratoma and yolk sac tumor (11, 12), the present data suggest instead that these malignancies evolved independently from a shared ancestral clone common to both malignancies (Supplemental Figure 1). For example, in one 24-year-old male with concurrent PM NSGCT and CMML, both cancers had i(12p) and KRAS G12S mutations, along with 8 additional shared truncal mutations of unknown functional significance (Figure 2, A–C). These alterations were then followed by the independent acquisition of a distinct set of mutations (including oncogenic mutations) and copy number alterations (CNAs) arising after a branching of their disease evolution, thereby indicating that both conditions developed from a shared precursor (as opposed to dedifferentiation or histologic transformation of mature elements in the GCT into the subsequent hematologic malignancy as has been speculated).
Figure 2. Genetic evolution of germ cell tumors (GCTs) and hematologic malignancies from a common ancestral clone.
Whole-exome sequencing was performed in 5 patients, 3 shown here and 2 shown in Figures 3 and 4. (A) Shown are the somatic mutations (known oncogenic and variants of undetermined significance [VUS]) and their fraction of tumor cells mutated (cancer cell fraction [CCF]), the (B) allelic DNA copy number, and (C) inferred evolutionary relationships of a GCT and associated myelodysplastic syndrome (MDS) in a 24-year-old male with both diagnoses. An ancestral clone with shared isochromosome 12p and a KRAS G12S mutation (among other shared mutations) was detected followed by independent acquisition of numerous distinct mutations. (D–F) Similar CCF (D), allele copy number (E), and evolutionary relationships (F) in a 28-year-old with shared GCT and AML. In this case, both tumors derived from a TP53 R248L–mutant ancestral precursor that underwent copy-neutral loss of heterozygosity (CNLOH). (G–J) In another patient, a KRAS G12D–mutant precursor gave rise to GCT and MDS, each of which later independently acquired TP53 hotspot mutations with CNLOH. (I) Aligned TP53 reads over the regions of mutations, highlighting mutual exclusivity of each mutation across the GCT and MDS samples. AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia.
Figure 3. Genomic evolution of multiple clinically and genetically distinct hematologic malignancies arising in the setting of a clonally related germ cell tumor (GCT).
(A) Cancer cell fraction (CCF) of mutations, (B) allele copy number (middle), (C) evolutionary relationship, and (D) histopathology (hematoxylin and eosin stain) of a 19-year-old patient successively diagnosed with a GCT, histiocytic sarcoma, chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML). Scale bars: 200 μm (black) and 1 μm (white). The timeline of acquisition of each diagnosis is shown at the top of A. In this case, TP53 Q144Rfs*26 mutation undergoing copy-neutral loss of heterozygosity defined the ancestral precursor with a PIK3CD mutation that gave rise to all 4 malignancies. This was followed by divergent evolution of 1 intermediate precursor that gave rise to the GCT and AML and another that resulted in development of the histiocytic sarcoma and CMML. MDS, myelodysplastic syndrome; CNLOH, copy-neutral loss of heterozygosity.
Figure 4. Lack of clonal hematopoiesis in autologous stem cell products from germ cell tumor (GCT) patients developing hematologic malignancies.
(A) Clinical course of a 25-year-old patient with metastatic primary mediastinal nonseminomatous GCT who was treated with multiple salvage chemotherapy regiments (VIP: etoposide, ifosfamide, cisplatin; TICE: paclitaxel, ifosfamide, followed by high-dose carboplatin plus etoposide) as well as autologous stem cell transplant (auto-SCT). This was followed by development of a myelodysplastic syndrome (MDS) 1 year after GCT diagnosis, treated by allogeneic SCT. A histiocytic sarcoma was then found 1 year after allogeneic SCT based on a PET/CT scan. (B) Histopathology (hematoxylin and eosin stain) of the yolk sac and teratoma component of the GCT as well as MDS and histiocytic sarcoma. Scale bars: 200 μm (black) and 1 μm (white). (C) Somatic mutations in TP53, BCOR, and NRAS illustrate the clonal relationships of the GCT, MDS, and histiocytic sarcoma versus the auto-SCT product. (D) The cancer cell fraction (CCF) of both shared and distinct somatic mutations in the GCT and MDS. (E) Copy number analysis of the GCT and MDS. (F) Evolutionary relationship of the GCT, MDS, and histiocytic sarcoma. (G) Aligned TP53 reads over the regions of mutations, highlighting mutual exclusivity of each mutation across the GCT and MDS samples compared with the histiocytic sarcoma sample. CNLOH, copy-neutral loss of heterozygosity; VAF, variant allele frequency; VUS, variants of undetermined significance.
In nearly every case, the common precursor for the GCT and blood cancer harbored either a p53 or RAS pathway alteration (Figure 2, D–J). For example, we identified one 28-year-old male with a PM NSGCT and a diagnosis of high-risk MDS wherein both malignancies developed from a shared KRAS G12D–mutant precursor bearing identical CNAs on chromosomes 13 and 17 (Figure 2, G–J). Each malignancy thereafter acquired distinct somatic mutations and CNAs including TP53 hotspot mutations, each of which underwent independent copy-neutral loss of heterozygosity (CNLOH), thus evolving from a shared common ancestor followed by divergent parallel evolution of each disease (Figure 2J).
Even in cases where patients harbored multiple temporally distinct blood cancers, each subtype of hematologic malignancy was found to have its own unique precursor shared with the GCT. For example, we studied a 19-year-old patient who developed successive diagnoses of histiocytic sarcoma, CMML, and AML within 18 months of GCT diagnosis (Figure 3). Tracing these 4 individual cancers via their somatic mutational profiles revealed that all were clonally related and derived from a common precursor bearing a TP53 Q144Rfs*28 mutation with CNLOH, a PI3KCD mutation, and shared CNAs on chromosomes 1, 2, and 5. From this shared precursor, the GCT and AML evolved from a daughter precursor bearing 1q and 21q gain, while the histiocytic sarcoma and CMML evolved from a separate daughter precursor. Further divergence between the CMML and histiocytic sarcoma is highlighted by a unique NRAS Q61H mutation in the histiocytic sarcoma (Figure 3, B and C). Interestingly, the inferred clonal relatedness of the individual malignancies did not match the order of diagnosis, supporting the hypothesis that all malignancies were derived from a common precursor clone, rather than serially transformed from GCT elements in the mediastinal tumor. Notably, this patient’s GCT did not have the i(12p) abnormality and thus might not have been identified as having clonally related GCT and leukemia based on karyotype alone.
High-dose chemotherapy and autologous stem cell transplantation (auto-SCT) is a treatment for relapsed GCTs (18), a clinical scenario encompassing some of the GCT patients in this study. Given recent recognition for the potential for contamination of hematopoietic stem cell transplant products with clonal hematopoiesis (CH) (19), we next sought to determine if there was CH in the G-CSF–mobilized stem cell products collected from individuals with high-risk PM GCTs. We therefore performed genetic analysis of G-CSF–mobilized hematopoietic stem cell products from 5 PM GCT patients who had undergone stem collection for potential autologous transplantation (Supplemental Table 3). Only 1 patient from this cohort developed a subsequent hematologic malignancy, despite undergoing high-dose chemotherapy with auto-SCT as treatment for their high-risk GCT; he developed a clonally related MDS shortly thereafter (1 year after GCT diagnosis) (Figure 4A). After a subsequent allogeneic SCT, the patient developed a second clinically and histologically distinct hematologic malignancy — a histiocytic sarcoma (Figure 4B). WES revealed that all 3 tumors were host derived and shared a somatic BCOR G943D mutation as well as an RRAS2 activating mutation with CNLOH (Figure 4, C–F). Every disease thereafter acquired TP53 mutations with CNLOH (Figure 4, D–G, and Supplemental Figure 4). Again, the acquisition of a different TP53 mutation and successive independent LOH event in the histiocytic sarcoma suggest secondary malignancies developing from a precursor clone that also gave rise to the GCT (Figure 4, D–G). To confirm, we sequenced the auto-SCT product, which was collected from the patient before being diagnosed with hematologic malignancies, only to find that the mobilized stem cell product did not contain any mutations (Figure 4C). As with this index case, none of 4 additional PM GCT patients with auto-SCT products sequenced had evidence of CH (Supplemental Table 3), which is consistent with the rarity of CH in GCT patients overall (20). Together, these data suggest that the hematologic malignancies seen in these patients did not arise from the hematopoietic system, but rather developed from the same cell of origin as the GCT.
Discussion
The development of clonally related somatic type cancers in the setting of GCTs has been recognized for nearly 30 years (5) and is marked by a very poor outcome, despite modern therapy for both cancer types. To date, the challenge of capturing both sets of malignancies in this aggressive clinical scenario combined with its relative rarity have limited efforts to understand this clinical phenomenon. Here we identify the precise clonal relationships of the tumors arising in these patients, the genetic features of PM GCTs in general, as well as the subset that develop secondary hematologic malignancies. These comprehensive maps of the somatic mutations and CNAs across solid and liquid malignancies in such individuals identify a common shared precursor in the germ cell lineage from which both the GCT and other malignancies develop. As such, the hematologic malignancies developing in these patients are genetically more similar to primary mediastinal GCTs than to de novo MDS or AML and lack many of the hallmark genomic drivers of myeloid malignancies. At the same time, the fact that hematologic malignancies bearing i(12p) do not develop unless accompanied by a GCT strongly suggests that hematologic malignancies present in this setting derive from a precursor along the GCT lineage.
All patients here who developed a hematologic malignancy clonally related to GCT had a PM NSGCT and we did not identify any patients with testicular or seminomatous GCTs that developed a clonally related hematologic malignancy. Although this is consistent with prior reports (7–10), the basis for the specific association between somatic type hematologic malignancies and PM NSGCTs remains unclear. Overall, our data are consistent with the model that PM NSGCTs are derived from transformed primordial germ cells lodged in the mediastinum, which have been reprogrammed to embryonal carcinoma cells and are believed to be the initiating cells of NSGCTs (21). Indeed, none of the hematologic malignancies here were preceded by a pure seminoma, making it unlikely that they can be derived directly from a primordial germ cell. Thus, we hypothesize that the precursor to both PM NSGCTs and clonally related hematologic malignancies in these individuals may be embryonal carcinoma cells, which have the potential to differentiate toward all components of an NSGCT as well as hematopoietic precursor cells. This is distinct from prior studies (11, 12), which postulated that hematologic malignancies in these cases develop from teratomas or yolk sac elements within the GCT.
The unique genetic features of hematologic malignancies in GCT patients along with the high frequency of TP53 alterations likely explains the frequent cisplatin resistance in this patient population as well the ineffectiveness of therapies used for high-risk MDS and AML. This clinical scenario is particularly unique given the co-occurrence of multiple, otherwise unrelated rare cancer types in a young adult population and the fact that this condition is invariably fatal. As such, future studies to evaluate for potential germline genetic predisposition for the development of hematologic malignancies in the setting of GCTs are warranted. Of note, 2 patients with PM NSGCT that developed hematologic malignancies here had clinical diagnoses of Klinefelter syndrome, consistent with prior data regarding increased incidence of PM NSGCT in patients with Klinefelter syndrome (22). Although none of the 41 additional PM NSGCT patients without hematologic malignancies had known Klinefelter syndrome, further germline genomic analysis would be needed to evaluate for potential mosaicism or subclinical Klinefelter syndrome in these individuals. Of note, we identify far more diverse forms of myeloid neoplasms developing in this setting than appreciated in prior studies and observed that multiple genetically distinct myeloid neoplasms often develop in individual patients affected by this condition. Overall, these findings argue that the development of blood cancers in patients with GCTs represents a unique clinical syndrome wherein the aggressive hematologic malignancies in these individuals do not arise from the hematopoietic system, like de novo hematologic malignancies, but rather develop from the same cell of origin as the GCT.
Methods
Pathology review.
All samples underwent detailed pathology review to confirm diagnosis by both specialized hematopathologists as well genitourinary pathologists, and cytogenetics of both GCT and hematologic malignancies were reviewed by dedicated cytogeneticists.
Genomic sequencing and mutation calling.
Next-generation sequencing was performed on DNA extracted from tumor and matched normal DNA samples from fingernails. Patient samples were sequenced using the Memorial Sloan Kettering IMPACT targeted sequencing panel, with somatic mutations (substitutions and small insertions and deletions), gene-level focal CNAs, and structural rearrangements detected with a clinically validated pipeline as previously described (23, 24). WES and mutational analysis was performed on a subset of cases as previously described (25). Depth coverage at the positions of nontruncal mutations in the tumors in which they were not detected was calculated as median 196 (IQR 112–312). Those loci with tumor depth less than 20 (the same depth cutoff used to filter called mutations) were excluded from all related samples. The OncoKB knowledge base was used to identify oncogenic alterations (26). For the post–allogeneic transplant histiocytic sarcoma sample from patient 2 (Figure 4), likely true somatic mutations were distinguished from donor-derived contamination using the variant allele frequency (VAF) of mutations previously identified in preallogeneic samples from the same patient along with the known zygosity of those mutations in said samples (Supplemental Figure 4). This sample was also excluded from copy number analysis. Patient 3’s NRAS mutation in their histiocytic sarcoma sample was initially identified in additional IMPACT sequencing, read support 2.05% (14 of 683), reaching the threshold for reporting oncogenic/hotspot mutations. Read support in the WES was 1.02% (2 of 197). The same NRAS mutation was not present in other the histologic components via IMPACT sequencing (GCT: 0 of 874; CMML: 1 of 358; AML: 0 of 574).
Allele-specific copy number analysis and cancer cell fractions.
Genome-wide total and allele-specific DNA copy number, purity, and ploidy were calculated for all exome data (FACETS version 0.5.6; ref. 27). Only clonal CNAs were considered, i.e., copy number segments with a cellular fraction having 80% or greater estimated purity. The expected number of copies for each mutation was generated based on observed VAF and local ploidy (28). Cancer cell fractions were calculated using a binomial distribution and maximum likelihood estimation normalized to produce posterior probabilities (29).
Phylogenetic reconstruction.
Clone trees were manually constructed based on the most parsimonious sequence of events using mutations and copy number events. Branch lengths represent the number of shared or unique alterations.
Data availability.
Whole-exome data were deposited at dbGap under accession ID phs2231.V1 and IMPACT mutation calls were deposited via the cBioPortal for Cancer Genomics (https://www.cbioportal.org/study/summary?id=gct_msk_2020).
Statistics.
Progression-free survival (PFS) 95% CIs were calculated using the R survival package (v.3.1-8).
Study approval.
These studies were approved by the Institutional Review Board of Memorial Sloan Kettering Cancer Center and conducted in accordance with the Declaration of Helsinki protocol (IRB protocols 15-021, 06-107). All patients provided written informed consent before solid tumor tissue, fingernails, or peripheral blood or bone marrow mononuclear cells were used.
Author contributions
JT, CH, KPD, DRF, and OAW designed the study. MTAD, PR, SSC, and BST performed genomic analyses. CH, KPD, HAA, AD, SKT, and VER performed histopathologic analyses; CH, KPD, and HAA performed detailed pathological review of all the samples. YZ and UA performed cytogenetic analyses of both GCT and hematologic malignancies. SAF, MH, RT, SG, MST, RKR, PB, RK, SK, GJB, DBS, CAID, and DRF helped with patient recruitment and sample collection. JT, MTAD, GJB, DRF, and OAW wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
We thank the patients, their families, and Barbara Solit for tireless help with patient recruitment through Cycle for Survival and the Memorial Sloan Kettering Cancer Center’s Make-An-IMPACT program. JT is supported by the Conquer Cancer Foundation of the American Society of Clinical Oncology, the American Association for Cancer Research, the American Society of Hematology, the Robert Wood Johnson Foundation, and the NIH/NCI (1K08CA230319-01). This work was also funded by Cycle for Survival grants to BST, DBS, DRF, and OAW, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology and the NIH/NCI Cancer Center Support Grant P30 CA008748.
Version 1. 09/08/2020
In-Press Preview
Version 2. 11/16/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Footnotes
Conflict of interest: SAF has received research support from AstraZeneca and Genentech/Roche, has served in a consulting or advisory role for Decibel, AstraZeneca, and Immunai, and has stock or other ownership interests in Urogen, Allogene Therapeutics, Neogene Therapeutics, Vaxigene, Kronos Bio, and Vida Ventures. RKR has received consulting fees from Constellation, Incyte, Celgene, Promedior, CTI, Jazz Pharmaceuticals, Blueprint, and Stemline, and research funding from Incyte, Constellation, and Stemline. DBS has served as a consultant for Pfizer, Loxo Oncology, Lilly Oncology, Q.E.D. Therapeutics, and Illumina. BST reports honoraria and research funding from Genentech and advisory board activities for Boehringer Ingelheim and Loxo Oncology, a wholly owned subsidiary of Eli Lilly. OAW has served as a consultant for H3B Biomedicine, Foundation Medicine Inc, Merck, and Janssen, and is on the Scientific Advisory Board of Envisagenics Inc; OAW has received prior research funding from H3B Biomedicine.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6668–6676.https://doi.org/10.1172/JCI139682.
See the related Commentary at Mediastinal germ cell tumors: many questions and perhaps an answer.
Contributor Information
Justin Taylor, Email: jtmed11@gmail.com.
Mark T.A. Donoghue, Email: donoghum@mskcc.org.
Caleb Ho, Email: hoc@mskcc.org.
Kseniya Petrova-Drus, Email: petrovak@mskcc.org.
Hikmat A. Al-Ahmadie, Email: alahmadh@mskcc.org.
Samuel A. Funt, Email: safunt@gmail.com.
Yanming Zhang, Email: zhangy1@mskcc.org.
Umut Aypar, Email: ayparu@mskcc.org.
Shweta S. Chavan, Email: chavans@mskcc.org.
Michael Haddadin, Email: michealmhm17@gmail.com.
Roni Tamari, Email: tamarir@mskcc.org.
Sergio Giralt, Email: giralts@mskcc.org.
Martin S. Tallman, Email: tallmanm@mskcc.org.
Raajit K. Rampal, Email: rampalr@mskcc.org.
Priscilla Baez, Email: baezp@mskcc.org.
Rajya Kappagantula, Email: kappagar@mskcc.org.
Satyajit Kosuri, Email: skosuri@medicine.bsd.uchicago.edu.
Ahmet Dogan, Email: dogana@mskcc.org.
Satish K. Tickoo, Email: tickoos@mskcc.org.
Victor E. Reuter, Email: reuterv@mskcc.org.
George J. Bosl, Email: boslg@mskcc.org.
Christine A. Iacobuzio-Donahue, Email: iacobuzc@mskcc.org.
David B. Solit, Email: solitd@mskcc.org.
Barry S. Taylor, Email: btaylor@loxooncology.com.
Darren R. Feldman, Email: Feldmand@MSKCC.ORG.
Omar Abdel-Wahab, Email: abdelwao@mskcc.org.
References
- 1.Cheng L, et al. Testicular cancer. Nat Rev Dis Primers. 2018;4(1):29. doi: 10.1038/s41572-018-0029-0. [DOI] [PubMed] [Google Scholar]
- 2.Hanna NH, Einhorn LH. Testicular cancer — discoveries and updates. N Engl J Med. 2014;371(21):2005–2016. doi: 10.1056/NEJMra1407550. [DOI] [PubMed] [Google Scholar]
- 3.No authors listed. International Germ Cell Consensus Classification: a prognostic factor-based staging system for metastatic germ cell cancers. International Germ Cell Cancer Collaborative Group. J Clin Oncol. 1997;15(2):594–603. doi: 10.1200/JCO.1997.15.2.594. [DOI] [PubMed] [Google Scholar]
- 4.Nichols CR, Hoffman R, Einhorn LH, Williams SD, Wheeler LA, Garnick MB. Hematologic malignancies associated with primary mediastinal germ-cell tumors. Ann Intern Med. 1985;102(5):603–609. doi: 10.7326/0003-4819-102-5-603. [DOI] [PubMed] [Google Scholar]
- 5.DeMent SH, Eggleston JC, Spivak JL. Association between mediastinal germ cell tumors and hematologic malignancies. Report of two cases and review of the literature. Am J Surg Pathol. 1985;9(1):23–30. doi: 10.1097/00000478-198501000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Hartmann JT, et al. Hematologic disorders associated with primary mediastinal nonseminomatous germ cell tumors. J Natl Cancer Inst. 2000;92(1):54–61. doi: 10.1093/jnci/92.1.54. [DOI] [PubMed] [Google Scholar]
- 7.Chaganti RS, et al. Leukemic differentiation of a mediastinal germ cell tumor. Genes Chromosomes Cancer. 1989;1(1):83–87. doi: 10.1002/gcc.2870010113. [DOI] [PubMed] [Google Scholar]
- 8.Ladanyi M, et al. Cytogenetic and immunohistochemical evidence for the germ cell origin of a subset of acute leukemias associated with mediastinal germ cell tumors. J Natl Cancer Inst. 1990;82(3):221–227. doi: 10.1093/jnci/82.3.221. [DOI] [PubMed] [Google Scholar]
- 9.Oshrine BR, et al. Acquired isochromosome 12p, somatic TP53 and PTEN mutations, and a germline ATM variant in an adolescent male with concurrent acute megakaryoblastic leukemia and mediastinal germ cell tumor. Cancer Genet. 2014;207(4):153–159. doi: 10.1016/j.cancergen.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Lu C, et al. A common founding clone with TP53 and PTEN mutations gives rise to a concurrent germ cell tumor and acute megakaryoblastic leukemia. Cold Spring Harb Mol Case Stud. 2016;2(1):a000687. doi: 10.1101/mcs.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, et al. Teratoma with malignant transformation: diverse malignant histologies arising in men with germ cell tumors. J Urol. 1998;159(1):133–138. doi: 10.1016/S0022-5347(01)64035-7. [DOI] [PubMed] [Google Scholar]
- 12.Orazi A, Neiman RS, Ulbright TM, Heerema NA, John K, Nichols CR. Hematopoietic precursor cells within the yolk sac tumor component are the source of secondary hematopoietic malignancies in patients with mediastinal germ cell tumors. Cancer. 1993;71(12):3873–3881. doi: 10.1002/1097-0142(19930615)71:12<3873::AID-CNCR2820711214>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagrodia A, et al. Genetic determinants of cisplatin resistance in patients with advanced germ cell tumors. J Clin Oncol. 2016;34(33):4000–4007. doi: 10.1200/JCO.2016.68.7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grinfeld J, et al. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379(15):1416–1430. doi: 10.1056/NEJMoa1716614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adra N, Abonour R, Althouse SK, Albany C, Hanna NH, Einhorn LH. High-dose chemotherapy and autologous peripheral-blood stem-cell transplantation for relapsed metastatic germ cell tumors: the Indiana University experience. J Clin Oncol. 2017;35(10):1096–1102. doi: 10.1200/JCO.2016.69.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson CJ, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol. 2017;35(14):1598–1605. doi: 10.1200/JCO.2016.71.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coombs CC, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374–382.e4. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oosterhuis JW, Looijenga LHJ. Human germ cell tumours from a developmental perspective. Nat Rev Cancer. 2019;19(9):522–537. doi: 10.1038/s41568-019-0178-9. [DOI] [PubMed] [Google Scholar]
- 22.Hasle H, Mellemgaard A, Nielsen J, Hansen J. Cancer incidence in men with Klinefelter syndrome. Br J Cancer. 1995;71(2):416–420. doi: 10.1038/bjc.1995.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsson P, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571(7766):576–579. doi: 10.1038/s41586-019-1382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravarty D, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017:PO.17.00011. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dentro SC, Wedge DC, Van Loo P. Principles of reconstructing the subclonal architecture of cancers. Cold Spring Harb Perspect Med. 2017;7(8):a026625. doi: 10.1101/cshperspect.a026625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7(283):283ra54. doi: 10.1126/scitranslmed.aaa1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-exome data were deposited at dbGap under accession ID phs2231.V1 and IMPACT mutation calls were deposited via the cBioPortal for Cancer Genomics (https://www.cbioportal.org/study/summary?id=gct_msk_2020).