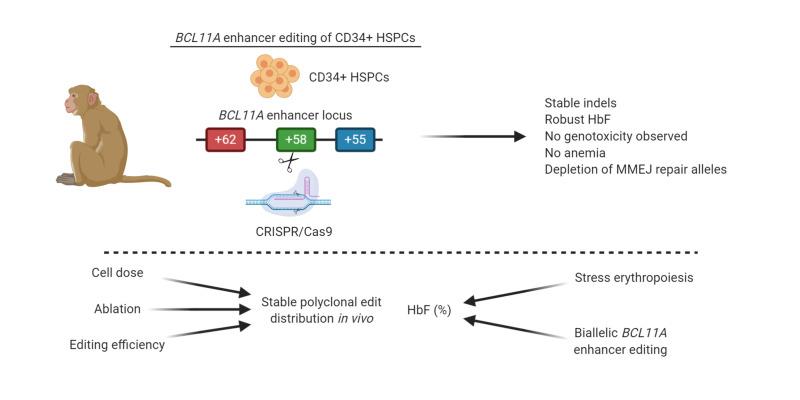
Abstract
Gene editing of the erythroid-specific BCL11A enhancer in hematopoietic stem and progenitor cells (HSPCs) from patients with sickle cell disease (SCD) induces fetal hemoglobin (HbF) without detectable toxicity, as assessed by mouse xenotransplant. Here, we evaluated autologous engraftment and HbF induction potential of erythroid-specific BCL11A enhancer–edited HSPCs in 4 nonhuman primates. We used a single guide RNA (sgRNA) with identical human and rhesus target sequences to disrupt a GATA1 binding site at the BCL11A +58 erythroid enhancer. Cas9 protein and sgRNA ribonucleoprotein complex (RNP) was electroporated into rhesus HSPCs, followed by autologous infusion after myeloablation. We found that gene edits persisted in peripheral blood (PB) and bone marrow (BM) for up to 101 weeks similarly for BCL11A enhancer– or control locus–targeted (AAVS1-targeted) cells. Biallelic BCL11A enhancer editing resulted in robust γ-globin induction, with the highest levels observed during stress erythropoiesis. Indels were evenly distributed across PB and BM lineages. Off-target edits were not observed. Nonhomologous end-joining repair alleles were enriched in engrafting HSCs. In summary, we found that edited HSCs can persist for at least 101 weeks after transplant and biallelic-edited HSCs provide substantial HbF levels in PB red blood cells, together supporting further clinical translation of this approach.
Keywords: Hematology, Transplantation
Keywords: Genetic diseases, Hematopoietic stem cells, Stem cell transplantation
Introduction
The β-hemoglobin (HBB) disorders, including sickle cell disease (SCD) and β-thalassemia, are the most common single-gene inherited disorders, resulting in severe morbidity and mortality worldwide. The only curative therapeutic modality for these conditions is allogeneic hematopoietic stem cell transplantation (HSCT), which is limited by a shortage of matched donors (1) and may be associated with graft-versus-host disease. Induction of fetal hemoglobin (HbF) in adults with SCD has proven clinically beneficial for patients with severe disease because γ-globin inhibits hemoglobin (Hb) polymerization under deoxygenated conditions by incorporating into tetramers and decreasing sickle hemoglobin (HbS) concentration in red blood cells (RBCs) (2). In addition, a high level of HbF in patients with β-thalassemia is associated with amelioration of disease severity as γ-globin improves α/β chain imbalance and HbF replaces deficient adult hemoglobin (HbA) (3). Genome-wide association studies recognized BCL11A as a significant repressor of γ-globin expression (4, 5), and multiple lines of evidence have validated its negative regulatory activity (6, 7). In addition to its HbF silencing role, BCL11A is required for normal B cell development and hematopoietic stem cell (HSC) function. We and others previously identified the intronic erythroid-specific BCL11A enhancer, itself subject to HbF-associated common genetic variation, as a favorable therapeutic target (8–12). We recently demonstrated that CRISPR/Cas9 disruption of the enhancer by indels (insertions and deletions introduced to the genome by DNA repair after targeted cleavage) induces HbF, inhibiting sickling and restoring globin chain balance in erythroid cells derived from hematopoietic stem and progenitor cells (HSPCs) from SCD and patients with β-thalassemia respectively, without detectable genotoxicity or adverse effects on HSC function (12). Xenotransplantation of HSCs may not fully evaluate the self-renewal and peripheral blood (PB) repopulation potential required for an effective cellular therapy. In particular, xenotransplant assays may emphasize the function of progenitors over HSCs and do not support normal PB reconstitution. Since HbF is a recent evolutionary adaptation originating in a shared ancestor of humans and old-world nonhuman primates, its endogenous regulation cannot be modeled easily in nonprimate cells. Therefore, nonhuman primate autologous hematopoietic cell transplantation may be a powerful experimental system where various gene manipulations may be investigated for their feasibility, safety, and efficacy, facilitating successful clinical development (13–15). To more fully investigate the clinical potential of autologous HSC gene editing, here we evaluated engraftment and HbF induction potential of erythroid-specific BCL11A enhancer editing using a nonhuman primate transplantation model (Rhesus macaque) in which Hb switching is conserved (16). Advantages of the rhesus macaque autologous HSCT model include similarities between rhesus macaques and humans in genomic profile, hematopoiesis, and erythropoiesis, as well as the possibility of measuring indels and HbF in PB and bone marrow (BM) for extended time periods (17–19).
Results
Editing the BCL11A erythroid enhancer in rhesus CD34+ HSPCs induces γ-globin.
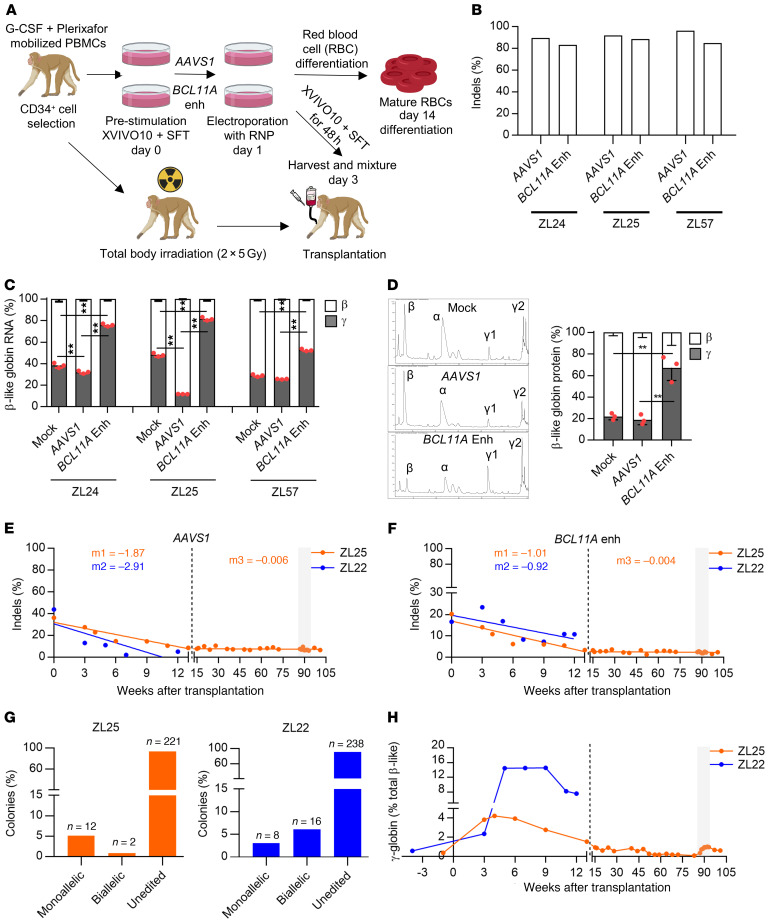
We previously employed pooled sgRNA screening in human erythroid precursors to identify a core GATA1 binding site in the BCL11A +58 DNAse I hypersensitive site that is required for γ-globin repression (9). Subsequently, we observed that electroporation of a nuclear localization signal (NLS) sequence modified form of SpCas9 (3×NLS-SpCas9) and chemically modified synthetic guide RNA #1617 producing cleavage directly within the GATA1 binding motif produced highly efficient indels that disrupted erythroid BCL11A expression and prevented HbF silencing in healthy donor and β-hemoglobinopathy patient cells without detectable genotoxicity or impairment of HSC function under xenotransplantation conditions (12). The protospacer and protospacer adjacent motif (PAM) sequences are identical at the human and rhesus #1617 target sequences within the BCL11A +58 enhancer. Since we began these rhesus studies before identifying 3×NLS-SpCas9 as a highly efficient RNP complex for HSPC editing, in our preliminary studies we delivered 2×NLS-SpCas9:sgRNA RNP to rhesus CD34+ HSPCs by electroporation. We observed efficient editing at the BCL11A enhancer (using sgRNA #1617), with 81%–85% indels, and at a control locus AAVS1, with 89%–96% indels (Figure 1, A and B). Following erythroid differentiation of rhesus CD34+ HSPCs, substantial γ-globin RNA induction was detected with BCL11A enhancer editing, with 51%–83% γ-globin in BCL11A enhancer edited cells compared with 27%–49% and 11%–33% in nonelectroporated and AAVS1 control locus–edited cells, respectively (P < 0.01, Figure 1C). Consistent with the RNA profiles, elevated γ-globin protein levels (54%–77%, P < 0.01) were observed in BCL11A-edited cells compared with nonelectroporated (19%–25%) and AAVS1-edited cells (15%–24%), respectively (Figure 1D and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI140189DS1), indicating that BCL11A enhancer editing derepresses HbF expression in rhesus erythroid cells. No significant difference in RBC enucleation efficiency (44%–47%) following erythrocyte differentiation and viability in electroporated cells (56%–68%) was observed between groups (Supplemental Figure 1, B and C). Furthermore, the colony-forming unit (CFU) capacity of the cells was comparable between the BCL11A-edited and control groups (Supplemental Figure 1D).
Figure 1. Durable autologous engraftment following BCL11A enhancer gene editing.
(A) Schematic representation of electroporation of rhesus CD34+ HSPCs with ribonucleoprotein (RNP) complex composed of 2×NLS SpCas9 or 3×NLS SpCas9 protein, and either BCL11A enhancer targeting (#1617) or AAVS1 targeting sgRNA. The cells are either used for ex vivo analysis or autologous transplantation. (B) Editing efficiency measured by Sanger sequencing with TIDE analysis, and (C) β-like globin RNA expression by RT-qPCR normalized to α-globin expression in nonedited (Mock), AAVS1-, and BCL11A-edited rhesus CD34+ HSPCs in small scale (ZL24 and ZL25, 5 × 104 cells, 200 pmol for both SpCas9 and sgRNAs) and large scale (ZL57, 5 × 106 cells, 1000 pmol for both SpCas9 and sgRNAs) electroporation conditions (n = 3, 1-way ANOVA followed by the Tukey’s post hoc test, **P < 0.01). (D) β-like globin protein expression by RP-HPLC (n = 3, 1-way ANOVA followed by the Tukey’s post hoc test, ** P < 0.01). ZL25 and ZL22 were transplanted with AAVS1- and BCL11A enhancer–edited cells (1:1). The gray rectangle represents the phlebotomy course. Editing frequencies in granulocytes for (E) AAVS1 and (F) BCL11A enhancer in transplanted rhesus macaques. Slopes were calculated separately for the first 13 weeks of transplantation (early progenitor phase) and later time points (HSC phase) as indicated by the dashed line. (G) Distribution of monoallelic and biallelic edited colonies collected from methylcellulose plates for bone marrow mononuclear cells of ZL25 at 100 weeks and ZL22 at 13 weeks after transplantation. (H) γ-globin protein expression percentage in ZL25 and ZL22.
To test the editing frequency in HSC and progenitor fractions, we sorted CD34+CD38–CD90+CD45RA– (HSC-enriched population) and CD34+CD38+ (committed progenitors, HPCs) from CD34+ HSPCs 2 hours after RNP electroporation. Indel sequence analysis of 5-day cultured fractions revealed modestly reduced editing frequency in the HSC-enriched population (56%) compared with HPCs (72%) (Supplemental Figure 2). In addition, the 15-bp deletion, a predicted microhomology-mediated end joining (MMEJ) product (20), was particularly reduced in the HSC-enriched population as compared with HPCs (from 8% to 2%) while the predominant nonhomologous end-joining (NHEJ) repair product, a +1 insertion, was similar in the 2 populations (33% and 34%, Supplemental Figure 2).
Edited HSPCs produce long-term engraftment in rhesus macaques.
To assess engraftment and HbF induction potential of BCL11A enhancer editing, we used 2 cohorts of macaques, with 2 animals per cohort. In cohort 1, we performed competitive engraftment of BCL11A enhancer– and AAVS1–edited CD34+ HSPCs to test the feasibility of the approach and long-term reconstitution of edited cells. Mobilized rhesus CD34+ HSPCs were electroporated with RNP composed of 2×NLS SpCas9 protein and either BCL11A enhancer–targeting (#1617) or AAVS1-targeting sgRNA, followed by ex vivo culture for 48 hours. Cells were harvested and freshly infused into rhesus monkeys following preparative conditioning with 2 × 5 Gy total body irradiation (Figure 1A). Using a small aliquot of cells reserved for ex vivo studies, marked induction in γ-globin protein level was observed in BCL11A-edited erythroid cells (33%–45%) compared with control groups (12%–27%) as determined by reverse-phase high-pressure liquid chromatography (RP-HPLC) and Hb electrophoresis, without a significant change in cell numbers among the BCL11A enhancer– and AAVS1-edited groups (Supplemental Figure 3, A–F).
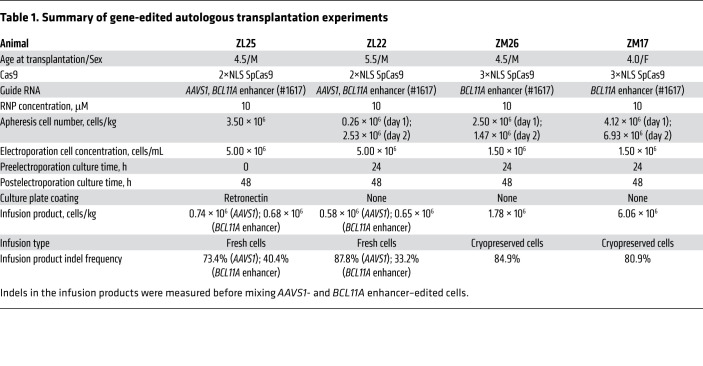
After CD34+ HSPC infusion (n = 2, ZL25 and ZL22, 1.23 × 106–1.42 × 106 CD34+ HSPCs/kg) (Table 1), animals engrafted with typical autologous reconstitution kinetics, i.e., 2–3 weeks as evidenced by blood cell counts and Hb levels in PB (Supplemental Figure 4). There was a moderate reduction in indel frequencies over the first 12 weeks after transplantation (3%–15%) compared with indels in the input cell product (17%–44%). However, after 12 weeks, stable indel frequencies were detected for both BCL11A enhancer (~3%–6%) and AAVS1 (~7%–11%) in PB-derived granulocyte and lymphocyte fractions of the edited animals (Figure 1, E and F, and Supplemental Figure 5, A and B). Stable AAVS1 and BCL11A enhancer editing frequencies were found in ZL22 and ZL25 for up to 101 weeks (Supplemental Figure 5, C and D), suggesting the absence of selective difference between BCL11A enhancer–edited, AAVS1-edited, and nonedited HSCs. Overall, these results appear consistent with previous observations that after ex vivo HSPC gene editing, indel frequencies in short-term engrafting progenitors often exceed those found in long-term engrafting HSCs (12, 21, 22).
Table 1. Summary of gene-edited autologous transplantation experiments.
By performing sequence analysis on individual BM mononuclear cell–derived (MNC-derived) hematopoietic colonies, we found that the BCL11A enhancer indels in ZL22 were distributed as approximately 2 of 3 in biallelic and approximately 1 of 3 in monoallelic edited colonies, whereas in ZL25 the BCL11A enhancer indels were distributed as approximately 15% biallelic and approximately 85% monoallelic edited colonies (Figure 1G). Blood cell counts returned to the normal range after autologous reconstitution (Supplemental Figure 4). In addition, gene editing did not change the frequency or distribution of BM MNC-derived colonies from ZL25 and ZL22 as compared with an unedited control animal (Supplemental Figure 5, E–G).
To analyze γ-globin induction in edited animals, we performed RP-HPLC from PB over time. There was a substantial induction of γ-globin during the first 12 weeks after transplantation (4.3%–14.6%) that gradually decreased and stabilized at pretransplantation levels (~0.5%) by around 20 weeks after transplantation (Figure 1H). Unfortunately, ZL22 was diagnosed with radiation pneumonitis at 11 weeks after transplantation and euthanized at 13 weeks after the transplantation, at which point γ-globin level was 7.5%. Transient γ-globin induction has been reported for conditions resulting in stress erythropoiesis such as recovery from anemia due to transient erythroblastopenia of childhood (23), BM transplantation (24), and phlebotomy (25, 26). Autologous hematopoietic transplantation of rhesus macaques produces transient expression of γ-globin up to a maximum of 0.99%–2.53% at 4–9 weeks of transplantation that stabilizes at pretransplantation levels 20 weeks after transplantation (27), suggesting that most of the γ-globin induction observed in ZL22 and ZL25 was due to BCL11A editing.
Stable engraftment with biallelic-edited HSCs after low-dose autologous HSPC infusion.
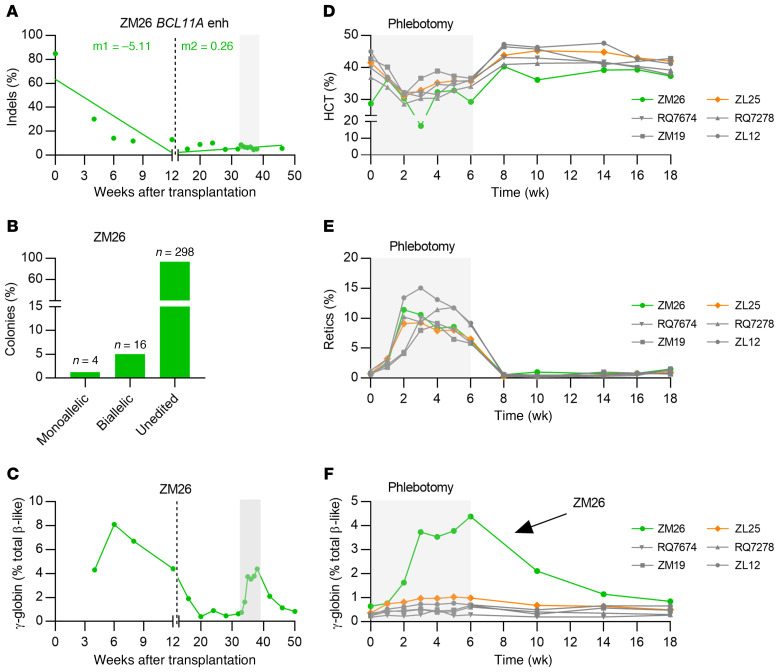
To further investigate γ-globin induction potential of BCL11A editing in rhesus monkeys, we transplanted 2 monkeys with HSPCs edited solely at the BCL11A enhancer (n = 2, ZM26 and ZM17) as cohort 2. We used 3×NLS-SpCas9, which in previous human HSPC studies was associated with highly efficient indel generation in engrafting HSCs (12). Furthermore, we cryopreserved HSPCs before infusion to more fully mimic an anticipated clinical protocol. Consistent with cohort 1 animals, these animals engrafted with typical kinetics, and rescued cell counts and Hb levels at 2–3 weeks after transplantation, similar to control transplanted animals (Supplemental Figure 4). Despite having high indel rates (84.9%) in the infusion product of ZM26, and robust ex vivo γ-globin induction (65.9% vs. 24.7% in nonedited control), there was a significant drop in the indel frequencies (~25%–30%) at 4 weeks after transplantation that stabilized to about 5%–16% at 28 weeks after transplantation (Figure 2A and Supplemental Figure 6, A–C). For prior efficient autologous reconstitution by gene-marked HSCs after lentiviral gene transfer, we infused cell products of at least approximately 3 × 106 CD34+ HSPCs/kg (28, 29). The relatively low infused HSPC number of 1.78 × 106 CD34+ HSPCs/kg could have contributed to inefficient autologous engraftment of the edited cells (Table 1). We observed that the BCL11A enhancer indels were mainly distributed in biallelic-edited BM MNC-derived colonies (~80%) as compared with monoallelic edited colonies (~20%) (Figure 2B). γ-globin expression in ZM26 peaked at 8.1% at 6 weeks after transplantation but decreased to levels indistinguishable from control animals (~0.5%) 20 weeks after transplantation (Figure 2C). The clonogenic capacity of BM MNCs of ZM26 at 38 weeks after transplant was similar to other edited and nonedited animals (Supplemental Figure 5, E–I).
Figure 2. Stress erythropoiesis cooperates with BCL11A enhancer editing to amplify γ-globin induction.
(A) Editing frequencies in granulocytes of ZM26 transplanted with BCL11A-edited CD34+ HPSCs. Slopes were calculated separately for the first 13 weeks of transplantation (early progenitor phase) and later time points (HSC phase) as indicated by the dashed line. (B) Distribution of monoallelic and biallelic edited colonies collected from methylcellulose plates for bone marrow mononuclear cells of ZM26 at 38 weeks after transplantation. (C) γ-globin protein expression in ZM26. The gray rectangle represents the phlebotomy course. (D) Hematocrit (HCT), (E) reticulocyte, and (F) γ-globin protein percentages in phlebotomized nontransplanted (ZL12 and ZM19), lentivirus-transduced (GFP vector–transduced) CD34+ HSPC-transplanted (RQ7278 and RQ7674), and BCL11A enhancer–edited animals (ZM26 and ZL25).
Erythropoiesis stress induces substantial γ-globin levels in edited animals.
As γ-globin induction peaked in the first 12 weeks following transplantation, we hypothesized that stress erythropoiesis might be reinforcing BCL11A editing–related γ-globin induction. To test this idea, we phlebotomized edited animals (ZL25 and ZM26) along with nontransplanted (ZL12 and ZM19) and lentivirally transduced (GFP vector) CD34+ HSPC transplanted (RQ7278 and RQ7674) control animals twice a week (20% of total blood volume) for 6 weeks. Following phlebotomy, we observed anemia and elevated reticulocyte counts, with peak reticulocytosis 2–5 weeks after onset of phlebotomy (Figure 2, D and E), consistent with accelerated erythropoiesis kinetics. ZM26, the animal with approximately 6% indel frequency with indels mainly distributed within biallelic-edited colonies (Figure 2B) demonstrated substantial γ-globin induction (from 0.6% to 4.4% after 6 weeks of phlebotomy) (Figure 2F). In contrast, ZL25, the animal with approximately 3% indel frequency, with indels mainly distributed within monoallelically edited colonies (Figure 1G) showed more modest γ-globin induction (from 0.4% to 1% after 6 weeks of phlebotomy). Control nontransplanted and marker vector–transduced transplanted animals showed minimal γ-globin induction (from ~0.2%–0.3% to ~0.5%–0.7% after 6 weeks of phlebotomy). Editing frequencies in ZL25 and ZM26 for both AAVS1 and BCL11A enhancer did not change during the phlebotomy experiment (Supplemental Figure 6, D–F), indicating that stress erythropoiesis did not preferentially drive hematopoiesis from edited compared with nonedited cells. After terminating phlebotomy, γ-globin expression returned to pre-phlebotomy levels for all animals, along with stable indel frequencies (Figure 2F). Together, the phlebotomy and autologous reconstitution results suggest a positive interactive effect between biallelic BCL11A enhancer editing and stress erythropoiesis with regard to γ-globin induction.
Efficient engraftment of BCL11A-edited HSPCs.
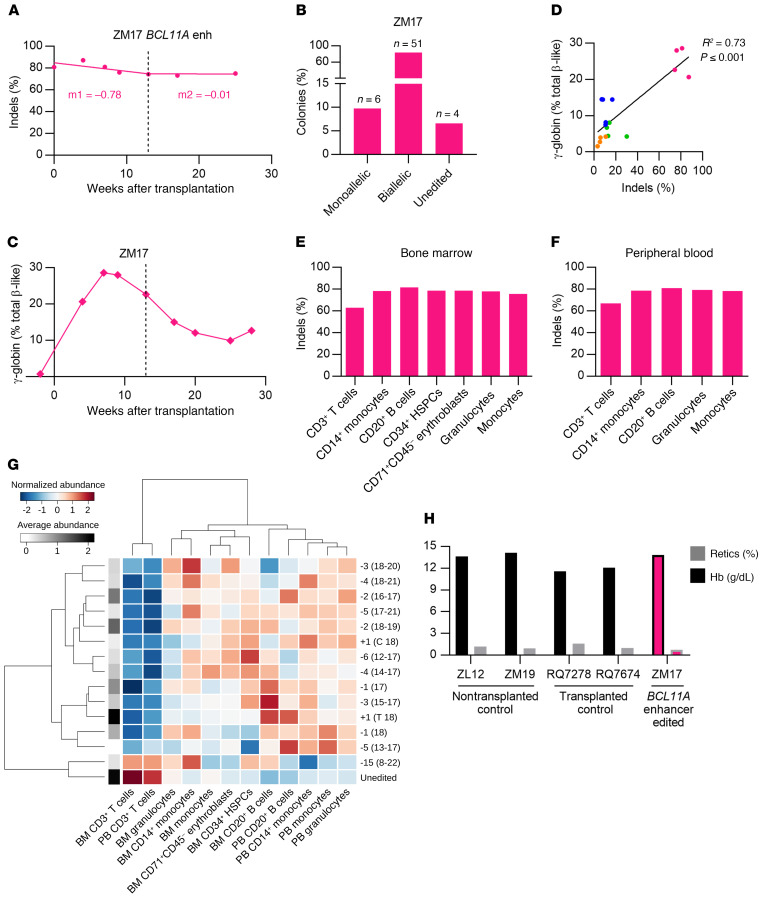
The indel frequency in the input cell product for ZM17, the second animal in cohort 2, was 80.9%. A higher HSPC dose of 6.06 × 106 CD34+ HSPCs/kg was infused to ZM17 (3.4-fold greater than infused to ZM26). Stable indel frequencies were detected in both PB granulocyte (75.1%) and lymphocyte (79.0%) fractions at 25 weeks after transplantation (Figure 3A and Supplemental Figure 7A). The animal ZM17 developed severe radiation pneumonitis and was euthanized at 28 weeks. In edited BM MNC colonies, 83.6% had biallelic indels, 9.8% had monoallelic indels, and 6.6% were unedited (Figure 3B). γ-globin expression in PB reached up to 28.6% at 7 weeks after transplantation (Figure 3C). Like the other animals, γ-globin in ZM17 decreased after autologous transplant, nadiring at 9.9% at 25 weeks and then 12.7% at 28 weeks after transplantation. Comparing all 4 edited animals, there was a strong correlation between BCL11A enhancer indel frequencies and peak γ-globin levels in peripheral blood during the first 13 weeks following transplantation (R2 = 0.73, P < 0.001, Figure 3D).
Figure 3. Robust BCL11A enhancer editing and γ-globin induction.
(A) Editing frequencies in granulocyte fraction in ZM17. Slopes were calculated separately for the first 13 weeks of transplantation (early progenitor phase) and later time points (HSC phase) as indicated by the dashed line. (B) Distribution of monoallelic and biallelic edited colonies collected from methylcellulose plates for bone marrow mononuclear cells of ZM17 at 28 weeks after transplantation. (C) γ-globin protein expression by RP-HPLC in peripheral blood of ZM17. (D) Correlation of editing frequencies in all transplanted animals (4–13 weeks after transplantation) with γ-globin protein expression by RP-HPLC. (E) Editing frequencies in BM-derived CD3+ T cells, CD14+ monocytes, CD20+ B cells, CD34+ HSPCs, CD71+ CD45– erythroblasts, and granulocytes and monocytes sorted using FSC A and SSC A. (F) Editing frequencies in PB-derived granulocytes, monocytes, CD3+, CD14+, and CD20+ cells. (G) Heatmap representation for normalized alleles abundance values among BM and PB lineages of ZM17 at 28 weeks after transplantation. The allele-specific average abundance value (log) is represented in grayscale on the left side of the heatmap. Both columns (lineages) and rows (alleles) are ordered according to their similarity measured by Ward distance. (H) Hemoglobin (Hb) and reticulocyte count percentages in nontransplanted (ZL12 and ZM19), lentivirus-transduced (GFP vector–transduced) CD34+ HSPC-transplanted (RQ7278 and RQ7674), and BCL11A enhancer–edited animals (ZM17).
BCL11A plays critical roles in B lymphocyte maturation and HSC self-renewal (30–32). The rationale of editing the BCL11A erythroid enhancer is to minimize impact on nonerythroid hematopoietic functions. To test any selective impact of BCL11A enhancer editing on various hematopoietic lineages, different cell fractions from both PB and BM MNCs of ZM17 at 28 weeks after transplantation were sorted and subjected to deep sequencing. The BCL11A enhancer indel frequencies ranged from 78%–81% across all PB and BM lineages (excluding CD3+ T cells with 63% indels), including CD20+ B lymphoid, CD14+ myeloid cells, CD34+ HSPCs, and CD71+ CD45– erythroblasts, suggesting no selective toxicity to any cell lineage tested (Figure 3, E and F).
We further analyzed the subset of specific gene edit alleles for which the frequency was at least 1% in one lineage, comprising 14 indels plus the unedited allele. Overall, the distribution of gene edits showed a very similar pattern among all hematopoietic lineages, suggesting the absence of strong lineage-specific selection pressure (Supplemental Figure 8A). In order to measure the similarity among cell subpopulations, we performed an unsupervised hierarchical clustering analysis (using Ward distance) on normalized (z-scored) edit frequencies (Figure 3G). The lineages were grouped in 3 main clusters: CD3+ T cells in BM and PB; all other PB lineages plus BM CD20+ B cells; and all other BM lineages. Multiple factors might contribute to this clustering configuration, such as the hierarchical nature of hematopoietic differentiation and heterogeneity in the life span of lineage-biased HSPCs. We also tested the correlation (Pearson’s coefficient) among all possible pairs of lineages using the edit frequency data (Supplemental Figure 8B and Supplemental Tables 1 and 2). BM and PB CD3+ T cells showed a significant positive correlation with each other and negative association with PB CD14+ monocytes and PB granulocytes. Positive association was also measured among BM CD14+ monocytes and BM granulocytes; BM monocytes and CD71+ CD45– erythroblasts; and all PB lineages (besides CD3+ T cells).
Peripheral blood counts, including Hb level and reticulocyte counts, of ZM17 at 6 months after transplantation were not different from those of untransplanted controls (ZL12 and ZM19) and GFP-expressing lentivirus-transduced CD34+ HSPC transplanted animals (RQ7278 and RQ7674) (Figure 3H and Supplemental Figure 4). Furthermore, similar clonogenicity of BM MNCs collected at 28 weeks after transplantation was observed as compared with other edited animals and the control nontransplanted animal (Supplemental Figure 5, E–I). Together, these data do not indicate apparent hematologic or erythroid effects of BCL11A enhancer editing aside from γ-globin induction.
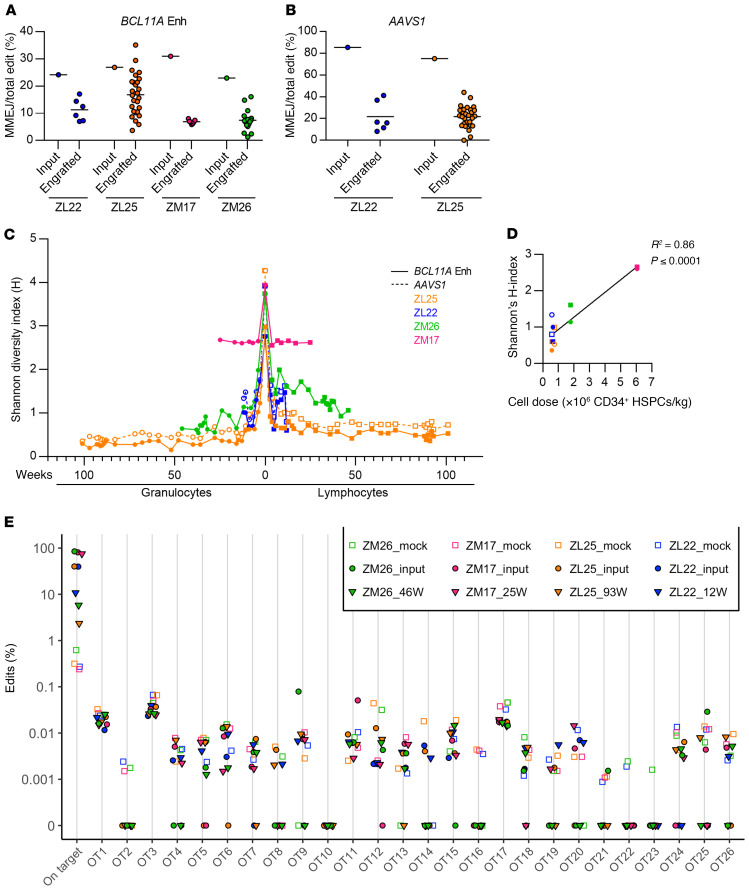
The most common alleles in rhesus HSPCs after BCL11A enhancer editing were a 1-bp T insertion, produced by NHEJ repair, followed by a –15 bp deletion, produced by MMEJ repair (Supplemental Figures 9 and 10). These predominant edits were the same as observed in human HSPCs after BCL11A enhancer editing with the identical sgRNA (12). Previously, we observed that engrafting human hematopoietic cells in mouse xenografts preferentially carried NHEJ repair alleles as compared with MMEJ repair alleles. To evaluate the engraftment potential of HSPCs repaired by NHEJ as compared with MMEJ, we analyzed individual gene edit alleles in input HSPCs and engrafting granulocytes and lymphocytes (Supplemental Figure 9, A–F). We observed that overall edits were variably reduced in the 4 animals comparing the input cell products to engrafting cells (Supplemental Figures 9 and 11). In each animal, we observed that the fraction of MMEJ alleles at the BCL11A enhancer was selectively reduced (1.6-fold to 4.5-fold) in engrafted cells as compared with the input cell product (Figure 4A and Supplemental Figure 11). Similar selective reduction of MMEJ repair alleles in engrafting cells (3.4-fold to 3.9-fold) was also observed after AAVS1 editing (Figure 4B, Supplemental Figure 9, A and C, and Supplemental Figure 11).
Figure 4. Gene editing dynamics after BCL11A enhancer editing.
Relative loss of engrafted edits repaired by MMEJ in transplanted animals for (A) BCL11A enhancer and (B) AAVS1 alleles. Deletions of at least 8 bp were categorized as MMEJ, while all other indels were categorized as NHEJ. (C) Shannon diversity index (H) for edit distribution in input cell products and engrafted samples. Granulocytes and lymphoid lineage data are plotted symmetrically, with the former on the left (for animals ZL22 and ZL25, both BCL11A enhancer and AAVS1 data are reported; for ZM17 and ZM26, only BCL11A enhancer data are reported). (D) Correlation of Shannon diversity index (H) with infusion cell dose in all transplanted animals (12–13 weeks after transplantation). Granulocytes (circles), lymphocytes (squares), BCL11A enhancer (solid shapes), AAVS1 (open shapes). (E) Off-target analysis of granulocytes in edited animals transplanted with BCL11A enhancer–edited CD34+ HSPCs. Using CIRCLE-Seq, 26 potential genomic off-target sites for BCL11A enhancer guide #1617 were identified. Editing frequencies at the 26 sites were evaluated through amplicon deep sequencing and analysis. The samples include the infusion product (input) and granulocyte fraction of a later peripheral blood collection for each rhesus macaque. Each point represents a single replicate.
We calculated the population diversity of gene edit alleles (excluding the unedited allele) as measured by the Shannon H index (33) in each input cell product and engrafted granulocyte and lymphocyte sample. We found that for each animal and sgRNA, there was a consistent reduction in the diversity in the engrafted cells with respect to the input cell product (Figure 4, C and D, and Supplemental Table 3). For ZM17, after the initial drop, the diversity of the edited cell population stabilized around H = 2.6, similar for both granulocytes and lymphocytes. This suggests a polyclonal distribution in which (a) a considerable number of cells bearing different edits engrafted (~250 alleles); (b) the number of alleles with a significant abundance was high; (c) the composition of this population was constant over time; and (d) any in vivo selection acts similarly in the 2 lineages. For all other animals, the clonality index decreased over time, approaching H < 1, suggesting that the edited cell populations lost more substantial complexity and heterogeneity, tending to an oligoclonal distribution, relative to ZM17. The number of detected edits in the engrafted population reduced over time to approximately 10 to 40 alleles, with few alleles having high relative abundance. In lymphocytes, Shannon’s H index decreased more slowly as compared with granulocytes, perhaps due to differences in the life span of lineage-biased progenitors in the input product or to a slower turnover of lymphocytes. For animals ZL22 and ZL25, the AAVS1 and BCL11A enhancer edits showed similar clonality dynamics. The H-index at week 12 was positively correlated to infused cell number, consistent with observations from human hematopoietic gene therapy that infused cell number is a key determinant of polyclonal engraftment of gene marked cells (R2 = 0.86, Figure 4D) (34).
We performed CIRCLE-seq (35) to empirically define rhesus genomic sites susceptible to off-target in vitro RNP cleavage. Then we performed amplicon deep sequencing of the top 26 candidate off-target sites (based on CIRCLE-seq read counts) in each of the 4 input BCL11A enhancer–edited input cell products as well as in engrafted hematopoietic cells. Despite detecting robust on-target BCL11A enhancer editing in each sample, we did not observe off-target gene editing at any of these 26 sites in any of the samples, at a limit of detection of 0.1% allele frequency (Figure 4E).
Discussion
Developing innovative autologous hematopoietic cell transplantation protocols in rhesus macaques is especially complex in that the procedures and reagents for HSC mobilization, collection, gene modification, culture, and reengraftment are largely adapted from protocols initially optimized for human cells. One challenge we encountered was to achieve consistently safe and effective myeloablation (36, 37), with 2 animals succumbing to radiation pneumonitis. Furthermore, we obtained variable recovery from mobilized peripheral blood cell apheresis. Three of the 4 infusion cell products included fewer than the typical target of 3 × 106 CD34+ HSPCs/kg (Table 1), which might disadvantage the engraftment of the cell product as compared with residual HSCs. Despite these challenges, by investigating just 4 animals, we observed improved gene-editing efficiency from approximately 30%–40% to approximately 80%–85% indels in the input cell product at scale to support autologous hematopoietic transplantation following myeloablative conditioning therapy. We demonstrated a proof-of-principle for therapeutic gene editing with approximately 70%–80% gene modifications in engrafting hematopoietic cells after 28 weeks, mainly distributed as biallelic edits. This result is the most efficient autologous hematopoietic gene editing achieved in nonhuman primates to our knowledge. Prior reports demonstrated substantially fewer gene edits in engrafting hematopoietic cells (14, 21). In a previous NHP setting, editing of the CCR5 gene dropped from 40% in the infusion product to 3%–5% at 6 months (22). A recent study aiming to induce HbF by creating hereditary persistence of fetal hemoglobin mutation in rhesus macaques showed a decrease in indels from approximately 70% in the input cell product to about 15% in engrafting cells (21). These results suggest that at intermediate editing frequencies, indels in unfractionated autologous HSPCs also may overestimate those in engrafting HSCs (12). A nonmutually exclusive possibility could be incomplete hematopoietic ablation with partial reconstitution by residual nonablated HSCs. These results encourage the development of gene editing protocols to maximize both the total number and fraction of biallelically modified HSCs.
We observed selective reduction of MMEJ repair alleles in engrafting compared with input cells. This result suggests that HSCs, a rare subset within HSPC cell products, preferentially repair by NHEJ. NHEJ repair with short indels appears sufficient to disrupt BCL11A enhancer function and support HbF induction in erythroid progeny of edited HSCs. We found that BCL11A enhancer editing yielded robust induction of γ-globin during steady-state erythropoiesis, with an approximately 18-fold increase from unedited steady-state level in the same animal (Figure 3C and Supplemental Figure 7B). In addition, we observed a substantial positive interaction between BCL11A enhancer editing and stress erythropoiesis associated with hematopoietic repopulation, with an additional 2.8-fold to 6.8-fold increase as compared with the steady-state levels in the edited animals. During phlebotomy-induced erythroid stress, we observed HbF induction only in ZM26, the animal with BCL11A enhancer edits mainly distributed as biallelic indels, also consistent with a positive interaction between erythroid stress and BCL11A modulation. Future studies appear warranted to further explore this interaction. In the setting of HbF induction for β-hemoglobinopathy patients as compared with healthy individuals, we anticipate there may be additional contributions from both disease-associated stress erythropoiesis as well as survival advantage of HbF-expressing erythrocytes and precursors. Together, we predict these effects, plus more pancellular than heterocellular distributions of gene edits, could further magnify the impact of HbF induction following BCL11A enhancer editing in patients with β-hemoglobinopathy.
For all animals and guide RNAs (including targeting both AAVS1 and BCL11A enhancer), we observed a reduction of the diversity of gene edits in the engrafted cells with respect to the input cell product. The starting cell product was relatively complex, with edits more evenly distributed. The engrafted cells had fewer edits, suggesting a bottleneck in the initial engrafting cell population, that then stabilized over extended examination. Cell products with greater HSPC numbers were associated with more polyclonal engraftment. These dynamics appear reminiscent of gene therapy with HSCs marked by lentiviral insertions.
Under efficient editing conditions, we observed a similar distribution of BCL11A enhancer indels across all PB and BM lineages (with the exception of modestly lower frequency of gene editing in long-lived CD3+ T lymphocytes), suggesting no skewed hematopoietic contributions of BCL11A enhancer gene-edited cells. Moreover, we observed normal blood counts, including no anemia or hemolysis following efficient BCL11A enhancer editing, consistent with intact hematopoiesis and erythropoiesis. Off-target genotoxicity is one possible concern of therapeutic gene editing. We did not observe any off-target gene edits attributable to BCL11A enhancer editing, although large deletions and rearrangements were not specifically evaluated (38). These findings are similar to the absence of apparent toxicity of BCL11A enhancer editing with 3×NLS-SpCas9 and sgRNA #1617 in human HSPCs (12).
In summary, we evaluated the clinical potential of autologous BCL11A erythroid enhancer editing in rhesus macaques. BCL11A enhancer–edited HSCs can persist for at least 101 weeks after transplant and provide potentially therapeutic levels of HbF in PB RBCs without anemia or other apparent hematologic toxicity. These results emphasize that gene editing efficiency and repair mode, input CD34+ HSPC number, and conditioning therapy are each critical variables that influence gene edits in engrafted hematopoietic cells following autologous HSCT. Overall, these findings support BCL11A erythroid enhancer genome editing as a promising strategy for HbF induction for β-hemoglobinopathies.
Methods
Detailed methods are provided in the Supplemental Methods. All raw sequence data have been deposited at NCBI Sequence Read Archive (SRA) under the accession number of PRJNA655555.
Statistics.
Standard errors of the mean are given as error bars in all figures. The data were statistically analyzed with 1-way analysis of variance (ANOVA) followed by the Tukey’s post hoc test using GraphPad Prism 7 software. Data were considered significantly different at P less than 0.05. Clonality index and lineage data analysis were performed by means of R statistical software (39) and packages vegan and ggplots.
Study approval.
Rhesus macaques (Macaca mulatta) were housed and handled in accordance with the guidelines set by the Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council, and all animal protocols were approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute (approved protocol H-0136R4).
Author contributions
SD and JZ designed, performed, and analyzed experiments, prepared the figures, and wrote the manuscript. AHS helped with figures and analysis. DP, SNP, and SMPM performed the clonality analyses. YW, NU, JG, MY, CD, JB, JJHM, AL, and TN helped with experiments. ACB, AEK, NSL, TE, and RED performed animal care, transplantation, and sample derivation. KL and SAW produced the 3×NLS SpCas9 protein. CRL, SQT, and MJW performed and analyzed off-target assays. DEB and JFT designed and analyzed experiments and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank the NIH National Heart, Lung, Blood Institute (NHLBI) Biochemistry core facility for the HPLC service. DEB was supported by NHLBI (P01HL053749), St. Jude Children’s Research Hospital Collaborative Research Consortium, and Burroughs Wellcome Fund. KL and SAW were supported in part by NIH grants F31HL147482 and R01GM115911.
Version 1. 09/08/2020
In-Press Preview
Version 2. 11/09/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Funding Statement
D.E.B. was supported by NHLBI (P01HL053749), St. Jude Children’s Research Hospital Collaborative Research Consortium, and Burroughs Wellcome Fund.
K.L. and S.A.W. were supported in part by NIH grants F31HL147482 and R01GM115911.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6677–6687.https://doi.org/10.1172/JCI140189.
Contributor Information
Selami Demirci, Email: selami.demirci@nih.gov.
Jing Zeng, Email: Jing.Zeng@childrens.harvard.edu.
Yuxuan Wu, Email: yuxuanwu2018@hotmail.com.
Naoya Uchida, Email: uchidan@nhlbi.nih.gov.
Anne H. Shen, Email: ahshen@mit.edu.
Danilo Pellin, Email: Danilo_Pellin@DFCI.HARVARD.EDU.
Jackson Gamer, Email: j14gamer@gmail.com.
Morgan Yapundich, Email: yapundichm@gmail.com.
Claire Drysdale, Email: clairemd@live.unc.edu.
Jasmine Bonanno, Email: jasminejbonanno@gmail.com.
Aylin C. Bonifacino, Email: bonifaca@nhlbi.nih.gov.
Theresa Engels, Email: theresa.engels@nih.gov.
Robert E. Donahue, Email: donahuer@nhlbi.nih.gov.
Juan J. Haro-Mora, Email: juanjharo@gmail.com.
Alexis Leonard, Email: alexis.leonard@nih.gov.
Tina Nassehi, Email: tina.nassehi@nih.gov.
Kevin Luk, Email: Kevin.Luk@umassmed.edu.
Shaina N. Porter, Email: Shaina.Porter@STJUDE.ORG.
Cicera R. Lazzarotto, Email: Cicera.Lazzarotto@STJUDE.ORG.
Shengdar Q. Tsai, Email: Shengdar.Tsai@STJUDE.ORG.
Shondra M. Pruett-Miller, Email: Shondra.Miller@STJUDE.ORG.
Scot A. Wolfe, Email: Scot.Wolfe@umassmed.edu.
Daniel E. Bauer, Email: daniel.bauer@childrens.harvard.edu.
John F. Tisdale, Email: johntis@intra.niddk.nih.gov.
References
- 1.Tisdale J. Improvements in haploidentical transplantation for sickle cell disease and β-thalassaemia. Lancet Haematol. 2019;6(4):e168–e169. doi: 10.1016/S2352-3026(19)30045-6. [DOI] [PubMed] [Google Scholar]
- 2.Eaton WA, Bunn HF. Treating sickle cell disease by targeting HbS polymerization. Blood. 2017;129(20):2719–2726. doi: 10.1182/blood-2017-02-765891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thein SL. The emerging role of fetal hemoglobin induction in non-transfusion-dependent thalassemia. Blood Rev. 2012;26(suppl 1):S35–S39. doi: 10.1016/S0268-960X(12)70011-5. [DOI] [PubMed] [Google Scholar]
- 4.Menzel S, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39(10):1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 5.Uda M, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of β-thalassemia. Proc Natl Acad Sci U S A. 2008;105(5):1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 7.Sankaran VG, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460(7259):1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith EC, et al. Strict in vivo specificity of the Bcl11a erythroid enhancer. Blood. 2016;128(19):2338–2342. doi: 10.1182/blood-2016-08-736249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vierstra J, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015;12(10):927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med. 2019;25(5):776–783. doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiem HP, et al. Pigtailed macaques as a model to study long-term safety of lentivirus vector-mediated gene therapy for hemoglobinopathies. Mol Ther Methods Clin Dev. 2014;1:14055. doi: 10.1038/mtm.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MY, et al. Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell. 2018;173(6):1439–1453.e19. doi: 10.1016/j.cell.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida N, et al. Development of a forward-oriented therapeutic lentiviral vector for hemoglobin disorders. Nat Commun. 2019;10(1):4479. doi: 10.1038/s41467-019-12456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson RM, et al. Humans and old world monkeys have similar patterns of fetal globin expression. J Exp Zool. 2000;288(4):318–326. doi: 10.1002/1097-010X(20001215)288:4<318::AID-JEZ4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 17.Donahue RE, Dunbar CE. Update on the use of nonhuman primate models for preclinical testing of gene therapy approaches targeting hematopoietic cells. Hum Gene Ther. 2001;12(6):607–617. doi: 10.1089/104303401300057289. [DOI] [PubMed] [Google Scholar]
- 18.Koelle SJ, et al. Quantitative stability of hematopoietic stem and progenitor cell clonal output in rhesus macaques receiving transplants. Blood. 2017;129(11):1448–1457. doi: 10.1182/blood-2016-07-728691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepherd BE, et al. Hematopoietic stem-cell behavior in nonhuman primates. Blood. 2007;110(6):1806–1813. doi: 10.1182/blood-2007-02-075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae S, Kweon J, Kim HS, Kim JS. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods. 2014;11(7):705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 21.Humbert O, et al. Therapeutically relevant engraftment of a CRISPR-Cas9-edited HSC-enriched population with HbF reactivation in nonhuman primates. Sci Transl Med. 2019;11(503):eaaw3768. doi: 10.1126/scitranslmed.aaw3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson CW, et al. Long-term multilineage engraftment of autologous genome-edited hematopoietic stem cells in nonhuman primates. Blood. 2016;127(20):2416–2426. doi: 10.1182/blood-2015-09-672337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papayannopoulou T, Vichinsky E, Stamatoyannopoulos G. Fetal Hb production during acute erythroid expansion. I. Observations in patients with transient erythroblastopenia and post-phlebotomy. Br J Haematol. 1980;44(4):535–546. doi: 10.1111/j.1365-2141.1980.tb08707.x. [DOI] [PubMed] [Google Scholar]
- 24.Ferster A, et al. Transplanted sickle-cell disease patients with autologous bone marrow recovery after graft failure develop increased levels of fetal haemoglobin which corrects disease severity. Br J Haematol. 1995;90(4):804–808. doi: 10.1111/j.1365-2141.1995.tb05199.x. [DOI] [PubMed] [Google Scholar]
- 25.DeSimone J, Biel SI, Heller P. Stimulation of fetal hemoglobin synthesis in baboons by hemolysis and hypoxia. Proc Natl Acad Sci U S A. 1978;75(6):2937–2940. doi: 10.1073/pnas.75.6.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonagh KT, et al. Hydroxyurea-induced HbF production in anemic primates: augmentation by erythropoietin, hematopoietic growth factors, and sodium butyrate. Exp Hematol. 1992;20(10):1156–1164. [PubMed] [Google Scholar]
- 27.Demirci S, et al. Fetal hemoglobin and F-cell variance in mobilized CD34+ cell-transplanted rhesus monkeys. Exp Hematol. 2019;75:21–25.e1. doi: 10.1016/j.exphem.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans ME, Kumkhaek C, Hsieh MM, Donahue RE, Tisdale JF, Uchida N. TRIM5α variations influence transduction efficiency with lentiviral vectors in both human and rhesus CD34(+) cells in vitro and in vivo. Mol Ther. 2014;22(2):348–358. doi: 10.1038/mt.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchida N, et al. Busulfan combined with immunosuppression allows efficient engraftment of gene-modified cells in a rhesus macaque model. Mol Ther. 2019;27(9):1586–1596. doi: 10.1016/j.ymthe.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4(6):525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 31.Luc S, et al. Bcl11a deficiency leads to hematopoietic stem cell defects with an aging-like phenotype. Cell Rep. 2016;16(12):3181–3194. doi: 10.1016/j.celrep.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brendel C, et al. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J Clin Invest. 2016;126(10):3868–3878. doi: 10.1172/JCI87885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14(4):306–317. [PubMed] [Google Scholar]
- 34.Six E, et al. Clonal tracking in gene therapy patients reveals a diversity of human hematopoietic differentiation programs. Blood. 2020;135(15):1219–1231. doi: 10.1182/blood.2019002350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, Joung JK. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14(6):607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayakawa J, Hsieh MM, Uchida N, Phang O, Tisdale JF. Busulfan produces efficient human cell engraftment in NOD/LtSz-Scid IL2Rgamma(null) mice. Stem Cells. 2009;27(1):175–182. doi: 10.1634/stemcells.2008-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh MM, Langemeijer S, Wynter A, Phang OA, Kang EM, Tisdale JF. Low-dose parenteral busulfan provides an extended window for the infusion of hematopoietic stem cells in murine hosts. Exp Hematol. 2007;35(9):1415–1420. doi: 10.1016/j.exphem.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.https://www.R-project.org/. Accessed September 26, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.