Abstract
BACKGROUND
Current methods for the detection and surveillance of bladder cancer (BCa) are often invasive and/or possess suboptimal sensitivity and specificity, especially in early-stage, minimal, and residual tumors.
METHODS
We developed an efficient method, termed utMeMA, for the detection of urine tumor DNA methylation at multiple genomic regions by MassARRAY. We identified the BCa-specific methylation markers by combined analyses of cohorts from Sun Yat-sen Memorial Hospital (SYSMH), The Cancer Genome Atlas (TCGA), and the Gene Expression Omnibus (GEO) database. The BCa diagnostic model was built in a retrospective cohort (n = 313) and validated in a multicenter, prospective cohort (n = 175). The performance of this diagnostic assay was analyzed and compared with urine cytology and FISH.
RESULTS
We first discovered 26 significant methylation markers of BCa in combined analyses. We built and validated a 2-marker–based diagnostic model that discriminated among patients with BCa with high accuracy (86.7%), sensitivity (90.0%), and specificity (83.1%). Furthermore, the utMeMA-based assay achieved a great improvement in sensitivity over urine cytology and FISH, especially in the detection of early-stage (stage Ta and low-grade tumor, 64.5% vs. 11.8%, 15.8%), minimal (81.0% vs. 14.8%, 37.9%), residual (93.3% vs. 27.3%, 64.3%), and recurrent (89.5% vs. 31.4%, 52.8%) tumors. The urine diagnostic score from this assay was better associated with tumor malignancy and burden.
CONCLUSION
Urine tumor DNA methylation assessment for early diagnosis, minimal, residual tumor detection and surveillance in BCa is a rapid, high-throughput, noninvasive, and promising approach, which may reduce the burden of cystoscopy and blind second surgery.
FUNDING
This study was supported by the National Key Research and Development Program of China and the National Natural Science Foundation of China.
Keywords: Genetics, Oncology
Keywords: Cancer, Molecular diagnosis, Urology
Introduction
Bladder cancer (BCa) is the most common malignancy of the urinary system, with an estimated roughly 549,393 new cases and approximately 199,922 deaths worldwide each year (1). Approximately 75% of patients present with non–muscle-invasive disease (NMIBC), and 70% of these tumors will recur, while 15% will progress in stage and grade (2). Therefore, patients diagnosed with NMIBC undergo frequent treatment and monitoring, resulting in BCa achieving the highest lifetime treatment cost per patient among all cancers (3). The current gold standard for the monitoring of BCa recurrence involves the use of cystoscopy and cytology (4). Cystoscopy is highly sensitive but is invasive, costly, and often associated with discomfort, while urine cytology is highly specific but lacks sensitivity (25%–35%), especially for low-grade BCa (4%–15%) (4–6). The UroVysion fluorescence in situ hybridization (FISH) has a higher sensitivity of 60%–80% and is widely used in the routine clinical detection of BCa, but it shows low sensitivity in low-grade or small tumors (5, 6). In addition, repeated transurethral resection of bladder tumor (Re-TURBT) is recommended for patients with high-grade and T1 tumors (4). However, we still lack effective means to estimate if the patient has residual tumors. Therefore, there is an urgent need to develop effective methods for the detection of early-stage, minimal, residual, and recurrent tumors, which in turn, may improve disease management.
DNA methylation is a key epigenetic regulator of gene expression that usually causes defective gene expression (7). Increased methylation of tumor suppressor genes is an early event in many tumors, and altered DNA methylation patterns could be one of the first detectable neoplastic changes associated with tumorigenesis (8, 9). Therefore, DNA methylation markers were widely used in the diagnosis and prognosis of common cancers (10–13). Several studies have also shown that methylation CpG sites in urine can be promising markers to detect or monitor BCa (14–16). In a multicenter study, Bladder EpiCheck, a commercial application in Europe, used a panel of 15 methylation markers to monitor recurrence in patients with NMIBC undergoing surveillance, and its overall sensitivity was 68.2% and specificity was 88.0% (17). Another multicenter study found that a 3-gene methylation classifier showed overall sensitivity of 89.6% and specificity of 30.5% for monitoring BCa (18). However, the performance of these assays still needs to be improved and validated in multicenter and large-scale cohorts in Asia. Hence, these assays have not been completely adopted in routine clinical practice in Asia.
In this study, we developed an efficient method for the detection of urine tumor DNA (utDNA) methylation at multiple regions by MassARRAY, termed utMeMA. Importantly, we applied it to a retrospective cohort and least absolute shrinkage and selection operator (LASSO) to build a 2-marker–based diagnostic model of BCa, and performed further validation in a prospective multicenter cohort. Furthermore, we systematically evaluated the performance of the utMeMA in the diagnoses of early-stage, minimal, residual, and recurrent tumors of BCa, in comparison with routine urine cytology and FISH.
Results
Discovery of DNA methylation markers to distinguish BCa from normal tissue.
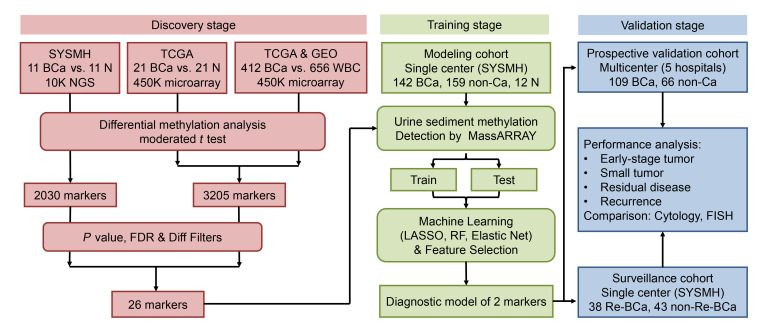
The design and implementation of this study are shown in detail in Figure 1. To investigate specific DNA methylation markers in the detection of BCa, we first performed DNA methylation profiling by high throughput DNA bisulfite targeted sequencing in 11 pairs of BCa and normal adjacent tissue (NAT) from the Sun Yat-sen Memorial Hospital (SYSMH) cohort. Next, we analyzed DNA methylation data of 21 pairs of BCa and NATs from The Cancer Genome Atlas (TCGA) cohort. It is well known that urine is considered as the best sample to noninvasively diagnose BCa. However, the leukocytes are common in the urine of urinary diseases and an interference factor to distinguish malignant and benign diseases. To eliminate the influence of leukocyte DNA in urine, we further analyzed DNA methylation profiles of 412 BCa tissues from TCGA and 656 normal blood samples from a Gene Expression Omnibus (GEO) data set (GSE40279) (19). Through differential methylation analysis, 2030 markers in the SYSMH cohort and 3205 markers in the combined TCGA and GEO cohorts were markedly changed between BCa and normal tissue (Supplemental Figure 1). Furthermore, we applied a series of statistical filters to reduce the number of markers and sought the most important and specific markers of BCa. Finally, we identified 26 markers that displayed high and stable methylation in tumors, but remained at very low levels in normal tissue and leukocytes (Figure 2, A and B, Supplemental Figure 2). These data suggested that DNA methylation markers could be used to distinguish BCa.
Figure 1. Workflow indicating study design.
SYSMH, Sun Yat-sen Memorial Hospital; TCGA, the Cancer Genome Atlas; BCa, bladder cancer; FDR, false discovery rate; LASSO, the least absolute shrinkage and selection operator; RF, random forest.
Figure 2. Discovery of DNA methylation markers to distinguish BCa and normal tissue.
(A) Unsupervised hierarchical clustering of 26 methylation markers differentially methylated between NAT (n = 21) and BCa tumor tissue (n = 412) in the TCGA cohort. (B) Box plot presenting the β-value distribution of cg21472056 among BCa tumor tissue samples (n = 412), NAT (n = 21), and normal blood WBCs (n = 656). A β-value of zero represents no methylation, whereas 1 represents full methylation. The data are shown as median with the interquartile range. Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s tests. (C) Unsupervised hierarchical clustering of 26 methylation markers differentially methylated among NAT (n = 21), BCa tumor tissue (n = 21), and matched urine (n = 18) in the SYSMH cohort. The unavailable value is shown in gray. (D) Box plot presenting the β-value distribution of cg21472056 among BCa tumor tissue (n = 21), matched urine (n = 18), and NAT (n = 21), which was detected by TOF-MS. The data are shown as median with the interquartile range. Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s tests. (E) The Spearman correlation analysis of cg21472056 methylation level between the tumor tissue and matched urine in 18 patients. Pearson’s χ2 test was used to analyze statistical significance. **P < 0.01 and ***P < 0.001.
Development of a novel urine DNA methylation assay for BCa detection.
To detect multiple markers in a fast, cost-effective, and high throughput way in clinics, we developed an efficient method called urine tumor DNA methylation MassARRAY (utMeMA) to diagnose BCa, which allows simultaneous multiplex quantification of CpG sites from various genomic regions at a low methylation frequency with high resolution. To validate whether 26 markers could be used to distinguish BCa from normal tissue, we performed utMeMA to detect the methylation levels of 21 pairs of BCa and NATs, and 18 matched urine samples (Figure 2C). There were 25 markers that showed high methylation levels in cancer tissue and urine, but low methylation levels in NATs, except cg12350762 (Figure 2D, Supplemental Figure 3). The methylation levels of tissues both in NMIBC and MIBC were higher than those in NATs, suggesting that these 25 markers may be used for the detection both of NMIBC and MIBC (Supplemental Figure 4). Furthermore, correlation analysis showed that 23 of 26 markers in urine were significantly and positively correlated with matched cancer tissue, such as cg21472506 which had the highest R2 of 0.625. Three markers (cg12350762, cg23180938, cg06782686) were the exceptions. These findings indicated that urine DNA methylation could represent cancer tissue methylation levels using utMeMA, and these 23 markers could be used as diagnostic markers in BCa (Figure 2E, Supplemental Figure 5).
Construction and validation of a urine diagnostic model to detect BCa in 3 cohorts by using 2 markers.
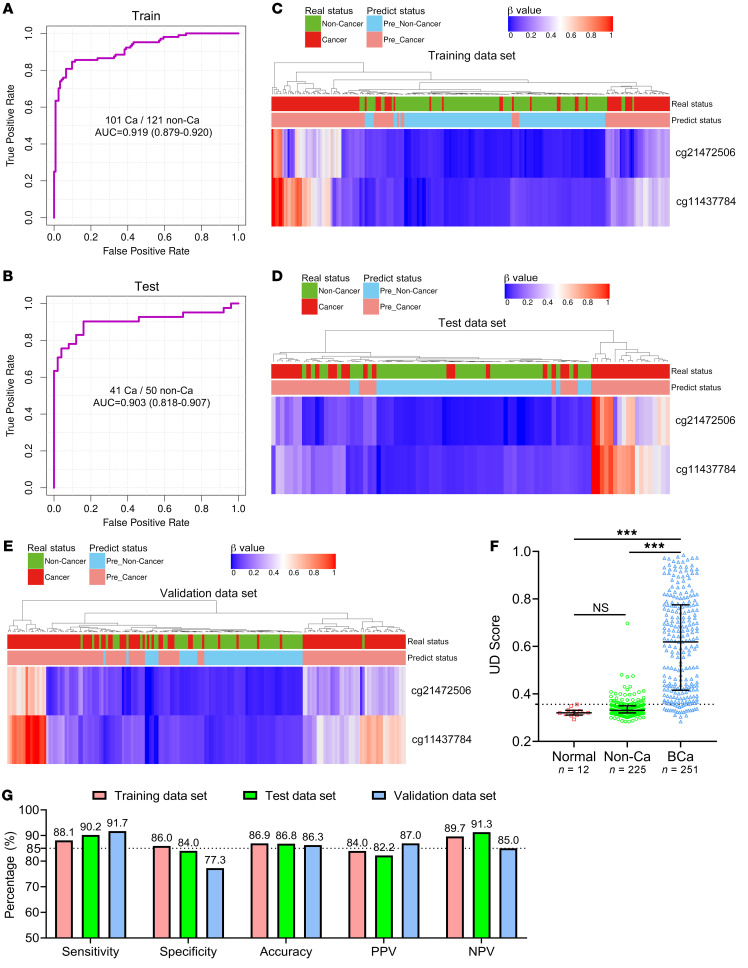
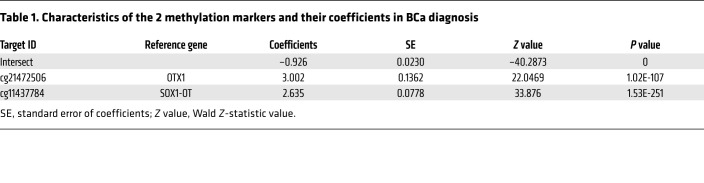
To build the diagnostic model, we enrolled 142 patients diagnosed with BCa, 159 non–cancer patients with benign diseases of the urinary system, and 12 healthy participants from the SYSMH cohort. We analyzed the methylation status of 23 markers by utMeMA, and used LASSO for marker selection and model development. We achieved an excellent performing model with only 2 CpG markers (cg21472506 and cg11437784), which exhibited high AUCs of 0.919 and 0.903, respectively, in the training and test data sets (Figure 3, A and B, Table 1, Supplemental Figure 6, A and B). Remarkably, we observed a high consistency between predicted results and pathological diagnosis results in both the training and test data sets using this model (Figure 3, C and D).
Figure 3. Construction and validation of a urine diagnostic model to detect BCa in 3 cohorts by using 2 markers.
(A and B) ROC curves and the associated AUCs of the diagnostic prediction model using urine DNA methylation analysis in the training (A) and testing (B) cohorts. (C–E) Unsupervised hierarchical clustering of 2 methylation markers that were differentially methylated between the DNA of BCa and non–cancer subjects in the training (C, n = 222), testing (D, n = 91), and independent prospective validation (E, n = 175) cohorts. Each row represents an individual patient and each column is a CpG marker. The real disease status and prediction status by model are shown ahead. (F) The UD score of healthy participants (n = 12), non–cancer patients (n = 225), and patients with BCa (n = 251) are shown. The dotted line shows the cutoff value (0.3564) to distinguish BCa from non–cancer cases. The data are shown as median with the interquartile range. Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s tests. (G) The sensitivity, specificity, accuracy, PPV, and NPV of this model in the training, testing, and validation cohorts were determined by the cutoff value. ***P < 0.001.
Table 1. Characteristics of the 2 methylation markers and their coefficients in BCa diagnosis.
To further assess the performance of the utMeMA-based diagnostic model for clinical application, we performed a prospective, multicenter, blinded study. This independent validation cohort enrolled 109 patients diagnosed with BCa and 66 controls with benign diseases from 5 hospitals in China. Similarly, this model showed good concordance with pathological diagnosis (Figure 3E). We then assessed a urine diagnostic score (UD score) of the model for differentiating between BCa and benign diseases. The UD score was significantly high in cases with BCa, but very low in patients with benign diseases and healthy people (Figure 3F). Importantly, this model achieved high sensitivities of 88.1%, 90.2%, and 91.7% and specificities of 86%, 84%, and 77.3% in the training, test, and validation data sets, respectively. In addition, the value of accuracy, the positive predictive value (PPV) and negative predictive value (NPV) of this model were almost more than 85% and showed great performance (Figure 3G). The performance of this model was better than either cg21472506 or cg11437784 (Supplemental Figure 6, C–D). The utMeMA-based model showed high sensitivity and strong diagnostic power in the detection of BCa.
The performance of utMeMA to diagnose BCa in comparison with urine cytology and FISH.
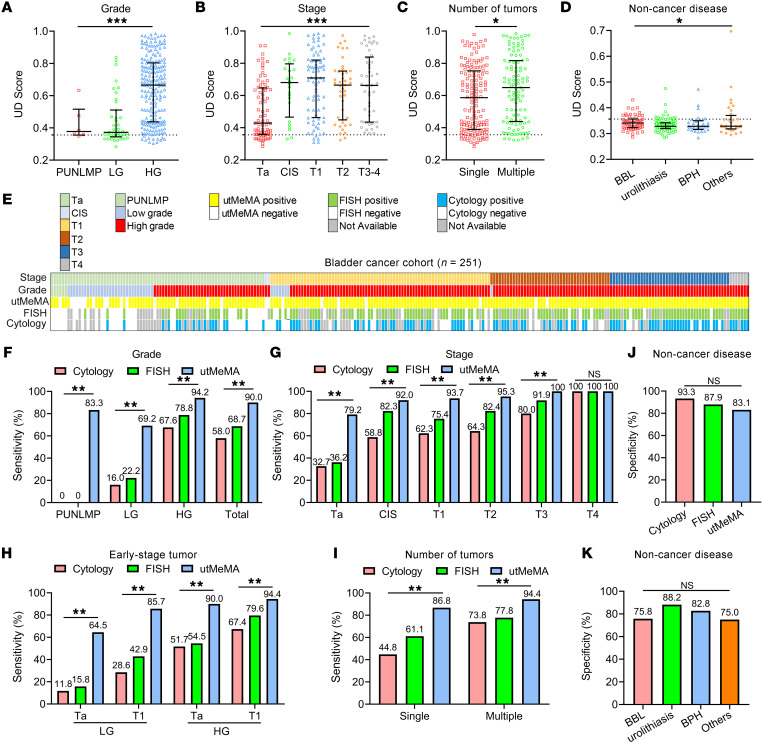
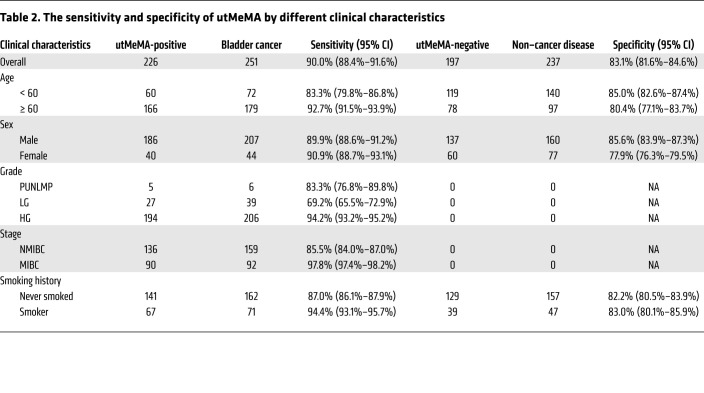
We found that the UD score was positively correlated with advanced grade, stage, number of tumor, and number of RBCs in urine, but no obvious difference in age, sex, smoking status, the type of non–cancer disease, and number of WBCs in urine (Figure 4, A–D, Supplemental Figure 7). From the integrated analysis of 488 cases in this study, this model showed an overall sensitivity of 90.0% and specificity of 83.1%. From further analysis of the sensitivity and specificity using various clinical characteristics, the sensitivity was markedly higher in patients with old age, high-grade, and MIBC, but no obvious difference was observed in sex and smoking status. In addition, the specificity showed no significant difference in age, sex, and smoking status (Table 2, Supplemental Figure 8).
Figure 4. The significantly improved sensitivity of utMeMA in the diagnosis of BCa in comparison with urine cytology and FISH.
(A–C) The UD score of patients with BCa in different grade (A), stage (B), and number (C) of tumors (n = 251). Carcinoma in situ (CIS) means all the cases that include CIS (n = 24). (D) The UD score of patients in 4 types of non–cancer diseases of the urinary system, including benign bladder lesions (BBLs), urolithiasis, benign prostatic hypertrophy (BPH), and other benign diseases (n = 237). The data are shown as median with the interquartile range. Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s tests (A, B, D) and unpaired t test (2-tailed, C). (E) Distribution of predicted diagnostic status using utMeMA across patients with BCa (n = 251) with associated tumor stage, grade, cytology, and FISH results. CIS means the cases which are CIS alone (n = 2). (F–I) The sensitivity of utMeMA in BCa patients with indicated grade (F), stage (G), early-stage (H), and number (I) of tumor, in comparison with urine cytology and FISH. CIS means all the cases that include CIS (n = 24). (J) The specificity of utMeMA in patients with non–cancer diseases in comparison with urine cytology and FISH. (K) The specificity of utMeMA in patients with 4 types of non–cancer diseases. Statistical significance was assessed by χ2 test (G–L). *P < 0.05, **P < 0.01, ***P < 0.001.
Table 2. The sensitivity and specificity of utMeMA by different clinical characteristics.
Urine cytology and UroVysion FISH were routine methods used in the detection of BCa (4–6). To compare the performance among the utMeMA-based model, urine cytology, and FISH, we included 251 patients with BCa for further analysis. The landscape of clinical characteristics and the diagnostic status of 3 methods is shown in Figure 4E. Surprisingly, utMeMA detected 5 of 6 patients (83.3%) with papillary urothelial neoplasm of low malignant potential (PUNLMP), but none of these patients were detected by the other 2 methods (Figure 4F). Furthermore, in patients with low-grade tumors, the sensitivity of utMeMA was 4-fold higher compared with cytology (69.2% vs. 16.0%) and 3-fold higher compared with FISH (69.2% vs. 22.2%). Remarkably, utMeMA achieved a great improvement in sensitivity over cytology and FISH in Ta (79.2% vs. 32.7%, 36.2%) and T1 (93.7% vs. 62.3%, 72.4%) stage patients (Figure 4G). In addition, the sensitivity of this model was also superior to cytology and FISH in high-grade, MIBC, and total patients, respectively (Figure 4, F and G). In the hardest-to-detect low-grade and Ta patients, the sensitivity of utMeMA was 5-fold higher compared with cytology (64.5% vs. 11.8%) and 4-fold higher compared with FISH (64.5% vs. 15.8%). The great advantage of utMeMA was also seen in other patients with early-stage tumors and single/multiple tumors (Figure 4, H and I). Although the specificity of cytology and FISH were higher than utMeMA, the difference was not statistically significant (Figure 4J). There were also no obvious differences among the 4 types of non–cancer diseases (Figure 4K). Similar results were also found in a multicenter validation cohort (Supplemental Figure 9). Collectively, utMeMA exhibited markedly improved sensitivity compared with urine cytology and FISH, particularly in patients with low-grade and early-stage tumors.
Application of utMeMA to detect minimal tumor in BCa.
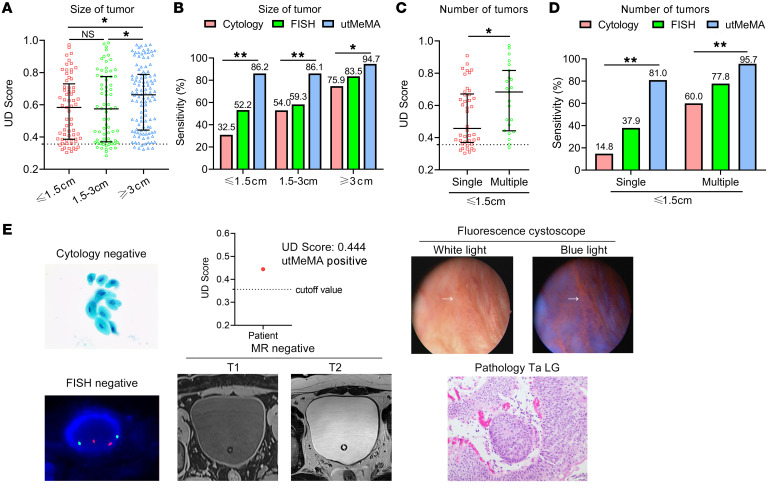
We then evaluated the performance of utMeMA in the size of tumors. The UD score and sensitivity were markedly increased in bigger tumors (≥ 3 cm), but were similar in tumors that were small and middle-sized (Figure 5, A and B). After dividing cases with small tumors into 2 groups, the UD score and sensitivity of small single tumors were lower than multiple tumors, which was consistent with tumor burden (Figure 5, C and D). The utMeMA achieved a great improvement in sensitivity over cytology and FISH in the above conditions, especially in small single tumors (81.0% vs. 14.8%, 37.9%) (Figure 5, B and D). The potential utility of this approach is highlighted by a case that was detected by utMeMA, but missed by cytology, FISH, MR imaging, and ordinary cystoscopy. The lesion was very flat and small, and not markedly abnormal in white light, but was later diagnosed as a low-grade and Ta tumor by fluorescence cystoscopy-guided TURBT (Figure 5E). Furthermore, a similar situation was observed in 3 other cases and the smallest tumor detected by utMeMA was 4 mm in diameter. These data strongly demonstrated the advantage of utMeMA in the detection of minimally sized tumors.
Figure 5. Application of utMeMA to detect minimal BCa tumor.
(A and B) The UD score and sensitivity of utMeMA in patients with different BCa tumor sizes, in comparison with urine cytology and FISH. Statistical significance was assessed using 1-way ANOVA followed by Dunnett’s tests (A) and χ2 test (B). (C and D) The UD score and sensitivity of utMeMA in patients with BCa with single or multiple small tumors, in comparison with urine cytology and FISH. Statistical significance was assessed using unpaired t test (2-tailed, C) and χ2 test (D). The data are shown as median with the interquartile range (A and C). (E) Example of a patient with minimal tumor detected by utMeMA, but missed by cytology, FISH, MR imaging, and ordinary cystoscopy, who was later diagnosed by fluorescence cystoscopy. The pathology of the tumor was Ta and low grade. *P < 0.05, **P < 0.01. The magnifications of the images in fluorescence cystoscopy, urine cytology, FISH and pathology were ×10, ×600, ×3000, ×400, respectively.
Application of utMeMA to detect residual tumor and monitor recurrence in BCa.
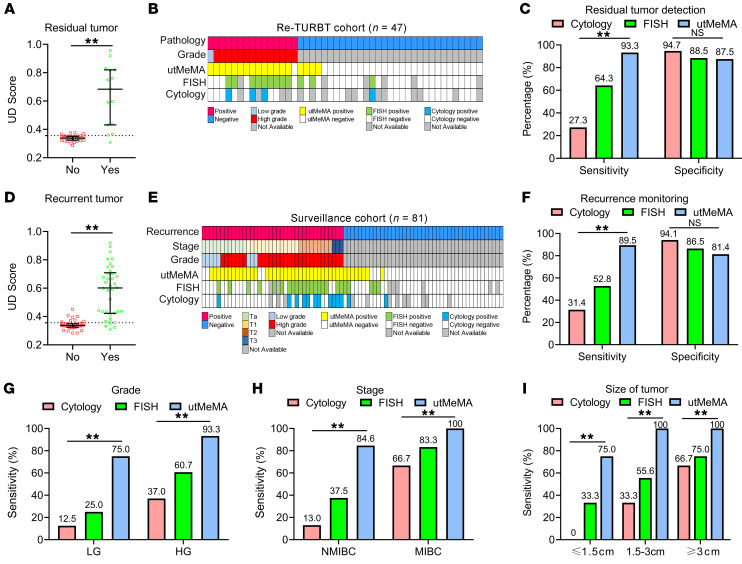
Re-TURBT is recommended for patients with high-grade and T1 tumors, but currently, we lack effective methods to estimate if the patient actually has residual tumors (4, 5). In our modeling and validation cohorts, 47 patients received Re-TURBT and the samples were collected before the surgery, when 15 patients had residual tumor but 32 patients did not. Interestingly, the UD score was significantly increased in patients with residual tumor compared with those without (Figure 6A). Importantly, utMeMA correctly diagnosed 14 of 15 (93.3%) patients with residual tumor, but cytology and FISH only diagnosed 3 of 11 (27.3%) and 9 of 14 (64.3%) of these patients, respectively (Figure 6, B and C). The specificity of utMeMA was 87.5%, which was similar with cytology and FISH (Figure 6C). These amazing findings suggested that utMeMA could be used to detect residual tumors and serve as a predictor to select patients for Re-TURBT.
Figure 6. Application of utMeMA to detect residual tumor and monitor the recurrence of BCa.
(A) The distribution of UD score in BCa patients with or without residual tumors (n = 47). Statistical significance was assessed using unpaired t test (2-tailed). The data are shown as median with the interquartile range. (B) The landscape of pathological characters and detection results in re-TURBT cohort, including 15 cases with residual tumor and 32 cases without tumor. (C) The sensitivity and specificity of utMeMA in the detection of residual tumor, in comparison with urine cytology and FISH (n = 47). (D) The distribution of UD score in patients with BCa with or without recurrent tumor. The data are shown as median with the interquartile range. Statistical significance was assessed using unpaired t test (2-tailed). (E) The landscape of pathological characteristics and detection results in surveillance cohort, including 38 cases with tumor recurrence and 43 cases without recurrence (n = 81). (F) The sensitivity and specificity of utMeMA in detection of recurrent tumor, in comparison with urine cytology and FISH (n = 81). (G–I) The sensitivity of utMeMA in patients with recurrent BCa with indicated grade (G), stage (H), and size (I) of tumor, in comparison with urine cytology and FISH (n = 38). Statistical significance was assessed using χ2 test (C, F, G–I). *P < 0.05, **P < 0.01.
Given the high recurrent rate of patients with NMIBC, it is important to develop a noninvasive and sensitive method to monitor recurrence (4). We observed a high consistency of UD score between first morning urine and random urine, suggesting that random urine was also suitable for the detection of BCa (Supplemental Figure 10, A and B). Next, we enrolled an additional 81 patients undergoing surveillance from SYSMH and collected urine samples before undergoing cystoscopy. Subsequently, 38 cases were found to have tumor recurrence and 43 cases did not. Interestingly, the UD score was markedly higher in patients with recurrence compared with patients without recurrence, and was positively correlated with tumor burden (Figure 6D Supplemental Figure 10, C–F). Importantly, utMeMA accurately detected 34 of 38 (89.5%) patients with recurrence, but cytology and FISH only detected 11 of 35 (31.4%) and 19 of 26 (52.8%) patients with recurrence, respectively (Figure 6, E and F). The specificity of utMeMA was 81.4%, which had no statistically significant difference from cytology and FISH (Figure 6F). The follow-up of patients with positive utMeMA results but no evidence of recurrence is ongoing and they will be reevaluated in a future study. In the subgroup analysis, utMeMA achieved a great improvement in sensitivity over cytology and FISH, especially in low-grade (75.0% vs. 12.5%, 25.0%), NMIBC (84.6% vs. 13.0%, 37.5%), and small and single tumors (75.0% vs. 0%, 33.3%) (Figure 6, G–I, Supplemental Figure 10G). utMeMA could serve as a noninvasive and highly sensitive approach to monitor the recurrence of BCa.
Discussion
Here, we first discovered the BCa-specific methylation markers by combined analyses of SYSMH, TCGA, and GEO cohorts. Then, we trained and tested the diagnostic model in the SYSMH cohort of 313 samples, and performed validation in a multicenter, prospective, independent cohort of 175 samples. This diagnostic model of BCa included only 2 CpG markers (cg21472506 and cg11437784), but exhibited an overall sensitivity of 90.0% and a specificity of 83.1%. The CpG site cg21472506, located on the 3′-untranslated region of OTX1, was previously reported as a useful marker to detect BCa in urine (20, 21). However, cg11437784, located in the intron of SOX1-OT, was first discovered as a tumor marker. The biological function and methylated mechanism of OTX1 and SOX1-OT remain largely unknown. A previous study used 4 methylation markers to identify bladder carcinoma with a sensitivity of 82% and a specificity of 53% (14). Bladder EpiCheck used a panel of 15 methylation markers to monitor recurrence in patients with NMIBC undergoing surveillance, and its overall sensitivity was 67% and specificity was 88% (22, 23). Our study showed that the overall sensitivity of NMIBC was 85.5% in modeling and validation cohorts, and it was 84.6% in the additional surveillance cohort. Our test also showed a higher sensitivity compared with EpiCheck in the monitoring of recurrence of low-grade (75% vs. 40%) and high-grade (93% vs. 89%) tumors (22, 23). Recent studies found that somatic mutation or combined DNA mutation and methylation were also promising markers to detect BCa (20, 21, 24). On the basis of this 2-marker test, it is worth exploring whether the performance could be improved by adding the detection of additional DNA mutations in the future.
The common methods to detect DNA methylation are methylation-specific PCR (MS-PCR) and genome bisulfite sequencing (14, 21, 24). MS-PCR is easy to use and cheap, but it fails to provide high resolution and specific detection of single CpG sites when multiple CpG sites are involved. The genome bisulfite sequencing enables high-throughput detection of large-scale methylation markers, but it is expensive and time consuming, which limits its clinical application. Our utMeMA method addresses these issues, allowing high-resolution and high-throughput quantification of multiple CpG sites even from samples with a low methylation frequency. Due to the superior sensitivity of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS), the methylation level of a single CpG site can be determined by single nucleotide amplification of the CpG site of interest without any normalization. The enhanced technical advantages therefore effectively improved assay detection sensitivity and the scale of samples being processed. This approach could analyze 300 samples at a time and provide clinical reports in 1 to 2 days. Thus, utMeMA is a fast, cost-effective, high-resolution, and high-throughput method to detect BCa in the clinical setting.
The early-stage, minimal, residual, and recurrent tumors of BCa were very difficult to diagnose, which was usually missed by urine cytology and FISH (4, 5). However, this method achieved a great improvement in sensitivity over cytology and FISH, serving as a promising solution in these conditions. Importantly, the UD score positively correlated with the grade, stage, size, and the presence of residual and recurrent tumors of BCa. These results make this method attractive for use in clinical decision-making across a variety of patients and situations, and could in turn reduce the current burden of repeated cystoscopy and blind Re-TURBT.
However, there are some limitations that need to be emphasized. First, the samples analyzed in Re-TURBT and the surveillance cohort were small, so the data need to be validated in a larger multicenter prospective study. Second, the performance of utMeMA in the monitoring of recurrence was a cross-sectional analysis and the long-term follow-up data are currently unavailable. Thus, we were unable to correlate false-positives with later recurrence.
In conclusion, we have developed an efficient utMeMA 2-marker–based test for the fast and noninvasive detection of BCa. Our approach achieved a great improvement in sensitivity over urine cytology and FISH, especially in the detection of early-stage, minimal, residual, and recurrent tumors. Therefore, it was adopted in the optional clinical detection of BCa by more than 10 hospitals in China. A large-scale, multicenter, and prospective clinical trial (NCT04314245) is ongoing to validate its clinical applicability in China.
Methods
Study design and participants.
In discovery stage, we identified the BCa-specific methylation markers by combined analyses of the SYSMH, TCGA, and GEO cohorts (19, 25). There were 32 paired BCa and NAT samples, and 18 matched urine samples from patients who underwent surgery at SYSMH between June 2016 and May 2017. The human methylation 450 K array data and clinical characteristics of 412 BCa tissue and 21 matched normal tissue samples were obtained from TCGA. The methylation profiles of 656 blood leukocyte samples of healthy control individuals were obtained from a GEO data set (GSE40279).
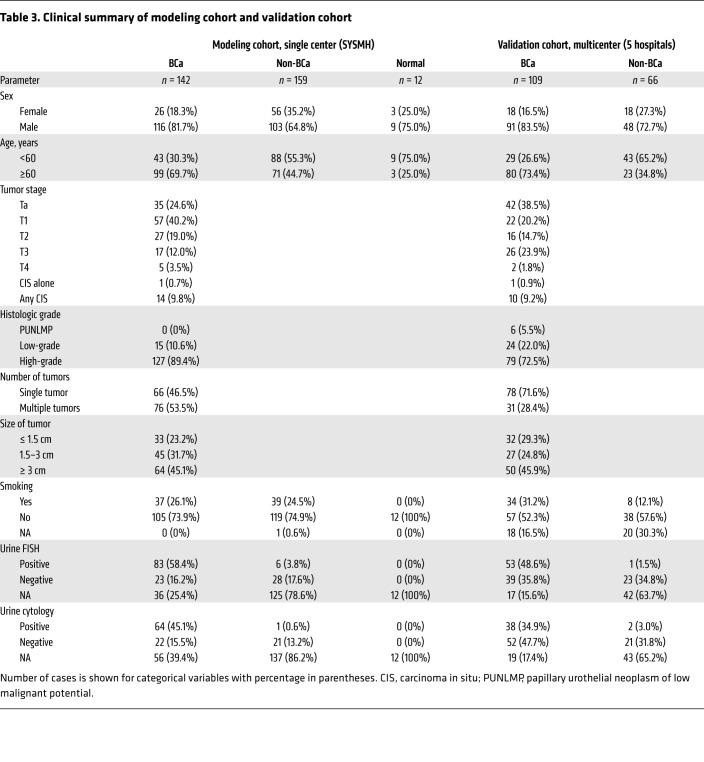
In the retrospective, single-center cohort (modeling cohort), we enrolled 142 patients with urothelial carcinoma of the bladder (UCB), 12 healthy participants, and 159 non–cancer controls from SYSMH between June 2017 and May 2019. In the multicenter, prospective, blinded cohort (validation cohort), we enrolled 109 patients with UCB and 66 non–cancer controls from 5 hospitals in China between August 2019 and December 2019. The multicenter validation cohort was collected from the SYSMH (n = 70), Zhujiang Hospital, Southern Medical University (n = 23) and Nanfang Hospital, Southern Medical University (n = 22) in Guangzhou, Tongji Hospital of Huazhong University of Science and Technology in Wuhan (n = 39), and the Affiliated Hospital/Clinical Medical College of Chengdu University in Chengdu (n = 21), China. Urine samples were collected from each hospital with written informed consent obtained from all patients. The non–cancer controls were diagnosed with benign urological diseases including benign bladder lesions (BBLs), urolithiasis, benign prostatic hypertrophy (BPH), and other benign diseases of the urinary system.
In the surveillance cohort, we enrolled 38 patients with tumor recurrence and 43 patients without recurrence from SYSMH. The samples with pathological diagnoses were reviewed by 2 independent pathologists. Flow of participant enrollment of these 3 cohorts is summarized in Supplemental Figures 11–13. The demographics and clinical characteristics of the participants are summarized in Table 3 and Supplemental Tables 1–3, respectively.
Table 3. Clinical summary of modeling cohort and validation cohort.
Sample processing.
Genomic DNA extraction from freshly frozen normal or cancer tissue was performed with the DNeasy Blood & Tissue Kit (Qiagen, catalog 69506) according to the manufacturer’s recommendations. Roughly 0.5 mg of tissue was used to obtain 5 μg of genomic DNA on average, which was stored at –80°C.
Voided urine (approximately 50–100 mL) was collected before surgery or cystoscopy, and immediately processed within an hour. The urine samples were centrifuged at 3000g for 10 minutes. The cell pellets were washed with 10 mL PBS twice and spun down for 10 minutes at 3000g. Then, cells were resuspended in 1 mL PBS, transferred to an Eppendorf vial, and centrifuged for another 5 minutes. The washed cell pellets were stored at –80°C. The DNA from urine cell pellets was isolated using the Quick-DNA Urine Kit (Zymo Research, catalog D3061) according to the manufacturer’s instructions. Ninety-five percent of the urine samples yielded more than 100 ng DNA, which was the required amount to perform all assays.
AnchorIRIS targeted methylation sequencing.
To discover differential methylation profiling of BCa, a targeted methylation sequencing of 100,000 CpG sites was performed by using the AnchorIRIS technologies as previously described (26). Detailed information is shown in Supplemental Methods. The raw sequence data reported in this study have been deposited in the Genome Sequence Archive in the Beijing Institute of Genomics (BIG) Data Center, Chinese Academy of Sciences, under GSA accession number CRA002787, and are publicly accessible at http://bigd.big.ac.cn/gsa
Methylation analysis by MALDI-TOF-MS.
Instead of using the typical EpiTYPER DNA methylation analysis technology which was limited to detect only one genomic region with a relatively large target fragment, we have adopted the SNP genotyping MassARRAY system to detect the methylation of multiple CpG sites from different genomic regions. By applying the bisulfite-converted target sequences on the Assay Design Suite software (Agena Bioscience), the amplification and extension primers for simultaneous multitarget methylation detections were designed and experimentally verified. Genomic DNA (100 ng) from each sample was treated with sodium bisulfite with the EZ DNA Methylation-Lightning Kits (Zymo Research, catalog D5030). A subsequent quantitative analysis of DNA methylation of selected methylation markers was carried out by the Agena MassARRAY platform with the iPLEX Pro reagent kit (Agena Bioscience, catalog 10217) according to the manufacturer’s instructions. All specific primers used in the utMeMA assay are listed in Supplemental Table 4. See Supplemental Methods for further details.
Identification of methylation markers discriminating between BCa and normal tissue.
To identify putative markers, we first compared the methylation data derived from paired BCa and normal urothelium samples from the SYSMH cohort (n = 11) and TCGA cohort (n = 21).Then, we compared the methylation data derived from 412 BCa tissue samples from the TCGA cohort and 656 healthy blood samples from a previous study (19). Group-wise (cancer vs. normal) moderated t test was used for the initial screening of markers from the above-mentioned data. Furthermore, we applied additional filters to increase the stringency of the screening to reduce potential false positives, including group-wise SE less than 0.1, mean β-value difference greater than 0.2, FDR less than 0.01 (450 K data) or less than 0.05 (targeted methylation sequencing data), mean β-value in normal or WBCs less than 0.17, mean β-value in cancer greater than 0.3.
Construction and validation of a urine diagnostic model to detect BCa.
In the modeling cohort, 313 cases were randomly subdivided into training (222 cases) and test (91 cases) sets, respectively, and samples were stratified against age, sex, smoking status, and pathological classes. The training and test sets were used for model building and the selection of CpG markers, and the 175 cases in the validation set were for independent testing of the selected model. We used LASSO to build the model and selected the best markers simultaneously by shrinking some coefficients to zero, which was equivalent to removing these markers from the model. The hyperparameter lambda in LASSO, which controlled the level of regularization, was selected from out-of-fold performance on 50 repetitions of 5-fold cross-validation analysis of the training data, and the metric for model selection was based on AUC scores in the cross-validation phase. The final model built from the whole training data set was used for testing and validation. The performance of the model was evaluated by AUC. UD scores were calculated based on the LASSO model as determined in the training data set. The formula for calculating the UD score is logistic (–0.926 + 3.002 × OTX1 + 2.635 × SOX1-OT), and the coefficients and intercepts and their statistical significances are listed in Table 1. The cutoff value (0.3564) on the UD score was determined by the method of Youden’s index on the model ROC, which maximizes the sum of sensitivities and specificities.
Statistics.
LASSO was fitted to build the UD score, and the ROC curve was adopted to assess the performance of the UD score–based model. The β-value and UD score distribution between clinical categories were presented as box plots with median and the interquartile range marks. Differences between 2 groups were analyzed with the unpaired/paired Student’s t test (2-tailed tests), and 1-way ANOVA followed by Dunnett’s multiple comparisons tests when more than 2 groups were compared. The sensitivity, specificity, accuracy, PPV, and NPV of utMeMA, cytology, and FISH in detecting BCa were obtained by comparison to pathology and are presented as univariate values in the figures. The positive and negative values for utMeMA were determined by the cutoff value (0.3564), while positive and negative values for cytology and FISH were determined by the clinical report. Pearson’s χ2 test was used to analyze the clinical variables on sensitivity and specificity. Spearman’s correlation analysis was performed to determine the correlation between 2 variables. All hypothesis testing was 2-sided with a P value less than 0.05 considered statistically significant. All statistical analyses and data visualizations were carried out in R (3.6.0) with R packages and Prism 8 (GraphPad Software).
Study approval.
This study was conducted in compliance with the principles of the 1975 Declaration of Helsinki and was approved by the Ethics Committees of the Sun Yat-sen Memorial Hospital, Sun Yat-sen University. Written informed consent was obtained from all patients or their legal representatives before their participation in the study.
Author contributions
TL, JH, JBF, and XC conceived, designed, and directed the study. WR, ZJ, JC, and ZC developed the methodology. XC, JZ, MH, CW, SW, ZL, CL, WT, JY, XZ, RX, and QZ acquired the data. XC, HW, and ZJ performed the analyses and interpretation of data. WR, JC, ZC, XL, DLI, PX, LC, and SX provided technical or material support. XC, TL, JH, and JBF wrote and critically reviewed the manuscript. All authors read and approved the final manuscript. The order of the co–first authors was assigned based on the relative contributions of the individuals.
Supplementary Material
Acknowledgments
We are grateful to the patients and families involved in this study, and to the Cellular & Molecular Diagnostics Center of SYSMH for preserving urine cytology and FISH data for this study. This study was supported by the National Key Research and Development Program of China (grants 2018YFA0902803 and 2017YFC1309002), the National Natural Science Foundation of China (grant 81825016, 81961128027, 81702523, 81772719, 81772728), The Key Areas Research and Development Program of Guangdong (grant 2018B010109006), Science and Technology Program of Guangzhou (grant 201804010041), Guangdong Basic and Applied Basic Research Foundation (grant 2020A1515010888), Science and Technology Planning Project of Guangdong Province, China (grant 2017B020226005), Guangdong Special Support Program (2017TX04R246), Project Supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (to TL), Scheme of Guangzhou for Leading Team in Innovation (grant 201909010010), Scheme of Guangzhou Economic and Technological Development District for Leading Talents in Innovation and Entrepreneurship (grant 2017-L152), Scheme of Guangzhou for Leading Talents in Innovation and Entrepreneurship (grant 2016007).
Version 1. 08/20/2020
In-Press Preview
Version 2. 10/26/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6278–6289.https://doi.org/10.1172/JCI139597.
Contributor Information
Xu Chen, Email: chenx457@mail.sysu.edu.cn.
Jingtong Zhang, Email: jtzhang930912@gmail.com.
Weimei Ruan, Email: weimei_ruan@anchordx.com.
Ming Huang, Email: 562457252@qq.com.
Chanjuan Wang, Email: luna0907@foxmail.com.
Hong Wang, Email: hong_wang@anchordx.com.
Zeyu Jiang, Email: zeyu_jiang@anchordx.com.
Shaogang Wang, Email: sgwangtjm@163.com.
Zheng Liu, Email: lz2013tj@163.com.
Chunxiao Liu, Email: liuchx888@hotmail.com.
Wanlong Tan, Email: twl@smu.edu.cn.
Jin Yang, Email: dr.jinyang@163.com.
Jiaxin Chen, Email: jiaxin_chen@anchordx.com.
Zhiwei Chen, Email: zhiwei_chen@anchordx.com.
Xia Li, Email: xia_li@anchordx.com.
Xiaoyu Zhang, Email: 1391032151@qq.com.
Peng Xu, Email: yihuixp88@hotmail.com.
Lin Chen, Email: clin360@gmail.com.
Ruihui Xie, Email: xrhsysu@foxmail.com.
Qianghua Zhou, Email: zhouqhsysu@163.com.
Shizhong Xu, Email: shizXu@foxmail.com.
Jian-Bing Fan, Email: jianbingfan1115@smu.edu.cn.
Jian Huang, Email: huangj8@mail.sysu.edu.cn.
Tianxin Lin, Email: lintx@mail.sysu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cumberbatch MGK, et al. Epidemiology of bladder cancer: a systematic review and contemporary update of risk factors in 2018. Eur Urol. 2018;74(6):784–795. doi: 10.1016/j.eururo.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Leal J, Luengo-Fernandez R, Sullivan R, Witjes JA. Economic burden of bladder cancer across the European Union. Eur Urol. 2016;69(3):438–447. doi: 10.1016/j.eururo.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Babjuk M, et al. European Association of Urology guidelines on non-muscle-invasive bladder cancer (TaT1 and carcinoma in situ) - 2019 update. Eur Urol. 2019;76(5):639–657. doi: 10.1016/j.eururo.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Dimashkieh H, Wolff DJ, Smith TM, Houser PM, Nietert PJ, Yang J. Evaluation of UroVysion and cytology for bladder cancer detection: a study of 1835 paired urine samples with clinical and histologic correlation. Cancer Cytopathol. 2013;121(10):591–597. doi: 10.1002/cncy.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin T, et al. Prospective evaluation of fluorescence in situ hybridization for diagnosing urothelial carcinoma. Oncol Lett. 2017;13(5):3928–3934. doi: 10.3892/ol.2017.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(9):a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, et al. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin Cancer Res. 2019;25(4):1389–1403. doi: 10.1158/1078-0432.CCR-18-1656. [DOI] [PubMed] [Google Scholar]
- 10.Yim JH, et al. Identification of tissue-specific DNA methylation signatures for thyroid nodule diagnostics. Clin Cancer Res. 2019;25(2):544–551. doi: 10.1158/1078-0432.CCR-18-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DD, et al. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J Clin Invest. 2019;129(1):223–229. doi: 10.1172/JCI121303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downs BM, et al. DNA methylation markers for breast cancer detection in the developing world. Clin Cancer Res. 2019;25(21):6357–6367. doi: 10.1158/1078-0432.CCR-18-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, et al. Loss of GATA6 expression promotes lymphatic metastasis in bladder cancer. FASEB J. 2020;34(4):5754–5766. doi: 10.1096/fj.201903176R. [DOI] [PubMed] [Google Scholar]
- 14.Guo RQ, et al. Detection of urothelial carcinoma, upper tract urothelial carcinoma, bladder carcinoma, and urothelial carcinoma with gross hematuria using selected urine-DNA methylation biomarkers: A prospective, single-center study. Urol Oncol. 2018;36(7):342.e15–342.e23. doi: 10.1016/j.urolonc.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Reinert T, et al. Comprehensive genome methylation analysis in bladder cancer: identification and validation of novel methylated genes and application of these as urinary tumor markers. Clin Cancer Res. 2011;17(17):5582–5592. doi: 10.1158/1078-0432.CCR-10-2659. [DOI] [PubMed] [Google Scholar]
- 16.Renard I, et al. Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur Urol. 2010;58(1):96–104. doi: 10.1016/j.eururo.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 17.Witjes JA, et al. Performance of the Bladder EpiCheck methylation test for patients under surveillance for non-muscle-invasive bladder cancer: results of a multicenter, prospective, blinded clinical trial. Eur Urol Oncol. 2018;1(4):307–313. doi: 10.1016/j.euo.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 18.van der Heijden AG, et al. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin Epigenetics. 2018;10:71. doi: 10.1186/s13148-018-0496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beukers W, et al. FGFR3, TERT and OTX1 as a urinary biomarker combination for surveillance of patients with bladder cancer in a large prospective multicenter study. J Urol. 2017;197(6):1410–1418. doi: 10.1016/j.juro.2016.12.096. [DOI] [PubMed] [Google Scholar]
- 21.van Kessel KE, et al. Validation of a DNA methylation-mutation urine assay to select patients with hematuria for cystoscopy. J Urol. 2017;197(3 Pt 1):590–595. doi: 10.1016/j.juro.2016.09.118. [DOI] [PubMed] [Google Scholar]
- 22.D’Andrea D, et al. Diagnostic accuracy, clinical utility and influence on decision-making of a methylation urine biomarker test in the surveillance of non-muscle-invasive bladder cancer. BJU Int. 2019;123(6):959–967. doi: 10.1111/bju.14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trenti E, Pycha S, Mian C, Schwienbacher C, Hanspeter E, Kafka M, et al. Comparison of 2 new real-time polymerase chain reaction-based urinary markers in the follow-up of patients with non-muscle-invasive bladder cancer. Cancer Cytopathol. 2020;128(5):341–347. doi: 10.1002/cncy.22246. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z, et al. Noninvasive detection and localization of genitourinary cancers using urinary sediment DNA methylomes and copy number profiles. Eur Urol. 2020;77(2):288–290. doi: 10.1016/j.eururo.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Robertson AG, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(3):540–556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang W, et al. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA) Theranostics. 2019;9(7):2056–2070. doi: 10.7150/thno.28119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.