We developed mice expressing Tlr4 SNPs homologous to those expressed in humans and associated with infectious and inflammatory diseases. Cellular and molecular analyses of TLR4 signaling in wild-type vs. TLR4-SNP mice revealed novel mechanisms of LPS hyporesponsiveness and altered disease susceptibility.
Abstract
Two cosegregating single-nucleotide polymorphisms (SNPs) in human TLR4, an A896G transition at SNP rs4986790 (D299G) and a C1196T transition at SNP rs4986791 (T399I), have been associated with LPS hyporesponsiveness and differential susceptibility to many infectious or inflammatory diseases. However, many studies failed to confirm these associations, and transfection experiments resulted in conflicting conclusions about the impact of these SNPs on TLR4 signaling. Using advanced protein modeling from crystallographic data of human and murine TLR4, we identified homologous substitutions of these SNPs in murine Tlr4, engineered a knock-in strain expressing the D298G and N397I TLR4 SNPs homozygously, and characterized in vivo and in vitro responses to TLR4 ligands and infections in which TLR4 is implicated. Our data provide new insights into cellular and molecular mechanisms by which these SNPs decrease the TLR4 signaling efficiency and offer an experimental approach to confirm or refute human data possibly confounded by variables unrelated to the direct effects of the SNPs on TLR4 functionality.
Introduction
Initiation of host-mediated inflammation by Gram-negative LPS is triggered by TLR4 through a relatively complex process that transmits signaling across the membrane following oligomerization of the TLR4 protein upon LPS binding. TLR4 does not bind LPS directly. A serum protein, LPS-binding protein, extracts an LPS monomer from micelles and transfers it to soluble or cell-associated CD14. In turn, CD14 transfers the LPS monomer to MD-2, a TLR4 coreceptor present in soluble form or preassociated with TLR4 on the cell surface. Interaction of LPS with the TLR4/MD-2 complex results in formation of TLR4/MD-2 heterotetramers (Latty et al., 2018), creating an intracellular “TLR4 signaling platform” that facilitates recruitment of adapters, MyD88 or TIR domain–containing adaptor-inducing IFN-β (TRIF), leading to MyD88- or TRIF-dependent signaling pathways, respectively (reviewed in Fitzgerald and Kagan, 2020). Other structurally unrelated, MD-2–dependent TLR4 agonists have been described, including respiratory syncytial virus (RSV) fusion protein (Kurt-Jones et al., 2000), Chlamydia Hsp 60 (Bulut et al., 2002), and host-derived high-mobility group box 1 (HMGB1; Yang et al., 2013), among others.
Two nonsynonymous, cosegregating single-nucleotide polymorphisms (SNPs) in human TLR4, an A896G transition at SNP rs4986790 (D299G) and a C1196T transition at SNP rs4986791 (T399I), have been associated with LPS hyporesponsiveness, as well as differential susceptibility to many infectious and noninfectious diseases (reviewed in Schröder and Schumann, 2005; Ferwerda et al., 2007; Medvedev, 2013; Mukherjee et al., 2019). Arbour et al. (2000) first demonstrated that inheritance of these relatively common SNPs resulted in hyporesponsiveness to inhaled LPS and decreased LPS sensitivity of airway epithelial cells. Michel et al. (2003) reported that these TLR4 SNPs were associated with systemic inflammatory hyporesponsiveness to inhaled LPS. However, the mechanistic basis for altered LPS responsiveness in cells expressing these SNPs has been highly controversial. In several studies, ectopic expression of the TLR4 SNPs was associated with significantly decreased surface expression compared with WT TLR4, suggesting that reduced surface TLR4 expression accounted for the observed LPS hyporesponsiveness in epithelial cell cultures (Arbour et al., 2000; Tulic et al., 2007). While Tulic et al. (2007) also observed reduced LPS-induced cytokine responses in peripheral blood mononuclear cells from individuals with these SNPs, Ferwerda et al. (2007) reported an increase in TNF-α in LPS-stimulated whole blood derived from D299G-expressing African individuals but no altered induction of TNF-α in LPS-stimulated blood samples from a Dutch population expressing both D299G and T399I SNPs. Transgenic mice expressing varying copy numbers of genes encoding human WT or TLR4 SNP variants exhibited LPS responses that were more dependent upon the relative extent of TLR4 surface expression rather than expression of the TLR4 SNPs per se (Hajjar et al., 2017). Yet, under conditions of apparently equivalent levels of ectopic expression of WT and mutant TLR4, diminished signaling and recruitment of intracellular adaptor molecules in response to LPS and other TLR4 agonists was reported (Rallabhandi et al., 2006; Figueroa et al., 2012). Prohinar et al. (2010) reported that the affinity of interaction of complexes composed of MD-2 and the TLR4 ectodomain with LPS-CD14 or of TLR4 ectodomain with LPS-MD-2 was not detectably affected by the presence of the TLR4 SNPs, yet accumulation of both extracellular TLR4 ectodomain and full-length surface TLR4 was significantly reduced in the presence of the human TLR4 SNPs.
Since all genetic analyses of inheritance of these TLR4 SNPs with human disease states are correlative and have the potential to be significantly impacted by genetic differences not directly related to TLR4 haplotype, we undertook studies to model the homologous human TLR4 SNPs in murine TLR4, based on published crystallographic analyses. Using a CRISPR/Cas9 approach, mice were derived onto a C57BL/6J background that expresses the Tlr4 SNPs homozygously to permit a rigorous and unambiguous evaluation of LPS sensitivity and susceptibility to infectious insults in mice and their macrophages. We report herein that C57BL/6J mice harboring nucleotide substitutions in the Tlr4 locus that encode D298G/N397I (TLR4-SNP mice) and their macrophages are LPS hyporesponsive, in contrast to LPS-unresponsive TLR4−/− mice, and exhibit increased susceptibility to Gram-negative infection and RSV but increased resistance to influenza, infections previously associated with TLR4. Our results further provide novel insights into underlying signaling mechanisms that dictate the observed altered responses of TLR4-SNP mice.
Results
Based on the crystal structures of WT and SNP-expressing human TLR4 proteins (Kim et al., 2007; Ohto et al., 2012), we modeled murine TLR4 with the homologous SNPs D298G and N397I (Fig. 1 A). Neither residue is at the TLR dimerization interface or contacts MD-2 or LPS in the cocrystal structures. In the five available murine TLR4 structures, the WT D298 side chain varies in its orientation, while the N397 side chain is invariant (Kim et al., 2007; Ohto et al., 2012; Wang et al., 2016). The leucine-rich repeats 10–13, which include D299 (human)/D298 (mouse), are close enough to LPS and MD-2 in the human TLR4 crystal structure (∼10 Å) that one might predict downstream effects on TLR4’s affinity for LPS or other ligands; however, no such differences were observed in affinity for the human proteins (Prohinar et al., 2010). As TLR activation is dependent upon ligand binding–induced dimerization, small alterations in binding kinetics could have profound downstream effects, as has been reported for T cell receptor signaling (Gagnon et al., 2006).
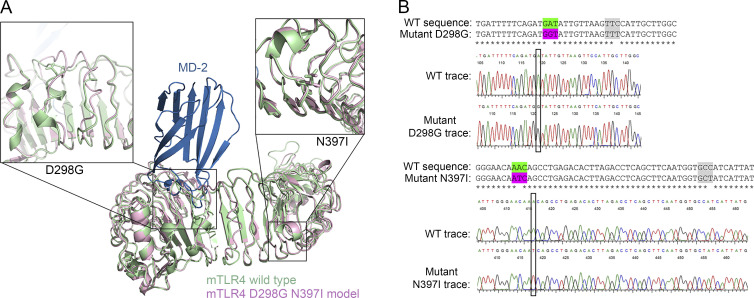
Figure 1.
Homology model of murine TLR4 D298G/N397I. (A) Superimposed models of murine WT vs. TLR4 D298G N397I. The x-ray crystal structure of murine TLR4 (green) with MD-2 (blue) is superimposed onto a model of the double mutant (pink). Inset panels show details around the two SNP sites, D298G (left) and N397I (right). (B) Alignment and DNA sequences of WT and homozygous mutant mice expressing both the D298G and N397I TLR4 SNPs (TLR4-SNP mice).
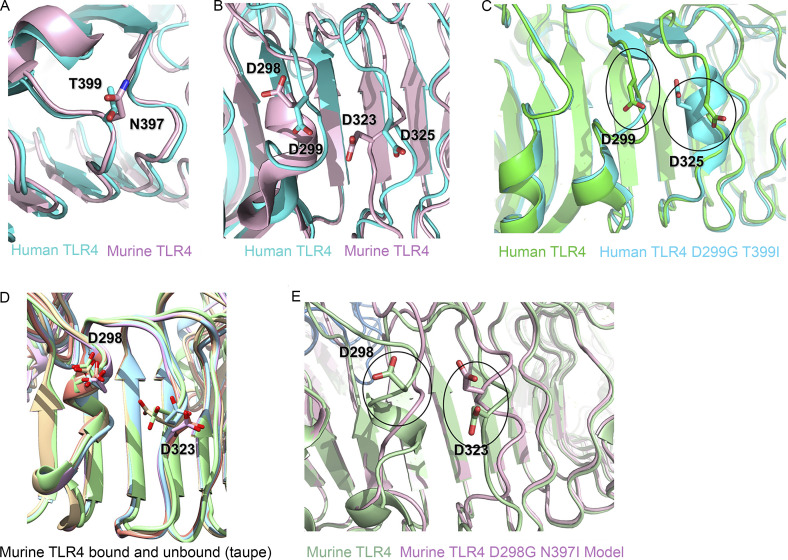
Alignment of the human and murine crystal structures revealed that the loop containing T399 (human) and N397 (mouse) is highly conserved (Fig. S1 A), and our model predicts that, as in the human mutation, the murine N397I is unlikely to alter the overall fold, as the native asparagine side chain makes no contact with other residues in the protein (Fig. S1 A). However, like the human T→I mutation, the electrostatic alteration caused by replacing a polar side chain with an aliphatic one may have consequences for TLR4 association with other membrane proteins such as CD14 (Fergestad et al., 2001; Zanoni et al., 2011). The structural effects of the murine D298G mutation are somewhat more difficult to predict, because this region is less well conserved between the human and murine proteins (Fig. S1 B). In the human D299G mutant, this region is substantially rearranged; both the loop containing 299 and an adjacent loop (containing D325) adopt altered conformations (Fig. S1 C), which Ohto et al. (2012) attribute, in part, to the loss of a hydrogen bond between the D299 side chain and the amide nitrogen of residue 302. Based on our modeling results, we suggest instead that the loss of electrostatic repulsion between the human D299 and D325 side chains allows for the conformational shift that moves the D325 side chain 5 Å closer to the 299 backbone (Fig. S1 C). The murine D298 and D323 side chains are conformationally heterogeneous in the available crystal structures (Kim et al., 2007; Ohto et al., 2012; Wang et al., 2016), but the distance between the aspartate side chains (8 Å) is approximately conserved in all of the bound structures and only slightly reduced in the unbound structure (Fig. S1 D; compare taupe [unbound murine TLR4] to green, blue, mauve, and orange [bound]). Our model (Fig. 1 A, left panel) predicts a rearrangement similar to that in human TLR4 but is somewhat less extended than the human equivalent because of the shorter loop length in the murine protein, with D323 moving 3 Å closer to the 298 backbone (Fig. S1 E). As in human TLR4, our model does not predict any structural changes to the regions of murine TLR4 responsible for dimerization or at the MD-2–LPS interface.
Figure S1.
Structural analysis of the D298 region in murine TLR4 D298G/N397I model. (A) Superimposed models of human T399 (blue) and murine N397 SNPs (pink). (B) Superimposed models of WT human D299 (blue) and murine D298 (pink) to show the relative distance from D325 and D323, respectively. (C) Superimposed models of human WT D299 and human TLR4 D299G SNP and their relative proximity to D325. (D) Superimposition of five models showing differences in the orientation of murine TLR4 D298. (E) Superimposed models of murine WT (green) vs. murine TLR4 D298G (pink).
Based on this structural model for murine TLR4, TLR4-SNP mice homozygously expressing TLR4 with cosegregating D298G and N397I mutations were engineered onto a C57BL/6J background (see Materials and methods; Fig. 1 B) and compared in vivo and in vitro for their responses to LPS and infectious agents previously associated with TLR4 signaling. The following results provide the first direct evidence in a mouse model for the significant impact of these two TLR4 SNPs in response to LPS and in three models of infection.
TLR4-SNP mice exhibit hyporesponsiveness to LPS in vivo
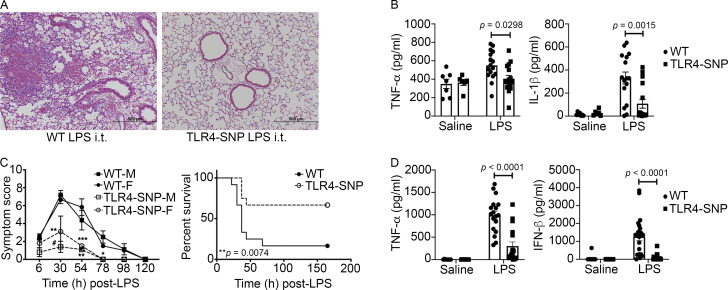
Arbour et al. (2000) originally reported an association of the inheritance of the human TLR4 SNPs D299G and T399I with hyporesponsiveness to inhaled LPS. To assess the responses of WT and TLR4-SNP (D298G and N397I) mice to LPS in the lung, we administered saline or highly purified, protein-free Escherichia coli LPS (5 µg/mouse) intratracheally (i.t.) and assessed the lung inflammatory response 18 h later (Feng et al., 2013). Based on the combined pathology scores from three separate experiments, lung sections of saline-treated WT and TLR4-SNP mice revealed no significant inflammation (data not shown; pathology scores of 0 ± 0 and 0.17 ± 0.11, respectively; n = 6 mice/strain). As expected, lung sections from LPS-treated WT mice show significantly greater inflammation, composed primarily of neutrophils and lymphocytes surrounding the airways and in the alveolar spaces (Fig. 2 A, left; and Fig. S2 A). In contrast, lungs of LPS-treated TLR4-SNP mice (Fig. 2 A, right) showed minimal inflammation (WT vs. TLR4-SNP pathology scores: 1.41 ± 0.35 vs. 0.34 ± 0.15 [n = 16 mice/strain]; P = 0.0123). Consistent with the differential inflammatory response of WT and TLR4-SNP mice to i.t. LPS, levels of the inflammatory cytokines TNF-α and IL-1β (Fig. 2 B), as well as Tnf, Il1b, and Cxcl10 mRNA (Fig. S2 B), were significantly lower in lung homogenates of LPS-treated TLR4-SNP mice compared with WT controls at 18 h after treatment.
Figure 2.
TLR4-SNP mice exhibit LPS hyporesponsiveness in vivo. (A) TLR4-SNP mice exhibit reduced lung pathology in response to i.t. LPS (5 µg/mouse). Representative H&E-stained lung sections from WT vs. TLR4-SNP mice 18 h after LPS treatment (n = 16 LPS-treated/strain in three separate experiments; 100× magnification, scale bars = 500 µm). In this and all subsequent figures, age- and sex-matched WT (C57BL/6J) mice were purchased from the Jackson Laboratory; TLR4-SNP mice were engineered onto a C57BL/6J background, and TLR4−/− mice were backcrossed onto a C57BL/6J background for >12 generations (see Materials and methods). (B) TNF-α and IL-1β protein levels in lung homogenates of mice treated as in A. Each point represents an individual mouse; each column represents the mean ± SEM. Data were analyzed by one-way ANOVA with Tukey’s post hoc test. For LPS-treated (n = 16/strain, combined data from three independent experiments) WT vs. TLR4-SNP mice, TNF-α, P = 0.0298; IL-1β, P = 0.0015. (C) TLR4-SNP mice exhibit reduced symptoms in response to lethal i.p. LPS. Left: Age-matched male (M) and female (F) WT and TLR4-SNP mice (n = 5 per sex/strain) were injected with LPS (600 µg/mouse i.p.) and cumulative symptom scores recorded at the indicated time points (mean ± SEM). Data are representative of three separate experiments. Differences between males and females of the same strain were not significant. Comparison of WT vs. TLR4-SNP mice revealed the following significant differences by sex as analyzed pairwise by nonparametric Mann–Whitney U test: at 30 h, male, #, P = 0.0079; female, not significant (N.S.); at 54 h, male, **, P = 0.0159; female, ***, P = 0.0286; at 78 h, male, *, P = 0.0079; female, not applicable (single survivor in the WT group). Right: WT and TLR4-SNP mice were injected i.p. with LPS (30 mg/kg) and survival monitored. This experiment is representative of three independent experiments; n = 10 mice (5 males, 5 females)/strain; P = 0.0074 (log-rank Mantel-Cox test). (D) Serum TNF-α and IFN-β protein 2 h after nonlethal LPS administration (25 µg/mouse i.p.). Each point represents an individual mouse: WT saline (n = 8); TLR4-SNP saline (n = 7), WT LPS (n = 18), and TLR4-SNP LPS (n = 17; derived from three separate experiments). Each column represents the mean ± SEM. Data were analyzed by one-way ANOVA with Tukey’s post hoc test. For LPS-treated WT vs. TLR4-SNP mice, TNF-α, P < 0.0001; IFN-β, P < 0.0001.
Figure S2.
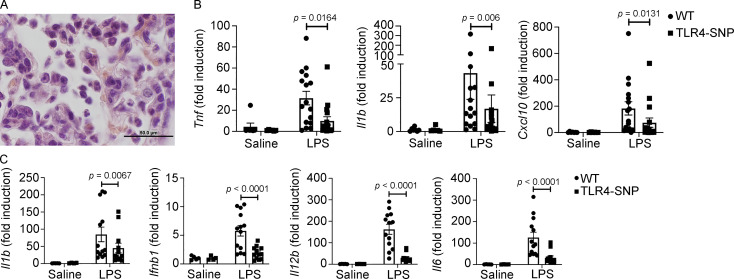
TLR4-SNP mice exhibit reduced lung pathology and cytokine mRNA in response to i.t. administration of LPS (5 µg/mouse). (A) Representative H&E-stained lung section from the WT mouse shown in Fig. 2 A showing neutrophil and lymphocyte infiltration at higher magnification (1,000×; scale bar = 50 µm). (B) Lung RNA was derived from the same mice described in Fig. 2 A, and cytokine mRNA was measured by qRT-PCR. Each point represents an individual mouse, and data are combined from three separate experiments. Each column represents the mean ± SEM. Data were analyzed by one-way ANOVA with Tukey’s post hoc test. LPS-stimulated WT vs. TLR4-SNP: Tnf, P = 0.0164; Il1b, P = 0.006; Cxcl10, P = 0.0131. (C) TLR4-SNP mice exhibit reduced liver cytokine mRNA in response to i.p. administration of LPS (25 µg/mouse). Liver RNA was derived from the same mice described in Fig. 2 D, and cytokine mRNA was measured by qRT-PCR. Each point represents the response of an individual mouse, and data are combined from three separate experiments. Each column represents the mean ± SEM. Data were analyzed by one-way ANOVA with Tukey’s post hoc test. LPS-stimulated WT vs. TLR4-SNP: Il1b, P = 0.0087; Ifnb1, P < 0.0001; Il12b, P < 0.0001; Il6, P < 0.0001.
WT and TLR4-SNP mice were also treated i.p. with doses of LPS that induce lethality in WT mice. In response to LPS (600 µg/mouse; Fig. 2 C, left), WT mice exhibited significantly greater symptoms than TLR4-SNP mice. No significant differences between male (M) and female (F) mice were observed within each strain. Further, when mice were challenged i.p. with LPS (30 mg/kg), to adjust for minor dose differences due to differing weights of each mouse, we observed that a much higher proportion of the WT mice (83%) succumbed to LPS than the TLR4-SNP mice (33%; Fig. 2 C, right). In contrast, TLR4−/− mice challenged with this same dose of LPS showed no symptoms of endotoxicity or lethality (data not shown). When WT and TLR4-SNP mice were challenged i.p. with a nonlethal dose of LPS (25 µg/mouse), levels of LPS-induced TNF-α protein (encoded by Tnf, an MyD88-dependent gene) and IFN-β protein (encoded by Ifnb1, a TRIF-dependent gene) in sera collected 2 h after LPS administration were significantly greater in WT mice (Fig. 2 D). These data were confirmed at the level of Il1b, Ifnb1, Il12b, and Il6 mRNA (encoding IL-1β, IFN-β, IL-12 p40, and IL-6, respectively) in the liver (Fig. S2 C). These results indicate that expression of the murine TLR4 D298G and N397I SNPs render mice LPS hyporesponsive.
TLR4-SNP macrophages exhibit hyporesponsiveness to LPS in vitro
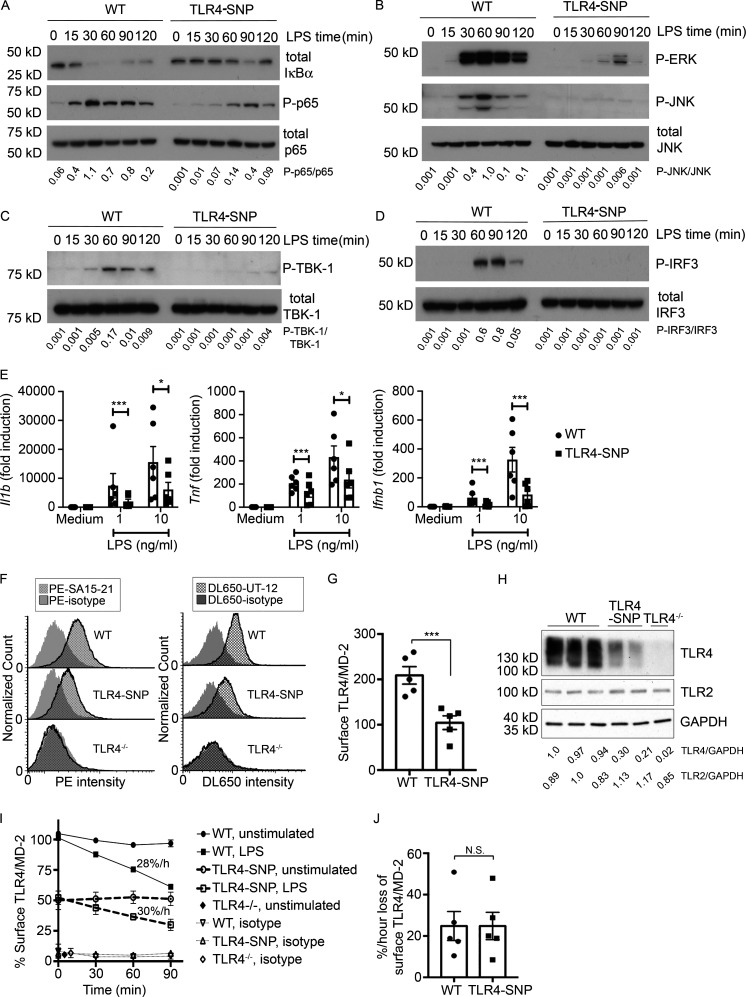
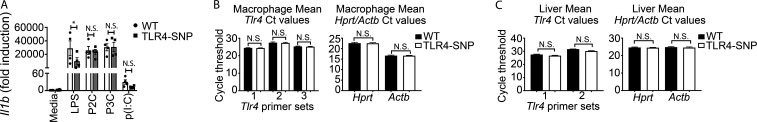
To compare the signaling capacities in response to LPS, thioglycollate-elicited WT and TLR4-SNP macrophages were treated with LPS and whole-cell lysates subjected to Western analysis for signaling intermediates over a 2-h period (Fig. 3, A–D; densitometric analysis is shown below each blot). Activation of NF-κB, as evidenced by rapid degradation of IκBα, and phosphorylation of the p65 subunit (Fig. 3 A) were observed in WT macrophages but were markedly less robust in TLR4-SNP macrophages. This was also observed for activation of both p-ERK 1,2 and p-JNK (Fig. 3 B). LPS-induced TRIF-dependent activation of TBK1 and IRF-3 was similarly reduced in TLR4-SNP macrophages (Fig. 3, C and D). Thus, TLR4-mediated activation of both MyD88 and TRIF signaling pathways were compromised by the presence of the D298G and N397I SNPs. Consistent with these findings, induction of MyD88-dependent gene expression (Il1b and Tnf mRNA) and TRIF-dependent expression of Ifnb1 mRNA was significantly lower in LPS-stimulated TLR4-SNP macrophages (Fig. 3 E), and these findings were confirmed in bone marrow–derived macrophages (data not shown). There was no significant difference in WT vs. TLR4-SNP macrophages stimulated with either TLR2 or TLR3 agonists (Fig. S3 A).
Figure 3.
TLR4-SNP macrophages are hyporesponsive to LPS in vitro. (A–D) Attenuated LPS-dependent signal transduction in TLR4-SNP mice. WT and TLR4-SNP thioglycollate-elicited macrophages were stimulated with LPS (10 ng/ml in A–C or 100 ng/ml in D) for the indicated times and whole-cell lysates probed by Western blot using antibodies directed against the indicated proteins. Densitometric analysis is shown below each lane. Images and normalized densitometric measurements are representative of n > 4 independent experiments with similar outcomes. (E) Cytokine gene expression of Il1b, Tnf, and Ifnb1 relative to expression of the housekeeping gene Hprt in macrophages treated with medium only or LPS (1 or 10 ng/ml). Each point represents the average responses of pooled macrophages from six independent experiments. Each column represents the mean ± SEM. Data were analyzed by two-way ANOVA with Tukey’s post hoc test; P values were adjusted for multiple comparison. Il1b: 1 ng/ml LPS, ***, P < 0.0001; 10 ng/ml LPS, *, P = 0.0204. Tnf: 1 ng/ml LPS, ***, P < 0.0001; 10 ng/ml LPS, *, P = 0.0104. Ifnb1: 1 ng/ml LPS, ***, P < 0.0001; 10 ng/ml LPS, ***, P < 0.0001. (F) Flow cytometric analysis of surface TLR4 on WT, TLR4-SNP, and TLR4−/− macrophages. The histograms on the left show a representative experiment (n = 5) illustrating the fluorescent intensity of WT or TLR4-SNP macrophages stained with anti-mouse mAb Sa15-21, and the histograms on the right show a representative experiment (n = 3) for WT or TLR4-SNP macrophages stained with mAb UT-12. TLR4−/− macrophages were included as a negative control. (G) Expression of surface TLR4 (median fluorescence intensity [MFI]) in WT vs. TLR4-SNP macrophages (mean ± SEM) in five separate experiments using mAb Sa15-21. Data were analyzed by paired Student’s t test. ***, P = 0.0008. (H) Western analysis of WT, TLR4-SNP, and TLR4−/− macrophage whole-cell lysates for TLR4 and TLR2 expression. Each lane represents a separate macrophage pool. Representative of two independent experiments. Densitometric ratios are shown below the blot. (I) LPS-induced internalization of TLR4 in WT vs. TLR4-SNP macrophages. This graph shows the results of a single representative experiment. Rate of internalization was calculated by linear regression (WT, slope −28%/h, R2 = 0.9938; TLR4-SNP, slope = −30%/h relative to its own expression level in absence of LPS, R2 = 0.9820); WT vs. TLR4-SNP, not significant (N.S.). (J) Rates of TLR4 internalization were calculated from five independent experiments (mean ± SEM) performed as shown in I. Data were analyzed by paired Student’s t test. WT vs. TLR4-SNP, N.S.
Figure S3.
TLR4-SNP macrophages exhibit normal responses to TLR2 and TLR3 agonists, and steady-state Tlr4 mRNA is expressed equivalently in WT and TLR4-SNP macrophages and liver samples. (A) Responses of WT (open bars) vs. TLR4-SNP (gray bars) macrophages to TLR2 (P2C and P3C, 1 µg/ml; 4 h) and TLR3 (p(I:C), 50 µg/ml; 6 h) agonists are not significantly different, in contrast to TLR4 (LPS, 10 ng/ml; 2 h). Results represent the mean ± SEM of four separate experiments and were analyzed by two-way ANOVA with Sidak’s multiple comparison post-test. *, P = 0.046. (B and C) Steady-state levels of Tlr4 mRNA are not significantly different in WT vs. TLR4-SNP macrophage or liver RNA. The data represent Tlr4 mRNA levels from untreated pooled macrophage cultures from the six individual experiments analyzed in Fig. 3 B (B) and livers of 9 WT vs. 10 TLR4-SNP mice (C). qRT-PCR was performed using distinct primer sets for Tlr4 mRNA and two housekeeping genes, Hprt and Actb mRNA, to ensure equivalent RNA loading. Data were analyzed by one-way ANOVA with Sidak multiple comparison post-test. N.S., not significant.
TLR4-SNP macrophages exhibit reduced TLR4 surface expression in vitro
Several studies have suggested that surface expression of human TLR4 expressing the D299G and T399I SNPs was lower than in WT epithelial cells (Arbour et al., 2000; Tulic et al., 2007), although this finding has been controversial (Rallabhandi et al., 2006; Figueroa et al., 2012). Fig. 3 F (left) is a representative flow cytometry histogram showing that cell surface staining of murine TLR4 with mAb Sa15-21, which binds to the N-terminal half of murine TLR4 (Akashi-Takamura et al., 2006) independently of MD-2 interaction (Visintin et al., 2006), was greater in primary WT macrophages than in TLR4-SNP macrophages. Staining of TLR4−/− macrophages was superimposable on that of the isotype control antibody. Based on five separate experiments (Fig. 3 G), the mean fluorescence intensity of TLR4 stained with Sa15-21 on WT macrophages was more than twice that of the TLR4-SNP macrophages. These findings were confirmed using a second mAb, UT-12 (Ohta et al., 2006; Fig. 3 F, right), that binds to transfectants expressing the TLR4/MD-2 complex, but not to cells expressing TLR4 or MD-2 alone (Bahrun et al., 2007). Lastly, Western analysis of whole-cell lysates from pools of WT, TLR4-SNP, and TLR4−/− macrophages was performed using a rabbit anti-recombinant mouse TLR4 mAb. Consistent with the finding by FACS analyses that TLR4-SNP macrophages expressed decreased surface TLR4 expression, levels of total TLR4 protein were also reduced in TLR4-SNP macrophage lysates compared with WT and were undetectable in the TLR4−/− macrophage lysates. In contrast, levels of TLR2 in these same preparations were equivalent (Fig. 3 H; densitometric analysis is provided below the blot). Despite the apparent difference in TLR4 expression on WT vs. TLR4-SNP macrophages, steady-state levels of Tlr4 mRNA were not significantly different (Fig. S3, B and C) when measured in either macrophage or liver mRNA using distinct murine Tlr4 primer sets to ensure complete coverage of the expressed mRNA.
Given the profound effect of the TLR4 SNPs on activation of the TRIF-dependent pathway (Fig. 3, C–E) and the fact that internalization of TLR4/MD-2/CD14 is required for TRIF activation (Kagan et al., 2008), macrophages were treated with LPS and loss of surface TLR4 quantified over 90 min (Kagan et al., 2008; Richard et al., 2019). While basal levels of macrophage WT TLR4 were approximately twice that of the TLR4-SNP macrophages (Fig. 3, F–H), WT and TLR4-SNP macrophages responded to LPS with comparable rates of internalization of the TLR4/MD-2/CD14 receptor complex, findings confirmed in five separate experiments (Fig. 3, I and J).
TLR4-SNP macrophages exhibit reduced LPS sensitivity for cell metabolic regulation
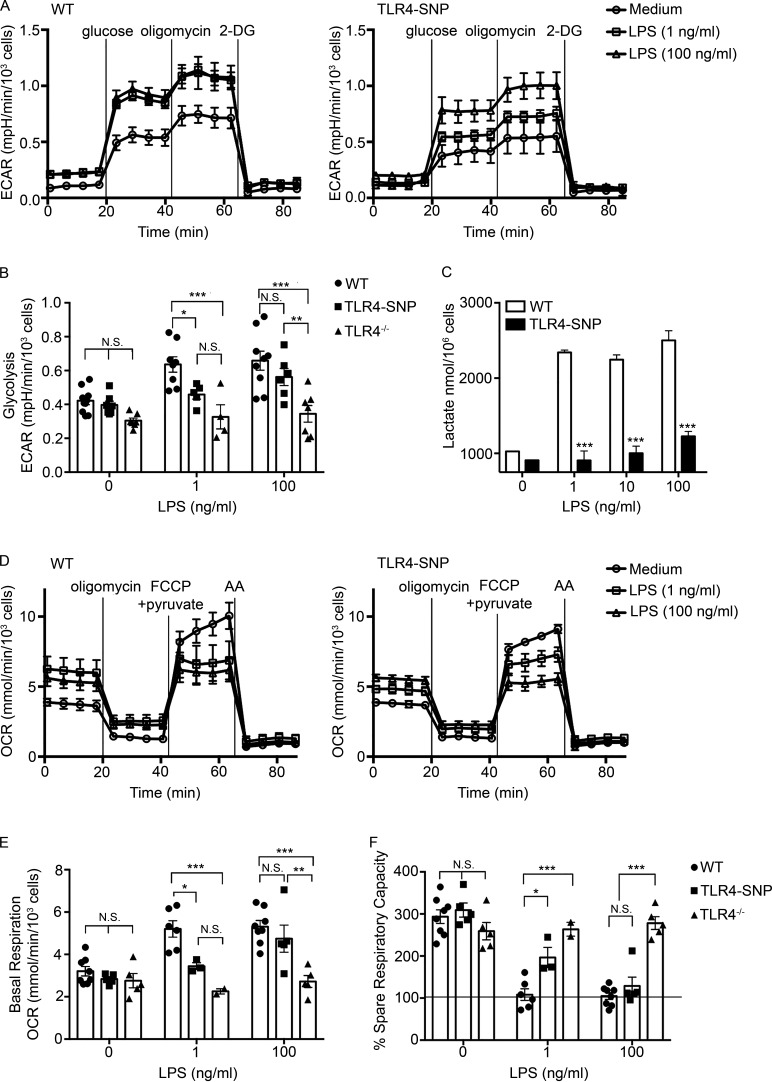
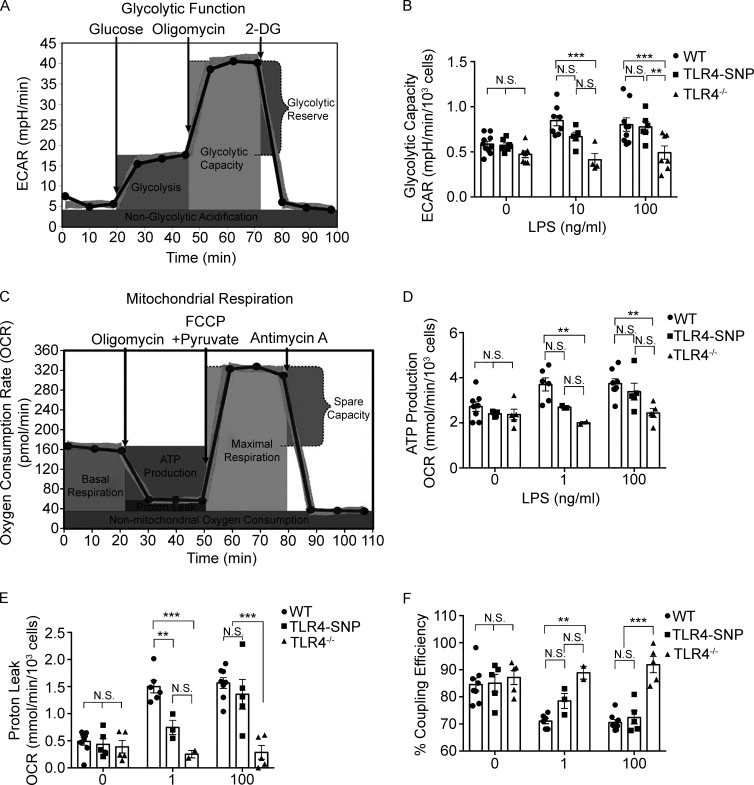
Previous studies have shown that LPS stimulation of macrophages activates glycolysis in order to support M1 (classically activated) macrophage activation (Mills et al., 2017; Hughes and O’Neill, 2018). Standard glycolytic stress tests were performed to compare WT and TLR4-SNP macrophage responses (Fig. 4 A), and glycolytic parameters (e.g., rates of glycolysis, glycolytic capacity, glycolytic reserve, and nonglycolytic acidification) were calculated (see diagram in Fig. S4 A). TLR4−/− macrophages were included as negative controls and did not respond to LPS for any of the measurements. Within 24 h of LPS treatment, WT macrophages consistently exhibited a modest increase in glycolysis (Fig. 4, A and B) and glycolytic capacity (Fig. S4 B) in response to both low (1 ng/ml) and high (100 ng/ml) LPS concentrations, whereas the glycolytic reserve and nonglycolytic acidification were essentially unchanged (data not shown). Compared with WT macrophages, TLR4-SNP macrophages exhibited reduced glycolysis after treatment with 1 ng/ml LPS, which was overcome at the higher LPS dose (Fig. 4, A and B). Reduced glycolytic capacity after low-dose LPS stimulation of the TLR4-SNP macrophages was also observed, although this trend did not reach statistical significance (Fig. S4 B). To confirm these data, lactate, a product of glycolysis, was measured in WT and TLR4-SNP macrophage cultures (Fig. 4 C). LPS-stimulated TLR4-SNP macrophages produced significantly less lactate than WT macrophages at 1, 10, and 100 ng/ml LPS.
Figure 4.
TLR4-SNP macrophages exhibit reduced metabolic activation in response to LPS. (A) Glycolytic stress test performed on thioglycollate-elicited macrophages derived from WT and TLR4-SNP mice. Results represent the mean ± SEM of a representative experiment shown as a Seahorse wave plot. (B) Glycolysis measurements ([ECAR with 10 mM glucose] − [ECAR after 1-h glucose starvation]) from glycolytic stress tests were performed as in A, with some experiments including TLR4−/− macrophages as an additional negative control. Each data point shows the average response of two to five technical replicates of a separate pool of thioglycollate-elicited macrophages from a total of nine independent experiments (at 1 ng/ml LPS: WT, n = 8; TLR4-SNP, n = 5; TLR4−/−, n = 4; at 100 ng/ml LPS: WT, n = 9; TLR4-SNP, n = 6; TLR4−/−, n = 7; columns represent mean ± SEM). Data were analyzed by two-way ANOVA (α = 0.10), with Sidak’s multiple comparison post-tests to compare WT vs. TLR4-SNP responses: untreated, not significant (N.S.); 1 ng/ml LPS, P = 0.067; 100 ng/ml LPS, N.S.; WT vs. TLR4−/− responses: untreated, N.S.; 1 ng/ml LPS, P = 0.0003; 100 ng/ml LPS, P < 0.0001; TLR4-SNP vs. TLR4−/− responses: 100 ng/ml LPS, P = 0.0086 (adjusted for multiple comparisons). (C) Macrophages isolated from WT and TLR4-SNP mice were stimulated for 24 h with the indicated dose of LPS (0–100 ng/ml). Culture supernatants were assayed for the concentration of L-lactate using an enzymatic detection kit. The data represent the mean ± SD for a representative experiment of two separate experiments. Data were analyzed by one-way ANOVA, with Tukey’s post hoc test to compare WT vs. TLR4-SNP responses; ***, P < 0.0001 for each pair. (D) Mitochondrial stress test performed on macrophages derived from WT and TLR4-SNP mice. Results represent the mean ± SEM of a representative experiment shown as a Seahorse wave plot. (E) Basal respiration ([OCR before injections] − [OCR after inhibition of electron transport with AA]). Each data point shows the average response of two to five technical replicates of a separate pool of thioglycollate-elicited macrophages (at 1 ng/ml LPS: WT n = 6, TLR4-SNP n = 3, TLR4−/− n = 2; at 100 ng/ml LPS: WT n = 8, TLR4-SNP n = 4, TLR4−/− n = 5; from a total of seven independent experiments; columns represent mean ± SEM). Data were analyzed by two-way ANOVA (α = 0.10), with Sidak’s multiple comparison post-tests to compare WT vs. TLR4-SNP responses: untreated, N.S.; 1 ng/ml LPS, *, P = 0.032; 100 ng/ml LPS, N.S.; WT vs. TLR4−/− responses: untreated, N.S.; 1 ng/ml LPS, ***, P = 0.0006; 100 ng/ml LPS, ***, P < 0.0001; TLR4-SNP vs. TLR4−/− responses at 100 ng/ml LPS, **, P = 0.0026 (adjusted for multiple comparisons). (F) Percent SRC (100 × [(maximum OCR after uncoupling oxidative phosphorylation from the mitochondria with carbonyl cyanide 4-[trifluoromethoxy]phenylhydrazone and providing excess pyruvate) − (OCR before injections)] / [basal respiration]) from the same experiments as in E. Data were analyzed by two-way ANOVA (α = 0.10), with Sidak’s multiple comparison post-tests to compare WT vs. TLR4-SNP responses: untreated, N.S.; 1 ng/ml LPS, *, P = 0.021; 100 ng/ml LPS, N.S.; WT vs. TLR4−/− responses: untreated, N.S.; 1 ng/ml LPS, ***, P < 0.0001; 100 ng/ml LPS, ***, P < 0.0001; TLR4-SNP vs. TLR4−/− responses: 100 ng/ml LPS, ***, P < 0.0001 (adjusted for multiple comparisons). FCCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone.
Figure S4.
Metabolic analysis of WT and TLR4-SNP macrophages. (A) Diagram of glycolytic stress test measurements (reproduced with permission; courtesy of Agilent Technologies). (B) Glycolytic capacity ([ECAR after inhibition of mitochondrial ATP synthase with oligomycin forcing all cellular energy production to the glycolytic pathway] − [ECAR after inhibition of glycolysis with excess 2-deoxyglucose]) from the same experiments as in Fig. 4 B (combined data from nine separate experiments). Data were analyzed by two-way ANOVA (α = 0.10), with Sidak’s multiple comparison post-tests to compare WT and TLR4-SNP responses as follows: untreated, not significant (N.S.); 1 ng/ml LPS, N.S.; 100 ng/ml LPS, N.S.; WT and TLR4−/− responses: untreated, N.S.; 1 ng/ml LPS, ***, P = 0.0001; 100 ng/ml LPS, ***, P = 0.0011; TLR4-SNP and TLR4−/− responses: 100 ng/ml LPS, **, P = 0.010 (adjusted for multiple comparisons). (C) Diagram of mitochondrial stress test measurements (reproduced with permission; courtesy of Agilent Technologies). (D) ATP production ([OCR before any injection] − [OCR after injecting ATP synthase inhibitor oligomycin]) from the same experiments as in Fig. 4 (E and F; combined data from seven separate experiments). Data were analyzed by two-way ANOVA (α = 0.10), with Sidak’s multiple comparison post-tests to compare WT and TLR4-SNP responses as follows: untreated, N.S.; 1 ng/ml LPS, N.S.; 100 ng/ml LPS, N.S.; WT and TLR4−/− responses: untreated, N.S.; 1 ng/ml LPS, **, P = 0.0067; 100 ng/ml LPS, **, P = 0.0023; TLR4-SNP and TLR4−/− responses at 100 ng/ml LPS, N.S. (adjusted for multiple comparisons). (E) Proton leak ([OCR after ATP synthase inhibition by oligomycin] − [OCR after electron transport inhibition by AA]) from the same experiments as in Fig. 4 (E and F). Data were analyzed by two-way ANOVA (α = 0.10), with Sidak’s multiple comparison post-tests to compare WT vs. TLR4-SNP responses as follows: untreated, N.S.; 1 ng/ml LPS, **, P = 0.014; 100 ng/ml LPS, N.S.; WT vs. TLR4−/− responses: untreated, N.S.; 1 ng/ml LPS, ***, P = 0.0002; 100 ng/ml LPS, ***, P < 0.0001; TLR4-SNP vs. TLR4−/− responses at 100 ng/ml LPS, ***, P < 0.0001. (adjusted for multiple comparisons). (F) Mitochondrial coupling efficiency (100 × [ATP production rate]/[basal respiration rate]) from the same experiments as in Fig. 4 (E and F) (combined data from seven separate experiments). Data were analyzed by two-way ANOVA (α = 0.10), with Sidak’s multiple comparison post-tests to compare WT vs. TLR4-SNP responses: untreated, N.S.; 1 ng/ml LPS, N.S.; 100 ng/ml LPS, N.S.; WT vs. TLR4−/− responses: untreated, N.S.; 1 ng/ml LPS, **, P = 0.0025; 100 ng/ml LPS,***, P < 0.0001; TLR4-SNP vs. TLR4−/− responses at 100 ng/ml LPS, ***, P < 0.0001 (adjusted for multiple comparisons).
Mitochondrial stress tests were also performed with modifications described previously (Bordt et al. 2017; Fig. 4 D), and measurements of oxidative phosphorylation (e.g., rates of basal respiration, ATP production, proton leak, mitochondrial coupling efficiency, maximal respiration, spare respiratory capacity [SRC], and nonmitochondrial oxygen consumption) were calculated (see diagram in Fig. S4 C). Treatment of WT macrophages with LPS for 24 h resulted in modest but consistent increases in the basal respiration rate (Fig. 4, D and E) and, to a lesser extent, ATP production rate (Fig. S4 D). Basal respiration and ATP synthesis rates of TLR4-SNP macrophages stimulated with 1 ng/ml LPS were minimally increased over the basal respiration rates of all media-treated control cells or LPS-treated TLR4−/− macrophages but were increased to the level of WT macrophages upon treatment with 100 ng/ml LPS (Fig. 4, D and E; and Fig. S4 D). WT macrophages also exhibited an approximately threefold increase in proton leak in response to low- or high-dose LPS (Fig. S4 E) that resulted in significantly decreased mitochondrial coupling efficiency (Fig. S4 F). Again, TLR4-SNP macrophages exhibited an intermediate response to low-dose LPS that was not statistically different from LPS-treated TLR4−/− macrophages, but this deficit was overcome at the higher LPS dose (Fig. S4, E and F). SRC (approximately threefold higher than basal respiration in the absence of LPS) was decreased in WT macrophages at both the doses of LPS and completely eliminated in approximately half of the macrophage pools tested (Fig. 4 F; the horizontal line on the graph at 100% represents the basal respiration rate), indicating that LPS-stimulated WT macrophages are using oxidative phosphorylation at maximal levels. TLR4-SNP macrophages still exhibited the capacity to double their respiratory rate after low-dose LPS treatment and only exhibited significantly reduced SRC at the high LPS dose. Maximal respiration and nonmitochondrial respiration rates were not affected by LPS treatment (data not shown). Together, these data reveal that while TLR4−/− macrophages were completely refractory to LPS stimulation, TLR4-SNP macrophages exhibited partial responses to low-dose LPS treatment and near-WT responses to high-dose LPS treatment for all metabolic measurements that were LPS responsive.
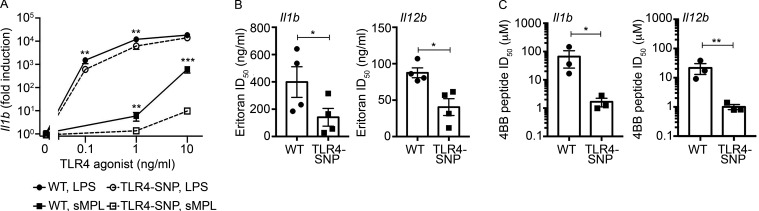
TLR4-SNP macrophages exhibit reduced responsiveness to weak TLR4 agonists and increased sensitivity to TLR4 antagonists
When stimulated by weak TLR4 agonists, the differential responses of human TLR4 D299G/T399I-expressing stable transfectants (Yamakawa et al., 2013) and primary human adherent white blood cells (Rallabhandi et al., 2008) were greater than observed when LPS was used as the stimulus. This was attributed to a SNP-induced conformational change that impaired TLR4 dimerization in transfectants (Yamakawa et al., 2013). Stimulation of murine WT and TLR4-SNP macrophages with the weak TLR4 agonist synthetic monophosphoryl lipid A (sMPL) also resulted in differences in gene induction between the two strains that were more disparate than seen in response to LPS stimulation (Fig. 5 A; plotted on a log10 scale to facilitate a direct comparison of the relative differences in LPS- vs. sMPL-stimulated gene expression). Since TLR4 expression levels were significantly lower in the TLR4-SNP macrophages (Fig. 3, F and G), we hypothesized that Eritoran, a synthetic TLR4 antagonist that competitively inhibits binding of LPS to MD-2, and thereby inhibits TLR4 dimerization (Kim et al., 2007), would more readily inhibit LPS-induced gene expression in TLR4-SNP macrophages. The dose of Eritoran required to inhibit LPS-induced Il1b and Il12b mRNA expression by ∼50% (ID50) was significantly lower in TLR4-SNP macrophages (Fig. 5 B). Finally, Toshchakov et al. (2011) demonstrated that 4BB, a cell-permeating inhibitory peptide based on the sequence of the BB loop of TLR4, disrupted TLR4 dimerization in response to LPS. Like Eritoran, the 4BB peptide more readily inhibited LPS-induced Il1b and Il12b gene expression in TLR4-SNP macrophages than in WT macrophages (Fig. 5 C).
Figure 5.
TLR4-SNP macrophages exhibit decreased responsiveness to weak TLR4 agonists and antagonists. (A) WT and TLR4-SNP macrophages were stimulated with LPS and sMPL at the indicated concentrations and Il1b mRNA quantified by qRT-PCR. The results represent the mean ± SEM of four separate experiments. Data were analyzed by two-way ANOVA and Tukey’s post hoc test. WT vs. TLR4-SNP: 0.1 ng/ml LPS, **, P = 0.005; 1.0 ng/ml LPS, **, P = 0.0079; 10 ng/ml LPS, P = not significant (N.S.); 1.0 ng/ml sMPL, **, P = 0.0032; 10 ng/ml sMPL, ***, P < 0.0001. (B) WT and TLR4-SNP macrophages were pretreated for 20 min with Eritoran (10-fold dilutions ranging from 100 pg/ml to 1 µg/ml) and then stimulated with LPS (10 ng/ml) for 2 h. The ID50 was calculated as described in Materials and methods. Results represent the mean ± SEM of four separate experiments. LogID50 data were analyzed by a paired Student’s t test; Il1b: *, P = 0.013; Il12b: *, P = 0.022. (C) WT and TLR4-SNP macrophages were pretreated for 30 min with 4BB peptide (dilutions ranging from 0.8 to 26 µM in experiment 1 or from 0.08 to ∼20 µM in experiments 2 and 3) and then stimulated with LPS (10 ng/ml) for 2 h. The ID50 for each genotype was calculated as described in Materials and methods. Results represent the mean ± SEM of three separate experiments. LogID50 data were analyzed by a paired Student’s t test; IL1b: *, P = 0.014; Il12b: **, P = 0.008.
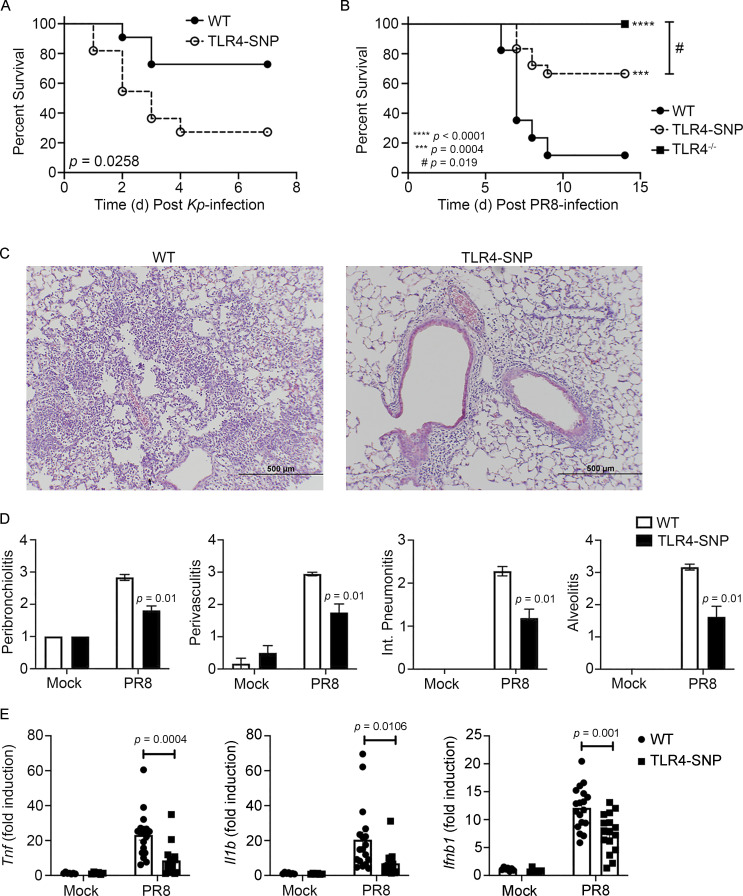
TLR4-SNP D298G/N397I mice exhibit increased sensitivity to Gram-negative infection
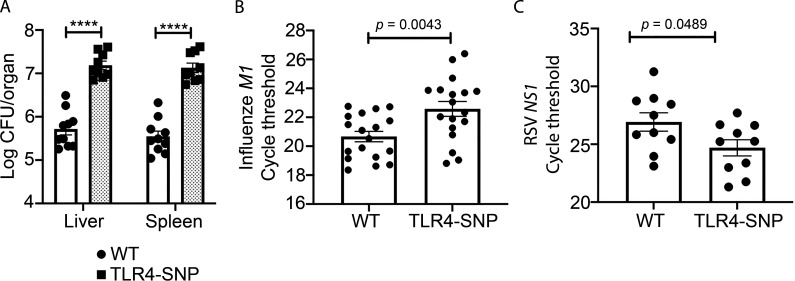
C3H/HeJ mice, shown to carry a point mutation in Tlr4 that renders them LPS unresponsive (Poltorak et al., 1998), as well as TLR4−/− mice, exhibited increased susceptibility to Gram-negative bacteria, including Salmonella and E. coli (O’Brien et al., 1980; Cross et al., 1989; Roger et al., 2009). Lorenz et al. (2002b) reported that the allele encoding TLR4 D299G was exclusively found in patients with septic shock, and Lorenz et al. (2002b) and Agnese et al. (2002) reported that septic shock patients expressing both SNPs had a higher prevalence of Gram-negative infection. More recently, Chatzi et al. (2018) reported that the human TLR4 SNPs D299G/T399I were associated with increased susceptibility to hospital-acquired pathogens such as Pseudomonas and Klebsiella, leading to increased hospital stay. To test the sensitivity of TLR4-SNP mice to Gram-negative infection, WT and TLR4-SNP mice were infected i.p. with Klebsiella pneumoniae (Kp) O1K2 strain B5055 (∼1,200 CFU/mouse) and monitored daily for survival. TLR4-SNP mice were significantly more susceptible to Kp infection than WT mice (Fig. 6 A), supporting the hypothesis that inheritance of the murine TLR4 D298G and N397I SNPs similarly increased sensitivity to Gram-negative bacterial infection. Increased susceptibility to Kp was accompanied by a significant increase in bacterial counts in the livers and spleens of TLR4-SNP mice 24 h after infection (Fig. S5 A).
Figure 6.
Altered susceptibility of TLR4-SNP mice to Kp and PR8 infections. (A) WT and TLR4-SNP mice were infected with Kp B5055 (∼1,200 CFU i.p.) and monitored daily for survival for 7 d. Data represent the combined results of two separate experiments (n = 11 mice/strain). Data analyzed by log-rank Mantel-Cox test. P = 0.0258. (B) WT, TLR4-SNP, and TLR4−/− mice were infected with influenza PR8 (∼7,500 TCID50 i.n.) and monitored daily for survival for 14 d after PR8 infection. Data represent the combined results of three separate experiments (WT, n = 15; TLR4-SNP, n = 18; TLR4−/−, n = 14). Data analyzed by log-rank Mantel-Cox test. TLR4−/− vs. TLR4-SNP: #, P = 0.019; WT vs. TLR4-SNP: ***, P = 0.0004; WT vs. TLR4−/−: ****, P < 0.0001. (C and D) WT and TLR4-SNP mice were infected as in B. On day 6 after infection, mice were euthanized and the lungs extracted, fixed, and sectioned for histology after H&E staining. (C) Representative sections from PR8-infected WT and TLR4-SNP mice are shown (magnification 100×; scale bars = 500 µm). (D) Lung histopathology scoring. Data from mock- or PR8-infected mice represent the mean ± SEM of two separate experiments (6 mice/strain for mock treatment; 18 mice/strain for PR8 infection). Data analyzed by two-way ANOVA with Tukey’s post hoc test. PR8-infected WT vs. TLR4-SNP: P = 0.01 for each histological parameter scored. (E) Gene expression analysis by qRT-PCR of lung RNA from WT and TLR4-SNP mice infected as in B. Each point represents the responses of individual mice from two separate experiments (identical to the mice in D). Columns represent the mean ± SEM. Data were analyzed by two-way ANOVA with Tukey’s post hoc test. PR8-infected WT vs. TLR4-SNP: Tnf, P = 0.0004; Il1b, P = 0.0106; Ifnb1, P = 0.001.
Figure S5.
Pathogen burden in WT- and TLR4-SNP–infected mice. (A) WT and TLR4-SNP mice were infected with Kp B5055 (∼1,500 CFU i.p.). Livers and spleens were harvested after 18 h for bacterial burden by CFUs for each organ. Data represent the combined results of two separate experiments (n = 10 WT; n = 9 TLR4-SNP). Data were analyzed by two-way ANOVA with Tukey’s post hoc test (****, P < 0.0001). (B) The identical samples from WT and TLR4-SNP mice shown in Fig. 6 E were analyzed by qRT-PCR for expression of the influenza M1 gene from two separate experiments. Data were analyzed by two-way ANOVA with Tukey’s post hoc test (P = 0.0043). (C) The WT and TLR4-SNP lung RNA samples shown in Fig. 7 A were used to measure RSV NS1 gene expression by qRT-PCR. Data were analyzed by two-way ANOVA with Tukey’s post hoc test (P = 0.0489).
TLR4-SNP 298G/N397I mice exhibit increased resistance to influenza infection
We previously reported that TLR4−/− mice were highly refractory to infection by mouse-adapted influenza A/PR/8/34 (PR8) and that therapeutic administration of Eritoran, or other TLR4-specific antagonists, protected WT mice from lethal infection and pathology (Shirey et al., 2013, 2016; 2020; Piao et al., 2015; Perrin-Cocon et al., 2017). Since there are no reports that have specifically examined the association of human TLR4 299/399 SNPs and influenza infection, we tested the hypothesis that the murine D298G/N397I TLR4 SNPs would confer resistance to PR8 infection, similar to TLR4−/− mice or WT mice treated therapeutically with Eritoran (Shirey et al., 2013). WT, TLR4−/−, and TLR4-SNP mice were infected i.n. with a 90% lethal dose (LD90) of PR8 previously determined in WT C57BL/6J mice (Shirey et al., 2013). Fig. 6 B shows that WT mice succumb to infection, while TLR4−/− mice are refractory to PR8 infection (P = 0.0001), as reported previously (Shirey et al., 2013). TLR4-SNP mice were significantly more resistant to PR8-induced lethality than WT mice (P = 0.0004) but less resistant than the TLR4−/− mice (P = 0.019).
Additional groups of WT and TLR4-SNP mice were infected with PR8 and euthanized on day 6 after infection, and the lungs were removed. Part of each lung was fixed and H&E stained for histopathology, and part was homogenized for analysis of gene expression by quantitative RT-PCR (qRT-PCR). Lung sections revealed extensive pathology and inflammation in PR8-infected WT mice (Fig. 6 C, left panel; pathology scores are shown in Fig. 6 D, open bars), whereas lung sections from PR8-infected TLR4-SNP mice exhibited significantly less inflammatory infiltrates and lung damage (Fig. 6 C, right panel; pathology scores in Fig. 6 D, closed bars). We previously reported that PR8-infected WT mice treated with the TLR4 antagonists also exhibited a significant reduction in inflammatory gene expression (Shirey et al., 2013, 2016, 2020; Piao et al., 2015; Perrin-Cocon et al., 2017). PR8-infected WT mice showed significantly greater Il1b, Tnf, and Ifnb1 mRNA expression than PR8-infected TLR4-SNP mice (Fig. 6 E). Finally, viral load, as measured by qRT-PCR analysis of lung RNA 6 d after infection, revealed significantly lower influenza M1 viral gene expression, as evidenced by increased Ct values, in TLR4-SNP mice (Fig. S5 B). These data support the conclusion that the TLR4-SNP mice are more resistant to influenza-induced disease.
TLR4-SNP 298G/N397I mice exhibit increased susceptibility to RSV infection
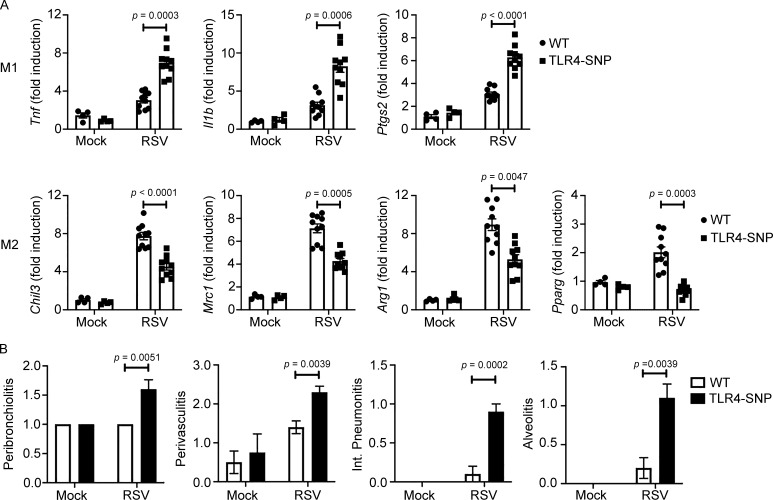
Previous studies associated the inheritance of TLR4 299/399 SNPs with RSV disease severity in infants and young children (Tal et al., 2004; Mandelberg et al., 2006; Awomoyi et al., 2007; Puthothu et al., 2006), although this too has been met with controversy (Douville et al., 2010; Kresfelder et al., 2011). In WT C57BL/6J mice, RSV-induced disease is associated with increased lung pathology, but not lethality, and resolution of RSV-induced lung pathology is dependent on IL-4Rα, IFN-β, and TLR4 for induction of alternatively activated (M2) macrophages (Shirey et al., 2010). In addition, RSV-infected TLR4−/− mice exhibited enhanced proinflammatory (M1) gene expression and reduced M2 gene expression in vivo and in RSV-infected macrophage cultures (Shirey et al., 2010). To evaluate the effect of inheritance of the homologous murine TLR4 298/397 SNPs on the host response to RSV infection, WT and TLR4-SNP mice were either mock or RSV infected i.n., and lungs were harvested for gene expression and pathology after 6 d. TLR4-SNP mice responded to RSV infection with significantly greater M1 cytokine gene expression (e.g., Tnf, Il1b, and Ptgs2) than WT mice (Fig. 7 A). Conversely, analysis of M2 gene expression (e.g., Chil3, Mrc1, and Arg1) revealed that each was expressed to a significantly reduced degree in TLR4-SNP mice (Fig. 7 A). The transcription factor PPARγ is necessary for M2 gene expression (Malur et al., 2009). Levels of Pparg mRNA were greatly reduced in RSV-infected TLR4-SNP lungs (Fig. 7 A), as observed for RSV-infected TLR4−/− lungs (Shirey et al., 2010). Consistent with these and previous findings using TLR4−/− mice, RSV-infected TLR4-SNP mice exhibited greater pathology than WT mice with significantly increased peribrochiolitis, perivasculitis, interstitial pneumonitis, and alveolitis (Fig. 7 B). Finally, qRT-PCR analysis of RSV NS1 viral gene expression in the lungs of RSV-infected mice revealed significantly increased viral load (i.e., decreased Ct values) in TLR4-SNP mice (Fig. S5 C). Together, these findings support the conclusion that TLR4-SNP mice are more susceptible than WT mice to RSV-induced lung damage secondary to enhanced M1 and reduced M2 gene expression.
Figure 7.
Altered susceptibility of TLR4-SNP mice to RSV infection. (A) WT and TLR4-SNP mice were infected with RSV A/Long (∼5 × 106 PFU i.n.). The lungs were harvested for M1 and M2 gene expression analysis by qRT-PCR on day 6 after infection. Each point represents the responses of individual mice from two separate experiments. Columns represent the mean ± SEM. Data were analyzed by two-way ANOVA with Tukey’s post hoc test. RSV-infected WT vs. TLR4-SNP: Tnf, P = 0.0003; Il1b, P = 0.0006; Ptgs2, P < 0.0001; Chil3, P < 0.0001; Mrc1, P = 0.0005; Arg1, P = 0.0047; Pparg, P = 0.0003. (B) WT and TLR4-SNP mice were infected with RSV A/Long as in A. The lungs were harvested for histopathology analysis on day 6 after infection. Each column represents the mean ± SEM of the combined results of two separate experiments. Data were analyzed by two-way ANOVA with Tukey’s post hoc test. RSV-infected WT vs. TLR4-SNP: peribronchiolitis, P = 0.0051; perivasculitis, P = 0.0039; interstitial pneumonitis, P = 0.0002; alveolitis, P = 0.0039.
Discussion
We engineered a knock-in mouse strain that homozygously expresses two nonsynonymous SNPs in the extracellular domain of TLR4 (D298G and N397I) that are positionally homologous to the human TLR4 SNPs D299G and T399I, expressed by ∼10% to 18% of Caucasians (Ferwerda et al., 2007). In humans, these often cosegregating TLR4 SNPs have been associated with decreased LPS responsiveness and increased or decreased susceptibility to a variety of infectious agents or immunological disorders (reviewed in Schröder and Schumann, 2005; Medvedev, 2013; Mukherjee et al., 2019; Balistreri et al., 2009). However, conclusions from studies of human cohorts have often been conflicting, although transfection studies generally support the conclusion that expression of mutant TLR4 proteins lead to reduced responsiveness to LPS as well as other TLR4 agonists, e.g., RSV F protein, chlamydia Hsp60, and MPL (Rallabhandi et al., 2006; Figueroa et al., 2012; Prohinar et al., 2010; Yamakawa et al., 2013); however, until now, there has been no experimental model with which to confirm or refute disease associations or to analyze underlying mechanisms by which these SNPs alter TLR4 functionality in primary cells.
After demonstrating the LPS hyporesponsiveness of the TLR4-SNP mice in vivo, the responses of primary TLR4-SNP macrophages to LPS were found to exhibit reduced MyD88- and TRIF-dependent signaling and cytokine gene/protein expression. In contrast to studies in human TLR4-SNP–expressing monocytes/macrophages (Hold et al., 2014), we observed no difference in basal expression of MyD88- or TRIF-dependent cytokine gene expression. Consistent with previous reports for human TLR4 SNPs (Arbour et al., 2000; Tulic et al., 2007), two different mAbs, Sa15-21 and UT12, were used to demonstrate that surface TLR4 expression in the TLR4-SNP macrophages was approximately half that of WT macrophages, observations confirmed by Western analysis of whole-cell lysates using a rabbit mAb made against recombinant murine TLR4 only. Differential receptor expression did not, however, affect the rate of internalization of the TLR4/MD-2/CD14 complex that initiates TRIF-dependent signaling (Kagan et al., 2008). Nagai et al. (2002) reported that MD-2 was required to chaperone the TLR4 to the cell surface, and Yamakawa et al. (2013) proposed that surface TLR4 expression is altered in human TLR4-SNP–expressing cells due to a conformational change that results in a failure of the mutant TLR4 to interact correctly with MD-2. In contrast, Visintin et al. (2006) used mAb Sa15-21 (the same mAb used in our studies) in FACS analysis to show that WT and MD-2−/− bone marrow–derived or thioglycollate-induced macrophages express comparable levels of TLR4 (i.e., MD-2 is not required for TLR4 expression). A possible conformational change in the structure of TLR4 caused by the proximity of the human D299G SNP to the TLR4 binding site for MD-2 has been proffered to underlie decreased responsiveness to LPS and, particularly, to weak TLR4 agonists (Yamakawa et al., 2013; Rallabhandi et al., 2008). Lipid A–stimulated Ba/F3 cells expressing the human TLR4 (D299G/T399I) SNPs exhibited decreased NF-κB activation compared with Ba/F3 cells expressing WT human TLR4; however, when stimulated with the weak TLR4 agonist, MPL (Henricson et al., 1992), NF-κB activation in both cell types was further reduced, with the response of the mutant cells more profoundly affected (Yamakawa et al., 2013). Indeed, the transcriptional responses of primary murine WT vs. TLR4-SNP macrophages to sMPL were much more divergent than the differential response induced by LPS.
Using purified proteins in solution, Yamakawa et al. (2013) also observed that compared with lipid A, native MPL elicited a much lower level of TLR4 dimerization in the presence of the WT TLR4 protein and that dimerization was further reduced when recombinant human TLR4(D299G/T399I)/MD-2 complexes were stimulated with lipid A or MPL. Consistent with this effect on purified protein–protein interactions, we observed that Eritoran, a lipid A analogue antagonist that blocks LPS-induced TLR4 signaling by competitively binding to MD-2 (Kim et al., 2007), and 4BB, a cell-permeating TLR4 BB loop peptide antagonist shown previously to block TLR4 dimerization (Toshchakov et al., 2011; Szmacinski et al., 2014), were significantly more effective at inhibiting TLR4-SNP than WT macrophage responses to LPS. These data provide compelling support for the notion that in addition to reduced cell surface expression, the TLR4-SNPs diminish TLR4 signaling by interfering with ligand-induced dimerization. These findings also support previous work in transfectants that express human WT or TLR4 SNP proteins comparably yet exhibit decreased signaling and recruitment of adaptor molecules to TLR4 upon LPS stimulation (Rallabhandi et al., 2006; Figueroa et al., 2012).
LPS-unresponsive C3H/HeJ and TLR4−/− mice are highly susceptible to infection with Gram-negative pathogens (O’Brien et al., 1980; Cross et al., 1989; Roger et al., 2009). The increased susceptibility of TLR4-SNP mice to Kp infection, accompanied by increased bacterial burden in the liver and spleen, further supports the hypothesis that a decreased ability to detect the LPS of Gram-negative pathogens permits the pathogen to evade detection, thereby facilitating systemic infection. Mechanistically, we observed that TLR4-SNP macrophages are less sensitive to LPS for regulating their metabolic profile. Efficient metabolic switching to glycolysis in response to M1 stimuli has previously been found to be a prerequisite for proinflammatory macrophage responses; glycolysis provides ATP for driving phagocytosis, proinflammatory cytokine production, and NADPH for the NADPH oxidase 2 enzyme to generate reactive oxygen species (Mills et al., 2017; Hughes and O’Neill, 2018). Thus, a reduced glycolytic response in TLR4-SNP macrophages possibly contributes to decreased antimicrobial capacity. In addition to increased glycolysis induced by LPS, WT macrophages exhibited increased oxidative phosphorylation following LPS stimulation, similar to the responses of resident peritoneal macrophages (Davies et al., 2017, 2019) and human monocytes (Lachmandas et al., 2016). The “macrophage metabolic switch” in which oxidative phosphorylation is suppressed while glycolysis is increased (Mills et al., 2017; Hughes and O’Neill, 2018) likely requires induction of nitric oxide (Bailey et al., 2019; Palmieri et al., 2020) and inhibition of the mechanistic target of rapamycin complex 1 (mTORC1), mediated by IFN-γ (Su et al., 2015). Activation cascades induced by LPS alone differ from that of LPS + IFN-γ in many regards (reviewed in Murray et al., 2014; Das et al., 2018). Stimulation with LPS alone led WT macrophages to increase mitochondrial respiration to maximum levels, as well as increase mitochondrial electron transport that is uncoupled from ATP synthesis (proton leak rate) and likely associated with production of reactive oxygen species for microbial killing (Chandel, 2015). For all observed LPS-induced metabolic changes, TLR4-SNP macrophages exhibited reduced sensitivity to LPS that may impact disease outcomes.
WT and TLR4-SNP mice were also challenged with influenza (PR8) or RSV, infections that have been shown to have a strong TLR4 involvement (Shirey et al., 2010, 2013). In response to influenza infection, the TLR4-SNP mice were significantly more resistant than WT mice (67% vs. ∼10% survival) yet significantly more sensitive to infection than the TLR4−/− mice. These findings were paralleled by significantly decreased viral burden, cytokine gene expression, and histopathology in the TLR4-SNP mice, similar to that seen in WT mice infected with PR8 and treated with the TLR4 antagonist Eritoran (Shirey et al., 2013, 2016). Eritoran blocks release of a host-derived “danger-associated molecular pattern” and TLR4 agonist, HMGB1, in influenza-infected mice, as well as HMGB1-induced TLR4 activation (Shirey et al., 2013, 2016). Conversely, therapeutic administration of a small molecule HMGB1 antagonist to PR8-infected mice was as protective as Eritoran (Shirey et al., 2016). It is interesting to note that a recent in silico molecular docking study revealed that the SARS-CoV-2 spike protein has the potential to bind to the extracellular domains of human TLR1, TLR4, and TLR6, with TLR4 being the strongest, and it was proposed that TLR4 antagonists might represent novel inhibitors of the virus (Choudhury and Mukherjee, 2020). It has also been proposed that HMGB1 may be a therapeutic target in severe pulmonary inflammation associated with COVID-19 (Andersson et al., 2020). This raises the intriguing possibility that individuals expressing the TLR4 299/399 SNPs may be more resistant to influenza, as well as SARS-CoV-2 infection, although to date, this has not been demonstrated.
In contrast to influenza, infection of WT C57BL/6 mice with RSV A/Long is not lethal, despite significant pathology that is resolved by M2 macrophages whose development is IL-4Rα, IFN-β, and TLR4 dependent (Shirey et al., 2010). RSV infection of TLR4-SNP mice was accompanied by increased histopathology and M1 macrophage gene expression compared with WT mice, confirming our previous report comparing WT and TLR4−/− mice (Shirey et al., 2010). Conversely, development of M2 macrophage markers, e.g., Chil3 (Ym1), Mrc1 (CD206), Arg1 (Arg-1), and Pparg (PPARγ) mRNA, were decreased in TLR4-SNP mice after RSV infection, supporting the concept that M2 macrophage development is necessary for amelioration of RSV-induced pathology. These data strongly support our earlier finding of an unexpected predominance of the D299G and T399I TLR4 SNPs in cohort of infants and children with documented severe RSV infection (Awomoyi et al., 2007). Several other reports similarly identified an association of the TLR4 299/399 SNPs with RSV severity in infants/children (Tal et al., 2004; Mandelberg et al., 2006; Puthothu et al., 2006), while others have failed to identify such an association (Douville et al., 2010; Kresfelder et al., 2011). The reasons for these differing observations are not obvious; however, expression of these TLR4 SNPs has been shown to differ in African, Asian, and European populations (Ferwerda et al., 2007). Moreover, Caballero et al. (2015) proposed that the severity of RSV was determined by a combined effect of TLR4 haplotype and environmental exposure to LPS in low vs. high socioeconomic populations. Notably, like the weak TLR4 agonist sMPL, which elicited a greater divergence in responses between WT and TLR4-SNP macrophages, the response to RSV F protein, also a weak TLR4 agonist, was significantly lower in HEK293T cells expressing both TLR4 SNPs than either SNP alone (Rallabhandi et al., 2006). It is important to note that prematurity, a predisposing factor for RSV, was shown in a Finnish cohort to be associated with inheritance of the TLR4 299/399 SNPs (Lorenz et al., 2002a).
In summary, our data reinforce the concept that the effects of the human TLR4 299/399 SNPs or the homologous murine SNPs are highly context dependent; an altered balance between innate detection and inflammation appears to underlie decreased LPS responsiveness, increased sensitivity to bacterial infection, resistance to influenza infection, and increased sensitivity to RSV infection. This engineered mouse strain represents the first model available for analysis of these common human TLR4-SNPs and will permit molecular and cellular analyses of the mechanisms underlying the importance of these TLR4 mutations in infection, as well as in other inflammatory diseases or immune processes in which TLR4 has been implicated (e.g., inflammatory bowel disease, atherosclerosis, Alzheimer’s disease, cancer, prematurity, arthritis, allergic airway disease, and adjuvanticity).
Materials and methods
Model of the murine TLR4 D298G/N397I mutant structure
Based on the existing x-ray crystal structures of human TLR4 (PDB ID: 3FXI; Park et al., 2009), human TLR4 D299G and T399I (PDB ID: 4G8A; Ohto et al., 2012), and murine TLR4 (PDB ID: 2Z64; Kim et al., 2007), we created a model of the equivalent murine TLR4 double mutant, D298G, N397I. After alignment of the human and murine TLR4 sequences (Clustal Omega; Sievers et al., 2011), the structural model was constructed through a combination of homology modeling (Phyre; Kelley et al., 2015) and ab initio loop sampling (Coot; Emsley et al., 2010; Fig. 1 A).
Generation of TLR4 D298G/N397I (TLR4-SNP) mice
WT C57BL/6J mice were purchased from the Jackson Laboratory. C57BL/6J mice harboring nucleotide substitutions in the endogenous Tlr4 locus (TLR4-SNP mice) were generated using CRISPR/Cas9-based targeting and homology-directed repair by Cyagen Biosciences. The knock-in allele is regulated by native elements present in the progenitor background. The amino acid residues to be mutated were selected from the original identification of two SNPs in human TLR4 that were associated with hyporesponsiveness to inhaled LPS (Arbour et al., 2000; i.e., the equivalent amino acid residues in humans are D299G and T399I). The small guide RNA (sgRNA) to the mouse Tlr4 gene, a donor vector containing nonsynonymous GAT→GGT (D298G) and AAC→ATC (N397I) nucleotide substitutions and synonymous TTC→TTT (F302F) and GCC→GCT (A409A) nucleotide substitutions in exon 3 of mouse Tlr4, and Cas9 were coinjected into zygotes to obtain targeted knock-in offspring. The sgRNAs selected for mouse generation had a minimum of three mismatches located predominantly in intergenic regions. For each sgRNA, only four potential exonic off-target sites were predicted, none of which ranked in the top 40 and each of which exhibited a minimum of four mismatches with a low probability score. Founder animals were identified by sequencing PCR product from genomic DNA. Two independent founder mice generated on a C57BL/6J background (Jackson Laboratory) were identified and crossed to WT C57BL/6J mice to test germline transmission. Four F1 heterozygotes were found to be positive for the knock-in allele. These heterozygotes were mated to derive distinct F2 populations that were subsequently intercrossed to obtain litters that were homozygous for the knock-in allele. Subsequent crosses of homozygous knock-in mice yielded healthy offspring that were used to study the responses presented in this report. DNA prepared from tail snips were sequenced from each litter to verify the presence of the knock-in allele in a homozygous state using the following sequencing primers: 5′-CAAAACCTGGCTGGTTTACACGTC-3′ (forward primer) and 5′-GTGTTAGTATAAGAGATGTCAAGG-3′ (reverse primer). An alignment of the nucleotides, followed by chromatogram traces, is shown in Fig. 1 B. WT and mutated codons (i.e., nonsynonymous) are highlighted in green and pink, respectively, while synonymous codons are highlighted in gray. As indicated in the text and figure legends, each treatment group (for in vivo studies) had at least five mice per treatment/experiment, and each experiment was repeated at least three times. In addition, most of our studies required that multiple litters be combined to obtain sufficient numbers of mice for both in vivo and in vitro studies. Age- (6–8 wk) and sex-matched WT C57BL/6J mice were used for all experiments. TLR4−/− (kindly provided by S. Akira; Osaka University, Osaka, Japan) and backcrossed n > 12 generations onto a C57BL/6J background) and TLR4-SNP mice described above were bred in the University of Maryland, Baltimore’s accredited animal facility. All animal experiments were performed with Institutional Animal Care and Use Committee approval from the University of Maryland, Baltimore.
Reagents
Protein-free E. coli K235 LPS was prepared by the hot phenol-water method as previously reported (McIntire et al., 1969). Murine TNF-α and IL-1β ELISA kits were purchased from R&D Systems and Invitrogen, respectively. sMPL (PHADTM) was purchased from Avanti Polar Lipids. N-palmitoyl-S-[2,3-bis(palmitoyloxy)-(2RS)-propyl]-(R)-cysteinyl-(S)-seryl-(S)-lysyl-(S)-lysyl-(S)-lysyl-(S)-Lys (P3C) and S-[2,3-bis(palmitoyloxy)-(2RS)-propyl]-(R)-cysteinyl-(S)-seryl-(S)-lysyl-(S)-lysyl-(S)-lysyl-(S)-Lys (P2C) and polyinosinic polycytidylic acid were purchased from Invivogen and reconstituted according to manufacturer’s instructions. Eritoran was kindly provided by Eisai (Andover, MA). The cell-permeating inhibitory peptide, 4BB, was synthesized as described previously (Toshchakov et al., 2011). Directly PE-conjugated anti-murine TLR4 (mAb Sa15-21) and isotype control (rat IgG2a,κ) antibodies were purchased from BioLegend. Purified UT-12 mAb (Ohta et al., 2006) was a kind gift from Drs. Robert Munford and Mingfang Lu (National Institutes of Health, Bethesda, MD). Both UT-12 and isotype control IgG2a,κ were conjugated to DyLight 650 using Lightening-Link rapid antibody labeling kit (Novus Biologicals) in parallel reactions following the manufacturer’s instructions. In addition to the antibodies previously described for Western analyses of signaling molecules (Perkins et al., 2018), rabbit mAbs specific for TLR2 (clone E1J2W), TLR4 (clone D8L5W), and GAPDH (clone 14C10) were purchased from Cell Signaling Technology.
qRT-PCR
qRT-PCR was performed on total RNA using an ABI Prism 7900HT Sequence Detection System with SYBR Green reagent (Applied Biosystems) and transcript-specific primers (Cole et al., 2006, 2008; Shirey et al., 2008). Levels of mRNA for specific genes are reported as relative gene expression normalized to that of WT mock-infected lungs, livers, or macrophages (“fold induction”; Livak and Schmittgen, 2001). The housekeeping gene encoding hypoxanthine-guanine phosphoribosyltransferase (Hprt) was used for normalization of RNA levels within each sample.
qRT-PCR was performed on total macrophage RNA as described above for lung and liver homogenates. In addition to previously published primer sequences for detection of cytokine gene expression, the following primer sets were used to detect Tlr4 and Actb mRNA: TLR4 primer set 1: forward, 5′-GGCAACTTGGACCTGAGGAG-3′; reverse, 5′-CATGGGCTCTCGGTCCATAG-3′; TLR4 primer set 2: forward, 5′-CCTGGCTGGGACTCTGATC-3′; reverse, 5′-ATTTCACACCTGGATAAATCC-3′; and TLR4 primer set 3: forward, 5′-CAAAACCTGGCTGGTTTACACGTC-3′; reverse, 5′-GTGTTAGTATAAGAGATGTCAAGG-3′; β-actin: forward, 5′-ACATTAAGGAGAAGCTGTGC-3′; reverse, 5′-TTCTGCATCCTGTCGGCAAT-3′.
Analysis of in vivo responses to LPS
Mice were administered saline or LPS (5 µg/mouse in 25 µl) i.t. (Feng et al., 2013). 18 h later, lungs were harvested for histochemical staining, mRNA analysis by qRT-PCR, and cytokine protein analysis of lung homogenates by ELISA as detailed previously (Shirey et al., 2013). LPS-induced pathology was scored on H&E-stained lung sections on a scale of 0–4 by a pathologist (where 0 is no inflammation and 4 is maximum inflammation). All histological images were taken using an Olympus BH-2 with light/fluorescence teaching microscope with an attached Olympus DP70 digital camera and Olympus DP Controller software.
In other experiments, age- and sex-matched mice were injected i.p. with saline or LPS (doses indicated in the text and figure legends). Mice were monitored for disease symptoms of endotoxicity (Divanovic et al., 2005) and survival every 12 h for 6–8 d. Each of the cardinal symptoms of endotoxicity (lethargy, piloerection, ocular discharge, and diarrhea) received a score from 0 to 3, with 0 (no symptoms), 1 (mild symptoms), 2 (moderate symptoms), and 3 (severe symptoms), and a cumulative score for each animal was recorded. In some experiments, mice were injected i.p. with saline or a nontoxic dose of LPS (25 µg/mouse), and 2 h later, sera and livers collected for cytokine protein (by ELISA) and cytokine mRNA (by qRT-PCR).
Analysis of in vitro responses to LPS
Thioglycolate-elicited peritoneal macrophages were obtained and cultured as previously described (Perkins et al., 2018; Cole et al., 2006, 2008; Shirey et al., 2008). Whole-cell lysates from macrophage cultures were prepared and subjected to Western blot and subsequent densitometric analyses for signaling molecules as described previously (Perkins et al., 2018).
Measurement of surface TLR4 was performed using a FACS Canto II Flow Cytometer (BD) using FACS Diva software (BD) in the University of Maryland Flow Cytometry Core Facility, Center for Innovative Biomedical Resources, collecting 104 single-cell events per sample as described elsewhere (Richard et al., 2019).
To calculate the ID50, 10-fold serial dilutions of Eritoran (0.1–1,000 ng/ml) were used to pretreat macrophages for 20 min or two- to threefold serial dilutions of 4BB were used to pretreat macrophages for 30 min, followed by a 2-h stimulation with LPS (10 ng/ml). The maximum inducible mRNA level was determined using the Prism 7 sigmoidal four-parameter best-fit algorithm, using log-transformed inhibitor concentrations on the x axis and −ΔCT (before anti-log transformation to obtain “fold induction”) on the y axis. The “minimum” response was defined as the average −ΔCT signal from unstimulated cells. For Eritoran, linear regression analysis was applied to the data points surrounding the half-maximal response on the linear portion of the curve to derive the ID50. For 4BB, the Prism 7 sigmoidal four-parameter best-fit algorithm was applied with comparison of the curves by extra sum-of-squares F test.
To measure macrophage glycolysis and oxidative phosphorylation, extracellular flux analyses (Seahorse) were performed. Briefly, macrophages were seeded at 1–1.5 × 105 cells/well in XF24 microplates (Agilent Technologies) or at 8 × 104 cell/well in XFe96 microplates (Agilent Technologies), and treated as indicated. Standard glycolytic stress tests and mitochondrial stress tests were performed on an XF24 or XFe96 Extracellular Flux Analyzer (Agilent Technologies) with the following optimizations (Bordt et al., 2017): XF media was supplemented with 0.4% fatty acid–free bovine serum albumin (#A7030; Sigma-Aldrich), pH was adjusted to 7.4 with sodium hydroxide, and the media was filter-sterilized (Steriflip; Millipore); compound delivery ports A, B, and C were loaded with 75 µl (XF24) or 20 µl (XFe96) of 10×, 11×, and 12× of the final drug concentrations, respectively; for the glycolytic stress test, the standard 10 mM L-glucose, 1 µM oligomycin, and 50 mM 2-deoxyglucose were used; for the mitochondrial stress tests, 0.6 µM oligomycin, 4 µM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone + 10 mM sodium pyruvate, and 1 µM antimycin A (AA) were used based on prior compound titration results; and after the standard temperature and pH equilibration, an additional 15-min equilibration step was added to the XF run protocol, consisting of three cycles of 3-min mix and 2-min wait before baseline measurements were performed. Data were normalized to cell counts on bright-field pictures taken of the center of each well using a Revolve light microscope (ECHO) before the Seahorse run, analyzed using the Wave for Desktop software suite (Agilent Technologies), and plotted in GraphPad Prism 7, whereby a scalar of 6.8 was applied to all extracellular acidification rate (ECAR) measurements performed on the XFe96 instrument based on the difference in sensitivity between this and the XF24 instrument used for prior experiments. Differences in oxygen consumption rate (OCR) were negligible between the two instruments. The tissue culture supernatants of macrophages treated for 24 h with medium alone or a range of LPS doses were assayed for the concentration of L-lactate using a colorimetric redox reaction (#MAK329; Sigma-Aldrich) that paired the oxidation of L-lactate to the reduction of a formazan reagent, forming a product that was detectable at 630 nm.
Analysis of in vivo susceptibility to bacterial and viral pathogens
Bacterial challenge studies
Kp O1K2 strain B5055 (kindly provided by Dr. Alan Cross, University of Maryland, School of Medicine, Baltimore, MD) was streaked onto trypticase soy agar plates and grown overnight at 37°C. Single-colony isolates were grown to log phase in trypticase soy broth. The optical density of the bacterial suspension (OD600) was determined. 6-wk-old male and female WT and TLR4-SNP mice were inoculated i.p. with Kp (∼1,000 CFU). Survival was monitored for 7 d after infection. To quantify bacterial burden, WT and TLR4-SNP mice were inoculated i.p. with Kp (∼1,500 CFU). After 18 h of infection, mice were euthanized. The spleen and medial lobe from the liver were removed and homogenized in sterile PBS and plated for colony counts.
Virus challenge studies
Mouse-adapted H1N1 influenza A/PR/8/34 virus (PR8; American Type Culture Collection) was kindly provided by Dr. Donna Farber (Columbia University, New York, NY). 6-wk-old male and female WT, TLR4−/−, and TLR4-SNP mice were infected with PR8 (∼7,500 50% tissue culture infectious dose [TCID50], i.n., 25 µl/nare). Mice were monitored daily for survival for 14 d. In some experiments, mice were euthanized 6 d after infection to harvest lungs for analysis of gene expression and lung pathology. Influenza M1 viral gene expression was measured to determine virus level in the lungs of WT and TLR4-SNP mice (Boukhvalova et al., 2020). The following primers were used: forward, 5′-GCAGAGACTTGAAGATGTCTTTGC-3′; reverse, 5′-GGGCATTTTGGACAAAGCGTCTAC-3′.
RSV Long strain (group A) was obtained from American Type Culture Collection and propagated as described previously (Richardson et al., 2005). 6-wk-old male and female WT and TLR4-SNP mice were mock-infected with saline or infected with RSV (5 × 106 PFU/mouse i.n., 25 µl/nare). Lungs were harvested 6 d after infection for analysis of gene expression and lung pathology as described previously (Prince et al., 1999). RSV NS1 viral gene expression was measured to determine virus level in the lungs of WT and TLR4-SNP mice (Boukhvalova et al., 2007). The following primers were used: forward, 5′-CACAACAATGCCAGTGCTACAA-3′; reverse, 5′-TTAGACCATTAGGTTGAGAGCAATGT-3′.
For both influenza and RSV infection, viral-induced lung histopathology was evaluated as follows. Fixed sections (10 µm) of paraffin-embedded lungs were stained with H&E. Four inflammatory parameters were scored independently from 0 to 4 for each section as described previously (Prince et al., 1999): peribronchiolitis (inflammatory cells, primarily lymphocytes surrounding a bronchiole), perivasculitis (inflammatory cells, primarily lymphocytes surrounding a blood vessel), alveolitis (inflammatory cells within alveolar spaces), and interstitial pneumonitis (increased thickness of alveolar walls associated with the infiltration of inflammatory cells). Slides of lung sections were randomized, read blindly, and scored for each parameter.
Statistics
All data were analyzed using GraphPad Prism software. Survival data were analyzed using a Wilcoxon log-rank test. All other data are presented as means with SEM, with individual responses indicated. Where relevant, the sample size is indicated in the text or figure legends. qRT-PCR data were analyzed using the nontransformed −ΔCt values after normalization to the housekeeping gene Hprt but are graphically presented as “fold-increase” relative to the response of untreated RNA preparations from WT mice or macrophages (2−ΔΔCt; Livak and Schmittgen, 2001) to ease interpretation of results. Statistical significance was determined using one- or two-way ANOVA, with differences between treatment groups evaluated by post hoc comparisons (e.g., Tukey or Sidak post hoc tests), extra sum-of-squares F test, a Student’s t test, or Mann–Whitney U test, as indicated in the figure legends. P values from post hoc analyses of ANOVAs were adjusted for multiple comparisons. P values are shown in the figures, figure legends, or both. A P value <0.05 was considered statistically significant, except in two-way ANOVA of metabolic measurements, where α = 0.10.
Online supplemental material
Fig. S1 shows various superimposed models of human and murine WT and TLR4 SNP proteins. Fig. S2 shows a high magnification of the lung section of a WT mouse administered LPS i.t., as well as lung and liver mRNA responses from WT and TLR4-SNP mice challenged with LPS by different routes. Fig. S3 shows that the TLR4-SNP macrophages have a reduced response to LPS but a normal response to TLR2 and TLR3 agonists and that equivalent levels of Tlr4 mRNA were found in macrophages and livers of WT and TLR4-SNP mice. Fig. S4 illustrates the glycolytic and mitochondrial stress test measurements taken and provides additional data on glycolytic capacity, ATP production, proton leak, and mitochondrial coupling efficiency in WT and TLR4-SNP macrophages stimulated with LPS. Fig. S5 shows measurements of pathogen burden in WT and TLR4-SNP mice infected with Kp, PR8, and RSV.
Acknowledgments
Flow cytometry and Seahorse metabolic analyses were performed at the Center for Innovative Biomedical Resources Core Facilities at the University of Maryland, School of Medicine. The authors thank Drs. Achsah Keegan, Tami Kingsbury, and Alan Cross for their careful review of the manuscript and thoughtful suggestions.
This work was supported in part by National Institutes of Health grants AI123371 (S.N. Vogel), S10 OD025101 (S.N. Vogel), AI125215 (S.N. Vogel and J.C.G. Blanco), AI082299 (V.Y. Toshchakov), AI123467 (K.H. Piepenbrink), and GM133126 (K.H. Piepenbrink).
Author contributions: K.H. Piepenbrink carried out crystallographic analyses of human TLR4 SNPs to define the homologous SNPs in mice; K. Richard, A. Gopalakrishnan, D.J. Prantner, D.J. Perkins, C. Feng, R. Fanaroff, V.Y. Toshchakov, and A. Vlk designed and carried out experiments to measure responses to LPS and other TLR agonists in vivo and in vitro; K. Richard carried out all flow cytometry analyses, D.J. Prantner carried out Western analysis for TLR2 and TLR4, and K. Richard and D.J. Prantner performed all metabolic studies; K.A. Shirey and W. Lai designed and carried out all infection experiments, and J.C.G. Blanco contributed to the design of virus infection studies and histopathology scoring; S. Nallar bred and genotyped all of the TLR4-SNP mice and provided significant guidance for analysis of specific Tlr4 transcripts; R. Fanaroff carried out histopathology scoring on lungs from LPS-treated mice; A.E. Medvedev (deceased) and S.N. Vogel designed the study; S.N. Vogel oversaw the execution of the study; and all authors except for A.E. Medvedev participated in the writing of the manuscript.
References
- Agnese, D.M., Calvano J.E., Hahm S.J., Coyle S.M., Corbett S.A., Calvano S.E., and Lowry S.F.. 2002. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J. Infect. Dis. 186:1522–1525. 10.1086/344893 [DOI] [PubMed] [Google Scholar]
- Akashi-Takamura, S., Furuta T., Takahashi K., Tanimura N., Kusumoto Y., Kobayashi T., Saitoh S., Adachi Y., Doi T., and Miyake K.. 2006. Agonistic antibody to TLR4/MD-2 protects mice from acute lethal hepatitis induced by TNF-α. J. Immunol. 176:4244–4251. 10.4049/jimmunol.176.7.4244 [DOI] [PubMed] [Google Scholar]
- Andersson, U., Ottestad W., and Tracey K.J.. 2020. Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19? Mol. Med. 26:42 10.1186/s10020-020-00172-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbour, N.C., Lorenz E., Schutte B.C., Zabner J., Kline J.N., Jones M., Frees K., Watt J.L., and Schwartz D.A.. 2000. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 25:187–191. 10.1038/76048 [DOI] [PubMed] [Google Scholar]
- Awomoyi, A.A., Rallabhandi P., Pollin T.I., Lorenz E., Sztein M.B., Boukhvalova M.S., Hemming V.G., Blanco J.C.G., and Vogel S.N.. 2007. Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J. Immunol. 179:3171–3177. 10.4049/jimmunol.179.5.3171 [DOI] [PubMed] [Google Scholar]
- Bahrun, U., Kimoto M., Tsukamoto H., Tsuneyoshi N., Kohara J., and Fukudome K.. 2007. Preparation and characterization of agonistic monoclonal antibodies against Toll-like receptor 4-MD-2 complex. Hybridoma (Larchmt.). 26:393–399. 10.1089/hyb.2007.0523 [DOI] [PubMed] [Google Scholar]
- Bailey, J.D., Diotallevi M., Nicol T., McNeill E., Shaw A., Chuaiphichai S., Hale A., Starr A., Nandi M., Stylianou E., et al. . 2019. Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep. 28:218–230.e7. 10.1016/j.celrep.2019.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balistreri, C.R., Colonna-Romano G., Lio D., Candore G., and Caruso C.. 2009. TLR4 polymorphisms and ageing: implications for the pathophysiology of age-related diseases. J. Clin. Immunol. 29:406–415. 10.1007/s10875-009-9297-5 [DOI] [PubMed] [Google Scholar]
- Bordt, E.A., Clerc P., Roelofs B.A., Saladino A.J., Tretter L., Adam-Vizi V., Cherok E., Khalil A., Yadava N., Ge S.X., et al. . 2017. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev. Cell. 40:583–594.e6. 10.1016/j.devcel.2017.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhvalova, M.S., Prince G.A., and Blanco J.C.G.. 2007. Respiratory syncytial virus infects and abortively replicates in the lungs in spite of preexisting immunity. J. Virol. 81:9443–9450. 10.1128/JVI.00102-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhvalova, M.S., Mortensen E., Mbaye A., McKay J., and Blanco J.C.G.. 2020. Effect of aging on immunogenicity and efficacy of inactivated influenza vaccines in cotton rats Sigmodon hispidus. Hum. Vaccin. Immunother. 2:1–13. 10.1080/21645515.2020.1766334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut, Y., Faure E., Thomas L., Karahashi H., Michelsen K.S., Equils O., Morrison S.G., Morrison R.P., and Arditi M.. 2002. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J. Immunol. 168:1435–1440. 10.4049/jimmunol.168.3.1435 [DOI] [PubMed] [Google Scholar]
- Caballero, M.T., Serra M.E., Acosta P.L., Marzec J., Gibbons L., Salim M., Rodriguez A., Reynaldi A., Garcia A., Bado D., et al. . 2015. TLR4 genotype and environmental LPS mediate RSV bronchiolitis through Th2 polarization. J. Clin. Invest. 125:571–582. 10.1172/JCI75183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel, N.S. 2015. Navigating Metabolism. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 248 pp. [Google Scholar]
- Chatzi, M., Papanikolaou J., Makris D., Papathanasiou I., Tsezou A., Karvouniaris M., and Zakynthinos E.. 2018. Toll-like receptor 2, 4 and 9 polymorphisms and their association with ICU-acquired infections in Central Greece. J. Crit. Care. 47:1–8. 10.1016/j.jcrc.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Choudhury, A., and Mukherjee S.. 2020. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 92:2105–2113. 10.1002/jmv.25987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, L.E., Elkins K.L., Michalek S.M., Qureshi N., Eaton L.J., Rallabhandi P., Cuesta N., and Vogel S.N.. 2006. Immunologic consequences of Francisella tularensis live vaccine strain infection: role of the innate immune response in infection and immunity. J. Immunol. 176:6888–6899. 10.4049/jimmunol.176.11.6888 [DOI] [PubMed] [Google Scholar]
- Cole, L.E., Santiago A., Barry E., Kang T.J., Shirey K.A., Roberts Z.J., Elkins K.L., Cross A.S., and Vogel S.N.. 2008. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J. Immunol. 180:6885–6891. 10.4049/jimmunol.180.10.6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, A.S., Sadoff J.C., Kelly N., Bernton E., and Gemski P.. 1989. Pretreatment with recombinant murine tumor necrosis factor alpha/cachectin and murine interleukin 1 alpha protects mice from lethal bacterial infection. J. Exp. Med. 169:2021–2027. 10.1084/jem.169.6.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A., Yang C.-S., Arifuzzaman S., Kim S., Kim S.Y., Jung K.H., Lee Y.S., and Chai Y.G.. 2018. High-Resolution mapping and dynamics of the transcriptome, transcription factors, and transcription co-factor networks in classically and alternatively activated macrophages. Front. Immunol. 9:22 10.3389/fimmu.2018.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, L.C., Rice C.M., Palmieri E.M., Taylor P.R., Kuhns D.B., and McVicar D.W.. 2017. Peritoneal tissue-resident macrophages are metabolically poised to engage microbes using tissue-niche fuels. Nat. Commun. 8:2074 10.1038/s41467-017-02092-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, L.C., Rice C.M., McVicar D.W., and Weiss J.M.. 2019. Diversity and environmental adaptation of phagocytic cell metabolism. J. Leukoc. Biol. 105:37–48. 10.1002/JLB.4RI0518-195R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divanovic, S., Trompette A., Atabani S.F., Madan R., Golenbock D.T., Visintin A., Finberg R.W., Tarakhovsky A., Vogel S.N., Belkaid Y., et al. . 2005. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat. Immunol. 6:571–578. 10.1038/ni1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville, R.N., Lissitsyn Y., Hirschfeld A.F., Becker A.B., Kozyrskyj A.L., Liem J., Bastien N., Li Y., Victor R.E., Sekhon M., et al. . 2010. TLR4 Asp299Gly and Thr399Ile polymorphisms: no impact on human immune responsiveness to LPS or respiratory syncytial virus. PLoS One. 5:e12087 10.1371/journal.pone.0012087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, C., Zhang L., Nguyen C., Vogel S.N., Goldblum S.E., Blackwelder W.C., and Cross A.S.. 2013. Neuraminidase reprograms lung tissue and potentiates lipopolysaccharide-induced acute lung injury in mice. J. Immunol. 191:4828–4837. 10.4049/jimmunol.1202673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergestad, T., Wu M.N., Schulze K.L., Lloyd T.E., Bellen H.J., and Broadie K.. 2001. Targeted mutations in the syntaxin H3 domain specifically disrupt SNARE complex function in synaptic transmission. J. Neurosci. 21:9142–9150. 10.1523/JNEUROSCI.21-23-09142.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferwerda, B., McCall M.B.B., Alonso S., Giamarellos-Bourboulis E.J., Mouktaroudi M., Izagirre N., Syafruddin D., Kibiki G., Cristea T., Hijmans A., et al. . 2007. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc. Natl. Acad. Sci. USA. 104:16645–16650. 10.1073/pnas.0704828104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, L., Xiong Y., Song C., Piao W., Vogel S.N., and Medvedev A.E.. 2012. The Asp299Gly polymorphism alters TLR4 signaling by interfering with recruitment of MyD88 and TRIF. J. Immunol. 188:4506–4515. 10.4049/jimmunol.1200202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, K.A., and Kagan J.C.. 2020. Toll-like receptors and the control of immunity. Cell. 180:1044–1066. 10.1016/j.cell.2020.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon, S.J., Borbulevych O.Y., Davis-Harrison R.L., Turner R.V., Damirjian M., Wojnarowicz A., Biddison W.E., and Baker B.M.. 2006. T cell receptor recognition via cooperative conformational plasticity. J. Mol. Biol. 363:228–243. 10.1016/j.jmb.2006.08.045 [DOI] [PubMed] [Google Scholar]
- Hajjar, A.M., Ernst R.K., Yi J., Yam C.S., and Miller S.I.. 2017. Expression level of human TLR4 rather than sequence is the key determinant of LPS responsiveness. PLoS One. 12:e0186308 10.1371/journal.pone.0186308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricson, B.E., Perera P.-Y., Qureshi N., Takayama K., and Vogel S.N.. 1992. Rhodopseudomonas sphaeroides lipid A derivatives block in vitro induction of tumor necrosis factor and endotoxin tolerance by smooth lipopolysaccharide and monophosphoryl lipid A. Infect. Immun. 60:4285–4290. 10.1128/IAI.60.10.4285-4290.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hold, G.L., Berry S., Saunders K.A., Drew J., Mayer C., Brookes H., Gay N.J., El-Omar E.M., and Bryant C.E.. 2014. The TLR4 D299G and T399I SNPs are constitutively active to up-regulate expression of Trif-dependent genes. PLoS One. 9:e111460 10.1371/journal.pone.0111460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, M.M., and O’Neill L.A.J.. 2018. Metabolic regulation of NLRP3. Immunol. Rev. 281:88–98. 10.1111/imr.12608 [DOI] [PubMed] [Google Scholar]
- Kagan, J.C., Su T., Horng T., Chow A., Akira S., and Medzhitov R.. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9:361–368. 10.1038/ni1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A., Mezulis S., Yates C.M., Wass M.N., and Sternberg M.J.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10:845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.M., Park B.S., Kim J.I., Kim S.E., Lee J., Oh S.C., Enkhbayar P., Matsushima N., Lee H., Yoo O.J., and Lee J.-O.. 2007. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 130:906–917. 10.1016/j.cell.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Kresfelder, T.L., Janssen R., Bont L., Pretorius M., and Venter M.. 2011. Confirmation of an association between single nucleotide polymorphisms in the VDR gene with respiratory syncytial virus related disease in South African children. J. Med. Virol. 83:1834–1840. 10.1002/jmv.22179 [DOI] [PubMed] [Google Scholar]
- Kurt-Jones, E.A., Popova L., Kwinn L., Haynes L.M., Jones L.P., Tripp R.A., Walsh E.E., Freeman M.W., Golenbock D.T., Anderson L.J., and Finberg R.W.. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398–401. 10.1038/80833 [DOI] [PubMed] [Google Scholar]
- Lachmandas, E., Boutens L., Ratter J.M., Hijmans A., Hooiveld G.J., Joosten L.A., Rodenburg R.J., Fransen J.A., Houtkooper R.H., van Crevel R., et al. . 2016. Microbial stimulation of different Toll-like receptor signalling pathways induces diverse metabolic programmes in human monocytes. Nat. Microbiol. 2:16246 10.1038/nmicrobiol.2016.246 [DOI] [PubMed] [Google Scholar]
- Latty, S.L., Sakai J., Hopkins L., Verstak B., Paramo T., Berglund N.A., Cammarota E., Cicuta P., Gay N.J., Bond P.J., et al. . 2018. Activation of Toll-like receptors nucleates assembly of the MyDDosome signaling hub. eLife. 7:e31377 10.7554/eLife.31377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lorenz, E., Hallman M., Marttila R., Haataja R., and Schwartz D.A.. 2002a Association between the Asp299Gly polymorphisms in the Toll-like receptor 4 and premature births in the Finnish population. Pediatr. Res. 52:373–376. 10.1203/00006450-200209000-00011 [DOI] [PubMed] [Google Scholar]
- Lorenz, E., Mira J.P., Frees K.L., and Schwartz D.A.. 2002b Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch. Intern. Med. 162:1028–1032. 10.1001/archinte.162.9.1028 [DOI] [PubMed] [Google Scholar]
- Malur, A., Mccoy A.J., Arce S., Barna B.P., Kavuru M.S., Malur A.G., and Thomassen M.J.. 2009. Deletion of PPAR gamma in alveolar macrophages is associated with a Th-1 pulmonary inflammatory response. J. Immunol. 182:5816–5822. 10.4049/jimmunol.0803504 [DOI] [PubMed] [Google Scholar]
- Mandelberg, A., Tal G., Naugolny L., Cesar K., Oron A., Houri S., Gilad E., and Somekh E.. 2006. Lipopolysaccharide hyporesponsiveness as a risk factor for intensive care unit hospitalization in infants with respiratory syncitial virus bronchiolitis. Clin. Exp. Immunol. 144:48–52. 10.1111/j.1365-2249.2006.03030.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire, F.C., Barlow G.H., Sievert H.W., Finley R.A., and Yoo A.L.. 1969. Studies on a lipopolysaccharide from Escherichia coli. Heterogeneity and mechanism of reversible inactivation by sodium deoxycholate. Biochemistry. 8:4063–4067. 10.1021/bi00838a024 [DOI] [PubMed] [Google Scholar]
- Medvedev, A.E. 2013. Toll-like receptor polymorphisms, inflammatory and infectious diseases, allergies, and cancer. J. Interferon Cytokine Res. 33:467–484. 10.1089/jir.2012.0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, O., LeVan T.D., Stern D., Dentener M., Thorn J., Gnat D., Beijer M.L., Cochaux P., Holt P.G., Martinez F.D., and Rylander R.. 2003. Systemic responsiveness to lipopolysaccharide and polymorphisms in the toll-like receptor 4 gene in human beings. J. Allergy Clin. Immunol. 112:923–929. 10.1016/j.jaci.2003.05.001 [DOI] [PubMed] [Google Scholar]
- Mills, E.L., Kelly B., and O’Neill L.A.J.. 2017. Mitochondria are the powerhouses of immunity. Nat. Immunol. 18:488–498. 10.1038/ni.3704 [DOI] [PubMed] [Google Scholar]
- Mukherjee, S., Huda S., and Sinha Babu S.P.. 2019. Toll-like receptor polymorphism in host immune response to infectious diseases: A review. Scand. J. Immunol. 90:e12771 10.1111/sji.12771 [DOI] [PubMed] [Google Scholar]
- Murray, P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., et al. . 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 41:14–20. 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, Y., Akashi S., Nagafuku M., Ogata M., Iwakura Y., Akira S., Kitamura T., Kosugi A., Kimoto M., and Miyake K.. 2002. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat. Immunol. 3:667–672. 10.1038/ni809 [DOI] [PubMed] [Google Scholar]
- O’Brien, A.D., Rosenstreich D.L., Scher I., Campbell G.H., MacDermott R.P., and Formal S.B.. 1980. Genetic control of susceptibility to Salmonella typhimurium in mice: role of the LPS gene. J. Immunol. 124:20–24. [PubMed] [Google Scholar]
- Ohta, S., Bahrun U., Shimazu R., Matsushita H., Fukudome K., and Kimoto M.. 2006. Induction of long-term lipopolysaccharide tolerance by an agonistic monoclonal antibody to the toll-like receptor 4/MD-2 complex. Clin. Vaccine Immunol. 13:1131–1136. 10.1128/CVI.00173-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto, U., Yamakawa N., Akashi-Takamura S., Miyake K., and Shimizu T.. 2012. Structural analyses of human Toll-like receptor 4 polymorphisms D299G and T399I. J. Biol. Chem. 287:40611–40617. 10.1074/jbc.M112.404608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, E.M., Gonzalez-Cotto M., Baseler W.A., Davies L.C., Ghesquière B., Maio N., Rice C.M., Rouault T.A., Cassel T., Higashi R.M., et al. . 2020. Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat. Commun. 11:698 10.1038/s41467-020-14433-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B.S., Song D.H., Kim H.M., Choi B.S., Lee H., and Lee J.O.. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 458:1191–1195. 10.1038/nature07830 [DOI] [PubMed] [Google Scholar]
- Perkins, D.J., Richard K., Hansen A.M., Lai W., Nallar S., Koller B., and Vogel S.N.. 2018. Autocrine-paracrine prostaglandin E2 signaling restricts TLR4 internalization and TRIF signaling. Nat. Immunol. 19:1309–1318. 10.1038/s41590-018-0243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin-Cocon, L., Aublin-Gex A., Sestito S.E., Shirey K.A., Patel M.C., André P., Blanco J.C., Vogel S.N., Peri F., and Lotteau V.. 2017. TLR4 antagonist FP7 inhibits LPS-induced cytokine production and glycolytic reprogramming in dendritic cells, and protects mice from lethal influenza infection. Sci. Rep. 7:40791 10.1038/srep40791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao, W., Shirey K.A., Ru L.W., Lai W., Szmacinski H., Snyder G.A., Sundberg E.J., Lakowicz J.R., Vogel S.N., and Toshchakov V.Y.. 2015. A decoy peptide that disrupts TIRAP recruitment to TLRs is protective in a murine model of influenza. Cell Rep. 11:1941–1952. 10.1016/j.celrep.2015.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak, A., He X., Smirnova I., Liu M.Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. . 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 282:2085–2088. 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- Prince, G.A., Prieels J.P., Slaoui M., and Porter D.D.. 1999. Pulmonary lesions in primary respiratory syncytial virus infection, reinfection, and vaccine-enhanced disease in the cotton rat (Sigmodon hispidus). Lab. Invest. 79:1385–1392. [PubMed] [Google Scholar]
- Prohinar, P., Rallabhandi P., Weiss J.P., and Gioannini T.L.. 2010. Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on coexpression of MD-2 than does wild-type TLR4. J. Immunol. 184:4362–4367. 10.4049/jimmunol.0903142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthothu, B., Forster J., Heinzmann A., and Krueger M.. 2006. TLR-4 and CD14 polymorphisms in respiratory syncytial virus associated disease. Dis. Markers. 22:303–308. 10.1155/2006/865890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallabhandi, P., Bell J., Boukhvalova M.S., Medvedev A., Lorenz E., Arditi M., Hemming V.G., Blanco J.C., Segal D.M., and Vogel S.N.. 2006. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J. Immunol. 177:322–332. 10.4049/jimmunol.177.1.322 [DOI] [PubMed] [Google Scholar]
- Rallabhandi, P., Awomoyi A., Thomas K.E., Phalipon A., Fujimoto Y., Fukase K., Kusumoto S., Qureshi N., Sztein M.B., and Vogel S.N.. 2008. Differential activation of human TLR4 by Escherichia coli and Shigella flexneri 2a lipopolysaccharide: combined effects of lipid A acylation state and TLR4 polymorphisms on signaling. J. Immunol. 180:1139–1147. 10.4049/jimmunol.180.2.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, K., Vogel S.N., and Perkins D.J.. 2019. Quantitation of TLR4 internalization in response to LPS in thioglycollate elicited peritoneal mouse macrophages by flow cytometry. Bio Protoc. 9:3369 10.21769/BioProtoc.3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, J.Y., Ottolini M.G., Pletneva L., Boukhvalova M., Zhang S., Vogel S.N., Prince G.A., and Blanco J.C.. 2005. Respiratory syncytial virus (RSV) infection induces cyclooxygenase 2: a potential target for RSV therapy. J. Immunol. 174:4356–4364. 10.4049/jimmunol.174.7.4356 [DOI] [PubMed] [Google Scholar]
- Roger, T., Froidevaux C., Le Roy D., Reymond M.K., Chanson A.L., Mauri D., Burns K., Riederer B.M., Akira S., and Calandra T.. 2009. Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proc. Natl. Acad. Sci. USA. 106:2348–2352. 10.1073/pnas.0808146106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, N.W.J., and Schumann R.R.. 2005. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect. Dis. 5:156–164. 10.1016/S1473-3099(05)01308-3 [DOI] [PubMed] [Google Scholar]
- Shirey, K.A., Cole L.E., Keegan A.D., and Vogel S.N.. 2008. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J. Immunol. 181:4159–4167. 10.4049/jimmunol.181.6.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey, K.A., Pletneva L.M., Puche A.C., Keegan A.D., Prince G.A., Blanco J.C.G., and Vogel S.N.. 2010. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R α-, TLR4-, and IFN-β-dependent. Mucosal Immunol. 3:291–300. 10.1038/mi.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey, K.A., Lai W., Scott A.J., Lipsky M., Mistry P., Pletneva L.M., Karp C.L., McAlees J., Gioannini T.L., Weiss J., et al. . 2013. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. 497:498–502. 10.1038/nature12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey, K.A., Lai W., Patel M.C., Pletneva L.M., Pang C., Kurt-Jones E., Lipsky M., Roger T., Calandra T., Tracey K.J., et al. . 2016. Novel strategies for targeting innate immune responses to influenza. Mucosal Immunol. 9:1173–1182. 10.1038/mi.2015.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey, K.A., Lai W., Brown L.J., Blanco J.C.G., Beadenkopf R., Wang Y., Vogel S.N., and Snyder G.A.. 2020. Select targeting of intracellular Toll-interleukin-1 receptor resistance domains for protection against influenza-induced disease. Innate Immun. 26:26–34. 10.1177/1753425919846281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, X., Yu Y., Zhong Y., Giannopoulou E.G., Hu X., Liu H., Cross J.R., Rätsch G., Rice C.M., and Ivashkiv L.B.. 2015. Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 16:838–849. 10.1038/ni.3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmacinski, H., Toshchakov V., and Lakowicz J.R.. 2014. Application of phasor plot and autofluorescence correction for study of heterogeneous cell population. J. Biomed. Opt. 19:046017 10.1117/1.JBO.19.4.046017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal, G., Mandelberg A., Dalal I., Cesar K., Somekh E., Tal A., Oron A., Itskovich S., Ballin A., Houri S., et al. . 2004. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J. Infect. Dis. 189:2057–2063. 10.1086/420830 [DOI] [PubMed] [Google Scholar]
- Toshchakov, V.Y., Szmacinski H., Couture L.A., Lakowicz J.R., and Vogel S.N.. 2011. Targeting TLR4 signaling by TLR4 Toll/IL-1 receptor domain-derived decoy peptides: identification of the TLR4 Toll/IL-1 receptor domain dimerization interface. J. Immunol. 186:4819–4827. 10.4049/jimmunol.1002424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulic, M.K., Hurrelbrink R.J., Prêle C.M., Laing I.A., Upham J.W., Le Souef P., Sly P.D., and Holt P.G.. 2007. TLR4 polymorphisms mediate impaired responses to respiratory syncytial virus and lipopolysaccharide. J. Immunol. 179:132–140. 10.4049/jimmunol.179.1.132 [DOI] [PubMed] [Google Scholar]
- Visintin, A., Halmen K.A., Khan N., Monks B.G., Golenbock D.T., and Lien E.. 2006. MD-2 expression is not required for cell surface targeting of Toll-like receptor 4 (TLR4). J. Leukoc. Biol. 80:1584–1592. 10.1189/jlb.0606388 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Su L., Morin M.D., Jones B.T., Whitby L.R., Surakattula M.M., Huang H., Shi H., Choi J.H., Wang K.W., et al. . 2016. TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS. Proc. Natl. Acad. Sci. USA. 113:E884–E893. 10.1073/pnas.1525639113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa, N., Ohto U., Akashi-Takamura S., Takahashi K., Saitoh S., Tanimura N., Suganami T., Ogawa Y., Shibata T., Shimizu T., and Miyake K.. 2013. Human TLR4 polymorphism D299G/T399I alters TLR4/MD-2 conformation and response to a weak ligand monophosphoryl lipid A. Int. Immunol. 25:45–52. 10.1093/intimm/dxs084 [DOI] [PubMed] [Google Scholar]
- Yang, H., Antoine D.J., Andersson U., and Tracey K.J.. 2013. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukoc. Biol. 93:865–873. 10.1189/jlb.1212662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni, I., Ostuni R., Marek L.R., Barresi S., Barbalat R., Barton G.M., Granucci F., and Kagan J.C.. 2011. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 147:868–880. 10.1016/j.cell.2011.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]