Abstract
Due to global shortages of flocked nasopharyngeal swabs and appropriate viral transport media during the COVID-19 pandemic, alternate diagnostic specimens for SARS-CoV-2 detection are sought. The accuracy and feasibility of saliva samples collected and transported without specialized collection devices or media were evaluated. Saliva demonstrated good concordance with paired nasopharyngeal swabs for SARS-CoV-2 detection in 67/74 cases (90.5%), though barriers to saliva collection were observed in long-term care residents and outbreak settings. SARS-CoV-2 RNA was stable in human saliva at room temperature for up to 48 h after initial specimen collection, informing appropriate transport time and conditions.
Keywords: COVID-19, SARS-CoV-2, Saliva, Nasopharyngeal, Transport
Introduction
Diagnostic testing is a cornerstone of the COVID-19 pandemic response strategy [1], yet the establishment of laboratory testing which is accurate, practical, and scalable to meet the demand for public health surveillance measures has been a considerable challenge. Nasopharyngeal swabs are the preferred specimen type over throat swabs due to superior sensitivity [2, 3], and over nasal aspirates due to lower risk of aerosol generation. Flocked nasopharyngeal swabs are designed to maximize mucosal contact and more efficiently release contents into the testing medium [4, 5]; however, due to global demand during the pandemic response, a reliable supply of high-quality, flocked swabs and appropriate viral transport medium has been difficult to procure. Furthermore, studies describe variable collection quality of nasopharyngeal specimens leading to diminished sensitivity and potential false-negative results for SARS-CoV-2[6–8]. Although samples from the lower respiratory tract such as sputum or bronchoalveolar lavage have been widely used [9–11], only a subset of patients under investigation for COVID-19 are able to expectorate sputum or undergo invasive bronchoscopic procedures.
Saliva has been described as an acceptable alternative diagnostic specimen for the detection of common respiratory viruses [12–15], and more recently, SARS-CoV-2 [6, 15–19]. Salivary gland epithelial cells have demonstrated high expression of ACE2 receptors [20, 21], which may enhance the replication of SARS-CoV-2 at this site. Patients may describe the collection of saliva as more comfortable or acceptable compared to nasopharyngeal swabs [22], and the collection requires minimal healthcare worker involvement. Collection techniques vary widely in the published medical literature and include the following: passively drooling into a 50-mL Falcon tube for 1 min [16]; coughing 0.5–1 mL of saliva from the back of the throat into a sterile container and adding 2 mL of viral transport media upon receipt in the laboratory [17, 23]; repeatedly spitting into a sterile container upon waking in the morning prior to eating, drinking, or brushing teeth [6]; or pooling 1–2 mL of saliva into a container with the addition of liquid Amies media in a 1:1 ratio upon receipt in the laboratory [18].
Saliva collection techniques which require transport media or specialized collection devices are problematic during a pandemic. Distribution of pre-packaged kits may be costly and impractical for community settings, and the reliable supply of necessary materials will continue to be a global challenge. Specialized saliva collection devices typically marketed for biochemical tests [24] may contain components such as cotton which can inhibit nucleic acid amplification. Furthermore, it is uncertain in the current medical literature whether the enzymatic properties of human saliva will readily degrade intact virus particles from infected patients, as opposed to naked viral RNA [25, 26]. Commercial RNA stabilization solutions have been marketed to preserve cellular RNA in various specimen types, yet have also been described to decrease the efficiency of nucleic acid extraction [27, 28]. Clinical diagnostic laboratories require immediate guidance for implementing practical and feasible methods of saliva collection, transport, and processing for SARS-CoV-2 detection. The purpose of the present study was to evaluate the feasibility of saliva collection from various patient populations, and to evaluate the optimal transport time of saliva samples without the use of transport media for SARS-CoV-2 detection.
Materials and methods
From March–May 2020, paired saliva and nasopharyngeal swabs were received from patients under investigation for COVID-19 from various clinical settings: inpatients, residents of long-term care (LTC) facilities, healthcare workers, and outpatients. Nasopharyngeal swabs were collected using COPAN Flocked Swabs with 2 mL of Universal Transport Medium™ (COPAN, Brescia, Italy) or 3 mL of BD™ Universal Viral Transport System (Becton Dickinson, Sparks, MD), depending on availability. After the nasopharyngeal swab was collected, patients were asked to provide approximately 1 mL of saliva by pooling saliva in the mouth and spitting into a sterile screw-top container (Starplex Scientific Inc., Etobicoke, Canada). Saliva samples were transported to the laboratory at room temperature without the addition of transport media, with routine transport times (< 24 h). Upon receipt in the laboratory, samples were diluted 1:2 with sterile phosphate-buffered saline (PBS) and vortexed with glass beads until liquid consistency was achieved. Processed saliva samples were stored at 4 °C until nucleic acid extraction could be performed (< 24 h from the time of specimen receipt). Nucleic acid extraction was completed using the MagNA Pure 96 System (Roche Molecular Diagnostics, CA, USA) with an extraction volume of 500 μL and elution volume of 50 μL. Molecular detection of the SARS-CoV-2 envelope (E) gene from saliva samples was performed with the LightMix® ModularDx SARS-CoV (COVID19) E-gene assay (TIB Molbiol, Germany) and LightCycler® Multiplex RNA Virus Master (Roche Molecular Diagnostics, CA, USA). In contrast, nasopharyngeal swabs did not undergo processing with PBS nor glass bead vortexing. An aliquot of the viral transport media from each sample was tested for SARS-CoV-2 as per standard laboratory procedures, using either the RT-PCR assay previously described or the cobas® SARS-CoV-2 Test (Roche Molecular Diagnostics, Laval, QC).
To investigate the optimal transport time of saliva samples without the use of transport media, the stability of SARS-CoV-2 RNA detectable in human saliva samples over time was evaluated.
Two patients known to be positive for SARS-CoV-2 and three healthy volunteers provided five saliva samples each. The saliva from volunteers was spiked with 50 μL of remnant viral transport media from the nasopharyngeal swabs of SARS-CoV-2-positive patients (cycle threshold [Ct] value 15–16 for E-gene). Saliva from the patients and volunteers was processed as previously described, but an aliquot of each sample was processed at delayed time points: 0, 12, 24, 36, and 48 h. In the interim, saliva was stored at room temperature to mirror typical transport conditions. SARS-CoV-2 detection was performed as previously described in triplicate for each sample. Mean Ct values were compared over time using the paired t test with a significance level of α = 0.05 (GraphPad™ QuickCalcs).
Results
One hundred and six (106) saliva specimens were received from patients under investigation for COVID-19. Thirty-one saliva specimens (29.2%) had negligible sample volume (< 0.5 mL). The majority of saliva samples with insufficient volume were from residents and healthcare workers of a LTC facility (30/31, 96.8%). One further specimen was excluded from the analysis due to its appearance as expectorated sputum. On average, the approximate time from saliva collection to the time of laboratory processing was 13 h for inpatients, 16 h for outpatients, and 19 h for samples from LTC facilities. The final analysis included 13/74 samples from symptomatic patients requiring admission to a tertiary acute care hospital (17.6%), 20/74 samples from LTC residents in outbreak settings (27.0%), 28/74 samples from healthcare workers (37.8%), and 13/74 samples from mildly symptomatic outpatients including household contacts of known positive cases (17.6%). The overall median age was 51 years (range: 22 to 95 years), with 43 (58.1%) female and 31 (41.9%) male patients. For the detection of SARS-CoV-2, saliva was concordant with paired nasopharyngeal swabs in 67/74 cases (90.5%). Of the 21 cases positive for SARS-CoV-2 by nasopharyngeal swab, 15 were positive by saliva. Of the 53 cases negative for SARS-CoV-2 by nasopharyngeal swab, one case was positive by saliva.
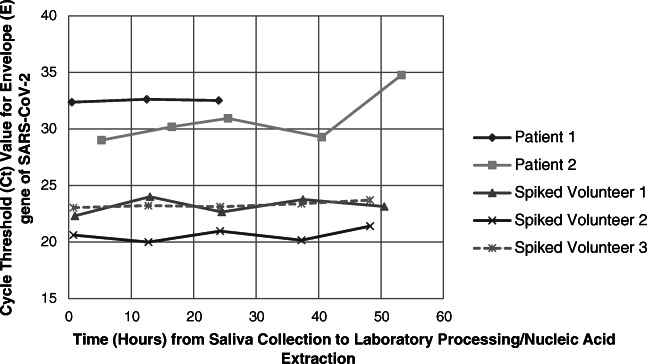
Stability of SARS-CoV-2 viral RNA in saliva with delayed processing up to 36 h was demonstrated (Fig. 1; mean Ct value for E-gene was 23.75 at time zero versus 24.16 at 36 h; p value 0.328, paired t test). The mean Ct value appeared to demonstrate a significant increase at the last reading near 48 h (25.76, p value 0.014 compared to time zero, paired t test), although this may be attributed to a single outlier sample (patient 2), which was collected from an outpatient and had the longest total transport time (53 h) of all samples.
Fig. 1.
Saliva samples from patients with known COVID-19 infection or from healthy volunteers spiked with viral transport media from the nasopharyngeal swabs of known positive COVID-19 cases were held in the laboratory at room temperature and processed at different time points. The stability of SARS-CoV-2 viral RNA in these samples is represented by the average detected cycle threshold (Ct) value of the Envelope (E) gene of SARS-CoV-2 tested in triplicate and plotted over time. Patient 1 was unable to provide sufficient sample volume for five readings and was only tested up to 24 h after the initial time of collection
Discussion
As a diagnostic specimen for SARS-CoV-2 detection, saliva performed well and demonstrated good general concordance with nasopharyngeal swabs. Although a minority of saliva samples (6/74, 8.1%) were discordantly negative for SARS-CoV-2 RNA, the paired nasopharyngeal swabs in these cases had evidence of low viral loads (late Ct values in the range of 31–39), suggesting the decrease in sensitivity may be attributable to viral loads near the limit of detection of the assay rather than the inherent properties of saliva.
Pre-analytical and analytical factors must be carefully considered before implementation of saliva as a routine diagnostic specimen. Nearly 30% of saliva specimens in this study were of insufficient volume. These specimens were rejected by the laboratory as they appeared to solidify, adhered to the sides of the collection container, and were not amenable to further laboratory processing. Although strategies to improve saliva collection have been described [6], LTC residents may face particular challenges due to xerostomia, inability to follow commands, physical barriers with dentures, or insufficient time provided in busy outbreak settings. The provision of careful collection instructions and training for healthcare providers is highlighted, as specimens in this study were received with heterogeneous consistency, and some were rejected due to their obvious appearance as expectorated sputum. Importantly, the findings from this study indicate saliva without transport media can be in transit at room temperature for up to 48 h prior to laboratory processing without losing diagnostic yield.
The amount of saliva processing required in the laboratory was significant. Manual labor by laboratory technologists is required to decrease the viscosity of saliva and ensure compatibility with laboratory instruments, which has added operational cost. The addition of 1:2 PBS dilutes the specimen and may affect diagnostic sensitivity. However, the treatment of saliva in this manner is necessary, as previous experiences in our laboratory revealed nearly all saliva samples demonstrated inhibition with the LightMix® assay when run neat, requiring repeat testing and delaying turn-around-time.
This study had several limitations. Firstly, due to a limited number of samples per each patient group, we are not able to make inferences on performance in any one particular setting. Samples were collected by various healthcare workers and submitted from diverse locations leading to variable transport times. However, these factors do provide a realistic view of the utility of saliva samples in clinical settings. The evaluation of the stability of SARS-CoV-2 viral RNA in human saliva samples without transport media was limited to 48 h by the available volume of saliva from patients and volunteers. However, the vast majority of samples received by our laboratory for SARS-CoV-2 detection are processed in less than 48 h, and thus these results have relevance to many clinical diagnostic laboratories.
Further study may include the establishment of a reliable method for saliva collection in patient populations with barriers (e.g. LTC residents, intubated patients in critical care units), and optimization of high-throughput automated laboratory instruments to accommodate these highly viscous specimens collected as part of mass surveillance measures.
Acknowledgments
We sincerely extend our gratitude to the patients and volunteers who contributed samples to this study as part of the COVID-19 response. We thank Dr. Michael Schwandt and Noah Reich for their contributions to this project. We are also indebted to our medical laboratory technologists who are highly committed to patient care and laboratory quality improvement.
Data availability
Included in the manuscript
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study received approval from the Providence Health Care Research Ethics Board.
Consent to participate
Verbal consent was obtained from patients and volunteers willing to provide saliva samples when not already part of their clinical care.
Consent for publication
Not applicable.
Code availability
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization (2020) 2019 Novel Coronavirus (2019-nCoV): strategic preparedness and response plan. 28. Available at: https://www.who.int/publications-detail/strategic-preparedness-and-response-plan-for-the-new-coronavirus. Accessed 4 May 2020
- 2.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Yang M, Yuan J, et al. Comparative sensitivity of different respiratory specimen types for molecular diagnosis and monitoring of SARS-CoV-2 shedding. Innovation (N Y) 2020;1:100061. doi: 10.1016/j.xinn.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daley P, Castriciano S, Chernesky M, Smieja M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J Clin Microbiol. 2006;44:2265–2267. doi: 10.1128/JCM.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron EJ, Miller JM, Weinstein MP, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the infectious diseases society of America (IDSA) and the American Society for Microbiology (ASM) Clin Infect Dis. 2013;57:e22–121. doi: 10.1093/cid/cit278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyllie AL, Fournier J, Casanovas-Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:13. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian Y, Zeng T, Wang H, et al. Safety management of nasopharyngeal specimen collection from suspected cases of coronavirus disease 2019. Int J Nurs Sci. 2020;7:153–156. doi: 10.1016/j.ijnss.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinloch N, Ritchie G, Brumme CJ, Dong W, Dong W, Lawson T, Jones RB, Montaner JSG, Leung V, Romney MG, Stefanovic A, Matic N, Lowe CF, Brumme Z. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J Infect Dis. 2020;222:899–902. doi: 10.1093/infdis/jiaa370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Y, Chang L, Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev Med Virol. 2020;30:e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US Centers for Disease Control and Prevention (CDC) (2020) Interim guidelines for collecting, handling, and testing clinical specimens from persons under investigation (PUIs) for Coronavirus Disease 2019 (COVID-19). Available at: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html. Accessed 5 May 2020
- 11.World Health Organization (2020) Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. Available at:https://www.who.int/publications/i/item/10665-331501. Accessed 30 March 2020
- 12.Robinson JL, Lee BE, Kothapalli S, Craig WR, Fox JD. Use of throat swab or saliva specimens for detection of respiratory viruses in children. Clin Infect Dis. 2008;46:e61–e64. doi: 10.1086/529386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartholomeusz A, Locarnini S. Associated with antiviral therapy. Antivir Ther. 2006;55:52–55. [Google Scholar]
- 14.Kim YG, Yun SG, Kim MY, Park K, Cho CH, Yoon SY, Nsm MH, Lee CK, Cho YJLC. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J Clin Microbiol. 2017;55:226–233. doi: 10.1128/JCM.01704-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To KK, Lu L, Yip CC et al (2017) Additional molecular testing of saliva specimens improves the detection of respiratory viruses. Emerg Microbes Infect 6:e49. 10.1038/emi.2017.35 [DOI] [PMC free article] [PubMed]
- 16.Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:E45–E50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.To KKW, Tsang OTY, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;3099:1–10. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58:e00776–e00720. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Lowe CF, Ritchie G et al. (2020) SARS-CoV-2 molecular testing for the diagnosis of COVID-19: One test does not fit all. J Med Virol. 10.1002/jmv.26532 [DOI] [PMC free article] [PubMed]
- 20.Xu J, Li Y, Gan F, Du Y, Yao Y. Salivary glands: potential reservoirs for COVID-19 asymptomatic infection. J Dent Res. 2020;99:989. doi: 10.1177/0022034520918518. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Ren B, Peng X, et al. Saliva is a non-negligible factor in the spread of COVID-19. Mol Oral Microbiol. 2020;35:141–145. doi: 10.1111/omi.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinloch N, Shahid A, Ritchie G, Dong W, Lawson T, Montaner JSG, Romney MG, Stefanovic A, Matic N, Brumme CJ, Lowe CF, Brumme ZL, Leung V (2020) Evaluation of nasopharyngeal swab collection techniques for nucleic acid recovery and participant experience: recommendations for COVID-19 diagnostics. Open Forum Infect Dis 1–14 [DOI] [PMC free article] [PubMed]
- 23.To KKW, Tsang OTY, Chik-Yan Yip C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenander-Lumikari M, Johansson I, Vilja P, Samaranayake L. Newer saliva collection methods and saliva composition: a study of two Salivette® kits. Oral Dis. 1995;1:86–91. doi: 10.1111/j.1601-0825.1995.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 25.Fábryová H, Celec P. On the origin and diagnostic use of salivary RNA. Oral Dis. 2014;20:146–152. doi: 10.1111/odi.12098. [DOI] [PubMed] [Google Scholar]
- 26.Park NJ, Li Y, Yu T, Brinkman BMN, Wong DT. Characterization of RNA in saliva. Clin Chem. 2006;52:988–994. doi: 10.1373/clinchem.2005.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez A, Vaneechoutte M (2019) Comparison of the efficiency of different cell lysis methods and different commercial methods for RNA extraction from Candida albicans stored in RNAlater. BMC Microbiol 19:1–10 [DOI] [PMC free article] [PubMed]
- 28.Jiang L, Li L, Kang P et al (2019) Inappropriateness of RNAlater to preserve Caenorhabditis elegans for RNA extraction. MethodsX 6:2460–2467. 10.1016/j.mex.2019.10.015 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Included in the manuscript