Mycobacterium abscessus is a highly antibiotic-resistant opportunistic pathogen causing clinically challenging infections in patients with preexisting lung diseases or under immunosuppression. Hence, reliable antibiotic susceptibility data are required for effective treatment. Aims of this study were to investigate (i) the congruence of genotypic and phenotypic antimicrobial susceptibility testing, (ii) the relationship between resistance profile and clinical course, and (iii) the phylogenetic relations of M. abscessus in a German patient cohort.
KEYWORDS: Mycobacterium abscessus, NTM, antibiotic susceptibility, nontuberculous mycobacteria, transmission
ABSTRACT
Mycobacterium abscessus is a highly antibiotic-resistant opportunistic pathogen causing clinically challenging infections in patients with preexisting lung diseases or under immunosuppression. Hence, reliable antibiotic susceptibility data are required for effective treatment. Aims of this study were to investigate (i) the congruence of genotypic and phenotypic antimicrobial susceptibility testing, (ii) the relationship between resistance profile and clinical course, and (iii) the phylogenetic relations of M. abscessus in a German patient cohort. A total of 39 isolates from 29 patients infected or colonized with M. abscessus underwent genotypic and phenotypic drug susceptibility testing. Clinical data were correlated with susceptibility data. Phylogenetic analysis was performed by means of whole-genome sequencing (WGS) and single-nucleotide polymorphism (SNP) analysis. Macrolide resistance was mainly mediated by functional Erm(41) methyltransferases (T28 sequevars) in M. abscessus subsp. abscessus (n = 25) and M. abscessus subsp. bolletii (n = 2). It was significantly associated with impaired culture conversion (P = 0.02). According to the core SNP phylogeny, we identified three clusters of closely related isolates with SNP distances below 25. Representatives of all circulating global clones (Absc. 1, Absc. 2, and Mass. 1) were identified in our cohort. However, we could not determine evidence for in-hospital interhuman transmission from clinical data. In our patient cohort, we identified three M. abscessus clusters with closely related isolates and representatives of the previously described international clusters but no human-to-human in-hospital transmission. Macrolide and aminoglycoside susceptibility data are critical for therapeutic decision-making in M. abscessus infections.
INTRODUCTION
Mycobacterium abscessus is a highly drug-resistant, rapidly growing nontuberculous mycobacterium (RGM and NTM) (1). It mainly causes pulmonary infections in patients with predisposing lung diseases, such as cystic fibrosis (CF) or non-CF bronchiectasis (2), but has also played an important role in soft-tissue infections in immunocompromised hosts or following surgical procedures (3–7). Recently, we and others have shown that the species consists of three distinct subspecies, namely, M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii (8).
Treatment of infections with M. abscessus remains highly difficult, as it is inherently resistant to several antibiotics by various mechanisms, including impermeability of the cell wall and antibiotic-modifying enzymes (9). In addition, there is a lack of treatment studies correlating in vitro susceptibility patterns with clinical outcome (1). For CF patients, M. abscessus infection is of high clinical impact, as in several centers it is considered a contraindication for lung transplantation and has been shown to be associated with a decline in pulmonary function and clinical deterioration (10).
Constitutional macrolide resistance is conferred by mutations in the rrl gene, whereas inducible resistance is mediated by the erm(41) gene, which codes for an erythromycin resistance methyltransferase that modifies the bacterial 23S rRNA target, resulting in high-level macrolide resistance (11). The erm(41) gene is truncated and, therefore, is nonfunctional in M. abscessus subsp. massiliense but usually is fully functional in M. abscessus subsp. abscessus or M. abscessus subsp. bolletii. High-level resistance to aminoglycoside antibiotics is conferred mainly by mutations in the rrs gene (12).
As the three resistance mechanisms are well characterized, molecular assays (Genotype NTM-DR; Hain Lifesciences, Nehren Germany) are available that allow for their rapid detection within a workday, thereby improving the workflow for clinical decision making (13).
M. abscessus has been suspected to be involved in interhuman transmission in CF patients, suggesting the global spread of isolates of particular clones (14–17). So far, this human-to-human as well as global spread remains controversial, as a route of transmission could not be identified, and those findings were mainly based on phylogenetic core genome analyses (18, 19). Until now, data from a German cohort have not been available.
To shed further light on M. abscessus antimicrobial resistance determinants, clinical characteristics, and population dynamics, we investigated (i) the congruence of genotypic and phenotypic antimicrobial susceptibility testing for macrolides and aminoglycosides, (ii) the relationship between resistance profile and clinical course, and (iii) the phylogenetic relations of 39 M. abscessus isolates from a cohort of 29 patients in a global context.
MATERIALS AND METHODS
Bacterial isolates.
For this study, 39 isolates from 29 patients were recovered from 2006 to 2018. They were cultured on Middlebrook 7H10 agar with oleic albumin dextrose catalase (Becton, Dickinson, Heidelberg, Germany) at 37°C until visible growth could be detected.
pDST.
For phenotypic drug susceptibility testing (pDST), all isolates were tested in the Sensititre RAPMYCO broth microdilution assay (Thermo Fisher Scientific, Waltham, MA) as recommended by the manufacturer. MICs were interpreted according to CLSI guidelines (20). Incubation was conducted at 30°C for 3 to 5 days for all antibiotics. If no resistance to clarithromycin was detected within that time, a total incubation period of 14 days was used to detect inducible resistance. Plates were read on days 3, 4, 5, 7, and 14. The first day with sufficient growth in the control well was chosen for MIC determination. Mycobacterium peregrinum ATCC 700686 was used as a quality control.
The assay includes, among others, the following antibiotic agents (abbreviation and ranges are in parentheses; antibiotics with available CLSI breakpoints are marked with an asterisk): clarithromycin (CLA*; 0.06 to 16 μg/ml), amikacin (AMI*; 1 to 64 μg/ml), moxifloxacin (MXF*; 0.25 to 8 μg/ml), ciprofloxacin (CIP*; 0.12 to 4 μg/ml), linezolid (LZD*; 1 to 32 μg/ml), cefoxitin (FOX*; 4 to 128 μg/ml), imipenem (IMI*; 2 to 64 μg/ml), trimethoprim-sulfamethoxazole (SXT*; 0.25/4.75 to 8/152 μg/ml), doxycycline (DOX*; 0.12 to 16 μg/ml), tigecycline (TGC; 0.015 to 4 μg/ml), and minocycline (MIN; 0.25 to 8 μg/ml).
gDST.
For genotypic drug susceptibility testing (gDST), DNA from all isolates was extracted using the Hain Genolyse kit (Hain Lifescience, Nehren, Germany) (21). DNA samples underwent a PCR program with primers provided by Genotype NTM-DR (Hain Lifescience, Nehren, Germany), and amplicons were then applied to the DNA strip test, as recommended by the manufacturer (13). The assay allows for differentiation of seven nontuberculous mycobacterial species and subspecies (M. avium, M. chimera, M. intracellulare, M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, M. abscessus subsp. massiliense, and M. chelonae) as well as the detection of inducible macrolide resistance [erm(41), C28T], inherent macrolide resistance (rrl gene, A2058C, A2058G, A2058T, A2059C, A2059G, and A2059T), and constitutional aminoglycoside resistance (rrs gene, A1408G, T1406A, and C1409T).
Clinical data and statistical analysis.
This study was approved by our local ethics committee under file number 386/18. Patient data were retrieved by retrospective chart review, collected in Libre Office Calc (The Document Foundation), and analyzed in R (22). All graphs were drawn using the ggplot package (23).
Mortality and culture conversion analyses were conducted by Cox regression within R’s survival and survminer packages (24, 25). Significance tests were performed using the log-rank test. To evaluate other clinical parameters, lung functions lying within temporal proximity to the first isolation of M. abscessus were analyzed (vital capacity, VC, and forced expiratory volume in 1 s, FEV1). For nonnormally distributed continuous data, significance tests were conducted using the Wilcoxon-Mann-Whitney test, and for categorical variables, tests were conducted using the Fisher exact test. All tests were performed two-sided at a significance level of alpha = 0.05.
Whole-genome sequencing (WGS).
Cetyltrimethylammonium bromide (CTAB)-chloroform DNA extraction was performed as previously described (26). Briefly, cells from solid cultures were dissolved in 400 μl of Tris-EDTA (TE) buffer and heat inactivated for 20 min at 80°C. Fifty microliters of 10-mg/ml lysozyme was added and incubated at 37°C for at least 1 h, and then 75 μl of proteinase K, sodium dodecyl sulfate, and 100 μl 5 M NaCl plus 100 μl CTAB-NaCl were added, each followed by 10 min of incubation at 65°C. Chloroform-isoamyl alcohol was used for DNA extraction. Supernatant was transferred into a fresh tube with 450 μl of isopropanol. After incubation for at least 30 min at –20°C, the sample was centrifuged and the precipitate was washed with 70% ethanol. Finally, the pellet was dried for 20 min at 60°C and DNA dissolved in 80 μl of TE buffer.
From extracted genomic DNA, next-generation sequencing libraries were generated using a modified Illumina Nextera library kit protocol (27), and libraries were sequenced in a 2× 150-bp paired-end run on the Illumina NextSeq 500 instrument (Illumina, San Diego, CA, USA).
Sequence data and network analysis.
Sequence quality and GC content of obtained sequence data were assessed using fastQC (Babraham Bioinformatics, Cambridge, United Kingdom). The prediction of macrolide and aminoglycoside resistance was performed using the mab-ariba database by Lipworth et al., with the addition of rrs gene mutations to detect aminoglycoside resistance (28).
For phylogenetic analysis, sequence data in the form of fastq files were processed by the MTBseq pipeline with default settings using the type strain M. abscessus subsp. abscessus ATCC 19977 as a reference genome (GenBank accession number NC_010397.1), yielding aligned core single-nucleotide polymorphism (SNP) positions, a respective pairwise SNP distance matrix, and groups of isolates determined by single linkage clustering (29).
For the detailed cluster analysis, a whole-genome de novo assembly was generated with SPAdes v. 3.11.1 for one isolate of the cluster (30), which was then used as the reference genome for the MTBseq analysis of the cluster members.
From concatenated SNP positions, phylogenetic trees were calculated with RaxML v. 8.2.12 with 100 bootstraps and GTRGAMMA as a substitution model (31). Those trees were midpoint rooted with the phangorn package v. 2.5.5 (32), visualized, and annotated with ggtree (33).
Patient hospital interaction was classified into four categories: (i) no documented interaction, (ii) same hometown, (iii) same day of hospital visit, and (iv) same day at the same ward/outpatient clinic. Patient visits and hospital stays were withdrawn from our local patient information system with a python script, and all patient-to-patient combinations (n = 409) were compared for the possibility of overlaps in hospital visits in R (22).
Data availability.
Sequences determined in the course of this work were deposited in the European Nucleotide Archive (ENA) (see Table S1 in the supplemental material).
RESULTS
Patient characteristics.
In total, 39 isolates from 29 patients were included in this study (29 primary and 10 follow-up isolates from 10 patients dating from 2006 to 2018). The majority of patients suffered from CF (n = 21) followed by other structural lung diseases (n = 3), no apparent predisposition (n = 2), HIV, malignoma, and immunosuppression after solid-organ transplantation (one patient each) (Tables 1 and 2). Isolates of M. abscessus subsp. abscessus and M. abscessus subsp. massiliense were recovered from respiratory specimens only, whereas a single case of lymphadenitis due to M. abscessus subsp. bolletii was included. Median age was 29.9 years (interquartile range [IQR], 18.7 years). Eighteen out of 29 primary isolates showed smooth-colony morphology, whereas 11 had a rough-colony morphotype. Twenty-seven patients were German, living within a 95-km radius around our tertiary care center, whereas one patient was from Croatia and one from Russia.
TABLE 1.
Baseline characteristics of patients included into this study: categorical variablesa (n = 29)
| Categorical variable | All patients (n = 29) |
Macrolide susceptibility |
P value | ||||
|---|---|---|---|---|---|---|---|
| Susceptible (n = 9) |
Resistant (n = 20) |
||||||
| n | % | n | % | n | % | ||
| Gender | |||||||
| Male | 13 | 44.8 | 6 | 66.7 | 7 | 35.0 | 0.2256 |
| Female | 16 | 55.2 | 3 | 33.3 | 13 | 65.0 | |
| M. abscessus subsp. | |||||||
| abscessus | 17 | 58.6 | 0 | 0.0 | 17 | 85.0 | <0.001 |
| massiliense | 10 | 34.5 | 9 | 100.0 | 1 | 5.0 | <0.001 |
| bolletii | 2 | 6.9 | 0 | 0.0 | 2 | 10.0 | 1 |
| Morphology | |||||||
| Smooth | 18 | 62.1 | 7 | 77.8 | 9 | 45.0 | 0.1296 |
| Rough | 11 | 37.9 | 2 | 22.2 | 11 | 55.0 | |
| Manifestation | |||||||
| NTM-PD/colonization | 28 | 96.6 | 9 | 100.0 | 19 | 95.0 | 1 |
| ATS criteria fullfilled | 20 | 71.4 | 5 | 55.6 | 15 | 78.9 | 0.3715 |
| Lymphadenitis | 1 | 3.4 | 0 | 0.0 | 1 | 5.0 | |
| Disposition | |||||||
| CF | 21 | 72.4 | 6 | 66.7 | 15 | 75.0 | 0.6749 |
| HIV | 1 | 3.4 | 0 | 0.0 | 1 | 5.0 | |
| SLD | 3 | 10.3 | 0 | 0.0 | 3 | 15.0 | |
| Lung transplantation | 1 | 3.4 | 1 | 11.1 | 0 | 0.0 | |
| Malignoma | 1 | 3.4 | 1 | 11.1 | 0 | 0.0 | |
| None | 2 | 6.9 | 1 | 11.1 | 1 | 5.0 | |
M. abscessus subsp. abscessus was the most frequent subspecies. All isolates except one originated from pulmonary samples. NTM-PD, NTM pulmonary disease; ATS, American Thoracic Society; CF, cystic fibrosis; HIV, human immunodeficiency virus; SLD, structural lung disease. P values in boldface are those that reached the significance level of α < 0.05.
TABLE 2.
Baseline characteristics of patients included in this study: continuous variablesa
| Continuous variable | All patients |
Macrolide susceptibility |
P value | ||||
|---|---|---|---|---|---|---|---|
| Susceptible |
Resistant |
||||||
| Median | IQR | Median | IQR | Median | IQR | ||
| Age (yr) | 29.9 | 18.7 | 36.4 | 29.4 | 29.9 | 14.15 | 0.8849 |
| Observation time (days) | 1,366 | 1,719 | 1,366 | 1,107 | 1,614.5 | 2,276 | 0.6268 |
| Lung function at baseline (%) | |||||||
| VC | 76.6 | 24.6 | 91.3 | 11.7 | 68.4 | 20.1 | 0.01499 |
| FEV1 | 62.8 | 27.3 | 73.7 | 11.7 | 59.4 | 27.1 | 0.09108 |
Vital capacity (VC) was significantly lower in patients with macrolide-resistant isolates (n = 20, one of those with an rrl mutation). FEV1, forced expiratory volume in one second; IQR, interquartile range. The P value in boldface is that which reached the significance level of α < 0.05
Phenotypic and genotypic drug susceptibility testing.
Thirty-one percent of the primary isolates were macrolide susceptible (confidence interval [CI], 16.0% to 51.0%; n = 9), whereas 69% (49.0% to 84.0%; n = 20) showed phenotypic resistance (Table 3). All M. abscessus subsp. abscessus and M. abscessus subsp. bolletii isolates were resistant to macrolides (n = 19). In contrast, this was the case for only one M. abscessus subsp. massiliense isolate, in which we could detect an rrl mutation. Aminoglycoside resistance was a rare event in this cohort, with only the first isolate being resistant to amikacin due to a mutation in the rrs gene (M. abscessus subsp. abscessus).
TABLE 3.
MIC50, MIC90, range, and susceptibility of isolates for all antibiotics, measured in our base cohort, excluding follow-up isolatesa (n = 29)
| Antibiotic agent | MIC50 (mg/ml) | MIC90 (mg/ml) | Range (mg/ml) | S |
I |
R |
|||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||||
| CLA | >16 | >16 | 0.06 to >16 | 9 | 31.0 | 0 | 0.0 | 20 | 69.0 |
| AMI | 8 | 16 | 2 to >64 | 27 | 93.1 | 1 | 3.4 | 1 | 3.4 |
| FOX | 32 | 128 | 3–128 | 3 | 10.3 | 22 | 75.9 | 4 | 13.8 |
| IMI | 16 | 32 | 4 to >64 | 2 | 6.9 | 19 | 65.5 | 8 | 27.6 |
| MXF | 8 | >8 | 2 to >8 | 0 | 0.0 | 3 | 10.3 | 26 | 89.7 |
| SXT | >8 | >8 | 0.25 to >8 | 5 | 17.2 | 0 | 0.0 | 24 | 82.8 |
| LZD | 8 | 16 | 1 to >32 | 18 | 62.1 | 9 | 31.0 | 2 | 6.9 |
| CIP | 4 | >4 | 2 to >4 | 0 | 0.0 | 3 | 10.3 | 26 | 89.7 |
| DOX | >16 | >16 | 0.25 to >16 | 1 | 3.4 | 1 | 3.4 | 27 | 93.1 |
| MIN | 16 | 16 | 1 to >8 | ||||||
| TGC | 0.5 | 0.5 | 0.25–1 | ||||||
S, susceptible; R, resistant; I, intermediate; CLA, clarithromycin; AMI, amikacin; FOX, cefoxitin; IMI, imipenem; MXF, moxifloxacin; SXT, trimethoprim-sulfamethoxazole; LZD, linezolid; CIP, ciprofloxacin; DOX, doxyxycline; MIN, minocycline; TGC, tigecycline; TOB, tobramycin.
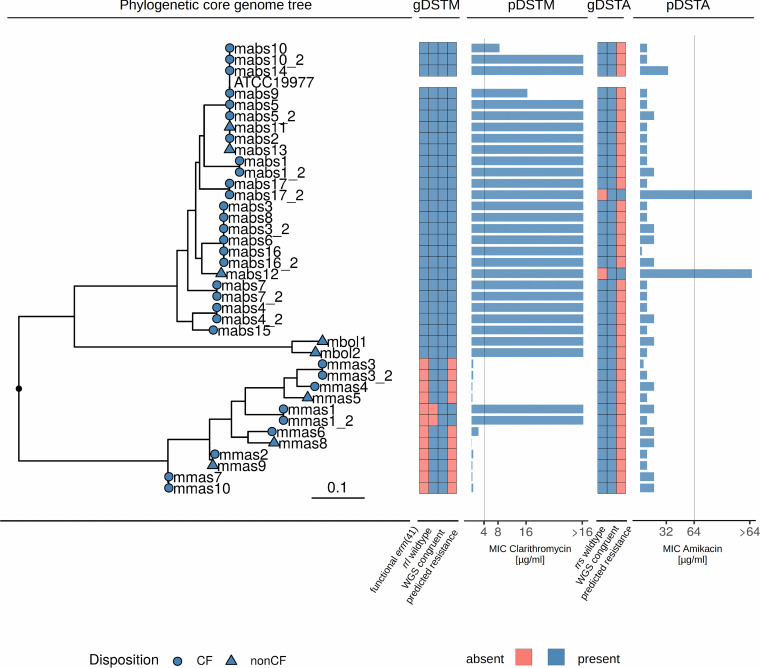
To assess the congruence between genotypic and phenotypic antimicrobial susceptibility for macrolides and aminoglycosides, primary and follow-up isolates from the same patients were included in the analysis (n = 39). The Genotype NTM-DR and the Mab-ariba prediction from whole-genome data of macrolide and aminoglycoside resistance confirmed pDST results in all cases, but in one patient with aminoglycoside resistance both the resistant and susceptible haplotype of the rrs gene were detected, indicating the possibility of a resistant subpopulation within this patient (Fig. 1; see also Table S2 in the supplemental material). Sensitivity and specificity using pDST as a gold standard were 100% for both methods.
FIG 1.
Phylogenetic core genome tree with all isolates from our cohort (n = 39), heatmap of present or absent resistance genes for macrolide resistance (gDSTM), pDST results for clarithromycin (pDSTM), heatmap of gDST results for aminoglycosides (gDSTA), and pDST results for amikacin (pDSTA). Macrolide resistance was due mainly to functional erm(41) genes in M. abscessus subsp. abscessus. One rrl mutation could be found in an M. abscessus subsp. massiliense isolate. Two isolates showed mutations in the rrs gene and accordingly high MICs in amikacin. All predicted resistance levels by gDST (NTM-DR and WGS) were confirmed by pDST. Vertical lines in pDST panels depict breakpoints, as given by the CLSI. mabs, M. abscessus subsp. abscessus; mmas, M. abcsessus subsp. massiliense; mbol, M. abscessus subsp. bolletii. The tree scale depicts SNPs/site.
No acquisition of macrolide or aminoglycoside resistance was observed in longitudinal isolates from individual patients, except for strain mabs17_2, where aminoglycoside resistance due to an rrs mutation was detected in an isolate that was cultivated 958 days after the first isolate. The patient had received aminoglycoside treatment in the meantime (4 weeks of gentamicin). The core SNP distance between those two isolates was 11 SNPs, rendering reinfection as the cause for the advent of resistance unlikely.
Correlation of clinical parameters and antibiotic susceptibility.
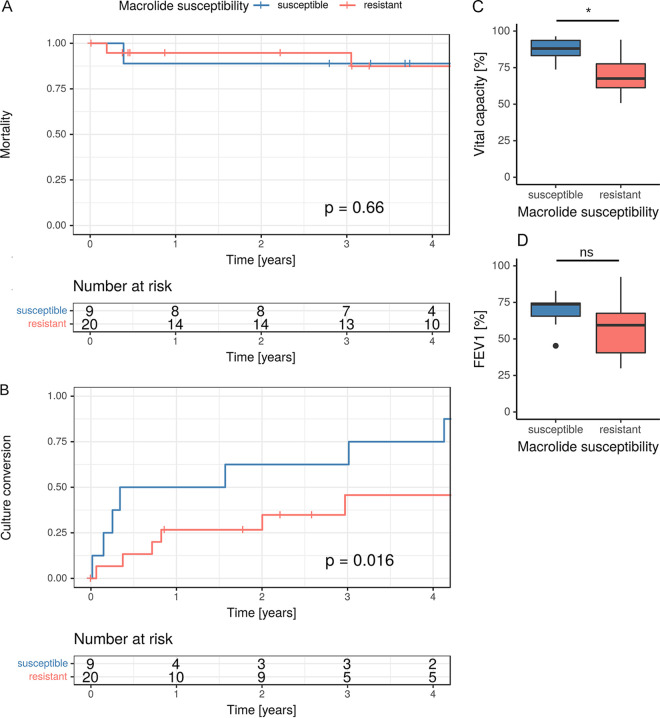
Between macrolide-resistant and macrolide-susceptible M. abscessus isolates, no significant difference in 5-year mortality was found (P = 0.66) (Fig. 2). Nevertheless, VC was significantly lower in patients who were infected or colonized with a macrolide-resistant M. abscessus isolate (68.4% versus 91.3%; P = 0.02). There was no significant difference in FEV1, but a trend toward lower FEV1s in patients with macrolide-resistant M. abscessus isolates (59.4% versus 73.7%; P = 0.09) was observed.
FIG 2.
(A) Kaplan-Meier curve showing mortality within the cohort differentiated by macrolide susceptibility in M. abscessus isolates. Mortality did not differ within the cohort (P = 0.66). (B) Kaplan-Meier curve showing culture conversion differentiated by macrolide susceptibility. Patients with macrolide-susceptible M. abscessus isolates did reach culture conversion significantly more often (P = 0.016). (C) Comparison of vital capacity in patients with macrolide-susceptible and -resistant isolates. Vital capacity was significantly impaired in patients with macrolide-resistant isolates (P = 0.015). (D) Comparison of FEV1 in patients with macrolide-susceptible and -resistant isolates. Here, differences were nonsignificant (P = 0.091).
Culture conversion was significantly more frequently reached in patients infected with a macrolide-susceptible isolate (P = 0.02). Cox regression showed no significant relationships between culture conversion and drug susceptibility to other antimycobacterial agents.
Phylogenetic analysis.
For comparison in an international context, we used all isolates from our cohort (n = 39) and 10 genome sequences of different patient isolates from each of the international described complexes, Absc. 1, Absc. 2, and Mass. 1, as well as 10 unclustered isolates of M. abscessus subsp. abscessus and M. abscessus subsp. massiliense from Bryant et al., which were randomly chosen (ENA accession code ERP001039) (14, 15).
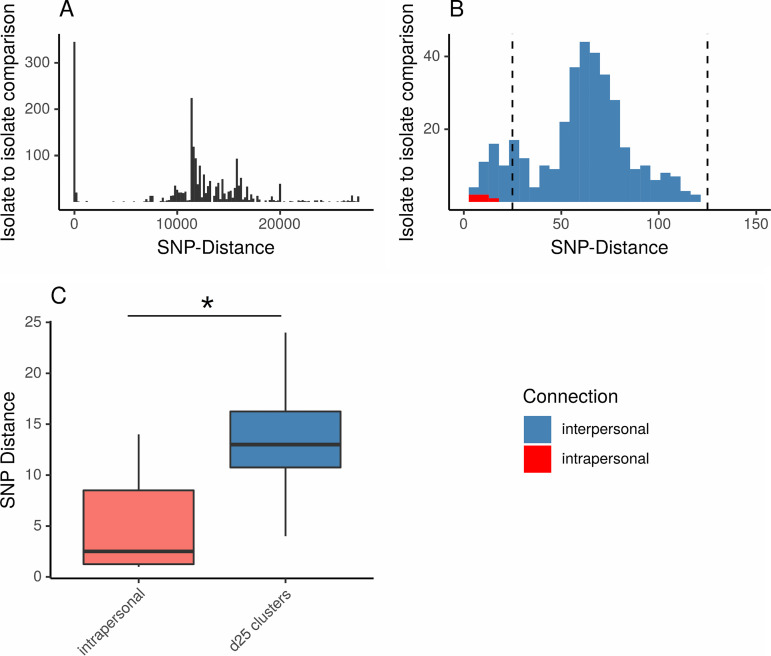
Analysis of this combined set of 89 isolates revealed two peaks in the pairwise SNP distance distribution, one below 25 and one below 125 SNPs (Fig. 3). Therefore, we chose 25 and 125 SNPs as thresholds for single linkage clustering, where SNP distances of <25 were considered indicative of recent close relationship (d25 clusters) and SNP distances of <125 indicated a closer genetic relation than the background diversity (d125 complexes). Intrapersonal isolates showed significantly smaller SNP distances than interpersonal isolates from d25 clusters (median of 2.5 versus 13 core genome SNPs; P < 0.01).
FIG 3.
(A and B) Histograms of pairwise SNP distance distribution in 89 isolates. Only pairwise SNP distances within a subspecies are shown. (A) All SNP distances. (B) Zoomed-out view of pairwise SNP distances between 0 and 150 SNPs. Horizontal lines mark 25 and 125 SNPs. (C) Box plot of pairwise SNP distances in intrapersonal isolates compared to interpersonal pairwise SNP distances within d25 clusters.
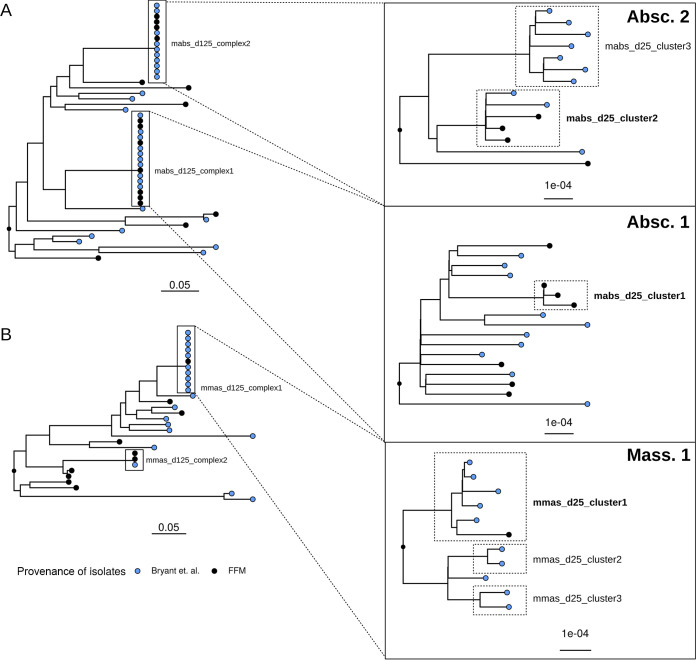
A phylogenetic tree was built, using one isolate per patient from our cohort (n = 29) and the 50 isolates from Bryant et al. as mentioned above. We identified four d125 complexes (2 in M. abscessus subsp. abscessus, 2 in M. abscessus subsp. massiliense) (Fig. 4). The first consisted of the 10 isolates from the international complex Absc. 1 and seven of our patients (mabs_d125_complex1), the second consisted of isolates from the international complex Absc. 2 and four of our patients (mabs_d125_complex2), the third consisted of isolates from the international complex Mass. 1 and one of our patients (mmas_d125_complex1), and, finally, the mmas_d125_complex2 was built from one international isolate and two from our patients. In total, representatives of all three dominant circulating clones were found in our cohort (Absc. 1, Absc. 2, and Mass. 1).
FIG 4.
Phylogenetic trees depicting 29 isolates from different patients in our cohort and 50 from Bryant et al. (14, 15). (A) Phylogenetic tree of M. abscessus subsp. abscessus isolates. Two d125 complexes with representatives of the international clusters Absc. 1 and Absc. 2 were identified. Within those, we found three d25 clusters. (B) Phylogenetic tree of M. abscessus subsp. massiliense isolates. Two d125 complexes could be identified as well, with one formed by the international Mass. 1 cluster and one of our patients. Tree scales depict SNPs/site. d25 clusters containing patients from our cohort are marked in boldface. Complexes found in our study were named in accordance with the method of detection: mabs_d125_complex1, mabs_d125_complex2, mmas_d125_complex1, and mmas_d125_complex2. FFM, Frankfurt/Main.
Within these d125 complexes, there were three M. abscessus subsp. abscessus clusters and three M. abscessus subsp. massiliense clusters with SNP distances of <25. Three of those contained isolates of patients from our center (mabs_d25_cluster_1, mabs_d25_cluster_2, and mmas_d25_cluster_1).
In the cluster analysis, discrimination resolution could be increased (Table S3 and Fig. S1). Overall, SNP distances increased with a median of 14 SNPs (minimum, 1 SNP; maximum, 44 SNPs) and a ratio of 1.59. Interestingly, in M. abscessus subsp. abscessus the SNP ratio was lower than that in M. abscessus subsp. massiliense (1.49 versus 2.36; P < 0.0001). In all clusters, more than 99% of the corresponding reference genomes were used for cluster SNP analysis (Table S3).
Network analysis.
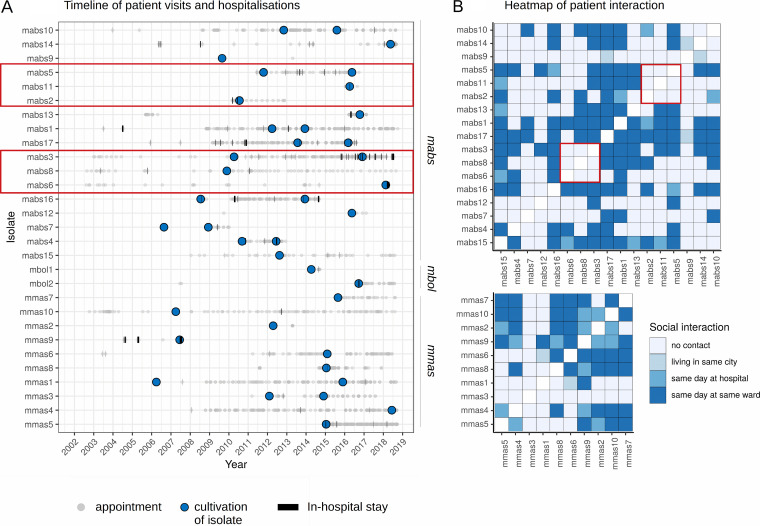
In total, the 29 patients attended 2,412 appointments and 96 stays in the hospital from June 2002 to October 2018 in our tertiary care center (Fig. 5). For 406 possible patient-to-patient combinations, 2,931,370 date-to-date combinations were analyzed. Out of those, 812 occasions on which patients attended our clinics on the same day (98 patient-to-patient combinations) were identified. After manual control of the results, 181 events in which patients were in the same ward or outpatient clinic on the same day remained (84 patient-to-patient combinations).
FIG 5.
(A) Timeline showing hospital visits and stays on wards by all patients. Gray dots represent hospital visits, time points of cultured M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense isolates are represented by blue dots, and hospital stays are represented by black bars. (B) Heatmaps showing social interaction in all patients within one subspecies. The timelines and heatmap for mabs_d25_cluster1 and mabs_d25_cluster2 are bordered in red. For those complexes, we could not identify concurrent hospital visits before cultivation of the respective isolates.
Interestingly, within the two clusters of patients containing more than one patient from our center (mabs_d25_cluster_1, mabs2, mabs5, and mabs11; mabs_d25_cluster_2, mabs3, mabs6, and mabs8), we did not identify concurrent hospital visits before the cultivation of the respective isolates (Fig. 5, red squares). Notably, in mabs_d25_cluster1 was a non-CF patient from Russia who supposedly had no social contacts with the other patients. In addition, we presume that our patients in d25 clusters never met the other patients within those international clusters.
DISCUSSION
In this study, the prediction of macrolide and aminoglycoside susceptibility by both the Genotype NTM-DR line probe assay and the mab-ariba WGS-based analysis was in full agreement with phenotypic testing methods. All mutations detected by the Genotype NTM-DR could be confirmed by WGS analysis. Therefore, we deem both methods viable options to infer resistance for those two antibiotic groups.
Even in our cohort with restricted case numbers, we could demonstrate a significant effect of macrolide resistance on culture conversion. Patients with macrolide-resistant isolates reached culture conversion less frequently than those with macrolide-susceptible isolates. This is consistent with similar studies from the past where significant differences in culture conversion between M. abscessus subsp. abscessus and M. abscessus subsp. massiliense were shown. Those were considered to be most likely related to macrolide resistance (34), as M. abscessus subsp. massiliense isolates are usually macrolide susceptible due to their truncated erm(41) gene.
One patient developed aminoglycoside resistance during the course of treatment, while the serial isolates showed little variation with regard to SNP analysis. This suggests the development of an rrs mutation leading to aminoglycoside resistance within the same bacterial population rather than reinfection with a new isolate. In total, we could not demonstrate reinfection in any of the 10 patients with a follow-up isolate. Thus, the M. abscessus population within one patient seems to be stable. On the other hand, in patients colonized by or infected with M. avium complex, reinfection was shown to be a frequent event (35, 36).
When analyzing the set of patient isolates together with representative data from a recent international study (14, 15), we detected four groups of related isolates (d125 complexes). Three of these correlate with the previously described dominant internationally circulating clones (Absc. 1, Absc. 2, and Mass. 1), with one additional d125 complex detected. Strikingly, all four d125 complexes contain isolates from our patients and from the international study. Whether the existence of those d125 complexes is caused by a global spread of dominant circulating clones, for example, via inanimate surfaces or liquids, or is inherent to the genomic structure of M. abscessus remains unclear. It is, however, quite intriguing to find representatives of all three previously described international complexes in our study collection.
Using a threshold of 25 SNPs, we identified six clusters of very closely related isolates within the d125 complexes that likely stem from a recent common ancestor. However, according to our network analysis, patients within mabs_d25_cluster1 and mabs_d25_cluster2 have never met in our hospital. In addition, we presume that patients grouping in global clusters did not have social contacts with any of the international patients. SNP distances between intrapersonal isolates were still significantly lower than those between interpersonal cluster isolates, even in cases where intrapersonal isolates were cultivated several years apart. Taking these findings together, it seems unlikely that the d25 clusters represent direct transmission events.
It is important to note that, similar to previous genomic comparisons in M. abscessus, the joint analysis of the 79 isolates was limited to the core genome. Therefore, we have also tried to increase discrimination resolution by conducting cluster-specific analyses, but cluster isolates kept close genetic relations.
Our study has several limitations. First, it is monocentric. Second, with 29 patients, the number of investigated isolates is small. Third, we sequenced at most two isolates from each patient; thus, the genetic variance over time could not be fully assessed. Fourth, we did not conduct environmental sampling in our hospital. Thus, a possible transmission in clusters by inanimate sources, such as surfaces, sinks, bronchoscopes, or even spirometer machines, independent of personal contact, cannot be fully excluded.
Conclusions.
This study delivers a first characterization of the genomic landscape of M. abscessus isolates in a German tertiary care center and sets it in an international context. In summary, we demonstrate that in the investigated M. abscessus isolates, both Genotype NTM-DR and WGS were excellent methods for the prediction of macrolide and aminoglycoside susceptibility and that macrolide resistance is a risk factor for impaired culture conversion. We identified clusters with closely related isolates and representatives of all three dominant circulating international clones, but we found no cases of likely in-hospital transmission. Whether these findings can only be related to our hospital or are applicable to the whole of Germany has to be answered in the future with more extensive studies that include M. abscessus isolates from other centers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Josephine Bryant for forwarding the strain IDs and accession codes to conduct phylogenetic analysis with isolates from the three international clusters: Absc. 1, Absc. 2, and Mass. 1. In addition, we thank K. Gebreamlack, R. Räder, N. Avemaria, V. Mohr, F. Boysen, and T. Ubben for excellent technical advice and support.
This study was partially supported by a financial grant from Mukoviszidose Institut gGmbH, Bonn, Germany, the research and development arm of the German Cystic Fibrosis Association Mukoviszidose e.V. (grant 2003), grants from the German Center for Infection Research Federal Ministry of Education and Research, Germany, and from Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany's Excellence Strategy (EXC 22167-390884018), and grants from the Leibniz Science Campus EvoLUNG. Nils Wetzstein received a research grant from the Deanery of Education and Research, University Hospital Frankfurt, Goethe University Frankfurt.
S.A., V.A.J.K., T.A.K., A.L., T.G.S., H.S., N.W., T.A.W., and T.W. have no conflict of interest to report. C.B. received research funding from Gilead Sciences outside the submitted work. M.H. received honoraria for consulting (Chiesi GmbH) and lectures (Chiesi GmbH/Horizon Pharma/Thieme Publisher Group), all outside the submitted work. C.H. has received speaker honoraria from Boehringer Ingelheim and PulmonX, reimbursement of congress fees and travel expenses as a speaker honorarium from Muko e.V., and reimbursement of congress fees and travel expenses from Novartis, Gilead, and Insmed, all outside the submitted work. F.P.M. reports grant support by the Joachim Herz Foundation during the study. S.N. reports grants from the German Center for Infection Research, grants from Excellenz Cluster Precision Medicine in Chronic Inflammation EXC 2167, and grants from Leibniz Science Campus Evolutionary Medicine of the LUNG (EvoLUNG) during the conduct of the study. M.J.G.T.V. has served at the speakers' bureau of Akademie für Infektionsmedizin, Ärztekammer Nordrhein, Astellas Pharma, Basilea, Gilead Sciences, Merck/MSD, Organobalance, and Pfizer, received research funding from 3M, Deutsches Zentrum für Infektionsforschung, IMI, Evonik, Glycom, Astellas Pharma, DaVolterra, Gilead Sciences, MaaT Pharma, Merck/MSD, Morphochem, Organobalance, Seres Therapeutics, and Aredypharm, and is a consultant to Alb-Fils Kliniken GmbH, Arderypharm, Astellas Pharma, Ferring, DaVolterra, MaaT Pharma, and Merck/MSD, all outside the submitted work.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 2.Swenson C, Zerbe CS, Fennelly K. 2018. Host variability in NTM disease: implications for research needs. Front Microbiol 9:2901. doi: 10.3389/fmicb.2018.02901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurer FP, Castelberg C, von Braun A, Wolfensberger A, Bloemberg GV, Böttger EC, Somoskovi A. 2014. Postsurgical wound infections due to rapidly growing mycobacteria in Swiss medical tourists following cosmetic surgery in Latin America between 2012 and 2014. Eurosurveillance 19:20905. doi: 10.2807/1560-7917.ES2014.19.37.20905. [DOI] [PubMed] [Google Scholar]
- 4.Henkle E, Winthrop K. 2015. Nontuberculous mycobacteria infections in immunosuppressed hosts. Clin Chest Med 36:91–99. doi: 10.1016/j.ccm.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson RM, Hasan NA, de Moura VCN, Duarte RS, Jackson M, Strong M. 2013. Phylogenomics of Brazilian epidemic isolates of Mycobacterium abscessus subsp. bolletii reveals relationships of global outbreak strains. Infect Genet Evol 20:292–297. doi: 10.1016/j.meegid.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnabel D, Gaines J, Nguyen DB, Esposito DH, Ridpath A, Yacisin K, Poy JA, Mullins J, Burns R, Lijewski V, McElroy NP, Ahmad N, Harrison C, Parinelli EJ, Beaudoin AL, Posivak-Khouly L, Pritchard S, Jensen BJ, Toney NC, Moulton-Meissner HA, Nyangoma EN, Barry AM, Feldman KA, Blythe D, Perz JF, Morgan OW, Kozarsky P, Brunette GW, Sotir M, Centers for Disease Control and Prevention. 2014. Notes from the field: rapidly growing nontuberculous Mycobacterium wound infections among medical tourists undergoing cosmetic surgeries in the Dominican Republic–multiple states, March 2013-February 2014. MMWR Morb Mortal Wkly Rep 63:201–202. [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte RS, Lourenço MCS, Fonseca LDS, Leão SC, Amorim EDLT, Rocha ILL, Coelho FS, Viana-Niero C, Gomes KM, da Silva MG, Lorena NSDO, Pitombo MB, Ferreira RMC, Garcia MHDO, de Oliveira GP, Lupi O, Vilaça BR, Serradas LR, Chebabo A, Marques EA, Teixeira LM, Dalcolmo M, Senna SG, Sampaio JLM. 2009. Epidemic of postsurgical infections caused by Mycobacterium massiliense. J Clin Microbiol 47:2149–2155. doi: 10.1128/JCM.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tortoli E, Kohl TA, Brown-Elliott BA, Trovato A, Leão SC, Garcia MJ, Vasireddy S, Turenne CY, Griffith DE, Philley JV, Baldan R, Campana S, Cariani L, Colombo C, Taccetti G, Teri A, Niemann S, Wallace RJ, Cirillo DM. 2016. Emended description of Mycobacterium abscessus Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii and designation of Mycobacterium abscessus subsp. massiliense comb. nov. Int J Syst Evol Microbiol 66:4471–4479. doi: 10.1099/ijsem.0.001376. [DOI] [PubMed] [Google Scholar]
- 9.van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. 2012. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat 15:149–161. doi: 10.1016/j.drup.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maurer FP, Castelberg C, Quiblier C, Bottger EC, Somoskovi A. 2014. Erm(41)-dependent inducible resistance to azithromycin and clarithromycin in clinical isolates of Mycobacterium abscessus. J Antimicrob Chemother 69:1559–1563. doi: 10.1093/jac/dku007. [DOI] [PubMed] [Google Scholar]
- 12.Prammananan T, Sander P, Brown BA, Frischkorn K, Onyi GO, Zhang Y, Böttger EC, Wallace RJ Jr.. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2‐deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis 177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 13.Hain Life Science. 2017. GenoType NTM-DR. Detection of NTM resistances. https://www.hain-lifescience.de/en/products/microbiology/mycobacteria/ntm/genotype-ntm-dr.html. Accessed 8 February 2018.
- 14.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, Esther CR, Noone PG, Giddings O, Bell SC, Thomson R, Wainwright CE, Coulter C, Pandey S, Wood ME, Stockwell RE, Ramsay KA, Sherrard LJ, Kidd TJ, Jabbour N, Johnson GR, Knibbs LD, Morawska L, Sly PD, Jones A, Bilton D, Laurenson I, Ruddy M, Bourke S, Bowler IC, Chapman SJ, Clayton A, Cullen M, Daniels T, Dempsey O, Denton M, Desai M, Drew RJ, Edenborough F, Evans J, Folb J, Humphrey H, Isalska B, Jensen-Fangel S, Jönsson B, Jones AM, Katzenstein TL, Lillebaek T, et al. . 2016. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tettelin H, Davidson RM, Agrawal S, Aitken ML, Shallom S, Hasan NA, Strong M, Calado Nogueira de Moura V, De Groote MA, Duarte RS, Hine E, Parankush S, Su Q, Daugherty SC, Fraser CM, Brown-Elliott BA, Wallace RJ, Holland SM, Sampaio EP, Olivier KN, Jackson M, Zelazny AM. 2014. High-level relatedness among Mycobacterium abscessus subsp. massiliense strains from widely separated outbreaks. Emerg Infect Dis 20:364–371. doi: 10.3201/eid2003.131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tortoli E, Kohl TA, Trovato A, Baldan R, Campana S, Cariani L, Colombo C, Costa D, Cristadoro S, Di Serio MC, Manca A, Pizzamiglio G, Rancoita PMV, Rossolini GM, Taccetti G, Teri A, Niemann S, Cirillo DM. 2017. Mycobacterium abscessus in patients with cystic fibrosis: low impact of inter-human transmission in Italy. Eur Respir J 50:1602525. doi: 10.1183/13993003.02525-2016. [DOI] [PubMed] [Google Scholar]
- 18.Harris KA, Underwood A, Kenna DTD, Brooks A, Kavaliunaite E, Kapatai G, Tewolde R, Aurora P, Dixon G. 2015. Whole-genome sequencing and epidemiological analysis do not provide evidence for cross-transmission of Mycobacterium abscessus in a cohort of pediatric cystic fibrosis patients. Clin Infect Dis 60:1007–1016. doi: 10.1093/cid/ciu967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson RM. 2018. A closer look at the genomic variation of geographically diverse Mycobacterium abscessus clones that cause human infection and disease. Front Microbiol 9:2988. doi: 10.3389/fmicb.2018.02988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical Laboratory Standards Institute. 2018. Susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes, 3rd ed. Document no. M24 Clinical Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 21.Hain Life Science. 2017. GenoLyse. Extraction of genomic bacterial DNA. Available at: https://www.hain-lifescience.de/en/products/dna-isolation/genolyse.html. Accessed 2 July 2019.
- 22.R Core Team. 2018. R: a language and environment for statistical computing. https://www.r-project.org/.
- 23.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY: https://ggplot2.tidyverse.org. [Google Scholar]
- 24.Therneau TM. 2015. A package for survival analysis in S. https://cran.r-project.org/package=survival.
- 25.Kassambara A, Kosinski M, Biecek P. 2019. survminer: drawing survival curves using ggplot2. https://cran.r-project.org/package=survminer.
- 26.de Almeida IN, da Carvalho WS, Rossetti ML, Costa ERD, de Miranda SS. 2013. Evaluation of six different DNA extraction methods for detection of Mycobacterium tuberculosis by means of PCR-IS6110: preliminary study. BMC Res Notes 6:561. doi: 10.1186/1756-0500-6-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipworth S, Hough N, Buchanan R, Smith EG, Robinson E, Alexander E, Peto T, Crook D, Walker T. 2019. Improved performance predicting clarithromycin resistance in Mycobacterium abscessus on an independent dataset. Antimicrob Agents Chemother 63:e00400-19. doi: 10.1128/AAC.00400-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohl TA, Utpatel C, Schleusener V, De Filippo MR, Beckert P, Cirillo DM, Niemann S. 2018. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 6:e5895. doi: 10.7717/peerj.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G, Smith D, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 34.Park J, Cho J, Lee C-H, Han SK, Yim J-J. 2017. Progression and treatment outcomes of lung disease caused by Mycobacterium abscessus and Mycobacterium massiliense. Clin Infect Dis 64:301–308. doi: 10.1093/cid/ciw723. [DOI] [PubMed] [Google Scholar]
- 35.Jhun BW, Kim S-Y, Moon SM, Jeon K, Kwon OJ, Huh HJ, Ki C-S, Lee NY, Shin SJ, Daley CL, Koh W-J. 2018. Development of macrolide resistance and reinfection in refractory Mycobacterium avium complex lung disease. Am J Respir Crit Care Med 198:1322–1330. doi: 10.1164/rccm.201802-0321OC. [DOI] [PubMed] [Google Scholar]
- 36.Wetzstein N, Kohl TA, Andres S, Schultze TG, Geil A, Kim E, Biciusca T, Hügel C, Hogardt M, Lehn A, Vehreschild MJGT, Wolf T, Niemann S, Maurer FP, Wichelhaus TA. 2020. Comparative analysis of phenotypic and genotypic antibiotic susceptibility patterns in Mycobacterium avium complex. Int J Infect Dis 93:320–328. doi: 10.1016/j.ijid.2020.02.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences determined in the course of this work were deposited in the European Nucleotide Archive (ENA) (see Table S1 in the supplemental material).