Fungal infections are being caused by a broadening spectrum of fungi, yet in many cases, identification to the species level is required for proper antifungal selection. We investigated the fungal intergenic spacer (IGS) sequence in combination with nanopore sequencing for fungal identification. We sequenced isolates from two Cryptococcus species complexes, C. gattii and C. neoformans, which are the main pathogenic members of this genus, using the Oxford Nanopore Technologies MinION device and Sanger sequencing.
KEYWORDS: DNA sequencing, diagnostic, fungal
ABSTRACT
Fungal infections are being caused by a broadening spectrum of fungi, yet in many cases, identification to the species level is required for proper antifungal selection. We investigated the fungal intergenic spacer (IGS) sequence in combination with nanopore sequencing for fungal identification. We sequenced isolates from two Cryptococcus species complexes, C. gattii and C. neoformans, which are the main pathogenic members of this genus, using the Oxford Nanopore Technologies MinION device and Sanger sequencing. There is enough variation within the two complexes to argue for further resolution into separate species, which we wanted to see if nanopore sequencing could detect. Using the R9.4.1 flow cell, IGS sequence identities averaged 99.57% compared to Sanger sequences of the same region. When the newer R10.3 flow cell was used, accuracy increased to 99.83% identity compared to the same Sanger sequences. Nanopore sequencing errors were predominantly in regions of homopolymers, with G homopolymers displaying the largest number of errors and C homopolymers displaying the least. Phylogenetic analysis of the nanopore- and Sanger-derived sequences resulted in indistinguishable trees. Comparison of average percent identities between the C. gattii and C. neoformans species complexes resulted in only a 74 to 77% identity between the two complexes. Sequencing using the nanopore platform could be completed in less than an hour, and samples could be multiplexed in groups as large as 24 sequences in a single run. These results suggest that sequencing the IGS region using nanopore sequencing could be a potential new molecular diagnostic strategy.
INTRODUCTION
Fungal infections, particularly systemic mycoses, continue to be a major health care challenge. In 2017, the estimated cost of fungal infections in the United States was $7.2 billion, and there were more than 75,000 hospitalizations (1). Globally, there are an estimated 1.7 billion people infected, and 15 to 30% of the cases are serious (2–4). These infections result in approximately 1.6 million annual deaths, a rate equivalent to that of tuberculosis and more than that of malaria (5). One of the most important tools for reducing fungal morbidity and mortality is accurate and rapid diagnosis. In fact, based on autopsy reports, it has been estimated that 50% of fungal infections may be undiagnosed (6).
Although the literature typically cites 300 to 500 species as pathogenic for humans (7–10), this number is an underestimation, based on case reports alone. Since 1995, the number of new fungal pathogens of animals, plants, and humans has increased almost 10-fold (11). Our own diagnostic program has found more than 1,500 unique species capable of infecting humans (12). Most of these species are opportunistic pathogens that take advantage of immunosuppressed hosts, with most of them rarely, if ever, seen by the majority of clinical microbiology laboratories. Because of the ability of modern medicine to sustain increasingly sicker patients, the continued expansion of the number of fungal species capable of causing infection is to be expected. This expansion will necessitate new fungal diagnostic strategies to identify these organisms. However, a confounding problem with this need is that because so many human fungal pathogens are so rarely encountered in clinical specimens, it is not possible or financially worthwhile to include them in assays requiring specific analytes to be targeted with specific probes, antibodies, etc., for each potential species. Consequently, virtually all FDA-cleared fungal diagnostic assays focus on a single fungal pathogen or a very limited scope of the most common fungal pathogens (12), with most assays directed toward a few Candida species. The reasons are both epidemiological and economic. Candida spp. are among the most common causes of systemic mycoses, and because getting a diagnostic assay all the way through FDA clearance is so challenging, it is difficult to incentivize the development of assays that target organisms that may be only rarely, if ever, encountered. However, when viewed in regard to the morbidity and mortality costs of undiagnosed, slowly diagnosed, or misdiagnosed infections, there is a strong argument for developing assays that are panfungal. This strategy would allow an assay to identify the fungi commonly encountered in the clinic but would also enable the identification of rare fungal pathogens.
Assay development arguably requires choosing one of two paths: an assay that targets one or a few fungi or an assay that is potentially panfungal. Targeted assays require two levels of suspicion before deployment; a patient has a fungal infection, and the infecting agent can be identified by the targeted assay. Panfungal assays require only a suspicion that an infection is fungal in nature since the assay is “one size fits all.” However, panfungal assays are dependent on having the identity of the infecting fungus in the assay database, which is interrogated by the assay output. Matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) is one potentially panfungal assay; however, this assay requires a biocurated database, which is attached to the instrument by the vendor and is proprietary, although it is possible for laboratories to add new species to research-use-only (RUO) databases under certain conditions. Sequenced-based identification, because of the vast amount of data in public databases (more than 500,000 sequence records from more than 5,000 fungi [12]), is generally unlimited in species scope since sequences from even newly discovered species are quickly added to GenBank as part of the formal species description (13).
DNA sequencing, ribosomal DNA (rDNA) sequencing in particular, is the gold standard of fungal molecular identification for many reasons. First, fungal ribosomal genes are multicopy in nature, which increases the sensitivity of detection by PCR. Second, the organization of these loci in fungi places multiple conserved ribosomal subunit genes (18S, 5.8S, and 28S) in close proximity, which offers conserved PCR primer sites that span variable regions. In fact, the conserved nature of the subunits and their primer annealing sites enables virtually any unknown fungus to be amplified with universal primers targeted to these regions (14, 15). Third, the overall organization of this region confers variability due to the fact that the informative regions, the internal transcribed spacer (ITS) regions (comprised of ITS1 and ITS2), are short enough to be covered in a single Sanger sequencing run. For this reason, the ITS region serves as the universal barcode for fungi (16). A third variable region, called the D1/D2 region (∼700 bp in length), exists within the large 28S ribosomal subunit and was commercialized as a diagnostic kit (MicroSEQ; Thermo Fisher, Waltham, MA) many years ago, but it is not widely used due to lack of sensitivity. The fourth variable region is the intergenic spacer (IGS) sequence (made up of IGS1 and IGS2), which is not transcribed and is the most variable region within the ribosomal repeat (∼3 kb to 8 kb in length) (17). However, the region is too long to sequence quickly and efficiently using Sanger sequencing. Finally, sequence data can be used to search public databases, such as GenBank, using the Web-based BLASTn algorithm (18). The breadth and depth of these data are unmatched as a diagnostic tool. However, sequence instrumentation is expensive, and the procedure requires specific expertise and takes a day or longer to obtain results. Furthermore, while the ITS region can be species specific for many fungi, other fungi, including Aspergillus, Penicillium, Alternaria, and Fusarium species, require additional sequences from other genes with greater specificity, which may delay results substantially, particularly if the first sequence identity is required for selection of the second sequence. Consequently, while reference or research laboratories with the proper expertise can identify virtually any fungus by sequencing, this arrangement is suboptimal or not possible in most clinical microbiology laboratories. If throughput is low, purchase and maintenance of a sequencing platform may not be justified no matter how well it is operated. Therefore, while DNA sequencing arguably has the greatest potential for specificity, crucial assay factors such as turnaround time, cost, and expertise are the main barriers blocking it from being a viable option as the main fungal diagnostic platform.
To overcome these hurdles, another jump in sequencing technology was needed. This jump came in the recent development of fourth-generation sequencing technology called nanopore sequencing. Nanopore sequencing technology generates sequences based on detecting changes in electrical conductivity which occur as DNA molecules are threaded through a biological pore that is embedded in a solid-state film (19). While the basic technology has been known for more than 20 years, commercial platforms were only recently developed by Pacific Biosciences Inc. and Helicos Biosciences. Subsequently, Oxford Nanopore Technologies (Oxford, UK) introduced their commercial version of this technology in 2015 as a small desktop device (20). This technology has numerous advantages over first (Maxam-Gilbert)-, second (Sanger)-, and third (next generation)-generation methods. Depending on the application, there is no library preparation required, reads can be a megabase in length (21), and unique technician skills are not needed. Importantly, depending on the platform, sequencing can be done on the benchtop and data can be captured and analyzed with a laptop.
In this study, we used nanopore sequencing to obtain complete IGS sequences from multiple genotypes of Cryptococcus neoformans and Cryptococcus gattii and compared these sequences to Sanger sequences of the same regions. Samples could be multiplexed using barcoding and sequencing completed in less than an hour, depending on the number of samples. Nanopore-derived sequences were, on average, ≥99.8% identical to Sanger-derived sequences of the same region. These results suggest that nanopore sequencing of the IGS region could be a viable fungal identification strategy.
MATERIALS AND METHODS
Strains and media.
YPD consisted of 1% yeast extract, 2% peptone, and 2% dextrose and was solidified with 2% agar (all obtained from Thermo Fisher Scientific, Waltham, MA) as needed. C. neoformans and C. gattii strains were chosen from the Wickes culture collection and were stored at −70°C in 15% glycerol–YPD. Genotype reference cultures (WM prefix) were supplied by June Kwon-Chung (Table 1). Hybrids, such as serotype AD, were excluded from the study. Strains were revived from −70°C by plating onto YPD plates and incubating at 30°C for 1 to 2 days. Individual colonies from each strain were then subcultured onto a new YPD plate and grown for 24 h at 30°C prior to nucleic acid extraction.
TABLE 1.
Strains
| Strain | Alias | Species | Serotype | Genotype | Contributor |
|---|---|---|---|---|---|
| W10 | NIH B-4508 | C. gattii | B | VGI | Jeffrey Edman |
| W85 | NIH 435 | C. gattii | B | VGI | K. J. Kwon-Chung |
| W3482 | WM 179 | C. gattii | B | VGI | Weiland Meyer |
| W432 | USC#1014 | C. gattii (C. deuterogattii) | B | VGII | Vishnu Chaturvedi |
| W530 | Thai 37-141-141 | C. gattii (C. deuterogattii) | B | VGII | Natteewan Poonam |
| W3481 | WM 178 | C. gattii (C. deuterogattii) | B | VGII | Weiland Meyer |
| W84 | NIH 198 | C. gattii (C. bacillisporus) | B | VGIII | K. J. Kwon-Chung |
| W83 | NIH 189 | C. gattii (C. bacillisporus) | B | VGIII | K. J. Kwon-Chung |
| W15 | NIH 34 | C. gattii (C. bacillisporus) | C | VGIII | Jeffrey Edman |
| W16 | NIH 191 | C. gattii (C. bacillisporus) | C | VGIII | Jeffrey Edman |
| W87 | NIH 312 | C. gattii (C. bacillisporus) | C | VGIII | K. J. Kwon-Chung |
| W89 | NIH 139 | C. gattii (C. bacillisporus) | C | VGIII | K. J. Kwon-Chung |
| W90 | NIH 113 | C. gattii (C. bacillisporus) | C | VGIII | K. J. Kwon-Chung |
| W3479 | WM 161 | C. gattii (C. bacillisporus) | C | VGIII | Weiland Meyer |
| W3480 | WM 779 | C. gattii (C. tetragattii) | C | VGIV | Weiland Meyer |
| W393 | H99 | C. neoformans var. grubii | A | VNI | John Perfect |
| W72 | NIH 288 | C. neoformans var. grubii | A | VNI | K. J. Kwon-Chung |
| W473 | IUM 96-2828 | C. neoformans var. grubii | A | VNI | Marianna Vivianni |
| W516 | IFM-46660 | C. neoformans var. grubii | A | VNI | Reiko Tanaka |
| W1047 | IUM 993617-3 | C. neoformans var. grubii | A | VNI | Marianna Vivianni |
| W3477 | WM 148 | C. neoformans var. grubii | A | VNI | Weiland Meyer |
| W3478 | WM 626 | C. neoformans var. grubii | A | VNII | Weiland Meyer |
| W21 | JEC21 | C. neoformans var. neoformans (C. deneoformans) | D | VNIV | Jeff Edman |
| W77 | NIH 433 | C. neoformans var. neoformans (C. deneoformans) | D | VNIV | K. J. Kwon-Chung |
| W78 | NIH 430 | C. neoformans var. neoformans (C. deneoformans) | D | VNIV | K. J. Kwon-Chung |
| W79 | NIH 52 | C. neoformans var. neoformans (C. deneoformans) | D | VNIV | K. J. Kwon-Chung |
| W3484 | WM 629 | C. neoformans var. neoformans (C. deneoformans) | D | VNIV | Weiland Meyer |
DNA extraction.
DNA extraction was done by subculturing cells from a 2- to 3-day-old YPD culture onto fresh YPD and incubating at 30°C for 20 h. Approximately 107 cells were removed from the subculture and prepared for DNA extraction as previously described (22).
Primers.
PCR and sequencing primers were obtained from Eurofins Genomics (Louisville, KY) and are shown in Table 2. Sequences of primers that were used in sequencing reactions (P55 to P122) were stored as MacVector files (MacVector, Inc., Apex, NC) and placed into their own folder, which was used by the MacVector software for mapping primer walks.
TABLE 2.
Primers
| Name | Sequence |
|---|---|
| P1 | 5'-GCTGGGGCGGCACATCTGTT-3' |
| P2 | 5'-TGAGCCATTCGCAGTTTCACAGT-3' |
| URA5.F | 5'-ATGTCCTCCCAGCCCTCGACTCCG-3' |
| URS5.R | 5'-TTAAGACCTCTGAACACCGTACTC-3' |
| P55 | 5'-CTGTCTCACGACGGTCTAAACC-3' |
| P56 | 5'-GGGCTTTACGCATTCGCATAAC-3' |
| P57 | 5'-ACACTGCCAACTTGCGATGG-3' |
| P58 | 5'-GTCGTGGGGGACTTTGGTAATG-3' |
| P59 | 5'-CCCACAATGAGCAAGAGAAGTGAC-3' |
| P60 | 5'-GACCTTGGGTGACAAAAAATCGG-3' |
| P61 | 5'-TAGCCTTCATACAGCACCCTGC-3' |
| P62 | 5'-GGTGCTGTATGAAGGCTATGGC-3' |
| P63 | 5'-TTTGGTCTACTGGACTTGCGTC-3' |
| P64 | 5'-CGTGGTAGGCTCAAACACTCTC-3' |
| P65 | 5'-GAGAGAGTGTTTGAGCCTACCAC-3' |
| P66 | 5'-CTTCTCTGAAAACACTTGGAGG-3' |
| P67 | 5'-CGCACCTCCAAGTGTTTTCAG-3' |
| P68 | 5'-CGGATTACTTTTCGTAACGCC-3' |
| P69 | 5'-CAGAACAAGACAAGTAGGGAAG-3' |
| P70 | 5'-AACAAGGGCTTAGCCTCAG-3' |
| P71 | 5'-TTTTACCCTACTGATGGAGTGTCG-3' |
| P72 | 5'-TGCCTGTCTTTCTAGCTGGGTG-3' |
| P73 | 5'-CAATCACCAAGAATTGCCCGAG-3' |
| P74 | 5'-ACACACAGTCTCATCAGTCTTCAG-3' |
| P75 | 5'-GAAAGAATGTTTGAGCCTACCACG-3' |
| P76 | 5'-CTACCAAAGTCCCCCACGAC-3' |
| P77 | 5'-TGGTTCACAGCCGAAGCC-3' |
| P78 | 5'-GCTCATTGTGGGTCCAGTCTTC-3' |
| P79 | 5'-CTCACATCACATACTCACCTGGG-3' |
| P80 | 5'-TGTGCTAAGTTGAGTTGAAACGC-3' |
| P81 | 5'-ATAGGCTTCGGCTGTGAACC-3' |
| P82 | 5'-CTTTCTTACAAACTCGGATGGTGC-3' |
| P83 | 5'-TGACTTAGAGGCGTTCAGCC-3' |
| P84 | 5'-GCAAGATCCACTGGCTTATAGTGTC-3' |
| P85 | 5'-TGTGCGGGACCAAAATCGTC-3' |
| P86 | 5'-ATCAGTCCGTCATTTCAGC-3' |
| P87 | 5'-GCAAATGAACAACCTAGCG-3' |
| P88 | 5'-GTAAGTAGGCTCTGAATGACGGG-3' |
| P89 | 5'-ATCCACTGAGGCTAAGCCCTTG-3' |
| P90 | 5'-TCACCCTTGGGTCAATTATTCCTC-3' |
| P91 | 5'-TTATCGCAAGTTGGGCAG-3' |
| P92 | 5'-GATCTTTCAAACTGGACATGCTGC-3' |
| P93 | 5'-TACGGGTACTAGAGACCACTTGGC-3' |
| P94 | 5'-CGCAACATGGTTCTCGATCAGG-3' |
| P95 | 5'-TCCGATCTGCGAAGTCAAGC-3' |
| P96 | 5'-ACGTTTGCTTGACCAGCCTATTAG-3' |
| P97 | 5'-GGATTCAGCGTGTTCTTTGGC-3' |
| P98 | 5'-AATTACCAGCCCGACCTCTCTC-3' |
| P99 | 5'-TGCAGAAAGGGTGAGAAGAACG-3' |
| P100 | 5'-CATTCATTTCGCCCAAGTCCAC-3' |
| P101 | 5'-GCTCAAGTACGAGGAGCAGTAG-3' |
| P102 | 5'-TGAAAACTGGACCCACAGTGG-3' |
| P103 | 5'-TCTGGCGTATGATAGCTTCGC-3' |
| P104 | 5'-CCGCATTGCCAACTACATGAAAG-3' |
| P105 | 5'-GGTGTCTCCTTCATTCCCTTTTC-3' |
| P106 | 5'-ACGTGGTAGGCTCAAACACTC-3' |
| P107 | 5'-AATCAGTAGTATGCCCAGTGCAG-3' |
| P108 | 5'-TCCCATTCAAGTCGTCTGC-3' |
| P109 | 5'-CTTCCTCCTCCCTTTCATTC-3' |
| P110 | 5'-CACAGGGATAACTGGCTTG-3' |
| P111 | 5'-GGATGGATGGAAGAGAAGC-3' |
| P112 | 5'-CCCCCACGACCATAAAATC-3' |
| P113 | 5'-GACTTACCTTTTCTTCCTCCTCCC-3' |
| P114 | 5'-TGCGGCAAGTAGAGTCAACCAG-3' |
| P115 | 5'-TATCCAAACGGGCAAAGGCG-3' |
| P116 | 5'-GACACCGCCCCAACTTTTTG-3' |
| P117 | 5'-ATGGGGGACTTTGATAGTGGTTG-3' |
| P118 | 5'-TTGGCTACTGGGTGCTTGTGTTGC-3' |
| P119 | 5'-AGCTGAAGACTGATGAGACTGTG-3' |
| P120 | 5'-AAATGTGGTATGGATGGTGAGAGG-3' |
| P121 | 5'-CAGTCTGCAATGTTGGAAAAGTGG-3' |
| P122 | 5'-TCCACTGTGGCTCTGATACCAG-3' |
PCR.
The IGS region was prepared for sequencing as a PCR product using 1 μl of extracted DNA as the PCR template. The PCR mixture consisted of 12.5 μl of 2× KOD Xtreme Buffer, 5.0 μl of 2 μM deoxynucleoside triphosphate (dNTP) mix, 4.5 μl of H2O, 0.75 μl of both primers (primers P1 and P2, 10 mM stock), and 0.5 μl of KOD Xtreme HotStart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO). Initial denaturation was done at 94°C for 2 min, followed by 36 cycles of denaturation at 98°C for 1 s, annealing at 60°C for 10 s, and amplification at 68°C for 4 min and a final extension at 68°C for 2 min in a SimpliAmp thermal cycler (Thermo Fisher Scientific). The PCR amplicons were electrophoresed on a 1% agarose gel (Bio-Rad Laboratories, Inc., Hercules, CA) in Tris-borate-EDTA (TBE) buffer to confirm amplification and then purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany). DNA concentrations were determined with a NanoDrop (Thermo Fisher).
Genotyping was performed with the URA5 gene sequence (23). Reaction mixtures contained 1 μl of template DNA, 2.5 μl of 10× buffer, 2.0 μl of 2.5 mM dNTP mix, 16.0 μl of H2O, 1.0 μl of both primers (URA5.F and URA5.R, 10 mM stock), 1 μl of dimethyl sulfoxide, and 0.5 μl EconoTaq DNA polymerase (Lucigen, Middleton, WI). Initial denaturation was done at 94°C for 2 min 20 s, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 61°C for 30 s, and amplification at 72°C for 45 s and a final extension at 72°C for 5 min. The PCR amplicons were electrophoresed on a 1% agarose gel in TBE buffer to check for amplification and then purified and sequenced as described above or digested with Sau96I and HhaI (New England Biolabs, Beverly, MA). Sequences were analyzed by MacVector to place Sau96I and HhaI restriction enzyme sites or run on a 2% Nusieve gel (Lonza Group, Ltd., Basel, Switzerland) and were also used in BLASTn searches to determine genotype (18). The sizes of the internal fragments after mapping or restriction enzyme digestion were noted for genotype pattern. Internal fragments were defined as fragments produced by two or more cuts which were flanked by a restriction site. The flanking fragments were ignored because they contained one restriction site and the end of the amplicon, which could vary in size if sequencing did not proceed all the way through the primer. With this strategy, sequencing with only a single forward or reverse primer, which will miss some of the amplicon ends, still can be used to genotype. For example, an amplicon that has two Sau96I sites would result in three fragments after restriction enzyme digestion, but only the Sau96I-Sau96I fragment size was noted.
Sanger sequencing.
Sanger sequencing of the IGS region was done by primer walking. Sequencing primers were selected with MacVector software from the primer folder using the Align To Folder command. The software maps all primers in the primer folder with identity to the target sequence in both the forward and reverse orientations. Sequencing primers that were 600 to 800 bp apart were selected and used to obtain double-stranded sequences of PCR templates covering the entire IGS region, including primers. After the initial sequencing runs with the two primers used for amplification (P1 and P2), new primers were designed using MacVector, if needed, to extend the sequence until it was double stranded or to close gaps and then added to the primer folder. Sequencing was performed by Eurofins Genomics (Louisville, KY). Sequencing of the URA5 gene was done using the same primers used to amplify the template, to yield a double-stranded sequence.
Nanopore sequencing.
To prepare our IGS PCR amplicons for nanopore sequencing (Oxford Nanopore Technologies [ONT], Oxford, UK), we followed the Native Barcode Expansion 1–12 and 13–24 protocols (EXP-NBD104 and EXP-NBD114 kits; ONT). Individual IGS amplicons were adjusted to 440 ng of template DNA in 48 μl of distilled water (dH2O). The manufacturer’s instructions for end repair using FFPE DNA repair and Ultra II End preparation enzyme mixes (New England Biolabs) were followed. After cleanup, samples were quantified with the QuantiFluor One double-stranded DNA (dsDNA) system (Promega, Madison, WI) and adjusted to 220 ng in 22.5 μl of dH2O. Each sample was then barcoded using the Native Barcode Expansion kits. After cleanup, samples were again quantified and pooled in equimolar amounts for a total of 400 ng in 65 μl. Adapter ligation was performed using the ligation sequencing kit (SQK-LSK109) with adapter mix II (from the Native Barcode Expansion kit) (ONT). The NEBNext quick ligation kit (New England Biolabs) was replaced with T4 ligase and buffer (New England Biolabs) for this step. Ligation reaction mixtures were incubated for 20 min instead of 10 min at room temperature, and then, without heat killing, Ampure XP beads (Beckman Coulter, Indianapolis, IN) were added to clean samples. After a 5-min incubation, the beads were washed twice with long-fragment buffer (from the ligation sequencing kit) (ONT). The samples were then resuspended in 15 μl elution buffer, incubated at 37°C for 10 min, and recovered after removal of beads. The samples were quantified a final time with QuantiFluor dye.
The MinION sequencer (ONT) loaded with an R9.4.1 or R10.3 flow cell was connected to a MacBook Pro 2018 laptop with 1 Tb of memory (Apple, Cupertino, CA) running MinKNOW software version 19.12.5 (ONT). After hardware and flow cell checks, the kit type was selected (SQK-LSK109) along with the corresponding barcode kits (EXP-NBD104 and/or EXP-NBD114), high-accuracy basecalling, output format (FASTq, with FAST5 if additional basecalling was done later), and 10,000 reads per file. The flow cell was prepared using the flow cell priming kit according to manufacturer’s instructions (ONT). Twelve microliters of the sample pool containing 100 ng of the barcode pool was prepared for loading by mixing with sequencing buffer and loading beads from the ligation sequencing kit (ONT). The sample was then loaded according to the manufacturer’s instructions. Once started, total barcode reads and individual barcode read numbers were monitored during the run to guide run duration. The sequencing run was stopped after ∼35 to 200 Mb of total bases were collected, depending on the number of barcoded samples present in the run, to yield ∼700 to 1,700 reads per barcode. After the sequencing run was complete, the flow cell was cleaned using the flow cell wash kit (EXP-WSH003) according to manufacturer’s instructions (ONT) and stored at 4°C. The MinION device was disconnected from the computer, and basecalling was allowed to continue if needed.
Data analysis.
At the completion of basecalling, edited and trimmed sequences were distributed by the software into individual pass, fail, and unclassified read folders as FASTq and FAST5 files (if selected), with each folder labeled according to barcode (e.g., Barcode01, Barcode02, etc.) by the software. A Perl script was used to produce consensus sequences from basecalled FASTq files in pass folders using CANU (24), which is a single-molecule sequence assembly program for data generated by nanopore platforms. The script was deposited at Github and is available at <https://github.com/embunnik/nanopore-amplicon-consensus>. In order to set up and run the program, nanopore sequence data from demultiplexed FASTq pass folders labeled Barcode01, Barcode02, etc., were copied into a subdirectory called Barcode_first_run located within a directory called NANOPORE_DATA, which contained the script README file. Each barcode subdirectory will contain one or more FASTq files of raw sequence data; however, there is no need to rename any of the FASTq files in these directories (nanopore-amplicon-consensus/<run>/<barcodesxx>/<files.fastq>). Runs were initiated with the command sh nanopore-amplicon-consensus.sh <name_of_run_directory> <minimum_amplicon_length> <maximum_amplicon_length>. The program first removes any existing TEMP or result folders that may have been generated during a previous execution of the script. Next, multiple FASTq files from the same barcode directory, if they exist, will be merged and filtered for minimum and maximum amplicon length. It is recommended to use a target amplicon length of +100 to 200 nucleotides. Prior to the sequencing run start, we set the number of reads to 10,000 to reduce the likelihood of multiple FASTq files being produced from the same run. Merged and filtered files are next used by CANU to generate consensus sequences, which are copied to the output folder “consensus_seqs” as FASTa files. The output directory and directories with merged, filtered, and consensus FASTa files created in the process will be removed when the script is executed again on the same run directory.
After runs were completed, consensus files were opened directly with MacVector and used for alignments, GenBank searches, and annotation. Annotation was done using RFAM (25), which identified the 28S ribosomal subunit, 5S ribosomal subunit, and 18S ribosomal subunit sequences and boundaries. The IGS1 and IGS2 regions were identified from the boundaries of these three subunits. The 28S and 18S subunits were partial sequences, based on primer location, and the IGS1, 5S, and IGS2 sequences were complete sequences.
IGS analysis.
Phylogenetic analysis was performed on 28 IGS sequences, including the outgroup, Cryptococcus wingfieldii. The Cryptococcus wingfieldii IGS region was recovered from the CBS7118 chromosome 14 genome sequence after a BLASTn search using the corresponding H99 sequence. The chromosomal region was downloaded, analyzed by RFAM, and then annotated for subunit boundaries as described above. All sequences were aligned using MUSCLE (26) as implemented in Sequencher version 5.4.6 Build 46289 (Gene Codes Corporation, Ann Arbor MI, USA) and manually checked in PAUP version 4.0a Build 167 (PAUP) (27). The alignment was analyzed using maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI). The maximum-likelihood tree was constructed in IQ-TREE (28) with 1,000 resampling of standard nonparametric bootstrapping (BS) implemented in IQ-TREE as UFBoot (29). TPM3u+G was the best-fit substitution model for maximum likelihood as determined by the ModelFinder implemented in IQ-TREE using the corrected Akaike information criterion (AICc) (30). Bayesian posterior probabilities (PP) were calculated using MrBayes 3.2.5 (31). The substitution model for Bayesian analysis was determined as described above. However, TPM3u+G is not implemented in MrBayes 3.2.5 and was replaced by the GTR+G as an alternative model (32). The analysis ran for 2 × 107 generations, two parallel runs with four chains, and every 1,000th tree was sampled until convergence was reached when the standard deviation of split frequency was <0.01. The first 25% of trees were discarded as burn-in, and the remaining trees were combined into a single tree with 50% majority rule consensus. Thresholds of ≥80% BT and ≥0.95 PP on nodes were considered significantly supported.
Percent identities were compared by generating a consensus sequence for strains within the same genotype and then creating a matrix of pairwise alignments of each consensus sequence. The average identities of the sequences within each genotype were then calculated and compared to all genotype averages.
Comparison of nanopore and Sanger sequences.
Nanopore sequence accuracy was determined by aligning nanopore sequences of the IGS regions (IGS1, 5S, and IGS2) with CLUSTAL (33) to the Sanger sequences of each strain. Percent identities were determined for each alignment to ascertain nanopore sequencing accuracy. Each Sanger and nanopore alignment was then examined for error location and type, including homopolymers, indels, and mismatches. Homopolymer definitions can be variable, such as runs of 2 or more (34) or 3 or more (35) of the same nucleotide. We used runs of 2 or more of the same nucleotide to identify a homopolymer.
Data availability.
Annotated IGS sequences obtained from Sanger sequencing were submitted to GenBank under BioProject record number PRJNA614507, with accession numbers MT712017 to MT712043.
RESULTS
Twenty-seven strains, including reference isolates representing seven genotypes, were analyzed by Sanger sequencing of the URA5 gene to determine genotype based on Sau96I and HhaI restriction enzyme sites (23) and BLASTn search results. We found that locating the restriction sites based on sequence instead of trying to determine sizes based on gel pattern eliminated the potential ambiguity of trying to determine exact fragment sizes, some of which were less than 100 bp. One strain, WSA3478, had a single Sau96I restriction enzyme site, and no strains had a single HhaI restriction enzyme site. Table 3 shows that the restriction enzyme fragments determined by sequence cluster in 100% agreement with VN-VG genotype.
TABLE 3.
Strain genotypes
| Strain | URA5 genotype | BLAST genotype |
Restriction enzyme fragment size (bp)
|
||||
|---|---|---|---|---|---|---|---|
| Sau96I | Sau96I-HhaI | Sau96I-Sau96I | HhaI-Sau96I | HhaI-HhaI | |||
| W3482 | VGI | VGI | 324 | ||||
| W10 | VGI | VGI | 324 | ||||
| W85 | VGI | VGI | 324 | ||||
| W530 | VGII | VGII | 128 | 138 | 58 | ||
| W3481 | VGII | VGII | 128 | 138 | 58 | ||
| W432 | VGII | VGII | 128 | 138 | 58 | ||
| W84 | VGIII | VGIII | 128 | ||||
| W87 | VGIII | VGIII | 128 | ||||
| W83 | VGIII | VGIII | 128 | ||||
| W3479 | VGIII | VGIII | 128 | ||||
| W15 | VGIII | VGIII | 128 | ||||
| W16 | VGIII | VGIII | 128 | ||||
| W89 | VGIII | VGIII | 128 | ||||
| W90 | VGIII | VGIII | 128 | ||||
| W3480 | VGIV | VGIV | 128 | 196 | |||
| H99 | VNI | VNI | 61 | ||||
| W3477 | VNI | VNI | 61 | ||||
| W473 | VNI | VNI | 61 | ||||
| W516 | VNI | VNI | 61 | ||||
| W72 | VNI | VNI | 61 | ||||
| W1047 | VNI | VNI | 61 | ||||
| W3478a | VNII | VNII | 269 | ||||
| JEC21 | VNIV | VNIV | 186 | 156 | |||
| W3484 | VNIV | VNIV | 186 | 156 | |||
| W77 | VNIV | VNIV | 186 | 156 | |||
| W78 | VNIV | VNIV | 186 | 156 | |||
| W79 | VNIV | VNIV | 186 | 156 | |||
Sau96I has a single cut site.
IGS region characteristics.
After annotating for ribosomal subunit position, IGS1 and IGS2 regions were identified and the sizes were determined (Table 4). The sizes of the entire IGS region (IGS1, 5S, and IGS2) within genotypes were consistent, with little variation (0 to 5 nucleotides over an IGS size range of 2,426 to 2,622 bp), depending on genotype. Individual regions also showed little variation. The 5S subunit was uniform in length across all genotypes and strains (119 bp). The IGS1 size varied from 1,229 to 1,418 bp, while the IGS2 size varied from 1,061 to 1,123 bp. The IGS1 region varied from 0 to 7 nucleotides and the IGS2 region varied from 0 to 12 nucleotides, depending on genotype. When the two species complexes were compared, for C. gattii, the IGS average length was 2,487.13 ± 15.44 bp, while the IGS1 average was 1,268± 2.83 bp, and the IGS2 average was 1,100.13 ± 12.91 bp. For C. neoformans, the IGS average length was 2,539.33 ± 98.48 bp, while the IGS1 average was 1,340.33 ± 95.83 bp, and the IGS2 average was 1,078.50 ± 8.39 bp. The C. gattii IGS, IGS1, and IGS2 length ranges were 2,482 to 2,514 bp, 1,265 to 1,274 bp, and 1,106 to 1,123 bp, respectively, with the IGS2 region being more variable. The C. neoformans IGS, IGS1, and IGS2 length ranges were 2,426 to 2,622 bp, 1,229 to 1,418 bp, and 1,061 to 1,085 bp, respectively, with the IGS2 region also being more variable. Interestingly, the C. neoformans IGS and IGS1 lengths were greater than the corresponding C. gattii lengths, while the C. gattii IGS2 length was greater than that of the corresponding C. neoformans region.
TABLE 4.
IGS analysis
| Strain | Genotype | IGS length (bp) |
|||
|---|---|---|---|---|---|
| Total | IGS1 | 5S | IGS2 | ||
| W3482 | VGI | 2,470 | 1,265 | 119 | 1,086 |
| W10 | VGI | 2,470 | 1,265 | 119 | 1,086 |
| W85 | VGI | 2,470 | 1,265 | 119 | 1,086 |
| Avg ± SD | 2,470 ± 0 | 1,265 ± 0 | 119 ± 0 | 1,086 ± 0 | |
| W530 | VGII | 2,514 | 1,272 | 119 | 1,123 |
| W3481 | VGII | 2,513 | 1,271 | 119 | 1,123 |
| W432 | VGII | 2,514 | 1,272 | 119 | 1,123 |
| Avg ± SD | 2,513.7 ± 0.6 | 1,271.7 ± 0.6 | 119 ± 0 | 1,123 ± 0 | |
| W84 | VGIII | 2,482 | 1,267 | 119 | 1,096 |
| W87 | VGIII | 2,483 | 1,267 | 119 | 1,097 |
| W83 | VGIII | 2,482 | 1,267 | 119 | 1,096 |
| W3479 | VGIII | 2,482 | 1,267 | 119 | 1,096 |
| W15 | VGIII | 2,482 | 1,267 | 119 | 1,096 |
| W16 | VGIII | 2,482 | 1,267 | 119 | 1,096 |
| W89 | VGIII | 2,482 | 1,267 | 119 | 1,096 |
| W90 | VGIII | 2,482 | 1,267 | 119 | 1,096 |
| Avg ± SD | 2,482.1 ± 0.4 | 1,267.0 ± 0 | 119 ± 0 | 1,096.1 ± 0.4 | |
| W3480 | VGIV | 2,499 | 1,274 | 119 | 1,106 |
| Avg ± SD | 2,499 ± 0 | 1,274 ± 0 | 119 ± 0 | 1,106 ± 0 | |
| H99 | VNI | 2,619 | 1,418 | 119 | 1,082 |
| W3477 | VNI | 2,619 | 1,418 | 119 | 1,082 |
| W473 | VNI | 2,618 | 1,418 | 119 | 1,082 |
| W516 | VNI | 2,622 | 1,418 | 119 | 1,085 |
| W72 | VNI | 2,619 | 1,418 | 119 | 1,082 |
| W1047 | VNI | 2,615 | 1,418 | 119 | 1,078 |
| Avg ± SD | 2,618.7 ±2.3 | 1,418 ± 0 | 119 ± 0 | 1,081.8 ± 2.2 | |
| W3478 | VNII | 2,621 | 1,417 | 119 | 1,085 |
| Avg ± SD | 2,621 ± 0 | 1,417 ± 0 | 119 ± 0 | 1,085 ± 0 | |
| JEC21 | VNIV | 2,426 | 1,236 | 119 | 1,061 |
| W3484 | VNIV | 2,428 | 1,229 | 119 | 1,081 |
| W77 | VNIV | 2,426 | 1,236 | 119 | 1,061 |
| W78 | VNIV | 2,431 | 1,229 | 119 | 1,083 |
| W79 | VNIV | 2,428 | 1,229 | 119 | 1,080 |
| Avg ± SD | 2,427.8 ± 2.0 | 1,231.8 ± 3.8 | 119 ± 0 | 1,073.2 ± 11.2 | |
IGS phylogeny.
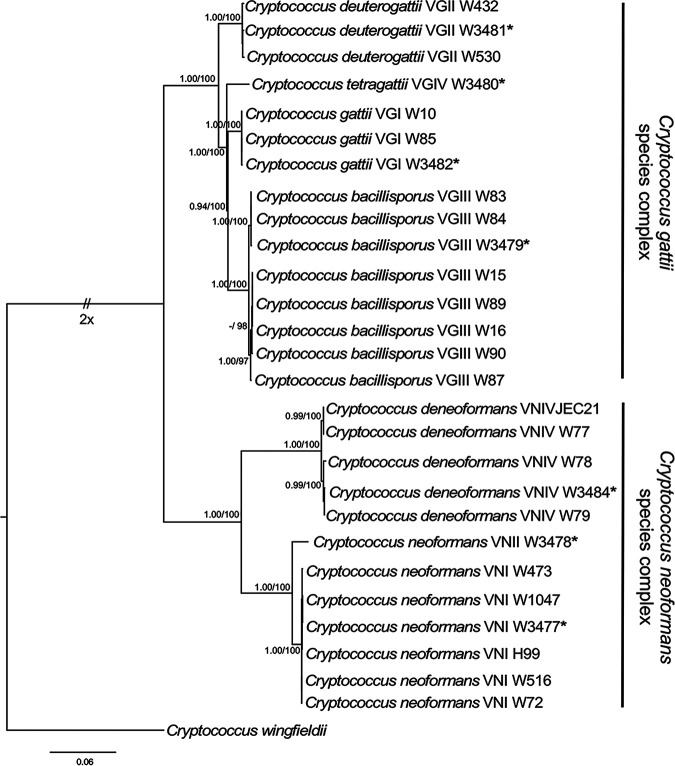
The IGS alignment was 2,872 characters with 594 (20%) parsimony-informative characters. Twelve most-parsimonious trees were generated from the maximum-parsimony analysis in PAUP, with the following tree scores: consistency index (CI), 0.924; retention index (RI), 0.976; homoplasy index (HI), 0.076. The topologies of the best-scoring MP (not shown) and ML trees were congruent with the BI tree (not shown). In the best-scoring ML tree, two main clades corresponding to the two species complexes were shown with high BI-PP/ML-BS support (1.00/100%) (Fig. 1). Strain identities included strain number, VN-VG genotype, and new names of the recent Cryptococcus neoformans/Cryptococcus gattii species complex (36) to determine if IGS sequences clustered by this new naming method. The 27 isolates of each species complex were resolved in well-supported subclades of strains with the same genotype as well as species designation for each genotype, which demonstrates that the sequences generated by nanopore sequencing match previous results generated by Sanger sequencing (36). A CLUSTAL alignment matrix of consensus sequences for each genotype was consistent with the phylogenetic tree (Table 5). Importantly, from a diagnostic perspective, the consensus IGS sequences of genotypes within a species complex differed from the other species complex substantially, displaying only 74.6 to 77.2% identity. Within a species complex, percent identities ranged from 92.3 to 96.0% for the C. gattii complex and from 83.6 to 98.1% for the C. neoformans complex.
FIG 1.
Phylogenetic relationships of strains belonging to the Cryptococcus gattii and C. neoformans species complexes inferred from maximum-likelihood analysis of IGS sequences. Bayesian posterior probabilities (PP) (left) and maximum-likelihood bootstrap support (BT) (right) are shown on the nodes. The tree is rooted with the corresponding IGS sequence from Cryptococcus wingfieldii strain CBS 7118. Asterisks represent reference strains.
TABLE 5.
Consensus VG-VN IGS alignment matrix
| Genotype | Identity (%) to genotype: |
||||||
|---|---|---|---|---|---|---|---|
| VGI | VGII | VGIII | VGIV | VNI | VNII | VNIV | |
| VGI | 100.0 | 93.6 | 96.0 | 94.7 | 75.5 | 75.4 | 77.2 |
| VGII | 93.6 | 100.0 | 93.1 | 92.3 | 74.6 | 74.6 | 76.2 |
| VGIII | 96.0 | 93.1 | 100.0 | 93.9 | 75.6 | 75.4 | 77.1 |
| VGIV | 94.7 | 92.3 | 93.9 | 100.0 | 75.5 | 75.5 | 77.0 |
| VNI | 75.5 | 74.6 | 75.6 | 75.5 | 100.0 | 98.1 | 83.6 |
| VNII | 75.4 | 74.6 | 75.4 | 75.5 | 98.1 | 100.0 | 83.6 |
| VNIV | 77.2 | 76.2 | 77.1 | 77.0 | 83.6 | 83.6 | 100.0 |
Nanopore IGS sequence analysis derived from R9.4.1 flow cells.
The same PCR products used for Sanger sequencing were used for nanopore sequencing in various barcoded combinations, with up to 24 barcoded samples in a single run. Run speed depended on a variety of factors, including the number of previous runs of the flow cell and number of barcoded samples in the run. Our amplicons ranged from ∼3.3 to 3.5 kb, and we typically barcoded 5 to 12 samples. We used the MinION flow cell, which has an enormous excess capacity for PCR amplicons in our size range. We were able to get almost 10 runs per flow cell, although each successive run depletes functioning pores. Sequencing speed was generally in the range of 500 bases/s, with only a slight drop-off toward the end of the flow cell life. With a new flow cell and a set of 12 barcodes, we were able to generate at least 1,500 reads per barcode within 20 to 30 min.
After base-called reads were demultiplexed by MinKNOW and processed by the nanopore amplicon consensus script to derive a consensus sequence, the sequence from each strain was aligned with its respective Sanger-derived sequence to identify mismatches, with the goal being to determine how close in percent identity the nanopore sequences were to Sanger sequences. Two of the Sanger sequences, H99 and JEC21, were 100% identical to sequences in GenBank derived from the genome sequence of each strain, which provided the rationale for using Sanger sequences as the baseline for measuring nanopore sequence accuracy. The nanopore sequences for each strain displayed an average identity of 99.57% ± 0.19% (range, 99.20% to 100%) to their respective Sanger-derived sequences.
Alignments of sequences derived from the R9.4.1 flow cell were then examined more closely for error type. Regions of homopolymers were most commonly prone to errors, with G homopolymers showing an average of 5.15 mismatches per IGS and C homopolymers showing the least, with an average of 0.04 mismatch per IGS (Table 6). All IGS sequences except one had at least one G homopolymer mismatch, while only one of the 27 IGS sequences had a C homopolymer mismatch. For every IGS sequence that displayed homopolymer errors in the nanopore sequence, all were deletions. No insertion errors were found in the homopolymer regions. Other types of errors were rarely seen. Out of the 27 IGS sequences, seven sequences displayed errors in repetitive motif regions (e.g., ATATATAT), one displayed a deletion error, and five displayed mismatch errors. No insertion errors were detected. When homopolymer errors were excluded from percent identity determinations, the nanopore sequences were 99.97 to 100% identical to Sanger sequences, with 17/27 sequences displaying 100% identity.
TABLE 6.
Sanger versus nanopore R9.4.1 flow cell sequence IGS mismatches
| Strain | Genotype | No. of errors |
% identityb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homopolymer mismatches |
Repetitive motifs | Deletionsa | Mismatches | |||||||
| A | T | G | C | Total | ||||||
| W3482 | VGI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 |
| W10 | VGI | 1 | 7 | 6 | 0 | 14 | 0 | 0 | 0 | 99.55 |
| W85 | VGI | 3 | 4 | 5 | 0 | 12 | 0 | 0 | 0 | 99.64 |
| W3481 | VGII | 4 | 4 | 4 | 0 | 12 | 0 | 0 | 0 | 99.52 |
| W432 | VGII | 1 | 5 | 4 | 0 | 10 | 0 | 0 | 0 | 99.58 |
| W530 | VGII | 5 | 5 | 5 | 0 | 15 | 2 | 0 | 0 | 99.38 |
| W3479 | VGIII | 0 | 4 | 5 | 0 | 9 | 0 | 0 | 0 | 99.73 |
| W15 | VGIII | 1 | 0 | 3 | 0 | 4 | 0 | 0 | 0 | 99.85 |
| W16 | VGIII | 3 | 5 | 5 | 0 | 13 | 0 | 0 | 1 | 99.58 |
| W83 | VGIII | 3 | 3 | 6 | 0 | 12 | 0 | 0 | 0 | 99.61 |
| W84 | VGIII | 0 | 4 | 2 | 0 | 6 | 0 | 0 | 0 | 99.76 |
| W87 | VGIII | 2 | 4 | 4 | 0 | 10 | 0 | 0 | 0 | 99.70 |
| W89 | VGIII | 2 | 3 | 4 | 0 | 9 | 0 | 0 | 0 | 99.70 |
| W90 | VGIII | 1 | 4 | 3 | 0 | 8 | 0 | 0 | 0 | 99.70 |
| W3480 | VGIV | 1 | 5 | 5 | 0 | 11 | 0 | 0 | 0 | 99.61 |
| W3477 | VNI | 6 | 3 | 8 | 0 | 17 | 0 | 0 | 0 | 99.48 |
| W72 | VNI | 5 | 2 | 5 | 0 | 12 | 0 | 0 | 0 | 99.63 |
| W473 | VNI | 6 | 4 | 6 | 0 | 16 | 0 | 0 | 1 | 99.51 |
| W516 | VNI | 5 | 4 | 7 | 0 | 16 | 5 | 0 | 6 | 99.20 |
| W1047 | VNI | 7 | 3 | 10 | 0 | 20 | 0 | 1 | 1 | 99.34 |
| H99 | VNI | 6 | 3 | 7 | 0 | 16 | 2 | 0 | 2 | 99.23 |
| W3478 | VNII | 7 | 3 | 9 | 0 | 19 | 0 | 0 | 0 | 99.34 |
| W3484 | VNIV | 1 | 4 | 8 | 0 | 13 | 2 | 0 | 0 | 99.48 |
| JEC21 | VNIV | 1 | 5 | 8 | 0 | 14 | 3 | 0 | 0 | 99.48 |
| W77 | VNIV | 3 | 4 | 4 | 0 | 11 | 0 | 0 | 0 | 99.54 |
| W78 | VNIV | 1 | 0 | 2 | 1 | 4 | 2 | 0 | 0 | 99.75 |
| W79 | VNIV | 2 | 3 | 4 | 0 | 9 | 2 | 0 | 0 | 99.60 |
| Total | 77 | 95 | 139 | 1 | 301 | 18 | 1 | 11 | ||
| Avg | 2.85 | 3.52 | 5.15 | 0.04 | 11.15 | 0.67 | 0.04 | 0.41 | 99.57 | |
Deletions in regions other than a homopolymer.
Based on comparison to Sanger sequence of the same region.
Nanopore IGS sequence analysis derived from R10.3 flow cells.
During this study, the next version of the MinION flow cell was released, designated R10.3. The flow cell utilizes a new nanopore that reads homopolymers with greater accuracy. The same strains that were sequenced with the R9.4.1 flow cell were sequenced with the R10.3 flow cell. The sequences were then compared to the Sanger and R9.4.1 sequences. Overall, the percent identity increased from 99.57% ± 0.19% to 99.83% ± 0.092% for the R10.3 flow cell compared to the R9.4.1 flow cell (Table 7). As expected, the accuracy of homopolymer regions improved, with a 57% decrease in errors. Errors for A, T, and G homopolymers all decreased, although C homopolymer errors increased from one to eight. Errors associated with repetitive motifs and deletions increased slightly, while mismatch errors decreased substantially (Table 8).
TABLE 7.
Comparison of sequences obtained with R9.4.1 and R10.3 flow cells to Sanger sequences
| Strain | Identity (%) to Sanger sequence |
||
|---|---|---|---|
| R9.4.1 flow cell sequence | R10.3 flow cell sequence | Change | |
| W3482 | 100 | 99.91 | −0.09 |
| W10 | 99.55 | 99.85 | +0.30 |
| W85 | 99.64 | 99.94 | +0.30 |
| W3481 | 99.52 | 99.73 | +0.21 |
| W432 | 99.58 | 99.82 | +0.24 |
| W530 | 99.38 | 99.70 | +0.32 |
| W3479 | 99.73 | 99.94 | +0.21 |
| W15 | 99.85 | 99.88 | +0.03 |
| W16 | 99.58 | 99.85 | +0.27 |
| W83 | 99.61 | 99.91 | +0.30 |
| W84 | 99.76 | 99.70 | −0.06 |
| W87 | 99.70 | 99.91 | +0.21 |
| W89 | 99.70 | 99.85 | +0.15 |
| W90 | 99.70 | 99.82 | +0.12 |
| W3480 | 99.61 | 99.94 | +0.33 |
| W3477 | 99.48 | 99.71 | +0.23 |
| W72 | 99.63 | 99.91 | +0.28 |
| W473 | 99.51 | 99.86 | +0.35 |
| W516 | 99.20 | 99.89 | +0.69 |
| W1047 | 99.34 | 99.91 | +0.57 |
| H99 | 99.23 | 99.83 | +0.60 |
| W3478 | 99.34 | 99.74 | +0.40 |
| W3484 | 99.48 | 99.60 | +0.12 |
| JEC21 | 99.48 | 99.87 | +0.39 |
| W77 | 99.54 | 99.70 | +0.16 |
| W78 | 99.75 | 99.79 | +0.04 |
| W79 | 99.60 | 99.88 | +0.28 |
| Avg | 99.57 | 99.83 | +0.26 |
TABLE 8.
Sanger versus nanopore R10.3 flow cell sequence IGS mismatches
| Strain | Genotype | No. of errors |
% identityb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Homopolymer mismatches |
Repetitive motifs | Deletionsa | Mismatches | |||||||
| A | T | G | C | Total | ||||||
| W3482 | VGI | 0 | 0 | 2 | 1 | 3 | 0 | 0 | 0 | 99.91 |
| W10 | VGI | 1 | 1 | 3 | 0 | 5 | 0 | 0 | 0 | 99.85 |
| W85 | VGI | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 99.94 |
| W3481 | VGII | 0 | 2 | 6 | 1 | 9 | 0 | 0 | 0 | 99.73 |
| W432 | VGII | 0 | 1 | 5 | 0 | 0 | 6 | 0 | 0 | 99.82 |
| W530 | VGII | 1 | 1 | 6 | 0 | 8 | 0 | 2 | 0 | 99.70 |
| W3479 | VGIII | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 99.94 |
| W15 | VGIII | 1 | 0 | 3 | 0 | 4 | 0 | 0 | 0 | 99.88 |
| W16 | VGIII | 1 | 0 | 3 | 0 | 4 | 0 | 0 | 1 | 99.85 |
| W83 | VGIII | 0 | 1 | 2 | 0 | 3 | 0 | 0 | 0 | 99.91 |
| W84 | VGIII | 2 | 2 | 4 | 0 | 8 | 2 | 0 | 0 | 99.70 |
| W87 | VGIII | 0 | 1 | 2 | 0 | 3 | 0 | 0 | 0 | 99.91 |
| W89 | VGIII | 0 | 1 | 3 | 1 | 5 | 0 | 0 | 0 | 99.85 |
| W90 | VGIII | 2 | 1 | 3 | 0 | 6 | 0 | 0 | 0 | 99.82 |
| W3480 | VGIV | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 99.94 |
| W3477 | VNI | 0 | 1 | 7 | 0 | 8 | 2 | 0 | 0 | 99.71 |
| W72 | VNI | 1 | 0 | 1 | 1 | 3 | 0 | 0 | 0 | 99.91 |
| W473 | VNI | 2 | 0 | 2 | 0 | 4 | 0 | 0 | 1 | 99.86 |
| W516 | VNI | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 | 99.89 |
| W1047 | VNI | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 99.91 |
| H99 | VNI | 1 | 0 | 3 | 0 | 4 | 2 | 0 | 0 | 99.83 |
| W3478 | VNII | 5 | 0 | 4 | 0 | 9 | 0 | 0 | 0 | 99.74 |
| W3484 | VNIV | 6 | 0 | 3 | 0 | 9 | 2 | 0 | 2 | 99.60 |
| JEC21 | VNIV | 0 | 0 | 2 | 1 | 3 | 0 | 1 | 0 | 99.87 |
| W77 | VNIV | 6 | 0 | 2 | 0 | 8 | 2 | 0 | 0 | 99.70 |
| W78 | VNIV | 4 | 0 | 2 | 1 | 7 | 0 | 0 | 0 | 99.79 |
| W79 | VNIV | 1 | 0 | 2 | 1 | 4 | 0 | 0 | 0 | 99.88 |
| Total | 36 | 12 | 78 | 8 | 128 | 18 | 3 | 4 | ||
| Avg | 1.33 | 0.44 | 2.89 | 0.30 | 4.74 | 0.67 | 0.11 | 0.15 | 99.83 | |
Deletions in regions other than a homopolymer.
Based on comparison to Sanger sequence of the same region.
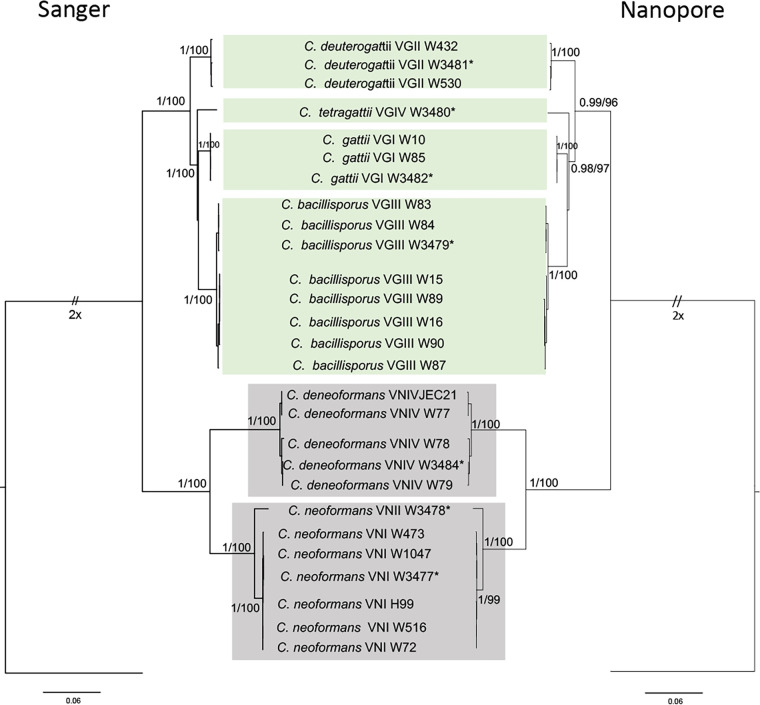
The phylogenetic tree of IGS sequences generated with this flow cell was compared to a tree of their respective Sanger sequences (Fig. 2). No differences were found with regard to clusters observed between the two trees. These results suggest that the R10.3 flow cell has sufficient accuracy to be used for IGS sequence-based identification.
FIG 2.
Phylogenetic relationships of species belonging to the C. gattii and C. neoformans species complexes inferred from maximum-likelihood analysis of IGS sequences using the nanopore (R10.3 flow cell) and Sanger methods. Bayesian posterior probabilities (PP) (left) and maximum-likelihood bootstrap support (BT) (right) are shown on the nodes. The tree is rooted with Cryptococcus wingfieldii CBS 7118. Asterisks represent reference strains.
DISCUSSION
The IGS region is a little-used area of the ribosomal cluster that has the potential to be extremely informative with regard to fungal identification. It has not been used extensively for fungal identification in the past because it is too long to cover efficiently using Sanger sequencing. In contrast, the fungal ITS region is short enough to be covered in a single Sanger sequence read and can be amplified with universal primers. However, for many fungi it does not have enough discriminatory power to yield a species-level identification independently, especially for closely related sibling species, which may necessitate identification and sequencing of additional loci. Sequencing of multiple loci may be problematic for clinical microbiology laboratories due to turnaround time, which can be extended further if the first sequence is needed before the second sequence is chosen. Because the IGS region is not transcribed and is much longer than the ITS region, it can vary extensively and can potentially solve the problem of yielding an identification in a single sequencing run; however, short of a whole genome sequence, there was no convenient way to sequence this region in a single run. The development of nanopore sequencing solves this problem since this type of sequencing can yield single reads that exceed 2 megabases in length (34, 37), so target sequence length is not a concern. When nanopore sequencing first became available in 2014, it was not accurate enough to be used for microbial sequence-based diagnosis (38). However, since its initial release, accuracy has improved greatly due to upgrades in instrumentation, chemistry, and base-calling algorithms, in addition to editing and assembly programs, to the point that nanopore sequencing is finding its way into clinical microbiology laboratories (39). Because of these improvements, we wanted to determine how useful nanopore sequencing could be for fungal identification using the IGS region as a target.
Diaz et al. performed a detailed analysis of the Cryptococcus IGS region a number of years ago by sequencing the IGS1-5S-IGS2 (partial) region from 107 isolates of Cryptococcus neoformans and Cryptococcus gattii, and they identified six genotypes and 12 different lineages (40, 41). Although those studies were completed prior to the publication of the species complex proposal (36), they demonstrated the specificity of this region with regard to species identification. The percent identity between the C. neoformans and C. gattii complex in those studies was ∼66 to 69%. Our results were similar, with a range of 74 to 77%, with the disparity due to the inclusion of the complete IGS2 region in our alignments. The studies by Diaz et al. and our study were able to resolve individual species within the species complex, suggesting that the IGS region is sufficiently sensitive for species-level identification. The utility of the IGS region as a potential molecular diagnostic target was the reason we investigated nanopore sequencing as a method for fungal identification, since it offers a number of advantages over Sanger sequencing. Importantly, in spite of its variability, the location between the 28S and 18S ribosomal subunits offers a number of options for universal PCR priming sites, analogous to the utility of the ITS region for sequence-based identification of unknown fungi. Consequently, it matches an important asset of ITS sequencing, which is that, due to universal PCR priming sites, unknown isolates can be identified with no preliminary information regarding suspected taxonomic placement. Based on the results of this study, it appears to have more discriminatory power than ITS sequencing, since it was able to resolve the C. neoformans and C. gattii complexes into individual clades, which cannot be done accurately with the ITS region. If this outcome also was observed for other, more complicated fungal taxonomies, such as Aspergillus and Fusarium, it could eliminate the need for a second locus to be sequenced from these fungi in order to yield the most accurate identity.
In addition to the ability of nanopore sequencing to easily and quickly generate complete IGS reads from PCR amplicons of the region, sequencing can be done in the laboratory using a sequencing device with a small footprint (the MinION) and a laptop computer to capture and analyze data. Sample preparation is easy and consists of end repair, barcoding, and adapter ligation, all of which are enzymatic manipulations done in a microcentrifuge tube. The barcode step is optional, as it is not needed for single samples or if the initial PCR step uses custom barcodes. In fact, we found that sequencing capacity of amplicons using a MinION flow cell was overkill, as the capacity in a single run is far more than the maximum barcode number using the two different 12 barcode kits from ONT. While we do not know the precise limits, we estimate that it might be possible to sequence 500 different amplicons or more if custom barcodes were used. With the ONT barcode kits providing a maximum of 24 barcodes per run, well in excess of 100 samples could be run on the same flow cell. There is a newer flow cell (Flongle) that is a single-use model capable of generating almost 2 gigabases of data, which is still excess capacity, yet it is 1/10 the cost of a MinION flow cell. Running multiple samples on this flow cell using custom barcodes would lower the cost further and make this strategy a realistic option, based on cost, for clinical microbiology laboratories, particularly with further improvements. Importantly, barcodes can be read in real time, and it is possible to run mixed microbial or other samples in a single run, which would further expand capability. Although the PCR step is not optimal for a diagnostic assay, it enables nanopore sequencing to be used directly on clinical specimens that have yeast or fungi in them if the target IGS region can be amplified directly from the sample. Nevertheless, compared to existing systems such as Vitek2 or MALDI-TOF MS, which may have lower costs per sample, nanopore sequencing using the MinION has a negligible capital investment ($1,000), no maintenance contract, and no required specialized technical expertise. While a biocurated database would need to be used for identification in a clinical laboratory, the spectrum of organisms identified by this system would arguably be much broader due to the availability of fungal DNA sequences in public databases. In our hands, some of the drawbacks included the need to purchase reagents from a third party (New England Biolabs) with no clear less-expensive alternative, which adds substantially to cost, and the requirement by ONT to order packages of 12 if the smaller module (Flongle) is purchased, which is quite expensive. We also found that our laptop computer handles one or a few samples quickly with regard to base calling and downstream data analysis, which can be done within minutes for a single sample, but can take longer than a day for a full set of 24 barcoded samples, depending on how large the FASTq files are. For a clinical or high-throughput laboratory, alternate computing resources or the Oxford Nanopore supporting computer system (MinIT), which can base call almost 10 times faster than our laptop, would be needed.
During this study the newest flow cell, R10.3, became available after we had analyzed IGS sequences generated with the previous flow cell version. Repeating the experiments using the R10.3 flow yielded improved results. Average percent identity compared to the corresponding Sanger sequence increased to 99.83%, with much of the increased accuracy due to better recognition of homopolymer regions. Homopolymer errors were reduced an average of 57%, and phylogenetic trees constructed with the nanopore sequences produced from the R10.3 flow cell compared to the corresponding Sanger sequences were indistinguishable, both of which supported previous phylogenetic trees produced by a variety of other methods (36, 42). In these investigations, VGI and VGIII were most closely related, VGIV was basal, and VGIII was the most distantly related to these genotypes in C. gattii (43). In the C. neoformans species complex, VNI and VNII were most closely related, while VNIV was most distant (43). The accuracy of nanopore sequencing compared to Sanger sequencing appears to be sufficient to allow this sequencing method and the IGS sequencing target to serve as a novel molecular identification method for fungal identification. The specificity of this region was sufficient to reproduce the recently proposed clades of the C. neoformans and C. gattii species complexes. It would be interesting to see how the IGS region performs on more-challenging taxonomic problems, such as the Fusarium species complexes or the Aspergillus sections. Technically, only basic molecular skills are required to use nanopore sequencing, since templates are PCR amplicons and preparation of these amplicons for sequencing consists only of end preparation to add adapters and barcodes (if used). Postrun processing is done in part by ONT software, and while we wrote our own scripts to generate consensus sequences, ONT has internal software programs that can be used to establish other pipelines. Importantly, for clinical laboratories, almost all steps have been automated, with future configurations incorporating dockable components (extraction, PCR, and sequencing) into a single system. The speed of sequencing and small footprint make nanopore sequencing of the IGS region an option to consider for sequence-based identification of fungi.
REFERENCES
- 1.Benedict K, Jackson BR, Chiller T, Beer KD. 2019. Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis 68:1791–1797. doi: 10.1093/cid/ciy776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi (Basel) 3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denning DW. 2017. Calling upon all public health mycologists: to accompany the country burden papers from 14 countries. Eur J Clin Microbiol Infect Dis 36:923–924. doi: 10.1007/s10096-017-2909-8. [DOI] [PubMed] [Google Scholar]
- 4.Havlickova B, Czaika VA, Friedrich M. 2008. Epidemiological trends in skin mycoses worldwide. Mycoses 51(Suppl 4):2–15. doi: 10.1111/j.1439-0507.2008.01606.x. [DOI] [PubMed] [Google Scholar]
- 5.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 6.Dignani MC. 2014. Epidemiology of invasive fungal diseases on the basis of autopsy reports. F1000Prime Rep 6:81. doi: 10.12703/P6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Solache MA, Casadevall A. 2010. Global warming will bring new fungal diseases for mammals. mBio 1:e00061-10. doi: 10.1128/mBio.00061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, Villarreal Ruiz L, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Poldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Partel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, et al. 2014. Fungal biogeography. Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 9.Taylor LH, Latham SM, Woolhouse ME. 2001. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal M. 2018. Morbidity and mortality due to fungal infections. J Appl Microbiol Biochem 1:1–3. doi: 10.21767/2576-1412.100002. [DOI] [Google Scholar]
- 11.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickes BL, Wiederhold NP. 2018. Molecular diagnostics in medical mycology. Nat Commun 9:5135. doi: 10.1038/s41467-018-07556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seifert KA, Rossman AY. 2010. How to describe a new fungal species. IMA Fungus 1:109–116. doi: 10.5598/imafungus.2010.01.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5' end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35:1216–1223. doi: 10.1128/JCM.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White TJ, Bruns TD, Lee SB, Taylor JW. 1990. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. Academic Press, New York, NY. [Google Scholar]
- 16.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium, Fungal Barcoding Consortium Author List. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mekha N, Sugita T, Makimura K, Poonwan N, Sawanpanyalert P, Ikeda R, Nishikawa A. 2010. The intergenic spacer region of the ribosomal RNA gene of Penicillium marneffei shows almost no DNA sequence diversity. Microbiol Immunol 54:714–716. doi: 10.1111/j.1348-0421.2010.00270.x. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, Zhang Y, Ying C, Wang D, Du C. 2015. Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics 13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain M, Olsen HE, Paten B, Akeson M. 2016. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Midha MK, Wu M, Chiu KP. 2019. Long-read sequencing in deciphering human genetics to a greater depth. Hum Genet 138:1201–1215. doi: 10.1007/s00439-019-02064-y. [DOI] [PubMed] [Google Scholar]
- 22.Romanelli AM, Fu J, Herrera ML, Wickes BL. 2014. A universal DNA extraction and PCR amplification method for fungal rDNA sequence-based identification. Mycoses 57:612–622. doi: 10.1111/myc.12208. [DOI] [PubMed] [Google Scholar]
- 23.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E, IberoAmerican Cryptococcal Study Group. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis 9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4 b10. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 28.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lecocq T, Vereecken NJ, Michez D, Dellicour S, Lhomme P, Valterova I, Rasplus JY, Rasmont P. 2013. Patterns of genetic and reproductive traits differentiation in mainland vs. Corsican populations of bumblebees. PLoS One 8:e65642. doi: 10.1371/journal.pone.0065642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, Tyson JR, Beggs AD, Dilthey AT, Fiddes IT, Malla S, Marriott H, Nieto T, O'Grady J, Olsen HE, Pedersen BS, Rhie A, Richardson H, Quinlan AR, Snutch TP, Tee L, Paten B, Phillippy AM, Simpson JT, Loman NJ, Loose M. 2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol 36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zascavage RR, Thorson K, Planz JV. 2019. Nanopore sequencing: an enrichment-free alternative to mitochondrial DNA sequencing. Electrophoresis 40:272–280. doi: 10.1002/elps.201800083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T. 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Payne A, Holmes N, Rakyan V, Loose M. 2019. BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 35:2193–2198. doi: 10.1093/bioinformatics/bty841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noakes MT, Brinkerhoff H, Laszlo AH, Derrington IM, Langford KW, Mount JW, Bowman JL, Baker KS, Doering KM, Tickman BI, Gundlach JH. 2019. Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage. Nat Biotechnol 37:651–656. doi: 10.1038/s41587-019-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen LM, Martin IW, Moschetti WE, Kershaw CM, Tsongalis GJ. 2019. Third-generation sequencing in the clinical laboratory: exploring the advantages and challenges of nanopore sequencing. J Clin Microbiol 58:e01315-19. doi: 10.1128/JCM.01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz MR, Boekhout T, Theelen B, Fell JW. 2000. Molecular sequence analyses of the intergenic spacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast. Syst Appl Microbiol 23:535–545. doi: 10.1016/S0723-2020(00)80028-4. [DOI] [PubMed] [Google Scholar]
- 41.Diaz MR, Boekhout T, Kiesling T, Fell JW. 2005. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res 5:1129–1140. doi: 10.1016/j.femsyr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR, Bicanic TA, Castaneda E, Chang YC, Chen J, Cogliati M, Dromer F, Ellis D, Filler SG, Fisher MC, Harrison TS, Holland SM, Kohno S, Kronstad JW, Lazera M, Levitz SM, Lionakis MS, May RC, Ngamskulrongroj P, Pappas PG, Perfect JR, Rickerts V, Sorrell TC, Walsh TJ, Williamson PR, Xu J, Zelazny AM, Casadevall A. 2017. The case for adopting the “species complex” nomenclature for the etiologic agents of Cryptococcosis. mSphere 2:e00357-16. doi: 10.1128/mSphere.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You M, Xu J. 2018. The effects of environmental and genetic factors on the germination of basidiospores in the Cryptococcus gattii species complex. Sci Rep 8:15260. doi: 10.1038/s41598-018-33679-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Annotated IGS sequences obtained from Sanger sequencing were submitted to GenBank under BioProject record number PRJNA614507, with accession numbers MT712017 to MT712043.